Abstract
BACKGROUND
Fluoroquinolones and second-line injectable drugs are the backbone of treatment regimens for multidrug-resistant tuberculosis, and resistance to these drugs defines extensively drug-resistant tuberculosis. We assessed the accuracy of an automated, cartridge-based molecular assay for the detection, directly from sputum specimens, of Mycobacterium tuberculosis with resistance to fluoroquinolones, aminoglycosides, and isoniazid.
METHODS
We conducted a prospective diagnostic accuracy study to compare the investigational assay against phenotypic drug-susceptibility testing and DNA sequencing among adults in China and South Korea who had symptoms of tuberculosis. The Xpert MTB/RIF assay and sputum culture were performed. M. tuberculosis isolates underwent phenotypic drug-susceptibility testing and DNA sequencing of the genes katG, gyrA, gyrB, and rrs and of the eis and inhA promoter regions.
RESULTS
Among the 308 participants who were culture-positive for M. tuberculosis, when phenotypic drug-susceptibility testing was used as the reference standard, the sensitivities of the investigational assay for detecting resistance were 83.3% for isoniazid (95% confidence interval [CI], 77.1 to 88.5), 88.4% for ofloxacin (95% CI, 80.2 to 94.1), 87.6% for moxifloxacin at a critical concentration of 0.5 μg per milliliter (95% CI, 79.0 to 93.7), 96.2% for moxifloxacin at a critical concentration of 2.0 μg per milliliter (95% CI, 87.0 to 99.5), 71.4% for kanamycin (95% CI, 56.7 to 83.4), and 70.7% for amikacin (95% CI, 54.5 to 83.9). The specificity of the assay for the detection of phenotypic resistance was 94.3% or greater for all drugs except moxifloxacin at a critical concentration of 2.0 μg per milliliter (specificity, 84.0% [95% CI, 78.9 to 88.3]). When DNA sequencing was used as the reference standard, the sensitivities of the investigational assay for detecting mutations associated with resistance were 98.1% for isoniazid (95% CI, 94.4 to 99.6), 95.8% for fluoroquinolones (95% CI, 89.6 to 98.8), 92.7% for kanamycin (95% CI, 80.1 to 98.5), and 96.8% for amikacin (95% CI, 83.3 to 99.9), and the specificity for all drugs was 99.6% (95% CI, 97.9 to 100) or greater.
CONCLUSIONS
This investigational assay accurately detected M. tuberculosis mutations associated with resistance to isoniazid, fluoroquinolones, and aminoglycosides and holds promise as a rapid point-of-care test to guide therapeutic decisions for patients with tuberculosis. (Funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and the Ministry of Science and Technology of China; ClinicalTrials.gov number, NCT02251327.)
Multidrug-resistant tuberculosis (defined by resistance to isoniazid and rifampin) is a consequence of ineffective treatment, both at the individual level, when antibiotics are improperly selected or taken, and at the programmatic level, when the use of standard regimens is based on algorithms that may not include drug-susceptibility testing. Incorrect diagnosis and treatment of drug-resistant tuberculosis is associated with morbidity, mortality, and ongoing transmission of infection. For uncomplicated multidrug-resistant tuberculosis, the World Health Organization (WHO) recently endorsed a treatment regimen of 9 to 12 months, as a potential alternative to conventional regimens of 18 to 24 months.1 Fluoroquinolones and second-line injectable drugs — aminoglycosides and capreomycin — are core components of this shortened regimen. Rapid methods for the detection of susceptibility and resistance to fluoroquinolones and second-line injectable drugs are needed to identify patients who are microbiologically eligible for the shortened regimen. Detection of Mycobacterium tuberculosis drug resistance with the use of conventional culture-based phenotypic drug-susceptibility testing is slow and biohazardous and requires substantial laboratory infrastructure and training.
Xpert MTB/RIF (Cepheid) is an integrated, automated, cartridge-based system, used with GeneXpert instrumentation, for the rapid molecular detection of M. tuberculosis and mutations associated with rifampin resistance. Xpert MTB/RIF is widely used in tuberculosis programs and has contributed to the global increase in detection of rifampin-resistant tuberculosis.2 However, for patients with rifampin-resistant tuberculosis, Xpert MTB/RIF provides no further information to guide the selection of appropriate antibiotics or to promptly identify and triage to equipped health care centers those patients who have extensively drug-resistant tuberculosis (defined as multidrug-resistant tuberculosis that is additionally resistant to fluoroquinolones and second-line injectables). M. tuberculosis resistance to fluoroquinolones, aminoglycosides, and isoniazid is associated with approximately 25 mutations in six genes and promoter regions.3–10 We recently described a new cartridge, for use with the GeneXpert platform, for the rapid molecular detection of these mutations.11 This assay can provide results from unprocessed sputum samples in just over 2 hours, with minimal hands-on technical time. Here, we describe a clinical study to assess the diagnostic accuracy of this investigational assay for the detection of resistance to fluoroquinolones, aminoglycosides, and isoniazid.
METHODS
STUDY DESIGN AND OVERSIGHT
We conducted a blinded, multicenter, prospective diagnostic accuracy study. The investigational assay was the index test, and phenotypic culture-based drug-susceptibility testing and DNA sequencing, considered separately, were the reference standards for resistance detection. The primary objective of the study was to determine the sensitivity and specificity of the investigational assay for the detection of M. tuberculosis resistance to isoniazid, moxifloxacin, ofloxacin, amikacin, and kanamycin.
The study was designed, implemented, and supervised by the Tuberculosis Clinical Diagnostics Research Consortium (TB-CDRC), which is made up of academic investigators at participating sites. Members of the TB-CDRC performed statistical analyses, wrote the manuscript, and made the decision to submit the manuscript for publication. All the authors vouch for the accuracy and completeness of the data and analyses and for the fidelity of the study to the protocol, which (with the statistical analysis plan) is available with the full text of this article at NEJM.org.
The investigational assay cartridges, Xpert MTB/RIF cartridges, and 10-color GeneXpert instruments were donated by Cepheid. Cepheid personnel had no role in study design, implementation, data analysis, manuscript writing, or the decision to submit the study findings for publication.
STUDY POPULATION
Adults in Seoul, South Korea, and Zhengzhou, China, who had symptoms of pulmonary tuberculosis were enrolled in the study. Participants were enrolled prospectively into one of two groups — the drug-resistance-risk group and the case-detection group. The inclusion and exclusion criteria for each group are provided in Table S1 in the Supplementary Appendix, available at NEJM.org. All participants provided written informed consent. The ethics committees at the enrolling sites and Johns Hopkins University approved this study.
STUDY PROCEDURES
For each participant, one investigational assay and one Xpert MTB/RIF test were performed directly on the same sputum specimen; in addition, one smear microscopy test, one mycobacterial liquid culture, and one solid culture were performed after digestion and decontamination of sputum. Participants provided up to two expectorated sputum specimens 1 hour to 4 days apart. If the volume of the first specimen exceeded 3.5 ml, it was homogenized with glass beads (Fisher Scientific) and split into two portions: 1.5 ml for the investigational assay and Xpert MTB/RIF and the remainder for smear and cultures. If the volume of the first specimen was between 1.5 and 3.5 ml, it was used for the investigational assay and Xpert MTB/RIF, and the second sputum specimen was used for smear and culture. Full details of the study design and procedures are provided in the protocol.
XPERT MTB/RIF AND INVESTIGATIONAL ASSAY
The investigational assay cartridge was assembled by Cepheid, using components synthesized or sourced by Cepheid. Sample reagent was added to sputum (in a 2:1 dilution), and 2.0 ml of the resulting mixture was added to one Xpert MTB/RIF and one investigational cartridge.12 Xpert MTB/RIF and investigational assays were performed with the use of standard four-module GeneXpert instruments. For the investigational assay, the instrument was calibrated to detect 10 fluorescence channels, with the use of research-version software that enabled 10-color melting-temperature analysis. For each probe targeting a resistance-associated genetic region, the melting temperature or temperatures were determined and interpreted as wild-type, mutant (if the measured temperature was different from the known wild-type melting temperature), or heteroresistant (if both wild-type and mutant melting temperatures were present).11 A specimen was considered to be resistant to a drug if a mutant melting temperature was detected in any of the gene targets associated in high frequency with resistance — namely, katG and the inhA promoter for isoniazid, gyrA for the fluoroquinolones ofloxacin and moxifloxacin, and rrs for kanamycin and amikacin.3,4,8,11,13–18 A specimen with a wild-type or uninterpretable melting temperature in a high-frequency gene target was considered to be susceptible or of indeterminate susceptibility, respectively, to the corresponding drug, unless there was a mutant melting temperature in the low-frequency gene target — namely, gyrB for fluoroquinolones and the eis promoter for kanamycin.3,4,8,11,13,15,19,20 The personnel performing the investigational assay were unaware of the other test results. The investigational assay was conducted in real time, but the results were not released to clinicians and therefore did not affect decisions regarding treatment.
MYCOBACTERIAL CULTURES
Sputum was digested with N-acetyl-L-cysteine and sodium hydroxide (final concentration, 2%) and processed with the use of standard methods.21 Smear microscopy was performed with Ziehl–Neelsen staining on the concentrated pellet.22 A volume of 0.5 ml of the resuspended pellet was inoculated into liquid culture (BACTEC MGIT, BD Microbiology Systems), and 0.2 ml was inoculated onto Löwenstein–Jensen medium. Cultures that were positive for growth of acid-fast bacilli underwent confirmation of the presence of M. tuberculosis complex by means of paranitro-benzoic acid (PNB) and thiophen-2-carboxylic acid hydrazide (TCH) testing,22 MPT64 antigen detection (Capilia TB, Tauns Laboratories), or polymerase chain reaction (MolecuTech MTB-ID V3, YD Diagnostics).
PHENOTYPIC DRUG-SUSCEPTIBILITY TESTING
Indirect drug-susceptibility testing was performed from the first positive M. tuberculosis culture with the use of the BACTEC MGIT 960 system. The critical concentrations used for each drug were 0.1 μg per milliliter for isoniazid, 0.5 and 2 μg per milliliter for moxifloxacin, 2 μg per milliliter for ofloxacin, 1 μg per milliliter for amikacin, and 2.5 μg per milliliter for kanamycin.23
DNA SEQUENCING
The molecular drug resistance of cultured isolates was characterized by Sanger DNA sequencing (Table S2 in the Supplementary Appendix). DNA was prepared from the same culture that was used for phenotypic drug-susceptibility testing. Each of the six gene targets included in the assay was sequenced to detect mutations in target and nontarget regions. Sequencing was the reference standard for the detection of heteroresistance, defined as the presence, in a single sample, of both wild-type and mutant sequences for a given gene locus.11
STATISTICAL ANALYSIS
The primary objective of the study was to determine the sensitivity and specificity of the investigational assay for the detection of resistance to isoniazid, fluoroquinolones (ofloxacin and moxifloxacin), and aminoglycosides (kanamycin and amikacin). In the primary analyses, the reference standards for phenotypic and molecular drug-susceptibility testing were considered separately, and sensitivity and specificity were calculated for the detection of resistance to each drug.24 The main analysis population for drug-susceptibility testing, which was made up of participants who were culture-positive for M. tuberculosis and who had interpretable results of reference-standard drug-susceptibility testing, included participants who were prospectively enrolled into the drug-resistance-risk and case-detection groups. Secondary analyses of diagnostic accuracy for tuberculosis case detection included only participants in the case-detection group. To explore the diagnostic accuracy of the investigational assay for the testing of patients who are already known to have rifampin-resistant tuberculosis, we conducted a post hoc analysis of a “reflex-test population” that was made up of participants who were included in the main analysis of drug-susceptibility testing and in whose sputum the Xpert MTB/RIF test indicated the presence of rifampin-resistant M. tuberculosis. Reference-standard drug-susceptibility testing was performed independently and regardless of the investigational-assay results. Invalid or indeterminate investigational-assay results were enumerated but not included in sensitivity and specificity calculations. The binomial exact method was used to calculate 95% confidence intervals.
RESULTS
PARTICIPANTS
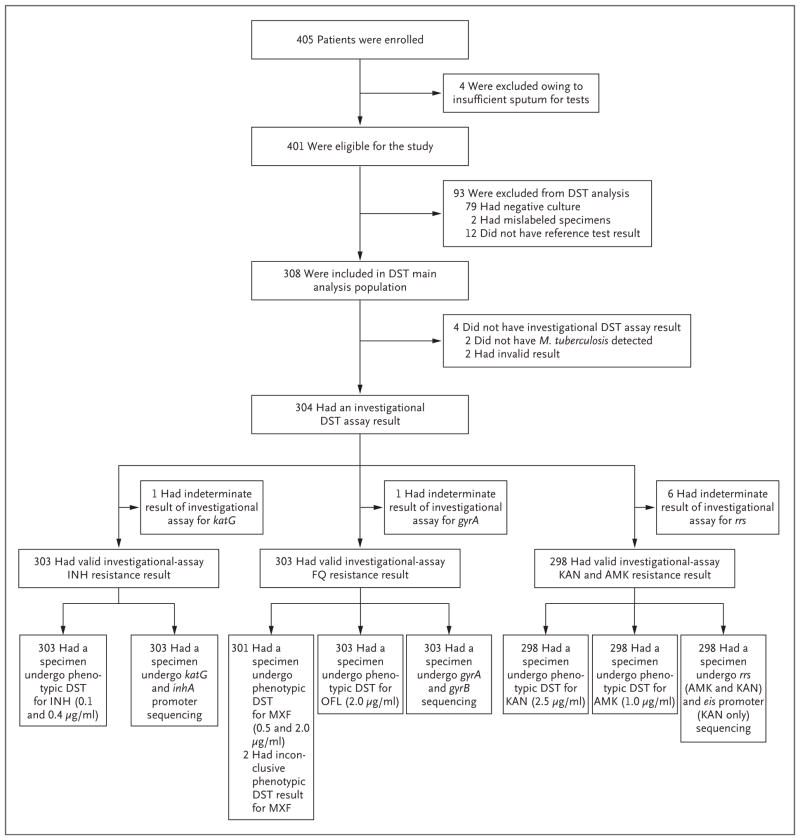
A total of 405 participants were enrolled in the study from June 2014 through June 2015 (Fig. 1). Four enrolled participants produced insufficient sputum for tests and were excluded late in the study. Thus, 401 participants were eligible for inclusion in the study. A total of 93 participants were excluded from the analyses of drug-susceptibility testing, including 79 for whom cultures were negative for M. tuberculosis, 2 for whom specimens were mislabeled, 10 for whom M. tuberculosis DNA was of insufficient quality or quantity for sequencing, and 2 for whom MGIT drug-susceptibility testing cultures were contaminated. Thus, 308 culture-positive participants were included in the main analysis population for drug-susceptibility testing.
Figure 1. Participant Enrollment and Testing in the Main Analysis Population.
Among the 12 patients who were excluded from the analysis of drug-susceptibility testing (DST) because of a lack of a reference-test result, 10 had DNA that was of insufficient quality or quantity for sequencing, and 2 had uninterpretable MGIT phenotypic DST results because of contamination. Complete DST reference-standard results were not achievable for 4.3% of participants (14 of 322) whose culture was positive for Mycobacterium tuberculosis. Among the 308 participants in the main analysis population for DST, 152 were excluded from the reflex-test analysis population (146 with an Xpert MTB/RIF result indicating that rifampin resistance was not detected, 4 with an Xpert MTB/RIF result indicating that M. tuberculosis was not detected, and 2 with an indeterminate Xpert MTB/RIF result with regard to rifampin resistance); the reflex-test analysis population therefore included 156 participants. AMK denotes amikacin, FQ fluoroquinolone, INH isoniazid, KAN kanamycin, MXF moxifloxacin, and OFL ofloxacin.
DISTRIBUTION OF PHENOTYPIC AND GENOTYPIC RESISTANCE
Phenotypic drug-susceptibility testing identified 194 of 308 participants (63.0%) as having infections that were resistant to one or more drugs, including 55 participants with multidrug-resistant tuberculosis, 54 with tuberculosis that was multidrug-resistant in addition to being resistant to fluoroquinolones or aminoglycosides, and 39 with extensively drug-resistant tuberculosis (Table 1). DNA sequencing identified 25 different mutations among the six gene targets; 15 of the mutations occurred in isolates from both China and South Korea (Table S3 in the Supplementary Appendix). The most common mutations were katG S315T, gyrA D94G, rrs A1401G, inhA C(-15)T, and gyrA A90V.
Table 1.
Demographic and Clinical Characteristics of Participants at Enrollment, and Drug-Resistance Status Based on Phenotypic Drug-Susceptibility Testing.
| Characteristic | Case-Detection Group* | Drug-Resistance-Risk Group† | All Participants |
|---|---|---|---|
| Demographic and clinical characteristics‡ | |||
| Enrolled in China — no./total no. (%) | 86/111 (77) | 196/290 (68) | 282/401 (70) |
| Enrolled in South Korea — no./total no. (%) | 25/111 (23) | 94/290 (32) | 119/401 (30) |
| Male sex — no./total no. (%) | 72/111 (65) | 230/290 (79) | 302/401 (75) |
| Median age (range) — yr | 48 (21–82) | 47 (18–85) | 47 (18–85) |
| Previous tuberculosis — no./total no. (%) | 0/111 (0) | 184/290 (63) | 184/401 (46) |
| Receiving tuberculosis treatment at enrollment — no./total no. (%) | 96/111 (86) | 285/290 (98) | 381/401 (95) |
| All study tests performed from one sputum specimen — no./total no. (%) | 72/111 (65) | 228/290 (79) | 300/401 (75) |
| Results of testing for Mycobacterium tuberculosis — no./total no. (%) | |||
| Study culture positive | 99/111 (89) | 223/290 (77) | 322/401 (80) |
| Smear microscopy positive for acid-fast bacilli | 67/111 (60) | 196/290 (68) | 263/401 (66) |
| Smear microscopy negative for acid-fast bacilli | 32/111 (29) | 27/290 (9) | 59/401 (15) |
| Study culture negative | 12/111 (11) | 67/290 (23) | 79/401 (20) |
| Drug-resistance status — no./total no. (%)§ | |||
| Isoniazid resistance only | 13/98 (13) | 14/210 (7) | 27/308 (9) |
| Rifampin resistance only | 0 | 2/210 (1) | 2/308 (0.6) |
| Fluoroquinolone resistance only | 2/98 (2) | 4/210 (2) | 6/308 (2) |
| Aminoglycoside resistance only | 1/98 (1) | 1/210 (0.5) | 2/308 (0.6) |
| Multidrug resistance only | 2/98 (2) | 53/210 (25) | 55/308 (18) |
| Multidrug resistance with fluoroquinolone resistance, aminoglycoside susceptibility | 0 | 46/210 (22) | 46/308 (15) |
| Multidrug resistance with aminoglycoside resistance, fluoroquinolone susceptibility | 1/98 (1) | 7/210 (3) | 8/308 (2.5) |
| Extensively drug resistant | 0 | 39/210 (19) | 39/308 (13) |
| Other polyresistance | 2/98 (2) | 7/210 (3) | 9/308 (3) |
| No resistance to isoniazid, rifampin, fluoroquinolones, or aminoglycosides | 77/98 (79) | 37/210 (18) | 114/308 (37) |
Participants in this group had suspected or confirmed new pulmonary tuberculosis and had received antituberculosis drugs for less than 3 days.
Participants in this group either had confirmed pulmonary tuberculosis with documented rifampin resistance and had received antituberculosis drugs for 31 days or less or had a history of tuberculosis plus ongoing signs or symptoms of pulmonary tuberculosis and suspected drug resistance.
Demographic and clinical characteristics are reported for the 401 study-eligible participants.
Drug-resistance status is reported for the 308 participants in the main analysis population for drug-susceptibility testing.
OPERATIONAL CHARACTERISTICS OF THE INVESTIGATIONAL ASSAY
The investigational assay provided no drug-susceptibility testing information for 4 of 308 participants (1.3%) (2 had invalid results, and 2 had no M. tuberculosis detected) (Table S4 in the Supplementary Appendix). Among the remaining 304 participants, the investigational-assay results were indeterminate for 18 (1.0%) of 1824 total gene targets. The assay sensitivities for tuberculosis case detection were 96 of 98, or 98.0% (95% confidence interval [CI], 92.8 to 99.8), for the investigational assay and 97 of 99, or 98.0% (95% CI, 92.9 to 99.8), for Xpert MTB/RIF.
INVESTIGATIONAL ASSAY VERSUS PHENOTYPIC DRUG-SUSCEPTIBILITY TESTING
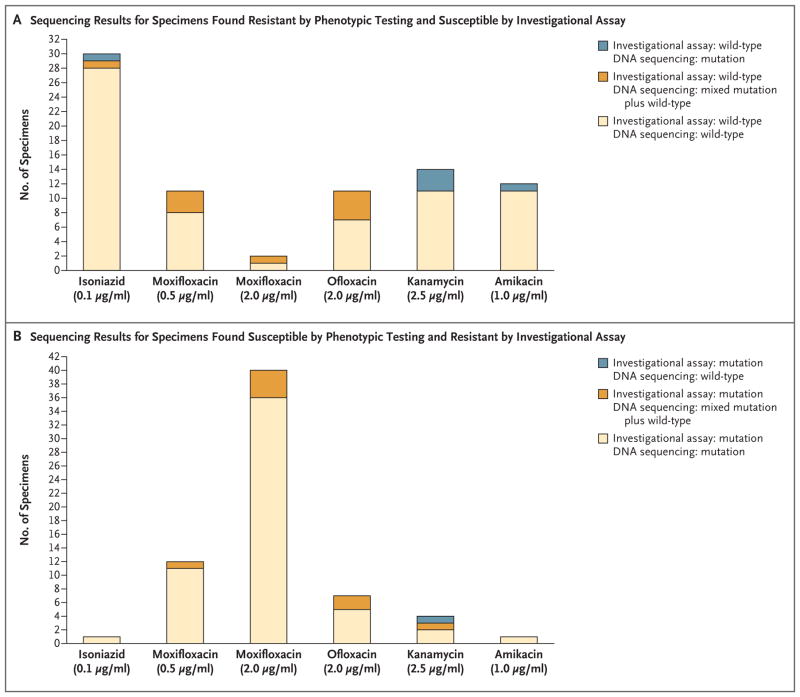
The sensitivity and specificity, respectively, of the investigational assay for the detection of phenotypic resistance were 83.3% and 99.2% for isoniazid, 88.4% and 96.6% for ofloxacin, 87.6% and 94.3% for moxifloxacin at a critical concentration of 0.5 μg per milliliter, 96.2% and 84.0% for moxifloxacin at a critical concentration of 2.0 μg per milliliter, 71.4% and 98.4% for kanamycin, and 70.7% and 99.6% for amikacin (Table 2). Almost all isolates that were found by phenotypic drug-susceptibility testing to be resistant but found by the investigational assay to be susceptible (i.e., to have a wild-type melting temperature) were also found to be wild-type by sequencing (Fig. 2A). All but one of the isolates that were found by phenotypic drug-susceptibility testing to be susceptible but were found by the investigational assay to be resistant (i.e., to have a mutant melting temperature detected) were found by DNA sequencing to have corresponding resistance mutations (Fig. 2B).
Table 2.
Sensitivity and Specificity of the Investigational Assay, with Phenotypic Drug-Susceptibility Testing as the Reference Standard, in the Main Analysis Population for Drug-Susceptibility Testing.
| Drug | Investigational-Assay Result + Phenotypic Drug-Susceptibility Test Result* | Sensitivity | Specificity | |||||
|---|---|---|---|---|---|---|---|---|
| R+R | R+S | S+R | S+S | |||||
| no. of specimens | no./total no. | % (95% CI) | no./total no. | % (95% CI) | ||||
| Isoniazid† | 150 | 1 | 30 | 122 | 150/180 | 83.3 (77.1–88.5) | 122/123 | 99.2 (95.6–100.0) |
|
| ||||||||
| Ofloxacin‡ | 84 | 7 | 11 | 201 | 84/95 | 88.4 (80.2–94.1) | 201/208 | 96.6 (93.2–98.6) |
|
| ||||||||
| Moxifloxacin, 0.5 μg/ml‡§ | 78 | 12 | 11 | 200 | 78/89 | 87.6 (79.0–93.7) | 200/212 | 94.3 (90.3–97.0) |
|
| ||||||||
| Moxifloxacin, 2.0 μg/ml‡ | 51 | 40 | 2 | 210 | 51/53 | 96.2 (87.0–99.5) | 210/250 | 84.0 (78.9–88.3) |
|
| ||||||||
| Kanamycin¶ | 35 | 4 | 14 | 245 | 35/49 | 71.4 (56.7–83.4) | 245/249 | 98.4 (96.0–99.6) |
|
| ||||||||
| Amikacin¶ | 29 | 1 | 12 | 256 | 29/41 | 70.7 (54.5–83.9) | 256/257 | 99.6 (97.9–100.0) |
The numbers of specimens with each combination of results for the investigational assay and the MGIT drug-susceptibility test are shown. R+R indicates specimens found resistant by both methods, R+S specimens found resistant by the investigational assay and susceptible by phenotypic drug-susceptibility testing, S+R specimens found susceptible by the investigational assay and resistant by phenotypic drug- susceptibility testing, and S+S specimens found susceptible by both methods.
One specimen was excluded because of an indeterminate investigational-assay result for katG (it was found to be susceptible to isoniazid by phenotypic drug-susceptibility testing).
One specimen was excluded because of an indeterminate investigational-assay result for gyrA (it was found to be susceptible to ofloxacin and moxifloxacin [0.5 μg per milliliter] by phenotypic drug-susceptibility testing).
Two specimens were excluded because of missing moxifloxacin (0.5 μg per milliliter) phenotypic drug-susceptibility testing results (one was found resistant and one was found susceptible by the investigational assay).
Six specimens were excluded because of indeterminate investigational-assay results for rrs (by phenotypic drug-susceptibility testing, five were found susceptible and one was found resistant to amikacin and kanamycin).
Figure 2. Sequencing Analysis of Isolates with Discrepant Investigational Assay and Phenotypic DST Results.
Panel A shows the results of sequencing analysis of isolates that were found by the investigational assay to be susceptible (i.e., to have a wild-type melting temperature) and were found by phenotypic DST to be resistant. Panel B shows the results of sequencing analysis of isolates that were found by the investigational assay to be resistant (i.e., to have a mutant melting temperature detected) and were found by phenotypic DST to be susceptible. A full list of the genotypes found by DNA sequencing is provided in the Supplementary Results section in the Supplementary Appendix.
INVESTIGATIONAL ASSAY VERSUS DNA SEQUENCING
The sensitivity and specificity, respectively, of the investigational assay for the detection of resistance mutations were 98.1% and 100.0% for isoniazid, 95.8% and 100.0% for fluoroquinolones, 92.7% and 99.6% for kanamycin, and 96.8% and 100.0% for amikacin (Table 3, and Table S5 in the Supplementary Appendix). Among the 13 specimens that had mutations that were missed by the investigational assay, 10 (76.9%) were identified as heteroresistant by DNA sequencing. The assay successfully detected the mutant population in 16 of 26 (61.5%) sequencing-confirmed heteroresistant bacillary populations, the majority of which were found to be resistant by phenotypic drug-susceptibility testing (Table S6 in the Supplementary Appendix). The diagnostic accuracies of the investigational assay in the reflex-test analysis population are provided in Table S7 in the Supplementary Appendix, and the diagnostic accuracies according to sputum smear microscopy status and enrollment site are provided in Tables S8 and S9 in the Supplementary Appendix.
Table 3.
Sensitivity and Specificity of the Investigational Assay, with DNA Sequencing as the Reference Standard, in the Main Analysis Population for Drug-Susceptibility Testing.*
| Drug | Investigational-Assay Result + DNA Sequencing Result† | Sensitivity | Specificity | |||||
|---|---|---|---|---|---|---|---|---|
| M+M | M+NM | NM+M | NM+NM | |||||
| no. of specimens | no./total no. | % (95% CI) | no./total no. | % (95% CI) | ||||
| Isoniazid‡ | 151 | 0 | 3 | 149 | 151/154 | 98.1 (94.4–99.6) | 149/149 | 100.0 (97.6–100.0) |
|
| ||||||||
| Fluoroquinolones§ | 91 | 0 | 4 | 208 | 91/95 | 95.8 (89.6–98.8) | 208/208 | 100.0 (98.2–100.0) |
|
| ||||||||
| Kanamycin¶ | 38 | 1 | 3 | 256 | 38/41 | 92.7 (80.1–98.5) | 256/257 | 99.6 (97.9–100.0) |
|
| ||||||||
| Amikacin¶ | 30 | 0 | 1 | 267 | 30/31 | 96.8 (83.3–99.9) | 267/267 | 100.0 (98.6–100.0) |
There are no known silent mutations that occur within the gene regions tested by the investigational assay for resistance. In the study, no silent mutations in these regions were detected either by the investigational assay or by sequencing.
The numbers of specimens with each combination of results for the investigational assay and DNA sequencing are shown. M+M indicates specimens found to have a mutation by both methods, M+NM specimens found by the investigational assay to have a mutation and found by DNA sequencing to have no mutation, NM+M specimens found by the investigational assay to have no mutation and found by DNA sequencing to have a mutation, and NM+NM specimens found by both methods to have no mutation.
One specimen was excluded because of an indeterminate investigational-assay result for katG (also found indeterminate for katG by sequencing).
Ofloxacin and moxifloxacin are grouped as fluoroquinolones, since gyrA and gyrB mutations confer resistance to both drugs. One specimen was excluded because of an indeterminate investigational-assay result for gyrA (no gyrA or gyrB mutation was detected by sequencing).
Six specimens were excluded because of an indeterminate investigational-assay result for rrs (five with no rrs mutation found by sequencing and one with an rrs mutation found by sequencing).
IDENTIFICATION OF PATIENTS ELIGIBLE FOR THE SHORTENED TREATMENT REGIMEN
One potential use of the investigational assay would be to determine the eligibility of patients for a shortened treatment regimen for multidrug-resistant tuberculosis. Among patients with multidrug-resistant tuberculosis, the investigational assay correctly identified 48 of 53 patients (90.6%; 95% CI, 79.3 to 96.9) who had tuberculosis without phenotypic resistance either to fluoroquinolones or to aminoglycosides (i.e., patients who were microbiologically eligible for the shortened regimen) and 81 of 92 patients (88.0%; 95% CI, 79.6 to 93.9) who had tuberculosis with phenotypic resistance to fluoroquinolones, aminoglycosides, or both (i.e., patients who were not microbiologically eligible for the shortened regimen). Accordingly, the predictive value of a positive investigational-assay result (resistance detected) for microbiologic unsuitability for the shortened regimen was 81 of 86 (94.2%; 95% CI, 87.0 to 98.1). The predictive value of a negative investigational-assay result (no resistance detected) for microbiologic eligibility for the shortened regimen was 48 of 59 (81.4%; 95% CI, 69.1 to 90.3).
DISCUSSION
In this study, we assessed the clinical diagnostic accuracy of a cartridge-based, automated assay for the rapid detection, directly from sputum specimens with the use of the GeneXpert platform, of M. tuberculosis mutations that are associated with resistance to isoniazid, fluoroquinolones, and aminoglycosides. This assay identified known mutations in genetic regions associated with resistance to these drugs with reasonable certainty; the sensitivity estimates were lower when phenotypic culture-based drug-susceptibility testing was used as the reference standard. Among patients with rifampin-resistant tuberculosis, the predictive value of a positive investigational-assay result for resistance to fluoroquinolones, aminoglycosides, or both was 94%. Although we did not test this strategy, we speculate that a positive test result could be used to triage patients away from the new shortened treatment regimen for multidrug-resistant tuberculosis and toward treatment centers with the capacity for comprehensive drug-susceptibility testing and with experience treating highly drug-resistant tuberculosis.
The WHO has set targets for the diagnostic sensitivity and specificity of next-generation molecular drug-susceptibility tests, with sequencing used as the reference standard, of 95% and 98%, respectively.24 The investigational assay described here met the sensitivity target for isoniazid, fluoroquinolones, and amikacin and missed the sensitivity target for kanamycin by approximately 2 percentage points. The investigational assay met the specificity target for all tested drugs.
Not surprisingly, the sensitivity of the investigational genotypic assay to detect resistance was lower when culture-based phenotypic drug-susceptibility testing was considered as the reference comparator. Most isolates that were found to be resistant by phenotypic drug-susceptibility testing but were found to be wild-type (and thus susceptible) by the investigational assay were also found to be wild-type by DNA sequencing. Our results are consistent with those reported in studies in which other molecular assays were used for M. tuberculosis drug-susceptibility testing.25–30 There are at least two potential causes of the phenotypic–genotypic discrepancies — alternative molecular mechanisms of resistance, many of which are still unknown, and limitations of the critical-concentration methods used for phenotypic testing, such that up to 5% of wild-type M. tuberculosis strains are categorized as drug-resistant.31
Phenotypic–genotypic discrepancy was the driver of the lower-than-ideal predictive value of a negative investigational test (no resistance detected) for microbiologic eligibility for the shortened regimen. If the investigational assay were used as the sole test for triaging patients with rifampin-resistant tuberculosis onto or away from the shorter treatment regimen for multidrug-resistant tuberculosis, approximately 20% of patients who have tuberculosis with phenotypic resistance to fluoroquinolones, aminoglycosides, or both would be inappropriately triaged to receive the shorter regimen. However, this is predicted to be a shortcoming of any molecular test that interrogates the set of common resistance-associated genetic loci targeted by the investigational assay. Currently, there are no other rapid, point-of-care tests to facilitate evidence-based treatment selection for patients with rifampin-resistant tuberculosis.
We identified sequencing-confirmed resistance mutations in some phenotypically susceptible isolates. This phenomenon was most commonly observed for fluoroquinolones, especially moxifloxacin, and resulted in the investigational assay having a lower specificity than culture when the moxifloxacin critical concentration of 2.0 μg per milliliter was used. This situation was almost always associated with gyrA A90V or S91P mutations, which confer low-level moxifloxacin resistance, with minimum inhibitory concentrations (MICs) of 0.25 to 1.0 μg per milliliter.16,32 Although these MICs are higher than those of wild-type M. tuberculosis, isolates with gyrA A90V or S91P mutations are potentially treatable with increased moxifloxacin dosing,33,34 and these mutations can be distinguished from other gyrA mutations with the use of the investigational assay.11 Heteroresistance was detected by the investigational assay in some phenotypically susceptible isolates. Mixed populations of susceptible and resistant bacilli can be clinically relevant, since the unmasking of drug resistance can occur during treatment.35,36 Thus, molecular diagnostics can provide important information for optimizing treatment that is not revealed by dichotomous results of critical concentration–based phenotypic drug-susceptibility testing and does not become available in a timely manner when agar-proportion-method testing is used.
Our study has several limitations. First, most participants with culture-confirmed tuberculosis had high sputum bacillary burdens, as evidenced by the high proportion of specimens that were found to be positive on smear microscopy; the results of our study will need to be confirmed in more patients with smear-negative tuberculosis and with a reference standard of multiple cultures. However, the completion of all tests from one sputum specimen for most participants minimized any potential effect of between-specimen heterogeneity. Second, the geographic representation of participants and M. tuberculosis strains was limited. However, DNA sequencing confirmed that the infecting organisms in the study population contained a diverse set of mutations that are prevalent in other geographic areas. Third, although we did not evaluate capreomycin, a cyclic peptide second-line injectable drug, the rrs A1401G mutation detected by the investigational assay accounts for the majority of capreomycin resistance when a molecular mechanism is identified.8 Finally, phenotypic drug-susceptibility testing was performed only at critical concentrations; MICs were not established, and therefore the full breadth of phenotypic–genotypic relationships was not assessed.
Our results compare favorably with published results for the newest versions of the Genotype MTBDR line probe assays (Hain Lifescience), although we did not perform Genotype MTBDR assays.28,30,37 The investigational assay and Genotype MTBDRsl, version 2.0, share most genetic targets for the detection of fluoroquinolone and aminoglycoside resistance.28,37 The line-probe assays, which are recommended by the WHO to assess candidacy for the shortened treatment regimen for multidrug-resistant tuberculosis, require a minimum of 4 to 6 hours before a result is obtained, which typically precludes same-day therapeutic decision-making; are largely confined to reference centers that meet laboratory-infrastructure and assay training requirements; and are inadequately sensitive when used to test smear-negative sputum specimens from patients with tuberculosis.30,38 In contrast, a result of the investigational assay can be obtained in 2 hours, the assay was found to preserve sensitivity when applied to smear-negative sputum, and it incorporates the same single-step specimen processing as the Xpert MTB/RIF, which can be implemented with minimal staff training and biosafety requirements.2 Existing GeneXpert instruments have the potential to be upgraded to run both the investigational assay and the Xpert MTB/RIF assay by upgrading software and performing a 10-color calibration, allowing both assays to be run contemporaneously with one instrument. These features should permit the investigational assay to be used in peripheral sectors of the global health care system, where more rapid identification of extended drug resistance may improve therapeutic decision-making.
Supplementary Material
Acknowledgments
Supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services (contract HHSN2722000900050C and K24AI104830, to Dr. Dorman), the Intramural Research Program of the NIAID (support to Dr. Barry), and a grant from the Ministry of Science and Technology of China (2014DFA30340).
We thank Mark Perkins (FIND, Geneva); David Hom (Boston Medical Center) and Ying Cai (NIAID) for logistic support; Drs. Wang Wei, Gao Xia, Yang Rui, and Guolong Zhang for study leadership and participant recruitment and enrollment in China; and Dr. Hongjo Choi for study recruitment and enrollment in South Korea.
APPENDIX
The authors’ full names and academic degrees are as follows: Yingda L. Xie, M.D., Soumitesh Chakravorty, Ph.D., Derek T. Armstrong, M.H.S., Sandra L. Hall, M.P.H., Laura E. Via, Ph.D., Taeksun Song, Ph.D., Xing Yuan, M.D., Xiaoying Mo, Ph.D., Hong Zhu, M.D., Peng Xu, Ph.D., Qian Gao, Ph.D., Myungsun Lee, M.D., Jongseok Lee, Ph.D., Laura E. Smith, M.S., Ray Y. Chen, M.D., Joon Sung Joh, M.D., YoungSoo Cho, M.D., Xin Liu, M.D., Xianglin Ruan, M.D., Lili Liang, M.D., Nila Dharan, M.D., Sang-Nae Cho, D.V.M., Ph.D., Clifton E. Barry III, Ph.D., Jerrold J. Ellner, M.D., Susan E. Dorman, M.D., and David Alland, M.D.
The authors’ affiliations are as follows: the Tuberculosis Research Section, Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda (Y.L.X., L.E.V., R.Y.C., C.E.B.), and Johns Hopkins University School of Medicine, Baltimore (D.T.A., S.E.D.) — both in Maryland; the Center for Emerging and Re-Emerging Pathogens, Rutgers New Jersey Medical School, Newark (S.C., L.E.S., N.D., D.A.); Boston Medical Center and Boston University School of Medicine, Boston (S.L.H., J.J.E.); the International Tuberculosis Research Center, Changwon (T.S., M.L., J.L., S.-N.C.), and the National Medical Center (J.S.J.), Seoul Metropolitan Seobuk Hospital (Y.C.), and the Department of Microbiology, College of Medicine, Yonsei University (S.-N.C.), Seoul — all in South Korea; Henan Provincial Chest Hospital (X.Y., X.M., X.L., X.R., L.L.) and Sino–U.S. Tuberculosis Research Collaboration (H.Z.), Zhengzhou, and Key Laboratory of Medical Molecular Virology of Ministries of Education and Health, School of Basic Medical Science, Fudan University, Shanghai (P.X., Q.G.) — all in China; and the Institute of Infectious Disease and Molecular Medicine and Department of Pathology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa (C.E.B.).
Footnotes
The authors’ full names, academic degrees, and affiliations are listed in the Appendix.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.WHO treatment guidelines for drug-resistant tuberculosis: 2016 update. Geneva: World Health Organization; 2016. ( http://www.who.int/tb/areas-of-work/drug-resistant-tb/MDRTBguidelines2016.pdf) [PubMed] [Google Scholar]
- 2.Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377:1495–505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui Z, Wang J, Lu J, Huang X, Hu Z. Association of mutation patterns in gyrA/B genes and ofloxacin resistance levels in Mycobacterium tuberculosis isolates from East China in 2009. BMC Infect Dis. 2011;11:78. doi: 10.1186/1471-2334-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazbón MH, Brimacombe M, Bobadilla del Valle M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50:2640–9. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi R, Zhang J, Li C, Kazumi Y, Sugawara I. Emergence of ofloxacin resistance in Mycobacterium tuberculosis clinical isolates from China as determined by gyrA mutation analysis using denaturing high-pressure liquid chromatography and DNA sequencing. J Clin Microbiol. 2006;44:4566–8. doi: 10.1128/JCM.01916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh P, Jain A, Dixit P, et al. Prevalence of gyrA and B gene mutations in fluoroquinolone-resistant and -sensitive clinical isolates of Mycobacterium tuberculosis and their relationship with MIC of ofloxacin. J Antibiot (Tokyo) 2015;68:63–6. doi: 10.1038/ja.2014.95. [DOI] [PubMed] [Google Scholar]
- 7.Wang JY, Lee LN, Lai HC, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis isolates: associated genetic mutations and relationship to antimicrobial exposure. J Antimicrob Chemother. 2007;59:860–5. doi: 10.1093/jac/dkm061. [DOI] [PubMed] [Google Scholar]
- 8.Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, Rodwell TC. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One. 2012;7(3):e33275. doi: 10.1371/journal.pone.0033275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maus CE, Plikaytis BB, Shinnick TM. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49:3192–7. doi: 10.1128/AAC.49.8.3192-3197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Li D, Zhao L, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. 2013;45:1255–60. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 11.Chakravorty S, Roh SS, Glass J, et al. Detection of isoniazid-, fluoroquinolone-, amikacin-, and kanamycin-resistant tuberculosis in an automated, multiplexed 10- color assay suitable for point-of-care use. J Clin Microbiol. 2016;55:183–98. doi: 10.1128/JCM.01771-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xpert MTB/RIF. Sunnyvale, CA: Cepheid; Feb, 2015. (package insert) [Google Scholar]
- 13.Slayden RA, Barry CE., III The genetics and biochemistry of isoniazid resistance in mycobacterium tuberculosis. Microbes Infect. 2000;2:659–69. doi: 10.1016/s1286-4579(00)00359-2. [DOI] [PubMed] [Google Scholar]
- 14.Xu C, Kreiswirth BN, Sreevatsan S, Musser JM, Drlica K. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J Infect Dis. 1996;174:1127–30. doi: 10.1093/infdis/174.5.1127. [DOI] [PubMed] [Google Scholar]
- 15.Takiff HE, Salazar L, Guerrero C, et al. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–80. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kambli P, Ajbani K, Sadani M, et al. Correlating Minimum Inhibitory Concentrations of ofloxacin and moxifloxacin with gyrA mutations using the genotype MTBDRsl assay. Tuberculosis (Edinb) 2015;95:137–41. doi: 10.1016/j.tube.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki Y, Katsukawa C, Tamaru A, et al. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J Clin Microbiol. 1998;36:1220–5. doi: 10.1128/jcm.36.5.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kambli P, Ajbani K, Nikam C, et al. Correlating rrs and eis promoter mutations in clinical isolates of Mycobacterium tuberculosis with phenotypic susceptibility levels to the second-line injectables. Int J Mycobacteriol. 2016;5:1–6. doi: 10.1016/j.ijmyco.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 20.Gikalo MB, Nosova EY, Krylova LY, Moroz AM. The role of eis mutations in the development of kanamycin resistance in Mycobacterium tuberculosis isolates from the Moscow region. J Antimicrob Chemother. 2012;67:2107–9. doi: 10.1093/jac/dks178. [DOI] [PubMed] [Google Scholar]
- 21.Kent PT, Kubica GP. Public health mycobacteriology — a guide for the level III laboratory. Atlanta: Centers for Disease Control; 1985. [Google Scholar]
- 22.Laboratory services in tuberculosis control: microscopy. Part 2. Geneva: World Health Organization; 1998. [Google Scholar]
- 23.Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. Geneva: World Health Organization; 2014. ( http://www.who.int/tb/publications/pmdt_companionhandbook/en/) [PubMed] [Google Scholar]
- 24.High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. Geneva: World Health Organization; 2014. ( http://www.who.int/tb/publications/tpp_report/en/) [Google Scholar]
- 25.Molina-Moya B, Lacoma A, Prat C, et al. Diagnostic accuracy study of multiplex PCR for detecting tuberculosis drug resistance. J Infect. 2015;71:220–30. doi: 10.1016/j.jinf.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Said HM, Kock MM, Ismail NA, et al. Evaluation of the GenoType MTBDRsl assay for susceptibility testing of second-line anti-tuberculosis drugs. Int J Tuberc Lung Dis. 2012;16:104–9. doi: 10.5588/ijtld.10.0600. [DOI] [PubMed] [Google Scholar]
- 27.Brossier F, Veziris N, Aubry A, Jarlier V, Sougakoff W. Detection by GenoType MTBDRsl test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2010;48:1683–9. doi: 10.1128/JCM.01947-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tagliani E, Cabibbe AM, Miotto P, et al. Diagnostic performance of the new version (v2. 0) of GenoType MTBDRsl assay for detection of resistance to fluoroquinolones and second-line injectable drugs: a multicenter study. J Clin Microbiol. 2015;53:2961–9. doi: 10.1128/JCM.01257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillemann D, Rüsch-Gerdes S, Richter E. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2009;47:1767–72. doi: 10.1128/JCM.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomasicchio M, Theron G, Pietersen E, et al. The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Sci Rep. 2016;6:17850. doi: 10.1038/srep17850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ängeby K, Juréen P, Kahlmeter G, Hoffner SE, Schön T. Challenging a dogma: antimicrobial susceptibility testing breakpoints for Mycobacterium tuberculosis. Bull World Health Organ. 2012;90:693–8. doi: 10.2471/BLT.11.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirgel FA, Warren RM, Streicher EM, Victor TC, van Helden PD, Böttger EC. gyrA Mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother. 2012;67:1088–93. doi: 10.1093/jac/dks033. [DOI] [PubMed] [Google Scholar]
- 33.Poissy J, Aubry A, Fernandez C, et al. Should moxifloxacin be used for the treatment of extensively drug-resistant tuberculosis? An answer from a murine model. Antimicrob Agents Chemother. 2010;54:4765–71. doi: 10.1128/AAC.00968-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigouts L, Coeck N, Gumusboga M, et al. Specific gyrA gene mutations predict poor treatment outcome in MDR-TB. J Antimicrob Chemother. 2016;71:314–23. doi: 10.1093/jac/dkv360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Rie A, Victor TC, Richardson M, et al. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med. 2005;172:636–42. doi: 10.1164/rccm.200503-449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hingley-Wilson SM, Casey R, Connell D, et al. Undetected multidrug-resistant tuberculosis amplified by first-line therapy in mixed infection. Emerg Infect Dis. 2013;19:1138–41. doi: 10.3201/eid1907.130313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brossier F, Guindo D, Pham A, et al. Performance of the new version (v2. 0) of the GenoType MTBDRsl test for detection of resistance to second-line drugs in multi-drug-resistant Mycobacterium tuberculosis complex strains. J Clin Microbiol. 2016;54:1573–80. doi: 10.1128/JCM.00051-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs. Geneva: World Health Organization; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.