Abstract
OXA-48-like beta-lactamase producing bacteria are now endemic in several European and Mediterranean countries. Among this carbapenemase family, the OXA-48 and OXA-181 variants predominate, whereas other variants such as OXA-204 are rarely reported. Here, we report the molecular epidemiology of a collection of OXA-204-positive enterobacterial isolates (n = 29) recovered in France between October 2012 and May 2014. This study describes the first outbreak of OXA-204-producing Enterobacteriaceae in Europe, involving 12 isolates of an ST90 Escherichia coli clone and nine isolates of an ST147 Klebsiella pneumoniae clone. All isolates co-produced the cephalosporinase CMY-4, and 60% of them co-produced the extended-spectrum beta-lactamase CTX-M-15. The bla OXA-204 gene was located on a 150-kb IncA/C plasmid, isolated from various enterobacterial species in the same patient, indicating a high conjugative ability of this genetic vehicle.
Keywords: carbapenems, outbreak, OXA-48-like, antibiotic resistance, endoscope, nosocomial
Introduction
Since the 2000s, the carbapenem-hydrolysing beta-lactamase OXA-48 has rapidly and widely disseminated and is now endemic in several European and Mediterranean countries [1-4]. Since its discovery, eleven variants of OXA-48 have been reported, classified into two main groups [1,5-13]. The first group contains variants with significant carbapenemase activity, such as OXA-48, OXA-162, OXA-181 or OXA-204 [5-7]. Some variants, such as OXA-232 and OXA-244, possess a hydrolytic profile similar to that of OXA-48 but have a lower capacity to hydrolyse imipenem and temocillin [8-10]. The second group of OXA-48-like variants includes beta-lactamases with extended-spectrum hydrolysis properties and without any significant carbapenemase activity because of deletions in the active site of the enzyme, such as OXA-163, OXA-247, or OXA-405 [11-13]. In most of these cases, the bla OXA-48-like genes are plasmid-borne and are associated with insertion sequences involved in their mobilisation and expression [1,7,9].
OXA-204 was first identified in 2012 in a Klebsiella pneumoniae isolate from Tunisia [7]. In this strain, the bla OXA-204 was co-located with a bla CMY-4 gene on a conjugative IncA/C-type plasmid [7]. The bla OXA-204 gene was part of a transposon Tn2016 that consisted of one copy of insertion sequence (IS) ISEcp1, disrupted by an ISKpn15 element, and a truncated lysR transcriptional regulator [7]. Since that report, OXA-204 has only been identified in two other K. pneumoniae isolates and a single Escherichia coli isolate, all recovered in Tunisia [14,15]. In those two strains, the bla OXA-204 gene was associated with the ISEcp1 element, in one of the two cases truncated by another IS element [14,15].
Our study aimed to compare the genetic features of OXA-204 beta-lactamase-producing strains recovered in France by analysing a collection of 29 bla OXA-204-positive enterobacterial isolates recovered from October 2012 to May 2014. The genetic context and the location of the bla OXA-204 gene were investigated. Finally, a clonal relationship analysis allowed us to identify a regional outbreak in France possibly related to an endoscope.
Methods
Bacterial isolates
We investigated a total of 29 OXA-204 beta-lactamase-producing enterobacterial isolates. All isolates had been recovered from clinical specimens and had been received between October 2012 and May 2014 at the National Reference Centre (NRC) for Antibiotic Resistance (division of carbapenemase-producing Enterobacteriaceae), France. The distribution of clinical samples was as follows: 12 rectal swabs, 12 urine samples, four bile samples and one pus specimen. Isolates were identified using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass-spectrometry (Maldi Biotyper, Bruker Daltonics, France).
Susceptibility testing
Antimicrobial susceptibilities were determined by disk diffusion method on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France) and interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [16]. In addition, minimal inhibitory concentrations (MICs) were determined for carbapenems (imipenem, meropenem, ertapenem) and tigecycline using E-tests (bioMérieux, Marcy-l’Etoile, France), and for colistin using broth microdilution according to EUCAST recommendations.
PCR and sequencing of beta-lactamase-encoding genes
Whole-cell DNA was extracted using the QiaAmp DNA minikit (Qiagen, Courtaboeuf, France). All isolates were screened by PCR for the Ambler class A, B and D carbapenemase-encoding genes bla KPC, bla IMP, bla VIM, bla NDM and bla OXA-48-like as previously described [6,17,18]. Detection of other beta-lactamase genes such as bla CTX-M and bla AmpC-like was performed with internal primers, as described previously [17]. PCR products were analysed on agarose gel. In case of positive signal, the full-length genes (basically bla CTX-M and bla CMY) were amplified and sequenced by using the amplification primers with an automated sequencer (ABI PRISM 3100; Applied Biosystems) as previously described [17]. The nucleotide and deduced protein sequences were analysed with software from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov).
Strain typing
Multilocus sequence typing (MLST) with seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB and tonB) was performed for K. pneumoniae isolates according to Diancourt et al. [19]. Allele sequences and sequence types (STs) were verified at the Institut Pasteur’s whole genome MLST database [20]. Fragments of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA) were amplified and sequenced for E. coli isolates as described on EnteroBase [21]. A different allele number was given to each distinct sequence within a locus, and a distinct ST number was attributed to each distinct combination of alleles.
Clonality analysis using repetitive element palindromic PCR (rep-PCR)
To evaluate their clonal relationship, all E. coli and K. pneumoniae isolates were subjected to Diversilab, a semi-automated rep-PCR (bioMérieux, Marcy-L’Etoile, France). As recommended by the manufacturer, a cut-off for similarity of 95% defined a cluster.
Plasmid DNA analysis and mating-out assays
Plasmid DNAs were extracted using the Kieser method [22], and analysed by agarose gel electrophoresis using the E. coli NCTC50192 strain that harbours four plasmids of 154, 66, 48 and 7 kb as plasmid size marker. Direct transfer of the carbapenem resistance markers was attempted by liquid mating-out assays at 37 °C using sodium azide-resistant E. coli J53 as recipient, as previously described [23]. Selection was performed on agar plates supplemented with ertapenem (0.5 µg/mL) and sodium azide (100 µg/mL).
Replicon and transposon typing
PCR-based replicon typing of the main plasmid incompatibility groups reported in Enterobacteriaceae was performed as previously described [24]. Genetic structures surrounding the bla OXA-204 gene were determined using the primers listed in Table 1.
Table 1. Primers used for Tn2016 PCR mapping.
| Primer | Sequence (5’ to 3’) | PCR product size (bp) |
|---|---|---|
| ISEcp1A | TGCAGGTCTTTTTCTGCTCC | 1,099 |
| ISKpn15–5’ext | CTGCGTGGCTATGTGCTCTG | |
| ISKpn15-for | GGTGTTCGGTGACGAGATTAGC | 1,955 |
| OXA-48–5’ext | ATTCCAGAGCACAACTACGC | |
| ISEcp1P + | TGCTCTGTGGATAACTTGCA | 998 |
| OXA48B | GAGCACTTCTTTTGTGATGGC |
Results
Bacterial isolates
A total of 29 OXA-204-producing enterobacterial isolates were received at the NRC for Antibiotic Resistance from October 2012 to May 2014. These isolates were sent to the NRC because they exhibited decreased susceptibility to carbapenems and/or because they were isolated from a patient who was identified in epidemiological investigations around an infected or colonised patient. These isolates included 11 K. pneumoniae, 15 E. coli, one Proteus mirabilis, one Citrobacter freundii and one Serratia marcescens (Table 2). The 29 strains were isolated from 24 patients. Of these 29 OXA-204 producers, 27 were isolated from 22 patients (n = 22) located in the same geographical area (Paris area, Ile-de-France).
Table 2. Phenotypic and genetic features associated with OXA-204-beta-lactamase producing Enterobacteriaceae, France, 2012–14 (n = 29).
| Species /clone | Number of isolates | Sequence type | Beta-lactams MIC range (µg/mL) |
Genetic location of blaOXA-204 | Incompatibility group of blaOXA-204-carrying plasmid |
Non-beta-lactam-associated resistance (number of strains) | Associated broad-spectrum beta-lactamasesa (number of strains) | Close genetic environment of the blaOXA-204 gene | ||
|---|---|---|---|---|---|---|---|---|---|---|
| ERT | IMP | MER | ||||||||
| Escherichia coli | ||||||||||
| A | 12 | ST90 | 0.38–32 | 0.19–12 | 0.094–6 | Plasmid | IncA/C | Gm (12), Tm (12), Q (12), Tet(12), Sxt (10) | CMY-4 (12), CTX-M-15 (9) | Tn2016 |
| B | 1 | ST104 | 0.5 | 0.38 | 0.25 | Plasmid | IncA/C | Tet | CMY-4 | ISEcp1 |
| C | 1 | ST617 | 1 | 0.25 | 0.25 | Plasmid | IncA/C | Gm, Tm, Q, Tet, Sxt | CMY-4 | Tn2016 |
| D | 1 | ST949 | 0.75 | 0.5 | 0.19 | Plasmid | IncA/C | Q, Tet, Sxt | CMY-4 | ISEcp1 |
| Klebsiella pneumoniae | ||||||||||
| A | 9 | ST147 | 3– >32 | 1–12 | 1– >32 | Plasmid | IncA/C | Ak (1), Tm (8), Gm (6), Q (9), Tet (9), Sxt (8), Fos (7) | CMY-4 (9), CTX-M-15 (9) | ISEcp1 |
| B | 1 | ST1683b | 3 | 0.25 | 0.19 | Plasmid | IncA/C | Gm, Tm, Tet | CMY-4 | Tn2016 |
| C | 1 | ST1709b | 1 | 0.25 | 0.12 | Plasmid | IncA/C | Gm, Tm, Tet, Tig | CMY-4 | Tn2016 |
| Proteus mirabilis | 1 | ND | 1.5 | 0.38 | 0.094 | Plasmid | IncA/C | Gm, Tm, Sxt | CMY-4 | ISEcp1 |
| Citrobacter freundii | 1 | ND | 3 | 1 | 0.5 | Plasmid | IncA/C | Gm, Tm, Q, Tet, Tig | CMY-4 | Tn2016 |
| Serratia marcescens | 1 | ND | 0.75 | 0.75 | 0.5 | Plasmid | IncA/C | Gm, Tm, Tet, Tig | CMY-4 | Tn2016 |
Ak: amikacin; ERT: ertapenem; Fos: fosfomycin; Gm: gentamicin; IMP: imipenem; MER: meropenem; MIC: minimum inhibitory concentration; ND: not determinable; Q: quinolones; Sxt: sulfamethoxazole-trimethoprim; Tet: tetracycline; Tig: tigecycline; Tm: tobramycin.
a Resistance markers co-harboured on the bla OXA-204-carrying plasmid are underlined.
b ST1683 ad ST1709 are new sequence types.
Susceptibility to beta-lactams and related beta-lactamase genes
According to the EUCAST guidelines, 21 isolates were susceptible to imipenem and meropenem. Of the 15 E. coli isolates, 14 were susceptible to imipenem and meropenem whereas only four of the 11 K. pneumoniae isolates were susceptible to those antibiotics (Table 2). By contrast, 21 of the 29 OXA-204 producers had a decreased susceptibility (intermediate or resistant) to ertapenem (7/15 of the E. coli and 11/11 of the K. pneumoniae isolates). All P. mirabilis, C. freundii and S. marcescens isolates were susceptible to imipenem and meropenem and had decreased susceptibility to ertapenem (Table 2). Regarding the broad-spectrum cephalosporins, all isolates were resistant to ceftazidime and cefotaxime. Since OXA-204 has no hydrolytic activity towards broad-spectrum cephalosporins, we searched for the expression of additional beta-lactamases (extended-spectrum beta-lactamases (ESBLs) and/or cephalosporinases). As expected, all OXA-204-producing isolates co-produced an AmpC-type beta-lactamase, CMY-4 (Table 2). In addition, 18 isolates were of intermediate susceptibility or resistant to cefepime (nine E. coli isolates and nine K. pneumoniae isolates). CMY-4 and OXA-204 are not able to hydrolyse cefepime, but co-production of the CTX-M-15 ESBL was found in all those isolates (Table 2).
Susceptibility to non-beta-lactams antibiotics
Four antibiotics were active against the majority of the isolates. Except for the P. mirabilis and S. marcescens isolates that are intrinsically resistant to polymyxins, all other OXA-204 producers were susceptible to colistin. In addition, 28, 26 and 22 of the 29 OXA-204 producers were susceptible to amikacin, tigecycline and fosfomycin, respectively (Table 2). Conversely, 22 OXA-204 producers were resistant to sulfamethoxazole-trimethoprim, and 24 were resistant to ciprofloxacin and gentamicin (Table 2).
Multilocus sequence typing
ST90 was the most commonly observed ST for the E. coli isolates, accounting for 12 of 15 isolates. The three remaining single isolates belonged to ST104, ST617 and ST949 (Table 2). Among the 11 OXA-204-positive K. pneumoniae isolates, nine isolates belonged to ST147. The remaining two isolates belonged to the new ST1683 and ST1709 (Table 2).
Genetic support of the bla OXA-204 gene
Using mating-out assays, transconjugants harbouring the bla OXA-204 and the bla CMY-4 genes were obtained for all 29 strains. Plasmid DNA analysis of those transconjugants revealed a single plasmid (ca 150 kb), which was identified as an IncA/C-type plasmid (Table 2).
Close genetic environment of the blaOXA-204 gene
The genetic environment of the bla OXA-204 gene was analysed using specific primers designed from the plasmid p204-B of K. pneumoniae 204 (Table 1). For 17 strains, the bla OXA-204 gene was part of transposon Tn2016, where ISEcp1 was disrupted by insertion of ISKpn15. This transposon was identified in the 12 ST90 E. coli strains and in five other single isolates (Table 2). In the 12 remaining isolates (nine ST147 K. pneumoniae isolates and three single isolates), ISEcp1 was not disrupted by ISKpn15 element (Table 2).
Endoscopy-related outbreak
Rep-PCR analysis confirmed the MLST results and showed that 12 of the 15 OXA-204-producing E. coli isolates (all ST90 E. coli) and nine of the 12 OXA-204-producing K. pneumoniae isolates (all ST147 K. pneumoniae) were clonally related (Figure 1).
Figure 1.
Rep-PCR analysis of OXA-204-producing Enterobacteriaceae, France, 2012–14 (n = 21)
Dendrogram and computer-generated image of rep-PCR banding patterns of OXA-204-producing Klebsiella pneumoniae (A) and OXA-204-producing E. coli (B) isolates. Epidemic clones (ST90 Escherichia coli and ST147 K. pneumoniae are highlighted in bold. The red similarity line at 95% shows the cut-off to separate different clone.
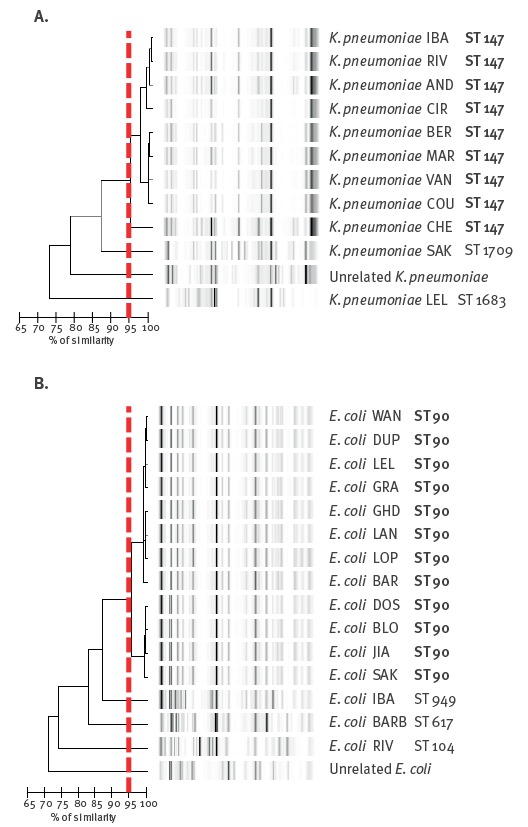
The results of this analysis led us to do an epidemiologically investigation of this dual outbreak (ST90 E. coli and ST147 K. pneumoniae). An endoscope was identified as the possible source of the outbreak in that the investigation showed that 17 patients had direct contact with the endoscope, while five (Patients 10, 11, 13, 14 and 16) were considered as secondary cases through patient-to-patient transmission on a clinical ward (Figure 2).
Figure 2.
Synoptic curve of patients involved in the endoscope-related outbreak caused by OXA-204-producing Enterobacteriaceae, France, 2012–14 (n = 22)
Patient numbers are indicated on the left of each time line. Numbers in parenthesis indicate patients who were not in direct contact with the suspectedly contaminated endoscope. Crosses indicate the date of endoscopy with the suspectedly contaminated endoscope and coloured boxes correspond to hospitalisations. Black arrows represent direct patient-to-patient transmission of the OXA-204-producing K. pneumoniae ST147.
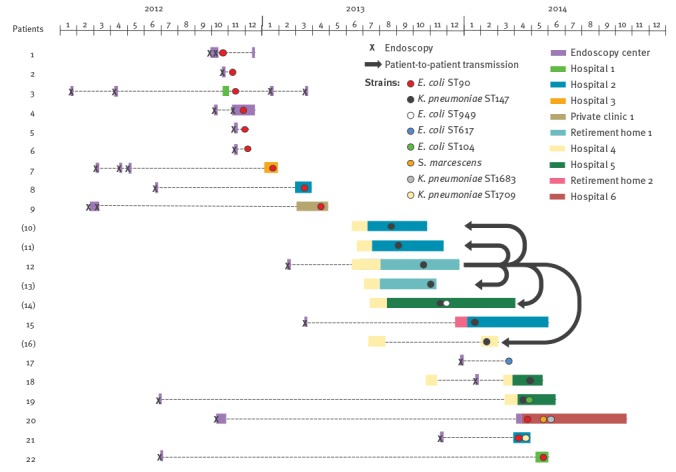
Of note, retrospective screening of all patients who had endoscopy with the suspectedly contaminated endoscope but were not hospitalised identified two colonised patients who underwent endoscopy as outpatients (Patients 6 and 17) (Figure 2). In addition, for four patients, transmission of a bla OXA-204-carrying plasmid from one strain to another was observed (several enterobacterial isolates of different species carrying the same plasmid with the same close genetic environment for bla OXA-204 were isolated from patients 14, 19, 20 and 21; Figure 2 and Table 2). Finally, 14 patients were infected (four biliary infections, one hepatic abscess and nine urinary tract infections) and 12 patients were colonised. For one patient (Patient 19), the acquired biliary tract infection resulted in fatal septicaemia. Overall, this outbreak spread in 10 health institutions including one endoscopy centre where patients received the endoscopy with the suspected endoscope, six hospitals, one private clinic and two retirement homes (Figure 2).
In February 2014, the sequestration of the endoscope immediately stopped further detection of colonised patients, confirming that the endoscope was probably the source of this outbreak. However, audit of the reprocessing procedures that were performed in accordance with the manufacturer guidelines and French recommendations [25,26] did not reveal any dysfunction. In microbiological investigations of the incriminated endoscope in February 2014, no OXA-204-producing strain was recovered from the device. However, polymorphic human flora was cultured from the endoscope and three additional reprocessing procedures were needed until the device was clean enough to conform with the French recommendations [26].
Discussion
We analysed different features of the OXA-204-positive enterobacterial isolates collected between October 2012 and May 2014 at the NRC for Antibiotic Resistance (division of carbapenemase-producing Enterobacteriaceae), France.
According to the EUCAST guidelines, 21 of the 29 OXA-204 producers remained susceptible to imipenem and meropenem, complicating their detection. Similar phenotypical characteristics have already been reported for OXA-48-producers [27]. OXA-204-producing E. coli seem to be more susceptible to those two carbapenems than OXA-204-producing K. pneumoniae. By contrast, ertapenem appeared to be the best carbapenem to detect those strains since 21 of the 29 isolates were resistant. All strains were resistant to extended-spectrum cephalosporins because of the production of the beta-lactamase CMY-4, thus limiting therapeutic options. In addition, 18 of the 29 OXA-204 producing isolates also produced a CTX-M-15-type ESBL, compromising the efficiency of cefepime. However, most of the isolates (27/29) remained susceptible to colistin, tigecycline, amikacin and fosfomycin.
We investigated the clonal distribution of OXA-204-positive isolates and identified two main STs. Twelve of the 15 E. coli isolates belonged to ST90. To our knowledge, this ST has not been reported to be associated with OXA-48-like-producing E. coli. However, the occurrence of other carbapenemases such as NDM-1 had been reported twice in ST90 E. coli isolates, and a link with India was demonstrated [28,29]. Nine of 11 the K. pneumoniae isolates belonged to ST147, which was the predominant ST in our study. The ST147 K. pneumoniae clone is linked to the worldwide spread of different carbapenemases (OXA-48, OXA-204, NDM-1, NDM-5, VIM-1, KPC-2) and ESBL (SHV-12, CTX-M-15) [15,27,30-33]. Additional single STs were identified, namely ST617, ST104 and ST949 for E. coli and two new STs (ST1683 and ST1709) for K. pneumoniae, supporting the hypothesis that a single bla OXA-204-positive plasmid is spreading among various genetic backgrounds (Figure 2). One OXA-204-positive ST617 E. coli strain was identified in Tunisia in 2015 [14]. Of note, ST617 is a widespread ST type, associated with various beta-lactamase-encoding genes (CTX-M-15, NDM-1) [34,35]. Interestingly, ESBL-producing ST617 E. coli were recently recovered from companion and farm animals in Tanzania and from water samples in Tunisia [36,37].
In this study, between October 2012 and May 2014, OXA-204 ST90 E. coli strains were regularly identified. Concomitantly, OXA-204-producing ST147 K. pneumoniae isolates were identified in the same area (Paris and its suburbs) between August 2013 and May 2014. Although microbiological investigations were not conclusive, our results strongly suggest that the endoscope may have been contaminated with at least two OXA-204 producing isolates: ST90 E. coli that co-harboured a bla CTX-M-15-carrying plasmid and ST147 K. pneumoniae (Figure 2). Interestingly, following this outbreak the manufacturer released several notes concerning the reprocessing procedures of the endoscope [38] and updated in June 2015 the manual for reprocessing procedures which now includes reference to a novel brush (MAJ-1888/MyBrush) [39]. This dual outbreak was controlled in May 2014 after the sequestration of the endoscope suspected to be the source of the outbreak. Gastrointestinal endoscopy has previously been identified as a risk factor for infection and colonisation with carbapenemase-producing Enterobacteriaceae [40].
Four patients were colonised or infected with more than one OXA-204-producing enterobacterial species. Three patients were colonised with one OXA-204-positive E. coli plus one OXA-204-producing K. pneumoniae, and one patient with one OXA-204-positive E. coli plus one OXA-204-producing K. pneumoniae and one additional OXA-204-positive S. marcescens. Those results suggest a high conjugative ability of the OXA-204-IncA/C-type plasmids. As shown for OXA-48 [41], OXA-204 is associated with an efficient genetic vehicle, thus promoting the interspecies spread of bla OXA-204-carrying plasmids among various enterobacterial species in the same patient. In addition, as previously reported, endoscopy-associated transmission of carbapenemase-producing Enterobacteriaceae (CPE) might result in long-term carriage of the acquired CPE [42] that poses a risk of further secondary outbreaks from primary infected or colonised patients. Indeed, these patients are often at high risk of recurrence of a hepato-biliary infection that needs to be treated by endoscopic procedures. To decrease the risk of secondary outbreaks, we propose making a note in these these CPE patients’ record and systematically screening them before performing any endoscopy.
The transposon Tn2016 was identified in 17 of the 29 isolates. The truncation of ISEcp1 by another IS may have stabilised this genetic structure on the IncA/C plasmid by disrupting the ISEcp1 transposase activity. However, in the 12 remaining isolates, the ISEcp1 copy was intact and we can therefore speculate that the transposon made of ISEcp1 and bla OXA-204 is functional. ISEcp1 is known to be an efficient genetic vehicle for spreading clinically significant beta-lactamases such as CMY or CTX-M-15 [43,44]. The association of bla OXA-204 with an intact copy of ISEcp1 on the one hand and an IncA/C broad host range plasmid on the other might increase the capability of the bla OXA-204 to disseminate among various genetic elements (plasmids, chromosome, etc) and among various bacterial species.
Acknowledgements
We thank the platform Genotyping of Pathogens and Public Health (Institut Pasteur, Paris, France) for coding MLST alleles and profiles and making them available at www.pasteur.fr/mlst.
Funding: This work was supported by grants from French Ministry of Health and The French Public Health Agency (Santé publique France).
Conflict of interest: None declared.
Authors’ contributions: AP, LD, LP and PN designed the study. AP, SB, GC and LD performed the experiments and recorded the data. AP, LD, PN, LP, TN, HB and VP contributed to the writing of the manuscript.
PN was head of the Associated French NRC for Antibiotic Resistance (division of carbapenemase-producing Enterobacteriaceae) from 2012 to July 2013.
References
- 1. Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67(7):1597-606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 2. Dortet L, Cuzon G, Nordmann P. Dissemination of carbapenemase-producing Enterobacteriaceae in France, 2012. J Antimicrob Chemother. 2014;69(3):623-7. 10.1093/jac/dkt433 [DOI] [PubMed] [Google Scholar]
- 3. Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) working group Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015;20(45):30062. 10.2807/1560-7917.ES.2015.20.45.30062 [DOI] [PubMed] [Google Scholar]
- 4. Vaux S, Carbonne A, Thiolet JM, Jarlier V, Coignard B, RAISIN and Expert Laboratories Groups Emergence of carbapenemase-producing Enterobacteriaceae in France, 2004 to 2011. Euro Surveill. 2011;16(22):19880. 10.2807/ese.16.22.19880-en [DOI] [PubMed] [Google Scholar]
- 5. Oueslati S, Nordmann P, Poirel L. Heterogeneous hydrolytic features for OXA-48-like β-lactamases. J Antimicrob Chemother. 2015;70(4):1059-63. 10.1093/jac/dku524 [DOI] [PubMed] [Google Scholar]
- 6. Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2011;55(10):4896-9. 10.1128/AAC.00481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Potron A, Nordmann P, Poirel L. Characterization of OXA-204, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013;57(1):633-6. 10.1128/AAC.01034-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oteo J, Hernández JM, Espasa M, Fleites A, Sáez D, Bautista V, et al. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J Antimicrob Chemother. 2013;68(2):317-21. 10.1093/jac/dks383 [DOI] [PubMed] [Google Scholar]
- 9. Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. 2013;41(4):325-9. 10.1016/j.ijantimicag.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 10. Potron A, Poirel L, Dortet L, Nordmann P. Characterisation of OXA-244, a chromosomally-encoded OXA-48-like β-lactamase from Escherichia coli. Int J Antimicrob Agents. 2016;47(1):102-3. 10.1016/j.ijantimicag.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 11. Poirel L, Castanheira M, Carrër A, Rodriguez CP, Jones RN, Smayevsky J, et al. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 2011;55(6):2546-51. 10.1128/AAC.00022-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gomez S, Pasteran F, Faccone D, Bettiol M, Veliz O, De Belder D, et al. Intrapatient emergence of OXA-247: a novel carbapenemase found in a patient previously infected with OXA-163-producing Klebsiella pneumoniae. Clin Microbiol Infect. 2013;19(5):E233-5. 10.1111/1469-0691.12142 [DOI] [PubMed] [Google Scholar]
- 13. Dortet L, Oueslati S, Jeannot K, Tandé D, Naas T, Nordmann P. Genetic and biochemical characterization of OXA-405, an OXA-48-type extended-spectrum β-lactamase without significant carbapenemase activity. Antimicrob Agents Chemother. 2015;59(7):3823-8. 10.1128/AAC.05058-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Charfi K, Mansour W, Khalifa AB, Mastouri M, Aouni M, Mammeri H. Emergence of OXA-204 β-lactamase in Tunisia. Diagn Microbiol Infect Dis. 2015;82(4):314-7. 10.1016/j.diagmicrobio.2015.04.003 [DOI] [PubMed] [Google Scholar]
- 15. Grami R, Mansour W, Ben Haj Khalifa A, Dahmen S, Chatre P, Haenni M, et al. Emergence of ST147 Klebsiella pneumoniae producing OXA-204 carbapenemase in a University Hospital, Tunisia. Microb Drug Resist. 2016;22(2):137-40. 10.1089/mdr.2014.0278 [DOI] [PubMed] [Google Scholar]
- 16.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0. Växjö: EUCAST; 2016. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf
- 17. Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55(11):5403-7. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119-23. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 19. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178-82. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klebsiella sequence typing. Paris: Institut Pasteur. [Accessed: 15 Jan 2017). Available from: http://bigsdb.pasteur.fr/klebsiella/klebsiella.html
- 21.EnteroBase. Escherichia coli MLST database. Warwick: EnteroBase. [ Accessed: 15 Jan 2017). Available from: http://mlst.ucc.ie/mlst/dbs/Ecoli
- 22. Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12(1):19-36. 10.1016/0147-619X(84)90063-5 [DOI] [PubMed] [Google Scholar]
- 23. Potron A, Poirel L, Nordmann P. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob Agents Chemother. 2014;58(1):467-71. 10.1128/AAC.01344-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219-28. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 25.Circulaire n°2003-591 du 17/12/2003 relative aux modalités de traitement manuel pour la désinfection des endoscopes non autoclavables dans les lieux de soins. [Circular no. 2003-591 of 17 Dec 2003 relating to the manual treatment modalities for the disinfection of non-autoclavable endoscopes in places of care]. Paris: Ministere de la Sante, de la Famille et des Personnes handicapees; 2003. French. Available from: http://circulaires.legifrance.gouv.fr/pdf/2009/04/cir_8584.pdf
- 26.Eléments d'assurance qualité en hygiène relatifs au contrôle microbiologique des endoscopes et à la traçabilité en endoscopie. [Elements of quality assurance in hygiene relating to microbiological control of endoscopes and traceability in endoscopy]. Paris: Ministère des Affaires sociales et de la Santé; 2007. Franch. Available from: http://social-sante.gouv.fr/IMG/pdf/microbio_endoscopes-2.pdf
- 27. Potron A, Poirel L, Rondinaud E, Nordmann P. Intercontinental spread of OXA-48 β-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 2013;18(31):20549. 10.2807/1560-7917.ES2013.18.31.20549 [DOI] [PubMed] [Google Scholar]
- 28. Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, et al. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother. 2011;66(9):2002-5. 10.1093/jac/dkr226 [DOI] [PubMed] [Google Scholar]
- 29. Österblad M, Kirveskari J, Hakanen AJ, Tissari P, Vaara M, Jalava J. Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008-11). J Antimicrob Chemother. 2012;67(12):2860-4. 10.1093/jac/dks299 [DOI] [PubMed] [Google Scholar]
- 30. Gamal D, Fernández-Martínez M, Salem D, El-Defrawy I, Montes LA, Ocampo-Sosa AA, et al. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int J Infect Dis. 2016;43:17-20. 10.1016/j.ijid.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 31. Shin J, Baek JY, Cho SY, Huh HJ, Lee NY, Song JH, et al. bla NDM-5-bearing IncFII-type plasmids of Klebsiella pneumoniae sequence type 147 transmitted by cross-border transfer of a patient. Antimicrob Agents Chemother. 2016;60(3):1932-4. 10.1128/AAC.02722-15 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Rodrigues C, Machado E, Ramos H, Peixe L, Novais Â. Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIK). Int J Med Microbiol. 2014;304(8):1100-8. 10.1016/j.ijmm.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 33. Bathoorn E, Tsioutis C, da Silva Voorham JM, Scoulica EV, Ioannidou E, Zhou K, et al. Emergence of pan-resistance in KPC-2 carbapenemase-producing Klebsiella pneumoniae in Crete, Greece: a close call. J Antimicrob Chemother. 2016;71(5):1207-12. 10.1093/jac/dkv467 [DOI] [PubMed] [Google Scholar]
- 34. Aibinu I, Odugbemi T, Koenig W, Ghebremedhin B. Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect. 2012;18(3):E49-51. 10.1111/j.1469-0691.2011.03730.x [DOI] [PubMed] [Google Scholar]
- 35. Torres-González P, Bobadilla-Del Valle M, Tovar-Calderón E, Leal-Vega F, Hernández-Cruz A, Martínez-Gamboa A, et al. Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-β-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob Agents Chemother. 2015;59(11):7080-3. 10.1128/AAC.00055-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seni J, Falgenhauer L, Simeo N, Mirambo MM, Imirzalioglu C, Matee M, et al. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front Microbiol. 2016;11;7:142. [DOI] [PMC free article] [PubMed]
- 37. Ben Said L, Jouini A, Alonso CA, Klibi N, Dziri R, Boudabous A, et al. Characteristics of extended-spectrum β-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci Total Environ. 2016;550:1103-9. 10.1016/j.scitotenv.2016.01.042 [DOI] [PubMed] [Google Scholar]
- 38.Information de sécurité. Ecouvillonnage de l’érecteur du Duodénoscope TJF Q180V. [Safety information. Swabbing the TJF Q180V duodenoscope erector]. Rungis Cedex: OLYMPUS France S.A.S; 2014. French. Available from: http://ansm.sante.fr/content/download/65965/844017/version/2/file/mes-140807-Duodenoscope-Olympus.pdf
- 39.Mise à jour du mode d’emploi et nouvelles de nettoyage pour le duodénoscope OLYMPUS TJF - Q180V. Guide de nettoyage et écouvillons de nettoyage MAJ-1888/MyBrush. [Update of the user manual and cleaning news for the OLYMPUS TJF - Q180V duodenoscope - MAJ-1888. MyBrush cleaning guide and cleaning swabs]. Rungis Cedex: OLYMPUS France S.A.S; 2015. French. Available from: http://ansm.sante.fr/content/download/78191/991251/version/1/file/mes_150619_olympus.pdf
- 40. Muscarella LF. Risk of transmission of carbapenem-resistant Enterobacteriaceae and related "superbugs" during gastrointestinal endoscopy. World J Gastrointest Endosc. 2014;6(10):457-74. 10.4253/wjge.v6.i10.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arana DM, Saez D, García-Hierro P, Bautista V, Fernández-Romero S, Ángel de la Cal M, et al. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin Microbiol Infect. 2015;21(2):148.e1-4. 10.1016/j.cmi.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 42. Dortet L, Naas T, Boytchev I, Fortineau N. Endoscopy-associated transmission of carbapenemase-producing Enterobacteriaceae: return of 5 years’ experience. Endoscopy. 2015;47(6):561. 10.1055/s-0034-1392098 [DOI] [PubMed] [Google Scholar]
- 43. Nakano R, Okamoto R, Nagano N, Inoue M. Resistance to gram-negative organisms due to high-level expression of plasmid-encoded ampC β-lactamase blaCMY-4 promoted by insertion sequence ISEcp1. J Infect Chemother. 2007;13(1):18-23. 10.1007/s10156-006-0483-6 [DOI] [PubMed] [Google Scholar]
- 44. Poirel L, Decousser JW, Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) β-lactamase gene. Antimicrob Agents Chemother. 2003;47(9):2938-45. 10.1128/AAC.47.9.2938-2945.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]


