Abstract
Liver injury resulting from exposure to drugs and chemicals is a major health problem. Autophagy is an important factor in a wide range of diseases, such as cancer, liver disease, muscular disorder, neurodegeneration, pathogen infection, and aging, and emerging evidence indicates that autophagy makes a substantial contribution to the pathogenesis of drug- and chemical-induced liver toxicity. In this review, we summarize current knowledge on autophagy triggered by toxicants/toxins, the protective role of autophagy in liver toxicity, and the underlying molecular mechanisms. We also highlight experimental approaches for studying autophagy.
Keywords: autophagy, drug-induced liver toxicity
1. Introduction to drug-induced liver toxicity
Acute liver failure is defined as a rapid deterioration of liver function in a patient without obvious preexisting chronic liver disease. Some acute liver failure can be reversed when the insults are withdrawn; however, a liver transplant is needed for certain severe situations.
There are geographic variations in the etiology of acute liver failure. Although hepatitis virus diseases, particularly hepatitis B, are prevalent in the Mediterranean region, drug-induced liver injury (DILI) is the most frequent cause of acute liver failure in the United States and Europe [1], accounting for over 50% of reported cases of acute liver failure [2]. In China, DILI has replaced hepatitis B as the leading cause of acute liver failure [3].
It is estimated that, worldwide, the annual incidence rate of DILI ranges from 13.9 to 24.0 per 100,000 inhabitants [4]. A variety of drugs has been reported to cause DILI, with different degrees of toxicity; these include antibacterial, antiviral, antifungal, anti-inflammatory, antigout, antidepressant, antiulcer, antidiabetic, antiarrhythmic, antineoplastic, vasoprotective, analgesic, and laxative drugs [5]. There is a difference in the prevalence of DILI between countries; in the United States, intentional and unintentional acetaminophen (APAP) overdose is the main reason for liver toxicity, far exceeding the other causes [6], whereas traditional herbal medication predominates as the cause of DILI in Asian countries such as China and Korea [3,4]. DILI is also a leading cause of drug failure in clinical trials and drug withdrawals from the market and it becomes a major burden to pharmaceutical companies and also poses a great challenge to regulatory authorities [2,7,8].
The liver has multiple functions, so injury responses and mechanisms are diverse, and they also depend on the nature of the drug or chemical exposure. If the toxicity is idiosyncratic (an infrequent reaction occurring in from 1 in 1000 patients to 1 in 100,000 patients), other factors such as genetic variations of patients, preexisting diseases, and environmental factors also have an impact on the complexity of the injury. Increasing knowledge of molecular events is leading to a better understanding of the pathogenesis of DILI, and emerging new technologies, such as next-generation genome sequencing, help in identifying genetic risk factors.
Numerous mechanisms of DILI have been identified, among which metabolism-mediated toxicity is well-recognized. Although some parent compounds directly damage specific organelles such as mitochondria, reactive electrophilic metabolites and free radicals generated via hepatic drug metabolizing enzymes often produce toxicity. The toxic metabolites covalently bind to proteins, lipids, and DNA and cause adverse consequences such as oxidative stress, lipid peroxidation, DNA damage, glutathione depletion, and eventually cell death. Toxic metabolites also provoke immune responses and sensitize the liver to toxicity [9]. Recently, mitochondrial dysfunction has emerged as one of the important underlying mechanisms in DILI, because several lines of evidence have revealed that mitochondria are the target of some drugs [10–12]. Different modes of cell death have been described in DILI studies, with apoptosis and necrosis being the two general modes [12]. Apoptosis, also called “programmed cell death”, involves various cellular events and signaling pathways leading to a series of morphological changes, such as cell shrinkage, chromatin condensation, and nuclear fragmentation. Apoptotic cells are eventfully broken down into small fragments, which will be cleared by neighboring cells or professional phagocytes (phagocytosis). If the phagocytosis is incomplete, cell death (secondary necrosis) follows. Necrosis is characterized as cell swelling, loss of integrity of nuclear and cell membranes, and leakage of cellular contents. The type of cell death is determined by the population and the type of cells affected and the intensity and duration of the exposure. The mechanisms of apoptosis and necrosis in drug hepatotoxicity are summarized by Gunawan and Kaplowitz [9] and Kaplowitz [13]. In recent years, besides apoptosis and necrosis, alternative cell death modes have been receiving attention. One of these is autophagy, considered to be an important cell death mechanism. It is classified as type 2 cell death mode, whereas apoptosis is classified as type 1 and necrosis is type 3 [14,15]. Apoptosis and autophagy are regulated and controlled processes, whereas necrosis is considered a passive, unregulated cell death. Most recently, a regulated necrosis, also called “programmed necrosis” or “necroptosis”, was described. Accumulated evidence indicates that necroptosis participates in the pathogenesis of various diseases [16]. Although a deep understanding of necroptosis in liver toxicity has not yet been achieved, studies have shown that necroptosis is involved in toxicant- and toxin-induced liver toxicity [17–19].
It should be mentioned that the regulation of cell death is a complicated process, with the involvement of multiple signaling pathways, and the balance between “repair/adaptation” and “cell death” decides the fate of cells. Prior to deterioration, organisms initiate repair and adaptive responses to counteract the progress of toxicity. Prolonged stress or toxicity exceeding cellular adaptive capacity then causes cell death. The regenerative capacity of the liver is much greater than that of other organs, so adaptation is an important mechanism for protecting against cellular damage. For instance, activation of drug metabolizing enzymes, especially the Phase II conjugation enzymes, detoxifies the effects of many drugs. In response to oxidative stress, antioxidant genes can be activated through the Keap1-nuclear factor erythroid-related factor 2-antioxidant-responsive element (Keap1-Nrf2-ARE) signaling pathway, which plays a critical role in protecting cells from endogenous and exogenous stresses [20,21]. Under normal physiological conditions, the transcription factor Nrf2 localizes in the cytoplasm and interacts with Keap1. Upon oxidative stress, Nrf2 is released from Keap1 and translocates to the nucleus; it subsequently activates its various downstream target genes by interacting with ARE, which is a regulatory element in the promoter region of the target. The target genes include: NAD(P)H (nicotinamide adenine dinucleotide phosphate-oxidase) dehydrogenase, quinone 1 (NQO1); carboxylesterase (CES)1; CES2; and glutathione S-transferase (GST). These target genes have a wide spectrum of functions to enhance cell survival, such as inactivating oxidants, increasing the levels of glutathione, and enhancing toxin export via transporters [20].
A different type of adaptive response to stress is endoplasmic reticulum (ER) stress. ER function can be disturbed by stimuli leading to the accumulation of unfolded proteins in the ER. To reestablish ER function, an adaptive mechanism termed the unfolded protein response (UPR) is activated to upregulate the expression of genes involved in protein folding and quality control. Three UPR branches, protein kinase RNA-like ER kinase (PERK), inositol-requiring enzyme 1 α (IRE1α), and activation of transcription factor 6 (ATF6), are activated when cells are under ER stress [22–24]. The role of ER stress in liver disease and DILI has been summarized in review articles [25–28].
Autophagy is not only involved in cell death, but also plays a protective role, functioning in either a prosurvival or a prodeath capacity (as will be discussed in the following sections). The role of autophagy has been described for various diseases and its importance in DILI has started to be recognized. In this article, we summarize the literature on autophagy in drug- or chemical-induced liver toxicity and highlight research approaches in autophagy studies.
2. Introduction to autophagy
Autophagy is a cellular process in eukaryotes involved in the degradation of cytoplasmic components by lysosomes. Autophagic cell death has been observed in multiple tissues during the pathogenesis of diseases [29,30]. The term “autophagy” was invented by Christian de Duve, who earned the 1974 Nobel Prize in “Physiology and Medicine” for his discovery of lysosomes and peroxisomes. Keith Porter and Thomas Ashford first observed autophagy in 1962, by electron microscopy of perfused rat liver when glucagon was added to the perfusing fluid [31]. Following this observation, the de Duve laboratory performed pioneering work on autophagy, clearly demonstrating the fusion between autophagosomes and lysosomes at the cellular level [32]. Thirty years later, the molecular mechanisms of autophagy have just started to be elucidated owing to the use of yeast genetics. To date, highly conserved genes named autophagy-related genes (Atg 1–10, Atg 12–14, Atg 16, and Atg 18) have been identified as required for autophagy [33].
Autophagy has been classified into three types according to the route of delivery of the cytoplasmic material to the lysosome: (1) chaperone-mediated autophagy; (2) micro-autophagy; and (3) macroautophagy. Chaperone-mediated autophagy involves the direct lysosomal import of unfolded proteins across the lysosomal membrane. Microautophagy involves the engulfing of cytoplasmic material by the lysosome by membrane rearrangement. Macroautophagy is considered to be the central process during nutrient starvation and physiological stress stimuli, and often is referred to simply as “autophagy” (as will be done hereafter in this review) [34]. Autophagy begins with the sequestration of cytoplasm into double-membrane cytosolic vesicles called autophagosomes. Subsequently, autophagosomes fuse with lysosomes to form autolysosomes, followed by degradation by acidic lysosomal hydrolases [35]. Autophagy is a dynamically regulated process composed of several sequential steps, including induction, autophagosome formation, autolysosome fusion, and degradation.
Studies of autophagy have gained great popularity because of its essential role in physiological and pathological processes [36]. The role of autophagy in human health and disease is complex, because it plays a role in both cell survival and cell death. The prosurvival function of autophagy is an adaptive response to most forms of cellular stress, including nutrient and growth factor deprivation, ER stress, development, microbial infection, and protein aggregate accumulation [37–39]. A well-demonstrated prosurvival function of autophagy is the starvation response during starvation, which happens in organisms from yeast to humans, in which cells degrade “self” amino acids by autophagy to maintain energy homeostasis and stay alive [40,41].
Several regulatory molecular components have been reported to be involved in the modulation of autophagy activity. The best characterized regulator of autophagy is the mammalian target of rapamycin (mTOR) which is involved in multiple cellular processes; the inactivation of mTOR is essential for the induction of autophagy [33]. Other signaling pathways, including c-Jun N-terminal kinase (JNK), calcium/ calmodulin-dependent protein kinase kinase (CaMKK), liver kinase B (LKB), protein kinase B (AKT), adenosine monophosphate-activated protein kinase (AMPK), and p-53 mediated signaling, are also involved in autophagy activation (reviewed elsewhere [15]).
As a result of progress in our knowledge of autophagy processes and the identification of signaling pathways that regulate autophagy, methods for detection and manipulation of autophagy have greatly increased. Methods for detecting autophagy are summarized in the following section.
3. Methods for monitoring autophagy
Within the past decade, there has been an increased interest in studying autophagy in various research areas, and numerous methods have thus been developed. In general, there is no single marker or gold standard for defining pathogenesis and determining mechanisms; this is also the case for autophagy, especially considering the complexity of its physiology and pathogenesis [42–44].
Because autophagy is a highly dynamic and multistep process, a number of methods have been applied to follow the progress of autophagy, detecting either the induction (autophagosome formation) or the dynamic process (autophagic flux) of autophagosome synthesis. Mizushima et al [45] summarized autophagy methods and an autophagy consortium published guidelines for standardizing autophagy research in 2008 and updated it in 2012 [42]. Here, we briefly list the most commonly used in vitro experimental assays for monitoring autophagy.
3.1. Detection of autophagosome formation
3.1.1. LC3 conversion
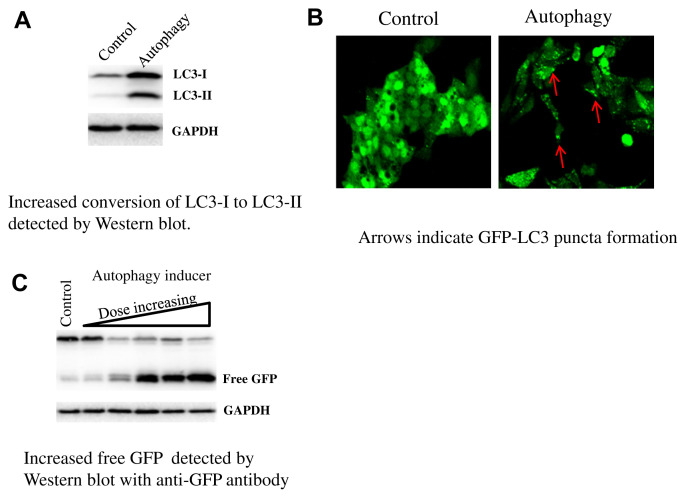
LC3 (Microtubule-associated protein 1A/1B light chain 3), the most commonly monitored autophagy marker, has four isoforms, of which LC3B is the one most often used. The amount of LC3-II correlates well with the number of autophagosomes [46]. Standard Western blots can be used to detect an increase in the conversion of endogenous LC3-I to LC3-II when there is autophagosome induction, as shown in Fig. 1A. Although LC3-II accumulation is a marker for autophagosome induction, the accumulation of LC3-II can also be seen if the dynamic progress of autophagy is inhibited, i.e., the degradation of LC3-II is blocked. For instance, inhibition of autophagy caused by interrupting the autophagosome-lysosome fusion step (with bafilomycin A1) or by inhibiting lysosome-mediated proteolysis (with chloroquine) leads to an increase in LC3-II accumulation; in these cases, the increase in expression of LC3-II does not prove increased autophagy.
Fig. 1.
Methods for monitoring autophagy.
3.1.2. GFP-LC3 puncta formation assay
A fusion protein of LC3 with a fluorescent protein such as GFP attached to the N terminus (GFP-LC3) has been used to monitor autophagosome formation by fluorescence microscopy, to determine the increase in puncta and to determine the accumulation of LC3 (Fig. 1B). The use of stable GFP-LC3 transformants is recommended to avoid the problem of artificial GFP-LC3 aggregation, which may be caused by transient transfection.
3.2. Monitoring autophagic flux
3.2.1. GFP-LC3 cleavage assay
The GFP-LC3 can be used as an autophagy indicator detected by fluorescence microscopy as described above, and it can be used to follow autophagic flux by immunoblotting as well. The GFP-LC3 cleavage assay is based on the idea that free GFP, after being cleaved from GFP-LC3 in the autolysosomes, is relatively resistant to degradation by hydrolysis. Therefore, the appearance of free GFP detected by immunoblotting with an anti-GFP antibody can be used to monitor the process of lysosomal degradation of LC3, as shown in Fig. 1C.
3.2.2. LC3 and SQSTM1/p62 degradation
Detection of LC3 degradation is based on the quantification of GFP-LC3 reduction by measuring fluorescence reduction. Although the amount of LC3-II transiently increases during autophagy induction, the total GFP-LC3 fluorescence decreases upon LC3 degradation in autolysosomes. This reduction of GFP fluorescence can be monitored with a flow cytometer or spectrometer. Also, the best studied autophagy substrate, SQSTM1/p62, can be used as a monitor of autophagic flux. The p62 protein has both LC3 and ubiquitin binding domains and thus links LC3 and ubiquitinated substrates. The p62 incorporates into the completed autophagosome and is degraded in autolysosomes. The p62 cellular expression level is negatively regulated by autophagic activity and can serve as a readout of autophagic degradation.
4. Autophagy inhibitors and activators
The use of modulators (activators and inhibitors) of autophagy is helpful in understanding autophagy activity. Although no highly specific autophagy inhibitors or activators are available to date, a couple of chemicals are currently used for autophagy studies [42,45]. Here, we list the frequently used autophagy modulators.
4.1. Inhibitors
4.1.1. PI3-kinase inhibitors
3-Methyladenine (3-MA) and LY294002 block the formation of autophagosomes and inhibit LC3-II conversion; the drawback is that these inhibitors affect lysosomal acidification, glycogen metabolism, or endocytosis.
4.1.2. Lysosomotropic reagents
Chloroquine and bafilomycin A1 block LC3-II degradation by inhibiting endosomal or lysosomal acidification; the drawback is that they also affect other cellular processes such as mitosis and endocytosis [47,48].
4.2. Activators
4.2.1. Starvation
Withdrawing amino acids for 1 hour can induce autophagy in most cell lines. The drawback is that it is not applicable for certain tumor cell lines.
4.2.2. Rapamycin
Rapamycin is an inhibitor of mTOR, which is a potent suppressor of autophagy. The drawback is that rapamycin affects other cellular processes besides autophagy, such as protein synthesis and cellular metabolism.
Although various methods and modulators can thus be considered in autophagy studies, it should kept in mind that each method has its own limitations and current autophagy modulators lack specificity; thus, multiple independent experiments should be conducted and two or more modulators should be used in order to avoid misinterpretation of the results.
5. Case studies
5.1. APAP
APAP is widely used as a pain and fever relief agent. APAP overdose is the leading cause for acute liver failure in the United States [6], with about 44,000 APAP overdose-related hospital visits per year [49]. It has been well documented that APAP-induced hepatotoxicity is initiated by its reactive metabolite, N-acetyl-p-benzoquinoneimine (NAPQI), which is generated by liver cytochrome P450, with CYP2E1 being the major form [50]. NAPQI depletes glutathione, an intracellular antioxidant, and disturbs cellular redox homeostasis. When glutathione is sufficiently depleted, NAPQI reacts with cellular proteins to form protein adducts and impair multiple cellular liver functions. N-acetylcysteine, which acts as a precursor for glutathione, has been used as a clinical antidote in the early stage of APAP toxicity. Studies of APAP toxicity have been going on for decades, and continue because of the complex of mechanisms [51]. Recent studies have demonstrated that APAP causes autophagy induction both in vivo and in vitro [52–54] and that the activation of autophagy is a defense mechanism in APAP-induced liver toxicity [53]. Mitochondrial damage is a well-defined mechanism of APAP’s toxicity, as demonstrated by mitochondrial protein adducts, inhibition of mitochondrial respiration, ATP depletion, opening of mitochondrial membrane permeability transition pores, and the formation of reactive oxygen species (ROS) [55]. Autophagy removes damaged mitochondria and is a main mitochondrial turnover mechanism (mitophagy), so that it is not entirely a surprise that induction of autophagy protects against APAP-induced liver toxicity. A study by Ni et al [53] showed that pharmacological induction of autophagy by rapamycin protects against APAP-induced liver injury in mice, but does not affect glutathione depletion, and interestingly, rapamycin administration after APAP metabolism and after the occurrence of hepatic glutathione depletion still significantly reduces APAP’s toxicity. These results suggest that in addition to treatment with N-acetylcysteine during the early stage of APAP toxicity, pharmacological induction of autophagy can be considered as a potential therapeutic approach for the treatment of APAP overdosed patients, particularly those who have passed the metabolic phase and have not received the appropriate treatment [41].
5.2. Efavirenz
Efavirenz is the most commonly used non-nucleoside analogue reverse transcriptase inhibitor administrated in the combined pharmacological treatment of human AIDS. Although generally considered safe, evidence indicates that the use of efavirenz has been related to several side effects, such as disrupting lipid metabolism [56], inducing liver fibrosis [57,58], inhibiting hepatic cell proliferation, and inducing apoptosis [59,60]. Apostolova et al [61] reported that mitochondrial dysfunction is a mechanism responsible for efavirenz-induced liver damage. Another study by the same group, focusing on autophagy induction, was conducted because mitochondrial damage is one of the main autophagy inducers [59]. In their study, the conversion of LC3-I to LC3-II (a hallmark of autophagy) was detected and autophagic degradation of mitochondria (double-membrane mitochondria-containing vacuoles) was also observed in efavirenz-exposed human hepatic cells. In addition, co-treatment with the autophagy inhibitor 3-MA enhanced the toxic effect of efavirenz, indicating that efavirenz-induced autophagy is a rescue mechanism to promote cell survival [59,62].
5.3. Usnic acid
Usnic acid is a prominent metabolite of the Usnea species of lichen [63,64]. Both crude extracts of usnic acid and pure usnic acid have been marketed in the United States as dietary supplements to aid in weight loss; however, liver injury has been documented with the ingestion of pure usnic acid and usnic acid-containing products [65–68]. The proposed mechanisms of usnic acid-induced liver toxicity include mitochondrial impairment and oxidative stress [67,69–73].
We recently studied the usnic acid associated autophagy induction in vitro using a series of morphological and biochemical assays. In our study, we showed that usnic acid caused apoptosis, as demonstrated by increased caspase 3/7 activity and an increased ratio of sub-diploid nuclei; we also showed that autophagy was induced following usnic acid treatment as demonstrated by LC3 conversion, p62 degradation, and puncta formation, all of which are autophagy indicators (Chen et al, unpublished). The use of autophagy inhibitors enhanced usnic acid-elicited apoptosis, implying that autophagy is cytoprotective against usnic acid’s hepatotoxicity.
5.4. Alcohol
Liver is the main organ metabolizing alcohol and alcohol abuse causes acute and chronic liver damage. The common features of acute and chronic alcoholic liver injury are mitochondrial damage, ROS generation, and steatosis [74–76]. Although great effort has been put into studying alcohol-induced liver damage, the underlying molecular mechanisms are still not fully understood. Recently, autophagy has been reported to play a role in alcohol-induced liver damage in both cell and animal models [77,78].
Ding et al [77] reported that alcohol induces autophagy in mouse hepatic cells, both in vivo and in vitro. In vivo studies using transgenic GFP-LC3 mice demonstrated that both acute and chronic alcohol exposure induces a significant elevation of autophagosome formation. Similarly, alcohol treatment of primary hepatocytes caused dose-dependent increases in autophagosome formation and autophagic flux [77]. Autophagy inhibitors or small interfering RNAs increased hepatic apoptosis and exacerbated liver toxicity, whereas autophagy activators reduced liver toxicity associated with ethanol exposure, demonstrating a protective mechanism of autophagy in ethanol’s liver toxicity [77]. In addition, ethanol treatment inhibited the mTOR signaling pathway which, as a main modulator, negatively regulates autophagy activity. In mouse primary hepatocytes, the inhibition of mTOR was evidenced by the reduction of two major downstream targets of mTOR, phosphorylated p70 S6 kinase and 4E-binding protein 1 [77].
It is well known that ethanol is metabolized by alcohol dehydrogenase (ADH) and CYP2E1 to generate metabolites such as acetaldehyde, which effectively initiates ROS production. In the study by Ding et al [77], 4-methylpyrozole, an inhibitor of ADH and CYP2E1, or N-acetylcysteine, an antioxidant, suppressed ethanol-induced autophagy in primary hepatocytes, indicating that the process of autophagy is dependent on ethanol metabolism and ROS. This was confirmed in VL-17A cells, a HepG2 derived cell line overexpressing ADH and CYP2E1; alcohol treatment significantly enhanced autophagosome content in VL-17A cells compared with the parental HepG2 cells (which lack the ability to metabolize ethanol). Ding et al [77] concluded that autophagy protects against alcohol-induced liver toxicity, possibly by removing damaged mitochondria and accumulated lipid droplets.
Another study, by Wu et al [79], investigated the role of CYP2E1 and the impact of autophagy in ethanol-induced steatosis using HepG2 E47 cells, which express CYP2E1, and HepG2 C34 cells, which do not express CYP2E1, as an in vitro model; wild type, CYP2E1 knockin and knockout mice were used for their in vivo study. Interestingly, they found that treatment of HepG2 E47 cells with ethanol increased fat accumulation and oxidative stress but decreased autophagy, whereas alcohol had no effect in HepG2 C34 cells [79]. The results of Wu et al [79] seem to contradict to the results of Ding et al [77], in which autophagy was induced in cells expressing CYP2E1. Wu et al [79] also reported that in wild type and CYP2E1 knockin mice, a 4 day alcohol treatment impaired autophagy, as demonstrated by a decreased ratio of LC3-II to LC3-1 and reduced total LC-3 compared to CYP2E1 knockout mice. In addition, alcohol treatment activated the mTOR signaling pathway, as shown by increases in pAKT/ AKT and mTOR. Wild type and CYP2E1 knockin mice induced steatosis compared to CYP2E1 knockout mice. Use of an autophagy inhibitor increased alcohol liver toxicity, whereas stimulation with an autophagy activator reduced liver steatosis in CYP2E1 knockin but not CYP2E1 knockout mice. These results led the authors to conclude that impaired autophagy promoted ethanol-induced toxicity via CYP2E1 [79]. It is worth noting that both studies, those by Ding et al [77] and by Wu et al [79], described the involvement of autophagy in alcohol’s toxicity and they shared a similar outcome, that is, inhibition of autophagy exacerbated liver toxicity and induction of autophagy reduced liver toxicity. However, Ding et al [77] showed elevated autophagy and inhibited mTOR signaling; by contrast, Wu et al [79] demonstrated lowered autophagy and activated mTOR. The cause of this discrepancy is not clear, but it may be due to the difference in treatment durations between the two studies. Alcohol initially activates autophagy as a protective mechanism for an 18 hour exposure as shown by Ding et al [77]. When induction of autophagy fails to be maintained by a longer alcohol treatment (4 days), autophagy impairment follows, with a loss of protection against liver toxicity as shown by Wu et al [79].
6. Conclusion
Drug-induced liver toxicity is one of the leading causes of acute liver failure. An increasing number of studies have reported that herbal dietary supplements also cause acute liver damage [3,4,63,80–82]. The causes and mechanisms of liver damage vary, particularly for idiosyncratic liver toxicity. Understanding the molecular mechanisms of liver damage is fundamentally important for learning how to prevent and cure liver injury caused by drugs and chemicals.
The role of autophagy in drug-induced liver toxicity is beginning to be recognized, although a deep insight into the mechanisms remains elusive. From our study of usnic acid (Chen et al, unpublished) and studies on APAP [53], efavirenz [61], and alcohol [77,79], autophagy appears to be a protective mechanism, so maintaining or inducing autophagy may be important in drug- and chemical-induced liver toxicity and may have potential therapeutic implications.
Acknowledgments
Si Chen and Yuanfeng Wu were supported by appointments to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science Education, through an interagency agreement between the US Department of Energy and the US Food and Drug Authority (FDA). This article is not an official guidance or policy statement of the US FDA. No official support or endorsement by the US FDA is intended or should be inferred.
Footnotes
Conflicts of interest
All authors declare no conflicts of interest.
REFERENCES
- 1. Ichai P, Samuel D. Epidemiology of liver failure. Clin Res Hepatol Gastroenterol. 2011;35:610–7. doi: 10.1016/j.clinre.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 2. Lee WM. Drug-induced hepatotoxicity. N Engl J Med. 2003;349:474–85. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- 3. Zhao P, Wang C, Liu W, et al. Causes and outcomes of acute liver failure in China. PLoS One. 2013;8:e80991. doi: 10.1371/journal.pone.0080991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suk KT, Kim DJ. Drug-induced liver injury: present and future. Clin Mol Hepatol. 2012;18:249–57. doi: 10.3350/cmh.2012.18.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen M, Vijay V, Shi Q, et al. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov Today. 2011;16:697–703. doi: 10.1016/j.drudis.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 6. Larson AM, Polson J, Fontana RJ, et al. Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology. 2005;42:1364–72. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 7. Kaplowitz N. Drug-induced liver disorders: implications for drug development and regulation. Drug Saf. 2001;24:483–90. doi: 10.2165/00002018-200124070-00001. [DOI] [PubMed] [Google Scholar]
- 8. Senior JR. Drug hepatotoxicity from a regulatory perspective. Clin Liver Dis. 2007;11:507–24. vi. doi: 10.1016/j.cld.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 9. Gunawan BK, Kaplowitz N. Mechanisms of drug-induced liver disease. Clin Liver Dis. 2007;11:459–75. v. doi: 10.1016/j.cld.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 10. Nadanaciva S, Will Y. The role of mitochondrial dysfunction and drug safety. IDrugs. 2009;12:706–10. [PubMed] [Google Scholar]
- 11. Nadanaciva S, Will Y. Current concepts in drug-induced mitochondrial toxicity. Curr Protoc Toxicol. 2009;Chapter 2(Unit 2):15. doi: 10.1002/0471140856.tx0215s40. [DOI] [PubMed] [Google Scholar]
- 12. Russmann S, Kullak-Ublick GA, Grattagliano I. Current concepts of mechanisms in drug-induced hepatotoxicity. Curr Med Chem. 2009;16:3041–53. doi: 10.2174/092986709788803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplowitz N. Drug-induced liver disease. Zug. Switzerland: Informa Healthcare; 2007. Mechanisms of cell death and relevance to drug hepatotoxicity; pp. 85–95. [Google Scholar]
- 14. Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 15. Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–14. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 17. Park EJ, Lee AY, Park S, et al. Multiple pathways are involved in palmitic acid-induced toxicity. Food Chem Toxicol. 2014;67:26–34. doi: 10.1016/j.fct.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 18. Roychowdhury S, McMullen MR, Pisano SG, et al. Absence of receptor interacting protein kinase 3 prevents ethanol-induced liver injury. Hepatology. 2013;57:1773–83. doi: 10.1002/hep.26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang YF, He W, Zhang C, et al. Role of receptor interacting protein (RIP)1 on apoptosis-inducing factor-mediated necroptosis during acetaminophen-evoked acute liver failure in mice. Toxicol Lett. 2014;225:445–53. doi: 10.1016/j.toxlet.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 20. Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 21. Ramos-Gomez M, Kwak MK, Dolan PM, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–5. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 23. Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–84. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 24. Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–64. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen S, Xuan J, Wan L, et al. Sertraline, an antidepressant, induces apoptosis in hepatic cells through the mitogen-activated protein kinase pathway. Toxicol Sci. 2014;137:404–15. doi: 10.1093/toxsci/kft254. [DOI] [PubMed] [Google Scholar]
- 26. Dara L, Ji C, Kaplowitz N. The contribution of endoplasmic reticulum stress to liver diseases. Hepatology. 2011;53:1752–63. doi: 10.1002/hep.24279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaplowitz N, Than TA, Shinohara M, et al. Endoplasmic reticulum stress and liver injury. Semin Liver Dis. 2007;27:367–77. doi: 10.1055/s-2007-991513. [DOI] [PubMed] [Google Scholar]
- 28. Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740–3. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 33. Pyo JO, Nah J, Jung YK. Molecules and their functions in autophagy. Exp Mol Med. 2012;44:73–80. doi: 10.3858/emm.2012.44.2.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118(Pt 1):7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yorimitsu T, Nair U, Yang Z, et al. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–88. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams A, Jahreiss L, Sarkar S, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 40. Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 41. Czaja MJ, Ding WX, Donohue TM, Jr, et al. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131–58. doi: 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klionsky DJ. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 44. Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 45. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Quintart J, Leroy-Houyet MA, Trouet A, et al. Endocytosis and chloroquine accumulation during the cell cycle of hepatoma cells in culture. J Cell Biol. 1979;82:644–53. doi: 10.1083/jcb.82.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McSheehy PM, Troy H, Kelland LR, et al. Increased tumour extracellular pH induced by Bafilomycin A1 inhibits tumour growth and mitosis in vivo and alters 5-fluorouracil pharmacokinetics. Eur J Cancer. 2003;39:532–40. doi: 10.1016/s0959-8049(02)00671-8. [DOI] [PubMed] [Google Scholar]
- 49. Mitka M. FDA asks physicians to stop prescribing high-dose acetaminophen products. JAMA. 2014;311:563. doi: 10.1001/jama.2014.716. [DOI] [PubMed] [Google Scholar]
- 50. James LP, Mayeux PR, Hinson JA. Acetaminophen-induced hepatotoxicity. Drug Metab Dispos. 2003;31:1499–506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- 51. Uzi D, Barda L, Scaiewicz V, et al. CHOP is a critical regulator of acetaminophen-induced hepatotoxicity. J Hepatol. 2013;59:495–503. doi: 10.1016/j.jhep.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 52. Igusa Y, Yamashina S, Izumi K, et al. Loss of autophagy promotes murine acetaminophen hepatotoxicity. J Gastroenterol. 2012;47:433–43. doi: 10.1007/s00535-011-0500-0. [DOI] [PubMed] [Google Scholar]
- 53. Ni HM, Bockus A, Boggess N, et al. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222–32. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ni HM, Boggess N, McGill MR, et al. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–50. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jaeschke H, McGill MR, Ramachandran A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: lessons learned from acetaminophen hepatotoxicity. Drug Metab Rev. 2012 Feb;44(1):88–106. doi: 10.3109/03602532.2011.602688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tashima KT, Bausserman L, Alt EN, et al. Lipid changes in patients initiating efavirenz- and indinavir-based antiretroviral regimens. HIV Clin Trials. 2003;4:29–36. doi: 10.1310/f2v7-3r46-vx6j-241r. [DOI] [PubMed] [Google Scholar]
- 57. Jones M, Nunez M. Liver toxicity of antiretroviral drugs. Semin Liver Dis. 2012;32:167–76. doi: 10.1055/s-0032-1316472. [DOI] [PubMed] [Google Scholar]
- 58. Loko MA, Bani-Sadr F, Winnock M, et al. Impact of HAART exposure and associated lipodystrophy on advanced liver fibrosis in HIV/HCV-coinfected patients. J Viral Hepat. 2011;18:e307–14. doi: 10.1111/j.1365-2893.2010.01417.x. [DOI] [PubMed] [Google Scholar]
- 59. Apostolova N, Gomez-Sucerquia LJ, Gortat A, et al. Compromising mitochondrial function with the antiretroviral drug efavirenz induces cell survival-promoting autophagy. Hepatology. 2011;54:1009–19. doi: 10.1002/hep.24459. [DOI] [PubMed] [Google Scholar]
- 60. Bumpus NN. Efavirenz and 8-hydroxyefavirenz induce cell death via a JNK- and BimEL-dependent mechanism in primary human hepatocytes. Toxicol Appl Pharmacol. 2011;257:227–34. doi: 10.1016/j.taap.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 61. Apostolova N, Gomez-Sucerquia LJ, Moran A, et al. Enhanced oxidative stress and increased mitochondrial mass during efavirenz-induced apoptosis in human hepatic cells. Br J Pharmacol. 2010;160:2069–84. doi: 10.1111/j.1476-5381.2010.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Apostolova N, Gomez-Sucerquia LJ, Gortat A, et al. Autophagy as a rescue mechanism in efavirenz-induced mitochondrial dysfunction: a lesson from hepatic cells. Autophagy. 2011;7:1402–4. doi: 10.4161/auto.7.11.17653. [DOI] [PubMed] [Google Scholar]
- 63. Guo L, Shi Q, Fang JL, et al. Review of usnic acid and Usnea barbata toxicity. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2008;26:317–38. doi: 10.1080/10590500802533392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ingolfsdottir K. Usnic acid. Phytochemistry. 2002;61:729–36. doi: 10.1016/s0031-9422(02)00383-7. [DOI] [PubMed] [Google Scholar]
- 65. Durazo FA, Lassman C, Han SH, et al. Fulminant liver failure due to usnic acid for weight loss. Am J Gastroenterol. 2004;99:950–2. doi: 10.1111/j.1572-0241.2004.04165.x. [DOI] [PubMed] [Google Scholar]
- 66. Favreau JT, Ryu ML, Braunstein G, et al. Severe hepatotoxicity associated with the dietary supplement LipoKinetix. Ann Intern Med. 2002;136:590–5. doi: 10.7326/0003-4819-136-8-200204160-00008. [DOI] [PubMed] [Google Scholar]
- 67. Pramyothin P, Janthasoot W, Pongnimitprasert N, et al. Hepatotoxic effect of (+)usnic acid from Usnea siamensis Wainio in rats, isolated rat hepatocytes and isolated rat liver mitochondria. J Ethnopharmacol. 2004;90:381–7. doi: 10.1016/j.jep.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 68. Yellapu RK, Mittal V, Grewal P, et al. Acute liver failure caused by ’fat burners’ and dietary supplements: a case report and literature review. Can J Gastroenterol. 2011;25:157–60. doi: 10.1155/2011/174978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Han D, Matsumaru K, Rettori D, et al. Usnic acid-induced necrosis of cultured mouse hepatocytes: inhibition of mitochondrial function and oxidative stress. Biochem Pharmacol. 2004;67:439–51. doi: 10.1016/j.bcp.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 70. Liu Q, Zhao X, Lu X, et al. Proteomic study on usnic-acid-induced hepatotoxicity in rats. J Agric Food Chem. 2012;60:7312–7. doi: 10.1021/jf2046834. [DOI] [PubMed] [Google Scholar]
- 71. Sahu SC, Amankwa-Sakyi M, O’Donnell MW, Jr, et al. Effects of usnic acid exposure on human hepatoblastoma HepG2 cells in culture. J Appl Toxicol. 2012;32:722–30. doi: 10.1002/jat.1721. [DOI] [PubMed] [Google Scholar]
- 72. Sahu SC, O’Donnell MW, Jr, Sprando RL. Interactive toxicity of usnic acid and lipopolysaccharides in human liver HepG2 cells. J Appl Toxicol. 2012;32:739–49. doi: 10.1002/jat.2768. [DOI] [PubMed] [Google Scholar]
- 73. Sonko BJ, Schmitt TC, Guo L, et al. Assessment of usnic acid toxicity in rat primary hepatocytes using (1)(3)C isotopomer distribution analysis of lactate, glutamate and glucose. Food Chem Toxicol. 2011;49:2968–74. doi: 10.1016/j.fct.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Massey VL, Arteel GE. Acute alcohol-induced liver injury. Front Physiol. 2012;3:193. doi: 10.3389/fphys.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med. 2002;32:11–6. doi: 10.1016/s0891-5849(01)00769-9. [DOI] [PubMed] [Google Scholar]
- 76. Donohue Jr TM. Alcohol-induced steatosis in liver cells. World J Gastroenterol. 2007;13:4974–8. doi: 10.3748/wjg.v13.i37.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ding WX, Li M, Chen X, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–52. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu D, Cederbaum AI. Inhibition of autophagy promotes CYP2E1-dependent toxicity in HepG2 cells via elevated oxidative stress, mitochondria dysfunction and activation of p38 and JNK MAPK. Redox Biol. 2013;1:552–65. doi: 10.1016/j.redox.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wu D, Wang X, Zhou R, et al. Alcohol steatosis and cytotoxicity: the role of cytochrome P4502E1 and autophagy. Free Radic Biol Med. 2012;53:1346–57. doi: 10.1016/j.freeradbiomed.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chen S, Wan L, Couch L, et al. Mechanism study of goldenseal-associated DNA damage. Toxicol Lett. 2013;221:64–72. doi: 10.1016/j.toxlet.2013.05.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fu PP, Xia Q, Guo L, et al. Toxicity of kava kava. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2008;26:89–112. doi: 10.1080/10590500801907407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo L, Fuscoe J, Fu P, Mei N. Application of DNA microarray in studies of herbal dietary supplements. In: Casciano DA, Sahu SC, editors. Handbook of systems toxicology. Chichester, UK: John Wiley & Sons, Ltd; 2010. pp. 407–18. [Google Scholar]