Abstract
Post-transcriptional modifications of ribosomal RNAs (rRNAs) are involved in ribosome biogenesis and fine-tuning of translation. 5-Hydroxycytidine (ho5C), a modification of unknown biogenesis and function, is present at position 2501 of Escherichia coli 23S rRNA. We conducted a genome-wide screen in E. coli to identify genes required for ho5C2501 formation, and found a previously-uncharacterized gene, ydcP (renamed rlhA), iron-sulfur cluster (isc) genes, and a series of genes responsible for prephenate biosynthesis, indicating that iron-sulfur clusters and prephenate are required for ho5C2501 formation. RlhA interacted with precursors of the 50S ribosomal subunit, suggesting that this protein is directly involved in formation of ho5C2501. RlhA belongs to a family of enzymes with an uncharacterized peptidase U32 motif and conserved Cys residues in the C-terminal region. These elements were essential for ho5C2501 formation. We also found that the frequency of ho5C2501 is modulated by environmental iron concentration. Together, our results reveal a novel biosynthetic pathway for RNA hydroxylation and its response to iron.
INTRODUCTION
RNA molecules contain a wide variety of modifications that are introduced enzymatically after transcription. Over 130 different types of chemical modifications have been identified in RNA molecules across all domains of life (1). These modifications play critical roles in modulating RNA stability and function (2–6).
During ribosome biogenesis, precursors of ribosomal (r)RNAs undergo post-transcriptional processing, including cleavage and trimming of the leader and trailer sequences, as well as chemical modifications such as methylation, pseudouridylation and base-modification (7). These rRNA modifications are clustered in the functional sites of the ribosome, including the peptidyl transferase center (PTC), nascent polypeptide chain exit tunnel, subunit interface, and binding sites for mRNA and tRNAs (8,9); collectively, they are required to fine-tune ribosomal function for optimal cellular fitness (10,11). Genetic and biochemical studies have functionally characterized many rRNA modifications, revealing their roles in fidelity of translation (12–14), efficient rRNA processing (15–19), subunit assembly (20,21), antibiotic resistance (22), virulence of pathogenic bacteria (23–25), and evasion of innate immunity in eukaryotic hosts (26).
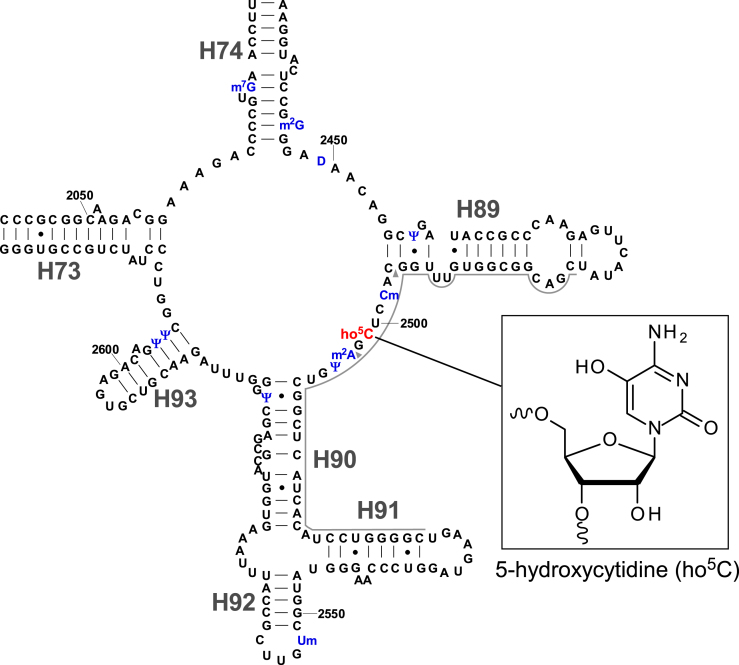
In Escherichia coli, 17 species of known modified nucleosides are present at 36 positions in the 16S and 23S rRNAs. Over the last two decades, the biogenesis of these rRNA modifications has been studied extensively, and most of the responsible methyltransferases and pseudouridylases have been identified (7,27). However, the function and biogenesis of 5-hydroxycytidine at position 2501 (ho5C2501) (Figure 1) and dihydrouridine at position 2449 (D2449) (Figure 1) in 23S rRNA remain to be elucidated (27).
Figure 1.
Secondary structure of domain V of E. coli 23S rRNA with post-transcriptional modifications. Inset shows the chemical structure of 5-hydroxycytidine (ho5C). The other modifications (blue) in this region are 7-methylguanosine (m7G) at position 2069, N2-methylguanosine (m2G) at position 2445, dihydrouridine (D) at position 2449, pseudouridine (Ψ) at positions 2457, 2504, 2580, 2604 and 2605, 2’-O-methylcytidine (Cm) at position 2498, 2-methyladenosine (m2A) at position 2503, and 2’-O-methyluridine (Um) at position 2552. Helix and position numbers are indicated. Two arrowheads indicate the RNase T1 cleavage sites for the ho5C2501-containing fragment.
Partial modification at position 2501 in E. coli 23S rRNA was first reported in 1993 (28). In 2011, the chemical structure of this modification was determined to be ho5C (29). ho5C2501 localizes at the PTC and forms a base triple with C2063 and A2451, which interacts with A76 of the P-site tRNA (30). Given that C2501 is universally conserved in all domains of life and essential in E. coli (31), this residue is likely to play a critical role in PTC function and recognition of P-site tRNA. Although the phylogenetic distribution of ho5C2501 is unclear, this modification is also present in Deinococcus radiodurans (29), which belongs to a phylum evolutionarily distant from E. coli, suggesting that it is widely distributed among bacteria. ho5C2501 is a sub-stoichiometric modification, and its frequency varies in a growth phase-specific manner (32), suggesting that the frequency of ho5C2501 is regulated in response to environmental conditions. Previously, however, the molecular function and physiological roles of ho5C2501 remained completely unknown.
To identify genes responsible for RNA modifications, we developed a method called the ‘ribonucleome analysis’, which entails a genome-wide reverse-genetic screen combined with liquid chromatography-mass spectrometry (LC/MS) (33). Using this approach, we have discovered a number of genes responsible for RNA modifications in tRNAs (34–42) and rRNAs (12,20). Here, we used this approach to identify genes required for ho5C2501 formation in E. coli. The screen revealed the previously-uncharacterized gene ydcP (renamed rlhA), whose deletion resulted in complete loss of ho5C2501. RlhA interacted with precursors of 50S subunit, strongly suggesting that RlhA is directly involved in ho5C2501 formation. RlhA belongs to a family of enzymes with a peptidase U32 motif and conserved Cys residues in the C-terminal region. These elements were essential for ho5C2501 formation. We also found that a series of genes required for iron–sulfur (Fe–S) cluster biogenesis or prephenate biosynthesis are necessary for ho5C2501 formation. Moreover, the frequency of ho5C2501 was altered in response to iron, suggesting that this rRNA modification is metabolically modulated by environmental iron availability.
MATERIALS AND METHODS
Construction of E. coli strains and media
A series of E. coli genomic-deletion strains (OCL/R-series) derived from MG1655 sp (MG1655 rpsL polA12 Zih::Tn10) was kindly provided by Dr Jun-ichi Kato (43). Strains ME5100 (MG1655sp ΔycjD-ydfJ::kanr) and ME5046 (MG1655sp ΔynhA-b1695::kanr) lack the regions containing ydcP and aroD, respectively. Single-deletion strains with kanamycin-resistance markers (Keio collection) (44) were obtained from the Genetic Stock Research Center, National Institute of Genetics, Japan. Escherichia coli DY330 strains [W3110, ΔlacU169, gal490, pgl8, λc1857(cro-bioA)] for expression of YdcP(RlhA)-SPA, MiaB-SPA, RlmN-SPA and DnaK-SPA were obtained from Thermo Scientific Open Biosystems. To construct strains that lacked subregions of the deletion spanning between ycjD and ydfJ, as well as ΔtyrA/ΔpheA double knockout strain, E. coli K-12 strain BW25113 (lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78) was subjected to one-step inactivation of chromosomal genes (45). The chloramphenicol resistance cassette derived from vector pBT was used to replace the knockout region. To construct the ΔtyrA/ΔpheA/ΔcmoA triple knockout strain, ΔcmoA::kanr was introduced into the ΔtyrA/ΔpheA double knockout strain by P1 transduction (46). The disruption was confirmed by colony PCR. Primers used for construction of E. coli strains in this study are listed in Supplementary Table S1. To rescue ho5C2501 formation, ydcP gene expressed from a mobile plasmid (pNT3-ydcP) was transferred to the Δ(ycjD-ydfJ) strain by conjugation (47). Escherichia coli strains were grown in LB medium or M9 medium with or without 0.2 mM FeCl3 at 37°C.
Plasmid construction
The ORF of YdcP-SPA with flanking promoter region was PCR-amplified from a genomic DNA of DY330 YdcP-SPA and cloned into the NdeI and BamHI sites in pBR322 to generate pBR-RlhA. Point mutations were introduced into pBR-RlhA by site-directed mutagenesis. To generate PheA with only CM activity [pMW-pheA(CM)], the ORF of pheA with the promoter region was PCR-amplified from genomic DNA of E. coli BW25113 and the product cloned into pMW118 (pMW-pheA), followed by introduction of the T278A mutation into the prephenate dehydratase (PDT) domain by site-directed mutagenesis. This mutation decreased PDT activity by 10000-fold while retaining 98% of CM activity (48). Primers used for plasmid construction are listed in Supplementary Table S1.
RNA preparation
For ribonucleome analysis, E. coli strains were grown in 1 ml of LB medium in 96-deep well plates at 37°C overnight. Total RNA was extracted from each strain using the RNeasy kit (Qiagen). For RNA segment analysis, the 48-mer RNA segment containing ho5C2501 (C2480–C2527) was carved out of E. coli 23S rRNA using a procedure involving complementary oligodeoxynucleotides (21). To lyse cells, 5 ml cultures of E. coli were suspended in 200 μl TE buffer [10 mM Tris–HCl, 1 mM EDTA (pH 8.0)] containing 1 mg/ml lysozyme and incubated on ice for 5 min. The lysates were then mixed with 600 μl of Trizol LS reagent (ThermoFisher Scientific), followed by two rounds of freezing and thawing. An aliquot (800 μl) was mixed with 160 μl chloroform, and total RNA was recovered. The resultant total RNA (100 μg), dissolved in 323 μl of a buffer consisting of 60 mM HEPES–KOH (pH 7.6) and 115 mM KCl, was mixed with 2 μl (200 pmol) of 48-mer synthetic DNA (C2480–C2527) (Supplementary Table S1) and incubated at 90°C for 5 min, followed by gradual cooling to room temperature to allow the DNA probe to hybridize to the 23S rRNA. Next, the sample was mixed with 0.5 μg RNase A and 500 unit RNase T1 and incubated on ice for 1 h. The protected RNA-DNA heteroduplex was recovered by phenol-chloroform extraction and ethanol precipitation, and resolved by denaturing 15% polyacrylamide gel electrophoresis. The band was excised, and the heteroduplex was eluted from the gel fragment and isolated.
For Figure 6D, RNA was prepared as above with several modifications. RBS buffer [20 mM HEPES–KOH (pH7.6), 200 mM NH4Cl, 0.5 mM Mg(OAc)2, 6 μM 2-mercaptoethanol] was used for resuspension of the cells. Total RNA (20 μg) dissolved in 200 μl of a buffer consisting of 50 mM HEPES–KOH (pH 7.6) and 100 mM KCl was mixed with 200 pmol of 50-mer synthetic DNA (A2478–C2527) (Supplementary Table S1). 0.05 μg RNase A and 50 unit RNase T1 was used for RNA digestion.
Figure 6.
Growth phase and iron dependencies of ho5C2501 formation. (A) Frequencies of ho5C2501 in E. coli DY330 strain for YdcP(RlhA)-SPA cultured in LB medium and harvested at different growth phases. Circles and squares represent growth of the cell culture (left vertical axis) and ho5C2501 frequency (right vertical axis), respectively, at each time point. (B) Frequencies of ho5C2501 in E. coli DY330 strain for YdcP(RlhA)-SPA cultured in M9 medium supplemented with (open symbols) or without (filled symbols) FeCl3, and harvested at different growth phases. Circles and squares represent growth of the cell culture (left vertical axis) and ho5C2501 frequency (right vertical axis), respectively, at each time point. Frequencies of ms2i6A (C) and ho5C2501 (D) in E. coli WT strain (BW25113) cultured in LB medium supplemented with FeCl3 and Dip at the indicated concentration. Data represent average values with s.d. of biological triplicate. Asterisks indicate the P-value of one-tailed t-test (*P < 0.05, ****P < 0.0001).
Phylogenic analysis
Amino acid sequences of peptidase U32 motif and full-length amino acid sequences of RlhA homologs were retrieved from Pfam (49) and Uniprot (50), respectively. Multiple sequence alignments and a Neighbor Joining tree were constructed using Clustal X2 (51) and displayed using Bioedit (52), NJ plot (53) and iTOL (54). Sources and accession numbers of peptidase U32 motifs are listed in Supplementary Table S2. Subfamilies of peptidase U32 proteins were categorized manually.
RNA mass spectrometry
RNA-MS was performed basically as described (21,33,55). Total RNA, the 48-mer-RNA segment (C2480–C2527) or 50-mer-RNA segment (A2478–C2527) of 23S rRNA were digested with 50 units of RNase T1 (Epicentre) in 20 mM NH4OAc (pH 5.3) at 37°C for 30 min or 1 h and then subjected to capillary LC/nano ESI-MS on an LTQ Orbitrap mass spectrometer (Thermo Scientific) with a nano-electrosprayer connected to a splitless nanoflow HPLC system (DiNa, KYA Technologies).
To measure ho5C frequency, we observe both modified and unmodified RNA fragments, and calculate from the ratio of extracted ion chromatogram (XIC) peak intensity of both fragments. Basically, in negative mode of ESI, ionization efficiencies of the RNA fragments bearing the same sequence but different modification do not differ largely, because ESI ionization relies mainly on numbers of phosphate groups, not on type of base modifications. Namely, modified and unmodified fragments mutually serve as internal standards in this analysis. Thus, we don’t need any standard curve for each modification, but reliably quantify ho5C frequency from their intensities of XICs even with a single measurement.
The methylthiolation level of ms2i6A was measured by total nucleosides analysis using LC/MS as described (56). Total RNA (0.1 μg) of E. coli WT cells harvested at late log phase was digested with 0.1 unit nuclease P1 (Wako Pure Chemical Industries), 0.35 unit Phosphodiesterase I (Worthington Biochemical Corporation) and 0.1 unit Bacterial Alkaline Phosphatase (BAP from E. coli C75, Wako Pure Chemical Industries). The enzymes used for nucleoside digestion were prepared as described (57). Proton adducts of nucleosides were scanned over the range of m/z 200–700. Frequency of ms2i6A can be measured from the proportion of the ms2i6A peak area to the total peak areas of ms2i6A and i6A, because the ionization efficiencies of these nucleosides were almost identical in our measurement.
Sucrose density gradient centrifugation
A 10 ml pre-culture of E. coli strain RlhA-SPA was inoculated into 1 l LB medium containing 50 μg/ml kanamycin and cultivated at 37°C until the OD600 reached 0.5. The harvested cell pellet was ground with alumina (Al2O3) and then dissolved in a buffer consisting of 20 mM HEPES-KOH (pH 7.6), 100 mM NH4Cl, 10 mM Mg(OAc)2, and 6 mM β-mercaptoethanol (β-ME). The lysate was cleared by ultracentrifugation at 22000 rpm in a 50Ti rotor for 45 min at 4°C. The supernatant was mixed with 5 U DNase I and ultracentrifuged at 40 000 rpm in an SW-28 rotor for 12 h at 4°C to obtain the crude ribosome fraction, which was then resuspended with the same buffer.
A 10–40% sucrose gradient was produced using a Gradient mate (BIOCOMP) with a buffer consisting of 20 mM HEPES–KOH (pH 7.6), 0.5 or 10 mM Mg(OAc)2, 100 mM NH4Cl, and 6 mM β-ME. The crude ribosome fractions (32 and 50A260 for the 0.5 and 10 mM Mg(OAc)2 conditions, respectively) were layered on the top of the sucrose gradient and ultracentrifuged at 20 000 rpm for 14 h at 4°C in a SW-28 rotor, followed by fractionation on a BIOCOMP fractionator. Ribosomal particles in each fraction were precipitated by trichloroacetic acid and subjected to western blotting.
Western blotting
For SDG fractions, SPA-tagged RlhA was detected using a mouse anti-FLAG M2 primary antibody (Sigma Aldrich) and an anti-mouse HRP secondary antibody (Dako). Chemical luminescence for HRP was detected using the ECL detection kit (GE healthcare). To examine the steady-state levels of SPA proteins in the presence or absence of Dip, E. coli DY330 strains (RlhA-SPA, MiaB-SPA, RlmN-SPA and DnaK-SPA) were cultured in 30 ml LB medium with or without 250 μM Dip and harvested at an OD600 of 0.4. Cell lysates of each strain were subjected to western blotting to detect the SPA-tagged protein using anti-FLAG-HRP antibody (Wako) and the ECL detection kit (GE Healthcare).
Reverse transcription quantitative PCR
Total RNA (1 μg) was treated with RNase-free DNase (RQ1 DNase, Promega) then converted to cDNAs through reverse transcription with Evoscript Reverse Transcriptase (Roche) and random N6 primer. cDNAs were mixed with KAPA SYBR (KAPA Biosystems) and 2 pmol primers listed in Supplementary Table S1. Quantitative PCR analysis was carried out with primers (Supplementary Table S1) using Light Cycler 480 (Roche) and KAPA SYBR (KAPA Biosystems). Detailed procedure is included in the checklist (Supplementary Table S3) according to MIQE guideline (58).
RESULTS
The ydcP gene is responsible for ho5C2501 formation
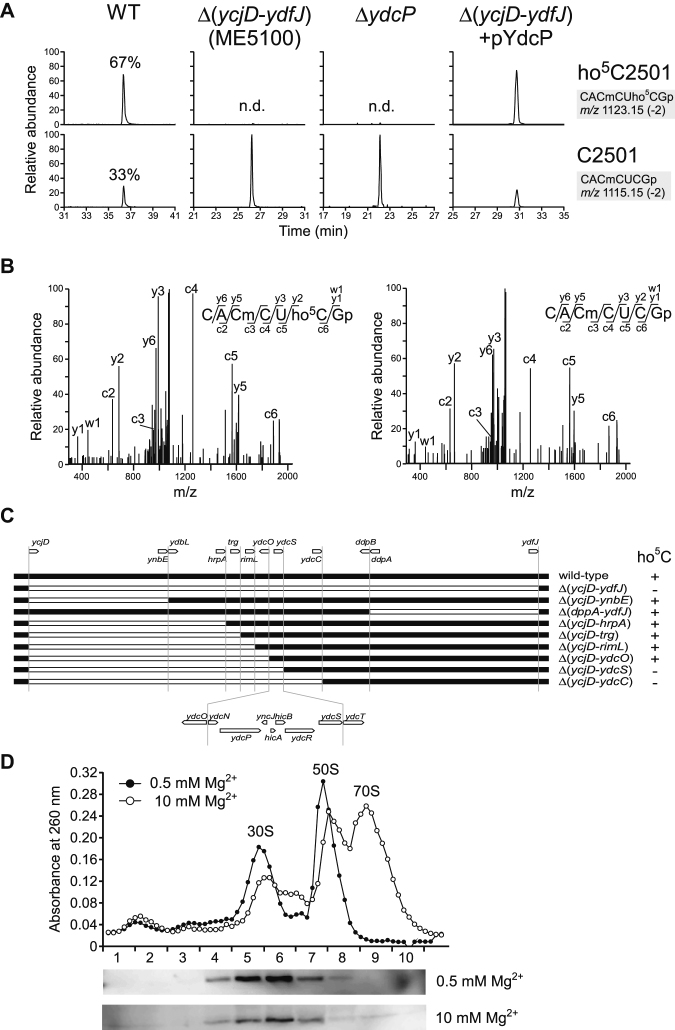
To detect ho5C2501 by RNA mass spectrometry (RNA-MS), we prepared the 48-mer segment of 23S rRNA containing ho5C2501 (C2480–C2527) (Figure 1) from the E. coli cells cultured in LB medium overnight using complementary DNA and RNase treatment (see Materials and Methods). The segment was then digested with RNase T1 and subjected to capillary liquid chromatography (LC) nano-electrospray ionization (nanoESI) MS to detect the heptamer RNA fragments containing C2501 (CACmCUCGp, m/z 1115.15) or ho5C2501 (CACmCUho5CGp, m/z 1123.15) (Figure 2A). The doubly-charged negative ion of each fragment was further probed by collision-induced dissociation (CID) to sequence the fragment and determined the exact positions of the RNA modifications (Figure 2B). Judging from the peak height ratio of the fragments, the modification frequency of ho5C2501 was 67%, confirming that the modification was sub-stoichiometric, as previously reported (29).
Figure 2.
RlhA/YdcP is responsible for ho5C2501 formation. (A) RNA-MS of 48-mer segments of 23S rRNA from WT (leftmost panels), ME5100 (left panels), ΔydcP (right panels) and ME5100 rescued by plasmid-encoded ydcP (rightmost panels). Shown are extracted mass chromatograms (XICs) for divalent negative ions of the RNase T1–digested heptamer fragments with (upper panels) or without (lower panels) ho5C2501. Frequencies of ho5C2501 are indicated. n.d., not detected. (B) Collision-induced dissociation (CID) spectra of the heptamer fragments with (left panel) or without (right panel) ho5C2501. A divalent ion for each fragment (m/z 1123.1 and 1115.1) was employed as the parent ion for CID. c, y, and w series product ions are assigned in each sequence. (C) E. coli genomic region from ycjD to ydfJ, and a series of deletion strains used to narrow down the gene responsible for ho5C2501 formation. The white bar represents a deleted region. The presence (+) or absence (–) of ho5C2501 is denoted in each strain. (D) Sucrose density gradient profiling of SPA-tagged RlhA in ribosomal fractions. Upper panel shows UV traces at 260 nm for the ribosomal fractions in 0.5 (filled circles) and 10 mM (open circles) Mg2+ conditions. Lower panels show western blotting to detect SPA-tagged RlhA protein in each Mg2+ concentration.
To identify the genes responsible for ho5C2501 formation, we implemented a reverse-genetic screen combined with RNA-MS, i.e. ribonucleome anaylsis (12,20,33). For the initial screen, we analyzed total RNA from 94 E. coli genomic-deletion strains, each of which lacked from 20 to 300 genes, covering almost 2000 genes in total. Among the complex mixture of RNA fragments produced by RNase T1 digestion of total RNA, we were able to successfully detected the heptamer RNA fragments containing ho5C2501 (m/z 1123.15) or C2501 (m/z 1115.15) by LC/MS analysis (Supplementary Figure S1). In two of the deletion strains, ME5100 and ME5046, no ho5C2501 was detected (Supplementary Figure S1). To validate this phenotype, we prepared the 48-mer RNA segments from these strains and confirmed lack of ho5C2501 in both (Figure 2A).
Next, we sought to extract a candidate gene responsible for ho5C2501 formation. Because the strain ME5100 lacks as many as 300 genes (from ycjD to ydfJ), we constructed eight strains containing smaller deletions within the large interval missing from the original strain and examined each for the presence or absence of ho5C2501 (Figure 2C). Eventually, we narrowed down the gene responsible for ho5C2501 formation to a genomic locus containing seven genes, from ydcN to ydcS (Figure 2C). Analysis of individual knockout strains of each gene in that locus revealed that ΔydcP completely lacked ho5C2501 (Figure 2A), indicating that ydcP is responsible for ho5C2501 formation. In support of this conclusion, ho5C2501 was restored when plasmid-encoded ydcP was introduced into strain ME5100 (Figure 2A).
YdcP co-localized with the precursor of ribosomal subunit
Assuming that YdcP is the dedicated enzyme for ho5C2501 formation, it should bind to a precursor of the ribosomal subunit. According to protein interactome studies in E. coli (59,60), YdcP interactes with several ribosomal proteins, as well as RlmN, which is the methyltransferase responsible for m2A2503 formation in 23S rRNA (Figure 1 and Supplementary Table S4). These facts imply that YdcP binds to an assembly intermediate of the 50S subunit. To explore this possibility, we performed sucrose density gradient ultracentrifugation (SDG) analysis to detect YdcP in ribosomal fractions. For this experiment, we used an E. coli strain in which endogenous YdcP was C-terminally sequential peptide affinity (SPA)-tagged (YdcP-SPA) (61). The crude ribosome fraction was subjected to SDG and fractionated in the presence of 0.5 or 10 mM Mg2+, followed by western blotting to detect YdcP-SPA in each fraction (Figure 2D). At both Mg2+ concentrations, YdcP-SPA was present in fractions mainly containing the 30S subunit. This sedimentation pattern is similar to those observed for other rRNA-modifying enzymes targeting 23S rRNA at the early stage of 50S subunit assembly (62). Moreover, this observation is consistent with the previous finding that ho5C2501 is introduced in an early stage of 50S biogenesis in the cell (63). Taken together with results of the interactome analyses (Supplementary Table S4), these data indicate that YdcP physically binds to an early-stage assembly intermediate of the 50S subunit, where C2501 of 23S rRNA is hydroxylated. Therefore, we renamed ydcP as rlhA (large subunit ribosomal RNA hydroxylation A).
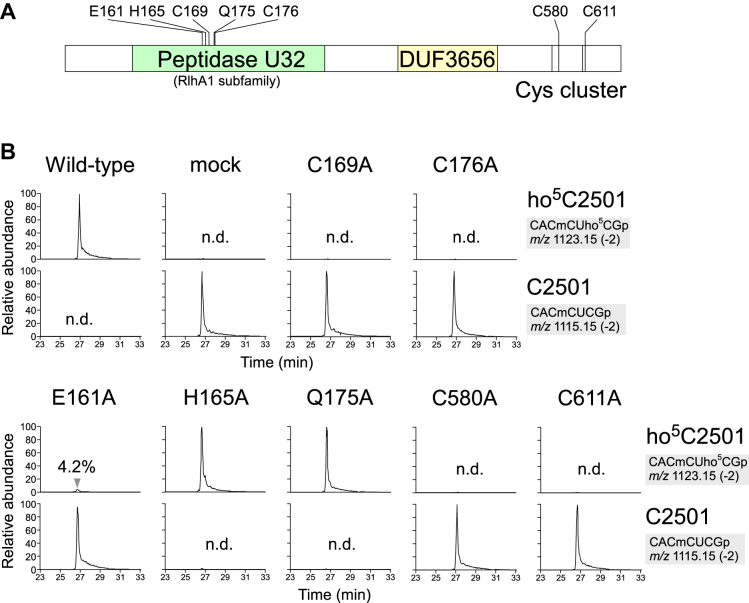
Conserved Cys residues are critical for RlhA function
RlhA possesses two conserved motifs, peptidase U32 and DUF3656 (Figure 3A). Peptidase U32 is categorized as a collagenase because some proteins containing this motif exhibit collagenolytic activity (64,65); however, the catalytic mechanism is unknown. In E. coli, peptidase U32 is present in four proteins, RlhA, YegQ, YhbU and YhbV, whereas DUF3656 is only present in RlhA, suggesting that the other three paralogs containing the peptidase U32 motif have functions distinct from those of RlhA. We performed multiple sequence alignments of 3521 peptidase U32 motifs (Supplementary Table S2) from 3066 proteins deposited in Pfam database and generated a phylogenetic tree (Supplementary Figure S2). On the basis of the tree, we picked up nine RlhA homologs, which were then subjected to multiple alignments (Supplementary Figure S3). This analysis revealed several conserved residues in the peptidase U32 motif (Figure 3A and Supplementary Figure S3). In addition, we noticed several cysteine residues clustered in the C-terminal region of the RlhA family proteins (Figure 3A and Supplementary Figure S3). To examine whether these conserved amino acids are important for ho5C2501 formation, we genetically complemented the ΔrlhA strain with multi-copy plasmids encoding wild-type rlhA or one of seven mutant variants, and then measured the modification frequency of each strain by LC/MS analysis. Introduction of wild-type rlhA fully restored ho5C2501 formation to even higher levels than in the wild-type strain, whereas no hydroxylation occurred in a mock-transformed negative control (Figure 3B). In five of the constructs, conserved residues in the peptidase U32 motif were individually replaced with Ala. Cys169 and Cys176 were completely essential for ho5C2501 formation, whereas a very small quantity of ho5C2501 was restored by the E161A mutant (Figure 3B). The H165A and Q175A plasmids complemented as efficiently as the wild type, indicating that these mutations had almost no effect. In the C-terminal region, mutations of two Cys residues at positions 580 and 611 totally abolished ho5C2501 formation, indicating that these cysteines are also essential for the modification (Figure 3B). Thus, these data revealed that four conserved Cys residues, two in the peptidase U32 motif and two in the C-terminal region, play critical roles in ho5C2501 formation.
Figure 3.
Mutation study of RlhA. (A) Schematic view of protein motifs in RlhA protein, including amino-acid residues mutated in this study. (B) RNA-MS of the 48-mer segments of 23S rRNA from a series of ΔrlhA strains harboring plasmids encoding wild-type or rlhA mutants. Shown are XICs for divalent negative ions of the RNase T1-digested heptamer fragments with (upper panels) or without (lower panels) ho5C2501 in each mutant. n.d., not detected.
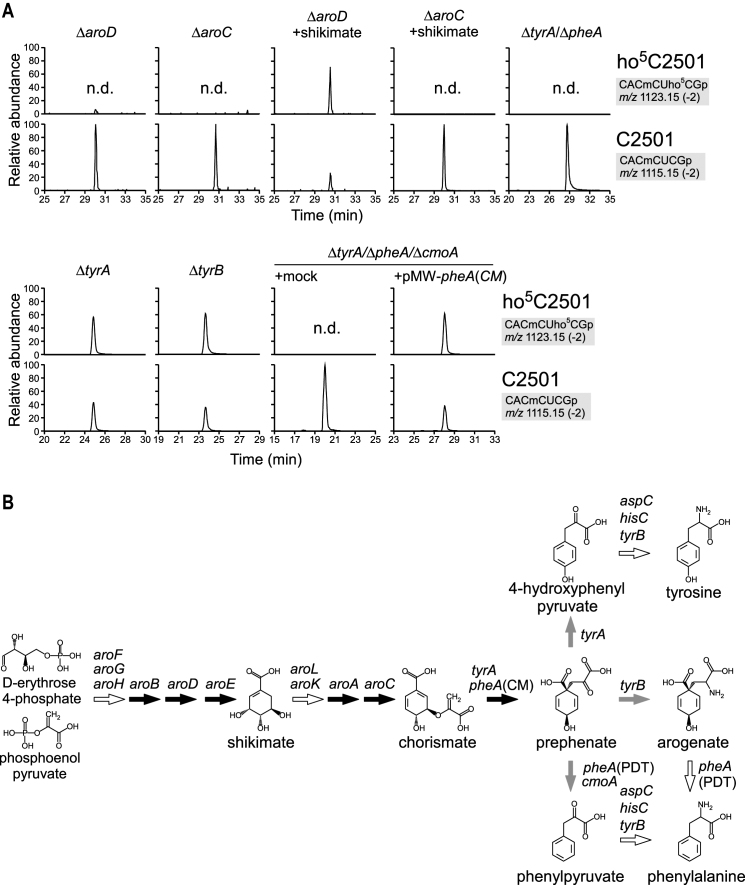
Prephenate synthesis is essential for ho5C2501 formation
In the ribonucleome analysis, we identified a second deletion mutant, strain ME5046, in which ho5C2501 was absent (Supplementary Figure S1). By analyzing a series of gene deletion strains, each of which lacked a single gene in the deleted region of ME5046, we found that aroD was essential for ho5C2501 formation (Figure 4A and Supplementary Figure S1).
Figure 4.
Prephenate synthesis and ho5C2501 formation. (A) RNA-MS of the 48-mer segments of 23S rRNA from a series of deletion strains related to prephenate synthesis. Shown are XICs for divalent negative ions of the RNase T1-digested heptamer fragments with (upper panels) or without (lower panels) ho5C2501 in each mutant. Frequencies of ho5C2501 are indicated. n.d., not detected. (B) Metabolic pathway of aromatic amino acid biogenesis. Genes responsible for each pathway are indicated. Black and gray arrows represent pathways examined in this study that were indispensable and dispensable for ho5C2501 formation, respectively. White arrows represent pathways not examined in this study.
aroD encodes 3-dehydroquinate dehydratase, a metabolic enzyme involved in the shikimate pathway. This pathway generates chorismate, a precursor of several metabolites including aromatic amino acids, quinones, folate and siderophores (66). We also confirmed the absence of ho5C2501 in several other knockout strains in the shikimate pathway, ΔaroB, ΔaroE, ΔaroA and ΔaroC (Figure 4AB and Supplementary Figure S4). Addition of shikimate to the culture medium rescued ho5C2501 formation in ΔaroD, but not in ΔaroC (Figure 4A), consistent with the fact that aroD and aroC are respectively upstream and downstream of shikimate in the pathway (Figure 4B). Chorismate produced by AroC is subsequently converted to prephenate by PheA and TyrA redundantly. ho5C2501 was absent in a ΔtyrA/ΔpheA double-knockout strain (Figure 4A), indicating that prephenate or its downstream metabolites (Figure 4B) are essential for ho5C2501 formation. Since ho5C2501 was present in both ΔtyrA and ΔtyrB strains (Figure 4A), we concluded that neither 4-hydroxyphenylpyruvate nor arogenate (Figure 4B) are involved in ho5C2501 synthesis. PheA is a metabolic enzyme bearing a chorismate mutase (CM) domain and a prephenate dehydratase (PDT) domain, which are responsible for synthesizing prephenate and phenylpyruvate, respectively (Figure 4B). To dissect these two reactions, we constructed a PheA variant with only CM activity [PheA(CM)] by introducing an active-site mutation in the PDT domain (48). To determine whether phenylpyruvate (Figure 4B) is responsible for ho5C2501 formation, we further deleted cmoA, which also converts prephenate to phenylpyruvate, from the ΔtyrA/ΔpheA strain to construct ΔtyrA/ΔpheA/ΔcmoA triple knockout strain, into which we introduced plasmid-encoded pheA(CM), resulting in accumulation of prephenate. As a result, ho5C2501 was rescued (Figure 4A), strongly suggesting that prephenate is required for ho5C2501 formation.
Iron-sulfur cluster biogenesis is required for ho5C2501 formation
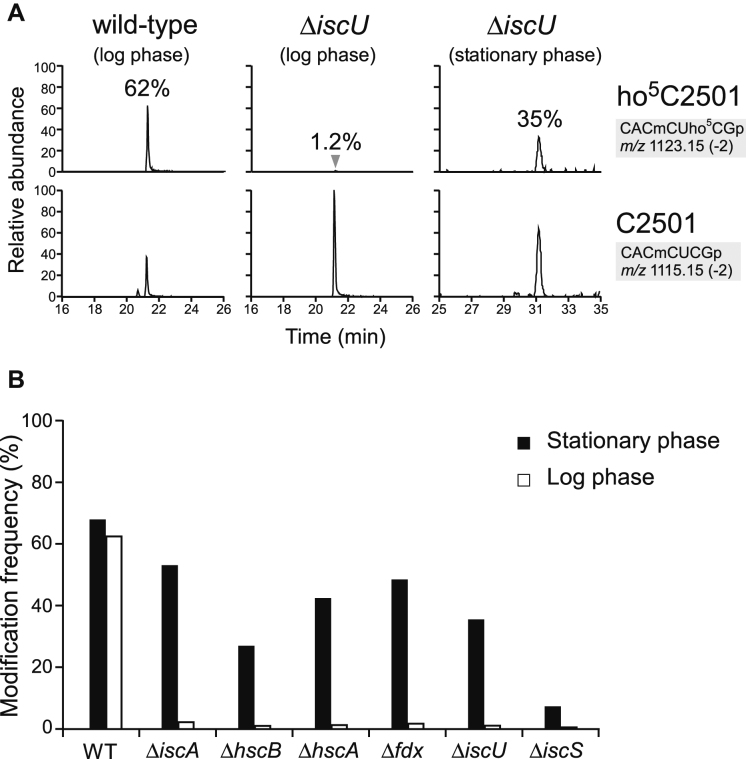
The importance of the four Cys residues in RlhA led us to speculate that Fe–S cluster formation is involved in ho5C2501 formation. Consistent with this hypothesis, we observed a profound reduction in the frequency of ho5C2501 upon knockout of iscU, which encodes a scaffold protein for Fe–S cluster formation in the ISC system (Supplementary Figure S1) (67). RNA segment analysis of ΔiscU revealed that the frequency of ho5C2501 was reduced to 1.2% in logarithmic (log) phase and 35% in stationary phase (Figure 5A). Next, we investigated whether five other genes involved in the ISC system are responsible for ho5C2501 formation. As in the ΔiscU strain, ho5C2501 frequency was reduced to 0.6–2.2% in log phase and 7–53% in stationary phase in deletion strains of iscA, iscS, hscB, hscA and fdx (Figure 5B). These results clearly demonstrate that Fe–S cluster biogenesis is involved in ho5C2501 formation. As in the ΔiscU strain, severe reduction of ho5C2501 was observed in log phase in all of these strains, indicating that the Fe–S cluster responsible for ho5C2501 formation is mainly generated by the ISC system in log phase. Studies of Fe–S-dependent tRNA modification (68) showed that in ΔiscS, the methylthio modification of 2-methylthio-N6-isopentenyladenosine (ms2i6A) and 2-thiocytidine (s2C) of tRNAs disappear in log phase, but are restored in stationary phase, probably due to redundant Fe–S cluster formation mediated by the other cysteine desulfurase (SufS and the SUF system) in stationary phase (69). Likewise, the robust ho5C2501 formation in stationary phase in E. coli strains lacking the ISC system can be explained by redundant Fe–S cluster biogenesis.
Figure 5.
Iron-sulfur cluster biogenesis and ho5C2501 formation. (A) RNA-MS of the 48-mer segments of 23S rRNA from WT and ΔiscU strains harvested in log and stationary phases. Shown are XICs for divalent negative ions of the RNase T1-digested heptamer fragments with (upper panels) or without (lower panels) ho5C2501 in each strain. Frequencies of ho5C2501 are indicated. (B) Frequencies of ho5C2501 in a series of knockout strains related to Fe–S cluster biogenesis, harvested at log (open bars) and stationary (filled bars) phases.
Iron-response alteration of ho5C2501 formation
According to a previous report (32), the modification frequency of ho5C2501 is higher in stationary phase than in log phase. To recapitulate this phenomenon, we analyzed ho5C2501 frequency in the wild-type strain cultured in rich medium and harvested at different growth phases (Figure 6A). ho5C2501 frequency was relatively low (33%) in early log phase (A600 = 0.18), but increased to 67% in late log or early stationary phase (A600 = 1.4), (Figure 6A). This observation is basically consistent with the results of the previous study (32). However, when the cells were cultured in minimal (M9) medium, the growth-phase dependent alteration of ho5C2501 frequency followed a different pattern. In this case, the modification frequency was 60% in log phase, but decreased markedly to 20% in stationary phase following overnight cultivation (Figure 6B). This drastic decrease in ho5C2501 formation might reflect the dearth of some nutrient in the minimum medium. In the case of methylthiolation of tRNA mediated by MiaB (a Fe–S protein), the modification frequency is regulated by iron starvation (70,71). Given that Fe–S cluster biogenesis is required for ho5C2501 formation, we asked whether the low level of ho5C2501 in stationary phase cells cultured in minimal medium could be restored by addition of excess iron. Indeed, when iron chloride (0.2 mM) was added to M9 medium, ho5C2501 frequency did not decrease during stationary phase, remaining at log-phase levels (∼60%) even after overnight cultivation (Figure 6B). This result strongly suggests that the severe reduction of ho5C2501 frequency in stationary phase was caused by iron depletion in minimal medium.
To confirm this finding we depleted iron from nutrient-rich medium by adding a strong iron chelator, 2,2’-dipyridyl (Dip) (71). As a positive control, we measured the methylthiolation level of ms2i6A in tRNAs, which is sensitive to iron depletion (70). As expected, the level of ms2i6A was significantly reduced to 3.6% in cells cultured in the presence of Dip (Figure 6C), indicating that Dip had effectively removed Fe2+ from the medium, as well as from cells. Under this condition, measurement of the ho5C2501 level by analysis of the corresponding rRNA segment revealed that the frequency of ho5C2501 decreased markedly, to 1.6% (Figure 6D). The reduced levels of ho5C2501 and ms2i6A were gradually restored by addition of increasing amounts of iron chloride (Figure 6CD), indicating that ho5C2501 frequency is altered by extracellular iron concentration.
The iron-sensitive alteration of ho5C2501 might be attributed to altered expression of RlhA. To explore this possibility, we analyzed steady-state levels of RlhA and two other Fe–S proteins, MiaB and RlmN, upon iron depletion. In this experiment, we used E. coli strains in which each endogenous protein was SPA-tagged. These strains were cultured in the presence or absence of Dip and subjected to western blotting to detect each protein. The level of RlhA decreased upon iron depletion (Supplementary Figure S5A). The MiaB level also decreased in the presence of Dip, whereas the level of RlmN did not decrease (Supplementary Figure S5A). We also measured steady-state level of mRNAs by RT-qPCR in each culture condition (Supplementary Figure S5B). The mRNA level of rlhA slightly increased upon iron depletion, whereas the mRNA level of miaB was not affected, denying transcriptional control of these proteins. Taken together, the protein levels of rlhA and miaB are controlled post-transcriptionally upon iron depletion via translational regulation or Fe–S protein instability by chelating Fe ion.
DISCUSSION
Hydroxylation of biological molecules is a fundamental enzymatic reaction involved in various biological events (72,73). In the context of the epitranscriptome, hydroxylation is a major modification that modulates RNA functions (73) and as such is involved in several tRNA modifications (34,74–77). N6-methyladenosine (m6A) and 1-methyladenosine (m1A) are demethylated via hydroxymethyl formation (78). The RNA hydroxylation events reported to date are catalyzed by Fe(II)- and 2-oxoglutarate (2-OG)-dependent oxygenases, including ALKBH family proteins (75,77,79), Tet family proteins (80,81), and JmjC-domain containing protein (34). Fe(II)/2-OG-dependent RNA oxygenases use molecular oxygen as a substrate for hydroxylation. RlhA does not have any of the characteristic domains and motifs conserved in these RNA oxygenases, suggesting that it represents a novel family of proteins responsible for RNA hydroxylation.
RlhA contains peptidase U32 and DUF3656 motifs. The former motif is present in bacterial collagenases, which in general contribute to bacterial infection (82–84). Porphyromonas gingivalis PrtC is a well-characterized peptidase U32 protein with proteolytic activity against a specific type of collagen, which allows the bacteria to exert virulence in gingival tissue (64). Helicobacter pylori HP0169, another peptidase U32 protein, is required for colonization of mouse gut by this bacterium (65). The collagenolytic activity of H. pylori HP0169 has been confirmed in vitro (65). However, the mechanisms underlying these protease activities remain largely unknown. The demonstration that RlhA is responsible for RNA hydroxylation reveals a novel function of peptidase U32 proteins, suggesting that this activity is involved in diverse cellular processes.
Phylogenetic analysis enabled us to classify peptidase U32 motifs into 12 subfamilies (Supplementary Figure S2 and Table S2). Four of these subfamilies are represented by four E. coli paralogs, YhbU, YhbV, YegQ and RlhA. Among all subfamilies, the co-occurrence of peptidase U32 and DUF3656 is unique to the RlhA subfamily. In proteins of the RlhA1 subfamily, about half of the homologs have a second peptidase U32 motif, named RlhA2a and RlhA2b, attached to their C-termini. Intriguingly, the RlhA1, RlhA2a and RlhA2b motifs are phylogenetically distant, indicating that they evolved independently. Two collagenases, P. gingivalis PrtC and H. pylori HP0169, are classified in subfamilies, named PepU32#4 and PepU32#2, respectively, which are clearly separated from the subfamilies of RlhA1, RlhA2a and RlhA2b. ho5C is present in D. radiodurans 23S rRNA (29). Therefore, D. radiodurans DR2130, a peptidase U32 protein in the RlhA1 subfamily, is likely involved in the biogenesis of ho5C.
Based on the distribution of the DUF3656 motif in bacterial and archaeal phyla (Supplementary Figure S6), RlhA homologs are abundant in γ-proteobacteria (16%), δ-proteobacteria (19%), clostridia (84%), bacteroidia (89%), cyanobacteria (17%) and some species of archaea. In particular, RlhA homologs are common in bacteroidia and clostridia, the main microbiota in human gut, suggesting that RlhA homologs and the ho5C2501 modification could contribute to bacterial survival in this environment.
In vivo complementation of the ΔrlhA strain revealed that four conserved Cys residues in the peptidase U32 motif and C-terminal region are essential for ho5C2501 formation, suggesting that RlhA is a Fe–S protein. Consistent with this, we demonstrated that Fe–S cluster biogenesis is involved in ho5C2501 formation. In a series of strains harboring knockouts in components of the ISC system, severe reduction of ho5C2501 frequency was observed in log phase, indicating that the Fe–S cluster of RlhA is mainly synthesized by the ISC system in log phase. However, in stationary phase, the level of ho5C2501 was restored in strains lacking the ISC system, likely because the SUF system can assist with Fe–S cluster biogenesis at this stage of growth.
Fe–S proteins are sensitive to oxidative stress and iron availability (67). In this study, we found that iron depletion decreased ho5C2501 frequency, suggesting that the Fe–S cluster of RlhA is sensitive to intracellular iron concentration. MiaB-mediated 2-methylthiolation of ms2i6A in tRNAs is also iron-sensitive, whereas m2A2503 formation mediated by RlmN, another Fe–S protein, is not, indicating that Fe–S proteins have different sensitivities to iron depletion. Bacteria often encounter iron-depleted environments, as in the case of pathogenic bacteria growing in animal host. Animal hosts express various types of iron-chelating proteins, in part to reduce the amount of extracellular iron available to bacteria (85). Under such conditions, the ho5C2501 frequency in 23S rRNA should be reduced. Pathogenic E. coli recovered from peritoneal cavities of lethally infected animals lacked the 2-methylthio group of ms2i6A (86), indicating that alteration of RNA modifications under low iron availability occurs physiologically. Given the strong iron sensitivity of ho5C2501, it is likely that ho5C2501 frequency is markedly reduced during colonization of an iron-restrictive host environment.
We also revealed that prephenate is required for ho5C2501 biogenesis. Prephenate is a precursor for aromatic amino acids (66), and also serves as a substrate for tRNA modification. To expand their decoding capacity, bacterial tRNAs responsible for family boxes contain 5-carboxymethoxyuridine (cmo5U) or its derivatives (87,88). CmoA and CmoB are the enzymes responsible for cmo5U formation (88,89). In this pathway, CmoA first synthesizes S-adenosyl-S-carboxymethyl-L-homocysteine (SCM-SAH or Cx-SAM) from AdoMet and prephenate (89,90). Then, CmoB transfers the carboxymethyl-group of SCM-SAH to the hydroxy group of 5-hydroxyuridine (ho5U) on tRNAs to yield cmo5U (89). In the first reaction, prephenate is converted to phenylpyruvate via elimination of a hydroxyl group and decarboxylation. Although we have not yet reconstituted ho5C formation, we speculate that RlhA utilizes prephenate as a hydroxyl donor for synthesis of ho5C. Further studies will be necessary to obtain mechanistic insight into ho5C formation.
In addition to iron availability, the intracellular concentration of prephenate could also affect ho5C2501 formation. Given that prephenate is a key metabolite involved in the synthesis of aromatic amino acids, dynamic changes in ho5C2501 frequency could regulate ribosomal function, leading to translational regulation associated with metabolic changes in shikimate pathway. Enterochelin, an iron-chelating metabolite, is synthesized from chorismate (66). Under the condition with little iron availability, EntC is overexpressed and catalyzes conversion of chorismate to isochorismate which is used for enterochelin biogenesis (91,92). Thus, cellular concentration of chorismate and prephenate would be reduced under iron starved condition, leading to much more severe reduction of ho5C2501.
In this study, we demonstrated that the novel gene rlhA is responsible for ho5C2501 formation. RlhA contains an uncharacterized peptidase U32 motif and does not belong to any known family of RNA hydroxylases. Although we do not still exclude a possibility that RlhA acts as a peptidase to activate an unknown proenzyme responsible for ho5C formation, we propose RlhA is directly involved in ho5C2501 formation. Fe–S cluster and prephenate were required for ho5C2501 formation, implying that RlhA is a Fe–S protein that catalyzes ho5C2501 formation using prephenate as a hydroxyl group donor. Consistent with this, the frequency of ho5C2501 is dynamically altered in response to environmental iron concentration.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the members of the Suzuki laboratory, in particular Yuriko Sakaguchi, for technical support and many insightful discussions. Special thanks are due to Kohji Seio (Tokyo Inst. Tech.) for providing materials.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan [to T.S.]; JSPS Fellowship for Japanese Junior Scientists [to S.K. and Y.S.]. Funding for open access charge: JSPS; Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. . MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duechler M., Leszczynska G., Sochacka E., Nawrot B.. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell. Mol. Life Sci. 2016; 73:3075–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 4. Li S., Mason C.E.. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014; 15:127–150. [DOI] [PubMed] [Google Scholar]
- 5. Helm M., Alfonzo J.D.. Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 2014; 21:174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki T. Grosjean H. Fine-Tuning of RNA Functions by Modification and Editing. 2005; 12:Springer-Verlag Berlin and Heidelberg GmbH & Co. KG; 23–69. [Google Scholar]
- 7. Sergiev P.V., Golovina A.Y., Prokhorova I.V., Sergeeva O.V., Osterman I.A., Nesterchuk M.V., Burakovsky D.E., Bogdanov A.A., Dontsova O.A.. Modifications of Ribosomal RNA: From Enzymes to Function. 2011; Vienna: Springer; 97–110. [Google Scholar]
- 8. Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A.. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015; 22:342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ge J., Yu Y.T.. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 2013; 38:210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sergeeva O.V., Bogdanov A.A., Sergiev P.V.. What do we know about ribosomal RNA methylation in Escherichia coli?. Biochimie. 2015; 117:110–118. [DOI] [PubMed] [Google Scholar]
- 12. Kimura S., Suzuki T.. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010; 38:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baxter-Roshek J.L., Petrov A.N., Dinman J.D.. Optimization of ribosome structure and function by rRNA base modification. PLoS One. 2007; 2:e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baudin-Baillieu A., Fabret C., Liang X.H., Piekna-Przybylska D., Fournier M.J., Rousset J.P.. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009; 37:7665–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lafontaine D., Vandenhaute J., Tollervey D.. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995; 9:2470–2481. [DOI] [PubMed] [Google Scholar]
- 16. Meyer B., Wurm J.P., Sharma S., Immer C., Pogoryelov D., Kotter P., Lafontaine D.L., Wohnert J., Entian K.D.. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res. 2016; 44:4304–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Connolly K., Rife J.P., Culver G.. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008; 70:1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ito S., Horikawa S., Suzuki T., Kawauchi H., Tanaka Y., Suzuki T., Suzuki T.. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J. Biol. Chem. 2014; 289:35724–35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito S., Akamatsu Y., Noma A., Kimura S., Miyauchi K., Ikeuchi Y., Suzuki T., Suzuki T.. A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem. 2014; 289:26201–26212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kimura S., Ikeuchi Y., Kitahara K., Sakaguchi Y., Suzuki T.. Base methylations in the double-stranded RNA by a fused methyltransferase bearing unwinding activity. Nucleic Acids Res. 2012; 40:4071–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arai T., Ishiguro K., Kimura S., Sakaguchi Y., Suzuki T., Suzuki T.. Single methylation of 23S rRNA triggers late steps of 50S ribosomal subunit assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4707–E4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilson D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014; 12:35–48. [DOI] [PubMed] [Google Scholar]
- 23. Garbom S., Forsberg A., Wolf-Watz H., Kihlberg B.M.. Identification of novel virulence-associated genes via genome analysis of hypothetical genes. Infect. Immun. 2004; 72:1333–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su J., Yang J., Zhao D., Kawula T.H., Banas J.A., Zhang J.R.. Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 2007; 75:3089–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kyuma T., Kimura S., Hanada Y., Suzuki T., Sekimizu K., Kaito C.. Ribosomal RNA methyltransferases contribute to Staphylococcus aureus virulence. FEBS J. 2015; 282:2570–2584. [DOI] [PubMed] [Google Scholar]
- 26. Oldenburg M., Kruger A., Ferstl R., Kaufmann A., Nees G., Sigmund A., Bathke B., Lauterbach H., Suter M., Dreher S. et al. . TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012; 337:1111–1115. [DOI] [PubMed] [Google Scholar]
- 27. Golovina A.Y., Dzama M.M., Osterman I.A., Sergiev P.V., Serebryakova M.V., Bogdanov A.A., Dontsova O.A.. The last rRNA methyltransferase of E. coli revealed: the yhiR gene encodes adenine-N6 methyltransferase specific for modification of A2030 of 23S ribosomal RNA. RNA. 2012; 18:1725–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bakin A., Ofengand J.. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993; 32:9754–9762. [DOI] [PubMed] [Google Scholar]
- 29. Havelund J.F., Giessing A.M., Hansen T., Rasmussen A., Scott L.G., Kirpekar F.. Identification of 5-hydroxycytidine at position 2501 concludes characterization of modified nucleotides in E. coli 23S rRNA. J. Mol. Biol. 2011; 411:529–536. [DOI] [PubMed] [Google Scholar]
- 30. Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 31. Sato N.S., Hirabayashi N., Agmon I., Yonath A., Suzuki T.. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15386–15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andersen T.E., Porse B.T., Kirpekar F.. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004; 10:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suzuki T., Ikeuchi Y., Noma A., Suzuki T., Sakaguchi Y.. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007; 425:211–229. [DOI] [PubMed] [Google Scholar]
- 34. Noma A., Ishitani R., Kato M., Nagao A., Nureki O., Suzuki T.. Expanding role of the jumonji C domain as an RNA hydroxylase. J. Biol. Chem. 2010; 285:34503–34507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noma A., Kirino Y., Ikeuchi Y., Suzuki T.. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006; 25:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ikeuchi Y., Shigi N., Kato J., Nishimura A., Suzuki T.. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006; 21:97–108. [DOI] [PubMed] [Google Scholar]
- 37. Sakai Y., Miyauchi K., Kimura S., Suzuki T.. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 2016; 44:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soma A., Ikeuchi Y., Kanemasa S., Kobayashi K., Ogasawara N., Ote T., Kato J., Watanabe K., Sekine Y., Suzuki T.. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003; 12:689–698. [DOI] [PubMed] [Google Scholar]
- 39. Ikeuchi Y., Kitahara K., Suzuki T.. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008; 27:2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miyauchi K., Kimura S., Suzuki T.. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013; 9:105–111. [DOI] [PubMed] [Google Scholar]
- 41. Kimura S., Miyauchi K., Ikeuchi Y., Thiaville P.C., Crecy-Lagard V., Suzuki T.. Discovery of the beta-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 2014; 42:9350–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noma A., Yi S., Katoh T., Takai Y., Suzuki T., Suzuki T.. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA. 2011; 17:1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kato J., Hashimoto M.. Construction of consecutive deletions of the Escherichia coli chromosome. Mol. Syst. Biol. 2007; 3:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:doi:10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomason L.C., Costantino N., Court D.L.. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007; doi:10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 47. Saka K., Tadenuma M., Nakade S., Tanaka N., Sugawara H., Nishikawa K., Ichiyoshi N., Kitagawa M., Mori H., Ogasawara N. et al. . A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 2005; 12:63–68. [DOI] [PubMed] [Google Scholar]
- 48. Zhang S., Wilson D.B., Ganem B.. Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli P-protein dehydratase domain. Biochemistry. 2000; 39:4722–4728. [DOI] [PubMed] [Google Scholar]
- 49. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. et al. . Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 52. Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999; 41:95–98. [Google Scholar]
- 53. Perriere G., Gouy M.. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996; 78:364–369. [DOI] [PubMed] [Google Scholar]
- 54. Letunic I., Bork P.. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016; 44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohira T., Suzuki T.. Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat. Chem. Biol. 2016; 12:648–655. [DOI] [PubMed] [Google Scholar]
- 56. Sakaguchi Y., Miyauchi K., Kang B.I., Suzuki T.. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol. 2015; 560:19–28. [DOI] [PubMed] [Google Scholar]
- 57. Kang B.I., Miyauchi K., Matuszewski M., D’Almeida G.S., Rubio M.A.T., Alfonzo J.D., Inoue K., Sakaguchi Y., Suzuki T., Sochacka E., Suzuki T.. Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res. 2017; 45:2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. . The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 59. Butland G., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N. et al. . Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005; 433:531–537. [DOI] [PubMed] [Google Scholar]
- 60. Rajagopala S.V., Sikorski P., Kumar A., Mosca R., Vlasblom J., Arnold R., Franca-Koh J., Pakala S.B., Phanse S., Ceol A. et al. . The binary protein-protein interaction landscape of Escherichia coli. Nat. Biotechnol. 2014; 32:285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zeghouf M., Li J., Butland G., Borkowska A., Canadien V., Richards D., Beattie B., Emili A., Greenblatt J.F.. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 2004; 3:463–468. [DOI] [PubMed] [Google Scholar]
- 62. Jiang M., Sullivan S.M., Walker A.K., Strahler J.R., Andrews P.C., Maddock J.R.. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J. Bacteriol. 2007; 189:3434–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Popova A.M., Williamson J.R.. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J. Am. Chem. Soc. 2014; 136:2058–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kato T., Takahashi N., Kuramitsu H.K.. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene, which expresses a novel collagenase activity. J. Bacteriol. 1992; 174:3889–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kavermann H., Burns B.P., Angermuller K., Odenbreit S., Fischer W., Melchers K., Haas R.. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 2003; 197:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M.. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014; 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Py B., Barras F.. Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 2010; 8:436–446. [DOI] [PubMed] [Google Scholar]
- 68. Lauhon C.T. Requirement for IscS in biosynthesis of all thionucleosides in Escherichia coli. J. Bacteriol. 2002; 184:6820–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Buhning M., Valleriani A., Leimkuhler S.. The role of SufS is restricted to Fe-S cluster biosynthesis in Escherichia coli. Biochemistry. 2017; 56:1987–2000. [DOI] [PubMed] [Google Scholar]
- 70. Griffiths E., Humphreys J.. Alterations in tRNAs containing 2-methylthio-N6-(delta2-isopentenyl)-adenosine during growth of enteropathogenic Escherichia coli in the presence of iron-binding proteins. Eur. J. Biochem. 1978; 82:503–513. [DOI] [PubMed] [Google Scholar]
- 71. Vecerek B., Moll I., Blasi U.. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007; 26:965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Katz M.J., Gandara L., De Lella Ezcurra A.L., Wappner P.. Hydroxylation and translational adaptation to stress: some answers lie beyond the STOP codon. Cell. Mol. Life Sci. 2016; 73:1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ploumakis A., Coleman M.L.. OH, the places you’ll go! hydroxylation, gene expression, and cancer. Mol. Cell. 2015; 58:729–741. [DOI] [PubMed] [Google Scholar]
- 74. Fu Y., Dai Q., Zhang W., Ren J., Pan T., He C.. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed. Engl. 2010; 49:8885–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kawarada L., Suzuki T., Ohira T., Hirata S., Miyauchi K., Suzuki T.. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017; 45:7401–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Persson B.C., Bjork G.R.. Isolation of the gene (miaE) encoding the hydroxylase involved in the synthesis of 2-methylthio-cis-ribozeatin in tRNA of Salmonella typhimurium and characterization of mutants. J. Bacteriol. 1993; 175:7776–7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Songe-Moller L., van den Born E., Leihne V., Vagbo C.B., Kristoffersen T., Krokan H.E., Kirpekar F., Falnes P.O., Klungland A.. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010; 30:1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao B.S., Roundtree I.A., He C.. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fedeles B.I., Singh V., Delaney J.C., Li D., Essigmann J.M.. The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J.f Biol. Chem. 2015; 290:20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Delatte B., Wang F., Ngoc L.V., Collignon E., Bonvin E., Deplus R., Calonne E., Hassabi B., Putmans P., Awe S. et al. . RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016; 351:282–285. [DOI] [PubMed] [Google Scholar]
- 81. Fu L., Guerrero C.R., Zhong N., Amato N.J., Liu Y., Liu S., Cai Q., Ji D., Jin S.G., Niedernhofer L.J. et al. . Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 2014; 136:11582–11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Navais R., Mendez J., Perez-Pascual D., Cascales D., Guijarro J.A.. The yrpAB operon of Yersinia ruckeri encoding two putative U32 peptidases is involved in virulence and induced under microaerobic conditions. Virulence. 2014; 5:619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhao H., Li X., Johnson D.E., Mobley H.L.. Identification of protease and rpoN-associated genes of uropathogenic Proteus mirabilis by negative selection in a mouse model of ascending urinary tract infection. Microbiology. 1999; 145:185–195. [DOI] [PubMed] [Google Scholar]
- 84. Zhao Y., Jansen R., Gaastra W., Arkesteijn G., van der Zeijst B.A., van Putten J.P.. Identification of genes affecting Salmonella enterica serovar enteritidis infection of chicken macrophages. Infect. immun. 2002; 70:5319–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Skaar E.P. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010; 6:e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Griffiths E., Humphreys J., Leach A., Scanlon L.. Alterations in the tRNA’s of Escherichia coli recovered from lethally infected animals. Infect. Immun. 1978; 22:312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Weixlbaumer A., Murphy F.V.t., Dziergowska A., Malkiewicz A., Vendeix F.A., Agris P.F., Ramakrishnan V.. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007; 14:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nasvall S.J., Chen P., Bjork G.R.. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004; 10:1662–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kim J., Xiao H., Bonanno J.B., Kalyanaraman C., Brown S., Tang X., Al-Obaidi N.F., Patskovsky Y., Babbitt P.C., Jacobson M.P. et al. . Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature. 2013; 498:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Byrne R.T., Whelan F., Aller P., Bird L.E., Dowle A., Lobley C.M., Reddivari Y., Nettleship J.E., Owens R.J., Antson A.A. et al. . S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA. Acta Crystallogr. D, Biol. Crystallogr. 2013; 69:1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Buss K., Müller R., Dahm C., Gaitatzis N., Skrzypczak-Pietraszek E., Lohmann S., Gassen M., Leistner E.. Clustering of isochorismate synthase genes menF and entC and channeling of isochorismate in Escherichia coli. Biochim. Biophys. Acta. 2001; 1522:151–157. [DOI] [PubMed] [Google Scholar]
- 92. Brickman T.J., Ozenberger B.A., McIntosh M.A.. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 1990; 212:669–682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.