Abstract
Most retroviruses preferentially integrate into certain genomic locations and, as a result, their genome-wide integration patterns are non-random. We investigate the epigenetic landscape of integrated retroviral vectors and correlate it with the long-term stability of proviral transcription. Retroviral vectors derived from the avian sarcoma/leukosis virus expressing the GFP reporter were used to transduce the human myeloid lymphoblastoma cell line K562. Because of efficient silencing of avian retrovirus in mammalian cells, only ∼3% of established clones displayed stable proviral expression. We analyzed the vector integration sites in non-selected cells and in clones selected for the GFP expression. This selection led to overrepresentation of proviruses integrated in active transcription units, with particular accumulation in promoter-proximal areas. In parallel, we investigated the integration of vectors equipped with an anti-silencing CpG island core sequence. Such modification increased the frequency of stably expressing proviruses by one order. The modified vectors are also overrepresented in active transcription units, but stably expressed in distal parts of transcriptional units further away from promoters with marked accumulation in enhancers. These results suggest that integrated retroviruses subject to gradual epigenetic silencing during long-term cultivation. Among most genomic compartments, however, active promoters and enhancers protect the adjacent retroviruses from transcriptional silencing.
INTRODUCTION
Retroviruses are unique in that their replication requires integration of proviral DNA into the host cell genome. This recombination event proceeds autonomously via the virus-encoded integrase; however, the functional structure and epigenetic features of the host cell genome as well as host-encoded factors are also important determinants of retrovirus integration. First, most retroviruses preferentially target certain chromatin segments so that, genome-wide, the patterns of retrovirus integration are skewed against random distribution. Second, proviral transcription can be efficiently controlled by adjacent cellular DNA and the state of chromatin at the site of integration. In general, transcriptionally active chromatin is permissive to provirus expression, whereas heterochromatin and intergenic regions promote provirus silencing.
Murine leukemia virus (MLV) integrates near active enhancers and transcription start sites (TSS) (1–3) that are favorable for provirus expression. However, when MLV was used as a vector in gene therapy trials, such provirus insertions have turned out to be genotoxic and have been shown to be prone to transactivation of adjacent proto-oncogenes (4). This distinct integration preference is directed by tethering of the bromodomain and extraterminal (BET) protein family members with MLV integrase, and abrogation of this interaction resulted in retargeting of MLV integration (5,6). MLV integration sites are enriched within BET binding sites (6), which have been identified within actively transcribed euchromatin and characterized by specific posttranslational histone modifications (7). Human immunodeficiency virus type 1 (HIV-1) was extensively studied from this point of view and its integration has displayed a bias towards transcriptionally active genes, gene-rich and GC-rich chromosomal regions, but not TSSs and CpG islands (8–10). Similarly to MLV, this bias has been shown to depend on HIV-1 integrase binding at the C-terminal domain of the lens epithelium-derived growth factor/p75 (LEDGF/p75) (11–14). The genome-wide profile of LEDGF/p75 binding is comprised of active transcription units (TU) downstream of TSS marked by H3/H4 acetylation and H3K4 monomethylation and to a great part overlaps with sites enriched by HIV-1 integration (15). As a proof of concept, MLV or HIV-1 integration can be redirected by hybrid targeting factors (5,16,17).
Avian sarcoma/leukosis viruses (ASLV), in contrast to gammaretroviruses and lentiviruses, have integration profiles that are closer to random distribution. Several studies have demonstrated that these viruses exhibit only a slight preference of integration for TUs but not for TSSs (18–20). Although FACT complex has recently been described to interact with ASLV integrase, no targeting effect was observed, hence, the slight preference for TUs might just be the effect of easier accessibility of the preintegration complex to active chromatin (21). An extreme example of randomly dispersed retrovirus integration has been represented by the mouse mammary tumor virus (MMTV) (22), which has been the apparent advantage of a recently established vector system derived from MMTV (23).
The aforementioned virus-specific integration profiles have been observed in non-selected cell cultures. However, this data tells us little about provirus distribution under real conditions during retrovirus infection or retrovirus-mediated gene therapy. The outcome of infection or gene therapy can be strongly affected by provirus silencing and the selection of a limited number of proviruses at certain integration sites. For example, latent HIV-1 copies that survive in resting memory cells and other reservoirs after combined antiretroviral therapy (cART) can be reactivated, hence providing a source of residual virus replication during the prolonged cART (24,25) or viremia rebound after therapy withdrawal (26). The resulting viral populations, however, are genetically less variable than before cART suggesting oligoclonal expansion from a limited number of proviral integrations (26). Clonal expansion was recently shown in multiple patients by deep sequencing of provirus integration sites. Furthermore, these expanded proviral clones are frequently mapped to cell division- and cancer-associated loci, which could have led to homeostatic proliferation and contributed to the persistence of latent HIV-1 (27,28).
Similar clonal expansion driven by MLV vector integration near the LMO2 oncogenic locus resulted in insertional leukemogenesis and compromised experimental gene therapy of X-linked severe immunodeficiency (4). An example of the benign clonal expansion of a lentiviral vector integrated into the HMGA2 gene was documented in a study, which successfully implemented β-thalassemia gene therapy (29). Even ASLV-derived vectors concentrate within genes and TSS when selected for long-term expression in sarcomas, which are induced by v-src-transducing vectors (30) or in cell clones bearing transcriptionally active and non-silenced proviruses (31).
Transcriptional silencing of integrated retroviruses and retroviral vectors is a general phenomenon, which has been observed in many experimental settings and gene transfer clinical trials (32,33). There are two examples of extremely efficient provirus silencing: first, MLV is transcriptionally suppressed in mouse embryonic cells (34) by embryo-specific zinc finger factor (35) and mutations in cis elements of LTR and leader release this block (36). Second, ASLV as well as ASLV-derived vectors are prone to epigenetic provirus silencing when integrated into the heterologous mammalian genome (37,38) whereas chicken host cells are permissive to ASLV productive infection (39). This silencing is mediated by the cellular protein, Daxx (40,41) and executed by epigenetic mechanisms, namely DNA methylation and histone modifications (31,41,42). Furthermore, previous studies suggest that genomic and epigenetic features at the site of integration determine the transcriptional activity of respective proviruses. Hence, TSS regions of broadly expressed genes that are enriched with histone 3 trimethylated lysine 4 (H3K4me3) represent loci, which accumulate the active ASLV proviruses (30,31). Thus, experimental ASLV-derived vectors serve as extremely sensitive markers of a repressive (epi)genomic environment, which might help to uncover genomic regions permissive to stable expression of retroviral vectors and possibly other de novo integrated elements.
The aim of this study was to investigate the epigenetic marks within the integration sites of long-term active proviruses. Therefore, we analyzed the retrovirus integration sites from single-cell clones selected for long-term provirus expression. This approach combines the clonal expansion with provirus silencing, eliminates the initial integration preference, and samples the host genome sequences facilitating stable retrovirus expression. Specifically, we employed replication-deficient ASLV-derived vectors with the GFP fluorescence reporter. In addition to the basic, silencing-prone vector, we also constructed a modified vector containing a CpG island core sequence that confers resistance to position effects and provides protection from de novo DNA methylation (43,44). The correlation between the reporter activity of both vectors as well as the characteristics of their integration sites suggests that stably active proviruses are preferentially found around TSSs of transcribed genes that are enriched in histone modifications, which in turn support transcription. The presence of a CpG island core sequence within the retroviral LTR releases dependence on TSS and partly protects proviruses from position effects.
MATERIALS AND METHODS
Construction of the retroviral vector
Construction of the pAG plasmid used for the AG retroviral vector propagation was described previously (31). pAG-2IE for AG-2IE vector propagation was derived from pAG by de novo creation of a unique KasI restriction site in the U3 region of 3′LTR (position –89 respective to the transcription start site) using the Transformer Site-Directed Mutagenesis Kit (Clontech). A tandem of two IEs from the hamster aprt CpG island was amplified from the previously described pRNIG2–2IE vector (44) and inserted into the KasI restriction site.
Cell culture and virus propagation
The packaging AviPack cell line (30) was maintained in D-MEM/F12 medium (Sigma) supplemented with 5% newborn calf serum, 5% fetal calf serum, 1% chicken serum (all Gibco BRL) and penicillin/streptomycin (100 mg/ml each, Sigma) in a 3% CO2 atmosphere at 37°C. The K562 human myeloid lymphoblastoma cell line was maintained in RPMI 1640 supplemented with 5% newborn calf serum, 5% fetal calf serum (all Gibco BRL) and penicillin/streptomycin (100 mg/ml each, Sigma) in a 5% CO2 atmosphere at 37°C. The AviPack packaging system was utilized for the virus propagation and pseudotyping with vesicular stomatitis virus protein G (VSV-G) as described in (30). Briefly, 107 AviPack cells plated on a 150 mm Petri dish were cultured and cotransfected with 50 μg of pAG3 and 10 μg of pVSV-G (Clontech) plasmids by calcium phosphate precipitation 24 h after plating. Fresh culture medium supplemented with 100 mM glucose was added 24 hours post transfection and viral stocks were collected at 48 and 72 h post transfection. These viral stocks were clarified by centrifugation at 200 × g for 10 min at 4°C, supernatants were collected and centrifuged at 23 000 rpm for 150 min at 4°C in a SW28 rotor, Beckman Optima100 (Beckman). The pellet was resuspended in medium containing 5% newborn calf serum, frozen and stored in –80°C. Titration of the infectious virus stock was performed by its serial dilution and subsequent infection of DF-1 cells. Two days post infection (dpi), the number of GFP-positive cells or cell clusters was counted. The titrated stock was used for infection of K562 cells.
Infection and subcloning of K562 cells
106 cells of the K562 cell line were collected and infected with the AG or AG-2IE replication-deficient retroviral vectors at MOI < 0.01. Prior to infection, viral stocks were passed through a 0.2 μm SFCA filter (Corning) and 600 μl of the suspension was applied and allowed to adsorb for 40 min at room temperature. After adsorption, 12 ml of fresh medium was added and cells were cultured at 37°C and 5% CO2. Three dpi, the percentage of GFP-positive cells was analyzed by flow cytometry and GFP-positive cells were sorted in a single-cell sort mode with an Influx cell sorter (Becton-Dickinson) into 96-well tissue culture plates to obtain single-cell clones. Expanded clones were subcultured and the percentage of GFP-positive cells was assessed at one-week intervals with an LSR II cytometer (Becton-Dickinson).
Cloning and sequencing of provirus integration sites
The provirus-cell DNA junction sequences were amplified using the splinkerette-PCR method (31,45). Genomic DNA was isolated by phenol–chloroform extraction from individual clones and cleaved with either DpnII or MseI restriction enzymes. The restriction fragments were ligated overnight at 15°C with a 10-fold molar excess of adaptors formed by annealing of HMspAa and HMspBb-Sau3AI or HMspBb-MseI oligonucleotides complementary to the particular cleavage site of the enzyme used for genomic DNA digestion. The ligation products were subsequently cleaved with Bsu36I to destroy undesirable products of adaptor ligation to the 3′LTRs. The resulting mixture of fragments was then purified by a High Pure PCR Cleanup Kit (Roche) and used as a template for nested PCR with primers specific for the retrovirus LTR and the splinkerette adaptor. Primary PCR was performed with primers Splink1 and spPCR-AG3-R or spAG3–2IEDD-R as follows: 94°C for 3 min, 2 cycles of 94°C 15 s, 68°C 30 s, 72°C 2 min and 31 cycles of 94°C 15 s, 62°C 30 s, 72°C 2 min and final polymerization 72°C for 5 min. The secondary PCR used primers Splink2 and spinPCR-AG3-R or spinAG3–2IEDD-R with the program setting: 94°C 3 min, 30 cycles of 94°C 15 s, 60°C 30 s, 72°C 2 min and final 72°C 5 min. The specific PCR products were sequenced and the resulting sequences adjacent to the 5′LTR were aligned to the Human Genome assembly version hg19.
Mapping and characterization of provirus integration sites
All junction sequences containing the end of 5′LTR and the unique cellular DNA sequence obtained from the splinkerette PCR were mapped to February 2009 human genome assembly (hg19) using BLAT from the UCSC Genome Browser website (http://genome.ucsc.edu/). Genomic coordinates of the LTR-proximal nucleotide of the obtained genomic sequences with unique score were considered as position of the integration sites. Further analysis of genomic features associated with the integration sites were performed with datasets obtained from UCSC golden path (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/) using MySQL Workbench 6.2 software.
Random integrations
As a control of random targeting of features for the low number of integration sites, we generated a set of 200 biologically unbiased genomic positions. Human chromosomes were virtually joined (from chromosome 1 to chromosome X) and 200 random positions in range 1–3 031 042 417 (chromosome Y was omitted) were generated. Genomic coordinates were then obtained by the mapping of random positions to chromosomal joint-genomic positions.
Transcriptional units
All integrations mapped into the RefSeq Genes were considered as intragenic. The absolute distance to TSS marks the distance to the closest TSS in the RefSeq Genes track. In the relative distance to TSS, the distance of intergenic integrations to TSS equals absolute distance to TSS. For integrations inside RefSeq Genes, the distance to the nearest TSS of a particular RefSeq Gene targeted by the integration was calculated.
RNA-seq data analysis
Seven RNA-seq datasets for the K562 cell line (ERR310212, SRR090233, SRR346063, SRR521457_1, SRR644784, SRR901899 and SRR901900) were obtained from Sequence Read Archive (SRA) at the NCBI website (http://www.ncbi.nlm.nih.gov/Traces/sra/). Reads were mapped to human genes using CLC Genomics Workbench 6.5.1 with default settings. RefSeq Genes were classified into groups by means of RPKM, with the NA group containing RefSeq Genes with the mean RPKM lower than 1 and RefSeq Genes with no match in the CLC Genomics Workbench 6.5.1 database and Q1–Q4 groups containing RefSeq Genes with the mean RPKM equal to/higher than 1 classified into the mean RPKM quartile groups with Q1 being the first quartile containing RefSeq Genes with the lowest RPKM.
Histone modification peaks
Histone modification peak datasets for the K562 cell line from the Broad Histone track were obtained from UCSC golden path. Peaks with signals higher than the median of the signal of a particular histone modification peak dataset were selected for further analysis. The distance to the nearest peak of the particular modification was calculated for each integration.
Chromatin segments
Chromatin segments for the K562 cell line were obtained from the UCSC Genome Segments track. Segments were grouped by the itemRgb field and the frequency of integrations into the grouped segments was calculated. For global analysis, we merged related segments to groups of active chromatin (Active Promoter, Promoter Flanking, Candidate Strong Enhancer, Candidate Weak Enhancer, Transcription Associated, Low Activity Proximal to Active States) and active regulatory elements (Active Promoter, Promoter Flanking, Candidate Strong Enhancer, Candidate Weak Enhancer).
Statistics
R software was used for statistical analysis. All statistical tests were performed at default settings.
RESULTS
The rate and kinetics of provirus silencing of ALSV-derived vectors in human cells
First, we quantitatively described the rate and kinetics of provirus silencing observed after infection of mammalian cells with ASLV-derived vectors. We compared the silencing of two replication-defective, GFP-transducing vectors derived from ASLV. Both vectors, AG and AG-2IE, differ solely by insertion of two internal elements (IE) from a CpG island into the U3 segment of AG-2IE (Figure 1A). The IE comprises the core sequence of CpG island from the hamster adenine phosphoribosyltransferase gene along with a tandem of high-affinity Sp1 binding sites. Each IE is 142 bp in length and comprises eight CpG dinucleotides (Figure 1B); the effect of this insertion has been previously described (44). The silencing of vectors was assessed in the human cell line K562, an ENCODE Tier1 cell line that has provided researchers with extensive data for subsequent analyses and represents a valuable gene therapy model.
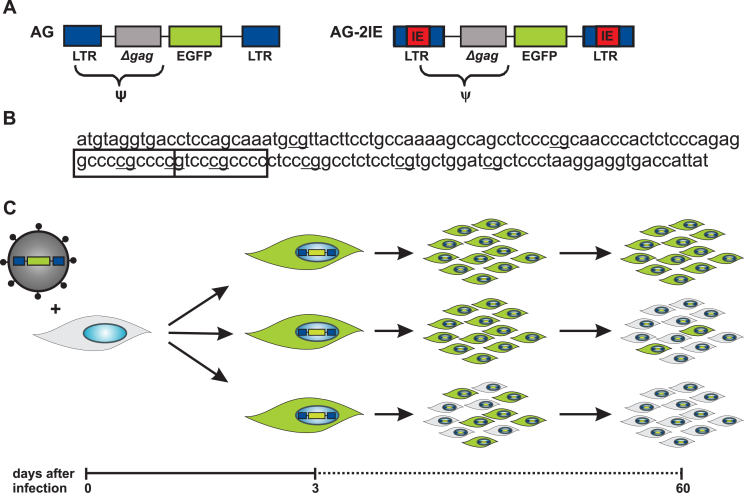
Figure 1.
Schematic representation of the experimental approach. (A) The scheme of replication-defective ASLV-derived retroviral vectors AG and AG-2IE used in this study. Both vectors consist of long terminal repeats (LTR, blue), part of the gag gene (Δgag, gray) and EGFP reporter gene (green). Tandems of CpG island core elements from hamster aprt gene (IE elements, red) are inserted in both LTRs of the AG-2IE vector. Encapsidation signal (Ψ) spans part of LTR and Δgag. (B) The nucleotide sequence of IE element with depicted CpG dinucleotides (bold) and highly affine Sp1 binding sites (boxed). (C) Human K562 cell line was infected with VSV-G pseudotyped vectors at low MOI (<0.01). Three dpi, GFP-positive cells were single-sorted and cellular clones were established. The expression of GFP in clones was measured weekly by flow cytometry up to 60 dpi. Three types of clones with different levels of provirus silencing are shown. The stable clone with cells maintaining the GFP expression over the period of 60 days is shown at the top.
K562 cells were infected with VSV-G-pseudotyped vectors at a low multiplicity of infection (MOI < 0.01), which was necessary to minimize the probability of multiple proviral integrations per cell. GFP-positive cells with transcriptionally active proviruses were separated by fluorescence-activated cell sorting (FACS) three dpi and single-cell clones were established. After expansion, the clones were cultivated for two months or longer with a FACS count of GFP-positive cells at 30 and 60 days (Figure 1C). Two sets of clones, 2128 clones, which contained AG proviruses and 558 clones with AG-2IE proviruses, were expanded and cultivated. Clonal analysis of the reporter expression confirmed the high rate of provirus silencing in the AG vector. At 60 dpi, only 3.5% (74 of 2128) of AG clones maintained stable provirus expression (at least 90% of cells in each clone were GFP-positive cells), whereas the majority of clones tended to undergo rapid silencing (Figure 2A). This behavior was in sharp contrast to much less effective and slow provirus silencing in AG-2IE clones with 29% (164 of 558) of the stable clones at 60 dpi. (Figure 2A). The striking contrast of silencing in AG and AG-2IE vectors was apparent already by 30 dpi (Figure 2A).
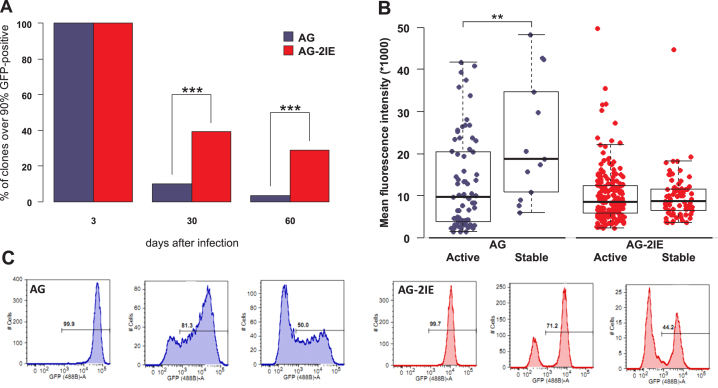
Figure 2.
Stability of expression and silencing of provirus transcription in single-cell clones. (A) The percentages of stable clones with ≥ 90% of GFP-positive cells 3, 30 and 60 dpi are shown for AG and AG-2IE virus vectors. Three dpi all clone-establishing cells were GFP-positive. In total, 2,128 and 558 clones with AG and AG-IE proviruses were established, respectively. Thirty dpi 10% of AG (210) and 39% of AG-2IE (218) clones kept stable expression of GFP. Sixty dpi 3.5% (74) and 29% (174) of AG and AG-2IE clones kept stable expression. (B) Mean fluorescence intensities (MFI) of AG and AG-2IE clones. Each dot represents one clone. Clones were established by sorting GFP-positive cells. MFIs of the clones with at least 20% GFP-positive cells at 21 dpi (active) and clones with at least 90% GFP-positive cells (stable) at 30 dpi were compared. (C) Histograms of GFP expression of representative clones. Silenced clones of AG show gradual decrease of GFP intensity, while AG-2IE clones silence with a bimodal profile.
In previous studies (43,44), we demonstrated that the anti-silencing effect of IE insertion lies in protection from DNA methylation. This insertion, however, did not increase LTR-driven expression as shown by the reporter activity assayed in multiple clones (43). Here, we compared the GFP fluorescence intensity in stable clones from both AG and AG-2IE groups. Stable clones, which contained AG proviruses, exhibited higher GFP fluorescence intensity in comparison with non-selected GFP-positive cells at the beginning of clonal expansion (Figure 2B). AG-2IE proviruses, however, exhibited lower variability and approximately the same fluorescence intensity in stable clones and non-selected cells (Figure 2B). This can be explained by the more autonomous expression of AG-2IE proviruses, which was less influenced by position effects. In contrast, the stability of expression of AG proviruses correlated with high level of expression, probably because this expression was mostly determined by position effects and only proviruses fortuitously integrated into the strongly supportive genetic environments could be selected as transcriptionally stable. Our clonal analysis independently suggested the anti-silencing but not transcription-enhancing effect of the IE insertion.
The higher autonomy of AG-2IE proviruses is also supported by the course of GFP expression silencing. Most AG clones proceeded to silencing through a gradual decrease of GFP intensity, and in the transient state, there was broad variability of fluorescence intensities in individual cells (Figure 2C). In contrast, most transcriptionally active AG-2IE proviruses in unstable clones simply switched to a silenced state and the distribution of fluorescence intensities was bimodal without intermediate states (Figure 2C).
We conclude that the insertion of the CpG island core sequence partly protected retroviral vectors from provirus silencing and releases the dependence of vectors on the local position effects towards vector expression.
Genome-wide mapping and characterization of provirus integration sites
We isolated genomic DNA from individual cellular clones with stably active AG or AG-2IE proviruses, digested them with specific restriction enzymes in order to release the proviruses together with flanking genomic sequences, and used them for cloning the junction DNA at the sites of vector integration utilizing the splinkerette PCR technique. In addition to individual clones that contained single proviral integrations, we cloned the proviral integration sites in en masse splinkerette PCR from cells that were infected with either AG or AG-2IE vectors without selection for GFP activity (non-selected control) or sorted for GFP-positivity 3 dpi (active 3 dpi controls). We sequenced the junction DNA fragments and identified integration sites using BLAT in the human genomic assembly GRCh37, version hg19. In total, after neglecting the small number of equivocal integrations, we identified 90 non-selected sites of AG proviruses and 82 non-selected sites of AG-2IE proviruses, 124 sites of active 3 dpi AG and 63 AG-2IE proviruses, and, finally, 46 sites of stable AG and 58 AG-2IE proviruses (Supplementary Table S1). For better comparison with previous studies, we generated 200 random integration sites by in silico targeting the human genome. This set of integration sites has been used in all subsequent analyses as a random control. The comparison of random with non-selected set of integrations also defines the possible bias of the technique given by the distribution of restriction recognition sequences. We further analyzed provirus integrations from the point of view of targeting TUs, transcriptionally active TUs in K562 cells, and TSSs.
We observed that the non-selected proviruses AG and AG-2IE had integrated within TUs at 57% and 48%, respectively, which is slightly higher than the 39% that was observed in random integration sites, corroborating the slight preference of ASLV integrase for genes as described previously in the studies of Narezkina et al (19) and Barr et al. (20). In the case of 3 dpi, the percentage of active proviruses found in TUs had increased to 65% and 64% for AG and AG-2IE proviruses, respectively. The enrichment of transcriptionally stable proviruses within TUs at 60 dpi was even higher, 74% and 79% for AG and AG-2IE proviruses, respectively (Figure 3A, Supplementary Table S1). This data demonstrates that insertion of the IE element into LTR does not influence integration preference and long-term selection of transcriptionally active proviruses increases the rate of genic/intergenic integration. We can assume that integration into TU increases the chance of the provirus to be transcriptionally active over time and resistant to epigenetic silencing. We also analyzed the proportion of insertions oriented in sense or anti-sense to the transcription of targeted TUs (Supplementary Figure S1). Although approximately equal at the beginning of the experiment, the selection for transcriptional activity of AG proviruses led to a slight but statistically insignificant preponderance of sense integrations. AG-2IE proviral integrations displayed approximately the same ratio of both orientations.
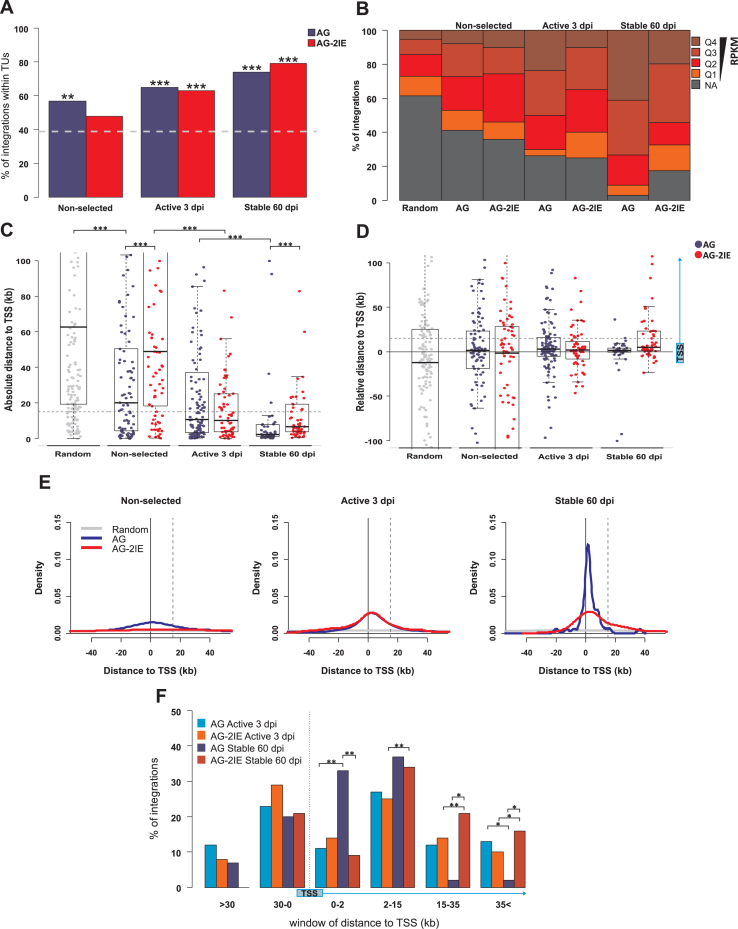
Figure 3.
Genomic features of integration sites of stably expressed proviruses. Four groups of integration sites were created representing the stages of selection for the expression stability presented in the paper: Random (200), Non-selected (90 AG, 82 AG-2IE), Active 3 dpi (124 AG, 63 AG-2IE) and Stable 60 dpi (46 AG, 58 AG-2IE). (A) The proportion of provirus integration into TUs represented by RefSeq Genes in the sets of AG proviruses and AG-2IE proviruses. The dashed line represents the percentage (39%) of TU targeting in the set of in silico generated random integration sites. (B) Frequency of proviruses integrated in TUs separated into categories by RPKM. Four quartiles of active TUs with Q4 being the most expressed group. NA represents the TUs with no detected or very low activity in the K562 cell line (RPKM < 1). (C) Absolute distance of proviruses to the closest transcriptional start site (TSS) of the TUs. Asterisks mark the P-value of Wilcoxon–Mann–Whitney Rank Sum Test. (D) Relative distance to TSS regarding the distribution of proviruses upstream and downstream to TSS. Positive values mark the distance to the nearest TSS of targeted TUs. Dashed line represents the distance of 15 kb inside TUs. (E) Density plots of the distance to TSS. Positive values mark the distance to TSS of proviruses inside TUs. Dashed line represents the distance of 15 kb inside TUs. (F) Barplots representing frequency of proviruses integrated within windows of distance to the TSS. Asterisks mark the P-value of Fisher's Exact Test for Count Data. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we analyzed the influence of transcription of targeted TUs. We combined transcription data of all TUs targeted by AG and AG-2IE proviruses from publicly available RNAseq data, which was retrieved from the Sequence Read Archive (SRA) for the K562 cell line. According to reads per kilobase per million (RPKM), TUs were divided into five groups at transcriptional level—a non-active (NA) group and four quartiles of active TUs (Q1–Q4) (Figure 3B). In the set of non-selected integration sites, the integrations into transcriptionally active TUs prevailed, which was in accordance with previous findings (18–20), showing preferential integration of retroviruses into active chromatin. We did not observe any striking difference between AG and AG-2IE proviruses. After short-term selection of active proviruses (active expression 3 dpi), the prevalence of transcriptionally active targeted TUs increased to ∼70%. Among the AG proviruses, there was an increase in integrations into Q4, the TUs with the strongest transcription intensity. In contrast, integrations into Q1, the group comprising weakly transcribed TUs, were underrepresented. This effect of short-term selection was not observed in AG-2IE proviruses (Figure 3B). Long-term selection for stable expression of AG proviruses led to strong overrepresentation of transcriptionally active TUs, particularly those of Q4 and Q3. Only 2% of stable AG proviruses resided in non-transcribed TUs and 40% in Q4 TUs. In summary, we have demonstrated that ongoing selection for transcriptional activity of proviruses leads to increased representation of proviruses integrated into transcribed TUs. Long-term stable proviruses are found almost exclusively in the transcribed TUs, particularly in those with highest transcription levels. This can be explained by a protective anti-silencing effect of the genomic environment in transcriptionally active TUs. Insertion of IE elements releases this dependence, in part, and some stable AG-2IE proviruses can also be found in non-transcribed or weakly transcribed TUs.
The epigenetic environment varies along the TUs, which may influence the stability of the provirus expression. Integration close to TSSs has already been documented for MLV (1) and correlate with the expression of ASLV proviruses (30,31). We therefore analyzed the position of integration sites within the targeted genes and in relation to the adjacent TSS in our set of AG and AG-2IE proviruses. First, we analyzed the absolute distance to the closest TSS (Figure 3C). Short-term selection of transcriptionally active proviruses concentrated integrations around the TSSs with the median distance around 10 kb in both AG and AG-2IE proviruses. After long-term selection, the stably expressed AG proviruses were found closer to TSSs with a median distance of 2 kb (P = 8.2e–06, Wilcoxon–Mann–Whitney Rank Sum Test). AG-2IE stably expressed proviruses were integrated at a median distance of 6.5 kb from TSSs (Figure 3C), resembling the integration pattern of shortly selected proviruses (P = 0.35, Wilcoxon–Mann–Whitney Rank Sum Test). Taking into account the distribution of integrations along targeted TUs, we calculated the relative distance to TSS (Figure 3D). Unlike the absolute distance to TSS, the relative distance indicates the distance to a particular TSS belonging to the targeted TU for intra-TU integrations. For inter-TU integrations, the relative distance is equal to the absolute distance to TSS. The relative distance to TSS showed that with further positive selection of expression, proviruses were concentrated around TSS with a mild bias for integrations within TUs downstream to TSSs. This bias is most striking for AG stably active proviruses (Figure 3D and E), where 70% of all integrations are found within 15 kb downstream of TSS from which one third of the proviruses (33%) are integrated within 2 kb downstream from TSS (Figure 3F). On the other hand, AG-2IE stably expressed proviruses that have spread into more distal parts downstream from TSS resembled the integration pattern that was observed with shortly-selected proviruses. We conclude that proviruses selected for stable transcription are predominantly found close to TSSs. Obviously, the vicinity to TSSs is favorable for provirus transcription and protects the proviruses from transcriptional silencing. Proviruses, which are equipped with protective IEs are less dependent on the need to be located within close proximity of TSSs.
CpG islands are an important part of the genomic and transcriptional landscape of vertebrate genomes. We analyzed the distribution of proviral integrations in relation to the closest CpG islands in a similar way as TSSs (Supplementary Figure S1). The random in silico integrations displayed a longer absolute distance to CpG islands than to TSSs (medians of 104 and 62 kb, respectively), which reflected the fact that CpG islands were less frequent in the genome. Furthermore, only ca. 60% of promoters, mostly promoters of housekeeping genes, which exhibit constitutive expression, are equipped with CpG islands (46). The distance to the CpG islands showed a pattern similar to that observed with distances to TSS, i.e., with further selection of expressional stability the proviruses of both vectors were found closer to the CpG island. The correspondence of the distances to TSS and CpG islands shows that most of TSS that are located close to active and stably active proviruses are associated with CpG islands (Supplementary Figure S1).
Active histone modifications at the sites of integration in stably active proviruses
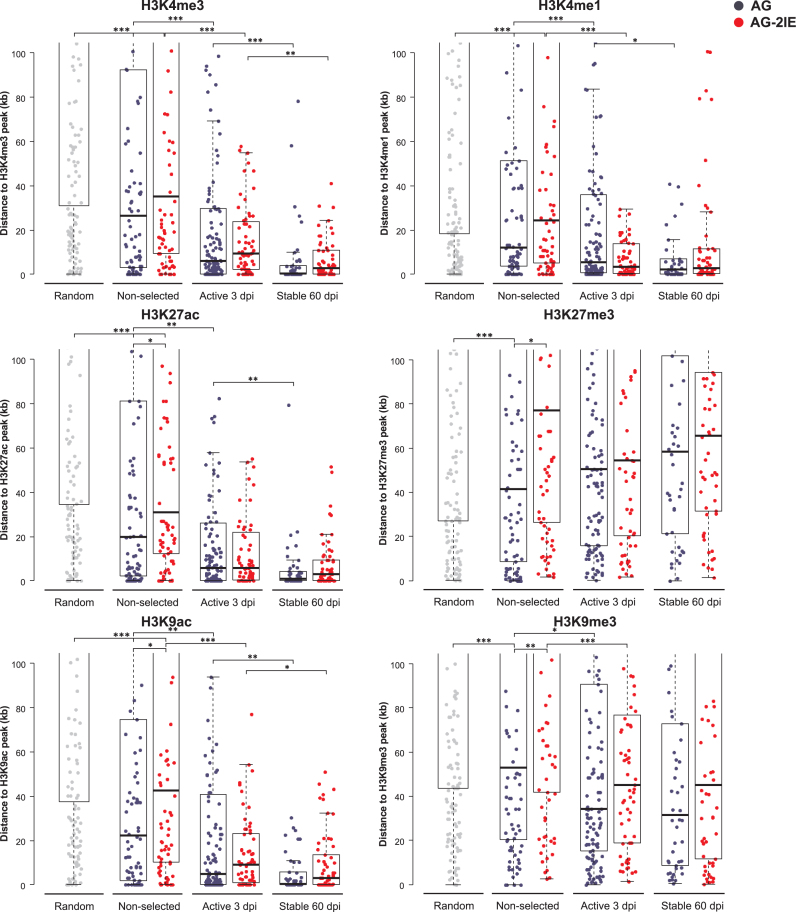
As described previously (31), active proviruses are overrepresented in chromatin regions which are enriched in trimethylation of the fourth lysine of histone H3 (H3K4me3). To obtain better insight into the epigenomic landscape of provirus integration and expression, we analyzed the distribution of our proviral integrations with respect to their distance to the peaks of different histone modifications. Because we obtained the epigenomic data from available database and not by experimental analysis of individual clones, we, technically speaking, correlate the transcriptional status of provirus with respective preintegration site characteristics in intact K562 cells. In Figure 4, we present results of analysis of histone modifications that are associated with both transcriptional activity and suppression, H3K4me3, H3K4me1, H3K9ac, H3K27ac, and H3K9me3, H3K27me3, respectively. As expected, transcriptionally active proviruses were found closer to peaks of H3K4me3, H3K4me1, H3K9ac, H3K27ac than to the peaks of H3K9me3, H3K27me3. The median distance of the AG active proviruses to the peaks of H3K4me3, H3K4me1, H3K9ac, H3K27ac ranged from 5 to 6 kb. Stable AG proviruses were found in close proximity to these peaks with the median distances between from 0.3 to 2 kb. The most significant shift of median distance was observed for the H3K4me3 mark (P = 1.3e–04, Wilcoxon-Mann–Whitney Rank Sum Test). The AG-2IE proviruses integrated slightly more distally to H3K4me3, H3K4me1, H3K9ac, H3K27ac peaks and maintained a more relaxed integration pattern even after long-term selection for transcriptional activity. The median distance of stable AG-2IE proviruses to those activation marks was around 3 kb. The peaks of suppressive histone modifications H3K9me3, H3K27me3 were found at a median distance of 40–50 kb from the AG proviral integrations and the long-term selection for transcriptional activity did not lead to any accumulation of proviruses closer to the H3K9me3, H3K27me3 peaks.
Figure 4.
Distance of proviruses to the peaks of histone modifications. Each dot represents a single provirus. The distance is measured as absolute distance to the nearest peak of a histone modification. More histone modifications evaluated in the study are depicted in Supplementary Figure S2. Asterisks mark the P-value of Wilcoxon–Mann-Whitney Rank Sum Test. *P < 0.05, **P < 0.01, ***P < 0.001.
A few more examples of correlation between histone modification enrichment and proviral integration are shown in Supplementary Figure S2. Dimethylation of lysine 79 in the H3 histone molecule (H3K79me2) is a lateral modification which is usually associated with transcriptional activation during cell differentiation (47) and aberrant gene expression in cancer (48). Although relatively non-selective for this histone modification at integration, after long-term selection for transcriptional activity, the AG and AG-2IE proviruses accumulated at a median distance to H3K79me2 peaks of 0 and 5 kb, respectively. Dimethylation of the fourth lysine in the H3 histone molecule (H3K4me2) is enriched in active enhancers (49) and transcriptionally active proviruses accumulate close to H3K4me2 peaks.
Other epigenomic features that correlate with transcription are histone isoforms, some of which are also enriched at specific and narrow genomic loci. In this study, we compared the distribution of proviral integrations and peaks of H2A.Z enrichment. H2A.Z has been found in association with active TSSs and enhancers occupied by various transcription factors (50). Both AG and AG-2IE proviruses integrated with a slight preference for H2A.Z enrichment at a mean distance of ca. 20 kb. Even short-term selection for proviral transcriptional activity accumulated proviral integrations of both vectors close to H2A.Z peaks (median of ca. 6 kb), but no decrease in distance was observed with further selection of expressional stability (Supplementary Figure S3). This suggests that not only integration within the proximity of H2A.Z-enriched areas might be important for the transcriptional activity of proviruses of either vector, but other features are important for the long-term transcriptional stability.
Proviral integration and transcriptional stability in functional chromatin segments
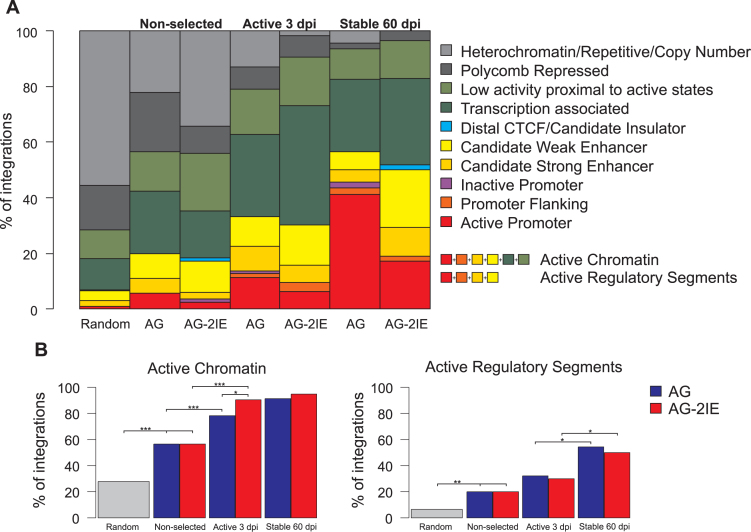
The integrative combination of epigenetic marks has been used for genome-wide annotation of functional chromatin states and non-coding functional elements in the human genome across multiple cell types. Two independent chromatin state annotation algorithms, ChromHMM (51,52) and Segway (53), based mostly on the results from ChIPseq assays, served for ENCODE-wide annotation of functional chromatin segments and regulatory elements (54). To describe proviral integration sites with regard to the function of their chromatin regions, we calculated the percentages of integrations within chromatin segments categorized by ChromHMM (Figure 5) and Segway (Supplementary Figure S4) databases.
Figure 5.
Integration into ChromHMM functional chromatin segments. (A) Barplots represent the percentage of proviruses integrated in one of ten types of chromatin segments. (B) Frequencies of provirus integrations into the merged active chromatin segments or merged active regulatory elements. Asterisks mark the P-value of Fisher′s Exact Test for Count Data: *P < 0.05, **P < 0.01, ***P < 0.001.
The randomly generated integration sites showed the approximate proportion of certain functional chromatin segments in the human genome with the majority being non-transcribed heterochromatin or polycomb-repressed chromatin. The proportion of targeted promoters and enhancers was found to be <10% in the random set of integration sites (Figure 5A). The experimentally-determined non-selected integration of both AG and AG-2IE proviruses preferred the merged active chromatin segments at the expense of non-transcribed ones (P = 6.1e–06 and P = 6.4e–05 for AG and AG-2IE, respectively, Fisher's Exact Test for Count Data, Figure 5B). Even the short-term selection of transcribed proviruses further increased the proportion of active chromatin among the targeted segments (P = 9.5e–04 and P = 2.7e–06 for AG and AG-2IE, respectively. Fisher's Exact Test for Count Data). Selection for stably active proviruses did not lead to any increase of proviruses located in active chromatin segments, but resulted in selection of proviruses in active regulatory segments, i.e. promoters and enhancers, where we observed more than 50% of stable integrations (P = 1.9e–04 and P = 4.0e–02 for AG and AG-2IE, respectively, Fisher's Exact Test for Count Data). The most striking observation was the enrichment of transcriptionally stable AG proviruses in promoters (ca. 40%, P = 4.0e–05, Fisher's Exact Test for Count Data). The described trends towards overrepresentation of transcribed chromatin and promoters/enhancers during the selection of transcribed proviruses were similar when the Segway database was used for chromatin segment categorization (Supplementary Figure S4). Here, the proportion of promoters/enhancers reached 70% after long-term selection for both AG and AG-2IE. In contrast to AG proviruses enriched in promoter-flanking segments, AG-2IE proviruses were found with particularly high frequency in strong enhancers (P = 2.6e–03, Fisher's Exact Test for Count Data).
DISCUSSION
The preference of retroviruses to integrate within certain genomic compartments such as transcriptionally active genes and promoters have been recently reviewed (55) and mechanistically explained by virus-specific cell-encoded factors that direct preintegration complex contacts with chromatin. The interplay of retroviral integrases and cellular factors is fixed by evolution and suggests that such integration bias is beneficial for retrovirus propagation. Intuitively, it might give the provirus a higher probability of access to transcriptional machinery in comparison to a random integration pattern [see also (56)]. However, the role of integration site selection in proviral expression is still not fully understood. Particularly, the stability of provirus expression in a given genomic context has not been studied in long-term experiments that involve selection. We combined retrovirus integration and selection using in silico genomic and epigenomic analysis of provirus integration sites in multiple parallel single-cell clones which were long-term selected for provirus expression. We have used this strategy in our previous work and described the accumulation of active proviruses in H3K4me3-enriched parts of genes (31).
Silencing of proviral expression is an important issue in the use of retroviral vectors and retroviral latency. For example, the reservoir of latently HIV-infected CD4+ T cells is the major obstacle to an HIV cure (57). Using the clonal approach presented in this study we observed extremely efficient silencing of the ASLV-based vector in human cells with only few percent of proviruses displaying strong and unsilenced expression from 3 to 60 dpi. Insertion of two copies of the aprt housekeeping gene CpG island IE into the LTR of the ASLV vector dramatically decreased the silencing intensity. This vector modification was designed in accordance with the well-defined anti-CpG methylation effect of CpG islands along with previously reported stable retroviral expression, even in a non-permissive genomic environment (44).
ASLV integration, in contrast to MLV or HIV, has not been associated with any significant preference for genomic features such as TUs or TSSs (18). Even in our work with a low number of integration sites, mild overrepresentation of proviral integrations close to the peaks of activating histone modifications and within active chromatin segments is apparent without selection for transcriptional activity (Figures 4 and 5). The next integration site selection is rapid; even short-term selection for transcriptional activity means substantial accumulation of proviruses close to the TSSs and sites enriched in H3K4me1, H3K4me3, H3K27ac and H3K9ac. The prolonged selection further highlights these trends. To our knowledge, this is the first observation of a significant integration bias of ASLV and further studies with larger sets of integration sites could help to validate our results. We can only speculate about the cause of this integration bias. In contrast to MLV and HIV, ASLVs have not been referred to as using cellular factors for preferential provirus integration into active chromatin. The first factor interacting with ALV integrase and specifically mediating ALV integration is the FACT protein complex (21). This complex stimulates the ALV integration activity in vitro and increases the frequency of provirus integration in infected cells. Although any FACT-dependent shift in ALV integration preference remains to be described, FACT has been characterized previously as histone chaperone acting in transcript elongation (58); hence its tethering to active chromatin is therefore understandable.
The presence of IE insertion in LTR relaxes the requirement for adjacent TSSs and enrichment in activating histone modifications. At the level of functional chromatin segments, we can see the accumulation of transcriptionally stable AG proviruses in promoters, whereas the modified proviruses AG-2IE preferentially accumulate in active enhancers. Nevertheless, if we merge both categories, the AG and AG-2IE vectors do not differ from each other with roughly 50% of proviruses accumulating in promoters and enhancers. Interestingly, when analyzing the genic and intergenic integrations separately, we observed no significant difference between AG and AG-2IE stably expressing proviruses integrated outside gene bodies. Conversely, distribution of stable AG-2IE proviruses integrated inside gene bodies differ from AG proviruses and exhibit a very flat distribution without strong preference for the vicinity of TSS. Such distribution strikingly resembles the distribution of AG proviruses which have integrated in DNMT3A–/– DNMT3B–/– cells (31), indicating the role of IE in the protection of proviruses from DNA methylation. The protective role of IE emerges in gene bodies, which have been shown to be the most densely methylated genomic compartment (59).
Differential distribution of stable AG and AG-2IE proviruses in gene bodies together with the lack of significant differences in distances of stable AG and AG-2IE proviruses to histone modifications marking mainly active TSSs and enhancers also suggest that the proximity of intragenic enhancers might play a role in the stability of AG-2IE proviral expression. This is supported by analysis of functional chromatin segments at the sites of integration. It is important to say that our analyses work with the linear genome sequence whereas enhancers can influence gene expression in the 3D genome. Therefore, the chromatin contact analysis could provide additional information. We, however, present significant evidence that the short-distance influence amenable to our ‘linear’ analysis correlate well with the proviral behavior. Second, we used exclusively in silico data on epigenetic marks at the site of integration. Provirus insertion, however, can affect the adjacent epigenetic landscape and we will analyze the actual epigenetic marks at individual proviruses in the future. We already provided an example of epigenetic events occurring at the site of proviral insertion and the stability of CpG methylation (60).
The epigenomic context of integrated retroviral vectors can be compared with distribution and transcriptional activity of endogenous retroviruses (ERV). Because of their abundance, insertional polymorphism, and different expression in various tissues, the murine ERVs of ETn/MusD and IAP families were studied from the point of view of ERV-induced heterochromatin spreading (61). ETn/MusD copies are less DNA methylated when near TSSs, but only in tissues where the adjacent gene is transcriptionally active (62). Furthemore, differences in DNA methylation of 5′ and 3′ LTRs reflect the vicinity to the nearby TSSs. The most striking parallel with our system is the presence of H3K4me3 enrichment in the flanking regions of non-methylated ERVs (62). Underrepresentation of these non-methylated copies indicates some kind of negative selection against methylated ERV insertions near genes, possibly because of the impact on host genes through heterochromatin spreading.
Our findings have important implications for the safety of gene therapy and genotoxicity of retroviral vectors. Such an approach not only demonstrates the possibility to protect vectors from repressive features at their integration site, but on the other hand, points to the accumulation of stably expressed vector integrations close to TSSs, i.e. in positions sensitive to genotoxic outcomes. The LTR modification just alleviates this trend. Our approach might also enable the discovery of highly repressive regions that would be non-permissive even for the protected vector.
CONCLUSIONS
We have provided a detailed analysis of retrovirus integration sites in single-cell clones long-term selected for the provirus transcriptional activity. Our analysis demonstrated that the simplified ASLV-based retroviral vectors integrate with mild preference for active chromatin and the subsequent selection leads to accumulation of proviruses integrated into transcriptionally active genes, close to TSSs, CpG islands, and regions enriched in active histone modifications H3K4me3, H3K4me1, H3K9ac and H3K27ac. Analyzing the functional chromatin elements defined as combinations of epigenetic marks, we found the proviruses with long-term transcriptional stability preferentially integrated within active promoters and enhancers. These trends are relaxed for vectors equipped with their own CpG island sequences, which are found at greater distance from TSS but still accumulate in enhancers. Collectively, these data suggest that active promoters and enhancers protect the adjacent retroviruses from transcriptional silencing.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Markéta Reinišová and Dana Kučerová for technical assistance and Daniel Elleder and Kateřina Trejbalová for helpful discussions and assistance with bioinformatic analyses. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Authors contributions: J.H. conceived the study and drafted the manuscript. F.S. constructed the vectors, performed most of the cell culture and FACS experiments, cloned the provirus integration sites, and analyzed the genomic data. DM cloned some of the provirus integration sites, analyzed the genomic data and developed the approaches for epigenomic analysis. M.A. contributed to the splinkerette cloning of integration sites at the beginning of the study. Both F.S. and D.M. helped to draft the manuscript. All authors carefully read and approved the final manuscript.
Footnotes
Present address: Miroslav Auxt, Pontificio Collegio Nepomuceno, Via Concordia 1, I-00183 Roma, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [14-34873S]; RVO [68378050]; NPU I [LO1419]. Funding for open access charge: Institute of Molecular Genetics, Czech Academy of Sciences, Prague, Czech Republic.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wu X., Li Y., Crise B., Burgess S.M.. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003; 300:1749–1751. [DOI] [PubMed] [Google Scholar]
- 2. De Ravin S.S., Su L., Theobald N., Choi U., Macpherson J.L., Poidinger M., Symonds G., Pond S.M., Ferris A.L., Hughes S.H. et al. Enhancers are major targets for murine leukemia virus vector integration. J. Virol. 2014; 88:4504–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LaFave M.C., Varshney G.K., Gildea D.E., Wolfsberg T.G., Baxevanis A.D., Burgess S.M.. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res. 2014; 42:4257–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008; 118:3132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Rijck J., de Kogel C., Demeulemeester J., Vets S., El Ashkar S., Malani N., Bushman F.D., Landuyt B., Husson S.J., Busschots K. et al. The BET family of proteins targets Moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013; 5:886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aiyer S., Swapna G.V.T., Malani N., Aramini J.M., Schneider W.M., Plumb M.R., Ghanem M., Larue R.C., Sharma A., Studamire B. et al. Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction. Nucleic Acids Res. 2014; 42:5917–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. LeRoy G., Chepelev I., DiMaggio P.A., Blanco M.A., Zee B.M., Zhao K., Garcia B.A.. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 2012; 13:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elleder D, Pavlíček A., Pačes J., Hejnar J.. Preferential integration of human immunodeficiency virus type 1 into genes, cytogenetic R bands and GC-rich DNA regions: insight from the human genome sequence. FEBS Lett. 2002; 517:285–286. [DOI] [PubMed] [Google Scholar]
- 9. Schroder A.R., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F.. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002; 110:521–529. [DOI] [PubMed] [Google Scholar]
- 10. Wang G.P., Ciuffi A., Leipzig J., Berry C.C., Bushman F.D.. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007; 17:1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z.. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003; 278:372–381. [DOI] [PubMed] [Google Scholar]
- 12. Cherepanov P., Devroe E., Silver P.A., Engelman A.. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004; 279:48883–48892. [DOI] [PubMed] [Google Scholar]
- 13. Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F.. A role for LEDGF/p75 in targeting HIV DNA integration. Nature Med. 2005; 11:1287–1289. [DOI] [PubMed] [Google Scholar]
- 14. Shun M.C., Raghavendra N.K., Vandegraaff N., Daigle J.E., Hughes S., Kellam P., Cherepanov P., Engelman A.. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007; 21:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Rijck J., Bartholomeeusen K., Ceulemans H., Debyser Z., Gijsbers R.. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 2010; 38:6135–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferris A.L., Wu X., Hughes C.M., Stewart C., Smith S.J., Milne T.A., Wang G.G., Shun M.C., Allis C.D., Engelman A.. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silvers R.M., Smith J.A., Schowalter M., Litwin S., Liang Z., Geary K., Daniel R.. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum. Gene Ther. 2010; 21:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitchell R.S., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J,R, Bushman F.D.. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004; 2:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Narezkina A., Taganov K.D., Litwin S., Stoyanova R., Hayashi J., Seeger C., Skalka A.M., Katz R.A.. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004; 78:11656–11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barr S.D., Leipzig J., Shinn P., Ecker J.R., Bushman F.D.. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 2005; 79:12035–12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winans S., Larue R.C., Abraham C.M., Shkriabai N., Skopp A., Winkler D., Kvaratskhelia M., Beemon K.L.. The FACT complex promotes avian leukosis virus DNA integration. J. Virol. 2017; 91:doi:10.1128/JVI.00082-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Faschinger A., Rouault F., Sollner J., Lukas A., Salmons B., Günzburg W.H., Indik S.. Mouse mammary tumor virus integration site selection in human and mouse genomes. J. Virol. 2008; 82:1360–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Konstantoulas C.J., Indik S.. Mouse mammary tumor virus-based vector transduces non-dividing cells, enters the nucleus via a TNPO3-independent pathway and integrates in a less biased fashion than other retroviruses. Retrovirology. 2014; 11:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bailey J.R., Sedaghat A.R., Kieffer T., Brennan T., Lee P.K., Wind-Rotolo M., Haggerty C.M., Kamireddi A.R., Liu Y., Lee J. et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+T cells. J. Virol. 2006; 80:6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kearney M.F., Spindler J., Shao W., Yu S., Anderson E.M., O'Shea A., Rehm C., Poethke C., Kovacs N., Mellors J.W. et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathogens. 2014; 10:e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Joos B., Fischer M., Kuster H., Pillai S.K., Wong J.K., Böni J., Hirschel B., Weber R., Trkola A., Günthard H.F. et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:16725–16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maldarelli F., Wu X., Su L., Simonetti F.R., Shao W., Hill S., Spindler J., Ferris A.L., Mellors J.W., Kearney M.F. et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014; 345:179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wagner T.A., McLaughlin S., Garg K., Cheung C.Y.K., Larsen B.B., Styrchak S., Huang H.C., Edlefsen P.T., Mullins J.I., Frenkel L.M.. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014; 345:570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010; 467:318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Plachy J., Kotab J., Divina P., Reinisova M., Senigl F., Hejnar J.. Proviruses selected for high and stable expression of transduced genes accumulate in broadly transcribed genome areas. J. Virol. 2010; 84:4204–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Senigl F., Auxt M., Hejnar J.. Transcriptional provirus silencing as a crosstalk of de novo DNA methylation and epigenomic features at the integration site. Nucleic Acids Res. 2012; 40:5298–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005; 16:1241–1246. [DOI] [PubMed] [Google Scholar]
- 33. Ellis J., Yao S.. Retrovirus silencing and vector design: relevance to normal and cancer stem cells?. Curr. Gene Ther. 2005; 5:367–373. [DOI] [PubMed] [Google Scholar]
- 34. Niwa O., Yokota Y., Ishida H., Sugahara T.. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983; 32:1105–1113. [DOI] [PubMed] [Google Scholar]
- 35. Wang C., Goff S.P.. Differential control of retrovirus silencing in embryonic cells by proteasomal regulation of the ZFP809 retroviral repressor. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Challita P.M., Skelton D., el-Khoueiry A., Yu X.J., Weinberg K., Kohn D.B.. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J. Virol. 1995; 69:748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Searle S., Gillespie D.A., Chiswell D.J., Wyke J.A.. Analysis of the variations in proviral cytosine methylation that accompany transformation and morphological reversion in a line of Rous sarcoma virus-infected Rat-1 cells. Nucleic Acids Res. 1984; 12:5193–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hejnar J., Svoboda J., Geryk J., Fincham V.J., Hak R.. High rate of morphological reversion in tumor cell line H-19 associated with permanent transcriptional suppression of the LTR, v-src, LTR provirus. Cell Growth Differ. 1994; 5:277–285. [PubMed] [Google Scholar]
- 39. Svoboda J., Hejnar J., Geryk J., Elleder D., Vernerova Z.. Retroviruses in foreign species and the problem of provirus silencing. Gene. 2000; 261:181–188. [DOI] [PubMed] [Google Scholar]
- 40. Poleshko A., Palagin I., Zhang R., Boimel P., Castagna C., Adams P.D., Skalka A.M., Katz R.A.. Identification of cellular proteins that maintain retroviral epigenetic silencing: evidence for an antiviral response. J. Virol. 2008; 82:2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shalginskikh N., Poleshko A., Skalka A.M., Katz R.A.. Retroviral DNA methylation and epigenetic repression are mediated by the antiviral host protein Daxx. J. Virol. 2013; 87:2137–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hejnar J., Plachy J., Geryk J., Machon O., Trejbalova K., Guntaka R.V., Svoboda J.. Inhibition of the Rous sarcoma virus long terminal repeat-driven transcription by in vitro methylation: different sensitivity in permissive chicken cells versus mammalian cells. Virology. 1999; 255:171–181. [DOI] [PubMed] [Google Scholar]
- 43. Hejnar J., Hajkova P., Plachy J., Elleder D., Stepanets V., Svoboda J.. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Senigl F., Plachy J., Hejnar J.. The core element of a CpG island protects avian sarcoma and leukosis virus-derived vectors from transcriptional silencing. J. Virol. 2008; 82:7818–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uren A.G., Mikkers H., Kool J., van der Weyden L., Lund A.H., Wilson C.H., Rance R., Jonkers J., van Lohuizen M., Berns A. et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 2009; 4:789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu J., He F., Hu S., Yu J.. On the nature of human housekeeping genes. Trends Genet. 2008; 24:481–484. [DOI] [PubMed] [Google Scholar]
- 47. Cattaneo P., Kunderfranco P., Greco C., Guffanti A., Stirparo G.G., Rusconi F., Rizzi R., Di Pasquale E., Locatelli S.L., Latronico M.V. et al. DOT1L-mediated H3K79me2 modification critically regulates gene expression during cardiomyocyte differentiation. Cell Death Differ. 2016; 23:555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deshpande A.J., Deshpande A., Sinha A.U., Chen L., Chang J., Cihan A., Fazio M., Chen C.W., Zhu N., Koche R et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014; 26:896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J.C., Ferrier P., Spicuglia S.. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011; 30:4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soboleva T.A., Nekrasov M., Pahwa A., Williams R., Huttley G.A., Tremethick D.J.. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat. Struct. Mol. Biol. 2012; 19:1076–1083. [DOI] [PubMed] [Google Scholar]
- 51. Ernst J., Kellis M.. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010; 28:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J.A., Noble W.S.. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012; 9:473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E. et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013; 41:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sultana T., Zamborlini A., Cristofari G., Lesage P.. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 2017; 18:292–308. [DOI] [PubMed] [Google Scholar]
- 56. Rouzine I.M., Raztoky B.S., Weinberger L.S.. Stochastic variability in HIV affects viral eradication. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13251–13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rasmussen T.A., Tolstrup M., Søgaard O.S.. Reversal of latency as part of a cure for HIV-1. Trends Microbiol. 2016; 24:90–97. [DOI] [PubMed] [Google Scholar]
- 58. Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D.. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998; 92:105–116. [DOI] [PubMed] [Google Scholar]
- 59. Suzuki M.M., Bird A.. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008; 9:465–476. [DOI] [PubMed] [Google Scholar]
- 60. Hejnar J., Elleder D., Hájková P., Walter J., Blažková J., Svoboda J.. Demethylation of host-cell DNA at the site of avian retrovirus integration. Biochem. Biophys. Res. Commun. 2003; 311:641–648. [DOI] [PubMed] [Google Scholar]
- 61. Rebollo R., Karimi M.M., Bilenky M., Gagnier L., Miceli-Royer K., Zhang Y., Goyal P., Keane T.M., Jones S., Hirst M. et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011; 7:e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rebollo R., Miceli-Rover K., Zhang Y., Farivar S., Gagnier L., Mager D.L.. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome Biol. 2012; 13:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.