Abstract
Crosstalk between growth factors (GFs) and steroid hormones recurs in embryogenesis and is co-opted in pathology, but underlying mechanisms remain elusive. Our data from mammary cells imply that the crosstalk between the epidermal GF and glucocorticoids (GCs) involves transcription factors like p53 and NF-κB, along with reduced pausing and traveling of RNA polymerase II (RNAPII) at both promoters and bodies of GF-inducible genes. Essentially, GCs inhibit positive feedback loops activated by GFs and stimulate the reciprocal inhibitory loops. As expected, no alterations in DNA methylation accompany the transcriptional events instigated by either stimulus, but forced demethylation of regulatory regions broadened the repertoire of GF-inducible genes. We report that enhancers, like some promoters, are poised for activation by GFs and GCs. In addition, within the cooperative interface of the crosstalk, GFs enhance binding of the GC receptor to DNA and, in synergy with GCs, promote productive RNAPII elongation. Reciprocally, within the antagonistic interface GFs hyper-acetylate chromatin at unmethylated promoters and enhancers of genes involved in motility, but GCs hypoacetylate the corresponding regions. In conclusion, unmethylated genomic regions that encode feedback regulatory modules and differentially recruit RNAPII and acetylases/deacetylases underlie the crosstalk between GFs and a steroid hormone.
INTRODUCTION
The human genome encodes two large clades of receptors pivotal for regulation of gene expression: the clade of 48 nuclear receptors (NRs) for steroid hormones and the group of 58 growth factor receptors (called receptor tyrosine kinases, RTKs) (1). However, the crosstalk between NRs and RTKs remains poorly understood. Unlike growth factors (GFs), which relay messages by binding to RTKs able to indirectly regulate transcription, the NRs directly regulate specific genes by acting as inducible transcription factors (TFs). Despite this difference, GFs and steroids often act simultaneously and maintain crosstalk. For example, during repair of cutaneous wounds, GFs accelerate healing by acting upon keratinocytes and immune cells, but the anti-inflammatory action of glucocorticoids (GCs) is inhibitory (2). Antagonistic interactions between steroids and GFs are similarly involved in lobuloalveolar morphogenesis of the mammary gland and in forelimb initiation (3).
Using a mammary cell system, we previously reported that the crosstalk between GCs and the epidermal growth factor (EGF) entails a group of feedback modifiers of signal transduction (4). In analogy, co-activation of the GC receptor (GR) and inflammation alters the repertoire of target genes (5). Because GR directly controls transcription (6) and several genes were co-regulated by EGF and GCs, it is plausible that epigenetic mechanisms involving both the respective promoters and specific classes of gene enhancer elements are involved (7,8). For example, dynamic acetylation of histone H3 at lysine 27 (H3K27Ac) marks active enhancers stimulated by the vascular endothelial growth factor (9) and epigenetic factors cooperate with steroid hormones to activate transcriptional programs in insects (10). Likewise, sites of GR repression utilize GRIP1’s corepressor function and display reduced histone acetylation (11). Another epigenetic mark, DNA methylation, may also be involved: although methylation marks rather stable developmental and differentiated states (12), recent observations raised the possibility that this modification underlays dynamic responses to certain hormones and neurotransmitters (13–15). Similar to histone and DNA (16) modifications, the ability of RNA polymerase II (RNAPII) to integrate multiple signals is yet another potential mechanism of the GC-to-GF crosstalk. Specifically, RNAPII elongation is increasingly recognized as a critical step. For example, GR represses pro-inflammatory genes by controlling a negative elongation factor (17), or by preventing phosphorylation of RNAPII at Serine 2 (18). Similarly, EGF permits productive elongation of immediate early genes by mobilizing molecular complexes able to release paused RNAPII (19), and transcriptional regulation by 17β-estradiol facilitates both recruitment of RNAPII and pause release (20). However, exactly how the concomitant input of opposing signals, such as some steroids and GFs, is reflected at the gene and chromatin levels has so far received limited attention.
To unravel the genetic and epigenetic bases of the crosstalk between RTKs and NRs, we selected MCF10A human mammary cells, which migrate in response to EGF, but their migration is strongly inhibited by simultaneous treatment with dexamethasone (DEX), a synthetic GC. We previously reported that the hormonal crosstalk involves several gene modules: GR represses EGFR’s positive feedback modifiers, while activating a negative feedback module (4). The present work investigated relationships between GFs and GCs by undertaking a genome-wide mapping of RNAPII binding, histone 3 acetylation and DNA methylation. These analyses revealed that master TFs, along with distinct patterns of RNAPII recruitment and pause release, underlie the hormonal crosstalk. In addition, EGF and DEX instigated rapid, but distinct, H3K27Ac changes at both transcription start sites (TSS) and putative enhancers. Altogether, our studies uncover the epigenetic mechanism underlying the crosstalk between GCs and GFs.
MATERIALS AND METHODS
Materials
Unless indicated, cells were obtained from the American Type Tissue Culture Collection (ATCC). MCF10A cells were cultured in DME:F12 medium (Gibco BRL, Grand Island, NY, USA) supplemented with insulin (10 μg/ml), cholera toxin (0.1 μg/ml), hydrocortisone (0.5 μg/ml), heat-inactivated horse serum (5%; Gibco BRL) and EGF (10 ng/ml). The following antibodies were used: anti-DUSP1 (clone EPR18884 from Abcam), polyclonal anti-EGR1 (C-19; Santa Cruz), anti-FOS antibody (H-125; Santa Cruz), an antibody specific to the K27 acetylated form of histone 3 (from Abcam), a rabbit anti-GR polyclonal antibody (PA1-511A; Thermo Fisher Scientific) and an antibody to the N-terminus of RNA polymerase II (from Cell Signaling). Protein content was estimated using the BCA kit (from Sigma) and HBEGF was assayed using an ELISA kit (DuoSet, from R&D Systems, Minneapolis, MN, USA).
Cell culture
MCF10A cells were maintained as described (4). For analysis of DNA methylation, 1.5 million cells were plated per 6-cm plate in full medium without EGF. Following 36 h of starvation, the cells were stimulated with EGF (10 ng/ml) and harvested. For H3K27Ac and RNAPII experiments, cells were starved overnight, and thereafter stimulated with EGF or DEX (100 nM). To measure the response of RNA, 625 000 cells were plated per well in 6-well plates and their RNA harvested for quantitative polymerase chain reaction (qPCR). Wild-type (WT) and Dnmt1/Dnmt3b double knockout HCT116 cells were the kind gift of Dr Bert Vogelstein (Johns Hopkins University School of Medicine). Both cell lines were grown in McCoy’s 5a medium supplemented with 10% fetal bovine serum (FBS). MCF7 cells were grown in Dulbecco's modified Eagle (DME) medium supplemented with 10% FBS.
Cell migration assays
Ibidi Culture-Inserts were placed in Nunc Lab-Teck 4-well chamber slides. A suspension of MCF10A cells (8 × 104) in 0.1 ml full medium (without EGF and hydrocortisone) was placed in both wells of the insert. After overnight incubation, the plastic barriers were removed and cells were washed twice with fresh medium to remove any floating or dead cells. Photographs were taken and labeled at time T = 0. The cells were then treated with starvation medium supplemented with EGF (10 ng/ml) or with DEX (100 nM). Additional photos were taken at 8 and 22 h. For Transwell migration assays, cells were counted and re-seeded on the upper face of Transwell migration chambers (Thermo Scientific). Cells (40 000–120 000 per chamber) were seeded in full medium and left at 37°C for 20 h. The same number of cells was seeded in parallel in 12-well plates and used as control for seeding variation. Cells were treated with EGF combined with IgG (control) or an antibody against HBEGF (21). Twenty hours later, cells were fixed in paraformaldehyde (3% in saline), washed and stained, using crystal violet. Cells attached to the upper face of the chamber were removed, and only the remaining migrating cells were imaged using a binocular. ImageJ was used for quantification of migration results.
Identification of transcription factors involved in cellular motility
Using gene expression profiling (22) and other criteria, we selected 182 TFs that are expressed in MCF10A cells. Each TF was singly knocked-down in biological triplicates using siRNAs, and effects on cell migration were assessed as previously described (23).
Analyses of histone acetylation, GR binding and RNA polymerase II pausing and traveling
Cell fixation, chromatin immunoprecipitation and nucleotide sequencing were performed as described (24). Reads obtained from chromatin immunoprecipitation followed by DNA sequencing (ChIP-Seq) were mapped to the human hg19 genome assembly using Bowtie2. To derive the H3K27Ac, GR and RNAPII data, ChIP-Seq tag reads were normalized according to the library size. All tag counts were expressed as reads/10 million reads. Tag counts of H3K27Ac and RNAPII data were tallied near promoter regions (±3 or −30 to +300 bp relative to the TSS, respectively) using the function ‘annotatePeaks’ of HOMER (http://biowhat.ucsd.edu/homer/). Resulting summary counts were incremented by one prior to log transformation. Histone acetylation (H3K27Ac), GR and RNA polymerase II signals for the genome browser views were calculated by approximation of ChIP fragment density at each position in the genome using ‘makeUCSCfile’ of HOMER (http://biowhat.ucsd.edu/homer/).
Differential peak calling for GR ChIP-Seq
For identification of DEX-induced GR binding sites to the genome we used the PePr program with default parameters, except that the peaktype parameter was defined as ‘sharp’. The genome alignment files corresponding to DEX induced cells (biological duplicates) were compared (using the ‘diff’ parameter) to alignments corresponding to untreated cells. Only GR peaks that increased by >2-fold and with a P-value < 0.001 upon treatment of mammary cells with DEX for 60 min were called. The identified peaks were used to measure the distance between module A and module B genes to the nearest GR binding site. To detect enhanced GR binding in response to EGF + DEX treatment, we compared, using similar parameters, GR ChIP sequencing reads derived from cells treated with EGF + DEX for 20 or 60 min to reads derived from cells treated with DEX alone. Only GR peaks that were enhanced by >2-fold by the combined treatment, and with a P-value < 0.001 compared to DEX alone treatment were called. The peaks called for the 20 and 60 min treatments were unified and used for measuring the distance between module A and random sets of genes to the nearest cooperatively induced GR binding sites.
qPCR analysis of gene expression
RNA was isolated using the PerfectPure kit from 5-Prime (Fisher Scientific) according to the manufacturer's instructions. cDNA was synthesized using the qScript kit from Quanta Biosciences (Gaithersburg, MD, USA). Real time qPCR analysis was performed on a StepOne instrument (Applied Biosystems) using Fast SYBR Green Master Mix. The following primers were used: GAPDH (control), F 5′-AGCCACATCGCTCAGACAC, R 5′-GCCCAATACGACCAAATCC; ERRFI1, F 5′-TCCATCTTCTACAGGCAGTCCT, R 5′-AGCCACACGTGGATTGTCTT (F, forward; R-reverse).
DNA methylation analysis
Genomic DNA from triplicate samples was harvested by first washing the plates with cell lysis solution (0.25% Triton X-100, 20 μg/ml RNase H, 10 mM Tris pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA)). Thereafter, nuclei were washed (200 mM NaCl, 20 μg/ml RNase H, 10 mM Tris pH 8.0, 1 mM EDTA), prior to lysis with nuclear lysis buffer (0.5% sodium dodecyl sulphate, 200 mM NaCl, 20 μg/ml RNase H, 10 mM Tris pH 8.0, 1 mM EDTA). The nuclear lysate was incubated at 37°C for 1 h. Proteinase K was added to 100 μg/ml and the samples were incubated overnight at 55°C. On the next day the samples were extracted twice using phenol-chloroform-isoamyl alcohol (PCIA, 25:24:1, pH 8.0, from Sigma) and washed twice with chloroform, prior to ethanol precipitation. The pellet was re-suspended in TE (10 mM Tris pH 8.0, 1 mM EDTA). CpG methylation analysis using Illumina 450K Bead Arrays was performed by Genome Quebec. DNA methylation of biological triplicates was measured using the Minfi package in-R (25) with the SWAN normalization method. Probes whose total intensity was less than 5000 were excluded. Custom routines written in Clojure were used to quantify DNA methylation at the TSS by finding, for each transcript, the closest CpG from the Illumina 450K array that lays within 100 bp.
Analysis of RNA-sequencing data
Seventy-five base-pair long, paired-end RNA-Seq reads were first filtered by using Bowtie2. The remaining reads were aligned to the human genome using TopHat and GENCODE. Cufflinks (26) was then used to assemble transcripts for each sample. Cuffquant and Cuffdiff were used to quantify and compare expression across samples for each TSS. For quantification of mRNAs in MCF7, HCT116 and IMR90 cells, 60-bp long, single-end, RNA-Seq reads were aligned to the human genome with STAR (27). Reads per gene were counted using the summarizeOverlaps function from the GenomicAlignments package in R with the Refseq transcriptome as reference. Reads per gene were normalized across samples using the DESeq2 package. All RNA-Seq experiments of MCF7, HCT116 and IMR90 cells were performed in biological duplicates.
Determination of RNAPII traveling ratio
RNAPII distribution across the gene body was estimated using the traveling ratio as an estimate of pause release. Data were generated as originally described (28). Briefly, ChIP-seq read density at the TSS (−30 to +300 bp) was divided by the read density over the rest of the gene body, plus an additional 1 kb beyond the transcription end site (TES). The traveling ratios for all genes were calculated using the hg19 Refseq isoform with the highest expression level according to our RNA-Seq data.
Selection of TSS for analyses of H3K27Ac, DNA methylation and expression
Of the 69 211 TSS identified by TopHat, 26 004 presented an Illumina 450K CpG measurement within 100 bp of the TSS. From these, we filtered out sites that lay adjacent (within ±4.5 kb) to another highly expressed TSS. Additionally, we filtered out TSS that displayed adjacent CpGs (±100 bp) with a methylation value that conflicted the methylation level associated with the specific TSS (i.e. if the methylation level associated with the TSS is 0.15 and the adjacent CpG has a methylation level >0.5). Following filtration we were left with 17 180 TSS.
Analysis of mRNA data and selection of responsive/unresponsive genes
Data of mRNA expression were previously generated in our lab (4). From the 17 126 genes represented on the microarray, we selected genes whose expression levels change (up or down) more than 0.6-fold (log2 scale), in two consecutive time points, in response to EGF or DEX (‘responsive genes’; 432 and 484 genes for EGF and DEX, respectively). To avoid mis-annotation of the TSS region and incorrect reading of DNA methylation and H3K27Ac levels, we used Refseq to filter out genes with multiple TSS (117 and 144, respectively). Of the selected genes, many have an Illumina 450K CpG measurement within 100 bp of the TSS (total of 236 and 252, for EGF and DEX responsive genes, respectively). Unresponsive genes were defined as genes whose expression levels did not change more than 0.1-fold (log2 scale) in all consecutive time points (817 and 524 genes for EGF and DEX, respectively). We filtered out all genes with multiple TSS (237 and 163 for EGF and DEX, respectively). Of the selected genes, a large fraction has an Illumina 450K CpG measurement within 100 bp of the TSS (total of 443 and 264 EGF and DEX unresponsive genes, respectively).
Analysis of DNA methylation and H3K27Ac at genomic regions distal to TSS
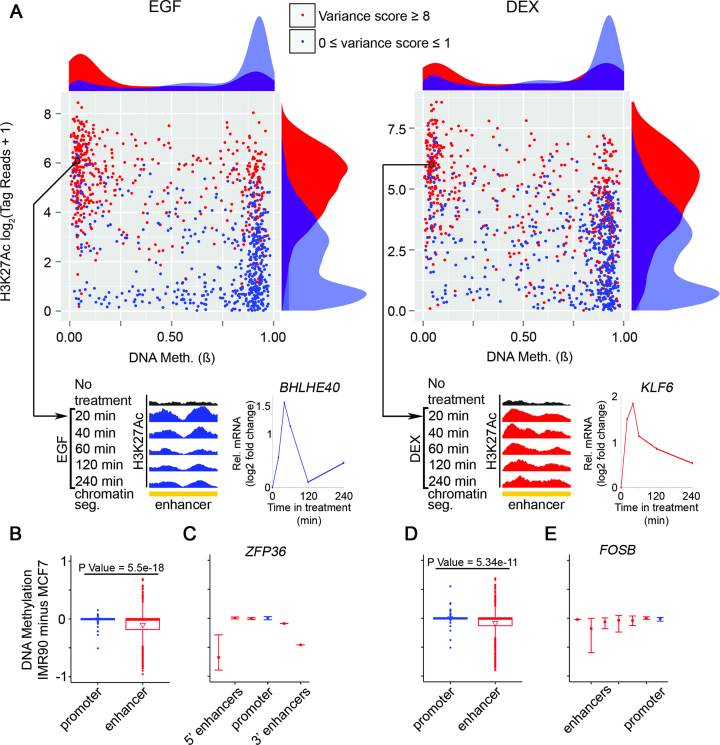
CpG sites that are more than 5 kb away (on each direction) from an actively transcribed TSS (164 251 of 446 796 CpGs) were selected. Their H3K27Ac occupancy variance score was calculated as previously described (9). Next, they were binned into highly dynamic sites (variance score ≥ 8) or relatively static sites (variance score < 1), according to responses to treatment with EGF (Figure 6A left panel; dynamic: n = 2183; static: n = 16 627) or DEX (Figure 6A right panel; dynamic: n = 826; static: n = 17 904).
Figure 6.
Lowly methylated regions distal to TSS are associated with dynamic acetylation, relatively high basal levels of H3K27Ac and enhanced responsiveness of genes to EGF stimulation. (A) Scatter plots presenting pre-stimulus levels of DNA methylation (ß) at the distal regions versus pre-stimulus H3K27Ac occupancy in MCF10A cells, reflecting either highly dynamic (red) or static (blue) sites (see ‘Materials and Methods’ section). Density plots are shown adjacent to the scatter plots. Dots are drawn for 500 randomly selected CpG sites. As examples, the lower panels show two genes: BHLHE40 and KLF6. The presented distal regions serve as enhancers (ENCODE/Broad chromatin state segmentation) that physically interact with the respective promoters. Shown are normalized H3K27Ac tracks corresponding to increasing treatment times with EGF or DEX. The graphs next to the browser views present alterations in RNA abundance. (B–E) MCF7 and IMR90 cells were serum starved overnight and then stimulated with EGF or DEX for 60 min. Whole genome 450K Illumina arrays of MCF7 and IMR90 cells were obtained from Encode (UCSC accession numbers: wgEncodeEH002180 and wgEncodeEH002194, respectively). (B and D) Boxplots showing average DNA methylation differences between IMR90 and MCF7 in TSS and enhancer regions of genes that are hyper-induced by EGF (B) or DEX (D) in IMR90 compared to MCF7 cells. (C and E) Shown are average DNA methylation differences between IMR90 and MCF7 cells in the TSS and associated enhancer regions of ZFP36 (C), which is hyper-induced by EGF in IMR90 compared to MCF7 cells, or FOSB (E), which is hyper-induced by DEX in IMR90 compared to MCF7 cells. Error bars represent the range of DNA methylation differences between the two cell lines in the corresponding regions.
Cell-lineage differential responsiveness to EGF and DEX
MCF7 and IMR90 cells were serum starved overnight and then stimulated for 60 min with EGF (10 ng/ml) or DEX (100 nM). RNA was isolated and subjected to sequencing. Reads were aligned to the Refseq annotation table, normalized and quantified using the DESeq2 package (29). Out of 111 genes that respond differentially to EGF in IMR90 compared to MCF7 cells, we selected 26 genes whose basal expression levels were similar in both cell lines (up to ±0.5-fold change on a log2 scale). For the selected group of genes, we calculated average DNA methylation differences between IMR90 and MCF7 cells in the TSS region (±500 bp) and in associated enhancer regions (±2 kb from enhancer center). These were identified using previously reported TSS–enhancer associations (30). Out of 80 genes that responded differentially to DEX in IMR90 compared to MCF7 cells, we selected 19 genes whose basal expression levels were similar in both cell lines (up to ±0.5-fold change on a log2 scale). For the selected group of genes, we calculated average DNA methylation differences between IMR90 and MCF7 cells as described above for EGF.
Sub-clustering of the cooperative and antagonistic modules corresponding to the GF-to-GC crosstalk
From module A we selected genes whose traveling ratio was decreased by more than 0.6 on a log2 scale after a combined treatment with EGF and DEX in at least one time point. Module B genes were clustered into two major groups: (i) type I, which is composed of nine genes whose traveling ratios decreased or increased more than 0.6 on a log2 scale in at least one time point (20 or 60 min) after EGF or DEX treatment, respectively, and (ii) type II, which is composed of 11 genes whose traveling ratios decreased more than 0.6 on a log2 scale after EGF treatment in at least one time point while DEX treatment caused a change in traveling ratio of <0.6 on log2 scale.
Statistical analysis
For statistical analysis of DNA methylation, the data were processed using the Minfi package (SWAN normalization) and Illumina’s custom algorithm. We used an False discovery rate (FDR) threshold of 5% for identification of genome-wide alterations in DNA methylation. For qPCR gene expression experiments we used one-way Anova with Tukey’s post-test. The Pearson’s Chi-squared Test was used for comparison of ratios of EGF responsive genes.
RESULTS
Dexamethasone robustly inhibits EGF-induced cell migration and alters expression of TFs and other genes controlling motility
Within short time of stimulation with EGF, MCF10A mammary epithelial cells initiate morphological alterations culminating in cell migration (31). This phenotype provides a model for the genome level action of GFs, because it is accompanied by time-dependent alterations in the abundance of multiple mRNAs (32). As demonstrated by ‘wound closure’ assays, incubation with EGF accelerated closure, but co-treatment with DEX inhibited the effect of EGF (Supplementary Figure S1A). Re-analysis of our previously reported gene expression data (4) showed that many of the >140 EGF-induced genes were suppressed when DEX was combined with EGF (Supplementary Figure S1B). Conversely, the EGF-induced downregulation of many genes was blocked by DEX (Supplementary Figure S1B). The ability of DEX to inhibit EGF-induced cell migration might be attributed to enhancement of DEX-inducible auto-inhibitors (module A genes; see a list in Supplementary Table S1), along with repression of auto-stimulators of the EGF-induced pathway (module B genes; Supplementary Table S2) (4). Importantly, ERRFI1, a module A gene, was shown to inhibit EGF-induced migration of mammary cells (4). In analogy to ERRFI1 and DUSP1, which are induced by GR and inhibit EGFR signaling, GR induces A20, an inhibitor of tumor necrosis factor (TNF) signaling, thereby restricts TNF-mediated inflammation (33).
While module A genes likely represent bone fide targets of GR, module B genes might be regulated by a plethora of GF-inducible TFs, whose interactions with GR may not be simple. To identify the corresponding TFs and also examine the possibility that they control motility, we undertook an RNA interference approach. First, we selected TFs that are regulated by EGF (84 TFs) and secondly, we enlisted TFs that are either relevant to breast cancer (88 TFs) or their chromatin binding patterns are well-characterized (52 TFs; altogether 182 TFs). Each TF was singly knocked-down in biological triplicates using siRNAs, and effects on collective cell migration were quantified using the software tool qCMA, which calculates the average migration distance (AMD) cells had moved during the measurement time (23). AMD values were normalized to the respective non-targeting control siRNAs and P-values were computed using two-sided unpaired t-test. This resulted in a list of 70 TFs significantly regulating EGF-induced collective cell migration in MCF10A cells. Using pscan (http://159.149.160.51/pscan/; Jaspar database) we selected from this list all TFs whose binding motif is enriched in the promoter regions of module B genes (Supplementary Table S3). As expected, the majority of these TFs positively regulate cell migration, but two factors of the list, TFAP2A and REL, may play regulatory roles. Altogether, the data presented in Supplementary Figure S1 and the three supplementary tables reveals that a complex transcriptional crosstalk enables DEX and GR to regulate EGF-induced mammary cell migration.
The antagonistic versus cooperative effects of DEX on EGF-induced genes are reflected in the dynamics of both H3K27Ac and RNAPII density
Assuming that RNAPII can be affected by signals generated by RTKs and NRs, we stimulated cells with EGF, DEX or the combination of both, and performed ChIP-Seq analyses using an antibody against the N-terminus of the largest subunit of RNAPII and an antibody recognizing the acetylated form of histone 3 (lysine 27). Focusing on module A and B genes, we analyzed temporal alterations in H3K27Ac occupancy at regions located ±3 kb relative to the TSS, along with RNAPII binding at 1 kb upstream to the TSS to 3 kb downstream to the TES. The results obtained are summarized in Figure 1 and Supplementary Figure S1C (module B genes), and in Figure 2 and Supplementary Figure S1D (module A genes).
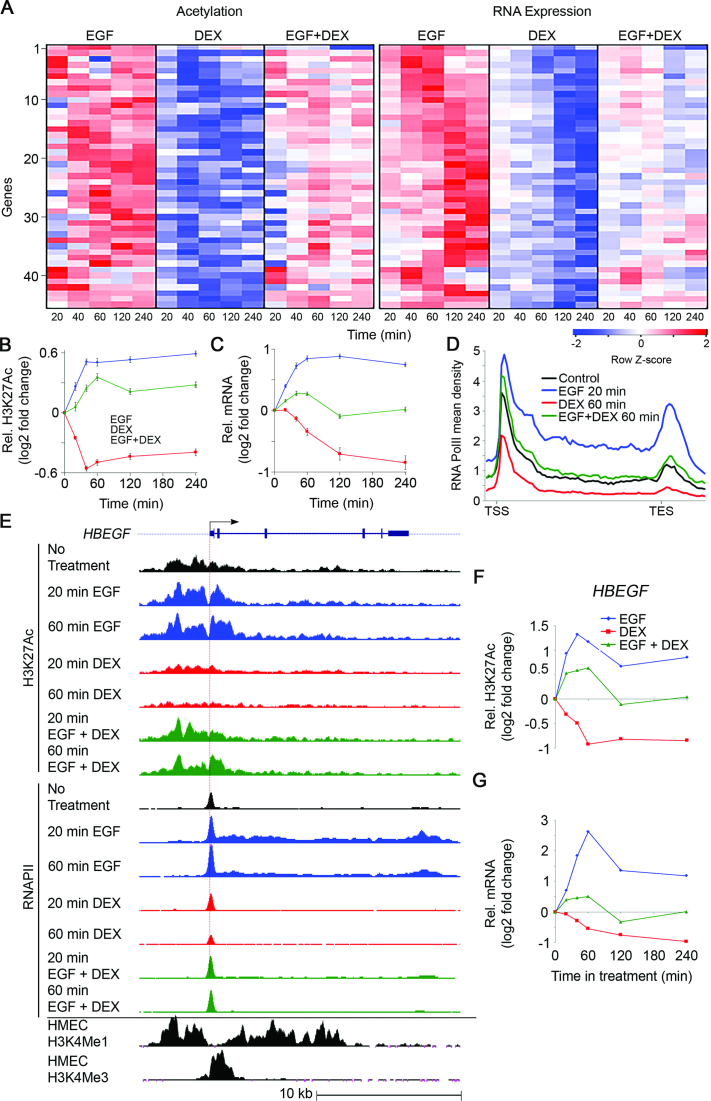
Figure 1.
The group of genes whose mRNAs are upregulated by EGF and downregulated by DEX (module B) displays concordant dynamic alterations of transcript abundance, H3K27Ac and RNA polymerase II occupancy. Serum-starved MCF10A cells were treated with EGF (10 ng/ml), DEX (100 nM) and the combination, for the indicated time intervals. (A) A heatmap showing alterations in H3K27Ac occupancy at promoter regions and RNA abundance of 45 Module B genes. (B and C) Time series showing average alterations in H3K27Ac occupancy (B) and mRNA abundance (C) of the 45 module B genes. (D) Shown are average profiles of RNAPII ChIP-Seq across all module B genes. Mean density was calculated as the average read number normalized to sequencing depth (total bin number: 85, from −1 kb upstream to the TSS to +3 kb after the TES). RNAPII profiles represent the average of biological duplicates. (E) The genomic structure and TSS of HBEGF are shown (top). Also shown are H3K27Ac and RNAPII tracks representing the normalized ChIP-seq read coverage. The lowermost two tracks show published ChIP-seq data of H3K4Me1 and H3K4Me3 for human mammary epithelial cells (wgEncodeEH000089 and wgEncodeEH000091, respectively). (F and G) Time series showing alterations in H3K27Ac occupancy (F) and transcript abundance (G) of the HBEGF gene. Bars represent standard errors of the mean of fold change.
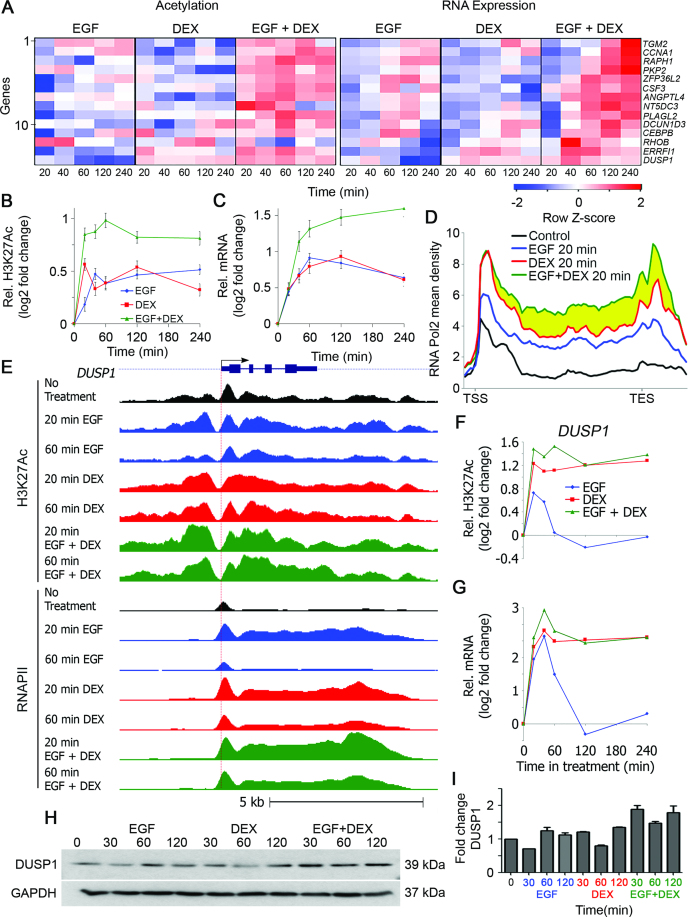
Figure 2.
Transcripts upregulated after stimulation with either EGF or DEX (module A genes) display additive abundance alterations, as well as synergistic H3K27Ac and RNAPII occupancy. MCF10A human mammary epithelial cells were grown and stimulated as in Figure 1. (A) A heatmap showing alterations in H3K27Ac occupancy and RNA expression of 14 module A (EGFUP/DEXUP) genes. (B and C) Time series showing average alterations in H3K27Ac occupancy (B) and transcript abundance (C) of the 14 module A genes. (D) Average profiles of RNAPII across 14 module A genes. Notably, the combined treatment further enhanced RNAPII occupancy in the gene body and not in the TSS region (yellow region). (E) The genomic structure and TSS (horizontal arrow and red vertical dotted line) of DUSP1, a module A gene, are shown. Also shown are H3K27Ac and RNAPII ChIP-Seq tag densities of cells treated with EGF, DEX and ‘EGF plus DEX’. (F and G) Time series showing alterations in H3K27Ac occupancy (F) and transcript abundance (G) corresponding to the DUSP1 gene. Bars represent standard errors of the mean of fold change. (H and I) MCF10A cells (1 × 106) grown in 6-well plates were serum starved for 24 h and then pretreated for 30 min with MG132 (5 μM). Thereafter, cells were stimulated with EGF (10 ng/ml), DEX (100 nM) and the combination of EGF and DEX. Following the indicated time periods, cells were extracted and processed for immunoblotting with antibodies to DUSP1 and GAPDH. Also shown is quantification of the DUSP1 protein levels of biological duplicates.
Notably, the temporal changes in mRNA levels well corresponded to the alterations in H3K27Ac and RNAPII signals. Thus, while EGF enhanced mRNA levels, as well as H3K27Ac and RNAPII signals corresponding to module B, DEX endorsed the opposite effects (Figure 1A–D). In the same vein, when separately applied, DEX and EGF induced highly similar effects on the cooperative module A genes (Figure 2A–D). Remarkably, the combined (EGF + DEX) treatment virtually abolished the induction by EGF of module B genes but cooperatively activated module A genes. As expected, nearly all module A and B genes displayed a peak of paused RNAPII at the promoter region, prior to their activation. Still, the inducible effects on RNAPII by both agents were also seen throughout the average gene locus, suggesting that both stimuli affect RNAPII recruitment, as well as pause release (Figures 1D and 2D). Surprisingly, the cooperative induction of module A genes by the combined EGF + DEX treatment affected mainly gene body density, suggesting that the simultaneous stimulation reaches maximal RNAPII density at promoter regions, while cooperatively inducing pause release and subsequent productive elongation (Figure 2D; yellow region).
To exemplify the genome-wide observations at the single gene level, we examined HBEGF, a module B gene (Figure 1E), encoding an EGFR agonist, along with a representative module A gene, DUSP1 (Figure 2E), which encodes an auto-inhibitory phosphatase. Methylation of histone 3 on lysine 4 (H3K4Me3) was used to identify the corresponding active promoters. Evidently, the temporal patterns of the selected transcripts precisely paralleled changes in H3K27Ac and RNAPII density (Figures 1F and G and 2F and G). Furthermore, as reflected by the RNAPII ChIP-seq tracks, both EGF and DEX transiently increased density of the enzyme at the DUSP1 promoter, but they oppositely affected the paused RNAPII at the TSS of HBEGF, in accordance with transcript levels. As expected, the combined EGF + DEX treatment additively upregulated abundance of the DUSP1 protein (Figure 2H and I) but almost nullified upregulation of HBEGF following stimulation with EGF (Supplementary Figure S2C). To test involvement of secreted HBEGF in inducible cell migration, we added a blocking anti-HBEGF antibody to the medium of EGF-stimulated MCF10A cells and observed strong inhibition of cell migration, but a control antibody exerted no effect on migration (Supplementary Figure S2). Presumably, inducible HBEGF sustains the effect of EGF on cell migration. In conclusion, the antagonistic GC-to-GF crosstalk displays modular and dynamic transcriptional changes, which are accompanied by rapid modifications of H3K27Ac occupancy, as well as RNAPII recruitment and elongation.
GR binding to DNA is enhanced following EGF treatment of mammary cells
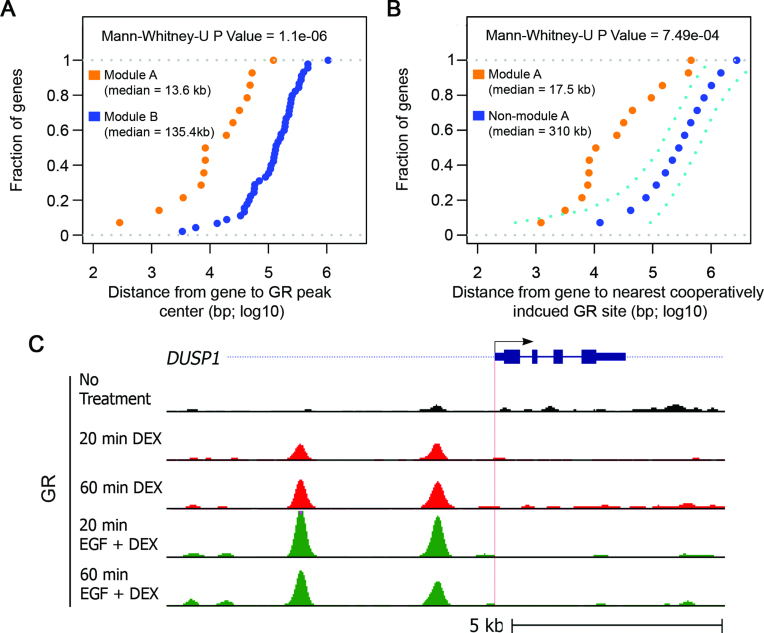
To gain deeper understanding of the crosstalk between growth factor signals and GC-controlled gene transcription, we performed GR ChIP-Seq. MCF10A cells were treated with DEX or DEX + EGF for 20 or 60 min and GR-binding to DNA was analyzed using nucleotide sequencing. The results we obtained confirmed that GR binding to chromatin strongly increases following stimulation with DEX. Specifically, we found that 2173 GR peaks increased by >2-fold already 20 min after treatment of mammary cells with DEX (P-value < 0.001). Interestingly, DEX-induced GR binding was found to be significantly closer to the TSS of module A genes than to the TSS of module B genes (Figure 3A), suggesting that DEX regulates module B genes indirectly, whereas induction of module A genes by DEX is mediated by GR binding proximal to the respective TSS. Notably, we also found that the combined treatment with EGF plus DEX, as compared to treatment with DEX alone, affected thousands of GR chromatin binding sites. Hence, we asked whether the corresponding sites are related to the cooperative module A, whose genes are additively upregulated by both EGF and DEX. Therefore, we compared the distance of loci presenting enhanced GR binding in response to EGF + DEX relative to the DEX alone treatment. This analysis found that the cooperatively induced module A genes are much closer to sites with enhanced GR binding than a set of randomly selected genes (Figure 3B). As an example, we present in Figure 3C the GR ChIP-Seq results for DUSP1: GR binding to two sites upstream to the TSS of DUSP1 was strongly enhanced already 20 min after the combined treatment with DEX and EGF. In summary, our new findings imply that the additive induction of module A genes by a growth factor and a GC is mediated, at least in part, by enhanced binding of GR to chromosomal regions proximal to these genes.
Figure 3.
In general, enhanced GR binding in response to EGF + DEX treatment is closer to module A genes than to a random set of genes. (A) Shown is the fraction of module A or module B genes as a function of the distance from the TSS to the identified nearest DEX-induced GR peak. The results were calculated based on data obtained using GR ChIP-Seq, and the distance was measured from the center of the peak. (B) Analyzed are two groups of genes: all module A genes and an equivalent number of randomly selected genes. For both groups of genes the dotted graphs present the fraction of genes as a function of the distance between the TSS and the nearest cooperatively induced GR binding site. The non-module A genes were randomly sub-sampled 1000 times from the whole population of genes. The mean, along with the upper 5% and lower 95% percentile, are shown in navy blue and cyan dashed lines, respectively. Distances were measured from the center of the respective peak of the GR binding site. (C) MCF10A cells were treated for 20 or 60 min with DEX, or EGF + DEX. Shown is the genomic structure and TSS (horizontal arrow and red vertical dotted line) of DUSP1 (top track). Also shown are normalized GR ChIP-Seq tag densities of cells treated with DEX, and ‘EGF plus DEX’. Note that the ChIP-Seq profiles shown represent the averages of biological duplicates.
Different stimuli dynamically influence RNAPII and chromatin acetylation, but DNA methylation remains unaltered
Because previous studies reported that DNA methylation dynamically changes at specific promoters in response to estrogen or during T-cell activation (13), we asked whether changes in DNA methylation play a role in the GC-to-GF crosstalk. To this end, we employed Illumina’s 450k arrays, which quantify methylation at more than 400 000 individual CpG sites located at the 5′ end of genes, gene bodies and enhancers (34). In addition to EGF and DEX, we selected two potent cellular agents, doxorubicin (Dox), a chemotherapeutic agent and phorbol 12-myristate 13-acetate (PMA), a strong activator of protein kinases. These stimuli were applied for 60 min, RNA and DNA were isolated, and thereafter we determined RNA abundance and DNA methylation levels. For gene-level analyses, we used qPCR and followed expression of ERRFI1, a module A gene. As expected, the transcript encoding ERRFI1 was strongly upregulated by DEX, PMA and EGF (Supplementary Figure S3A). However, while mRNA levels, H3K27Ac and both recruitment and elongation of RNAPII displayed highly dynamic and concordant patterns, all 13 cognate TSS and gene body CpGs from the Illumina arrays appeared unaffected by any of the diverse stimuli (Supplementary Figure S3B and C). We next asked if EGF (or DEX) causes changes in any of the more than 400 000 CpGs of the array. Plotting DNA methylation, before and after treatment with EGF (Supplementary Figure S4A) or DEX (Supplementary Figure S4B), demonstrated that methylation levels of the vast majority of the CpGs examined were extremely stable under treatment by either stimulus. Notably, changes in DNA methylation might require prolonged exposure (>60 min) to a challenge (13). However, we noted that methylation was stable over the examined 8-h interval, even for genes that are being strongly ‘turned on’ by EGF stimulation, such as CSF3 (>30-fold; Supplementary Figure S4C and D). Thus, while H3K27Ac and RNAPII signals displayed high dynamicity in response to EGF and DEX, DNA methylation appeared very stable throughout the genome, hence may not dynamically contribute to the steroid-GF crosstalk.
Forced demethylation of genomic regions distal to TSS is associated with hyper-responsiveness to EGF
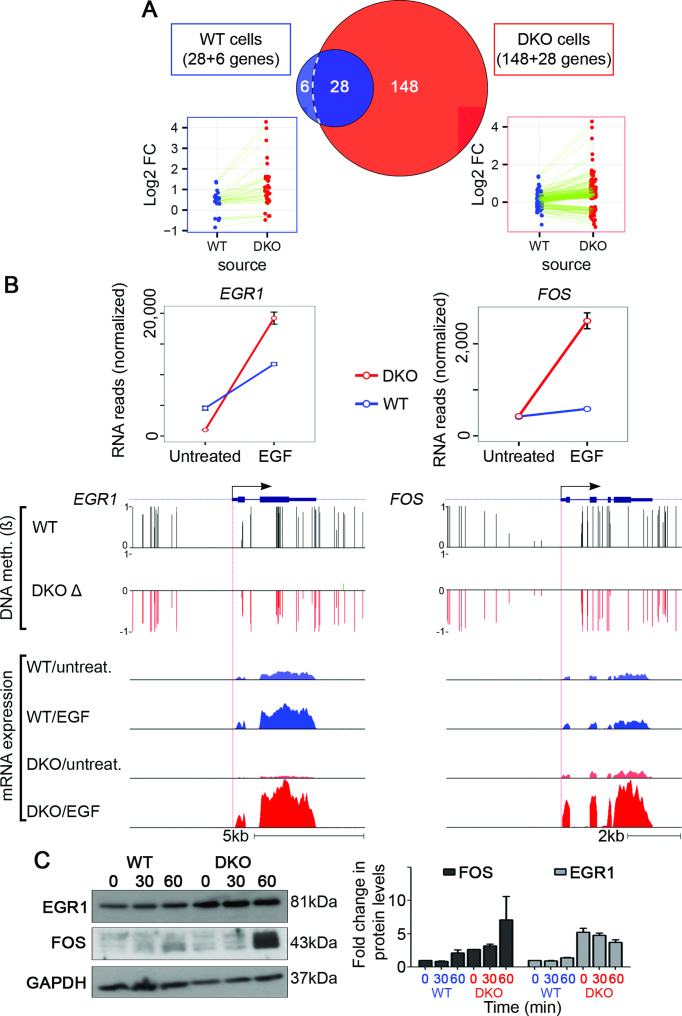
Because examining individual genes revealed that methylated regions of the genome are refractory to extracellular stimuli, we predicted that demethylation can broaden the repertoire of EGF-responsive genes. To test this, we made use of a WT colorectal cancer cell line, HCT-116, and a derivative that underwent disruption, by homologous recombination, of the genes encoding DNA methyltransferase 1 (DNMT1) and DNMT3b (double knockout, DKO) (35). Notably, knockout of both enzymes nearly eliminated (>95%) genomic methylation. Next, we stimulated WT and DKO cells with EGF for 60 min and performed RNA sequencing. Consistent with a previous report (36), we found that global demethylation only marginally altered basal transcription levels; however, it dramatically increased the number of genes that responded to EGF: from 34 genes in WT cells to 176 genes in DKO cells. Specifically, most EGF-inducible genes displayed by WT cells gained hyper-responsiveness to EGF in DKO cells (28 of 34 genes; hyper-responders; Figure 4A; left side), and another group (148 genes), responded to EGF only following global demethylation (de novo responders; Figure 4A; right side). Figure 4B contrasts a hyper-responder gene (EGR1) and a de novo-responder (FOS), the activation of which respectively increased by 7- and 4-fold in DKO cells. As shown, the combined effect of hypomethylation and EGF treatment strongly increased mRNA levels, relative to untreated DKO or WT cells (Figure 4B; lower tracks). Correspondingly, relatively high EGR1 and FOS protein levels were observed 60 min after stimulation of DKO cells with EGF (Figure 4C). Rather unexpectedly, the patterns of methylation displayed by EGR1 and FOS, along with all other genes we analyzed, disclosed hypomethylation of CpG sites located within gene bodies and distal regions, rather than promoter regions. In conclusion, although DNA methylation does not dynamically change in response to acute stimuli, basal methylation levels corresponding to both gene bodies and putative enhancers pre-determine responses to extracellular cues.
Figure 4.
Double-knockout of DNMTs both enhances responsiveness of EGF-inducible genes and exposes new genes to stimulation by EGF. (A) WT and DKO HCT116 cells were serum starved overnight and then stimulated with EGF (10 ng/ml; 60 min). Shown is a Venn diagram presenting genes responding to EGF treatment in WT HCT116 cells versus DKO cells. Note that in DKO cells, the expression of 176 genes was significantly altered following EGF treatment. Within this group, 28 genes were responsive to EGF in both cell lines but were hyper-induced by EGF in the DKO cells (hyper-responders), and 148 genes gained de novo responsiveness to EGF in the DKO cells (de novo responders). The last group comprises six genes whose expression levels changed significantly in response to EGF only in WT cells. (B) Shown are normalized RNA reads of FOS (a de novo-responder) and EGR1 (a hyper-responder), whose expression values were super-induced by EGF in DKO cells, as compared to WT cells. The browser views present WT DNA methylation levels (ß) and the relative change in DNA methylation levels (Δ) in cells depleted of two DNMTs. Also presented are normalized histograms of RNA reads corresponding to WT and DKO cells, treated (or not) with EGF. Bars represent standard errors of biological duplicates. Whole genome bisulfite sequencing of WT and DKO HCT116 cells were obtained from Gene Expression Omnibus (GEO) [GEO: GSE60106]. (C) WT and double DKO cells (1 × 106) were seeded in 6-well plates. Thereafter, cells were stimulated with EGF (10 ng/ml) for the indicated time intervals. Following the indicated time periods, cells were extracted and processed for immunoblotting with antibodies to EGR1, FOS, and GAPDH. Also shown is quantification of EGR1 and FOS protein levels in biological duplicates.
Relatively high RNAPII pausing and basal histone 3 acetylation, along with relatively low DNA methylation, poise large groups of genes for activation by either EGF or DEX
Although no rapid alterations in DNA methylation accompany inducible gene expression, we noticed that methylation levels of specific genes (e.g. ERRFI1, see Supplementary Figure S3) reciprocally correlated to basal mRNA levels, H3K27Ac occupancy and RNAPII density. To generalize this observation, we carried out genome-wide analyses of the relations between TSS methylation, transcript levels and H3K27Ac occupancy. RNA-Seq and GENCODE were used to measure transcription per each TSS and also locate the closest CpG. The resulting list of 26 004 TSS was divided into methylated and unmethylated sites. This analysis showed that while unmethylated TSS drive all levels of transcription, the methylated TSS rarely exhibited basal expression levels exceeding FPKM (Fragments Per Kilobase of transcript per Million) values of 1–3 (Supplementary Figure S5A and B). Next, we integrated methylation, transcript abundance and H3K27Ac data corresponding to 17 180 filtered TSS (see ‘Materials and Methods’ section). The results yielded a summary 3D scatter plot (Supplementary Figure S5C), which clearly demonstrates that genes, the TSS of which is methylated, are rarely expressed or associate with an acetylated histone 3 prior to stimulation by an extracellular cue.
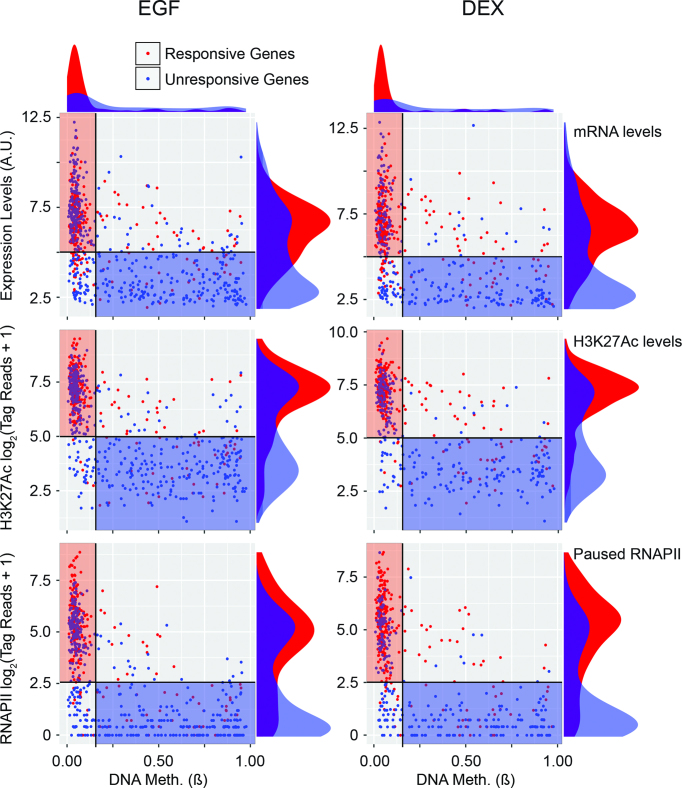
Because high basal (pre-stimulus) histone acetylation, RNAPII occupancy and low DNA methylation are typical to several module A and module B genes, we asked whether these attributes pre-determine responsiveness to both EGF and DEX. First, we generated lists of genes that are responsive or unresponsive to EGF or DEX (≤4 h of stimulation). Next, we measured the respective basal levels of mRNA, DNA methylation, H3K27Ac occupancy and RNAPII density at the respective TSS. The results are summarized in Figure 5. Remarkably, genes that respond to either ligand exhibited, prior to stimulation, very similar attributes: low DNA methylation, broad distributions of transcriptional activity, paused RNAPII and relatively high H3K27 acetylation (shown by red rectangles in the scatter plots). Interestingly, this four-dimension analysis similarly clustered unresponsive genes, but in a different area of each scatter plot (blue rectangles). These genes presented medium to high DNA methylation, along with low levels of mRNA expression, H3K27Ac and RNAPII signals. The relatively low methylation of genes responsive to either EGF or DEX was reflected by another type of analysis, which examined the respective cumulative fractions (Supplementary Figure S6A and B). Altogether, our data define a fraction of the genome, which is clearly poised for activation by two quite different extracellular ligands, namely EGF and DEX. Whether the rest of the genome of mammary cells is blocked to any extracellular stimulus is an intriguing question, which we exemplify in Supplementary Figure S6C. As shown, approximately a quarter of the mammary genome is non-responsive to EGF, whereas the vast majority of inducible genes belong to the unmethylated, relatively highly acetylated portion of the genome (rightmost green bar). These genome-wide conclusions are in line with the finding that primary response genes have pre-assembled RNAPII and positive histone modifications at their promoters in the basal state (37–39), and they highlight the role of poised chromatin in regulation of both the antagonistic as well as the cooperative gene modules.
Figure 5.
Prior to stimulation, promoter regions of genes that respond to treatment with either EGF or DEX display relatively low DNA methylation, high H3K27Ac and RNAPII occupancy, along with high mRNA expression, in comparison to unresponsive genes. Shown are scatter plots comparing TSS methylation (ß) and either abundance of the respective RNA (upper panels), H3K27Ac occupancy (middle panels) or RNAPII occupancy (lower panels). The data refer to two groups of genes: genes responsive to EGF (n = 236; left panels) and genes responsive to DEX (n = 252; right panels). Red and blue dots mark genes responsive to a treatment and unresponsive genes (EGF: n = 443; DEX: 264), respectively. Density plots are shown adjacent to the scatter plots. Rectangles in the scatter plots display possible clustering of the data that captures the majority of responsive and unresponsive genes (red and blue rectangles, respectively). To derive the H3K27Ac and RNAPII data, ChIP-Seq tag reads were normalized according to the library size and tallied near promoter regions. Note that all data correspond to untreated (control) MCF10A cells. A.U., arbitrary unit.
In similarity to inducible promoters, distal genomic regions respond to EGF or DEX and they are predisposed to activation
Recent reports have attributed to distal regulatory regions, including enhancers, critical roles in the modulation of gene expression under normal and pathologic conditions (40,41). Moreover, another study showed that GR binding to the chromatin is limited to distal preexisting foci of accessible chromatin (42). Hence, we examined relations between DNA methylation and dynamic H3K27Ac at distal loci (>5 kb from active TSS). Intersecting basal levels of CpG methylation and H3K27Ac occupancy revealed that, in similarity to promoter regions, distal sites with low methylation present relatively high and dynamic acetylation (Supplementary Figure S7A). This conclusion is exemplified by the CpG methylation probe cg18126802 (Supplementary Figure S7B). In the next step, we quantified the association between H3K27Ac dynamicity and low methylation, by utilizing the previously introduced ‘variance score’ (9). Using this score, we confirmed that high H3K27Ac variance, in response to either EGF or DEX, was associated with significantly lower DNA methylation at the corresponding loci of distal regions (Supplementary Figure S7C and D). This unexpected association emerged also from another type of analysis (see Figure 6A and B): Comparison of the group of distal regions with high H3K27Ac variance score (>8) to the group with low variance score (<1) revealed that the post-stimulus most dynamic group was endowed with very low basal DNA methylation. In contrast, distal regions with high methylation rarely exhibited H3K27Ac changes.
To address the epigenetic similarities between promoters and EGF/DEX responsive distal regions, we assumed that the distal regions loop onto the respective TSS and act as un-methylated enhancers of inducible genes. Analysis of enhancer–TSS association data from MCF7 mammary cells (30) supported this model: we identified an EGF-responsive distal region as a potential enhancer of BHLHE40. Likewise, we identified a DEX-responsive enhancer that physically interacts with the TSS of KLF6 (see the lower panels of Figure 6A). As expected, the normalized H3K27Ac tracks corresponding to the enhancers displayed dynamic patterns highly similar to the abundance of the BHLHE40 and KLF6 transcripts. Moreover, we found that acetylation signals of >25% of the EGF-responsive distal sites are affected (repressed or enhanced) by the concomitant addition of DEX, which identifies enhancers as an important site of the GC-to-GF crosstalk. In conclusion, hypomethylated and highly acetylated promoters, as well as distal, enhancer-containing regions, are poised to activation and contribute to the observed functional crosstalk between steroids and GFs.
Cell lineage-specific patterns of DNA methylation dictate responsiveness to extracellular cues
The hitherto described observations in epithelial cells indicated that unmethylated promoters and distal regions are predisposed to stimulation, but high methylation within such regions inhibits inducible activation. Because in general DNA methylation confers cell lineage-specific silencing (12), we assumed that treatment of non-epithelial cells with EGF or DEX would induce sets of genes distinct from those activated in epithelial cells. To test this, we stimulated both MCF7 epithelial cells and IMR90 fibroblasts with EGF and DEX, and then performed RNA-Seq. Focusing on genes sharing similar basal abundance in both cell lines, we found 26 and 19 hypomethylated genes that were hyper-induced by EGF and DEX, respectively, in IMR90 compared to MCF7 cells. Next, we used the FANTOM5 enhancer atlas (43) to associate enhancer regions to specific genes. Surprisingly, the promoter regions of these genes were lowly methylated in both cell lines and consequently exhibited strikingly small differences in DNA methylation between the two cells. In comparison, the associated enhancer sites exhibited much larger differences between fibroblasts and epithelial cells, as well as an overall hypomethylation (Figure 6B and D). To exemplify enhancer hypomethylation, we selected ZFP36 and FOSB, which were significantly hyper-induced by EGF or by DEX, respectively, in IMR90 compared to MCF7 cells (Figure 6C and E). Altogether, our analysis suggests that while promoter hypomethylation is a pre-requisite for gene induction by extracellular cues, the variation in enhancer methylation between different cell types plays a critical role in determining gene inducibility in response to EGF and DEX.
Differential recruitment and pause release of RNAPII underlay the genomic crosstalk between GCs and GFs
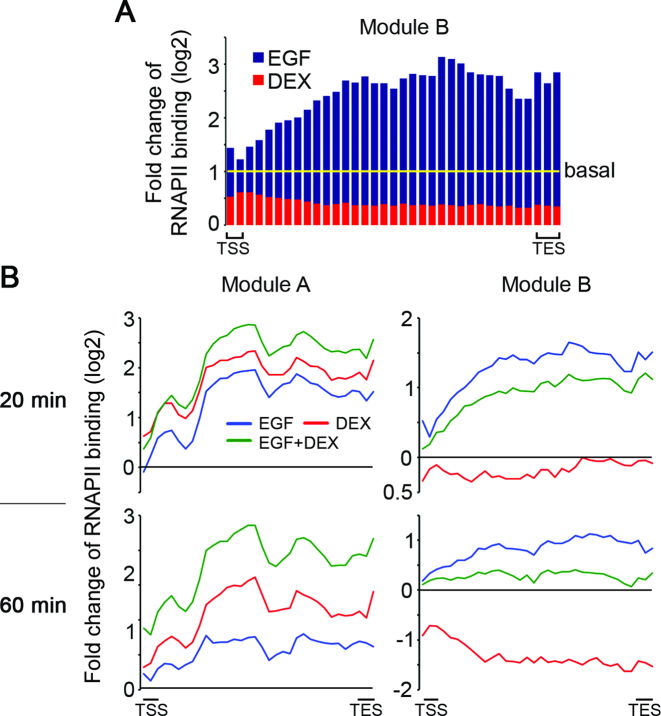
Because two main checkpoints, recruitment and pause release, bring about activation of RNAPII (16), we set out to examine the effects of EGF, DEX and the combined treatment on the dynamic patterns of RNAPII density. First, we examined the general effect of EGF (20 min of stimulation) and DEX (60 min) on the antagonistic, module B genes. Although EGF only slightly increased density of the already poised RNAPII at promoter regions, the very large increase in RNAPII density in gene bodies must be fed by enhanced recruitment in conjunction with an increased pause release (Figure 7A). In contrast, and in line with the antagonistic nature of module B, DEX treatment dramatically decreased RNAPII density at both the promoters and gene bodies of the 45 module B genes we examined (red histograms in Figure 7A; see tracks of HBEGF in Figure 1E). Further analyses supported a strong inhibitory effect of DEX on recruitment of RNAPII, and showed that the effect was stronger after 60 min of stimulation (Figure 7B, right panels). Unlike module B genes, we observed monotonic and similar effects of EGF and DEX on module A genes (cooperative): both stimuli enhanced RNAPII occupancy primarily inside gene bodies, suggesting that paused RNAPII is robustly released to allow productive elongation. Interestingly, the combined treatment with EGF and DEX enhanced RNAPII recruitment to the TSS and increased pause release, resulting in high and long-lasting (>60 min) density of RNAPII inside gene bodies (Figure 7B; left panels). In conclusion, GCs repress or enhance the EGF-induced effects on RNAPII recruitment and pausing, and these effects are in accord with the network function of each gene module.
Figure 7.
Stimulus-dependent RNAPII kinetics reflect the antagonistic and cooperative modules involved in the GC-to-GF crosstalk. (A) MCF10A cells were treated and ChIP-Seq of RNAPII was performed as in Figure 1. Shown is a bar graph presenting fold change of RNAPII binding of 45 module B genes following stimulation with EGF and DEX for 20 and 60 min, respectively, relative to starved cells. TSS and TES mark the TSS and end site, respectively. (B) Metagene graphs presenting fold change of RNAPII binding following stimulation with EGF, DEX and a combined stimulation (EGF + DEX) for 20 or 60 min. All module A (left side) and module B genes (right side) are presented.
Distinct patterns of RNAPII traveling disclose the identity of module-specific TFs
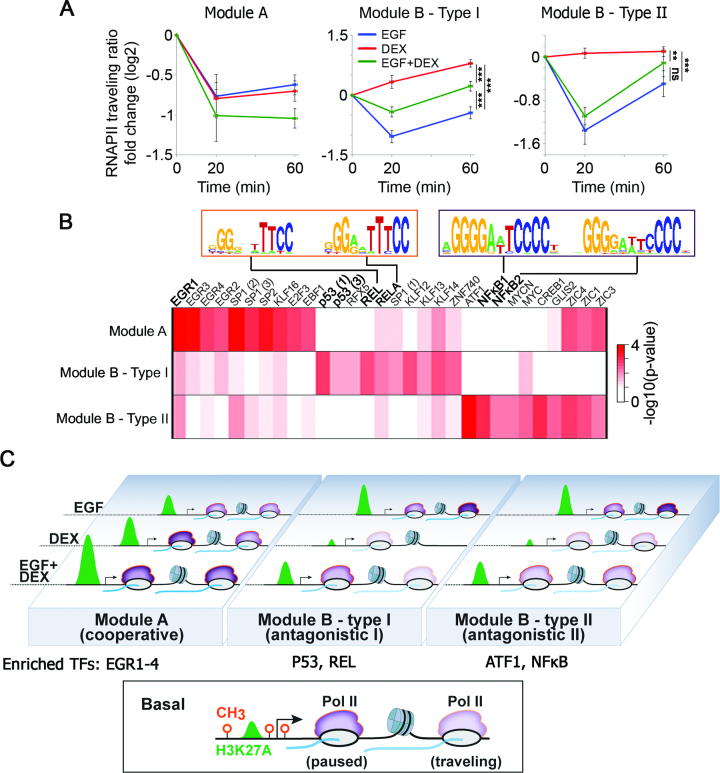
Because we found that DEX restrains the EGF-induced pause release of RNAPII at module B genes, we set to quantify the effect of the different treatments on RNAPII kinetics. To this end, we used the previously defined traveling ratio (28) (TR; RNAPII density in the promoter divided by the gene body density), a reliable measure of pause release. Applying this metric revealed that module B genes fall into two groups: (i) Genes like HBEGF, the RNAPII traveling ratio of which was increased by DEX, and (ii) genes like ARSJ, whose RNAPII occupancy was reduced by DEX throughout the promoter and gene body, therefore the traveling ratio was less affected. To resolve the mechanistic dichotomy, we clustered module B genes according to the fold change in their TRs in response to EGF, DEX and the combined treatment. This yielded two major groups of genes, whose average fold change in TRs in response to EGF and DEX is shown in Figure 8A. Surprisingly, although both groups belong to the antagonistic module, hence shared a strong EGF-induced release of RNAPII, they differed in the response to a concomitant DEX treatment: while in the first group (type I) simultaneous treatment with DEX blocked EGF-induced RNAPII pause release, in the second group (type II) DEX did not significantly affect RNAPII pause release following EGF treatment (Figure 8A and Supplementary Figure S8). Along this vein, the TRs of nine module A genes were significantly reduced (meaning enhanced RNAPII pause release) by the combined treatment (EGF + DEX). Taken together, these results show that GCs harness different mechanisms to repress the EGF-induced effects on RNAPII (module B) and enhance the RNAPII pause release in the cooperative module A.
Figure 8.
Kinetics of RNAPII recruitment and pause release in response to EGF and DEX exposes concealed subgroups of inducible genes and explains the transcriptional mechanism of the GF-to-GC crosstalk. (A) Average fold change of traveling ratios of RNAPII along different groups of genes before and after EGF, DEX and a combined stimulation. Shown are module A genes (nine genes) and module B genes, which were clustered into type I (nine genes whose traveling ratios decreased in response to EGF and increased in response to DEX) and type II (11 genes whose traveling ratios decreased after EGF treatment in at least one time point). **P < 0.01, ***P < 0.001 (two-way Anova). Non-significant differences to control are indicated (by ns). Bars represent standard errors of genes’ average. (B) A heatmap showing statistical enrichment of JASPAR motifs (67) among the different gene groups as defined in panel A. Pscan (http://159.149.160.51/pscan/; Jaspar 2016 database) was used to find over-represented TF binding sites. Only the top 10 enriched motifs from each group are shown (total of 30 motifs). Shown are the distinct NF-κB motifs enriched in the different gene groups. The respective P-values are minus log-transformed for easy presentation. (C) The scheme outlines the epigenetic processes governing the steroid-to-EGF crosstalk in mammary cells. The bottom box shows a typical inducible gene, which is poised for activation: the three methylation sites are unoccupied and moderate H3 acetylation occurs close to the TSS. The involved three gene modules (A, B-type I and B-type II), along with RNAPII molecules (in purple), are shown and their response to EGF, DEX or the combination (EGF + DEX) is outlined. Newly transcribed mRNA is shown in blue.
Since most promoters bind several TFs, and these may affect RNAPII, we assumed that different sets of TFs control the kinetics of RNAPII and determine each module’s genes. To examine this model, we used pscan (http://159.149.160.51/pscan/; Jaspar database) and searched for TF binding motifs that are over-represented in the promoter regions of the different subgroups. Next, we selected the resulting top 10 enriched TF motifs in each subgroup and compared their P-values to the other subgroups. The resulting log transformed heatmap is shown in Figure 8B. Although we detected no enrichment for the GR response element, implying indirect regulation by a tethered GR, we found that two response elements of p53, a direct binding partner of GR (44), are specifically enriched in the promoters of module B type I genes. Consistent with this observation, previous reports implicated p53 in RNAPII elongation (45). Likewise, DNA motifs of two distinct forms of NF-κB, another physical partner of GR, are enriched in the promoters of both type I and type II genes: whereas the REL motif was found mainly in type I genes, the NF-κB1 and NF-κB2 motifs are enriched in type II genes. In line with these observations, it has been reported that NF-κB binding to P-TEFb releases paused RNAPII (46).
The model presented in Figure 8C summarizes our findings: although steroid hormones, like GCs, and GFs like EGF remarkably differ, we found that the action of either ligand is confined to lineage-specific, un-methylated gene proximal as well as distal regions of the genome, which pre-exist in a poised state. The sign and kinetics of the induced transcriptional effects precisely correspond to another epigenetic mark, histone 3 acetylation. Analysis of RNAPII following treatment with either ligand, or with the combined treatment, revealed differential involvement of TFs like p53 and NF-κB, and consequent distinct recruitment and pause release of RNAPII. Altogether, our data portray harmonic interactions of epigenetic marks, RNA polymerase and specific gene modules, which collectively define the robust crosstalk between a steroid hormone and a growth factor.
DISCUSSION
Unlike bacterial operons, in which one promoter regulates multiple genes (47), mammalian genes are individually transcribed. This modular organization enables extracellular cues to specifically regulate individual target genes, as originally exemplified by the ‘classical’ binding of ER to the estrogen response element of the vitellogenin gene (48). Our study was motivated by the antagonistic effects induced by another steroid hormone receptor, GR, on cell migration (4,49), inflammation and wound healing (2), processes induced by GFs and cytokines. In a nutshell, our study attributes functional antagonism to a modular epigenetic program involving chromatin and DNA modifications, along with RNA polymerase pausing/elongation. These epigenetic processes are likely accompanied by post-translational modifications. For example, using MCF10A cells and a similar time frame, we previously reported pulses of protein degradation (50), along with dephosphorylation of the EGF-receptor by the inducible inhibitor ERRFI1/MIG6 (4). According to our results, GR might inhibit EGF induced genes (module B) through indirect pathways, while module A genes might be induced directly by GR binding to chromosome regions proximal to the TSS of these genes (see Figure 3). Consistent with a bi-directional crosstalk, relatively few genes are concordantly regulated by EGF and DEX (module A genes), and our GR ChIP data indicate that GF signals can enhance binding of GR molecules to cognate regulatory DNA elements (see Figure 3).
Genomic mechanisms shared between GFs and GCs
Despite the strong functional antagonism, we found that similar genomic mechanisms underlay the action of GFs and steroid hormones. First, the observed EGF- and DEX-induced effects on histone acetylation, RNAPII pausing and gene expression are strictly confined to un-methylated and relatively open regions of the genome. Our results are in line with previous reports that showed relatively high histone acetylation at active enhancers in different cell types and during different developmental stages (7,51–53). Unlike the relatively slow developmental course, our results highlight the role of histone acetylation in determining responses to external stimuli within dozens of minutes. Second, notwithstanding widespread transcriptional events, neither EGF nor DEX alter the state of genome methylation. This observation contrasts with the previously reported rapid, albeit local, changes in DNA methylation induced by other cues (13–15,50). Third, the chromosomal regions corresponding to both EGF- and DEX-inducible genes are occupied in cultured mammary cells by acetylated histone 3 and RNAPII prior to stimulation. Thus, in analogy to serum-induced immediate early genes (54), it seems that regulatory elements sensitive to either EGF or DEX are similarly poised for activation.
Enhancer hypomethylation predicts responsiveness to extracellular cues
Because we observed that gene demethylation confers de novo responsiveness to EGF, and GFs are frequently involved in cancer progression (55), it seems justified revisiting models associating hypo-methylation of non-coding regulatory regions with malignancy (40,56). Since, as we demonstrated, the state of gene methylation defines future responses to stimuli, different methylation profiles may define the trajectory of signal-induced tumor progression. It is notable that according to our data, prior to stimulation essentially all hyper-induced genes of cells defective in DNA methylation were actively transcribed and their promoters were hypomethylated. Hence, in addition to the respective promoters, the observed super sensitivity to EGF stimulation might be attributed to hypomethylation of distal regulatory regions, also known as gene enhancers. This conclusion is congruent with the observation that enhancer DNA methylation is significantly changed in cancers and is closely related to altered expression profiles of cancer genes (40,56). Steroid hormones, which play cardinal roles in breast cancer, may transiently increase local DNA methylation, and they are extensively involved in epithelial-mesenchymal transition (EMT), a GF-driven process essential for breast cancer progression (57). It remains to be seen if the epigenetic crosstalk we report herein is relevant to EMT and can be harnessed by attempts to inhibit cancer metastasis.
The role of RNAPII dynamics in the GFs-to-GCs crosstalk
RNAPII recruitment and elongation have been established as critical checkpoints of transcription modulation (16). Notably, several studies have shown that depletion of specific cofactors of RNAPII affects transcription initiation or elongation (37,58,59), along with responsiveness to extracellular cues (60). Previously, the kinetics of RNAPII has been examined in a cellular model stimulated by two factors (TNF-α and estrogen) (61,62). Our study examined the kinetics of RNAPII alterations in response to two different ligands, which were applied either separately or concurrently. The results identify pausing and release of RNAPII, along with recruitment of GR, chromatin modifying enzymes and TFs to both promoters and putative enhancers, as critical determinants of biological outcome. These principles are exemplified by the combined treatment with EGF and DEX: genes of the cooperative module (module A) are regulated primarily by EGR family TFs, but the antagonistic gene module (module B) may be divided into two different modes of action: type I is mediated by p53 and additional TFs, and it entails inhibition of RNAPII pause release, as well as subsequent reduction in elongation rates, whereas type II is mediated by NF-κB and its repression by DEX involves reduced recruitment, along with reduced RNAPII density in gene bodies. In conclusion, by comparing highly different routes of cellular activation, we gained modular resolution of the epigenetic mechanisms and putative TFs underlying differential gene regulation and cell behavior.
Progression of a promoter-proximal, paused RNAPII into productive elongation is considered a rate-limiting step in transcription of many mRNA-encoding genes (63), and accordingly GR represses pro-inflammatory genes by reducing RNAPII occupancy (17). Thus, our future studies will focus on the NF-κB-mediated module, which is characterized by reduced RNAPII recruitment in response to GR activation. Reduced RNAPII density inside the bodies of downregulated genes has previously been described (18,20). In addition, GR modulates the activity of several other TFs (64). For example, GR tethering to DNA-bound Stat3 results in transcriptional repression, whereas Stat3 tethering to GR results in synergism (65). Likewise, AP1 binding controls maintenance of baseline chromatin accessibility, which facilitates GR recruitment (66). Along with the study of downregulated transcription, our future studies will address the effect of combined treatment of EGF, which activates AP1, and DEX on additive GR binding proximal to genes of the cooperative module.
ACCESSION NUMBERS
Raw and processed ChIP-Seq, DNA methylation and RNA-Seq data were deposited in the Gene Expression Omnibus (GSE94063) database.
Reviewer private access link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=gdgpyymgbpoddid&acc=GSE94063
Supplementary Material
ACKNOWLEDGEMENTS
We thank Genome Quebec for methylation analyses. Ziv Machnes, Sebastian Alvarado and Claire Guillemin kindly provided valuable suggestions. We also thank Bert Vogelstein for the DKO cells.
Author Contributions: Y.E., M.F., M.S. and Y.Y. designed the study. Y.E. generated and analyzed ChIP-Seq data in MCF10A cells and RNA-Seq in MCF7 and HCT-116 cells. A.C. performed RNAPII ChIP-Seq experiments in MCF10A cells. M.F., M.J.S. and R.M. measured DNA methylation in MCF10A cells and the data were analyzed by M.F. S.S. and M.L. performed western blot analyses. K.S., C.K. and S.W. performed RNAi TF screening; A.S-C. performed and analyzed RNA-Seq data in MCF10A cells. All experiments in fibroblast cells were performed by Y.E. D.C. performed and analyzed DNA methylation in MCF7 cells. Analysis of DNA methylation at enhancer sites of MCF7 cells was performed by Y.Z. C.M. and M.L. performed and analyzed migration assays. YY, Y.E., M.F. and M.S. wrote the manuscript.
Footnotes
Present addresses:
Matthew J. Suderman, School of Social and Community Medicine, University of Bristol, Bristol, UK.
Kirti Shukla, School of Science, Monash University, Selangor Darul Ehsan, Malaysia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
M.D. Moross Institute for Cancer Research at the Weizmann Institute Collaborative Grant (to M.F., R.M.); Rosalind and Morris Goodman Cancer Research Centre at McGill University Collaborative Grant (to M.F., R.M.); Harold and Zelda Goldenberg Professorial Chair (to Y.Y.); Israel Science Foundation (ISF); Israel Cancer Research Fund (ICRF); Rising Tide Foundation; European Research Council; Dr Miriam and Sheldon G. Adelson Medical Research Foundation; Weizmann-McGill Joint Funding; Canadian Institute of Health Research [MOP-424111]; Deutsche Forschungsgemeinsachaft; Marvin Tanner Laboratory. Funding for open access charge: Dr Miriam and Sheldon G. Adelson Medical Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Heldin C.-H., Lu B., Evans R., Gutkind J.S.. Signals and receptors. Cold Spring Harb. Perspect. Biol. 2016; 8:a005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrientos S., Stojadinovic O., Golinko M.S., Brem H., Tomic-Canic M.. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008; 16:585–601. [DOI] [PubMed] [Google Scholar]
- 3. Cunningham T.J., Zhao X., Sandell L.L., Evans S.M., Trainor P.A., Duester G.. Antagonism between retinoic acid and fibroblast growth factor signaling during limb development. Cell Rep. 2013; 3:1503–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lauriola M., Enuka Y., Zeisel A., D’Uva G., Roth L., Sharon-Sevilla M., Lindzen M., Sharma K., Nevo N., Feldman M. et al. Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 2014; 5:5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rao N.A.S., McCalman M.T., Moulos P., Francoijs K.-J., Chatziioannou A., Kolisis F.N., Alexis M.N., Mitsiou D.J., Stunnenberg H.G.. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011; 21:1404–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weikum E.R., Knuesel M.T., Ortlund E.A., Yamamoto K.R.. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 2017; 18:159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zentner G.E., Tesar P.J., Scacheri P.C.. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011; 21:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why?. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang B., Day D.S., Ho J.W., Song L., Cao J., Christodoulou D., Seidman J.G., Crawford G.E., Park P.J., Pu W.T.. A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Res. 2013; 23:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirilly D., Wong J.J.L., Lim E.K.H., Wang Y., Zhang H., Wang C., Liao Q., Wang H., Liou Y.-C., Wang H. et al. Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila. Neuron. 2011; 72:86–100. [DOI] [PubMed] [Google Scholar]
- 11. Uhlenhaut N.H., Barish G.D., Yu R.T., Downes M., Karunasiri M., Liddle C., Schwalie P., Hubner N., Evans R.M.. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell. 2013; 49:158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007; 447:425–432. [DOI] [PubMed] [Google Scholar]
- 13. Bruniquel D., Schwartz R.H.. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat. Immunol. 2003; 4:235–240. [DOI] [PubMed] [Google Scholar]
- 14. Kangaspeska S., Stride B., Métivier R., Polycarpou-Schwarz M., Ibberson D., Carmouche R.P., Benes V., Gannon F., Reid G.. Transient cyclical methylation of promoter DNA. Nature. 2008; 452:112–115. [DOI] [PubMed] [Google Scholar]
- 15. Kersh E.N., Fitzpatrick D.R., Murali-Krishna K., Shires J., Speck S.H., Boss J.M., Ahmed R.. Rapid demethylation of the IFN-{gamma} gene occurs in memory but not naive CD8 T cells. J. Immunol. 2006; 176:4083–4093. [DOI] [PubMed] [Google Scholar]
- 16. Jonkers I., Lis J.T.. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015; 16:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gupte R., Muse G.W., Chinenov Y., Adelman K., Rogatsky I.. Glucocorticoid receptor represses proinflammatory genes at distinct steps of the transcription cycle. PNAS. 2013; 110:14616–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shang Y., Coppo M., He T., Ning F., Yu L., Kang L., Zhang B., Ju C., Qiao Y., Zhao B. et al. The transcriptional repressor Hes1 attenuates inflammation by regulating transcription elongation. Nat. Immunol. 2016; 17:930–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gardini A., Baillat D., Cesaroni M., Hu D., Marinis J.M., Wagner E.J., Lazar M.A., Shilatifard A., Shiekhattar R.. Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell. 2014; 56:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hah N., Danko C.G., Core L., Waterfall J.J., Siepel A., Lis J.T., Kraus W.L.. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011; 145:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindzen M., Lavi S., Leitner O., Yarden Y.. Tailored cancer immunotherapy using combinations of chemotherapy and a mixture of antibodies against EGF-receptor ligands. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:12559–12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kostler W.J., Zeisel A., Korner C., Tsai J.M., Jacob-Hirsch J., Ben-Chetrit N., Sharma K., Cohen-Dvashi H., Yitzhaky A., Lader E. et al. Epidermal growth-factor—induced transcript isoform variation drives mammary cell migration. PLoS One. 2013; 8:e80566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeisel A., Yitzhaky A., Koerner C., Lauriola M., Cohen-Dvashi H., Kostler W.J., Yarden Y., Wiemann S., Domany E.. qCMA: a desktop application for quantitative collective cell migration analysis. J. Biomol. Screen. 2013; 18:356–360. [DOI] [PubMed] [Google Scholar]
- 24. Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I.. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 2013; 8:539–554. [DOI] [PubMed] [Google Scholar]
- 25. Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A.. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014; 30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A.. c-Myc regulates transcriptional pause release. Cell. 2010; 141:432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012; 148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Debnath J., Muthuswamy S.K., Brugge J.S.. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003; 30:256–268. [DOI] [PubMed] [Google Scholar]
- 32. Zeisel A., Kostler W.J., Molotski N., Tsai J.M., Krauthgamer R., Jacob-Hirsch J., Rechavi G., Soen Y., Jung S., Yarden Y. et al. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Mol. Syst. Biol. 2011; 7:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kadiyala V., Sasse S.K., Altonsy M.O., Berman R., Chu H.W., Phang T.L., Gerber A.N.. Cistrome-based cooperation between airway epithelial glucocorticoid receptor and NF-kappaB orchestrates anti-inflammatory effects. J. Biol. Chem. 2016; 291:12673–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L. et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011; 98:288–295. [DOI] [PubMed] [Google Scholar]
- 35. Rhee I., Bachman K.E., Park B.H., Jair K.-W., Yen R.-W.C., Schuebel K.E., Cui H., Lengauer C., Kinzler K.W., Baylin S.B. et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002; 416:552–556. [DOI] [PubMed] [Google Scholar]
- 36. Blattler A., Yao L., Witt H., Guo Y., Nicolet C.M., Berman B.P., Farnham P.J.. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014; 15:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adelman K., Kennedy M.A., Nechaev S., Gilchrist D.A., Muse G.W., Chinenov Y., Rogatsky I.. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. PNAS. 2009; 106:18207–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hargreaves D.C., Horng T., Medzhitov R.. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009; 138:129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramirez-Carrozzi V.R., Braas D., Bhatt D.M., Cheng C.S., Hong C., Doty K.R., Black J.C., Hoffmann A., Carey M., Smale S.T.. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009; 138:114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aran D., Sabato S., Hellman A.. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013; 14:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., Wirbelauer C., Oakeley E.J., Gaidatzis D., Tiwari V.K. et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011; 480:490–495. [DOI] [PubMed] [Google Scholar]
- 42. John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A.. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2017; 43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sengupta S., Wasylyk B.. Physiological and pathological consequences of the interactions of the p53 tumor suppressor with the glucocorticoid, androgen, and estrogen receptors. Ann. N.Y. Acad. Sci. 2004; 1024:54–71. [DOI] [PubMed] [Google Scholar]
- 45. Balakrishnan S.K., Gross D.S.. The tumor suppressor p53 associates with gene coding regions and co-traverses with elongating RNA polymerase II in an in vivo model. Oncogene. 2008; 27:2661–2672. [DOI] [PubMed] [Google Scholar]
- 46. Nissen R.M., Yamamoto K.R.. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000; 14:2314–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jacob F., Jacob F.. The birth of the operon. Science. 2011; 332:767. [DOI] [PubMed] [Google Scholar]
- 48. Klein-Hitpass L., Schorpp M., Wagner U., Ryffel G.U.. An estrogen-responsive element derived from the 5′ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986; 46:1053–1061. [DOI] [PubMed] [Google Scholar]
- 49. D’Uva G., Lauriola M.. Towards the emerging crosstalk: ERBB family and steroid hormones. Semin. Cell Dev. Biol. 2016; 50:143–152. [DOI] [PubMed] [Google Scholar]
- 50. Miller C.A., Sweatt J.D.. Covalent modification of DNA regulates memory formation. Neuron. 2007; 53:857–869. [DOI] [PubMed] [Google Scholar]
- 51. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. PNAS. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009; 459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J.. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011; 470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lau L.F., Nathans D.. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Witsch E., Sela M., Yarden Y.. Roles for growth factors in cancer progression. Physiology (Bethesda). 2010; 25:85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009; 1:239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Voutsadakis I.A. Epithelial-mesenchymal transition (EMT) and regulation of EMT factors by steroid nuclear receptors in breast cancer: a review and in silico investigation. J. Clin. Med. 2016; 5:E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Balamotis M.A., Pennella M.A., Stevens J.L., Wasylyk B., Belmont A.S., Berk A.J.. Complexity in transcription control at the activation domain-mediator interface. Sci. Signal. 2009; 2:ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gilchrist D.A., Nechaev S., Lee C., Ghosh S.K.B., Collins J.B., Li L., Gilmour D.S., Adelman K.. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008; 22:1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fujita T., Piuz I., Schlegel W.. Negative elongation factor NELF controls transcription of immediate early genes in a stimulus-specific manner. Exp. Cell Res. 2009; 315:274–284. [DOI] [PubMed] [Google Scholar]
- 61. Danko C.G., Hah N., Luo X., Martins A.L., Core L., Lis J.T., Siepel A., Kraus W.L.. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell. 2013; 50:212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Franco H.L., Nagari A., Kraus W.L.. TNFalpha signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell. 2015; 58:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Min I.M., Waterfall J.J., Core L.J., Munroe R.J., Schimenti J., Lis J.T.. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011; 25:742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ratman D., Vanden Berghe W., Dejager L., Libert C., Tavernier J., Beck I.M., De Bosscher K.. How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Mol. Cell Endocrinol. 2013; 380:41–54. [DOI] [PubMed] [Google Scholar]
- 65. Langlais D., Couture C., Balsalobre A., Drouin J.. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol. Cell. 2012; 47:38–49. [DOI] [PubMed] [Google Scholar]
- 66. Biddie S.C., John S., Sabo P.J., Thurman R.E., Johnson T.A., Schiltz R.L., Miranda T.B., Sung M.-H., Trump S., Lightman S.L. et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011; 43:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mathelier A., Fornes O., Arenillas D.J., Chen C.-Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016; 44:D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.