ABSTRACT
Microbial communities are shaped by interactions among their constituent members. Some Gram-negative bacteria employ type VI secretion systems (T6SSs) to inject protein toxins into neighboring cells. These interactions have been theorized to affect the composition of host-associated microbiomes, but the role of T6SSs in the evolution of gut communities is not well understood. We report the discovery of two T6SSs and numerous T6SS-associated Rhs toxins within the gut bacteria of honey bees and bumble bees. We sequenced the genomes of 28 strains of Snodgrassella alvi, a characteristic bee gut microbe, and found tremendous variability in their Rhs toxin complements: altogether, these strains appear to encode hundreds of unique toxins. Some toxins are shared with Gilliamella apicola, a coresident gut symbiont, implicating horizontal gene transfer as a source of toxin diversity in the bee gut. We use data from a transposon mutagenesis screen to identify toxins with antibacterial function in the bee gut and validate the function and specificity of a subset of these toxin and immunity genes in Escherichia coli. Using transcriptome sequencing, we demonstrate that S. alvi T6SSs and associated toxins are upregulated in the gut environment. We find that S. alvi Rhs loci have a conserved architecture, consistent with the C-terminal displacement model of toxin diversification, with Rhs toxins, toxin fragments, and cognate immunity genes that are expressed and confer strong fitness effects in vivo. Our findings of T6SS activity and Rhs toxin diversity suggest that T6SS-mediated competition may be an important driver of coevolution within the bee gut microbiota.
KEYWORDS: Apis mellifera, Bombus, Rhs toxin, Snodgrassella alvi, coevolution
IMPORTANCE
The structure and composition of host-associated bacterial communities are of broad interest, because these communities affect host health. Bees have a simple, conserved gut microbiota, which provides an opportunity to explore interactions between species that have coevolved within their host over millions of years. This study examined the role of type VI secretion systems (T6SSs)—protein complexes used to deliver toxic proteins into bacterial competitors—within the bee gut microbiota. We identified two T6SSs and diverse T6SS-associated toxins in bacterial strains from bees. Expression of these genes is increased in bacteria in the bee gut, and toxin and immunity genes demonstrate antibacterial and protective functions, respectively, when expressed in Escherichia coli. Our results suggest that coevolution among bacterial species in the bee gut has favored toxin diversification and maintenance of T6SS machinery, and demonstrate the importance of antagonistic interactions within host-associated microbial communities.
INTRODUCTION
Host-associated microbiota are often complex communities comprised of hundreds of species. Bacteria that live in these communities employ diverse mechanisms of intercellular competition. Though first identified as pathogenicity factors (1), type VI secretion systems (T6SSs) are increasingly recognized for their role in mediating antagonistic interactions between bacteria (2). These multiprotein complexes participate in contact-dependent intercellular competition by driving needle-like structures, which can be loaded with a variety of toxins, through the membranes of nearby cells (3, 4). T6SSs have been shown to contribute to spatial organization of bacterial communities in vitro (5, 6). Furthermore, recent studies have shown that human gut microbes utilize T6SSs in interbacterial antagonism in vitro and in gnotobiotic mouse models; these findings suggest roles for these complexes in the cocolonization and persistence of bacterial species in the human gut (7–10). Although T6SSs are common—present in approximately 25% of sequenced genomes of Gram-negative bacteria (4, 11)—little is known about the role of T6SSs in the evolution of commensal communities.
The Western honey bee, Apis mellifera, has a highly conserved gut microbiota with properties comparable to those of mammalian gut communities, including high strain diversity, social transmission, and conferral of benefits to host health (12–15). The core gut community is comprised of nine bacterial species, which account for more than 95% of bacteria in the guts of healthy worker bees (16). We previously found T6SS and T6SS-associated effector genes in the genomes of some of these species, including the betaproteobacterium Snodgrassella alvi and gammaproteobacterium Gilliamella apicola (17). Therefore, T6SS-mediated competition among coevolved species may influence the structure and composition of the bee gut microbiota, as has been hypothesized for the gut communities of mammals based on bioinformatic analyses and on patterns of antimicrobial antagonism in vitro and in gnotobiotic mice (7–9, 11).
S. alvi is an abundant gut symbiont of honey bees (Apis spp.) and their close relatives, the bumble bees (Bombus spp.). It primarily colonizes the ileum section of the hindgut, where it forms biofilm-like layers with G. apicola (16, 18). Multiple S. alvi strains coexist within individual bees and bee hives, and strains differ among host species and geographic locations (13, 16, 19). How such strain diversity arises and is maintained in gut microbiomes is unclear (20). However, it is likely that interactions between members of the microbiota affect strain-level composition, which in turn may influence community-scale trends in stability, turnover, and diversity (21).
We hypothesize that T6SSs and T6SS-associated toxins mediate intraspecific competition among S. alvi strains in the bee gut, as well as interspecific competition between S. alvi and other gut microbes. We used transcriptome sequencing (RNA-Seq) to determine the conditions under which S. alvi T6SSs are expressed. To examine the diversity, prevalence, and evolution of T6SSs and their associated toxins in this gut symbiont, we isolated and sequenced the genomes of 28 S. alvi strains from diverse Apis and Bombus species. Finally, we provide evidence that T6SS-associated Rhs toxins have antibacterial activity in vivo and that extensive recombination and horizontal transfer of toxin/immunity genes between strains in the microbiota have resulted in tremendous diversity in their toxin repertoires. Our results support the view of gut microbiomes as exclusive assemblages whose membership is influenced by complex competitive interactions among coevolving species.
RESULTS
S. alvi upregulates T6SSs in vivo.
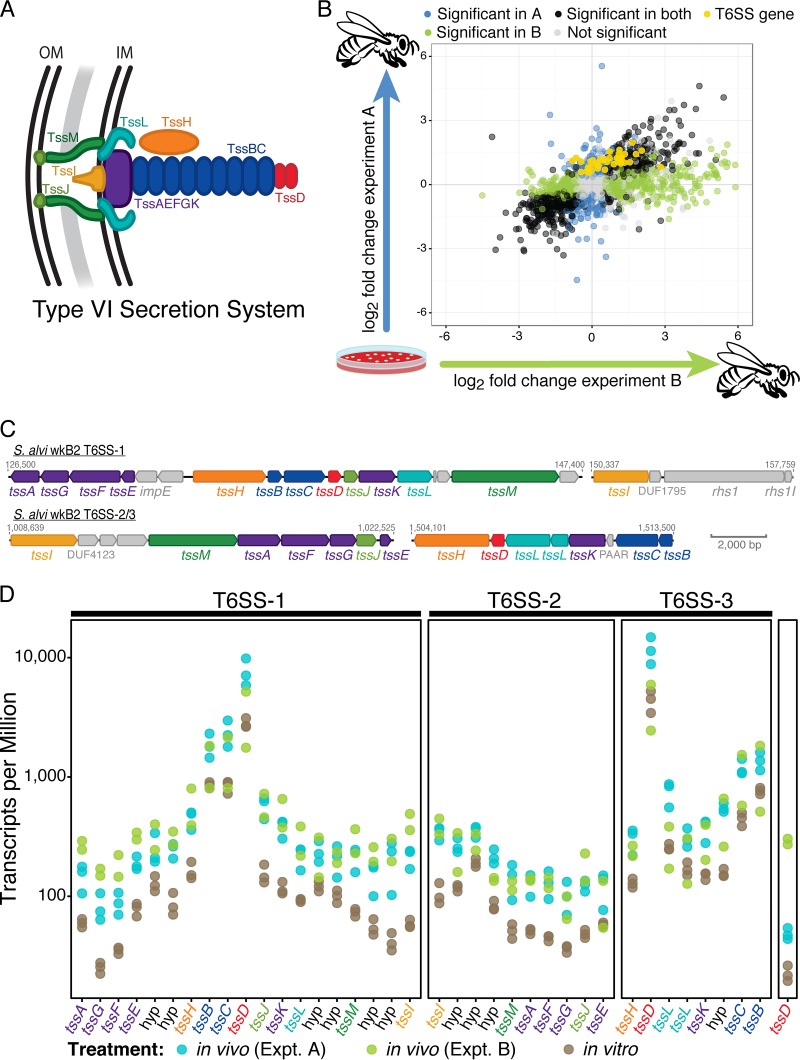
Inspection of the genome of the S. alvi type strain, wkB2, revealed 38 T6SS-associated genes clustered at three genomic loci. One locus (T6SS-1) contains 19 genes, including all 13 T6SS core components (17), while two other loci (T6SS-2 and T6SS-3 or, collectively, T6SS-2/3) contain complementary sets of 9 and 8 genes that together encode a second T6SS with very little amino acid sequence identity (17.3% average) to the genes in T6SS-1 (Fig. 1A and C). From this, we predict that two complete T6SS complexes are encoded in the S. alvi wkB2 genome. In addition to the core T6SS genes, the T6SS-1 locus contains impE and five hypothetical genes, while the T6SS-2/3 loci include four hypothetical genes, including a gene encoding a proline-alanine-alanine-arginine (PAAR) domain and a gene encoding a protein with a DUF4123 domain. This DUF4123 domain protein is likely to be an effector chaperone that facilitates interaction between an effector and the T6SS-2/3 VgrG (22).
FIG 1 .
Genes involved in intercellular competition are upregulated in vivo. (A) Diagram of the T6SS, showing the membrane complex (TssJ, TssL, and TssM; green), baseplate complex (TssA, TssE, TssF, TssG, and TssK; purple), a TssD needle (red) tipped with a TssI spike (yellow), the contractile sheath (TssB and TssC; blue), and an ATPase (TssH; orange). (B) In vivo fold change in each experiment for all genes. Axes depict comparisons between experimental treatments, with the in vitro condition serving as the control. Experiment A, from the work of Powell et al. (23); experiment B, from this study. T6SS-associated genes are highlighted in yellow. (C) ORF maps of the three T6SS loci in S. alvi wkB2; genes encoding core components are colored to match panel A, and gray numbers indicate the position of the diagrammed region within the genome. (D) Count-normalized gene expression of T6SS-associated genes for in vivo and in vitro replicates.
To determine whether the T6SSs identified in S. alvi wkB2 are expressed in vivo, we used whole-transcriptome sequencing (RNA-Seq) to measure gene expression of S. alvi wkB2 in gnotobiotic A. mellifera workers. To account for variation due to host genetic background, we also reanalyzed a published RNA-Seq data set—produced with the same experimental design—from the work of Powell et al. (23) and compared gene expression in each in vivo experiment to expression under standard laboratory culture. We identified 583 genes—including 19 T6SS genes—that were differentially expressed in the same direction in these two independent in vivo RNA-Seq experiments, relative to in vitro culture (Fig. 1B). Of the 19 T6SS genes that were upregulated in vivo in both experiments, 13 were from T6SS-1, 5 were from T6SS-2/3, and one (tssD) was not part of a T6SS locus (Fig. 1D). An additional 12 T6SS genes were significantly upregulated in only the data set of Powell et al. (see Table S1 in the supplemental material).
Differential expression of S. alvi wkB2 T6SS genes, Rhs toxin and putative immunity genes, and genes involved in iron and nitrogen metabolism. Download TABLE S1, DOCX file, 0.2 MB (160.6KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. alvi T6SSs are vertically inherited.
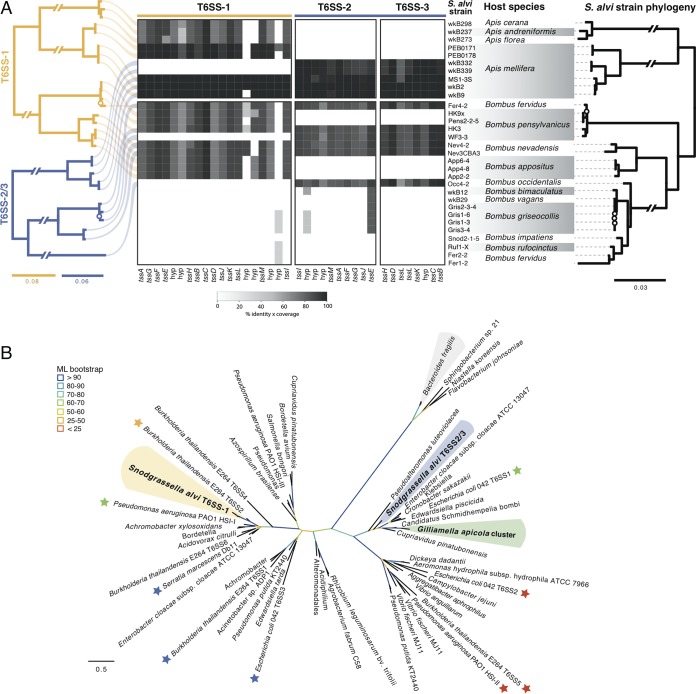
To better understand the evolution and ecology of T6SSs within S. alvi, we sequenced the genomes of 28 S. alvi strains isolated from diverse Apis and Bombus species collected from Southeast Asia and North America (Table S2). We determined whether T6SSs were present in these strains, as well as in three previously published strains (17), by screening for homologs of the T6SS genes in S. alvi wkB2. All seven S. alvi strains isolated from A. mellifera encoded at least one complete T6SS complex, and only the two strains from bees collected in Malaysia (wkB332 and wkB339) were missing T6SS-1 (Fig. 2A). S. alvi strains isolated from three honey bee species (Apis andreniformis, Apis cerana, and Apis florea) collected in Singapore also encoded T6SS-1. Both T6SS-1 and T6SS-2/3 were present in strains isolated from Bombus pensylvanicus, Bombus nevadensis, and Bombus appositus, although some strains lacked one or the other system. Finally, 10 strains isolated from seven different Bombus host species appear to have lost both T6SSs. To ensure that our homology-based searches had not missed any T6SS loci with low identity to T6SS-1 and T6SS-2/3, we searched for the 13 core T6SS genes in the genome annotations generated by the Rapid Annotation using Subsystems Technology (RAST) pipeline (24) for all 31 strains. We did not find any additional T6SS loci, confirming that only these two subfamilies of T6SSs are present in the entire set of S. alvi strains.
FIG 2 .
Two T6SSs are present in the bee gut symbiont S. alvi. (A) Presence of T6SS genes in S. alvi strains based on similarity to genes of the three T6SS loci of strain wkB2; the heat map shows the percent identity × coverage of the best match in each genome to the corresponding wkB2 gene. The S. alvi phylogeny (right) is reconstructed from ribosomal protein genes; nodes with less than 80% bootstrap support are marked with an open circle. The yellow and blue trees (left) show relationships between T6SS-1 and T6SS-2/3 loci, respectively. hyp, hypothetical open reading frame. (B) Maximum likelihood tree for TssB showing the relationship between T6SS-1 and T6SS-2/3. Sequences from T6SS with characterized functions are marked with stars: pathogenesis (red), intercellular competition (blue), Rhs-associated (yellow), and secretion of antimicrobials (green).
Bacterial strains and genomes used in this study. Download TABLE S2, DOCX file, 0.1 MB (133KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We reconstructed the phylogenetic relationships of the S. alvi strains using 37 single-copy ribosomal protein genes (Table S3), as well the relationships of the two T6SSs based on their constituent genes (Fig. 2A). The phylogenies of T6SS-1 and T6SS-2/3 were both highly congruent with the ribosomal protein-gene phylogeny, suggesting that evolution of S. alvi T6SSs occurs mainly by vertical descent and not through transfers between strains. Further, this phylogenetic congruence supports the acquisition of both T6SSs prior to the divergence of the S. alvi clades associated with Apis and Bombus hosts, with subsequent losses leading to the pattern of T6SS presence/absence observed across our strains (Fig. 2A and S1).
Ongoing deletion is eliminating the T6SS-2 locus in some S. alvi strains. The T6SS-2 locus in S. alvi strains PEB0171, wkB2, HK9, Gris1-3, and Fer1-2, which is bracketed by recO and integration host factor α in all but wkB2. The S. alvi strain phylogeny (left) shows the relationships between strains and supports the hypothesis that T6SS-2 has been lost multiple times in S. alvi. Download FIG S1, PDF file, 0.1 MB (91.7KB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. alvi genes used to reconstruct strain phylogeny and genes used for tests of positive selection. Download TABLE S3, DOCX file, 0.1 MB (93.4KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The phylogeny of the T6SS TssB protein was reconstructed to determine the relatedness of the S. alvi T6SSs to previously identified T6SSs. We found that TssB proteins associated with S. alvi T6SS-1 and T6SS-2/3 are phylogenetically distinct and have different evolutionary origins (Fig. 2B). TssB from S. alvi T6SS-1 clusters with proteins from Pseudomonas aeruginosa PAO1 HSI-I and Burkholderia thailandensis T6SS-2 and T6SS-6, while TssB from S. alvi T6SS-2/3 is more closely related to proteins from other Gram-negative bee symbionts, including G. apicola, Frischella perrara, and “Candidatus Schmidhempelia bombi,” and other gammaproteobacteria.
T6SS-1 likely mediates secretion of diverse Rhs toxins.
T6SSs secrete a variety of antibacterial proteins, including the Rhs family of toxins (25). We previously found numerous Rhs toxins encoded in the genomes of A. mellifera gut symbionts, including S. alvi wkB2 (17). Many of the newly sequenced genomes also contain large numbers of Rhs genes—up to 120 copies in some strains—but the function of these genes in the ecology of the bee gut is unclear.
We tested whether S. alvi Rhs toxins were associated with a particular T6SS locus and found that strains encoding T6SS-1 harbored significantly more Rhs genes than strains with no T6SS or only T6SS-2/3 (Fig. 3). In contrast, the number of Rhs genes per genome did not vary significantly with the presence or absence of T6SS-2/3. Among strains encoding T6SS-1, strains isolated from Bombus had more Rhs genes than did strains from Apis. However, the correlation between the presence of T6SS-1 and the number of Rhs toxins persisted when strains from Apis or Bombus hosts were examined separately (Fig. S2). Furthermore, the T6SS-1 tssI (vgrG) gene is located at the 3′ end of the T6SS locus, immediately upstream of one of the three Rhs loci in wkB2, while the T6SS-2/3 tssI gene occurs at the 5′ end of the locus, upstream of genes encoding proteins of unknown function (Fig. 1C). This strongly implicates a role for T6SS-1 in Rhs toxin secretion while suggesting that other, unidentified effectors are secreted through T6SS-2/3.
FIG 3 .

Rhs genes are associated with T6SS-1. The number of Rhs-family genes is significantly higher in genomes with the T6SS-1 locus, regardless of the presence or absence of the T6SS-2/3 locus (one-way ANOVA with Tukey’s HSD multiple-test correction; *, Padj < 0.05).
The association between S. alvi Rhs genes and T6SS-1 is consistent across host species. Statistical tests performed with ANOVA and Tukey’s HSD. *, Padj < 0.05; ***, Padj < 0.005. Download FIG S2, PDF file, 0.4 MB (386.9KB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rhs loci have a characteristic architecture.
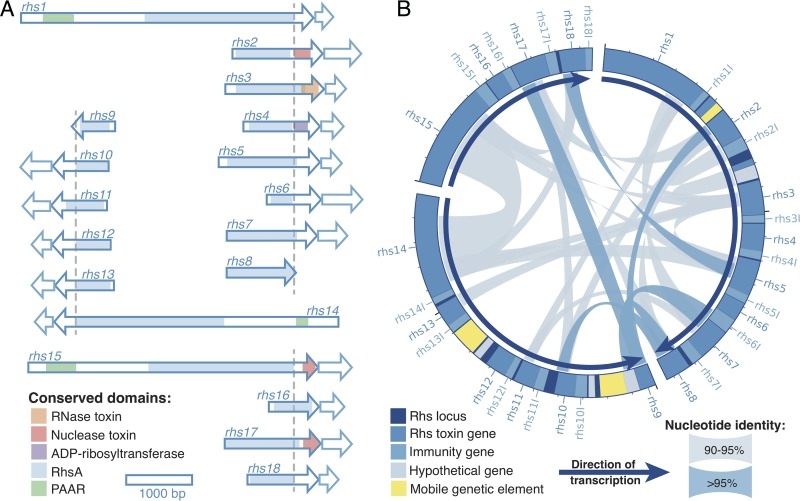
We examined the organization of Rhs loci in the completely closed genome of S. alvi wkB2 to better understand the structural diversity of these putative T6SS effectors. We identified 18 Rhs-family genes at three separate loci in wkB2. Rhs-family genes encode large, polymorphic proteins with variable C-terminal toxin domains and conserved core regions that contain the Rhs/YD repeats characteristic of this toxin family (26). Though many T6SSs secrete cargo effectors that form noncovalent associations with secreted components of the T6SS, Rhs toxins are often specialized effectors, fused to PAAR domains that interact with VgrG at the tip of the T6SS needle (25, 27). In contrast to cargo effectors, which are often classified by their cellular targets, the Rhs toxin family is defined by a core region containing characteristic Rhs repeats and includes proteins with variable C-terminal toxin domains that affect a variety of cellular targets within prokaryotes and eukaryotes (26). The highly conserved DPXG(18)DPXG motif, which is found at the end of the conserved core region (26), was used to predict the start of the C-terminal toxin domain in each Rhs gene, and the Conserved Domain Database (CDD) (28) was used to search for similarity to previously characterized toxin domains. Three genes (rhs1, rhs14, and rhs15) encode proteins with large N-terminal regions that contain complete RhsA domains (COG3209) and PAAR secretion domains (Fig. 4A). Each of these genes is found at the 5′ end of its respective Rhs locus and is followed by several relatively short Rhs genes with intact core and toxin domains, truncated RhsA domains, and no PAAR domain (Fig. 4B). Based on conserved motifs, Rhs8 and Rhs9, which are encoded at the 3′ end of their respective Rhs loci, appear to have truncated N-terminal domains and no C-terminal toxin domains. The C-terminal regions of five Rhs genes were similar to previously characterized toxin domains, including three domains associated with toxins of the HNH/endonuclease VII family, an RNase toxin domain, and an ADP-ribosyltransferase domain (Table S4), whereas the remaining 11 Rhs proteins may contain novel toxins.
FIG 4 .
S. alvi wkB2 has three Rhs loci with architectures congruent with extensive toxin domain replacement. (A) Size and structure of S. alvi wkB2 Rhs genes and cognate immunity genes, shown as arrows outlined in dark and light blue, respectively. A dashed line indicates the location of the conserved DPXG(18)DPXG motif, which marks the start of the C-terminal toxin domain in each toxin. PAAR, RhsA, and toxin domains were predicted through the NCBI CDD search (E value, <10−5). (B) Arrangement of Rhs loci in the wkB2 genome. Arrows show the 5′-to-3′ orientation of the loci, while dark and light blue ribbons connect >500-bp regions with >95% and >90% nucleotide identity, respectively. Rhs and cognate immunity genes are highlighted in dark and light blue, respectively; putative mobile genetic elements are yellow; and hypothetical genes are gray. Major tick marks occur every 10,000 bp; minor tick marks occur every 1,000 bp.
Conserved protein domains detected in S. alvi wkB2 Rhs toxin genes (E value, <0.00001) and immunity genes (E value, <0.001). Download TABLE S4, DOCX file, 0.1 MB (107.1KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rhs toxin genes have antibacterial function and are upregulated in vivo.
Like toxin-antitoxin systems, Rhs loci typically encode immunity proteins that protect the cell against the activity of their cognate toxins. These immunity genes generally have little sequence homology to each other, making it difficult to infer their activity from sequence data alone, but they are often found immediately downstream of their corresponding toxin (26, 29). We identified putative immunity genes immediately downstream of 16 Rhs toxin genes in the S. alvi wkB2 genome (Fig. 4A). Only rhs8 and rhs9, which do not have C-terminal toxin domains, lacked adjacent putative immunity genes.
To ascertain the functionality of the Rhs toxin-antitoxins in S. alvi, we reanalyzed data from a recent genome-wide transposon mutagenesis screen (Tn-Seq) of strain wkB2 (23) using our updated Rhs annotations. We found that three putative immunity genes were completely intolerant to disruption by transposon insertion, suggesting that these genes are essential (Fig. 5B; Table S5). Insertions in a further seven putative immunity genes were significantly detrimental to the fitness of S. alvi wkB2 in the bee gut, which demonstrates the importance of Rhs immunity genes for fitness within the host and indicates that the corresponding Rhs toxins in S. alvi do indeed have antibacterial activity. Some of the Rhs toxin genes whose cognate immunity genes do not contribute to fitness may not encode functional proteins. For example, rhs3 has been pseudogenized by multiple nonsense mutations, while rhs6 and rhs16 have truncated N-terminal regions that do not prevent their transcription but may interfere with their activity. Alternatively, these toxins may target species other than S. alvi.
FIG 5 .
Rhs toxins and immunity genes are upregulated and have antibacterial function in vivo. (A) Fold change in the abundance of S. alvi wkB2 mutants with transposon insertions in T6SS-associated genes, Rhs toxins, or putative Rhs immunity genes in vivo, relative to abundance in vitro. DESeq2 (55) was used to determine differential mutant fitness with a false discovery rate cutoff of 0.05. Green and red circles indicate mutants that are significantly more or less fit in vivo (Padj < 0.05), respectively. Gray circles indicate genes with a Padj of >0.05. *, three putative immunity genes that were essential in vivo are excluded. Essential genes were identified by comparing the frequency of observed transposon insertions for each gene to the frequency of insertion in a randomized data set, as described by Turner et al. (74). (B) Expression of Rhs toxin and immunity gene pairs, averaged across replicates, in in vitro culture and in two in vivo experiments: in vivo experiment A, from the work of Powell et al. (23); in vivo experiment B, from this paper. Gray arrows show the 5′-to-3′ organization of genes for each locus.
Fitness effects of transposon insertions in S. alvi wkB2 T6SS, Rhs toxin, and putative immunity genes. Download TABLE S5, DOCX file, 0.1 MB (139.8KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
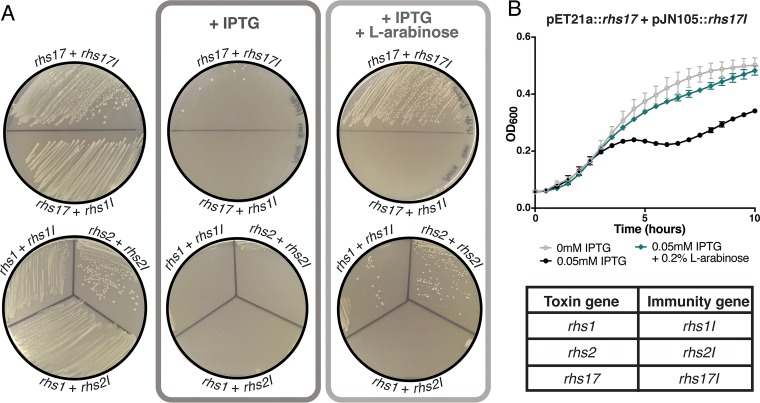
To further verify the function of these genes, we cloned three pairs of toxin and immunity genes into compatible expression vectors in Escherichia coli BL21(DE3). The toxin domains of rhs1, rhs2, and rhs17 were cloned into the pET21a expression vector under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible PT7 promoter. The putative immunity genes rhs1I, rhs2I, and rhs17I were cloned into expression vector pJN105 under the control of the l-arabinose-inducible PBAD promoter. Both rhs1I and rhs2I were identified as essential in the S. alvi wkB2 Tn-Seq analysis; rhs17I was important for fitness in vivo. E. coli cells containing pET21a::rhs17 and pJN105::rhs17I were able to grow on lysogeny broth (LB) agar without induction or LB containing 0.1 mM IPTG and 0.5% l-arabinose to induce expression of both toxin and immunity genes, but not on LB containing 0.1 mM IPTG alone, which induces expression of only the toxins (Fig. 6A). Induction of immunity gene expression with l-arabinose did not restore the growth of cells with pJN105::rhs1I instead of pJN105::rhs17I (Fig. 6A and S3B). Additionally, E. coli cells containing rhs17 and rhs17I demonstrate a growth defect in liquid cultures containing 0.05 mM IPTG that is counteracted by the addition of 0.2% l-arabinose (Fig. 6B). The observed growth defect is dependent on the presence of the toxin gene, while the restoration of growth with the addition of l-arabinose occurs only in cells with the cognate immunity gene (Fig. S3A). Similar growth patterns were observed for cells expressing rhs1 and rhs1I or rhs2 and rhs2I (Fig. 6A and S3C and D), though cells containing rhs1 exhibit much slower growth and often fail to grow, even in the absence of toxin gene induction.
FIG 6 .
Expression of cognate immunity genes restores growth in cells expressing Rhs toxins. (A) E. coli BL21(DE3) cells containing rhs17, rhs1, and rhs2 under the control of the IPTG-inducible promoter on pET21a grow on selective medium in the absence of induction (left) and do not grow when toxin expression is induced with 0.1 mM IPTG (center). Induction of the cognate immunity gene with 0.5% l-arabinose restores growth in the presence of IPTG (right). Induction of rhs1I does not restore growth in cells expressing rhs17, and rhs2I does not restore growth in cells expressing rhs1. (B) Induction of rhs17I with 0.2% l-arabinose (teal) restores growth in cells with induced toxin expression (black) in liquid culture.
Expression of the cognate immunity gene restores growth in cells expressing the rhs1 and rhs2 toxin genes. (A) l-Arabinose does not counteract the effects of toxin gene induction in the absence of the immunity gene. (B) Similarly, the presence of IPTG does not lead to reduced growth in the absence of the toxin gene. (C) Cells grown with 0.1 mM IPTG to induce rhs17 expression (dark teal circles and dark purple triangles) exhibit reduced growth relative to cells grown without IPTG (open circles and open triangles). Growth is restored by inducing immunity gene expression with 0.1% l-arabinose in cells containing rh17I (light teal circles) but not in cells with rhs1I (light purple triangles). (D and E) E. coli BL21(DE3) cells expressing rhs1 (D) or rhs2 (E) induced by IPTG demonstrate a severe growth defect in liquid culture that is at least partially alleviated by expression of the cognate immunity gene induced by l-arabinose. Download FIG S3, PDF file, 1 MB (1.1MB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As our RNA-Seq results indicated that T6SS expression was upregulated in vivo (Fig. 1D), we examined whether this also held true for Rhs toxin expression. Fifteen Rhs toxins were significantly upregulated in vivo in at least one experiment, and seven of these were significantly upregulated in both experiments (Fig. 5A; Table S1). We also observed a trend in expression, whereby—after normalizing for length—genes located at the 5′ end of the Rhs locus were more transcriptionally active than genes at the 3′ end (Fig. 5A). This trend was highly significant for the toxin genes of Rhs loci 1 and 2 (Spearman’s rank correlation ρ = −0.388, P < 0.005, and Spearman’s rank correlation ρ = −0.563, P < 0.001, respectively), although this trend was not observed for Rhs locus 3 (Spearman’s rank correlation ρ = 0.375, P = 0.034).
A large pool of interchangeable toxins drives Rhs ecology.
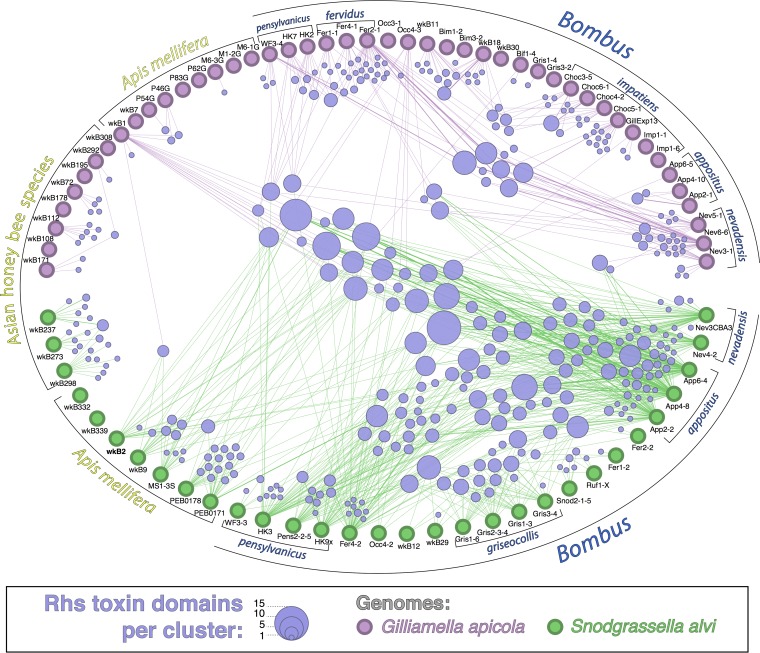
The large number of Rhs toxins encoded by S. alvi strains suggests that toxin diversity may be important for competitive ability among members of the bee gut microbiota. We examined the diversity and prevalence of these genes in all S. alvi strains, as well as in strains of a common coresident gut bacterium, G. apicola. In the 77 genomes analyzed (31 of S. alvi and 46 of G. apicola), we detected a total of 1,112 Rhs genes (813 from S. alvi and 299 from G. apicola), encoding potentially 364 distinct toxin domains. This reveals a tremendous pool of Rhs diversity accessible by, and possibly unique to, the bee gut microbiome (Fig. 7). Some toxins are found in both S. alvi and G. apicola, while others appear to be constrained to a single species or a set of closely related strains.
FIG 7 .
Members of the bee gut microbiome encode diverse Rhs toxins, which are shared within and between species. Each toxin node represents a unique C-terminal toxin domain derived by clustering at 90% nucleotide identity; size is proportional to number of sequences in a cluster. Lines connect toxin nodes to strains in which they are found. Strains are grouped and labeled according to host of origin. Spatial configuration of the Rhs clusters is to minimize overlap of lines and circles for visibility and does not correspond to any other metadata. Strain names are provided adjacent to each strain symbol.
In contrast to the T6SS structural genes, Rhs toxin distribution is not solely determined by vertical descent. Clustering toxin domains by cooccurrence reveals networks of dissemination that are likely governed by geography and shared host species. For instance, strains from Southeast Asia did not share any toxins with those from North America. Host relatedness does not pose a complete barrier, as S. alvi strains from both Apis and Bombus hosts carried many toxins in common. Toxins are mostly shared within a species (S. alvi or G. apicola) rather than between species, which may reflect more frequent gene transfer between conspecific bacterial strains or indicate that this toxin family is predominantly used for intraspecific, rather than interspecific, competition in these two gut symbionts.
Rhs toxin and immunity gene pairs are strongly linked.
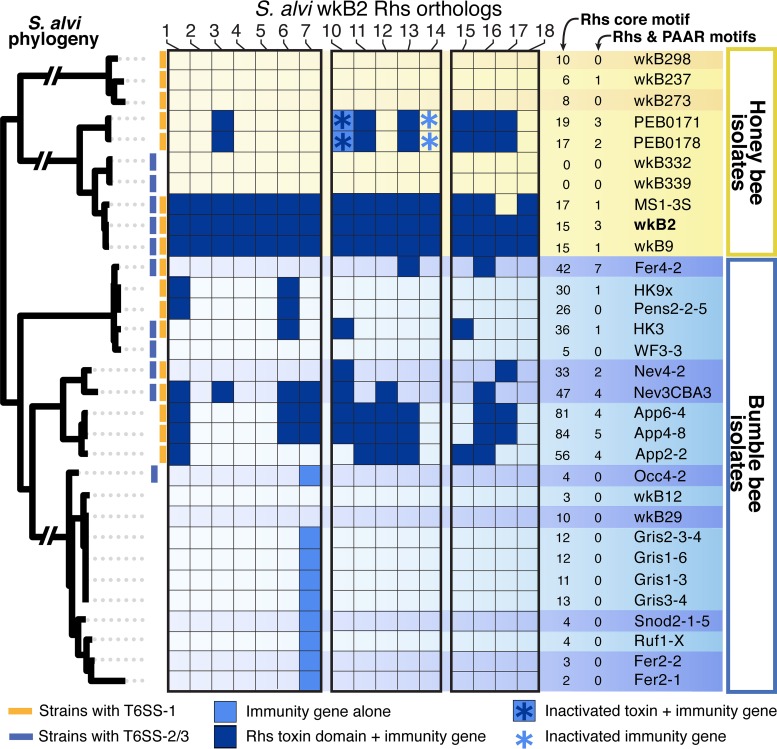
Frequent genetic transfers can break apart beneficial gene combinations, which would be highly detrimental in the case of a toxin and its immunity gene. The localization of Rhs immunity genes immediately adjacent to the C-terminal toxin domain is likely a mechanism that ensures against self-poisoning. We examine the linkage of Rhs toxin and immunity genes in S. alvi by searching the genomes of 20 S. alvi strains harboring T6SSs for the presence of S. alvi wkB2 toxins and their cognate immunity genes (Fig. 8). With the exception of isolates from Asian Apis species, every strain encoding T6SS-1 had at least one sequence homologous to a wkB2 toxin domain, and each of these also contained the associated immunity gene. In a few strains, immunity genes were detected without their associated toxin; however, the reverse was never true. The genomic context of these orphaned immunity genes suggests that they were originally acquired alongside their cognate toxin that has since been pseudogenized. Pseudogenization may also be the eventual fate of orphaned immunity genes in S. alvi, as several strains contain homologs to immunity genes from S. alvi wkB2 that have acquired nonsense mutations in the absence of the cognate toxin. Altogether, this illustrates both the importance of the immunity gene for the fitness of cells encoding the cognate Rhs toxin and the specificity of the toxin-immunity gene interaction.
FIG 8 .
Rhs toxins and cognate immunity genes cooccur and are tightly linked. The presence/absence of S. alvi wkB2 C-terminal toxin domains and their cognate immunity gene in other S. alvi strains is shown. rhs8 and rhs9 lack toxin domains and immunity genes and are excluded. S. alvi strains are grouped according to strain phylogeny (left).
Rhs toxins undergo slow sequence evolution but rapid recombination.
Although coevolving systems often impose strong selection upon particular genes, homologous Rhs toxins have very little sequence variation, suggesting that rapid de novo mutation is not a major source of toxin diversity in the bee gut microbiota. We tested Rhs core and toxin domains for positive selection and found that both regions were under purifying selection (ratio of nonsynonymous to synonymous changes per nucleotide site [dN/dS] of <1), albeit with more relaxed constraints relative to highly conserved housekeeping genes (Fig. S4; Table S3).
Rhs toxins are under purifying selection and affect S. alvi fitness. (A) dN/dS ratios for Rhs C-terminal toxin domains were not significantly different from those of the Rhs core. Both Rhs domains had dN/dS ratios of <1 but significantly higher dN/dS ratios than conserved housekeeping genes (one-way ANOVA with Tukey’s multiple comparisons of means, Padj < 0.005). Several homologous toxin domains were excluded from this analysis due to insufficient variation (i.e., only 0 to 5 polymorphisms). (B) Fold change in abundance of S. alvi wkB2 mutants with transposon insertions in Rhs toxins (top) and putative immunity genes (bottom) in vivo, relative to abundance in vitro (Padj < 0.05). E, gene essential in vitro; NS, nonsignificant change in mutant abundance in vivo, −, immunity gene not present in genome. Gray arrows show the 5′-to-3′ organization of genes for each locus. Download FIG S4, PDF file, 0.2 MB (247.3KB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
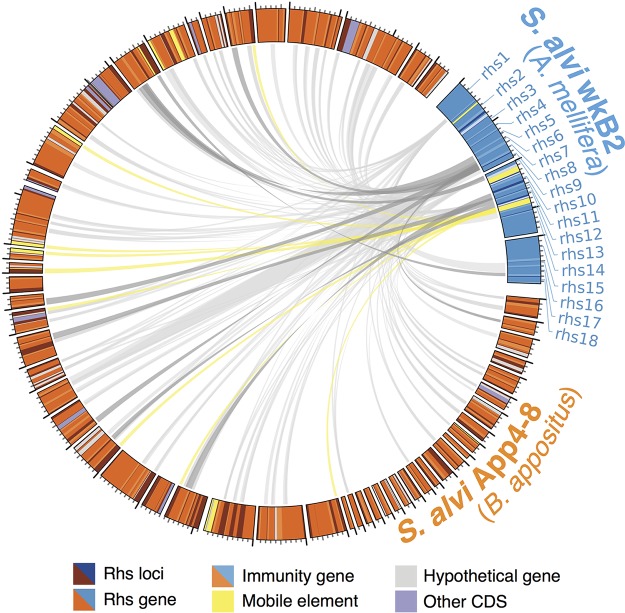
As de novo mutation is not the primary source of Rhs diversity between strains, it is likely that the horizontal acquisition of novel toxin alleles and functional diversification through toxin/core recombination provide the adaptive basis of T6SS/Rhs-mediated competition in the bee gut microbiota. Comparison of the 3 Rhs loci in S. alvi wkB2 and the 52 Rhs-encoding contigs in S. alvi App4-8 identified several shared Rhs genes and a similar integrase-like gene in proximity to Rhs genes in the two genomes (Fig. 9). This gene, at >80% amino acid identity, was found in 19 S. alvi and 11 G. apicola genomes, while another Rhs-associated integrase was detected in 13 and 22 genomes of S. alvi and G. apicola, respectively. Such genes might facilitate the transfer and recombinatorial capture of Rhs sequences across gut microbiota strains, but further work will be necessary to determine whether these integrases contribute to the transfer of toxin genes between bee gut microbes.
FIG 9 .
Rhs loci in divergent S. alvi strains share similar sequences. Dark and light gray ribbons show regions of >95% and >90% nucleotide similarity between S. alvi strains wkB2 and App4-8, respectively. Nine sequences within App4-8 Rhs loci have high identity (>95%) to 7 of the 18 Rhs genes in wkB2. Sequences similar to a putative integrase in wkB2 (yellow) are present in many App4-8 contigs. CDS, coding sequences.
DISCUSSION
T6SS-mediated competition is likely to be an integral component of the ecology of S. alvi in the bee gut. Two phylogenetically distinct T6SSs were upregulated by S. alvi wkB2 in gnotobiotic A. mellifera workers. We determined that the toxins associated with one of these systems, T6SS-1, have antibacterial properties, which is consistent with a role in intraspecific competition. S. alvi strains with T6SS-1 can carry dozens of Rhs genes; some Bombus-derived strains have more than 100. We discovered a correspondingly large pool of toxins in the bee “pan-microbiome,” with over 1,000 Rhs genes and up to 364 different toxin domains detected across S. alvi and G. apicola strains isolated from Apis and Bombus hosts.
Given these results, we were surprised to find that many S. alvi and G. apicola strains completely lacked T6SSs and/or Rhs genes (Fig. 2A and 7). It is possible that these strains utilize other toxins or mechanisms of intercellular antagonism that we have not yet identified. However, the absence of these genes may be due to fitness tradeoffs involved in maintaining large, energy-intensive T6SSs and toxins that can cause self-poisoning. In fact, inactivation of T6SS and Rhs toxin genes by transposon insertion generally increases fitness in vivo in an S. alvi monospecies community (Fig. 5B). This suggests that, under some circumstances, loss of expensive competitive machinery in favor of maximizing growth may be a viable route for success within the gut community.
The two T6SSs found in S. alvi are strictly vertically inherited, meaning that loss of one or both systems is likely to be permanent. Accordingly, few S. alvi strains contain partial T6SSs, indicating that once a T6SS becomes nonfunctional, the remaining genes are rapidly deleted. In one large clade of Bombus-specific S. alvi, both T6SSs appear to be permanently lost, with a few strains retaining only the ends of the T6SS-2 locus (Fig. 2A and S1). This stands in sharp contrast with T6SS evolution in bacteria such as Bacteroides and Salmonella, where horizontal gene transfers of entire systems, possibly via association with mobile genetic elements, appear to be common (8, 11, 30).
Neither S. alvi T6SS-1 nor S. alvi T6SS-2/3 is closely related to T6SSs known to participate in virulence (Fig. 2B). Instead, S. alvi T6SS-1 clusters with the prokaryote-targeting HSI-I of P. aeruginosa PAO1 (31) and three Burkholderia thailandensis T6SSs, two of which are encoded within loci containing Rhs genes (32). The diverse sets of Rhs toxin genes associated with T6SS-1 suggest that it may have an important role in mediating intraspecific competition in S. alvi. However, this does not eliminate the possibility that T6SS-1 is also used to antagonize other Gram-negative bee gut microbes, including G. apicola, or even opportunistic pathogens. Recent studies have found that the bee gut microbiota reduces colonization by some opportunistic bacterial pathogens (33, 34), which makes it tempting to speculate that the S. alvi T6SSs may play a role in colonization resistance in the bee gut. We have demonstrated that Rhs toxins from S. alvi wkB2 have an antibacterial function (Fig. 6), but this does not eliminate the possibility that these toxins may also be able to antagonize eukaryotic cells, as T6SS effectors in other systems are toxic to both prokaryotes and eukaryotes (2, 29). S. alvi T6SS-2/3 is more closely related to those of bee-associated gammaproteobacteria, including G. apicola and F. perrara. F. perrara is part of the core honey bee gut microbiota but appears to cause damage to host tissue, triggering melanization at the site of colonization (35). The function of S. alvi T6SS-2/3 is not yet known. It is likely to be antibacterial, like T6SS-1, but T6SSs can also be used for host interaction. For instance, T6SS effectors are used by Vibrio cholerae to kill eukaryotic cells (1), and Yersinia pestis requires a T6SS to enter and grow inside human macrophages (36). However, this T6SS is unlikely to be used in virulence, as S. alvi is a commensal member of the bee gut microbiota and is not known to harm its host.
In contrast to the vertically inherited T6SSs in S. alvi, we find evidence that Rhs toxins may be horizontally transferred between strains. Strains from different hosts can share the same toxins, and some toxins are found within both S. alvi and G. apicola. S. alvi harbors a greater diversity and abundance of Rhs genes than G. apicola, suggesting that S. alvi may be the predominant reservoir for these toxins in the bee gut community. Additionally, the Rhs-associated T6SS-1 system was not found in G. apicola, which may constrain the types of toxins that it can secrete. There is evidence that T6SS-associated toxins are shared among strains and species in other systems (37–40). Furthermore, the potential for gene exchange in the bee gut is supported by our finding of near-identical mobile elements in divergent bacteria (Fig. 9), as well as previous evidence from whole-genome analyses (17) and the presence of identical antibiotic resistance determinants in G. apicola and S. alvi (41).
Further sampling will be necessary to explain the observed toxin distribution pattern (Fig. 7), as it is not yet clear what factors allow for the exchange of toxin genes between strains. Geographic isolation could explain the small and nonoverlapping assortment of Rhs genes found in honey bees from Southeast Asia compared to North American samples; however, this is confounded by the fact that these Asian bees represent different Apis spp. and come from a region where Bombus does not occur. That North American S. alvi strains from A. mellifera and Bombus species encode similar toxins is also intriguing, as S. alvi strains are host specialized, and the same strains do not typically reside in both Apis and Bombus hosts (13, 17, 19). Most toxins are not shared between G. apicola and S. alvi, which is consistent with observations that some Rhs genes are primarily used for competition between closely related strains or species (7, 27). Barriers to cross-species toxin exchange may explain these patterns but do not eliminate the possibility of Rhs-mediated interspecies competition. Rhs toxins potentially influence the capability of strains or species to coexist in the same gut community, a competitive interaction that has been implied for other antibacterial effectors (8, 39, 42). Interestingly, G. apicola wkB1 and S. alvi wkB2, which were isolated from the same A. mellifera colony at the same time, share a large proportion of Rhs genes—11 toxins and 12 immunity genes (17).
Many coevolving systems impose strong selection on traits that enhance competitiveness, leading to ongoing evolutionary responses in interacting lineages. The two major types of antagonistic coevolution are fluctuating selection dynamics and arms race dynamics (43). Evolutionary arms races typically feature rapid sequence evolution of proteins mediating the interaction (43, 44). However, we observed that very similar Rhs toxins are shared among distantly related bacterial strains. Rhs evolution also appears to be dominated by purifying selection (dN/dS ratio of <1), suggesting that positive selection for amino acid replacements is not a major mode of toxin evolution in these genes. These observations weigh against the possibility that Rhs evolution is driven by an evolutionary arms race. Instead, our results are more congruent with a fluctuating selection dynamics model of coevolution, which involves frequency-dependent selection favoring rare types (43). Phylogenetic studies suggest that S. alvi has evolved within bee hosts for over 80 million years (14); the large pool of Rhs toxins that has accumulated over this time may reflect the ever-changing competitive dynamics at play in bee gut communities. Frequency-dependent selection may prevent rare toxins from being lost, while the metabolic expense of maintaining T6SSs and toxins is likely to constrain the diversity of competitive machinery encoded by a single cell, ensuring that fluctuating selection dynamics remain predominant (44).
As in several enterobacterial species (45), the Rhs loci of S. alvi are comprised of a large, complete Rhs toxin gene at the 5′ end, followed by its cognate immunity gene, and then a series of truncated Rhs genes, or “orphaned” toxin domains, along with their immunity genes. This genetic architecture likely arises from “C-terminal displacement,” the exchange of toxin domains through recombination between conserved core regions (45, 46), which can generate functional diversity by packaging novel combinations of C-terminal toxin domains and N-terminal secretion domains. This mechanism also results in the genomic accrual of remnant, presumably nonsecreted Rhs toxin domains, which can subsequently be rerecruited into action via recombination (45, 46). In contrast to Serratia (45), the orphaned toxin domains in S. alvi wkB2 are not all silenced by nonsense mutations or missing translational start sites. Instead, many of these genes are transcribed and appear to produce proteins with antibacterial function, as inhibition of growth is observed when these toxins are heterologously expressed in E. coli (Fig. 6 and S3), and inactivating transposon insertions in their cognate immunity genes are detrimental to the fitness of S. alvi (Fig. 5B). Expression of S. alvi toxin and immunity genes in E. coli reveals that the protective effect of each immunity gene is specific to its cognate toxin, which is consistent with what has been observed in other species (27, 47). We also observed a correlation between the gene expression and gene position in two of the three Rhs loci in S. alvi wkB2, with the highest expression at the 5′ end and the lowest expression at the 3′ end, consistent with the direction of transcription and probable promoter placement.
The accumulation of numerous, potentially functional Rhs genes in S. alvi stands in contrast to what is found in many previously characterized bacteria: Serratia marcescens has two Rhs toxins (27), Dickeya dadantii 3937 has five (47), and E. coli K-12 has eight, including orphans (45). The conditions that promote the toxin expansion seen in S. alvi are unclear. Possibly, specialized bacteria living in communities with fewer species or more constrained niches might experience greater pressure to acquire diverse toxin and immunity gene pairs to compete against close relatives. However, this does not seem to be universally true of host-associated bacteria, as a recent survey of T6SS effectors in human gut Bacteroidales identified relatively few toxin and immunity gene pairs within individual strains and metagenomes (10). Though single strains of Bacteroides fragilis dominate the human gut microbiome, multiple S. alvi strains coexist within the bee gut (13), and interactions between these strains may contribute to increased toxin diversification in S. alvi. Additionally, the organization of microbes within individual bees or across individuals within the hive may allow for maintenance of otherwise incompatible sets of toxin and immunity genes, ultimately allowing for greater toxin diversity. It would be interesting to determine whether the diversity of Rhs toxins or other effectors correlates with host characteristics or gut community diversity or variability. Particularly, it is curious why S. alvi and G. apicola strains from Bombus hosts tend to encode far more Rhs toxins than do their counterparts in Apis species. In contrast to Apis species, Bombus species—and their gut microbiota—go through an annual population bottleneck when individual queens found new colonies, resulting in lower S. alvi strain diversity per individual bee (13). Potentially, the Bombus life cycle may impose more random fluctuations in the intensity of T6SS-mediated competition within their gut communities. Such differences in host ecology may help to explain why strains associated with some host species have accumulated large numbers of toxin genes while strains from other hosts have lost T6SSs entirely.
Conclusion.
This study broadens our knowledge of the diversity of T6SSs and their effectors and highlights their potential role in shaping host-associated microbiomes. While the T6SS has been well studied in the decade since its discovery, the evolution and ecological role of these systems in naturally occurring polymicrobial communities have received relatively little attention until recently (48). We found that S. alvi, a resident member of the bee gut microbiota, encodes two T6SSs as well as numerous Rhs toxins, which are expressed in vivo and have antibacterial activities. These T6SSs were maintained during the diversification of S. alvi strains, suggesting that intercellular competition is important in the gut communities of diverse Apis and Bombus species. An enormous diversity of toxins was identified across the genomes of S. alvi and G. apicola, another bee gut resident. These toxins were often shared between distantly related strains, as well as between these two species, which represent different classes of Proteobacteria.
However, we also found that T6SS presence and toxin abundance vary among strains. The loss of T6SSs in some lineages indicates that participation in this mode of competition may not always be beneficial. Human gut bacteria with and without T6SSs have been shown to coexist in gnotobiotic mouse models (8). Clearly, T6SSs are only one component of bacterial competitiveness in gut communities. Other factors, such as metabolic tradeoffs, spatial distribution, and coevolutionary dynamics, will need to be considered to better understand the influence of T6SSs and associated effectors on microbial community structure, particularly over longer timescales. Furthermore, the bee gut microbiota has recently been shown to affect resistance to opportunistic bacterial pathogens that invade the body cavity through the gut (33, 34). Potentially, the T6SSs and Rhs toxins of S. alvi and G. apicola contribute to resistance to invasion by potential pathogens, such as strains of Serratia and other Enterobacteriaceae. The specialized gut microbiota of social bees presents a promising system in which to investigate these questions and will undoubtedly offer further insights into competition within coevolving bacterial communities.
MATERIALS AND METHODS
RNA sequencing.
For the in vitro samples, S. alvi wkB2 (49) was streaked in triplicate on heart infusion agar plates supplemented with 5% defibrinated sheep’s blood (HIA plus 5% SB) and incubated at 35°C in a 5% CO2 environment for 24 h. RNA was extracted from plated cells using TRIzol (Ambion), according to the manufacturer’s instructions.
For in vivo samples, microbiota-free bees were acquired as previously described (50). Briefly, pupae were extracted from brood frames of managed hives in New Haven, CT (experiment A), and Austin, TX (experiment B), and allowed to emerge under sterile conditions. Within 36 h of emergence, bees were fed 5 µl of a 20% sucrose–phosphate-buffered saline (PBS) solution containing approximately 106 S. alvi wkB2 cells (optical density at 600 nm [OD600] of 0.5) or sterile PBS. Inoculated bees were transferred to sterile cup cages in triplicate groups of 8 to 10 and fed a filter-sterilized 1:1 sucrose solution and gamma-irradiated pollen ad libitum.
Four (experiment A) or 5 (experiment B) days after inoculation, bees were frozen at −80°C and then dissected on ice. Guts were placed in RNAlater (Thermo Fisher) and stored at −80°C. In experiment A, each replicate was comprised of three pooled ileums, while replicates in experiment B consisted of ileums and rectums of individual bees. For experiment B, DNA and RNA were extracted using the bead-beating and RNA-Bee method described by Jorth et al. (51). The absolute numbers of S. alvi 16S rRNA (RNA) and rRNA gene (DNA) copies in experiment B samples were quantified in triplicate with a 109- to 103-copy standard curve using the Beta-1009-qtF, Beta-1115-qtR, Gamma1-459-qtF, and Gamma1-648-qtR primers described in reference 18 on an Eppendorf Mastercycler EP RealPlex as described in reference 50. The Thermo Scientific Verso cDNA kit was used to synthesize cDNA for quantitative PCR (qPCR) according to the manufacturer’s instructions. Eukaryotic and prokaryotic rRNA was depleted using the Ribo-Zero Gold rRNA removal kit (Epidemiology).
The University of Texas at Austin Genomic Sequencing and Analysis Facility prepared stranded Illumina libraries for in vitro and experiment B samples and performed single-end 50-bp sequencing with an Illumina HiSeq4000 sequencer. Extraction and sequencing for experiment A are described in reference 23.
Read processing and analysis.
RNA-Seq reads were trimmed with Flexbar (52) to remove Illumina adapters, and Bowtie2 (53) was used to map trimmed reads to the S. alvi wkB2 genome. HTSeq-count (54) was used to count the number of reads mapping to each gene in the RAST annotation. Differential expression analysis was done with DESeq2 (55), using a false discovery rate cutoff of 0.05. Transcripts per million (TPM) were calculated using a custom R script based on the calculation described in reference 56. One experiment B replicate had reads that mapped to other bacteria and was excluded from the analysis. Pearson’s coefficients for the relationship between Rhs toxin gene TPM and position for each Rhs locus were calculated using the corr.test function implemented in R (57).
Reanalysis of Tn-Seq data.
The T6SS and Rhs loci of the NCBI-annotated S. alvi wkB2 genome (CP007446.1) were reannotated by locating open reading frames (ORFs) and performing blastp searches of the predicted proteins. The reannotated genome was used as the reference on which Tn-Seq data from a previous study (23) were remapped. Scoring of essential genes and genes beneficial in vivo was performed as described previously (23).
Genome sequencing.
DNA was extracted from S. alvi cultures using the Qiagen blood and tissue kit and submitted for library preparation and sequencing using the Illumina MiSeq platform with paired-end, 2- by 250-bp or 2- by 300-bp reads. Genomes were assembled with Velvet (58) or MaSuRCA (59) and annotated using the RAST (24) pipeline.
Identification of T6SS loci and correlation between T6SS-1 and Rhs genes.
T6SS homologs were identified by amino acid similarity to the T6SS-associated genes of S. alvi wkB2 using tblastx best hits with an E value cutoff of ≤10−5, coverage of ≥50%, and ≥50% identity. A 70% identity cutoff was used for tssH. tblastx (E value, <0.01) was used to determine the average sequence similarity of the proteins encoded by the T6SS-1 and T6SS-2/3 loci of wkB2. Four genes (tssA, tssE, tssJ, and tssM) had no matches below this threshold. For the nine remaining core T6SS genes, percent identity for each match was normalized by the percentage of residues in the query sequence that aligned to the reference sequence, and the average of these values was taken. Open reading frame (ORF) maps for wkB2 T6SS loci were visualized using Geneious 10.1.3. HMMER 3.0 (60) was used to identify S. alvi protein coding genes containing the Rhs core motif (TIGR03696) and to determine the number of these proteins that also contain a PAAR motif (PF05488). Strains were then grouped based on the presence of T6SS-1, T6SS-2/3, both, or neither. Rhs gene counts were compared between groups using a one-way analysis of variance (ANOVA) with Tukey’s honestly significant difference (HSD) multiple-test correction.
Cloning Rhs toxin and immunity genes.
Standard restriction enzyme cloning methods were used to clone rhs1 and rhs2 into pET21a and rhs2I into pJN105, whereas Gibson assembly (61) was used to clone rhs17 into pET21a and rhs1I and rhs17I into pJN105. Genes were amplified with Phusion DNA polymerase (New England Biolabs) and the primers listed in Table S6 in the supplemental material. Digestions were performed using NdeI and XhoI (rhs1) or NdeI and NotI (rhs2), and T4 DNA ligase (New England Biolabs) was used to catalyze ligations. The rhs1 forward primer was designed to generate a truncated C-terminal toxin domain comprised of the last 146 amino acids of the toxin. Electroporation was used to transform ligation and Gibson assembly reaction mixtures into E. coli DH5α, and PCR was used to screen transformants for the presence of the correct insert. Inserts were verified by sequencing the cloning site of the purified plasmid. All insert sequences matched the S. alvi wkB2 genome exactly, except for rhs2, which has a nonsense mutation 99 nucleotides before the end of the gene.
Primers used to clone Rhs toxin and immunity genes into expression vectors. Download TABLE S6, DOCX file, 0.1 MB (80.7KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of Rhs toxin and immunity genes in E. coli.
Purified plasmids were transformed into E. coli BL21(DE3) via electroporation for induction assays. Cells containing either pET21a::toxin and pJN105::immunity, pET21a::toxin and pJN105 empty vector, or pJN105::immunity and pET21a empty vector were streaked out on LB plates containing 10 µg/ml gentamicin, 75 µg/ml ampicillin, and 1% glucose and incubated overnight at 37°C. Single colonies were picked from these plates and streaked out onto fresh 10-µg/ml gentamicin and 75-µg/ml ampicillin plates with and without 0.1 mM IPTG or 0.5% l-arabinose. Growth was observed after 24 h at 37°C.
Growth curves were constructed to measure the effect of toxin and immunity gene induction on growth over time. Cells containing pET21a with or without a toxin gene and pJN105 with or without the immunity gene were streaked out on LB plates containing 10 µg/ml gentamicin and 75 µg/ml ampicillin and incubated overnight. Single colonies were used to inoculate 4 ml LB with selection in a 14-ml culture tube, and these cultures were grown to mid-log phase at 37°C and 220 rpm, then diluted back to an OD600 of 0.01 and incubated for another 3 h, and then diluted to an OD600 of 0.2. Ten microliters of this culture was added to 190 µl LB broth with selection, with and without IPTG or l-arabinose, in a 96-well plate. A Tecan Spark 10 M plate reader was used to measure the OD600 of cultures every 30 min for 12 h.
Phylogenetic trees.
Concatenated nucleotide sequences of 37 ribosomal protein genes (Table S3) were used to construct a phylogenetic tree for 31 S. alvi strains. Sequences were aligned with MUSCLE 3.8.31 (62). Bayesian and maximum likelihood (ML) analyses were performed using MrBayes 3.2 (63) and RAxML v8 (64), respectively. Both Bayesian and ML analyses used a general time-reversible model of nucleotide substitution with a proportion of sites assumed to be invariable and the remaining sites drawn from a gamma distribution. The Bayesian analysis was run until the standard deviation of split frequencies dropped below 0.01. Bootstrapping was performed for the ML analysis (n = 1,000). Phylogenetic trees were visualized with FigTree v.1.4.2 (65).
Phylogenetic trees of S. alvi T6SS-1 and T6SS-2/3 were constructed using the same method. T6SS homologs were extracted from a local BLAST database of S. alvi protein-encoding genes based on amino acid similarity to T6SS-associated genes in wkB2. The only gene from the wkB2 T6SS-1 locus that had amino acid identity to its T6SS-2/3 homolog above our cutoff was tssC.
The TssB protein is well conserved and has previously been used to determine evolutionary relationships between distantly related T6SSs (32, 66). A phylogeny of the TssB protein was constructed using representative sequences from S. alvi and G. apicola, as well as reference sequences from the NCBI RefSeq database. The SecReT6 database (67) was used to identify proteins associated with previously identified T6SS subtypes. CD-HIT (68) was used to cluster S. alvi and G. apicola TssB sequences by 95% similarity, and MUSCLE was used to align representative sequences from each cluster with the reference sequences. Maximum likelihood analyses were performed with RAxML v8 using the WAG amino acid substitution model and assuming a gamma distribution of rates with a proportion of invariable sites and 1,000 bootstrap replicates.
Comparison of Rhs loci.
Conserved domains in the S. alvi wkB2 Rhs genes were identified using the NCBI’s conserved domain database (28) with a 10−5 E value cutoff. C-terminal toxin domains were identified based on the location of the conserved DPXG(18)DPXG motif in the translated sequence. Homologous toxin domains were detected by comparing translated sequences of the wkB2 C-terminal domains to all S. alvi genomes with the tblastx (69) tool and 70% coverage and 70% amino acid identity cutoffs.
Rhs loci were identified using RAST annotations and manually edited in Geneious R9 (70). Regions of high primary sequence homology between S. alvi Rhs loci were identified using blastn (69) with a 10−5 E value cutoff and visualized with Circos (71).
To obtain the Rhs toxin cooccurrence network, C-terminal toxin domains were extracted from a codon alignment of all S. alvi and G. apicola Rhs genes and clustered at 90% nucleotide identity with CD-HIT-est (56). Predicted C-terminal domains shorter than 40 bp were excluded. From this, 364 unique toxin domains were identified. A more conservative analysis, in which a more stringent match requirement for the DPXG motif and a 200-bp length cutoff were applied, revealed 258 unique C-terminal domains. Output was visualized in Gephi v0.9.1 (72).
Tests of selection.
Homologous conserved gene and Rhs sequences (Table S3) were identified in S. alvi genomes using tblastx, based on amino acid identity to S. alvi wkB2 genes. Alignments were constructed with MUSCLE and manually checked in Geneious R9. Omega (dN/dS) ratios were calculated using the PAML 4.8 codeml model 0 (73). A one-way ANOVA with Tukey’s HSD multiple-test correction was used to compare the dN/dS ratios of conserved genes, Rhs core domains, and Rhs toxin domains.
Accession number(s).
RNA-Seq data from experiment B are deposited under accession number SRP082731 in the NCBI Sequence Read Archive. Genome sequences are deposited under accession numbers MDUY00000000, MDUZ00000000, MDVA00000000, MDVB00000000, MDVC00000000, MDVD00000000, MDVE00000000, MDVF00000000, MDVG00000000, MDVH00000000, MDVI00000000, MDVJ00000000, MEII00000000, MEIJ00000000, MEIK00000000, MEIL00000000, MEIM00000000, MEIN00000000, MEIO00000000, MEIP00000000, MEIQ00000000, MEIR00000000, MEIS00000000, MEIT00000000, MEIU00000000, MEIV00000000, MEIW00000000, and MEIX00000000 in GenBank.
ACKNOWLEDGMENTS
We thank Kim Hammond for maintenance of honey bee hives, Philipp Engel for providing strains, Hauke Koch for providing strains and sequenced genomes, Drew Vander Wood for assembly and annotation of genomes, Eva Frederick and Tejashwini Gattu for assistance with cloning experiments, Rebecca Chong for assistance with codeml, Sean Leonard for assistance in analysis of Tn-Seq data, and members of the Moran and Whiteley labs for helpful discussions. Sequencing was performed by the University of Texas Genome Sequencing and Analysis Facility, and computational analyses used the resources of the Texas Advanced Computing Center.
This work was funded by the National Institutes of Health award RO1GM108477-02 and the National Science Foundation award DEB1415604 to N.A.M. and the National Science Foundation award DEB1701430 to M.I.S. M.I.S. was supported in part by the Rodeo Fund of the Department of Integrative Biology.
Footnotes
Citation Steele MI, Kwong WK, Whiteley M, Moran NA. 2017. Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. mBio 8:e01630-17. https://doi.org/10.1128/mBio.01630-17.
REFERENCES
- 1.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cascales E, Cambillau C. 2012. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci 367:1102–1111. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNally L, Bernardy E, Thomas J, Kalziqi A, Pentz J, Brown SP, Hammer BK, Yunker PJ, Ratcliff WC. 2017. Killing by type VI secretion drives genetic phase separation and correlates with increased cooperation. Nat Commun 8:14371. doi: 10.1038/ncomms14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong M, Liang X, Smart M, Tang L, Moore R, Ingalls B, Dong TG. 2016. Microbial herd protection mediated by antagonistic interaction in polymicrobial communities. Appl Environ Microbiol 82:6881–6888. doi: 10.1128/AEM.02210-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. 2014. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, Goodlett DR, Ochman H, Mougous JD, Goodman AL. 2016. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A 113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. 2016. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A 113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E. 2017. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 22:411–419.e4. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne MJ, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cariveau DP, Powell JE, Koch H, Winfree R, Moran NA. 2014. Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus). ISME J 8:2369–2379. doi: 10.1038/ismej.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell E, Ratnayeke N, Moran NA. 2016. Strain diversity and host specificity in a specialized gut symbiont of honey bees and bumble bees. Mol Ecol 25:4461–4471. doi: 10.1111/mec.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. 2017. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci U S A 114:4775–4780. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran NA, Hansen AK, Powell JE, Sabree ZL. 2012. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong WK, Engel P, Koch H, Moran NA. 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A 111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol 78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch H, Abrol DP, Li J, Schmid-Hempel P. 2013. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol 22:2028–2044. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- 20.Greenblum S, Carr R, Borenstein E. 2015. Extensive strain-level copy number variation across human gut microbiome species. Cell 160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang X, Moore R, Wilton M, Wong MJQ, Lam L, Dong TG. 2015. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci U S A 112:9106–9111. doi: 10.1073/pnas.1505317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. 2016. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc Natl Acad Sci U S A 113:13887–13892. doi: 10.1073/pnas.1610856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durand E, Cambillau C, Cascales E, Journet L. 2014. VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol 22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcoforado Diniz J, Coulthurst SJ. 2015. Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol 197:2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. 2013. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A 110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. 2009. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Zou Y, She P, Wu Y. 2015. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol Res 172:19–25. doi: 10.1016/j.micres.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raymann K, Shaffer Z, Moran NA. 2017. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol 15:e2001861. doi: 10.1371/journal.pbio.2001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong WK, Mancenido AL, Moran NA. 2017. Immune system stimulation by the native gut microbiota of honey bees. R Soc Open Sci 4:170003. doi: 10.1098/rsos.170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engel P, Bartlett KD, Moran NA. 2015. The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. mBio 6:e00193-15. doi: 10.1128/mBio.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JB, Telepnev MV, Zudina IV, Bouyer D, Montenieri JA, Bearden SW, Gage KL, Agar SL, Foltz SM, Chauhan S, Chopra AK, Motin VL. 2009. Evaluation of a Yersinia pestis mutant impaired in a thermoregulated type VI-like secretion system in flea, macrophage and murine models. Microb Pathog 47:243–251. doi: 10.1016/j.micpath.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salomon D, Klimko JA, Trudgian DC, Kinch LN, Grishin NV, Mirzaei H, Orth K. 2015. Type VI secretion system toxins horizontally shared between marine bacteria. PLoS Pathog 11:e1005128. doi: 10.1371/journal.ppat.1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. 2014. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirchberger PC, Unterweger D, Provenzano D, Pukatzki S, Boucher Y. 2017. Sequential displacement of type VI secretion system effector genes leads to evolution of diverse immunity gene arrays in Vibrio cholerae. Sci Rep 7:45133. doi: 10.1038/srep45133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian B, Fadhil NH, Powell JE, Kwong WK, Moran NA. 2012. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3:e00377-12. doi: 10.1128/mBio.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE. 2016. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio 7:e01055-16. doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet 32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 44.Hall AR, Scanlan PD, Morgan AD, Buckling A. 2011. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett 14:635–642. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 45.Jackson AP, Thomas GH, Parkhill J, Thomson NR. 2009. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics 10:584. doi: 10.1186/1471-2164-10-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koskiniemi S, Garza-Sánchez F, Sandegren L, Webb JS, Braaten BA, Poole SJ, Andersson DI, Hayes CS, Low DA. 2014. Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet 10:e1004255. doi: 10.1371/journal.pgen.1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. 2014. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. mBio 5:e01305-14. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwong WK, Moran NA. 2013. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order “Enterobacteriales” of the gammaproteobacteria. Int J Syst Evol Microbiol 63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 50.Powell JE, Martinson VG, Urban-Mead K, Moran NA. 2014. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol 80:7378–7387. doi: 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. 2014. Metatranscriptomics of the human oral microbiome during health and disease. mBio 5:e01012-14. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dodt M, Roehr JT, Ahmed R, Dieterich C. 2012. FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology 1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. 2015. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 57.R Core Team 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 58.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics 29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 62.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu K, Linder CR, Warnow T. 2011. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS One 6:e27731. doi: 10.1371/journal.pone.0027731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambaut A. 2014. FigTree. http://tree.bio.ed.ac.uk/software/figtree.
- 66.Förster A, Planamente S, Manoli E, Lossi NS, Freemont PS, Filloux A. 2014. Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes. J Biol Chem 289:33032–33043. doi: 10.1074/jbc.M114.600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J, Yao Y, Xu HH, Hao L, Deng Z, Rajakumar K, Ou HY. 2015. SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ Microbiol 17:2196–2202. doi: 10.1111/1462-2920.12794. [DOI] [PubMed] [Google Scholar]
- 68.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. Blast+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks, p 361–362. In Proceedings of the Third International ICWSM Conference. Association for the Advancement of Artificial Intelligence, Palo Alto, CA: http://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154. [Google Scholar]
- 73.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 74.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A 112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential expression of S. alvi wkB2 T6SS genes, Rhs toxin and putative immunity genes, and genes involved in iron and nitrogen metabolism. Download TABLE S1, DOCX file, 0.2 MB (160.6KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains and genomes used in this study. Download TABLE S2, DOCX file, 0.1 MB (133KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ongoing deletion is eliminating the T6SS-2 locus in some S. alvi strains. The T6SS-2 locus in S. alvi strains PEB0171, wkB2, HK9, Gris1-3, and Fer1-2, which is bracketed by recO and integration host factor α in all but wkB2. The S. alvi strain phylogeny (left) shows the relationships between strains and supports the hypothesis that T6SS-2 has been lost multiple times in S. alvi. Download FIG S1, PDF file, 0.1 MB (91.7KB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. alvi genes used to reconstruct strain phylogeny and genes used for tests of positive selection. Download TABLE S3, DOCX file, 0.1 MB (93.4KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The association between S. alvi Rhs genes and T6SS-1 is consistent across host species. Statistical tests performed with ANOVA and Tukey’s HSD. *, Padj < 0.05; ***, Padj < 0.005. Download FIG S2, PDF file, 0.4 MB (386.9KB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conserved protein domains detected in S. alvi wkB2 Rhs toxin genes (E value, <0.00001) and immunity genes (E value, <0.001). Download TABLE S4, DOCX file, 0.1 MB (107.1KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fitness effects of transposon insertions in S. alvi wkB2 T6SS, Rhs toxin, and putative immunity genes. Download TABLE S5, DOCX file, 0.1 MB (139.8KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of the cognate immunity gene restores growth in cells expressing the rhs1 and rhs2 toxin genes. (A) l-Arabinose does not counteract the effects of toxin gene induction in the absence of the immunity gene. (B) Similarly, the presence of IPTG does not lead to reduced growth in the absence of the toxin gene. (C) Cells grown with 0.1 mM IPTG to induce rhs17 expression (dark teal circles and dark purple triangles) exhibit reduced growth relative to cells grown without IPTG (open circles and open triangles). Growth is restored by inducing immunity gene expression with 0.1% l-arabinose in cells containing rh17I (light teal circles) but not in cells with rhs1I (light purple triangles). (D and E) E. coli BL21(DE3) cells expressing rhs1 (D) or rhs2 (E) induced by IPTG demonstrate a severe growth defect in liquid culture that is at least partially alleviated by expression of the cognate immunity gene induced by l-arabinose. Download FIG S3, PDF file, 1 MB (1.1MB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rhs toxins are under purifying selection and affect S. alvi fitness. (A) dN/dS ratios for Rhs C-terminal toxin domains were not significantly different from those of the Rhs core. Both Rhs domains had dN/dS ratios of <1 but significantly higher dN/dS ratios than conserved housekeeping genes (one-way ANOVA with Tukey’s multiple comparisons of means, Padj < 0.005). Several homologous toxin domains were excluded from this analysis due to insufficient variation (i.e., only 0 to 5 polymorphisms). (B) Fold change in abundance of S. alvi wkB2 mutants with transposon insertions in Rhs toxins (top) and putative immunity genes (bottom) in vivo, relative to abundance in vitro (Padj < 0.05). E, gene essential in vitro; NS, nonsignificant change in mutant abundance in vivo, −, immunity gene not present in genome. Gray arrows show the 5′-to-3′ organization of genes for each locus. Download FIG S4, PDF file, 0.2 MB (247.3KB, pdf) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used to clone Rhs toxin and immunity genes into expression vectors. Download TABLE S6, DOCX file, 0.1 MB (80.7KB, docx) .
Copyright © 2017 Steele et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.