Abstract
Isoprenoids are diverse natural compounds, which have various applications as pharmaceuticals, fragrances, and solvents. The low yield of isoprenoids in plants makes them difficult for cost-effective production, and chemical synthesis of complex isoprenoids is impractical. Microbial production of isoprenoids has been considered as a promising approach to increase the yield. In this study, we engineered the model cyanobacterium Synechocystis sp. PCC 6803 for sustainable production of a commercially valuable isoprenoid, limonene. Limonene synthases from the plants Mentha spicata and Citrus limon were expressed in cyanobacteria for limonene production. Production of limonene was two-fold higher with limonene synthase from M. spicata than that from C. limon. To enhance isoprenoid production, computational strain design was conducted by applying the OptForce strain design algorithm on Synechocystis 6803. Based on the metabolic interventions suggested by this algorithm, genes (ribose 5-phosphate isomerase and ribulose 5-phosphate 3-epimerase) in the pentose phosphate pathway were overexpressed, and a geranyl diphosphate synthase from the plant Abies grandis was expressed to optimize the limonene biosynthetic pathway. The optimized strain produced 6.7 mg/L of limonene, a 2.3-fold improvement in productivity. Thus, this study presents a feasible strategy to engineer cyanobacteria for photosynthetic production of isoprenoids.
Introduction
Recent studies have demonstrated the potential of using cyanobacteria as biological platforms to produce fuels and high-value chemicals1,2. Harnessing solar energy using the photosynthetic apparatus, atmospheric CO2 is fixed into sugars, which can be further converted to desired products by engineered cyanobacteria. Due to the recent development of genetic tools for model cyanobacteria3, expression of heterologous genes and pathways has become more feasible, thus facilitating the construction of engineered cyanobacteria for biotechnological applications. In this study, we engineered the model cyanobacterium Synechocystis sp. PCC 6803 (hereafter, Synechocystis 6803) for production of a commercially valuable isoprenoid, limonene.
Isoprenoids are one of the most diverse groups of natural products, with more than 55,000 compounds4. Isoprenoids have multiple commercial applications, including natural pharmaceuticals, nutraceuticals, solvents, and perfume components5,6. To date, commercially-used isoprenoids are mainly extracted from plants, but the low quantities of these naturally-produced chemicals have become an impediment for cost-effective production. Successful microbial production of valuable isoprenoids by engineered yeast and E. coli have been demonstrated7,8, whereas fewer researchers have studied production of isoprenoids by cyanobacteria. To improve photosynthetic production of isoprenoids, optimization of isoprenoid biosynthetic pathways in cyanobacteria is needed using metabolic engineering coupled with computational approaches.
Limonene is a 10-carbon isoprenoid produced by plants. (R)-limonene has a characteristic fragrance of orange, and commonly exists in the rinds of citrus fruits. It is commercially used as a fragrance in perfumes or a solvent in cleaning products. (S)-Limonene is a precursor for the biosynthesis of (S)-menthol, which is the major component of mint. Recently, limonene has been evaluated as a “drop-in” replacement for diesel9 and jet fuels10. The fully hydrogenated form of limonene was used as a diesel additive, exhibiting similar chemical properties compared to diesel fuel9. Moreover, the physical properties of limonene, such as viscosity, freezing point, and boiling point, are highly comparable to aviation fuel Jet A-110.
Cyanobacteria use the methylerythritol 4-phosphate (MEP) pathway to produce isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which are the building blocks for isoprenoid biosynthesis. The MEP pathway is a seven-step pathway that starts with glyceraldehyde 3-phosphate (GAP) and pyruvate, and ends with IPP and DMAPP. Further, IPP and DMAPP undergo a series of head-to-tail condensations to produce diphosphate substrates, which are then converted to isoprenoids by isoprenoid synthases. To increase isoprenoid production, the amounts of IPP and DMAPP need to be enhanced by increasing the carbon flux toward the MEP pathway.
Attempts have been made to engineer the MEP pathway for improving cyanobacterial limonene production. However, production titers are extremely low compared to other compounds such as ethanol11, butanol12, and free fatty acid13. Genes involving in the bottlenecks of the MEP pathway were overexpressed in Synechocystis 680314. The recombinant strain showed a 1.4-fold increase of limonene, and the final titer reached 1 mg/L after 30-day cultivation14. In addition, researchers used similar strategies to engineer the MEP pathway in the nitrogen-fixing cyanobacterium Anabaena sp. PCC 7120 for production of limonene15. The limonene yield increased up to 6.8-fold. However, the final titer remained low (0.5 mg/L over 12-day incubation)15.
A previous in vitro study suggested that isoprenoid production in Synechocystis 6803 is stimulated by compounds in the pentose phosphate (PP) pathway but not by substrates in the MEP pathway16. Using Synechocystis 6803 cell extracts, isoprenoid biosynthesis was significantly improved by supplying xylulose 5-phosphate (X5P) in the PP pathway, whereas providing substrates (GAP, pyruvate, and MEP) in the MEP pathway showed lower stimulation of isoprenoid production16. These results indicated a connection between the PP pathway and isoprenoid production in Synechocystis 6803.
In addition to experimental engineering approaches, computational strain design techniques can be useful to develop non-intuitive genetic interventions to achieve the desired level of production of a particular bioproduct. To this end, the OptForce procedure17 first characterizes the wild-type strain in the form of reaction flux ranges by utilizing the 13C MFA (Metabolic Flux Analysis) flux estimations as additional regulations. OptForce then contrasts the wild-type flux ranges with those in the overproducing phenotype. As a result, the algorithm identifies a set of genetic interventions (i.e., up/down-regulations and deletions) that must happen in the metabolic reaction network for a desired level of yield. Finally, OptForce pinpoints the minimal interventions (from these changes) that are directly related to achieving the desired yield. These strategies can then be tested in an experimental setting.
In this work, we engineered Synechocystis 6803 for photosynthetic limonene production (Fig. 1). To construct limonene-producing strains, genes encoding limonene synthase (lims) from Mentha spicata and Citrus limon were introduced into Synechocystis 6803. For generating computation-driven non-intuitive strain engineering strategies, we applied the OptForce algorithm17 on the genome-scale Synechocystis 6803 model iSyn73118 and also utilized 13C MFA flux estimations19 under photosynthetic wild-type condition. OptForce predicted the up-regulation of two PP pathway genes, ribose 5-phosphate isomerase (rpi) and ribulose 5-phosphate 3-epimerase (rpe), in limonene-producing strains in order to divert the carbon flux toward limonene production. Furthermore, based on the prediction made by OptForce to further improve limonene production, a geranyl diphosphate synthase (gpps) from Abies grandis was expressed to optimize the limonene production pathway. The final recombinant strain led to a 2.3-fold improvement in yield, producing 6.7 mg/L of limonene in 7 days. The metabolic engineering strategies used in this study demonstrate the feasibility of increasing limonene production in Synechocystis 6803 and can be applied to phototrophic production of other high-value isoprenoids.
Figure 1.
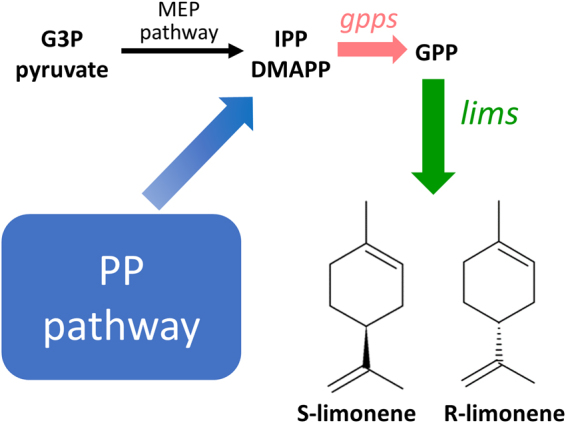
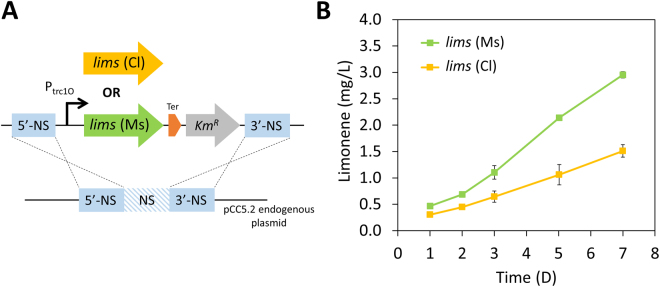
Schematic representation of engineering Synechocystis 6803 for production of limonene. Codon-optimized limonene synthases from Mentha spicata and Citrus limon were heterologously expressed in Synechocystis 6803 to produce S-limonene and R-limonene, respectively. The limonene biosynthetic pathway was optimized by overexpressing genes in the pentose phosphate (PP) pathway and a geranyl diphosphate synthase from Abies grandis. G3P, glyceraldehyde 3-phosphate; MEP, methylerythritol-4-phosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; lims, limonene synthase; gpps, geranyl diphosphate synthase.
Results
EngineeringSynechocystis6803 for production of limonene
Limonene is a C10 cyclic isoprenoid converted from geranyl diphosphate (GPP). Due to the complex nature of carbocation rearrangement from GPP to limonene, limonene synthase produces not only limonene but also other monoterpenes such as bicyclic α-pinene and acyclic mycene20. To avoid the production of other unwanted byproducts, we chose limonene synthases which have the highest specificity for limonene production. Based on previous studies, limonene synthase from Citrus limon and Mentha spicata produce limonene of high purity. Expression of each of these limonene synthases in E. coli showed that the former produces 99% pure (R)-limonene21, and the latter generates 94% of (S)-limonene22. The coding sequences of lims were codon optimized for Synechocystis 6803, and the plastid targeting sequences were removed23,24. The truncated enzyme is known to have better catalytic activity than the native protein25. Genes were cloned into a pCC5.2 neutral-site-targeting plasmid and driven by the trc1O promoter for higher level expression of lims (Fig. 2A). Expression of an enhanced yellow fluorescent protein (EYFP) from the pCC5.2 endogenous plasmid is 8 to 14 times higher than that on the chromosome26.
Figure 2.
Production of limonene by engineered Synechocystis 6803. (A) Introduction of limonene synthases into a neutral site on endogenous pCC5.2 plasmid to create limonene-producing mutants. (B) Time-course limonene production. Results were mean SD of three biological replicates. lims (Ms), limonene synthase from M. spicata; lims (Cl), limonene synthase from C. limon; Ptrc1O, trc1O promoter; NS, neutral site; Ter, terminator; KmR, kanamycin resistance cassette.
When the lims was cloned into a suicide plasmid and transformed into E. coli, we found that the gene accumulated random mutations in the E. coli host, leading to changes in amino acid residues or truncated proteins. This was presumably because the lims product is toxic to E. coli cells. To introduce a lims without mutations into Synechocystis 6803, we circumvented the E. coli cloning step by first cloning the lims into the suicide plasmid via Gibson assembly, and used the assembled product as template for PCR to amplify the lims cassette flanked by upstream and downstream homologous sequences of the neutral site in pCC5.226. Subsequently, the PCR product was directly used for natural transformation into Synechocystis 6803. The lims was introduced into Synechocystis 6803 genome via double homologous recombination (Fig. 2A). DNA sequencing results showed that the lims has no mutation in Synechocystis 6803 (data not shown). Mutants were fully segregated after re-streaking the cells several times on BG-11 plates with antibiotics.
Limonene production by engineered Synechocystis 6803 was tested by incubating cultures for 7 days. Because of the volatility of limonene, a dodecane overlay was applied on cultures to collect limonene in the organic layer. It has been reported that over 99% of limonene escapes from the cyanobacterial cultures14, and covering an organic overlay on cultures had little influence on growth in cyanobacteria23. The limonene yield by the strain expressing lims from M. spicata was two-fold higher than that by the strain expressing lims from C. limon (Fig. 2B). These results suggest that the limonene synthase from M. spicata exhibited better catalytic activity in Synechocystis 6803, and hence, the strain was used for further engineering.
Computational modeling
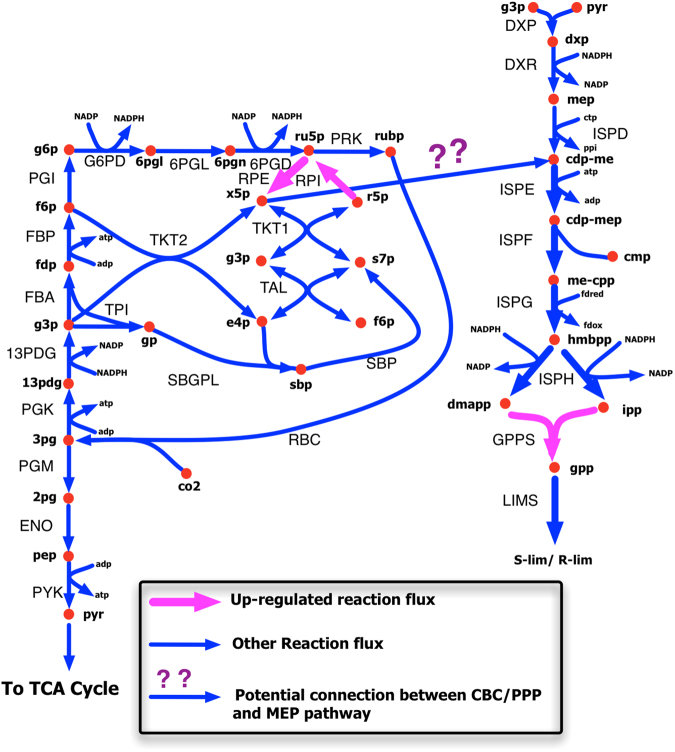
The iSyn731 metabolic model of Synechocystis 680318 was used to perform the computational strain designs using the OptForce algorithm17 for overproduction of limonene. Based on the current understanding as reported in literature16,27, a connection between Calvin Benson Cycle (CBC)/PP pathway and MEP pathway (Fig. 3) was included in the iSyn731 model. By superimposing the photoautotrophic flux measurements19 of 31 reactions of central carbon metabolism including the CBC and PP pathways of Synechocystis 6803 onto the iSyn731 model, the phenotypic space of the base strain was defined. All simulations were performed for a basis of 100 millimoles of CO2 plus H2CO3 uptake and unlimited photon supply19. The uptake fluxes for the remaining metabolites present in the BG11 medium was set to -1,000 and the non-growth associated ATP maintenance was set at 8.39 mmole/gDW-h. In addition, the biomass flux was fixed at the optimal value subject to the experimental flux measurements19. The upper bound of the fluxes of the remaining reactions was set to 1,000 mmole/gDW-h, whereas the lower bound was set to zero and -1,000 mmole/gDW-h for irreversible and reversible reactions, respectively.
Figure 3.
Metabolic interventions predicted by the OptForce algorithm. Up-regulation of rpe, rpi, and gpps (showed with pink arrows) leads to the improved production yield of limonene.
Similarly, the limonene overproducing phenotype was obtained by maximizing and minimizing each flux of the metabolic model iteratively subject to the network stoichiometry, uptake and medium conditions, regulatory constraints, and overproduction target. In this work, a minimum production yield of 85% of the theoretical maximum of limonene (i.e., 15.3 mmole/gDW-h) was set as the overproduction target, while the biomass flux was constrained to be at least 10% of its theoretical maximum (i.e., 0.021 h−1) with the basis of 100 millimoles of carbon fixed (i.e., CO2 plus H2CO3). The remaining parameter values including medium conditions and regulatory constraints were the same as those in the wild-type. By contrasting the maximal range of flux variability between the wild-type strain and the over-producing strain to meet the pre-specified yield of limonene, OptForce was used to identify the minimal set of genetic interventions (i.e., deletions and up-/down-regulations). In order to first explore non-intuitive interventions, reactions from the MEP and isoprenoid biosynthesis pathways were not considered as the candidates for any form of intervention. Integer cuts were used to identify alternative optimal solutions (i.e., alternative genetic intervention choices) to achieve the minimum production yield of limonene as specified earlier. The termination criterion for the OptForce procedure was set as either meeting a production yield of at least 85% of the theoretical maximum for limonene or exceeding the maximum allowable number of reaction interventions (i.e., three). Note that the OptForce procedure works at the reaction level, which is why the set of genetic manipulations can subsequently be identified by using gene-protein-reaction (GPR) associations from the iSyn731 model. Thus, the OptForce procedure identified up-regulation of rpi and rpe as the best possible solution, which can lead up to limonene yield at 89% of its theoretical maximum (i.e., 16.02 mmole/gDW-h). By up-regulating these two genes, OptForce suggested to force more flux from the CBC/PP pathway toward MEP pathway that can ultimately increase the production yield of limonene (Fig. 3). Once the set of non-intuitive interventions was obtained, as a next step, it was logical to explore if their combination with any of the intuitive one(s) from the MEP and isoprenoid biosynthesis pathways could further improve the limonene production yield that was otherwise not possible to achieve individually (i.e., by the non-intuitive candidates or by the intuitive ones). With a target of a minimum production yield of 90% of the theoretical maximum of limonene, the OptForce procedure identified the up-regulation of gpps, rpe, and rpi that could lead the limonene production yield to 16.56 mmole/gDW.h (i.e., 92% of its theoretical maximum). Thus, the proposed interventions combined the amplification (i.e., push) of flux from the CBB/PP pathway to MEP pathway with a similar increase (i.e., pull) in the flux of the limonene synthesis. As reported in the literature28, this kind of push-and-pull strategy can achieve the desired level of production yield with minimal effects caused by feedback inhibition.
Genetic interventions of the PP pathway to improve limonene production
Based on the prediction of the OptForce procedure, up-regulation of rpi and rpe genes in the PP pathway increases the flux toward limonene production. To test this hypothesis, the rpi and rpe genes driven by the Synechocystis 6803 native rbcL promoter were expressed on a replicating plasmid in the limonene-producing strain, resulting in 1.3-fold increase in limonene yield (3.7 mg/L) after 7 days of cultivation (Fig. 4). Furthermore, we introduced a gene encoding a specific GPP synthase (GPPS) to optimize the limonene biosynthetic pathway. In Synechocystis 6803, formation of GPP is catalyzed by a farnesyl diphosphate (FPP) synthase, CrtE. It performs consecutive condensation of IPP with DMAPP, and only synthesizes GPP as an intermediate29. Although the PP pathway was engineered to stimulate the limonene yield, it is possible that the native isoprenoids pathway in Synechocystis 6803 provides insufficient GPP for limonene production since the flux is diverted toward FPP formation for pigment synthesis. In addition, OptForce also predicted an increase (i.e., from 89% to 92% of maximum theoretical limonene yield) when up-regulation of rpe and rpi was combined with the up-regulation of gpps. It was reported that the GPPS 2 from Abies grandis specifically produces GPP30. Expressing this specific gpps with lims, the limonene yield increased 1.4-fold (4.1 mg/L) (Fig. 4). Finally, coexpression of rpi, rpe, gpps and lims resulted in a remarkable (2.3-fold) enhancement in productivity (6.7 mg/L) (Fig. 4).
Figure 4.
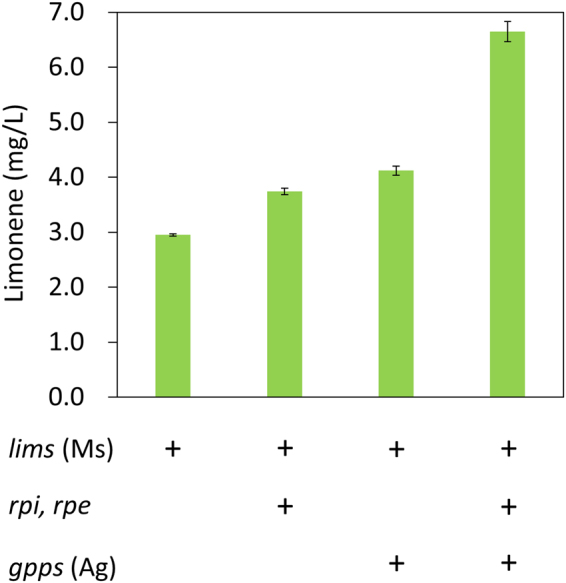
Increased limonene production by genetic modifications. Two genes in the PP pathway (rpi and rpe) were overexpressed in Synechocystis 6803 to divert carbon flux toward limonene production. The Abies grandis GPPS 2 that specifically produce GPP was expressed to ensure sufficient GPP for limonene production. Ms, Mentha spicata; Ag, Abies grandis. Results were mean SD of three biological replicates.
Pigment content in engineeredSynechocystis6803
Carotenoids and the phytol tail of chlorophyll, photosynthetic pigments, are derived from geranylgeranyl diphosphate (GGPP), a C20-intermediate for isoprenoid synthesis. Hence, production of limonene is expected to divert carbon flux away from pigment synthesis. To investigate the effect of limonene production on pigment content in engineered Synechocystis 6803, we extracted and quantified the chlorophyll and carotenoid contents. The chlorophyll content decreased over 30% in the gpps expression strains, whereas carotenoid levels were similar among the limonene-producing strains (Fig. 5). These results indicate that the specific GPPS diverts the carbon flux away from pigment synthesis.
Figure 5.
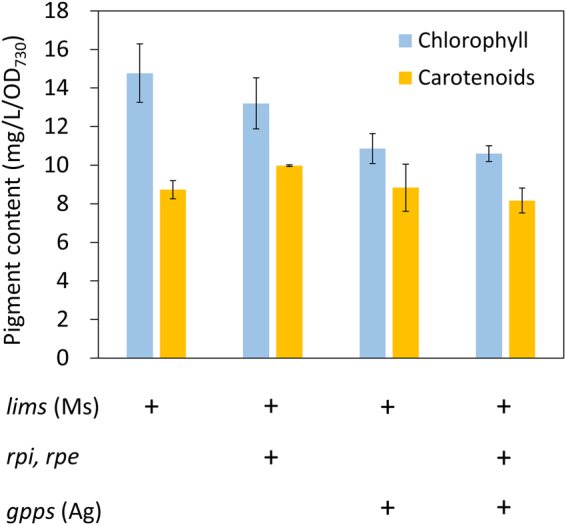
Pigment content of engineered Synechocystis 6803 strains. Chlorophyll and carotenoid contents of limonene-producing strains. Ms, Mentha spicata; Ag, Abies grandis. Results were mean ± SD of three biological replicates.
Discussion
In this study, we combined metabolic engineering with model-driven strain design strategies to engineer Synechocystis 6803 for enhanced limonene production. To generate limonene-producing Synechocystis 6803, we first constructed a suicide plasmid26 to engineer the lims into the neutral site on the pCC5.2 endogenous plasmid via double homologous recombination. This is the first time that the endogenous plasmid of Synechocystis 6803 has been used for enhanced production for the purpose of metabolic engineering. Expression of a gene on the pCC5.2 plasmid leads to higher expression level than that on the chromosome as well as the RSF1010 self-replicating plasmid26. Furthermore, during the stationary phase of cell growth, the copy numbers of the endogenous plasmids (pCA2.4, pCB2.4, pCC5.2) in Synechocystis 6803 are 3 to 7 per chromosome31. Using the endogenous plasmid to express the lims gene driven by the constitutive promoter trc1O allows high expression level at the stationary phase, decoupling growth and production, and thus leading to higher levels of production of limonene.
The higher yield with limonene synthase from M. spicata than that from C. limon may be due to the difference in enzyme kinetics of LIMS. Unfortunately, the kinetic parameters (both K m and k cat) are only available for the enzyme from M. spicata 25. In addition, it may be attributed to different protein expression levels. Although the same promoter was used to control the lims from two plant species, protein expression may vary because of different mRNA sequences and codon usage. To date, the highest reported limonene productivity in cyanobacteria was achieved by engineered Synechococcus sp. PCC 700223. In their study, only a lims from M. spicata was overexpressed, and the yield was over 4 mg/L in 4 days23. Our results also suggested that the LIMS from M. spicata performed better in limonene production (Fig. 2B). The doubling time of Synechococcus 7002 is shorter than Synechocystis 680332. Thus, the higher limonene yield from Synechococcus 7002 may be due to its faster growth rate. A recent study engineered Synechococcus elongatus PCC 7942 to produce limonene, achieving a 100-fold improvement in productivity33. However, it should be noted that such significant increase is due to the low productivity of the original strain, which produced merely 8.5 μg/L/OD/d of limonene. The best producing strain in this study, with a lims (M. spicata) controlled by the pea plant psbA promoter, produced 2.5 mg/L limonene in 5 days33.
Previously, researchers have engineered Synechocystis 6803 for limonene production by overexpressing genes involved in the bottleneck steps of the MEP pathway14. It is known that enzymes 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and isopentenyl diphosphate isomerase (IDI) catalyze the rate-limiting reactions in the MEP pathway34,35. With the introduction of an additional copy of endogenous dxs, idi, and gpps genes, the engineered Synechocystis 6803 produced 1.4-fold higher yield than that of the parent strain14. However, such improvement was less effective than the strategy used in the current study. As mentioned in the Results section, the endogenous gpps gene may not be suitable for enhancing the production of limonene. In addition, the MEP pathway is highly regulated at genetic and metabolic levels36. Expressing endogenous genes in the MEP pathway may be subject to native regulations, presenting a less effective engineering approach.
Instead of manipulating the MEP pathway, we took a systematic model-driven metabolic engineering approach for finding genetic interventions in order to increase the limonene production yield. As explained in the Materials and Methods section, the OptForce procedure finds the minimal interventions to reach a desired production target. To this end, we employed OptForce on our previously developed genome-scale model iSyn731 in order to ‘push’ more flux to MEP pathway and also to create better ‘pull’ for limonene synthesis (Fig. 3). From this in silico analysis, by up-regulating rpe and rpi, the metabolite pool of X5 P was found to be increased that, eventually, led to increased flux through the connection between the CBC/PP pathway and the MEP pathway. In addition, up-regulation of gpps created an improved ‘pull’ for limonene synthesis. Thus, the combination of this push-and-pull mechanism was proposed to be the best strategy to improve limonene yield by circumventing additional regulations (e.g. feedback inhibition). Interestingly, the same rationale could be applied to engineer cyanobacteria to produce other isoprenoid compounds.
Expression of the specific gpps modestly increased the limonene titer (Fig. 4), whereas the cellular chlorophyll content was greatly influenced (Fig. 5). Synthesis of limonene and the phytol tail of chlorophyll requires the same precursors, IPP and DMAPP. Table 1 compares the changes in chlorophyll and limonene contents between the strain with lims only and the two gpps-expressing strains. Compared to the lims-expressing strain, additional expression of gpps resulted in similar level of decrease in chlorophyll content in both strains. However, expression of rpi and rpe genes further led to 1.5-fold higher limonene productivity in the gpps-expressing strains (122 vs. 81 μM/OD730 isoprene unit). This result suggests that up-regulation of rpi and rpe enhances the carbon flux toward limonene synthesis.
Table 1.
Comparison of chlorophyll and limonene contents.
| Strains | lims | lims, gpps | lims, gpps, rpi, rpe |
|---|---|---|---|
| Chlorophyll* | 66 | 49 | 47 |
| Limonene* | 53 | 81 | 122 |
*Presented as μM/OD730 isoprenes.
Our results showed that overexpressing the genes in the PP pathway led to improved limonene production, suggesting an unidentified connection between the PP pathway and isoprenoids biosynthesis (Fig. 4). Our observation is consistent with previous in vitro study using Synechocystis 6803 cell lysate16. However, the connection between the PP pathway and isoprenoids biosynthesis remains to be elucidated. It was first shown that in vitro isoprenoid production increased significantly by providing substrates in the PP pathway16, while a recent study showed that increased production of isoprenoids by PP pathway substrates does not occur through the MEP pathway37. By removing the terminal enzyme of the MEP pathway in Synechocystis 6803 cell lysate, isoprenoid synthesis still increased by substrates in the PP pathway37. Taken together, it is still unclear how the PP pathway and isoprenoid production are connected in Synechocystis 6803. While our results made a strong argument for this connection, further investigation needs to be conducted to explore the details in terms of chemical conversions and genes/enzymes associated. From the modeling context, these details sometimes do not make much of a difference if they only involve aggregating linear reaction steps.
Conclusions
In this study, we engineered the model cyanobacterium Synechocystis 6803 to produce the isoprenoid, limonene. We applied computational strain design by using the OptForce procedure to identify minimal genetic interventions for improving limonene yield. Based on the prediction, the rpi and rpe genes in the PP pathway were overexpressed, and a specific gpps was introduced to optimize the limonene biosynthetic pathway. The final engineered strain produced 6.7 mg/L of limonene, which is a 2.3-fold improvement in productivity. The approach that we demonstrated can be applied to engineer cyanobacteria to produce other valuable isoprenoids.
Materials and Methods
Chemicals and reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. Phusion DNA polymerase were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Culture medium and condition
All strains were maintained in liquid BG-11 medium or on solid BG-11 plates with appropriate antibiotics at 30 °C continuous white light (50 μmoles photons m−2s−1).
DNA manipulations
Coding sequences of lims from Mentha spicata and Citrus limon were codon optimized for Synechocystis 6803 and synthesized by IDT (San Jose, CA, USA). The genes were cloned into a suicide plasmid, allowing gene insertion into the neutral site (NSP1) on the endogenous plasmid pCC5.2 in Synechocystis 680326. The constructed plasmids were directly used as templates for PCR to amplify a fragment which contains the lims and a kanamycin resistance cassette flanking by upstream and downstream homologous sequences of the NSP1 (Fig. 2A). The PCR product was then purified by DNA electrophoresis, and the linear DNA was transformed into Synechocystis 6803. The rpi, rpe, and gpps genes were cloned into a broad-host-range plasmid RSF1010 harboring a spectinomycin resistance cassette38. All the cloning works were done by Gibson isothermal DNA assembly method39.
Strains construction and transformation
The lims expression cassette was transformed into Synechocystis 6803 through homologous recombination. Cells at mid-log phase (OD730 of 0.4 to 0.6) were incubated with 600 ng of linear DNA overnight at 30 °C in the dark. Cells were then grown on BG-11 plates supplemented with 10 μg/mL of kanamycin for selection of transformants. Colonies were patched on BG-11 plates with 20 μg/mL of kanamycin for segregation. PCR was used to verify strain segregation. For the construction of rpi, rpe, and gpps expressing strains, self-replicating plasmids (600 ng per transformation) were transformed into the strain expressing lims. Transformants were selected by BG-11 plates with 2 μg/mL of spectinomycin and 5 μg/mL of kanamycin.
Limonene production by engineered cyanobacteria
Strains were inoculated in BG-11 medium with kanamycin (10 μg/mL) and spectinomycin (4 μg/mL) to mid-log phase at 30 °C with continuous white light (50 μmoles photons m−2s−1). Cells were collected by centrifugation at 7,000 x g, and washed by BG-11 medium to remove antibiotics. To test limonene production, the initial OD730 was adjusted to 0.34 (~0.5 g/L of biomass), and 50 mL of cell cultures were grown in 250-mL flasks at 30 °C with continuous white light (130 μmoles photons m−2s−1). A 10% (v/v) dodecane overlay was covered on top of cultures to trap evaporated limonene.
Quantification of limonene
Limonene samples were prepared by diluting 10 μL of dodecane overlay in 990 μL of ethyl acetate, and analyzed using a gas chromatography instrument with a flame ionization detector (Hewlett-Packard model 7890 A, Agilent Technologies, CA, USA) equipped with a 30 m DB5-MS column (J&W Scientific). The oven temperature program initiated at 60 °C, and increased at 12 °C/min to 300 °C. Limonene was quantified using a (R)-limonene standard.
Identification of engineering interventions via OptForce
We applied the OptForce algorithm17 on the genome-scale Synechocystis 6803 model iSyn73118. In order to characterize the wild-type phenotype, we utilized 13C MFA flux estimations19 under photosynthetic condition. Below is the step-by-step procedure that we followed:
Step 1
Identify the maximum biomass and limonene yields under photosynthetic condition.
Maximize v biomass or v ls
Subject to
| 1 |
| 2 |
| 3 |
Step 2
Characterize the wild-type phenotype
Maximize/Minimize reactions without experimental flux measurements
Subject to
| 1 |
| 2 |
| 3 |
| 4 |
Step 3
Characterize the limonene over-producing phenotype
Maximize/Minimizevj ∀ j ∈ 1, ……,m
Subject to
| 1 |
| 2 |
| 3 |
| 5 |
| 6 |
Step 4
Identify the MUST sets
In this step, fluxes ranging from step 2 and step 3 were compared to identify three different sets: reactions to be up-regulated (MUST U), down-regulated (MUST L), and deleted (MUST X).
Step 5
Identify the minimal engineering interventions
Maximize v j
(over MUST sets)
Subject to
Minimize v j
(over MUST sets)
Subject to
| 1 |
| 2 |
| 3 |
| 5 |
| 7 |
| 8 |
Here, S ij is the stoichiometric coefficient of metabolite i in reaction j and v j is the flux value of reaction j. Parameters v j,min and v j,max denote the minimum and maximum allowable fluxes for reaction j, respectively. V biomass and v ls represent biomass and limonene synthesis reactions under photosynthetic conditions, whereas v max biomass and v max ls represent the maximum theoretical yields of biomass and limonene under photosynthetic conditions. The minimal levels of biomass and the minimal target yield of limonene were set to be 10% of maximum biomass and 85% or 90% of maximum limonene yield, respectively. Finally, k represents the maximum number of interventions allowed.
Pigment content analysis
Cell cultures (1 mL) were collected by centrifugation at 16,000 x g for 7 min, and the supernatants were removed. To extract pigments in Synechocystis, pre-cooled methanol (1 mL) was added to the pellets, and mixed thoroughly by pipetting and vortexing. Samples were incubated at 4 °C for 20 mins, and centrifuged at 16,000 x g for 7 min. The supernatants were removed for a spectroph-otometer analysis to quantify the concentrations of carotenoids and chlorophyll. The following equations were used to calculate the pigment content: 40; 41.
Acknowledgements
Funding to support this work was provided by the Office of Science (BER), U. S. Department of Energy, to HBP. PCL was supported by a fellowship from the McDonnell International Scholars Academy at Washington University. We thank all members of the Pakrasi research group for collegial discussions.
Author Contributions
P.C.L. performed experiments; R.S. performed computational studies; All authors designed research, wrote and edited the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oliver JW, Atsumi S. Metabolic design for cyanobacterial chemical synthesis. Photosynthesis research. 2014;120:249–261. doi: 10.1007/s11120-014-9997-4. [DOI] [PubMed] [Google Scholar]
- 2.Angermayr SA, Gorchs Rovira A, Hellingwerf KJ. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends in biotechnology. 2015;33:352–361. doi: 10.1016/j.tibtech.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Berla BM, et al. Synthetic biology of cyanobacteria: unique challenges and opportunities. Frontiers in microbiology. 2013;4:246. doi: 10.3389/fmicb.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitmaier, E. Terpenes: flavors, fragrances, pharmaca, pheromones. (John Wiley & Sons, 2006).
- 5.George KW, Alonso-Gutierrez J, Keasling JD, Lee TS. Isoprenoid drugs, biofuels, and chemicals–artemisinin, farnesene, and beyond. Advances in biochemical engineering/biotechnology. 2015;148:355–389. doi: 10.1007/10_2014_288. [DOI] [PubMed] [Google Scholar]
- 6.Wang, G., Tang, W. & Bidigare, R. R. Terpenoids as therapeutic drugs and pharmaceutical agents. In Natural products pp. 197–227, Springer (2005).
- 7.Westfall PJ, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E111–118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 9.Tracy NI, Chen D, Crunkleton DW, Price GL. Hydrogenated monoterpenes as diesel fuel additives. Fuel. 2009;88:2238–2240. doi: 10.1016/j.fuel.2009.02.002. [DOI] [Google Scholar]
- 10.Chuck CJ, Donnelly J. The compatibility of potential bioderived fuels with Jet A-1 aviation kerosene. Applied Energy. 2014;118:83–91. doi: 10.1016/j.apenergy.2013.12.019. [DOI] [Google Scholar]
- 11.Gao Z, Zhao H, Li Z, Tan X, Lu X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy & Environmental Science. 2012;5:9857–9865. doi: 10.1039/C2EE22675H. [DOI] [Google Scholar]
- 12.Lan EI, Liao JC. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6018–6023. doi: 10.1073/pnas.1200074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Sheng J, Curtiss R., 3rd Fatty acid production in genetically modified cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6899–6904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyota H, Okuda Y, Ito M, Hirai MY, Ikeuchi M. Engineering of cyanobacteria for the photosynthetic production of limonene from CO2. Journal of biotechnology. 2014;185:1–7. doi: 10.1016/j.jbiotec.2014.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Halfmann C, Gu L, Zhou R. Engineering cyanobacteria for the production of a cyclic hydrocarbon fuel from CO2 and H2O. Green Chemistry. 2014;16:3175–3185. doi: 10.1039/C3GC42591F. [DOI] [Google Scholar]
- 16.Ershov YV, Gantt RR, Cunningham Jr FX, Jr., Gantt E. Isoprenoid biosynthesis in Synechocystis sp. strain PCC6803 is stimulated by compounds of the pentose phosphate cycle but not by pyruvate or deoxyxylulose-5-phosphate. Journal of bacteriology. 2002;184:5045–5051. doi: 10.1128/JB.184.18.5045-5051.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranganathan S, Suthers PF, Maranas CD. OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput Biol. 2010;6:e1000744. doi: 10.1371/journal.pcbi.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha R, et al. Reconstruction and comparison of the metabolic potential of cyanobacteria Cyanothece sp. ATCC 51142 and Synechocystis sp. PCC 6803. PloS one. 2012;7:e48285. doi: 10.1371/journal.pone.0048285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young JD, Shastri AA, Stephanopoulos G, Morgan JA. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metabolic engineering. 2011;13:656–665. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohlmann J, Steele CL, Croteau R. Monoterpene synthases from grand fir (Abies grandis). cDNA isolation, characterization, and functional expression of myrcene synthase, (−)-(4S)-limonene synthase, and (-)-(1S,5S)-pinene synthase. The Journal of biological chemistry. 1997;272:21784–21792. doi: 10.1074/jbc.272.35.21784. [DOI] [PubMed] [Google Scholar]
- 21.Lucker J, et al. Monoterpene biosynthesis in lemon (Citrus limon). cDNA isolation and functional analysis of four monoterpene synthases. European journal of biochemistry / FEBS. 2002;269:3160–3171. doi: 10.1046/j.1432-1033.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- 22.Colby SM, Alonso WR, Katahira EJ, McGarvey DJ, Croteau R. 4S-limonene synthase from the oil glands of spearmint (Mentha spicata). cDNA isolation, characterization, and bacterial expression of the catalytically active monoterpene cyclase. The Journal of biological chemistry. 1993;268:23016–23024. [PubMed] [Google Scholar]
- 23.Davies FK, Work VH, Beliaev AS, Posewitz MC. Engineering Limonene and Bisabolene Production in Wild Type and a Glycogen-Deficient Mutant of Synechococcus sp. PCC 7002. Frontiers in bioengineering and biotechnology. 2014;2:21. doi: 10.3389/fbioe.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonso-Gutierrez J, et al. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic engineering. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Williams DC, McGarvey DJ, Katahira EJ, Croteau R. Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry. 1998;37:12213–12220. doi: 10.1021/bi980854k. [DOI] [PubMed] [Google Scholar]
- 26.Ng AH, Berla BM, Pakrasi HB. Fine-Tuning of Photoautotrophic Protein Production by Combining Promoters and Neutral Sites in the Cyanobacterium Synechocystis sp. Strain PCC 6803. Applied and environmental microbiology. 2015;81:6857–6863. doi: 10.1128/AEM.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poliquin, K. et al. Inactivation of sll1556 in Synechocystis strain PCC 6803 impairs isoprenoid biosynthesis from pentose phosphate cycle substrates in vitro. J Bacteriol186, 4685-4693 (2004). [DOI] [PMC free article] [PubMed]
- 28.Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Hosfield DJ, et al. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. The Journal of biological chemistry. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 30.Burke C, Croteau R. Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Archives of biochemistry and biophysics. 2002;405:130–136. doi: 10.1016/S0003-9861(02)00335-1. [DOI] [PubMed] [Google Scholar]
- 31.Berla BM, Pakrasi HB. Upregulation of plasmid genes during stationary phase in Synechocystis sp. strain PCC 6803, a cyanobacterium. Applied and environmental microbiology. 2012;78:5448–5451. doi: 10.1128/AEM.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO(2) Scientific reports. 2015;5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14225–14230. doi: 10.1073/pnas.1613340113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estevez JM, Cantero A, Reindl A, Reichler S, Leon P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. The Journal of biological chemistry. 2001;276:22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- 35.Wang CW, Oh MK, Liao JC. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnology and bioengineering. 1999;62:235–241. doi: 10.1002/(SICI)1097-0290(19990120)62:2<235::AID-BIT14>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee A, Sharkey TD. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Natural product reports. 2014;31:1043–1055. doi: 10.1039/C3NP70124G. [DOI] [PubMed] [Google Scholar]
- 37.Poliquin, K., Cunningham, F. X., Gantt, R. R. & Gantt, E. In Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches (eds Thomas J. Bach & Michel Rohmer) 51–63 (Springer New York, 2013).
- 38.Taton A, et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 2014;42:e136. doi: 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 40.Porra R, Thompson W, Kriedemann P. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1989;975:384–394. doi: 10.1016/S0005-2728(89)80347-0. [DOI] [Google Scholar]
- 41.Wellburn AR. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of plant physiology. 1994;144:307–313. doi: 10.1016/S0176-1617(11)81192-2. [DOI] [Google Scholar]