Abstract
The unicellular photosynthetic organism, Chlamydomonas reinhardtii, represents a powerful model to study mitochondrial gene expression. Here, we show that the 5′- and 3′-extremities of the eight Chlamydomonas mitochondrial mRNAs present two unusual characteristics. First, all mRNAs start primarily at the AUG initiation codon of the coding sequence which is often marked by a cluster of small RNAs. Second, unusual tails are added post-transcriptionally at the 3′-extremity of all mRNAs. The nucleotide composition of the tails is distinct from that described in any other systems and can be partitioned between A/U-rich tails, predominantly composed of Adenosine and Uridine, and C-rich tails composed mostly of Cytidine. Based on 3′ RACE experiments, 22% of mRNAs present C-rich tails, some of them composed of up to 20 consecutive Cs. Polycytidylation is specific to mitochondria and occurs primarily on mRNAs. This unprecedented post-transcriptional modification seems to be a specific feature of the Chlorophyceae class of green algae and points out the existence of novel strategies in mitochondrial gene expression.
INTRODUCTION
Mitochondria, organelles with a bacterial origin, have preserved a remnant ancestral genome. In all organisms studied so far, mitochondrial (mt) genomes code for only a very small number of proteins. For instance, 32 protein genes are encoded in the mt DNA of the land plant Arabidopsis thaliana, 13 in human and only 8 in the yeast Saccharomyces cerevisiae. Most of these proteins are subunits of the respiratory chain or the adenosine triphosphate (ATP) synthase and their expression is essential. Despite the common prokaryotic origin of mitochondria, mechanisms allowing mt gene expression have diverged. Studies on mt gene expression in various organisms have highlighted the acquisition of a number of new features and this, in a species-specific manner (1–3). In plant mitochondria, post-transcriptional processes including RNA editing, splicing of introns, maturation of 5′- and 3′-ends of RNA transcripts and RNA degradation play an important role in gene expression (e.g. (4)). Still, many questions remain unresolved (5), for example concerning the identity of promoters, the mechanisms of transcript processing and of translation initiation.
The green alga Chlamydomonas reinhardtii is a prime model organism for photosynthesis and flagellar motility (6), but it is also the only photosynthetic organism where mt transformation is possible (7). Mutants impaired in mt respiration are viable in photoautotrophic conditions (8). The Chlamydomonas mt genome is linear, very compact, with short intergenic sequences and no introns (9). It codes for only eight proteins, three transfer RNAs (tRNAs) and the small (SSU) and large (LSU) ribosomal RNA (rRNA) subunits fragmented into numerous ‘modules’ (10), (Figure 1). Transcription starts in the short intergenic region between nad5 and cox1 genes and generates two divergent primary co-transcripts which are subsequently processed to generate mature RNAs (11,12). Previous analysis on mt mRNAs showed that their size is close to that of their coding sequence (CDS) indicating the existence of very short 5′- and 3′-UTRs (untranslated regions). This was confirmed for a few transcripts using S1 nuclease protection and primer extension experiments (13,14).
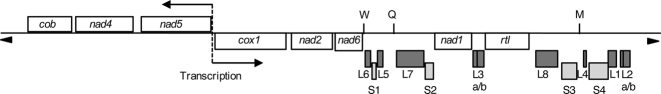
Figure 1.
Chlamydomonas reinhardtii mitochondrial genome. Schematic map of the C. reinhardtii mitochondrial genome. White boxes represent protein-coding genes: (cob) apocytochrome b of complex III; (nad1, 2, 4, 5 and 6) subunits of complex I; (cox1) subunit 1 of complex IV; (rtl) reverse transcriptase-like protein. The 3 tRNA genes are indicated with the one letter code (W, Q and M). The dark gray and light gray boxes correspond to the LSU (L1–L8) and SSU (S1–S4) RNAs fragments respectively. The bi-directional origin of transcription between nad5 and cox1 genes is represented by a dashed vertical line and two horizontal arrows. Telomeric regions are indicated by arrowheads.
In this work, the C. reinhardtii mt transcripts were analyzed using different methods (Supplementary Figure S1A): 5′ and 3′ RACE (Rapid Amplification of cDNA Ends), cRT-PCR (circular Reverse Transcription-Polymerase Chain Reaction) and three protocols based on Illumina single-end sequencing, namely sRNA-Seq (small RNA sequencing, TruSeq), directional WTSS (Whole Transcriptome Shotgun Sequencing, stranded TruSeq) and bi-directional WTSS (non-stranded TruSeq, public data retrieved from SRA). Our results shed light on two singular characteristics of the mRNA transcripts. First, all of them start at the AUG initiation codon of the CDS. Second, they carry post-transcriptionally added C-rich tails at the 3′-end of their short 3′-UTR. This unprecedented observation seems to be a specific feature of the Chlorophyceae class of green algae.
MATERIALS AND METHODS
Algae strains and growth conditions
Chlamydomonas reinhardtii strains CC-4351 (cw15–325 arg7–8 mt+) and CC-5101 (T222 nit1 nit2 mt+), Chlorella sorokiniana 211–32, Tetradesmus (Scenedesmus) obliquus 276–10 and Polytomella parva SAG 198.80 were grown on Tris-Acetate Phosphate (TAP) medium (15) supplemented with arginine (100 μg/ml) for CC-4351. Coccomyxa subellipsoidea C-16 was grown on Modified Bold’s Basal Medium (16). Culture conditions were 25°C under white light (10–50 μE m−2 s−1). The ‘Centre national de ressources biologiques marines’ (EMBRC France) provided us with Chondrus crispus. The Scandinavian Culture Collection of Algae and Protozoa (SCCAP) provided us with Cyanophora paradoxa (K-0262) and Pedinomonas minor (K-0264) strains. Physcomitrella patens Grandsen strain was provided by R. Resky (University of Freiburg, Germany). Ostreoccocus tauri OTTH0595 strain was provided by H. Moreau (Integrative Biology of Marine Organisms CNRS–UMR 7237). For all the organisms, strains and genome accessions are in Supplementary Table S1.
RNA analysis
Crude mt fractions were isolated from cell wall-less C. reinhardtii strain CC-4351 by digitonin treatment according to (17). Chlamydomonas reinhardtii RNA was prepared from mt fraction or whole cells using TRI Reagent® (Molecular Research Center) according to manufacturer’s instructions. For C. crispus, C. paradoxa, P. minor and P. patens, RNA was prepared from whole cells using TRI Reagent®. For C. subellipsoidea, C. sorokiniana, S. obliquus and P. parva, RNA was prepared from whole cells according to (18). For C. reinhardtii, cRT-PCR, 5′ and 3′ RACE experiments were performed with mt RNA, Illumina sequencing with whole-cell RNA. For the phylogenetic study of mt mRNA polycytidylation, 3′ RACE experiments were performed with whole-cell RNA from all species including C. reinhardtii.
Oligonucleotides are described in Supplementary Table S2. The cRT-PCR, 5′ and 3′ RACE analyses were performed using Sanger sequencing as in (19). Briefly, 3 μg of mt RNA or 4–6 μg of total RNA were ligated at the 3′-end to adaptor and cDNA was synthesized with SuperScript IV (Invitrogen) primed by the complementary RT primer. PCR was performed with gene-specific primers and RT primers. For some species, nested PCR were necessary to avoid non-specific amplifications. For 5′-RACE, the first-strand cDNA synthesized above was C-tailed with the terminal deoxynucleotidyl-transferase (Invitrogen) in the presence of dCTP. PCR was performed with a gene-specific forward primer and oligo(dG). For cRT-PCR, 4–6 μg of total RNA was circularized by T4 RNA ligase (New England Biolabs) prior to reverse transcription and PCR using gene-specific oligonucleotides. For RT-PCR analysis, 4 μg total RNA treated with DNAse RQ1 (Promega) was reverse transcribed in a 40 μl reaction, using 2,5 μM oligo(dG) primer or a mix of 2.5 µM oligo(dT) and 12 ng/μl random hexamer primer (Promega).
Sequence alignments were generated with MacVector software as described in more details in Supplementary Figure S1B. In order to monitor possible artifacts, 1 ng of synthetic transcript was incubated with 3,5 μg of total RNA and 3′ RACE analysis was performed. Analysis of this synthetic transcript showed that 1–2 additional nucleotides are observed at the 3′ extremity and in very few cases more than 3 nt (Supplementary Figure S1C). For 3′ RACE and cRT-PCR analysis, sequences with more than 3 nt added were therefore considered as bona fide 3′-tails.
Illumina sequencing
Single-end 50 nt directional WTSS and sRNA-Seq Illumina datasets (SRA accessions, resp.: SRX2725800 to SRX2725809 and SRX2725861 to SRX2725864) were used, along with ninety bi-directional WTSS datasets (single-end 100 nt) retrieved from SRA as described in (20). WTSS reads were mapped to the mt genome (EU306622) using bwa (mem). sRNA-Seq reads were mapped with either bwa (aln) or Bowtie2 to allow soft-clipping (21,22). Analysis of the mappings used bamtools, bedtools and the IGV browser (23–25).
Phylogenetic and prediction analysis
The maximum likelihood phylogenetic tree of Chlorophyta Class-II NTRs (RNA-nucleotidyl transferases) was generated with PhyML (http://www.atgc-montpellier.fr/phyml), (26) using a clustalW alignment modified to align catalytic residues and trimmed to the conserved regions. Organellar predictions were determined using TargetP (http://www.cbs.dtu.dk/services/TargetP/), (27) and Predalgo (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main), (28).
RESULTS
Reannotation of the Chlamydomonas mt genome
Combining various approaches, the mature 5′- and 3′-ends of all mt transcripts of C. reinhardtii were precisely determined. Our genome browser (http://chlamy-organelles.ibpc.fr) presents the data as well as a novel annotation that are proposed. Compared to the previous one (EU306622), one mis-sense error in the cob CDS is corrected, the start of nad2 is moved three codons upstream, we modify the ends of almost all rRNAs and reintroduce rrnL3a and rrnL2b present in the initial annotation (NC_001638). Therefore, the C. reinhardtii mt genome encodes 8 mRNAs, 3 tRNAs and 14 rRNA fragments (4 for the SSU, 10 for the LSU), (Figure 1).
Intercistronic maturation generates adjacent 3′- and 5′-termini for almost all RNAs
Concerning transcript processing, the near-absence of WTSS reads connecting successive cistrons indicates that the two primary transcripts that originate in the cox1_nad5 intergenic region are very efficiently processed into mono-cistronic RNAs (Figure 2). Gray et al. (12) previously showed that nad5, nad2 and cox1 mRNAs lack 5′-UTR. Here, 5′ RACE experiments carried out on all mRNAs extend this analysis and indicate that almost all mRNA molecules start at the A of the AUG start codon, meaning that they do not have a 5′-UTR (Figure 2A). In addition, the 5′-end of all mRNAs (except nad1 and cob) is marked by a cluster of sRNAs starting at the AUG initiation codon. A similar result was observed for most rRNAs, (Supplementary Figure S2). It is unknown whether these sRNAs result from protection by RNA-binding proteins, as observed in chloroplasts, or reveal an intrinsic stability of 5′-ends toward exonucleases (20,29). Their length varies greatly (the mode ranges from 16 to 33 nt) between mRNAs (Supplementary Figure S2) and at each locus heterogeneity is high compared to the cosRNAs (clustered organellar sRNAs) described in Arabidopsis organelles (29) and Chlamydomonas chloroplast (20). This suggests that they do not represent footprints of stalled ribosomes or 30S subunits. Whatever the origin of these sRNAs, their presence proved useful to map 5′-ends.
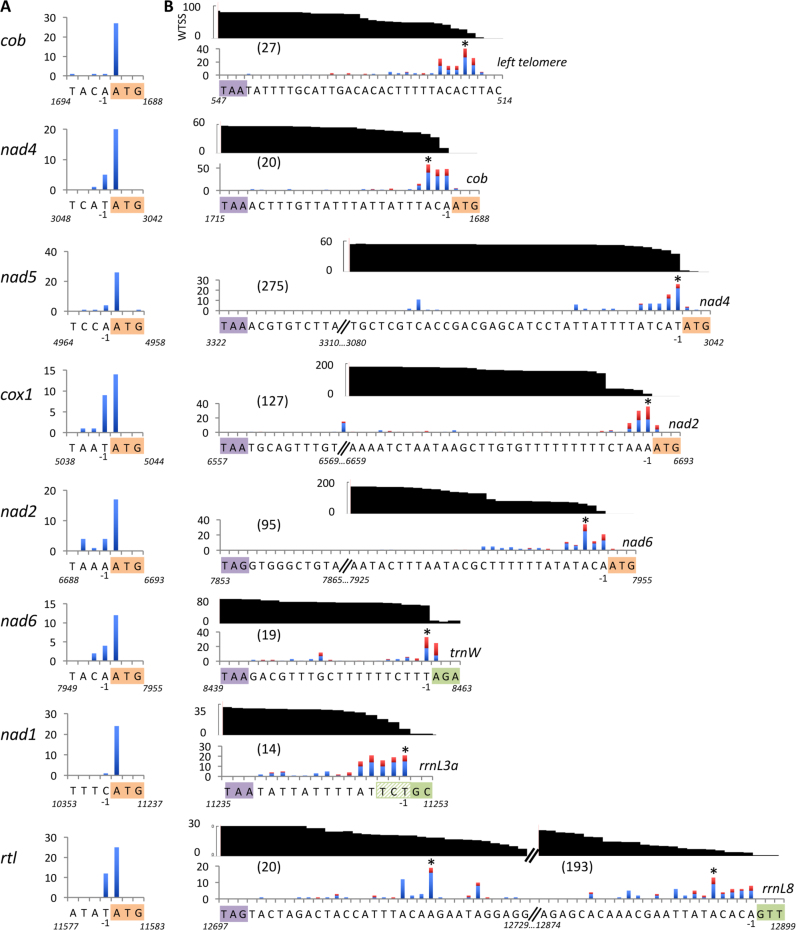
Figure 2.
Mapping of the mRNAs extremities. (A) Number of 5′ RACE clones for each 5′ nucleotide position. The start codon is highlighted and its coordinate on the genome indicated. (B) Number of 3′ RACE and cRT-PCR clones (respectively blue and red bars) for each position of the 3′ nucleotide. The asterisk (*) indicates the main position(s) of the 3′-end, used to calculate the size of the 3′-UTR (numbers in brackets). The UTR sequence is shown under the x-axis, with the stop codon and the first nucleotides of the next gene highlighted in colors. For rrnL3a, two start positions (TC or GC) were observed in 5′ RACE and sRNA data (Supplementary Figure S2) respectively. The chart in black indicates the coverage, expressed as reads per million, of directional WTSS reads starting inside the gene of interest. For cox1, the abrupt drop at the end of the poly-U stretch is an artefact due to the presence in most reads of a larger number of U’s, preventing mapping of the downstream nucleotides (see Supplementary Figure S3).
In contrast, all mRNAs had 3′-UTRs (Figure 2B) with sizes corresponding to the downstream intergenic regions (between 14 and 275 nt). The 3′-ends of nad4, nad5, cox1, nad2, nad6, nad1 mRNAs were mapped immediately upstream of the next stable transcript, or shortly before. This was true regardless of the nature of downstream genes, coding for a protein, a tRNA or a rRNA. Heterogeneity was slightly higher than at the 5′-end, suggesting that some molecules undergo trimming at the 3′-end after processing at the AUG. The cob mRNA, last in the leftward transcription unit and thus not followed by a gene, also harbors a 3′-UTR that ends just before the left telomere. The rtl mRNA has two major 3′-ends regions, shortly downstream of the stop codon and just upstream of rrnL8. For rRNAs as well, the main rule was that the mature 3′-ends lie at, or just a few nt upstream of the start of the next gene (Supplementary Figures S2 and 3). In addition to the previously described spacer regions separating rRNAs and tRNAs (30,31), a few other short sequences (sometimes as short as 3 nt) were found downstream tRNAs or rRNAs for which no transcripts have been observed (see bidirectional WTSS on genome browser: http://chlamy-organelles.ibpc.fr). In summary, our data show that the 3′-end of a mature RNA most of the time corresponds to the 5′-end of the downstream RNA.
A new type of tails at the 3′-end of mitochondrial transcripts
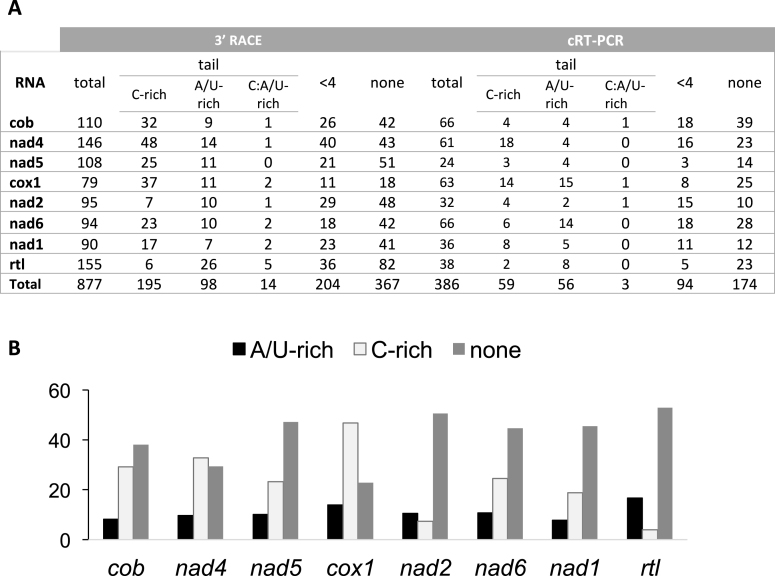
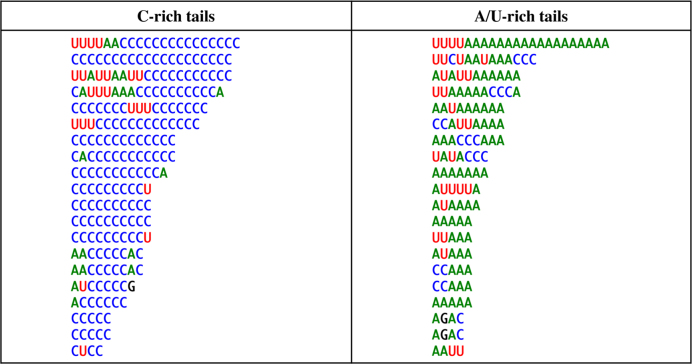
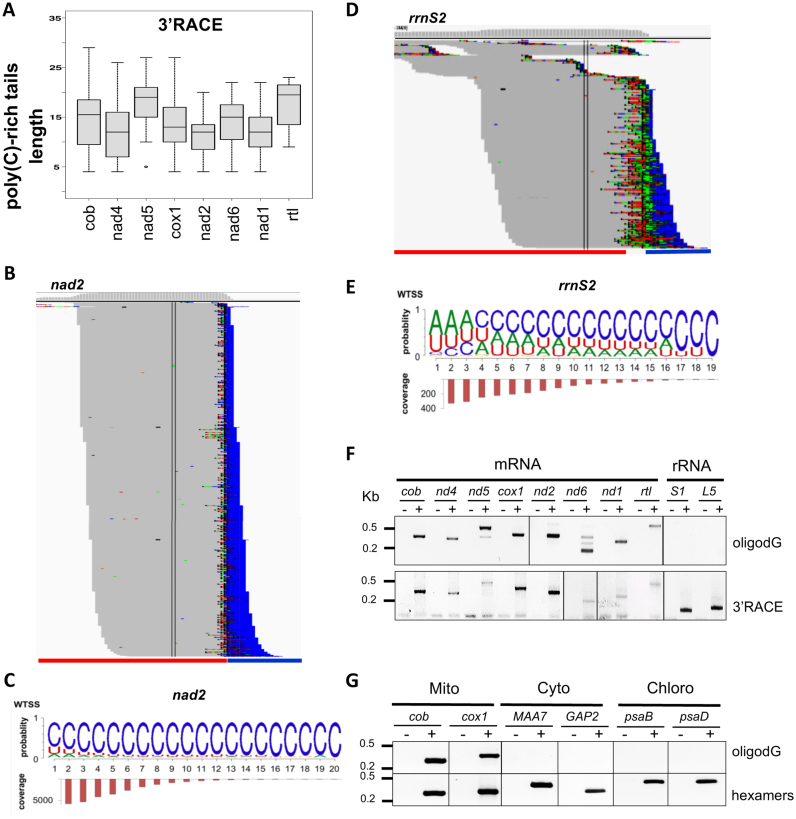
Sequencing also revealed unusual post-transcriptionally added tails at the 3′ extremities of all mRNAs (Supplementary Figure S3 and Figure 3). These tails are present in high proportions i.e. 35% of total transcripts analyzed using 3′ RACE. Interestingly, the nucleotide composition of these tails is distinct from that described in any other system: while containing certain proportions of Adenosine (20.1%) and Uridine (15.4%), they were strongly enriched in Cytidine (63.7%), a nucleotide hitherto not described in transcript tails. Figure 4 presents randomly selected examples of 3′-tails observed using 3′ RACE for all eight mRNAs. The tails have been partitioned between those predominantly composed of A and U and those composed mostly of C (Figure 3). Both types are equally represented for nad2 and the A/U-rich tails predominated for rtl, but for the six other mRNAs, the C-rich tails are by far the most abundant. Polycytidylation ratios (the fraction of mature mRNAs showing a C-rich tail) based on 3′ RACE clones vary between 3.2–46.8% among genes, with an average of 21.7%. The highest ratios were observed for nad4 and cox1. The length of the tails predominantly fell between 10–20 nt, with an average of 14, a maximum of 29 nt and substantial variations between mRNAs (Figure 5A). In some cases, homopolymeric tails comprising up to 20 consecutive cytidines were observed. Many ‘composite’ tails were also observed, defined here as tails starting with an A/U-rich sequence and ending in a C-rich sequence. For most genes, mRNAs with a complete 3′-UTR, (i.e. ending close to the start of the next transcript) were more likely to show C-rich tails than the truncated ones, where tails were fewer and shorter (Supplementary Figures S3 and 4). The analysis of WTSS data confirmed the presence of C-rich tails on mt mRNAs (Figure 5B–D). C prevailed at almost all positions of the tails (e.g. Figure 5C), even more than with other methods (Supplementary Figure S4), with a smaller proportion of A/U-rich and composite tails. WTSS provides deep coverage, but because it involves priming the reverse transcriptase with degenerate oligonucleotides, it rarely reaches the 3′-end and it is biased toward G/C rich sequences. The ligation-based methods (3′ RACE, cRT-PCR) were therefore favored to describe the composition and length of the tails.
Figure 3.
Summary of the 3′-UTR analysis of mRNAs. (A) For each method, the total number of sequences analyzed are indicated. Sequences were classified in three groups (see ‘Materials and Methods’ section): sequences without any added nt (none), sequences with 1–3 nt (<4) and sequences with a tail i.e. with more than 3 nt (tail). The tails were classified into three categories according to their nt content [(nt observed/nt tail length)*100]: tails with >50% of Cytidine (C-rich), tails with >50% of Adenosine/Uridine (A/U-rich) and tails with equal amounts of Cytidine and Adenosine/Uridine (C:A/U-rich). (B) Histogram summarizing the data obtained in (A) by the 3′ RACE approach and showing the percentage of transcripts with A/U-rich tail (black), C-rich tail (pale gray) and no tail (dark gray) for each mt gene.
Figure 4.
Randomly-selected C- (left) and A/U-rich (right) tails from 3′ RACE experiments.
Figure 5.
Analysis of C-rich tails. (A) Boxplot analysis of the length of C-rich tails observed by 3′ RACE. (B) and (D) 3′-tails at the end of nad2 and rrnS2, respectively (red, with a blue line marking the next gene). Browser view of WTSS 3′-soft-clipped reads; coverage is plotted in log scale above the reads and only soft-clipped bases are colored (C: blue, U: red; A: green; G: brown). (C) and (E) the corresponding proportion-based sequence logos and length profiles for the 3′-tails using WTSS are shown. (F) RT-PCR to detect C-rich tails on mt transcripts by oligo(dG) priming. The 3′ RACE experiment serves as a control. (G) RT-PCR showing absence of C-rich tails on cytosolic and chloroplast transcripts. First strand cDNA synthesis was primed with oligo(dG) (top) or random hexamers (bottom).
Because 3′-A-rich tails within transcripts are associated with their degradation in bacteria and chloroplasts (32), polycytidylation sites were searched inside transcripts. For this, a cox1 specific primer positioned far-upstream cox1 3′ end was used in 3′ RACE experiments. This analysis identified 61 transcripts truncated within the CDS, but none carried a C-rich tail, (Supplementary Figure S5A). WTSS revealed the presence of 3′-tails within the CDS of cox1 and cob, but the density was 33 and 805 times lower than over the 3′-UTRs, respectively. They were short and showed no enrichment in C (Supplementary Figure S5B).
Comparison of mRNA and rRNA tails
In order to determine whether C-rich tails are specific of mRNAs or not, the 3′-ends of the 14 rRNA fragments were analyzed by 3′ RACE. Out of 304 sequences obtained, only 18 tails were found (6%), all short and A/U-rich, and in only 9 of the rRNA modules (Supplementary Figure S3). Directional WTSS confirmed the enrichment of rRNA tails in A/U compared to mRNAs, but some C nucleotides were also found, especially toward tail ends (Figure 5D, E and Supplementary Figure S4). These results were confirmed using a standard 20-cycle PCR protocol on oligo(dG)-primed cDNA: amplification was observed for all mRNAs, but not rRNAs (Figure 5F), in spite of the abundance of the templates. So far, polycytidylation has been reported neither for plastidial nor for cytosolic mRNAs. Using oligo(dG)-priming, the absence of C-rich tails in plastidial and cytosolic mRNAs of Chlamydomonas was confirmed for two transcripts of each compartment (Figure 5G). In conclusion, C-rich tails in Chlamydomonas are only present in mitochondria, where they are added preferentially to mRNAs.
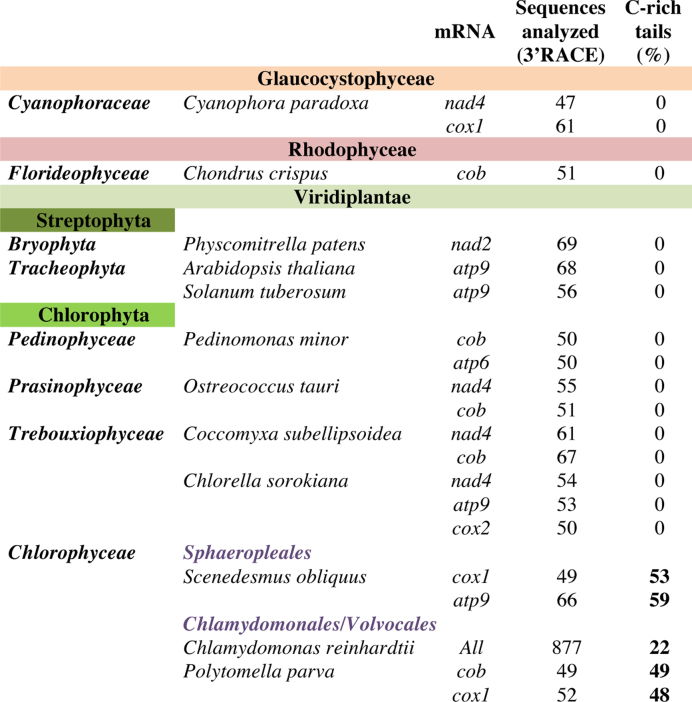
Polycytidylation is a hallmark of the Chlorophyceae
In order to describe the phylogenetic realm of mt mRNA polycytidylation, 3′ RACE analyses were performed on total RNA from a number of species belonging to the three clades of Archaeplastida, i.e. eukaryotes with primary plastids. They include the Glaucocystophyte C. paradoxa, the red alga C. crispus and representatives from the green lineage including three Streptophytes (the moss P. patens and two flowering plants A. thaliana and Solanum tuberosum) and five Chlorophyte algae ranging from the basal P. minor to the non-photosynthetic P. parva, a close relative of C. reinhardtii (33). For each species, up to 3 mt transcripts were analyzed by 3′ RACE (Figure 6 and Supplementary Figure S6). Polycytidylation was found in two other Chlorophyceae, namely P. parva and S. obliquus. This trait was not found in the other species examined, in particular in the green algae P. minor, O. tauri, Chlorella sorokiana and C. subellipsoidea. It therefore appears that the molecular process leading to C-rich tails is specific to the Chlorophyceae class.
Figure 6.
Analyzed organisms on the Archaeplastida lineage show the presence of C-rich tails in Chlorophyceae.
To further explore the Chlorophyceae, genome-wide WTSS datasets retrieved from the SRA or MMETSP databases were also analyzed (Supplementary Table S3). In the colonial alga Volvox carteri, complete intercistronic cleavage were found, mRNAs starting at AUG and C-rich tails at the end of the mRNAs, with rRNA tails enriched in As. The major difference with C. reinhardtii was that most 3′-UTR ended far upstream of the next gene. In P. parva, most mRNAs appeared to lack a 5′-UTR and showed C-rich tails, but the enrichment in A near the start was usually more pronounced than in C. reinhardtii, especially for mRNAs that were followed by a rRNA. In C. leiostraca and C. moewusii, two more distantly related Chlamydomonas species (34), polycytidylation of mRNAs was also found, even if the extent appeared lower than in C. reinhardtii.
DISCUSSION
The expression of the compact C. reinhardtii mt genome leads from two primary transcripts, to 3 tRNAs, 14 fragmented rRNA modules and 8 mRNAs containing no 5′-UTR and short 3′-UTR. If left and right telomeres are excluded, 905 nt (5.7%) are not part of a mature transcript. This organization is almost as compact as that of animal mt genomes, where a ‘tRNA punctuation’ model prevails for the generation of mature RNAs: primary transcripts are processed by endonucleolytic cleavage by RNase P and RNase Z respectively upstream and downstream of each tRNA to release mature rRNAs and mRNAs (35,36). In C. reinhardtii mitochondria as well, endonucleolytic cleavage is the major mode of RNA maturation. However, only one mRNA (nad6) has its 3′-end generated by PRORP, the enzyme that cleaves at the 5′ extremity of tRNAs in plants and algae (37) and no 5′-end coincides with the 3′-end of a tRNA, where RNase Z cleavage occurs. Thus, the identity of the endonuclease(s) cleaving at the AUG initiation codon of mRNAs and at the extremities of rRNAs in Chlamydomonas remains to be determined. In human mitochondria, FAST-Kinase Domain proteins have been shown to control, via their RAP domain, the maturation of transcripts not flanked by tRNAs (38). In Chlamydomonas, octotricopeptide repeat proteins with RAP domain such as those described in (39) may be involved, if targeted to the mitochondrion. A related question is how the cleavage sites are determined. Secondary structures within the 3′-UTR of mt mRNAs and palindromic repeats in the intergenic regions have been described (30,31) but they do not match the processing sites observed here. In addition, intervention of exonucleases is suggested not only by the ‘spacer regions’ not represented in mature RNAs (31) but also by the heterogeneity in length that we observe at the 3′-end of all mRNAs. The exonucleases identity is uncertain, because PNPase and the two RNase-II homologs in Chlamydomonas appear to function in the chloroplast (40,41).
The most intriguing feature of mRNA 3′-ends revealed by our study is the presence of non-templated post-transcriptionally added C-rich tails at the 3′-ends of mRNAs. In a study targeting all cell compartments, C was the only nucleotide not found to efficiently form tails (42). Thus, they potentially represent a novel way to regulate gene expression. Up to now, adenylation and uridylation were the two sole forms of nucleotide addition observed at the 3′-ends of mitochondrial RNAs. In animals, polyadenylation mostly stabilizes transcripts and completes the stop codon of several mRNAs, while in plants it serves as a transient signal for degradation (32,43). Accordingly, the 3 Streptophytes mt genes examined here by 3′-RACE showed only rare 1–2 nt additions, mostly As. In Myxomycetes, non-templated 3′ poly(U) tails were identified on edited mt mRNAs (44) while in Trypanosoma brucei, correctly edited mt transcripts are tailed with a poly(A/U) sequence of 200 to 300 nt which allows their recognition by the mt ribosomes and their translation activation (45).
C-rich tails were not observed in a previous study of Chlamydomonas mitochondria (46) which only retrieved A/U rich tails similar to those describe here, and long poly(A) tails (up to A32) that were not observed in this study. This is probably due to the use of oligo(dT) priming, rather than to strain differences, as C-rich tails were found here in various C. reinhardtii strains as well as in other Chlorophycean algae belonging to diverse genera (Chlamydomonas, Volvox, Polytomella, Scenedesmus). Trebouxiophyceae and Prasinophyceae had only A-rich tails, while a red alga (Chondrus) showed no tail and the Glaucophyte Cyanophora showed an unexpected U-enrichment in the tails of cox1. The emergence of polycytidylation is therefore a late event in the algal lineage, coinciding approximately with rRNA fragmentation (47).
What are the mechanisms and consequences of polycytidylation in Chlamydomonas mitochondria? It is first notable that rRNAs showed short tails with very little poly(C) and almost never exactly at the mature 3′-end. This could be due to their rapid folding preventing access of the polycytidylation enzyme. mRNAs truncated within the CDS also showed rare and short tails, with no particular C-enrichment, which suggests that polycytidylation could be restricted to translatable mRNAs. It may be significant that rtl, the less conserved, less abundant and probably less translated mRNA, showed the weakest C-enrichment in its tails. In UTRs, C-rich tails are more frequent at or near the initial cleavage site, suggesting that polycytidylation is an early maturation event. UTRs that have undergone trimming can also be tailed, but apparently with much lower efficiency. If the putative 3′-5′ exonuclease(s) have a lower affinity for 3′-C residues, C-tails could constitute a protection, stabilizing the longer transcript, while decay would occur once the A/U rich part of the tail or the genome-encoded UTR is reached. It is also noteworthy that many tails start being A/U rich before C-additions prevail. This can be interpreted in the framework either of two different RNA-NTRs acting sequentially, or a single one changing specificity as polymerization progresses.
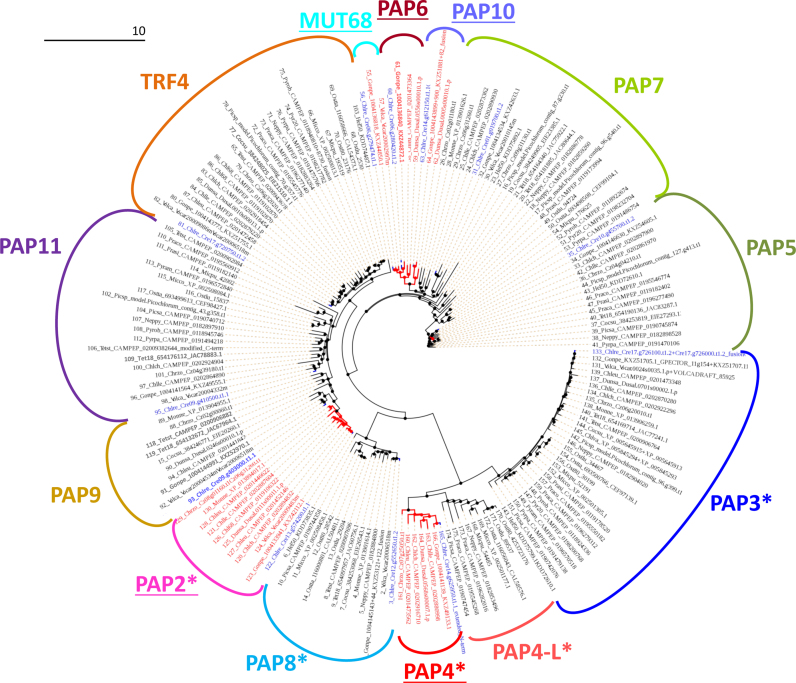
Since the lowly discriminating PNPase is believed to act mostly in the chloroplast (41), it is not a prime candidate for the generation of poly(C) tails. PAP1, a class-I NTR, is universally involved in cytosolic polyadenylation. The mt polycytidylation enzyme must therefore be sought among Class-II NTRs, even though the existence of still unknown classes of NTRs cannot be excluded. Among Chlamydomonas class-II NTRs (40), only PAP3 and PAP4 have been experimentally studied (46). As a first attempt to pinpoint the enzyme, we performed targeting predictions (Supplementary Table S4) and phylogenetic analysis (Figure 7) of Chlorophyta class-II NTRs, including a newly described isoform that we called PAP11. Among the 13 groups of NTRs that we identified, the majority was not predicted to reside in an organelle. This includes TRF4 and MUT68 which are known to act in the nucleolus or cytosol, respectively (48,49). Only PAP2, PAP3, PAP4, PAP4-L and PAP8 were consistently predicted to be organelle-targeted. Among them, PAP3 can be excluded as is has been shown to be a tRNA-CCA nucleotidyl transferase (46). PAP4-L, although related to Chlorophyceae-specific PAP4, is specific of Prasinophyceae which show A-rich tails. As to PAP8, it is present in all Chlorophytes and therefore not a prime candidate either. PAP4 and PAP2 thus appear as prime targets for future studies. The N-terminal sequence of CrPAP4 has been shown to address GFP to mitochondria in a heterologous system (46). This study also showed that CrPAP4 is able to elongate an A20 RNA substrate, but the nature of the added nucleotide(s) was not investigated. Among Chlamydomonas PAPs, CrPAP4 is the best hit of AGS1 which performs polyadenylation in Arabidopsis mitochondria (43). Note that the polymerization specificity of class-II NTRs can be low: in vitro, Escherichia coli poly(A) polymerase I can use all four triphosphate nucleotides with equal efficiency (50), which raises the question of whether the relative abundance of ATP and CTP plays a role in determining the composition of the tails. Further experiments will be required in order to identify the mitochondrial cytidyl transferase and shed light on the function of C-rich tails for Chlorophyceae mitochondrial gene expression processes.
Figure 7.
Phylogenetic tree of Chlorophyta class-II NTRs. Groups with Chlorophycean and non-Chlorphycean members are colored in red and black (branches and names), respectively. Chlamydomonas reinhardtii sequences are highlighted in blue. Groups are named based on the Chlamydomonas ortholog in Phytozome website (Chlamydomonas reinhardtii v5.5, http://phytozome.jgi.doe.gov/), those with only Chlorophycean members are underlined, those where most sequences are predicted organelle-targeted by TargetP or Predalgo are marked by an asterisk (Suplementary Table S4). Note that a PAP4 related family (PAP4-L) exists in Prasinophyceae. The abbreviations of the genus and species are listed in the Supplementary Table S4.
DATA AVAILABILITY
The data as well as a novel annotation proposed here for the Chlamydomonas mt genome can be visualized on our genome browser (http://chlamy-organelles.ibpc.fr) and downloaded by right-clicking on the tracks.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Gregorutti, M. Radoux and N. Cossemans for helpful technical assistance and J. Umen for sharing Volvox RNA-Seq data. A. Alioua is warmly acknowledged for sequencing work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (CNRS) in association with the Université de Strasbourg; Université Pierre et Marie Curie, Paris 06; Agence Nationale de la Recherche [ChloroRNP ANR-13-BSV7–0001-001]; ‘Initiative d’Excellence’ [DYNAMO ANR-11-LABX-0011–01, MITOCROSS ANR-11-LABX-0057]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mai N., Chrzanowska-Lightowlers Z.M., Lightowlers R.N.. The process of mammalian mitochondrial protein synthesis. Cell Tissue Res. 2017; 367:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ott M., Amunts A., Brown A.. Organization and regulation of mitochondrial protein synthesis. Annual Rev. Biochem. 2016; 85:77–101. [DOI] [PubMed] [Google Scholar]
- 3. Salinas T., Remacle C., Maréchal-Drouard L.. Duchêne AM. Mitochondrial translation in green algae and higher plants. Translation in Mitochondria and Other Organelles. 2013; Heidelberg; NY; Dordrecht; London: Springer; 181–206. [Google Scholar]
- 4. Binder S., Brennicke A.. Gene expression in plant mitochondria: transcriptional and post-transcriptional control. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2003; 358:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hammani K., Giegé P.. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014; 19:380–389. [DOI] [PubMed] [Google Scholar]
- 6. Merchant S.S., Prochnik S.E., Vallon O., Harris E.H., Karpowicz S.J., Witman G.B., Terry A., Salamov A., Fritz-Laylin L.K., Maréchal-Drouard L. et al. . The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007; 318:245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Remacle C., Cardol P., Coosemans N., Gaisne M., Bonnefoy N.. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:4771–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salinas T., Larosa V., Cardol P., Maréchal-Drouard L., Remacle C.. Respiratory-deficient mutants of the unicellular green alga Chlamydomonas: A review. Biochimie. 2013; 100:207–218. [DOI] [PubMed] [Google Scholar]
- 9. Grant D., Chiang K.S.. Physical mapping and characterization of Chlamydomonas mitochondrial DNA molecules: their unique ends, sequence homogeneity, and conservation. Plasmid. 1980; 4:82–96. [DOI] [PubMed] [Google Scholar]
- 10. Boer P.H., Gray M.W.. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell. 1988; 55:399–411. [DOI] [PubMed] [Google Scholar]
- 11. Duby F., Cardol P., Matagne R.F., Remacle C.. Structure of the telomeric ends of mt DNA, transcriptional analysis and complex I assembly in the dum24 mitochondrial mutant of Chlamydomonas reinhardtii. Mol. Gen. Genet. 2001; 266:109–114. [DOI] [PubMed] [Google Scholar]
- 12. Gray M.W., Boer P.H.. Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial DNA. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1988; 319:135–147. [DOI] [PubMed] [Google Scholar]
- 13. Boer P.H., Gray M.W.. Genes encoding a subunit of respiratory NADH dehydrogenase (ND1) and a reverse transcriptase-like protein (RTL) are linked to ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. EMBO J. 1988; 7:3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma D.P., Yang Y.W., King T.Y., Hasnain S.E.. The mitochondrial apocytochrome b gene from Chlamydomonas reinhardtii. Plant Mol. Biol. 1990; 15:357–359. [DOI] [PubMed] [Google Scholar]
- 15. Harris E. Chlamydomonas as a model organism. Annu. Rev. Plant Phys. Plant Mol. Biol. 2001; 52:363–406. [DOI] [PubMed] [Google Scholar]
- 16. Stein J. Handbook of Phycological methods: Culture methods and growth measurements. 1973; Cambridge; London; NY; New Rochelle; Melbourne; Sydney: Cambridge University Press. [Google Scholar]
- 17. Salinas T., Duby F., Larosa V., Coosemans N., Bonnefoy N., Motte P., Maréchal-Drouard L., Remacle C.. Co-evolution of mitochondrial tRNA import and codon usage determines translational efficiency in the green alga Chlamydomonas. PLoS Genet. 2012; 8:e1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loppes R., Radoux M.. Identification of short promoter regions involved in the transcriptional expression of the nitrate reductase gene in Chlamydomonas reinhardtii. Plant Mol. Biol. 2001; 45:215–227. [DOI] [PubMed] [Google Scholar]
- 19. Doublet V., Ubrig E., Alioua A., Bouchon D., Marcade I., Maréchal-Drouard L.. Large gene overlaps and tRNA processing in the compact mitochondrial genome of the crustacean Armadillidium vulgare. RNA Biol. 2015; 12:1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cavaiuolo M., Kuras R., Wollman FA., Choquet Y., Vallon O.. Small RNA profiling in Chlamydomonas: insights into chloroplast RNA metabolism. Nucleic Acids Res. 2017; doi:10.1093/nar/gkx668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barnett D.W., Garrison E.K., Quinlan A.R., Stromberg M.P., Marth G.T.. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011; 27:1691–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makaroff C.A., Apel I.J., Palmer J.D.. Characterization of radish mitochondrial atpA: influence of nuclear background on transcription of atpA-associated sequences and relationship with male-sterility. Plant Mol. Biol. 1990; 15:735–746. [DOI] [PubMed] [Google Scholar]
- 25. Quinlan A.R. BEDTools: the Swiss-Army Tool for genome feature analysis. Curr. Protoc. Bioinform. 2014; 47:11–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Criscuolo A. morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Mol. Phylogenet. Evol. 2011; 61:944–948. [DOI] [PubMed] [Google Scholar]
- 27. Emanuelsson O., Brunak S., von Heijne G., Nielsen H.. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007; 2:953–971. [DOI] [PubMed] [Google Scholar]
- 28. Tardif M., Atteia A., Specht M., Cogne G., Rolland N., Brugiere S., Hippler M., Ferro M., Bruley C., Peltier G. et al. . PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 2012; 29:3625–3639. [DOI] [PubMed] [Google Scholar]
- 29. Ruwe H., Wang G., Gusewski S., Schmitz-Linneweber C.. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016; 44:7406–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boer P.H., Gray M.W.. The URF 5 gene of Chlamydomonas reinhardtii mitochondria: DNA sequence and mode of transcription. EMBO J. 1986; 5:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boer P.H., Gray M.W.. Short dispersed repeats localized in spacer regions of Chlamydomonas reinhardtii mitochondrial DNA. Curr. Genet. 1991; 19:309–312. [DOI] [PubMed] [Google Scholar]
- 32. Lange H., Sement F.M., Canaday J., Gagliardi D.. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci. 2009; 14:497–504. [DOI] [PubMed] [Google Scholar]
- 33. Smith D.R., Lee R.W.. Nucleotide diversity of the colorless green alga Polytomella parva (Chlorophyceae, Chlorophyta): high for the mitochondrial telomeres, surprisingly low everywhere else. J. Eukaryot. Microbiol. 2011; 58:471–473. [DOI] [PubMed] [Google Scholar]
- 34. Pröschold T., Harris E.H., Coleman A.W.. Portrait of a species: Chlamydomonas reinhardtii. Genetics. 2005; 170:1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ojala D., Merkel C., Gelfand R., Attardi G.. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980; 22:393–403. [DOI] [PubMed] [Google Scholar]
- 36. Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. et al. . Sequence and organization of the human mitochondrial genome. Nature. 1981; 290:457–465. [DOI] [PubMed] [Google Scholar]
- 37. Bonnard G., Gobert A., Arrive M., Pinker F., Salinas-Giegé T., Giegé P.. Transfer RNA maturation in Chlamydomonas mitochondria, chloroplast and the nucleus by a single RNase P protein. Plant J. 2016; 87:270–280. [DOI] [PubMed] [Google Scholar]
- 38. Boehm E., Zaganelli S., Maundrell K., Jourdain A.A., Thore S., Martinou J.C.. FASTKD1 and FASTKD4 have opposite effects on expression of specific mitochondrial RNAs, depending upon their endonuclease-like RAP domain. Nucleic Acids Res. 2017; 45:6135–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boulouis A., Drapier D., Razafimanantsoa H., Wostrikoff K., Tourasse N.J., Pascal K., Girard-Bascou J., Vallon O., Wollman F.A., Choquet Y.. Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell. 2015; 27:984–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zimmer S.L., Fei Z., Stern D.B.. Genome-based analysis of Chlamydomonas reinhardtii exoribonucleases and poly(A) polymerases predicts unexpected organellar and exosomal features. Genetics. 2008; 179:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yehudai-Resheff S., Zimmer S.L., Komine Y., Stern D.B.. Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell. 2007; 19:1023–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chang H., Lim J., Ha M., Kim V.N.. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell. 2014; 53:1044–1052. [DOI] [PubMed] [Google Scholar]
- 43. Hirayama T., Matsuura T., Ushiyama S., Narusaka M., Kurihara Y., Yasuda M., Ohtani M., Seki M., Demura T., Nakashita H. et al. . A poly(A)-specific ribonuclease directly regulates the poly(A) status of mitochondrial mRNA in Arabidopsis. Nat. Commun. 2013; 4:2247–2255. [DOI] [PubMed] [Google Scholar]
- 44. Horton T.L., Landweber L.F.. Mitochondrial RNAs of myxomycetes terminate with non-encoded 3′ poly(U) tails. Nucleic Acids Res. 2000; 28:4750–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aphasizhev R., Suematsu T., Zhang L., Aphasizheva I.. Constructive edge of uridylation-induced RNA degradation. RNA Biol. 2016; 13:1078–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimmer S.L., Schein A., Zipor G., Stern D.B., Schuster G.. Polyadenylation in Arabidopsis and Chlamydomonas organelles: the input of nucleotidyltransferases, poly(A) polymerases and polynucleotide phosphorylase. Plant J. 2009; 59:88–99. [DOI] [PubMed] [Google Scholar]
- 47. Nedelcu A.M., Lee R.W., Lemieux C., Gray M.W., Burger G.. The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res. 2000; 10:819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sikorski P.J., Zuber H., Philippe L., Sement F.M., Canaday J., Kufel J., Gagliardi D., Lange H.. Distinct 18S rRNA precursors are targets of the exosome complex, the exoribonuclease RRP6L2 and the terminal nucleotidyltransferase TRL in Arabidopsis thaliana. Plant J. 2015; 83:991–1004. [DOI] [PubMed] [Google Scholar]
- 49. Ibrahim F., Rymarquis L.A., Kim E.J., Becker J., Balassa E., Green P.J., Cerutti H.. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl Acad. Sci. U.S.A. 2010; 107:3906–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yehudai-Resheff S., Schuster G.. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000; 28:1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data as well as a novel annotation proposed here for the Chlamydomonas mt genome can be visualized on our genome browser (http://chlamy-organelles.ibpc.fr) and downloaded by right-clicking on the tracks.