Abstract
The generation of reactive metabolites from therapeutic agents is one of the major mechanisms of drug-induced liver injury (DILI). In order to evaluate metabolism-related toxicity and improve drug efficacy and safety, we generated a battery of HepG2-derived cell lines that express 14 cytochrome P450s (CYPs) (1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5 and 3A7) individually using a lentiviral expression system. The expression/production of a specific CYP in each cell line was confirmed by an increased abundance of the CYP at both mRNA and protein levels. Moreover, the enzymatic activities of representative CYPs in the corresponding cell lines were also measured. Using our CYP-expressed HepG2 cells, the toxicity of three drugs that could induce DILI (amiodarone, chlorpromazine and primaquine) was assessed, and all of them showed altered (increased or decreased) toxicity compared to the toxicity in drug-treated wild-type HepG2 cells. CYP-mediated drug toxicity examined in our cell system is consistent with previous reports, demonstrating the potential of these cells for assessing metabolism-related drug toxicity. This cell system provides a practical in vitro approach for drug metabolism screening and for early detection of drug toxicity. It is also a surrogate enzyme source for the enzymatic characterization of a particular CYP that contributes to drug-induced liver toxicity.
Keywords: Drug-induced liver toxicity, HepG2, Drug metabolizing enzyme, Cytochrome P450s
1. Introduction
Drug-induced liver injury (DILI) is a leading cause of drug failures in clinical trials and the major reason of drug withdrawals from the market [1]. Identification of drugs that cause liver injury at the early stage of drug development poses a challenge to both the pharmaceutical industry and the U.S. Food and Drug Administration (FDA). Preclinical testing is one of the important approaches to early detect drug toxicity, and there is a constant need for the development of improved tools to facilitate toxicity assessment and risk identification.
The causes of DILI are multifactorial, including toxic effects caused by reactive metabolites, reactive oxygen species, inflammatory reactions, mitochondrial dysfunction, and imbalances between cellular damage and protective responses [1–5]. Metabolism-related toxicity is generally mediated by the generation of reactive metabolites from non-toxic parenteral compounds, particularly via cytochrome P450 (CYP) enzyme pathways [6]. The inter-individual variability in the expression of drug metabolizing genes predisposes certain individuals to increased susceptibility to DILI [7, 8]. Therefore, it is important to examine the roles of drug metabolizing enzymes and identify specific metabolizing enzymes that contribute to drug-induced liver toxicity.
Numerous in vitro models, such as recombinant enzymes, liver microsomes, liver cytosolic fractions, hepatic cells, liver slices and isolated perfused livers, have been used to examine drug-related hepatotoxicity [9]. Traditionally, cell-based assays have been performed using human primary hepatocytes, either freshly isolated or cryopreserved, to evaluate drug metabolism and drug–drug interactions [10, 11]. Indeed, the application of primary human hepatocytes in drug metabolism and toxicity studies is considered as a “gold standard”, because, under appropriate conditions, these cells retain functional activity of the major drug-metabolizing enzymes [12]. However, phenotypic instability, short life span, batch-to-batch variation and limited availability of primary human hepatocytes constrain their broad use.
Human hepatoma cell lines, such as HepG2, Hep3B, and Huh7, have been widely used in toxicity screening and mechanistic studies, owing to their high stability, unlimited life-span and ready availability. However, lower or no expression of the majority of drug-metabolizing genes is the most critical drawback associated with using these cell lines for drug metabolism and toxicity studies [13, 14]. As a strategy to overcome this limitation, genetically modified hepatic cell lines expressing human drug metabolizing genes have been developed and used for assessing drug metabolism and toxicity. For example, using adenoviral or lentiviral infection systems, cells that transiently or stably express individual CYPs, such as CYP1A1, CYP2C8, CYP2C9 or CYP3A4, have been generated [15–18]. These cells responded appropriately to known toxic chemicals, demonstrating their values for toxicity testing and mechanistic studies. However, not all of them are publicly available.
In this study, we aimed to develop a comprehensive set of cell lines that express the major human CYPs individually, to provide surrogate hepatic cell lines for the study of metabolism-mediated drug hepatotoxicity and the identification of specific CYP isoforms responsible for the metabolism of a drug. Toward this goal, using the lentiviral expression system, HepG2-derived cell lines expressing 14 individual CYPs (1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5 and 3A7) were generated, and the functionality of these CYPs was confirmed at the mRNA, protein, and enzymatic activity levels. In addition, three drugs that could cause metabolism-mediated DILI were examined to evaluate the utility of these cells in drug metabolism and toxicity screening.
2. Materials and methods
2.1. Chemicals and reagents
Dulbecco’s modified Eagle’s medium (DMEM), amiodarone hydrochloride, chlorpromazine hydrochloride, primaquine bisphosphate, proadifen (SKF-525A, SKF), alpha-naphthoflavone (ANF), ketoconazole (KET) and dimethyl sulfoxide (DMSO) were purchased from Sigma–Aldrich (St. Louis, MO). Fetal bovine serum (FBS) was from Atlanta Biologicals (Lawrenceville, GA). Blasticidin S hydrochloride and antibiotic-antimycotic were from Life Technologies (Grand Island, NY).
2.2. Cell culture
The human hepatocellular carcinoma cell line HepG2 was purchased from American Type Culture Collection (ATCC; Manassas, VA). HepG2 cells were routinely cultured in high-glucose DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin and 0.25 mg/ml fungizone. Stably transduced HepG2 cells were maintained in the above growth medium supplemented with 2.5 mg/ml blasticidin. The 293T cell line required for lentiviral packaging was obtained from Biosettia Inc. (San Diego, CA) and propagated in high-glucose DMEM supplemented with 10% FBS, 1% penicillin/streptomycin/fungizone, 1 mM sodium pyruvate and 0.1mM non-essential amino acids. All cells were grown at 37 °C in a humidified atmosphere with 5% CO2. Unless otherwise described, cells were seeded at a density of 3–5 × 105 cells/ml in 96-well tissue culture plates, 60-mm or 10-cm tissue culture dishes. Cells were allowed to adhere for 24 h prior to treatment with drugs or DMSO vehicle.
Primary human hepatocytes from 3 anonymous donors were obtained through the Liver Tissue Cell Distribution System (Pittsburgh, PA). Demographic information of all donors is shown in Supplemental Table 1. The isolation of human hepatocytes was conducted as described previously [19]. Hepatocytes were plated on collagen-coated T-25 flasks at a density of approximately 106 cells. Upon arrival, the shipping medium was replaced with serum-free hepatocyte maintenance medium supplemented with insulin and GA-1000 using an HMM SingleQuots kit (Lonza Walkersville, Inc., Walkersville, MD). Hepatocytes were allowed to recover for at least 12 h at 37 °C in a humidified atmosphere with 5% CO2 [14]. The use of primary human hepatocytes was approved by the Research Involving Human Subjects Committee of the U.S. Food and Drug Administration.
2.3. Construction of CYP-expressing HepG2 cell lines
A total of 14 human CYP cDNAs either amplified from cDNA libraries or prepared by gene synthesis were individually subcloned into the lentiviral expression vector pLV-EF1α-MCS-IRES-Bsd (Biosettia) along with an in-frame c-Myc extension added to the carboxyl terminus of each recombinant CYP protein. The identities and structures of each of the recombinant CYP expression vectors were confirmed by nucleotide sequencing. The sequencing primers used are shown in Table 1. EF1α-3′ and IRES-5′ are primers for 5′-and 3′-ends of the CYP gene inserts, respectively. For lentivirus production, 293T cells were seeded at a density of 1.0 × 106 cells per well in 6-well tissue culture plates and incubated overnight. On the following day, each of the 14 recombinant lentiviral CYP expression vectors as well as the lentiviral control vector were co-transfected with three lentiviral packaging vectors (pVSV-G, pRSV-Rev, pMDLg/pRRE) into the cultured 293T cells per well using lip-otransfectamine 2000 reagent (Life Technologies). After 48 h post-transfection, lentiviral supernatants were harvested from each well, filtered through 0.45 µm cellulose acetate filters and stored at −80 °C. Lentiviral titers were determined by infection of HepG2 cells with serial dilutions of lentiviral stocks followed by blasticidin selection. The titers ranged between 106 and 107 infectious units per milliliter. For lentiviral transduction, HepG2 cells were grown to 10–25% confluence in 6-well tissue culture plates, and infectious lentiviral particles were added to cell cultures at a multiplicity of infection of 10 with 4 µg/ml polybrene (Sigma). Transduced cells were selected in the presence of 10 µg/ml blasticidin to generate HepG2 cell lines expressing individual CYPs or the control vector.
Table 1.
Sequencing primers for recombinant CYP constructs.
| Target | Sequence |
|---|---|
| CYP1A1 | Forward: 5′-GGCCCGACCTCTACACCTTC-3′ |
| Reverse: 5′-TTGGATCTTTCTCTGTACCC-3′ | |
| CYP1A2 | Forward: 5′-CCTTCTCCATCGCCTCTGAC-3′ |
| Reverse: 5′-TCTTCCTCTGTATCTCAGGC-3′ | |
| CYP1B1 | Forward: 5′-CTTCACGCGCCAGCCGCGCAGC-3′ |
| Reverse: 5′-GCCAGGACATAGGGCAGGTTGG-3′ | |
| CYP2A6 | Forward: 5′-TCATCGACGCCCTCCGGGGC-3′ |
| Reverse: 5′-TTCATGAGCAGCAAGAAGCC-3′ | |
| CYP2B6 | Forward: 5′-GGCTCAGTGTCTGATAGAGG-3′ |
| Reverse: 5′-TGAGCATGAGCAGGAAGCCG-3′ | |
| CYP2C8 | Forward: 5′-TGGGGAAGAGGAGCATTGAG-3′ |
| Reverse: 5′-TGCTTCAGCAGGAGCAGGAG-3′ | |
| CYP2C9 | Forward: 5′-GACCGTGTTCAAGAGGAAGC-3′ |
| Reverse: 5′-TTCAGCAGGAGAAGGAGAGC-3′ | |
| CYP2C18 | Forward: 5′-CCTGGGCTGTGCTCCCTGCA-3′ |
| Reverse: 5′-TTCAGCAGGAGCAGGAGTCC-3′ | |
| CYP2C19 | Forward: 5′-CCAAGGCTTCACCCTGTGATC-3′ |
| Reverse: 5′-TTCAGCAGGAGAAGGAGAGC-3′ | |
| CYP2D6 | Forward: 5′-GGAGCAGTGGGTGACCGAGG-3′ |
| Reverse: 5′-GATCATGAGCAGGAGGCCCC-3′ | |
| CYP2E1 | Forward: 5′-GGAGGCCCACTTCCTGCTGG-3′ |
| Reverse: 5′-GGTATTTCATGAGAATCAGG-3′ | |
| CYP3A4 | Forward: 5′-TCTGAGGCGGGAAGCAGAGA-3′ |
| Reverse: 5′-GGACATCAGGGTGAGTGGCC-3′ | |
| CYP3A5 | Forward: 5′-TGGTGAGAAACTTGAGGCGG-3′ |
| Reverse: 5′-GGACATCAGGGTGAGTGGCC-3′ | |
| CYP3A7 | Forward: 5′-GTTGGTGAGAAATCTGAGGCGG-3′ |
| Reverse: 5′-CAGTATCATAGGTGGGTGGTGC-3′ | |
| EF1α-3′ | Forward: 5′-TTGGATCTTGGTTCATTCTC-3′ |
| IRES-5′ | Reverse: 5′-AATAACATATAGACAAACGC-3′ |
2.4. RNA extraction and cDNA synthesis
Total RNA was extracted from cells using the RNeasy Mini kit (Qiagen, Valencia, CA). The quantity and purity of RNA were measured with a NanoDrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE). RNA quality was assessed on an Agilent 2100 Bioanalyzer using the RNA 6000 Nano assay (Agilent Technologies, Palo Alto, CA). For mRNA quantification, cDNA was synthesized from 1 µg of total RNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions.
2.5. Quantitative real-time PCR
Quantitative real-time PCR was used to examine the expression of individual CYP enzymes in HepG2 cell lines at the mRNA level. Reactions were performed in a 10-µl volume according to the manufacturer’s protocol of FastStart Universal Probe Master (Rox) (Roche Applied Science, Indianapolis, IN) using a ViiA™ 7 Real-Time PCR system (Applied Biosystems) under the following conditions: 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. The following TaqMan probes (Applied Biosystems) were used: human CYP1A1 (Hs01054797_g1), human CYP1A2 (Hs00167927_m1), human CYP1B1 (Hs00164383_m1), human CYP2A6 (Hs00868409_s1), human CYP2B6 (Hs03044634_m1), human CYP2C8 (Hs02383390_s1), human CYP2C9 (Hs04260376_m1), human CYP2C18 (Hs00426403_m1), human CYP2C19 (Hs00426 380_m1), human CYP2D6 (Hs00164385_m1), human CYP2E1 (Hs00559368_m1), human CYP3A4 (Hs00604506_m1), human CYP3A5 (Hs00241417_m1), human CYP3A7 (Hs00426361_m1), and human β-actin (ACTB, Hs99999903_m1). The comparative threshold cycle (Ct) method [20] was used to determine the relative expression levels of target genes. Ct values were normalized to that of the endogenous control β-actin.
2.6. Western blotting
Western blotting was performed using whole-cell lysates prepared in RIPA buffer (150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 25 mM Tris–HCl pH 7.6) containing Halt protease inhibitor cocktail (Thermo Scientific). The protein concentrations of the lysates were measured using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA). Equivalent amounts of total protein were resolved by SDS-PAGE and immunoblotted with a mouse monoclonal anti-Myc-tag antibody (Cell Signaling Technology, Danvers, MA) followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG; Santa Cruz Biotechnology, Santa Cruz, CA) to confirm CYP protein expression. GAPDH (Santa Cruz Biotechnology) was used to verify equal protein loading.
2.7. CYP activity assay
CYP enzyme activities were measured using P450-Glo assays (Promega, Madison, WI). Briefly, lentivirus-transduced cells were seeded at a density of 5 × 104 cells per well in 96-well white wall/ clear bottom tissue culture plates and incubated overnight. Cells in triplicate wells were washed with PBS and incubated with the appropriate cell permeable luminogenic substrates (100 µM luciferin 6′-chloroethyl ether for CYP1A1 and CYP1B1, 6 µM luciferin-1A2 for CYP1A2, 3 µM luciferin-2B6 for CYP2B6, 150 µM luciferin 6′-methyl ether for CYP2C8, 100 µM 6′-deoxyluciferin for CYP2C9, 10 µM ethylene glycol ester of 6′-deoxyluciferin for CYP2C19, 30 µM ethylene glycol ester of luciferin 6′-methyl ether for CYP2D6, and 3 µM luciferin isopropyl acetal for CYP3A) prepared in 100 µl fresh media or PBS at 37 °C for 30 min-4 h (isoform dependent). A selective inhibitor of CYP1A, alpha-naphthoflavone (10 µM), was pre-incubated with cells prior to the addition of luciferin-ME, luciferin-H EGE and luciferin ME EGE, to reduce non-specific enzymatic reactions in appropriate cases. Luminescence was generated in separate 96-well white plates by addition of 25 µl luciferin detection reagent to an equal volume of supernatants collected from treated cells and incubation for 20 min at room temperature. Luminescent signals were measured with a Synergy 2 multi-mode microplate reader (BioTek, Winooski, VT). Background luminescence determined in a set of empty wells containing no cells was subtracted from sample measurements. The number of viable cells remaining in the original 96-well plates was determined by the CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI), and the luminescence measurements obtained using P450-Glo assays were normalized to the viable cell number to represent enzymatic activities.
2.8. Cytotoxicity assay
Cells were seeded at a density of 5 × 104 cells per well in 100 µl medium in 96-well white wall/clear bottom plates and incubated overnight. Two-fold serial dilutions of the test drugs were prepared and added to the culture medium. The maximum concentrations used in the cytotoxicity assay were at least 100-fold of the therapeutic Cmax (i.e., maximum therapeutic plasma concentration), which was considered a reasonable threshold to differentiate hepatotoxic drugs from non-hepatotoxic drugs. Lower concentrations were used if a cytotoxic effect was observed at the lowest concentration initially tested. In CYP inhibition assays, cells were pretreated with 20 µM SKF, 10 µM ANF or 10 µM KET for 1 h, followed by treatment with the test drugs for 24 h. Cell viability was determined at the end of a 24-h drug exposure period using the CellTiter-Glo assay (Promega). The luminescence was measured using a microplate reader (BioTek). DMSO was used as a vehicle control and the final DMSO concentration in the medium was ≤0.5% (v/v). The percentage of cell viability at each drug concentration was calculated using the formula: % cell viability = (luminescence of drug-treated cells/luminescence of vehicle-treated cells) × 100.
2.9. Statistical analysis
All data analyses were carried out using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Comparisons of two groups were performed by two-tailed unpaired Student’s t-test. The statistical significance level was set at p < 0.05.
3. Results and discussion
The central role of the liver in the metabolism and clearance of drugs makes it a major target of drug toxicity. A large number of drugs have been reported to cause hepatotoxicity, posing a profound challenge to the pharmaceutical industry and regulatory agencies [21, 22]. Although the mechanisms underlying drug-induced hepatotoxicity remain poorly understood, it has been widely accepted that in many cases the initial step triggering the progression of hepatotoxicity involves metabolism of a drug to chemically reactive metabolites, which covalently bind to cellular macromolecules (e.g., proteins, nucleic acids and lipids), resulting in irreversible chemical modification, adduct formation and functional impairment [23–25].
Human hepatoma cell lines are commonly used in vitro models in hepatotoxicity studies due to their ready accessibility, phenotypic stability and unlimited proliferative capacity. However, most hepatoma cell lines express low levels of drug-metabolizing enzymes, particularly CYP enzymes, hampering their use in drug metabolism studies. To confer metabolic competence to those cell lines, new cell models that transiently [15–17, 26–28] or stably [18] overexpress heterologous CYP enzymes have been developed and successfully used in toxicity studies. Of these models, HepG2 is the most widely used host cell line for transfection/transduction, since it retains many liver-specific biosynthetic functions, including those for metabolizing activities [29]. Importantly, HepG2 cells express considerable endogenous levels of NADPH-P450 reductase and cytochrome b5, which are electron transport components required for CYP activity [30]. Recombinant lentivirus is a widely used gene carrier, and the efficient delivery of transgenes via lentiviral vectors to HepG2 cells has been reported [31–33]. In the present study, we developed a panel of metabolically competent HepG2 cell lines by lentiviral-mediated transduction with expression vectors encoding individual human CYP isoforms. To our knowledge, this panel of hepatic cell lines expresses the most comprehensive set of individual human CYPs described thus far for a noncommercial source.
3.1. Establishment of HepG2-derived cells expressing CYPs
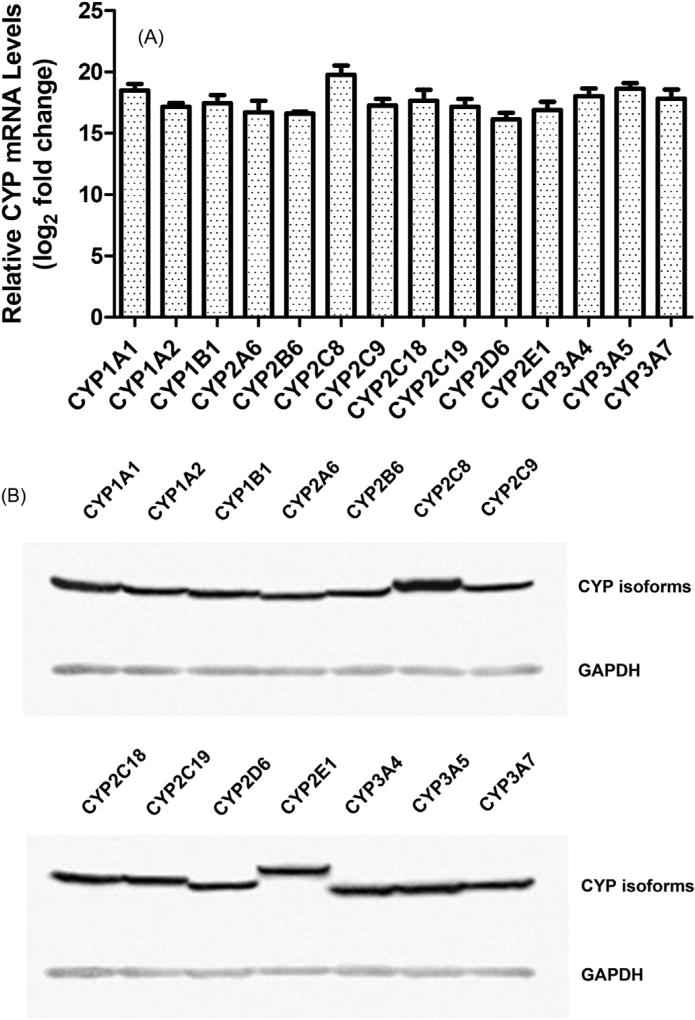
In this study, a recombinant lentivirus system was used to ensure efficient transduction and expression of CYP cDNAs. CYP gene expression was driven by the human EF1a promoter, a housekeeping gene whose promoter is insensitive to transcriptional silencing via DNA methylation [34]. In total, 15 stably transduced HepG2 cell lines were generated, including 14 cell lines transduced with individual CYP cDNAs and another containing an empty vector alone (referred to as EV controls). CYP expression in each HepG2 cell line was characterized. Significantly increased expression of each CYP mRNA in the corresponding cell line was detected by quantitative real-time PCR (Fig. 1A). The average increase in CYP mRNA levels after transduction is about 1.9 × 105 fold, with a ~12-fold difference between the highest and lowest fold changes. The expression of each CYP protein in the corresponding cell line was confirmed by western blotting (Fig. 1B). To examine the functionality of our expression system, the activities of representative CYP isoforms were measured by P450-Glo assays using luminogenic substrates. Compared with EV-transduced control cells, HepG2 cells transduced with CYP cDNAs showed significant increases in enzymatic activity. The fold changes in CYP activity are 92.5 ± 8.9 (CYP1A1), 87.7 ± 5.9 (CYP1A2), 77.6 ± 3.4 (CYP1B1), 71.0 ± 6.0 (CYP2B6), 56.9 ± 1.1 (CYP2C8), 67.9 ± 3.4 (CYP2C9), 47.6 ± 2.4 (CYP2C19), 52.9 ± 2.7 (CYP2D6), 95.9 ± 2.4 (CYP3A4), 68.1 ± 5.6 (CYP3A5), and 50.6 ± 3.4 (CYP3A7), respectively. These results indicate that we have developed a panel of HepG2-derived cell lines that express much higher levels of the introduced human CYPs than HepG2 parental cells. The cytotoxic effects of representative drugs were further assessed in the established cell lines.
Fig. 1.
Characterization of CYP expression in stably transduced HepG2 cells. (A) CYP mRNA expression. The mRNA levels of CYP isoforms in HepG2 cells stably transduced with individual CYP cDNAs or empty vector alone were measured by quantitative realtime PCR. Human β-actin was used as an internal control to normalize the amount of cDNA template. The fold expression of each CYP isoform relative to that in EV controls was determined by a comparative CT method. Data represent the mean ± SD of three independent experiments. (B) CYP protein expression. The protein levels of cDNA-expressed CYP isoforms that range in molecular weight between 45 and 60 kDa were detected by western blotting. GAPDH (37 kDa) was used as a loading control. Similar expression pattern was observed in three independent experiments.
The expression of CYP genes were also examined in primary human hepatocytes from 3 anonymous donors (Supplemental Fig. 1). Inter-individual variations were observed in CYP mRNA levels. The differences in CYP mRNA levels between CYP cell lines and human hepatocytes ranged from 11 to 6 × 104 fold (Supplemental Table 2). It is noteworthy that inter-individual variability of CYP expression can be wide. For example, CYP3A expression in human liver exhibits 10- to >500-fold inter-individual variation [35–37]. The hepatic expression of CYP2A6 varies 20- to >1000-fold among individuals [37–39]. Other CYPs (i.e., CYP1A2, CYP2B6, CYP2C8, CYP2C19 and CYP2D6) have also shown large inter-individual differences (>200-fold) in hepatic expression [37, 40–42]. Considering various environmental influences including chemical-mediated induction and inhibition of CYP expression, the range of such variations can be even wider. Therefore, our CYP cell lines may reflect CYP expression in human liver at certain levels. More importantly, our cell system is not confounded by inter-individual variability that hampers the use of human hepatocytes in the characterization of the contribution of individual CYP isoforms to the metabolism and toxicity of a drug.
3.2. Cytotoxic effects of amiodarone
Amiodarone, an iodinated benzofuran derivative, is one of the most commonly prescribed antiarrhythmic drugs [43]. Despite its effectiveness in the treatment of a broad spectrum of arrhythmias, amiodarone has numerous side effects, including pulmonary toxicity, thyroid toxicity, hepatotoxicity and other types of organ toxicity [44]. Hepatotoxicity, as manifested by liver enzyme abnormalities, has been reported in 14–82% of patients treated with amiodarone [45]. Although the hepatic side effects are generally mild and reversible, they frequently result in drug discontinuation [46]. Fatal cases have occasionally been reported [45, 47], and the prevalence of severe liver injury is estimated at 1–3% [45].
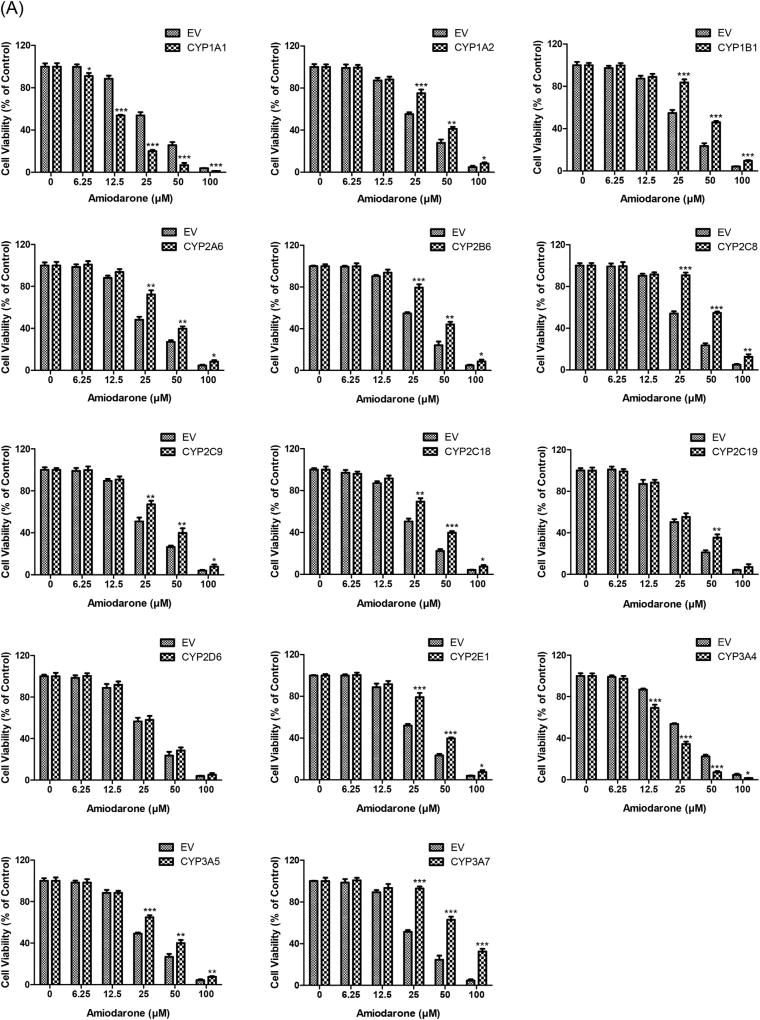
Amiodarone has been considered as a most-DILI-concern drug [48]. The maximum plasma concentration (Cmax) of amiodarone is about 0.85 µM at a therapeutic dose of 400 mg in healthy subjects [49]. In our study, significant cytotoxicity of amiodarone was observed in EV controls at 12.5 µM or higher concentrations (Fig. 2A). Cell viability was remarkably decreased by amiodarone exposure in HepG2 cells overexpressing CYP1A1 or CYP3A4 compared to EV controls, indicating cytotoxic effects due to metabolic activation of amiodarone by these two CYP enzymes. In contrast, cytotoxicity of amiodarone was significantly decreased by the overexpression of other CYP isoforms, except CYP2D6, suggesting that these metabolizing enzymes may contribute to the detoxification of amiodarone. Interestingly, CYP isoforms in the same subfamily may play opposite roles in the metabolism of amiodarone. While the overproduction of CYP1A1 enhanced the cytotoxic effect of amiodarone, the overexpression of CYP1A2 reduced its cytotoxic effect; likewise, whereas the overproduction of CYP3A4 increased the cytotoxic effect, CYP3A5 and CYP3A7 decreased such effect.
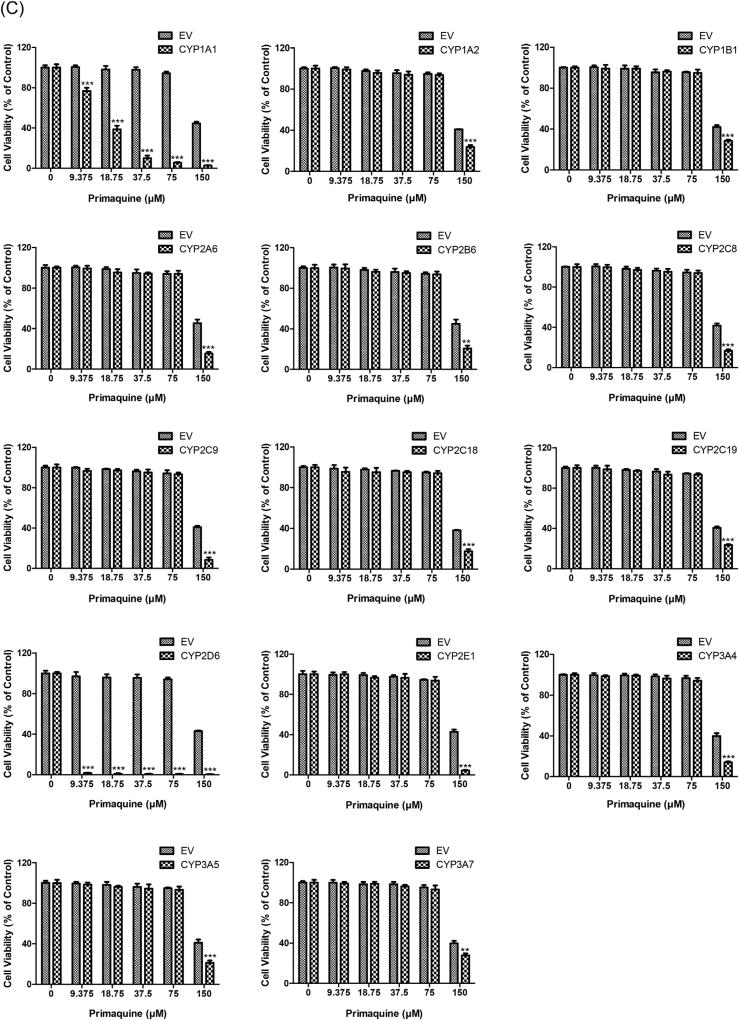
Fig. 2.
Cytotoxicity of amiodarone (A), chlorpromazine (B) and primaquine (C) in HepG2 cells expressing individual human CYPs. CYP cDNAs or EV-transduced HepG2 cells were treated with three drugs at the indicated concentrations for 24 h. Cell viability was determined by the CellTiter-Glo assay after drug exposure. Similar results were observed in three independent experiments. The bar graphs represent mean ± SD of triplicate determinations. Statistical significance compared with EV controls: *p < 0.05, **p < 0.01, and ***p < 0.001.
Amiodarone is primarily metabolized to mono-N-desethylamiodarone (DEA) in humans [50]. DEA is highly lipophilic and widely distributed in organ tissues, particularly in liver and lung [51]. It has been shown that DEA is more toxic to hepatocytes than amiodarone and may contribute to the amiodarone-associated hepatotoxicity, most likely by inhibiting the respiratory chain and producing oxidative stress [52, 53]. A high ratio of DEA/amino-darone in the plasma and liver is considered as a risk factor for hepatotoxicity [54]. There is evidence demonstrating that CYP1A1 and CYP3A4 are the most efficient enzymes transforming amiodarone to DEA and the former isoform has higher potency [55], which is consistent with the results of our cytotoxicity assays. Several other CYP isoforms, including CYP1A2, CYP2C8, CYP2C19, and CYP2D6, have also been involved in DEA production in humans, but we found that the overexpression of these metabolizing enzymes did not increase but even decreased the cytotoxicity of amiodarone, suggesting that they may contribute, even more significantly, in the detoxification of DEA. Recent studies have reported that the active metabolite DEA can be further transformed by hydroxylation to 3′OH–N-desethylamiodarone, dealkylation to di-N-desethylamiodarone and deamination to amiodarone-EtOH, and various CYP isoforms are involved in the elimination of DEA [56, 57]. Further studies are needed to elucidate whether these three minor metabolites of amiodarone are associated with detoxification pathways and to identify the CYPs involved in their formation.
3.3. Cytotoxic effects of chlorpromazine
Chlorpromazine, a dimethylamine derivative of phenothiazine, is one of the four antipsychotic drugs on the World Health Organization (WHO) essential medicine list [58]. Chlorpromazine is commonly associated with acute liver injury, with an incidence greater than 100 per 100,000 users [59–61]. The indication of chlorpromazine-induced liver injury is primarily cholestatic, but the hepatocellular injury marked by elevations in aminotransferase levels has also been reported in some chlorpromazine users [61, 62]. Although the mechanism of chlorpromazine-associated hepatotoxicity has not been elucidated, it has been suggested that reactive metabolites generated through CYP-mediated pathways may play a role [63].
Chlorpromazine has been classified as a less-DILI-concern drug with mild DILI severity [48]. The plasma level of chlorpromazine can reach up to 0.94 µM with a therapeutic dose of 400–800 mg/day [64]. As shown in Fig. 2B, in EV controls, cell viability was markedly decreased as the concentration of chlorpromazine increased to 12.5 µM and higher levels. Moreover, metabolic detoxification of chlorpromazine was apparently mediated by all tested CYP isoforms, as indicated by enhanced cell viability in those CYP-overexpressing HepG2 cells in comparison to EV controls, implicating the involvement of these CYP enzymes in the production of less- or non-toxic metabolites.
Chlorpromazine is known to be extensively metabolized to a number of metabolites, and various CYP enzymes are involved in the biotransformation of chlorpromazine [65, 66]. For instance, CYP2D6 and CYP1A2 mediate the 7-hydroxylation and N-oxidation of chlorpromazine, and the former is the major metabolic pathway for chlorpromazine in humans [67–69]. CYP1A2 and CYP3A4 primarily contribute to 5-sulfoxidation of chlorpromazine, whereas CYP2B6, CYP2C19 and CYP2D6 make a minor contribution [70]. CYP1A2 is also responsible for the mono- and di-N-demethylation of chlorpromazine, while CYP2C19, CYP2B6 and CYP3A4 play notable roles in the mono- and di-N-demethylation at high concentrations of the drug [70]. Some of these metabolites may contribute to chlorpromazine hepatotoxicity [71]. It has been suggested that the hydroxylated metabolites of chlorpromazine are more active than the sulfoxide metabolites in inducing jaundice [72]. It has also been shown that the mono- and di-demethylated metabolites of chlorpromazine are three and six times, respectively, more potent than the parent drug in causing the leakage of aspartate aminotransferase from isolated rat hepatocytes, while 7-and 8-hydroxychlorpromazine are slightly less potent than chlorpromazine, and sulfoxide metabolites are inactive [73]. Our data suggest that chlorpromazine may be inactivated by a series of CYP enzymes, and MacAllister et al. [63] noted that chlorpromazine detoxification in rat hepatocytes also required glutathione and glucuronide pathways. The toxicity of a drug is a result of functional balance among the activation, detoxification, and secretion processes that are regulated by a number of genes involved in phase I, phase II, and phase III metabolism and in drug transport. It is also important to develop a system that expresses the majority of drug metabolizing and transporter genes to investigate hepatic drug metabolism and toxicity as a whole, which is warranted for our future study.
3.4. Cytotoxic effects of primaquine
Primaquine, an 8-aminoquinoline, is the only drug licensed by the U.S. FDA for the radical cure of Plasmodium vivax infection [74]. Despite a unique role in the treatment of vivax malaria, the use of primaquine is limited due to its hemolytic toxicity, particularly in glucose-6-phosphate dehydrogenase (G6PD) deficient individuals [75]. In addition, recurrence of P. vivax occurs in up to 25% of patients following primaquine treatment, resulting from a lack of primaquine efficacy [76]. Although primaquine has been used since the 1950s, the mechanisms underlying its efficacy and toxicity remain elusive. It is thought that interference with the respiratory chain of parasite’s mitochondria or production of redox active metabolite(s) may be involved in the efficacy and toxicity of primaquine [77, 78].
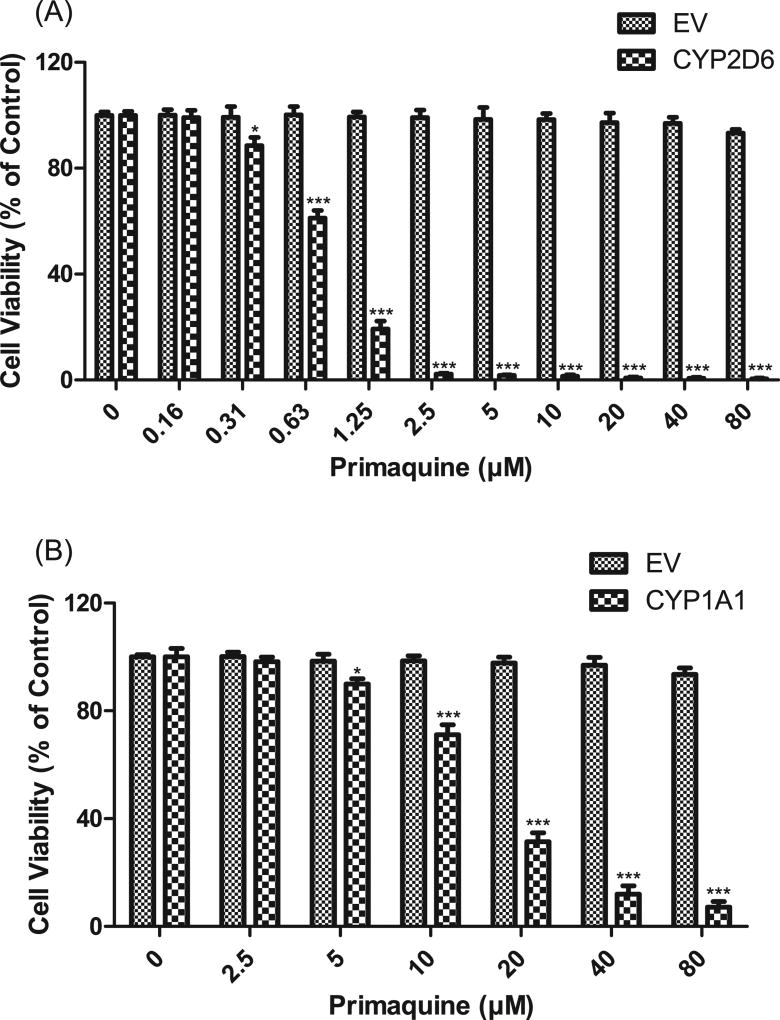
Primaquine has been designated as a no-DILI-concern drug due to the lack of associated DILI events [48]. The maximum plasma concentrations (Cmax) of primaquine achieved in healthy subjects after administration of a therapeutic dose are around 0.20 µM (15 mg base), 0.40 µM (30 mg base) and 0.68 µM (45 mg base) [79]. In our cytotoxicity assays, no toxic effects were observed at concentrations lower than 75 µM (>100-fold Cmax) primaquine in the majority of the HepG2-CYP cell lines (Fig. 2C). However, primaquine caused significant cytotoxicity in HepG2 cells overexpressing CYP2D6 and CYP1A1 in the range of concentrations tested. To determine the threshold concentrations for cytotoxicity in the two cell lines, lower concentrations of primaquine were subsequently employed. A significant decrease of cell viability was observed at as low as 0.31 µM (Fig. 3A) and 5 µM (Fig. 3B) primaquine in HepG2 cells overexpressing CYP2D6 and CYP1A1, respectively. The cytotoxicity of primaquine caused by the overexpression of CYP2D6 may be of pharmacological and toxicological significance since the treatment concentration is relevant to the concentration of the drug in the plasma, which is typically around 0.4 µM.
Fig. 3.
Effects of CYP2D6 (A) and CYP1A1 (B) overexpression on primaquine cytotoxicity. HepG2 cells expressing CYP2D6 or CYP1A1 enzymes and control vector were exposed to various concentrations of primaquine for 24 h. The data are shown as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with EV controls.
The metabolic profile of primaquine has been investigated for decades but is still not fully elucidated. Carboxyprimaquine has been identified as the major metabolite of primaquine in humans, but it lacks antimalarial activity and toxicity [80–82]. On the other hand, hydroxylated metabolites of primaquine, such as 5-hydroxyprimaquine, have been suggested to be responsible for the efficacy and toxicity of the drug, due to their capability of generating reactive oxygen species through redox cycling [78, 83–86]. The CYP system is implicated in the metabolism of primaquine. It has been shown that multiple CYP isoforms, including CYP3A4, CYP2D6, CYP2B6, CYP1A2 and CYP2E1, variably contributed to the oxidative toxicity of primaquine [87]. However, recent studies have demonstrated that CYP2D6 plays a major role in the production of hydroxylated metabolites and makes an essential contribution to the metabolic activation of primaquine [88, 89]. The study by Bennett et al. [90] has revealed that malaria relapse rate is increased with a decreased metabolism of primaquine in poor/intermediate CYP2D6 metabolizers, suggesting that primaquine efficacy may depend on CYP2D6 activity. The significant cytotoxic effect of primaquine mediated by CYP2D6, as observed in our study, provides further evidence that CYP2D6 may have a primary role in generating active metabolites that are responsible for the efficacy and toxicity of primaquine. It is noteworthy that CYP2D6 is highly polymorphic with a substantial inter-individual variability (>50-fold) in activity within a population [91]. The dependence of primaquine bioactivation via a CYP2D6-mediated metabolic pathway highlights the importance of stratifying CYP2D6 metabolizer phenotypes or genotypes prior to primaquine treatment.
Interestingly, we also observed a remarkable cytotoxicity of primaquine in HepG2 cells overexpressing CYP1A1, which might be related to an increased stability of CYP1A1 transcripts by the primaquine treatment. Several studies showed that primaquine upregulated CYP1A1 expression at transcriptional and post-transcriptional levels [92–94]. Notably, Werlinder et al. [93] demonstrated that primaquine inhibited the degradation of CYP1A1 in a dose-dependent manner. In their study, CYP1A1 degradation was dramatically inhibited by 10 µM or higher concentrations of primaquine, which is consistent with our observation that primaquine exhibited remarkable cytotoxic effects at concentrations higher than 10 µM in CYP1A1-overexpressing cells. The binding of primaquine at the active site of CYP1A1 has been postulated as a mechanism by which the drug protects CYP1A1 degradation. However, we did not observe the same effect for CYP1A2, suggesting that the regulatory mechanism may be specific for CYP1A1.
3.5. Effects of CYP inhibitors
The role of CYP inhibitors was examined in several CYP cell lines. SKF is a widely used, non-specific CYP inhibitor. ANF and KET are isoenzyme-specific CYP inhibitors for CYP1A and CYP3A, respectively. Pretreatment with ANF reduced the cytotoxicity of amiodarone in CYP1A1-overexpressing HepG2 cells as well as the cytotoxicity of primaquine in HepG2 cells expressing CYP1A1 or CYP1A2, while ANF-induced CYP inhibition enhanced the cytotoxicity of amiodarone in CYP1A2-overexpressing HepG2 cells (Supplemental Fig. 2A). Besides, KET-induced CYP inhibition alleviated the cytotoxicity induced by the overexpression of CYP3A4 and increased the cytotoxicity induced by the overproduction of CYP3A5 or CYP3A7 (Supplemental Fig. 2B). Moreover, pretreatment with SKF decreased the cytotoxicity induced by the overexpression of CYP2D6 at lower concentrations (Supplemental Fig. 2C). These results confirmed the involvement of CYP isoforms in the cytotoxicity induced by the test drugs.
4. Summary
In summary, we generated a metabolically competent cell system by stable transduction of CYP cDNAs individually. We also showed the potential of the system for in vitro screening of metabolism-related drug toxicity. Such an in vitro assay system represents a useful paradigm for the evaluation and early detection of drug toxicity. Considering the large variations in metabolic phenotypes observed within and among human populations that impact the use of normal human hepatocytes in pharmacological studies in vitro, the use of a panel of cells that allows the characterization of the role of a specific metabolizing enzyme in drug biotransformation could be critical to avoid confounding issues due to inter-individual variability. The battery of cell lines described in this report, which express CYPs separately, could be a promising tool for the screening of drug metabolism and the assessment of the contribution of a particular CYP to drug-induced hepatotoxicity. Our cell system is readily available for further drug metabolism and hepatotoxicity studies.
Supplementary Material
Acknowledgments
JX and SC were supported by appointments to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science Education through an interagency agreement between the U.S. Department of Energy and the U.S. FDA.
This article is not an official guidance or policy statement of the U.S. FDA. No official support or endorsement by the U.S. FDA is intended or should be inferred.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.cbi.2015.10.009.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.cbi.2015.10.009.
References
- 1.Kaplowitz N. Idiosyncratic drug hepatotoxicity, nature reviews. Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 2.Jaeschke H, McGill MR, Ramachandran A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: lessons learned from acetaminophen hepatotoxicity. Drug Metab. Rev. 2012;44:88–106. doi: 10.3109/03602532.2011.602688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ju C, Reilly T. Role of immune reactions in drug-induced liver injury (DILI) Drug Metab. Rev. 2012;44:107–115. doi: 10.3109/03602532.2011.645579. [DOI] [PubMed] [Google Scholar]
- 4.Yuan L, Kaplowitz N. Mechanisms of drug-induced liver injury. Clin. Liver Dis. 2013;17:507–518. vii. doi: 10.1016/j.cld.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S, Xuan J, Couch L, Iyer A, Wu Y, Li QZ, Guo L. Sertraline induces endoplasmic reticulum stress in hepatic cells. Toxicology. 2014;322:78–88. doi: 10.1016/j.tox.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunawan BK, Kaplowitz N. Mechanisms of drug-induced liver disease. Clin. Liver Dis. 2007;11:459–475. v. doi: 10.1016/j.cld.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Andrade RJ, Agundez JA, Lucena MI, Martinez C, Cueto R, Garcia-Martin E. Pharmacogenomics in drug induced liver injury. Curr. Drug Metab. 2009;10:956–970. doi: 10.2174/138920009790711805. [DOI] [PubMed] [Google Scholar]
- 8.Russmann S, Jetter A, Kullak-Ublick GA. Pharmacogenetics of drug-induced liver injury. Hepatology. 2010;52:748–761. doi: 10.1002/hep.23720. [DOI] [PubMed] [Google Scholar]
- 9.Huang SM, Strong JM, Zhang L, Reynolds KS, Nallani S, Temple R, Abraham S, Habet SA, Baweja RK, Burckart GJ, Chung S, Colangelo P, Frucht D, Green MD, Hepp P, Karnaukhova E, Ko HS, Lee JI, Marroum PJ, Norden JM, Qiu W, Rahman A, Sobel S, Stifano T, Thummel K, Wei XX, Yasuda S, Zheng JH, Zhao H, Lesko LJ. New era in drug interaction evaluation: US food and drug administration update on CYP enzymes, transporters, and the guidance process. J. Clin. Pharmacol. 2008;48:662–670. doi: 10.1177/0091270007312153. [DOI] [PubMed] [Google Scholar]
- 10.LeCluyse EL. Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur. J. Pharm. Sci. 2001;13:343–368. doi: 10.1016/s0928-0987(01)00135-x. [DOI] [PubMed] [Google Scholar]
- 11.Li AP. Evaluation of drug metabolism, drug-drug interactions, and in vitro hepatotoxicity with cryopreserved human hepatocytes. Methods Mol. Biol. 2010;640:281–294. doi: 10.1007/978-1-60761-688-7_15. [DOI] [PubMed] [Google Scholar]
- 12.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger J, Braeuning A, Budinsky RA, Burkhardt B, Cameron NR, Camussi G, Cho CS, Choi YJ, Craig Rowlands J, Dahmen U, Damm G, Dirsch O, Donato MT, Dong J, Dooley S, Drasdo D, Eakins R, Ferreira KS, Fonsato V, Fraczek J, Gebhardt R, Gibson A, Glanemann M, Goldring CE, Gomez-Lechon MJ, Groothuis GM, Gustavsson L, Guyot C, Hallifax D, Hammad S, Hayward A, Haussinger D, Hellerbrand C, Hewitt P, Hoehme S, Holzhutter HG, Houston JB, Hrach J, Ito K, Jaeschke H, Keitel V, Kelm JM, Kevin Park B, Kordes C, Kullak-Ublick GA, LeCluyse EL, Lu P, Luebke-Wheeler J, Lutz A, Maltman DJ, Matz-Soja M, McMullen P, Merfort I, Messner S, Meyer C, Mwinyi J, Naisbitt DJ, Nussler AK, Olinga P, Pampaloni F, Pi J, Pluta L, Przyborski SA, Ramachandran A, Rogiers V, Rowe C, Schelcher C, Schmich K, Schwarz M, Singh B, Stelzer EH, Stieger B, Stober R, Sugiyama Y, Tetta C, Thasler WE, Vanhaecke T, Vinken M, Weiss TS, Widera A, Woods CG, Xu JJ, Yarborough KM, Hengstler JG. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepa-totoxicity, cell signaling and ADME. Arch. Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donato MT, Lahoz A, Castell JV, Gomez-Lechon MJ. Cell lines: a tool for in vitro drug metabolism studies. Curr. Drug Metab. 2008;9:1–11. doi: 10.2174/138920008783331086. [DOI] [PubMed] [Google Scholar]
- 14.Guo L, Dial S, Shi L, Branham W, Liu J, Fang JL, Green B, Deng H, Kaput J, Ning B. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab. Dispos. Biol. Fate Chem. 2011;39:528–538. doi: 10.1124/dmd.110.035873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwamura A, Fukami T, Hosomi H, Nakajima M, Yokoi T. CYP2C9-mediated metabolic activation of losartan detected by a highly sensitive cell-based screening assay. Drug Metab. Dispos. Biol. Fate Chem. 2011;39:838–846. doi: 10.1124/dmd.110.037259. [DOI] [PubMed] [Google Scholar]
- 16.Zahno A, Brecht K, Morand R, Maseneni S, Torok M, Lindinger PW, Krahenbuhl S. The role of CYP3A4 in amiodarone-associated toxicity on HepG2 cells. Biochem. Pharmacol. 2011;81:432–441. doi: 10.1016/j.bcp.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Hosomi H, Fukami T, Iwamura A, Nakajima M, Yokoi T. Development of a highly sensitive cytotoxicity assay system for CYP3A4-mediated metabolic activation. Drug Metab. Dispos. Biol. Fate Chem. 2011;39:1388–1395. doi: 10.1124/dmd.110.037077. [DOI] [PubMed] [Google Scholar]
- 18.Yoshitomi S, Ikemoto K, Takahashi J, Miki H, Namba M, Asahi S. Establishment of the transformants expressing human cytochrome P450 subtypes in HepG2, and their applications on drug metabolism and toxicology. Toxicol. vitro Int. J. Publ. Assoc. BIBRA. 2001;15:245–256. doi: 10.1016/s0887-2333(01)00011-x. [DOI] [PubMed] [Google Scholar]
- 19.Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Methods Enzymol. 1996;272:388–401. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Lee WM. Drug-induced hepatotoxicity. N. Engl. J. Med. 2003;349:474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- 22.Gunawan B, Kaplowitz N. Clinical perspectives on xenobiotic-induced hep-atotoxicity. Drug Metab. Rev. 2004;36:301–312. doi: 10.1081/dmr-120034148. [DOI] [PubMed] [Google Scholar]
- 23.Antoine DJ, Williams DP, Park BK. Understanding the role of reactive metabolites in drug-induced hepatotoxicity: state of the science. Expert Opin. Drug Metab. Toxicol. 2008;4:1415–1427. doi: 10.1517/17425255.4.11.1415. [DOI] [PubMed] [Google Scholar]
- 24.Attia SM. Deleterious effects of reactive metabolites. Oxidative Med. Cell. Longev. 2010;3:238–253. doi: 10.4161/oxim.3.4.13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava A, Maggs JL, Antoine DJ, Williams DP, Smith DA, Park BK. Role of Reactive Metabolites in Drug-induced Hepatotoxicity. Handbook of Experimental Pharmacology. 2010:165–194. doi: 10.1007/978-3-642-00663-0_7. [DOI] [PubMed] [Google Scholar]
- 26.Bai J, Cederbaum AI. Adenovirus mediated overexpression of CYP2E1 increases sensitivity of HepG2 cells to acetaminophen induced cytotoxicity. Mol. Cell Biochem. 2004;262:165–176. doi: 10.1023/b:mcbi.0000038232.61760.9e. [DOI] [PubMed] [Google Scholar]
- 27.Vignati L, Turlizzi E, Monaci S, Grossi P, Kanter R, Monshouwer M. An in vitro approach to detect metabolite toxicity due to CYP3A4-dependent bioactivation of xenobiotics. Toxicology. 2005;216:154–167. doi: 10.1016/j.tox.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Yan QG, Shi JG, Zhang F, Zhao QT, Pang XW, Chen R, Hu PZ, Li QL, Wang Z, Huang GS. Overexpression of CYP2E1 enhances sensitivity of hepG2 cells to fas-mediated cytotoxicity. Cancer Biol. Ther. 2008;7:1280–1287. doi: 10.4161/cbt.7.8.6283. [DOI] [PubMed] [Google Scholar]
- 29.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez FJ, Korzekwa KR. Cytochromes P450 expression systems. Annu. Rev. Pharmacol. Toxicol. 1995;35:369–390. doi: 10.1146/annurev.pa.35.040195.002101. [DOI] [PubMed] [Google Scholar]
- 31.Cockrell AS, Kafri T. Gene delivery by lentivirus vectors. Mol. Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- 32.Zamule SM, Strom SC, Omiecinski CJ. Preservation of hepatic phenotype in lentiviral-transduced primary human hepatocytes. Chemico-Biological Interact. 2008;173:179–186. doi: 10.1016/j.cbi.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasri M, Karimi A, Allahbakhshian Farsani M. Production, purification and titration of a lentivirus-based vector for gene delivery purposes. Cytotechnology. 2014;66:1031–1038. doi: 10.1007/s10616-013-9652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skalamera D, Dahmer M, Purdon AS, Wilson BM, Ranall MV, Blumenthal A, Gabrielli B, Gonda TJ. Generation of a genome scale lentiviral vector library for EF1alpha promoter-driven expression of human ORFs and identification of human genes affecting viral titer. PLoS One. 2012;7:e51733. doi: 10.1371/journal.pone.0051733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thummel KE, Shen DD, Podoll TD, Kunze KL, Trager WF, Bacchi CE, Marsh CL, McVicar JP, Barr DM, Perkins JD, et al. Use of midazolam as a human cytochrome P450 3A probe: II. Characterization of inter- and intra-individual hepatic CYP3A variability after liver transplantation. J. Pharmacol. Exp. Ther. 1994;271:557–566. [PubMed] [Google Scholar]
- 36.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A–mediated metabolism. Adv. Drug Deliv. Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Price ET, Chang CW, Li Y, Huang Y, Guo LW, Guo Y, Kaput J, Shi L, Ning B. Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS One. 2013;8:e60368. doi: 10.1371/journal.pone.0060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamano S, Tatsuno J, Gonzalez FJ. The CYP2A3 gene product catalyzes coumarin 7-hydroxylation in human liver microsomes. Biochemistry. 1990;29:1322–1329. doi: 10.1021/bi00457a031. [DOI] [PubMed] [Google Scholar]
- 39.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 40.Rodriguez-Antona C, Donato MT, Pareja E, Gomez-Lechon MJ, Castell JV. Cytochrome P-450 mRNA expression in human liver and its relationship with enzyme activity. Arch. Biochem. Biophys. 2001;393:308–315. doi: 10.1006/abbi.2001.2499. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Tompkins LM. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr. Drug Metab. 2008;9:598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen MR, Farin FM, Omiecinski CJ. Quantification of multiple human cytochrome P450 mRNA molecules using competitive reverse transcriptase-PCR. DNA Cell Biol. 1998;17:231–238. doi: 10.1089/dna.1998.17.231. [DOI] [PubMed] [Google Scholar]
- 43.Vassallo P, Trohman RG. Prescribing amiodarone: an evidence-based review of clinical indications. J. Am. Med. Assoc. 2007;298:1312–1322. doi: 10.1001/jama.298.11.1312. [DOI] [PubMed] [Google Scholar]
- 44.Santangeli P, Di Biase L, Burkhardt JD, Bai R, Mohanty P, Pump A, Natale A. Examining the safety of amiodarone. Expert Opin. Drug Saf. 2012;11:191–214. doi: 10.1517/14740338.2012.660915. [DOI] [PubMed] [Google Scholar]
- 45.Lewis JH, Ranard RC, Caruso A, Jackson LK, Mullick F, Ishak KG, Seeff LB, Zimmerman HJ. Amiodarone hepatotoxicity: prevalence and clinicopatho-logic correlations among 104 patients. Hepatology. 1989;9:679–685. doi: 10.1002/hep.1840090504. [DOI] [PubMed] [Google Scholar]
- 46.Babatin M, Lee SS, Pollak PT. Amiodarone hepatotoxicity. Curr. Vasc. Pharmacol. 2008;6:228–236. doi: 10.2174/157016108784912019. [DOI] [PubMed] [Google Scholar]
- 47.Richer M, Robert S. Fatal hepatotoxicity following oral administration of amiodarone. Ann. Pharmacother. 1995;29:582–586. doi: 10.1177/106002809502900605. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Vijay V, Shi Q, Liu Z, Fang H, Tong W. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov. Today. 2011;16:697–703. doi: 10.1016/j.drudis.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Pourbaix S, Berger Y, Desager JP, Pacco M, Harvengt C. Absolute bioavail-ability of amiodarone in normal subjects. Clin. Pharmacol. Ther. 1985;37:118–123. doi: 10.1038/clpt.1985.22. [DOI] [PubMed] [Google Scholar]
- 50.Flanagan RJ, Storey GC, Holt DW, Farmer PB. Identification and measurement of desethylamiodarone in blood plasma specimens from amiodarone-treated patients. J. Pharm. Pharmacol. 1982;34:638–643. doi: 10.1111/j.2042-7158.1982.tb04692.x. [DOI] [PubMed] [Google Scholar]
- 51.Brien JF, Jimmo S, Brennan FJ, Ford SE, Armstrong PW. Distribution of amiodarone and its metabolite, desethylamiodarone, in human tissues. Can. J. Physiol. Pharmacol. 1987;65:360–364. doi: 10.1139/y87-062. [DOI] [PubMed] [Google Scholar]
- 52.Gross SA, Bandyopadhyay S, Klaunig JE, Somani P. Amiodarone and dese-thylamiodarone toxicity in isolated hepatocytes in culture, Proceedings of the Society for Experimental Biology and Medicine. Soc. Exp. Biol. Med. 1989;190:163–169. doi: 10.3181/00379727-190-42844. [DOI] [PubMed] [Google Scholar]
- 53.Waldhauser KM, Torok M, Ha HR, Thomet U, Konrad D, Brecht K, Follath F, Krahenbuhl S. Hepatocellular toxicity and pharmacological effect of amiodarone and amiodarone derivatives. J. Pharmacol. Exp. Ther. 2006;319:1413–1423. doi: 10.1124/jpet.106.108993. [DOI] [PubMed] [Google Scholar]
- 54.Ratz Bravo AE, Drewe J, Schlienger RG, Krahenbuhl S, Pargger H, Ummenhofer W. Hepatotoxicity during rapid intravenous loading with amiodarone: description of three cases and review of the literature. Crit. Care Med. 2005;33:128–134. doi: 10.1097/01.ccm.0000151048.72393.44. discussion 245–126. [DOI] [PubMed] [Google Scholar]
- 55.Elsherbiny ME, El-Kadi AO, Brocks DR. The metabolism of amiodarone by various CYP isoenzymes of human and rat, and the inhibitory influence of ketoconazole. J. Pharm. Pharm. Sci. Publ. Can. Soc. Pharm. Sci. Soc. Can. des Sci. Pharm. 2008;11:147–159. doi: 10.18433/j3sg66. [DOI] [PubMed] [Google Scholar]
- 56.Kozlik P. Metabolism of Amiodarone-biotransformation of Mono-N-Desethylamiodarone In-vitro. ETH Zurich; Switzerland: 2003. [Google Scholar]
- 57.Ha HR, Bigler L, Wendt B, Maggiorini M, Follath F. Identification and quantitation of novel metabolites of amiodarone in plasma of treated patients. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2005;24:271–279. doi: 10.1016/j.ejps.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 58.WHO. WHO Model List of Essential Medicines. (eighteenth) 2013 http://www.who.int/medicines/publications/essentialmedicines/en/
- 59.Garcia Rodriguez LA, Ruigomez A, Jick H. A review of epidemiologic research on drug-induced acute liver injury using the general practice research data base in the United Kingdom. Pharmacotherapy. 1997;17:721–728. [PubMed] [Google Scholar]
- 60.de Abajo FJ, Montero D, Madurga M, Garcia Rodriguez LA. Acute and clinically relevant drug-induced liver injury: a population based case-control study. Br. J. Clin. Pharmacol. 2004;58:71–80. doi: 10.1111/j.1365-2125.2004.02133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sabate M, Ibanez L, Perez E, Vidal X, Buti M, Xiol X, Mas A, Guarner C, Forne M, Sola R, Castellote J, Rigau J, Laporte JR. Risk of acute liver injury associated with the use of drugs: a multicentre population survey. Alimentary Pharmacol. Ther. 2007;25:1401–1409. doi: 10.1111/j.1365-2036.2007.03338.x. [DOI] [PubMed] [Google Scholar]
- 62.Munyon WH, Salo R, Briones DF. Cytotoxic effects of neuroleptic drugs. Psychopharmacology. 1987;91:182–188. doi: 10.1007/BF00217059. [DOI] [PubMed] [Google Scholar]
- 63.MacAllister SL, Young C, Guzdek A, Zhidkov N, O’Brien PJ. Molecular cytotoxic mechanisms of chlorpromazine in isolated rat hepatocytes. Can. J. Physiol. Pharmacol. 2013;91:56–63. doi: 10.1139/cjpp-2012-0223. [DOI] [PubMed] [Google Scholar]
- 64.Rivera-Calimlim L, Nasrallah H, Strauss J, Lasagna L. Clinical response and plasma levels: effect of dose, dosage schedules, and drug interactions on plasma chlorpromazine levels. Am. J. Psychiatry. 1976;133:646–652. doi: 10.1176/ajp.133.6.646. [DOI] [PubMed] [Google Scholar]
- 65.Turano P, Turner WJ, Manian AA. Thin-layer chromatography of chlor-promazine metabolites. Attempt to identify each of the metabolites appearing in blood, urine and feces of chronically medicated schizophrenics. J. Chromatogr. 1973;75:277–293. doi: 10.1016/s0021-9673(00)85556-9. [DOI] [PubMed] [Google Scholar]
- 66.Chetty M, Moodley SV, Miller R. Important metabolites to measure in pharmacodynamic studies of chlorpromazine. Ther. drug Monit. 1994;16:30–36. doi: 10.1097/00007691-199402000-00004. [DOI] [PubMed] [Google Scholar]
- 67.Hartmann F, Gruenke LD, Craig JC, Bissell DM. Chlorpromazine metabolism in extracts of liver and small intestine from guinea pig and from man. Drug Metab. Dispos. Biol. Fate Chem. 1983;11:244–248. [PubMed] [Google Scholar]
- 68.Cashman JR, Yang Z, Yang L, Wrighton SA. Stereo- and regioselective N-and S-oxidation of tertiary amines and sulfides in the presence of adult human liver microsomes. Drug Metab. Dispos. Biol. Fate Chem. 1993;21:492–501. [PubMed] [Google Scholar]
- 69.Yoshii K, Kobayashi K, Tsumuji M, Tani M, Shimada N, Chiba K. Identification of human cytochrome P450 isoforms involved in the 7-hydroxylation of chlorpromazine by human liver microsomes. Life Sci. 2000;67:175–184. doi: 10.1016/s0024-3205(00)00613-5. [DOI] [PubMed] [Google Scholar]
- 70.Wojcikowski J, Boksa J, Daniel WA. Main contribution of the cytochrome P450 isoenzyme 1A2 (CYP1A2) to N-demethylation and 5-sulfoxidation of the phenothiazine neuroleptic chlorpromazine in human liver-A comparison with other phenothiazines. Biochem. Pharmacol. 2010;80:1252–1259. doi: 10.1016/j.bcp.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 71.Selim K, Kaplowitz N. Hepatotoxicity of psychotropic drugs. Hepatology. 1999;29:1347–1351. doi: 10.1002/hep.510290535. [DOI] [PubMed] [Google Scholar]
- 72.Watson RG, Olomu A, Clements D, Waring RH, Mitchell S, Elias E. A proposed mechanism for chlorpromazine jaundice-defective hepatic sul-phoxidation combined with rapid hydroxylation. J. Hepatol. 1988;7:72–78. doi: 10.1016/s0168-8278(88)80508-7. [DOI] [PubMed] [Google Scholar]
- 73.Abernathy CO, Lukacs L, Zimmerman HJ. Adverse effects of chlorpromazine metabolites on isolated hepatocytes. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. 1977;155:474–478. doi: 10.3181/00379727-155-39833. [DOI] [PubMed] [Google Scholar]
- 74.Hill DR, Baird JK, Parise ME, Lewis LS, Ryan ET, Magill AJ. Primaquine: report from CDC expert meeting on malaria chemoprophylaxis I. Am. J. Trop. Med. Hyg. 2006;75:402–415. [PubMed] [Google Scholar]
- 75.Dern RJ, Beutler E, Alving AS. The hemolytic effect of primaquine. V. Pri-maquine sensitivity as a manifestation of a multiple drug sensitivity. J. Lab. Clin. Med. 1955;45:30–39. [PubMed] [Google Scholar]
- 76.John GK, Douglas NM, von Seidlein L, Nosten F, Baird JK, White NJ, Price RN. Primaquine radical cure of Plasmodium vivax: a critical review of the literature. Malar. J. 2012;11:280. doi: 10.1186/1475-2875-11-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlesinger PH, Krogstad DJ, Herwaldt BL. Antimalarial agents: mechanisms of action. Antimicrob. Agents Chemother. 1988;32:793–798. doi: 10.1128/aac.32.6.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vasquez-Vivar J, Augusto O. Hydroxylated metabolites of the antimalarial drug primaquine. Oxidation and redox cycling. J. Biol. Chem. 1992;267:6848–6854. [PubMed] [Google Scholar]
- 79.Mihaly GW, Ward SA, Edwards G, Nicholl DD, Orme ML, Breckenridge AM. Pharmacokinetics of primaquine in man. I. Studies of the absolute bioavailability and effects of dose size. Br. J. Clin. Pharmacol. 1985;19:745–750. doi: 10.1111/j.1365-2125.1985.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mihaly GW, Ward SA, Edwards G, Orme ML, Breckenridge AM. Pharmacokinetics of primaquine in man: identification of the carboxylic acid derivative as a major plasma metabolite. Br. J. Clin. Pharmacol. 1984;17:441–446. doi: 10.1111/j.1365-2125.1984.tb02369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bates MD, Meshnick SR, Sigler CI, Leland P, Hollingdale MR. In vitro effects of primaquine and primaquine metabolites on exoerythrocytic stages of Plasmodium berghei. Am. J. Trop. Med. Hyg. 1990;42:532–537. doi: 10.4269/ajtmh.1990.42.532. [DOI] [PubMed] [Google Scholar]
- 82.Link CM, Theoharides AD, Anders JC, Chung H, Canfield CJ. Structure-activity relationships of putative primaquine metabolites causing methemo-globin formation in canine hemolysates. Toxicol. Appl. Pharmacol. 1985;81:192–202. doi: 10.1016/0041-008x(85)90155-3. [DOI] [PubMed] [Google Scholar]
- 83.Vennerstrom JL, Eaton JW. Oxidants, oxidant drugs, and malaria. J. Med. Chem. 1988;31:1269–1277. doi: 10.1021/jm00402a001. [DOI] [PubMed] [Google Scholar]
- 84.Fletcher KA, Barton PF, Kelly JA. Studies on the mechanisms of oxidation in the erythrocyte by metabolites of primaquine. Biochem. Pharmacol. 1988;37:2683–2690. doi: 10.1016/0006-2952(88)90263-8. [DOI] [PubMed] [Google Scholar]
- 85.Bowman ZS, Oatis JE, Jr, Whelan JL, Jollow DJ, McMillan DC. Primaquine-induced hemolytic anemia: susceptibility of normal versus glutathione-depleted rat erythrocytes to 5-hydroxyprimaquine. J. Pharmacol. Exp. Ther. 2004;309:79–85. doi: 10.1124/jpet.103.062984. [DOI] [PubMed] [Google Scholar]
- 86.Bowman ZS, Morrow JD, Jollow DJ, McMillan DC. Primaquine-induced hemolytic anemia: role of membrane lipid peroxidation and cytoskeletal protein alterations in the hemotoxicity of 5-hydroxyprimaquine. J. Pharmacol. Exp. Ther. 2005;314:838–845. doi: 10.1124/jpet.105.086488. [DOI] [PubMed] [Google Scholar]
- 87.Ganesan S, Tekwani BL, Sahu R, Tripathi LM, Walker LA. Cytochrome P(450)-dependent toxic effects of primaquine on human erythrocytes. Tox-icol. Appl. Pharmacol. 2009;241:14–22. doi: 10.1016/j.taap.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 88.Pybus BS, Sousa JC, Jin X, Ferguson JA, Christian RE, Barnhart R, Vuong C, Sciotti RJ, Reichard GA, Kozar MP, Walker LA, Ohrt C, Melendez V. CYP450 phenotyping and accurate mass identification of metabolites of the 8-aminoquinoline, anti-malarial drug primaquine. Malar. J. 2012;11:259. doi: 10.1186/1475-2875-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pybus BS, Marcsisin SR, Jin X, Deye G, Sousa JC, Li Q, Caridha D, Zeng Q, Reichard GA, Ockenhouse C, Bennett J, Walker LA, Ohrt C, Melendez V. The metabolism of primaquine to its active metabolite is dependent on CYP 2D6. Malar. J. 2013;12:212. doi: 10.1186/1475-2875-12-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bennett JW, Pybus BS, Yadava A, Tosh D, Sousa JC, McCarthy WF, Deye G, Melendez V, Ockenhouse CF. Primaquine failure and cytochrome P-450 2D6 in Plasmodium vivax malaria. N. Engl. J. Med. 2013;369:1381–1382. doi: 10.1056/NEJMc1301936. [DOI] [PubMed] [Google Scholar]
- 91.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part I. Clin. Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 92.Fontaine F, Delescluse C, de Sousa G, Lesca P, Rahmani R. Cytochrome 1A1 induction by primaquine in human hepatocytes and HepG2 cells: absence of binding to the aryl hydrocarbon receptor. Biochem. Pharmacol. 1999;57:255–262. doi: 10.1016/s0006-2952(98)00304-9. [DOI] [PubMed] [Google Scholar]
- 93.Werlinder V, Backlund M, Zhukov A, Ingelman-Sundberg M. Transcriptional and post-translational regulation of CYP1A1 by primaquine. J. Pharmacol. Exp. Ther. 2001;297:206–214. [PubMed] [Google Scholar]
- 94.Bapiro TE, Andersson TB, Otter C, Hasler JA, Masimirembwa CM. Cytochrome P450 1A1/2 induction by antiparasitic drugs: dose-dependent increase in ethoxyresorufin O-deethylase activity and mRNA caused by quinine, primaquine and albendazole in HepG2 cells. Eur. J. Clin. Pharmacol. 2002;58:537–542. doi: 10.1007/s00228-002-0512-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.