Abstract
Misregulation of transcription factors, including signal transducer and activator of transcription (STAT) proteins, leads to inappropriate gene expression patterns that can promote tumor initiation and progression. Under physiologic conditions, STAT signaling is stimulus-dependent and tightly regulated by endogenous inhibitors, namely suppressor of cytokine signaling (SOCS) proteins, phosphatases, and protein inhibitor of activated STAT (PIAS) proteins. However, in tumorigenesis, STAT proteins become constitutively active and promote the expression of pro-growth and pro-survival genes. Although STAT activation has been widely implicated in cancer, therapeutic STAT inhibitors are still largely absent from the clinic. This review dissects the mechanisms of action of two families of endogenous STAT inhibitors, the SOCS and PIAS families, to potentially inform the development of novel therapeutic inhibitors.
Keywords: Cancer, STAT signaling, SOCS, PIAS
Transcriptional Misregulation in Cancer
Transcription factors regulate the expression of a cohort of genes, ultimately creating cell-specific gene expression patterns that define cellular function. Accordingly, misregulation of transcription factors, particularly those regulating genes controlling proliferation, survival, and self-renewal, leads to inappropriate expression patterns that can promote tumor initiation and progression in many types of human cancer [1]. Such mechanisms of misregulation include amplification, deletion, or mutation of genes encoding transcription factors, transcriptional cofactors, and chromatin remodelers. Furthermore, transcription factors serve as convergence points and terminal effectors of many oncogenic signal transduction cascades [2]. Thus, as functional mediators of many upstream oncogenic events, transcription factors are promising therapeutic targets for the treatment of cancer.
Unfortunately, transcription factors have proven difficult to target due to large, flat interaction surfaces and predominantly nuclear localization [3]. Thus, new therapeutic strategies are needed. One group that exemplifies these issues is the signal transducer and activator of transcription (STAT; see Glossary) family of transcription factors. Understanding the physiologic role and regulation of STATs can provide insight into cancer pathogenesis, as well as novel strategies for therapeutically targeting these proteins.
STAT Signaling in Cancer
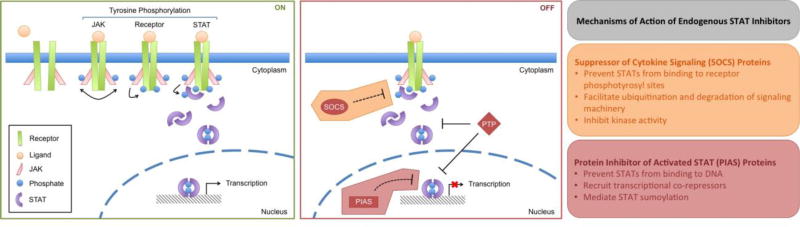
STAT target genes are involved in many cellular processes, including differentiation, immunity, inflammation, proliferation, and survival [4]. Due to the critical role these processes play in tumor initiation and progression, STAT signaling has been widely implicated in human cancer, including both hematologic malignancies and solid cancers [4,5]. Under physiologic conditions, STAT signaling is stimulus-dependent and tightly regulated by endogenous inhibitors, namely suppressor of cytokine signaling (SOCS; see Glossary) proteins, phosphatases, and protein inhibitor of activated STAT (PIAS; see Glossary) proteins (Figure 1, Key Figure). However, in cancer pathogenesis, STATs become constitutively active due to chronically elevated cytokine levels, loss of endogenous inhibitors, or activating mutations in tyrosine kinases. Such mutant kinases include gain-of-function JAKs, such as JAK2V617F and Tel-JAK2, constitutively active receptor tyrosine kinases, like epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), and FMS-like tyrosine kinase 3 (FLT3), and oncogenic non-receptor tyrosine kinases, including SRC and BCR-ABL [6,7].
Figure 1, Key Figure. JAK/STAT Signaling.
Under physiologic conditions, STAT signaling is stimulus-dependent and tightly regulated by endogenous inhibitors. Following receptor-ligand engagement, receptor oligomerization brings membrane-associated Janus kinases (JAKs) into close proximity and facilitates JAK transphosphorylation. Active JAKs then phosphorylate nearby tyrosine residues on the intracellular region of the receptor, creating docking sites for cytoplasmic STAT proteins. Following receptor docking, STATs are phosphorylated and activated by receptor-associated JAKs. Active STAT dimers then translocate to the nucleus where they regulate the expression of genes involved in differentiation, immunity, inflammation, proliferation, and survival. Three families of endogenous STAT inhibitors exist to effectively limit JAK/STAT signaling to a short pulse. Suppressor of cytokine signaling (SOCS) proteins are negative feedback regulators that prevent STAT activation, protein tyrosine phosphatases (PTPs) are endogenous inhibitors that promote STAT dephosphorylation, and protein inhibitor of activated STAT (PIAS) proteins are transcriptional regulators that inhibit STAT transcriptional activity.
Of the seven STAT family members, STAT1, STAT3, and STAT5 appear to play the most direct roles in tumorigenesis [8]. STAT1 primarily responds to interferons (IFNs), including IFNα, IFNβ, and IFNγ, and regulates the expression of genes involved in anticancer processes, including growth arrest, apoptosis, and immunosurveillance [4,9]. Accordingly, inactivation of STAT1 signaling has been shown to promote tumor growth in both colon cancer and MYC-driven prostate cancer [10,11]. However, persistent STAT1 activation has been seen in various cancer cell lines and primary disease, including multiple myeloma, leukemia, breast cancer, and head and neck cancer [5,7]. Moreover, STAT1 has been reported as a potential oncogene in serous papillary endometrial cancer [11]. Therefore, the commonly accepted role of STAT1 as a tumor suppressor is complex and likely tumor-specific.
In contrast, STAT3 predominantly responds to interleukins (ILs), namely IL-6, IL-10, IL-23, IL-21, IL-11, leukemia inhibitory factor (LIF), and oncostatin M (OSM), and regulates the expression of genes involved in pro-tumorigenic processes, including angiogenesis, inflammation, metastasis, proliferation, and survival. Such pro-tumorigenic STAT3 target genes include vascular endothelial growth factor (VEGF), matrix metalloproteinases (MMPs), p21, cyclin D1, MYC, BCL-2, BCL-XL, and survivin [4]. Furthermore, persistent STAT3 activation in tumor-associated immune cells has been shown to antagonize STAT1-mediated immunosurveillance and suppress both innate and adaptive immune responses [9,13]. Accordingly, constitutively active STAT3 has been reported in hematological malignancies, including lymphomas and multiple myeloma, as well as various solid tumors, including breast, melanoma, prostate, head and neck, lung, pancreatic, and ovarian [4,14]. Thus, across multiple malignancies, STAT3 is primarily oncogenic.
Although STAT1 and STAT3 tend to respond to distinct signals and play opposing roles in oncogenesis, cross-regulatory mechanisms between STAT1 and STAT3 have been described. For example, several groups have demonstrated that some cytokines can activate both STAT1 and STAT3. However, the majority of these studies have been performed in STAT-deficient systems in which only STAT1 or STAT3 is active [15]. Thus, the extent and significance of STAT1 and STAT3 co-activation, including the biological role of STAT1-STAT3 heterodimers, remain poorly understood. Additionally, Nivarthi and colleagues have recently demonstrated that the ratio of expression of STAT1 to STAT3 is a key determinant of progression in colorectal carcinoma and may be more prognostic of clinical outcome than STAT1 or STAT3 levels alone [16]. Together, these findings suggest that understanding the functional crosstalk between STAT1 and STAT3 will be critical for assessing whether STAT inhibitors will be advantageous or detrimental in certain tumor contexts.
STAT5 largely responds to IL-2, granulocyte macrophage CSF (GM-CSF), IL-15, IL-7, IL-3, IL-5, and prolactin (PRL) and regulates the expression of pro-growth and pro-survival genes [4,9]. The tumorigenic role of STAT5 has been most strongly established in FLT3 internal tandem duplication (FLT3-ITD)-containing acute myelogenous leukemia (AML) and BCR-ABL-driven chronic myelogenous leukemia (CML). In CML specifically, STAT5 is indispensable for disease initiation and maintenance [17] and capable of mediating both therapeutic resistance and disease persistence [18,19]. STAT5 activation has also been shown to play a role in breast, prostate, uterine, and head and neck cancers [4,20]. However, the pro-tumorigenic role of STAT5 in these solid tumors appears to be more complicated than that of STAT3. For example, in breast cancer, STAT5 has been shown to decrease proliferation and increase sensitivity to cell death when activated simultaneously with STAT3 [21]. Therefore, STAT5 is primarily oncogenic in hematologic malignancies, but its role in solid tumors may depend on the activation state of other STAT members, including STAT3.
Targeting STATs in Cancer
Although transcription factors have been notoriously difficult to target therapeutically, STAT inhibition remains an intriguing strategy for the treatment of cancer. First, transformed cells depend on STAT3 and STAT5, and to some extent STAT1, for growth and survival, whereas non-transformed cells do not. This discrepancy in dependence creates an ideal therapeutic window [22], potentially limiting therapeutic toxicities. Second, STATs act at the intersection of many upstream oncogenic signals [23], suggesting that STAT-specific inhibition may prevent resistance associated with the activation of parallel signaling pathways. Due to these two intriguing properties, many STAT inhibitors have been proposed,, developed, and tested for the treatment of cancer (reviewed in [4,24,25]). However, very few STAT inhibitors have shown clinical efficacy.
Some of the main limitations of current STAT inhibitors are insufficient specificity, potency, bioavailability, and delivery, as well as unknown mechanisms of action [26]. One potential strategy to overcome these limitations is to use endogenous STAT inhibitors as a model for the development of therapeutic inhibitors. Recent chemical biology approaches for identifying STAT inhibitors [27,28] have revealed drugs such as pimozide that decrease STAT tyrosine phosphorylation, yet are not direct kinase inhibitors [29,30]. These findings raise the possibility that such compounds are working through the activation of negative regulators of STAT function. Thus, a more comprehensive understanding of the biology of endogenous STAT inhibitors may reveal novel strategies for the design and development of therapeutic STAT inhibitors.
Endogenous STAT Inhibitors
Suppressor of Cytokine Signaling (SOCS) Proteins
The SOCS family consists of eight proteins, namely cytokine-inducible SH2-containing protein (CIS), SOCS1, SOCS2, SOCS3, SOCS4, SOCS5, SOCS6, and SOCS7. Although the biological roles of SOCS4–7 remain poorly understood, CIS and SOCS1–3 are well-characterized as cytokine-inducible negative feedback regulators of JAK/STAT signaling [31,32]. Accordingly, the expression levels of these four proteins are generally low in the absence of STAT-activating cytokines or growth factors. However, following an appropriate stimulus, the JAK/STAT signaling cascade becomes activated and newly phosphorylated STAT dimers enter the nucleus where they induce the expression of target genes, including CIS and SOCS1–3. Newly expressed SOCS proteins then inhibit the JAK/STAT cascade, limiting STAT-dependent transcription to a relatively short pulse. Following SOCS protein degradation, the cellular system returns to a pre-stimulus state and is primed for another round of activation.
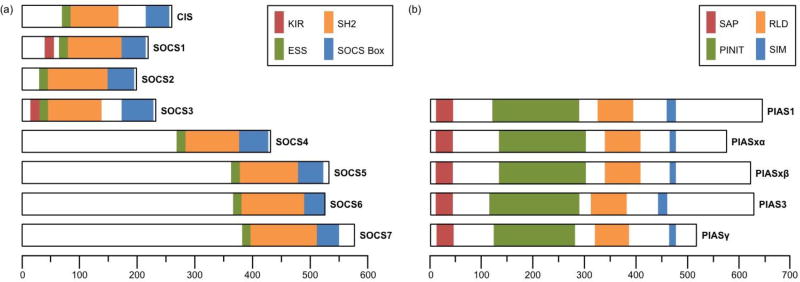
SOCS proteins are characterized by four functional domains (Figure 2A). Each member contains an SH2 domain, which recognizes tyrosine-phosphorylated substrates, preceded by an extended SH2 subdomain (ESS), which stabilizes SH2-phosphotyrosyl binding interactions. SOCS proteins also have a carboxyl-terminal SOCS box domain that recruits E3-ubiquitin ligase machinery. In brief, the first half of the SOCS box binds elongin C and the second half binds cullin 5, which is a scaffold protein responsible for recruiting RING-box protein 2 (RBX2) and forming the active E3 ubiquitin ligase complex. In addition to these two domains, CIS and SOCS1–3 have short amino-terminal domains, whereas SOCS 4–7 have long amino-terminal domains. These termini are thought to facilitate substrate binding, potentially for SOCS-mediated protein degradation [32]. Lastly, SOCS1 and SOCS3 have a kinase inhibitory region that enables them to inhibit the kinase activity of JAK1–2 and TYK2, but not JAK3 [31].
Figure 2. Domain structure of the SOCS and PIAS protein families.
(a) The suppressor of cytokine signaling (SOCS) family consists of eight proteins. All eight members share a Src homology 2 (SH2) domain preceded by an extended SH2 subdomain (ESS), as well as a carboxyl-terminal SOCS box domain. In brief, the SH2 domain recognizes and binds tyrosine-phosphorylated sites on SOCS substrates while the ESS domain stabilizes these interactions. The SOCS box domain recruits the E3-ubiquitin ligase machinery required for SOCS-mediated substrate degradation. SOCS1 and SOCS3 also have a kinase inhibitory region (KIR) that allows for direct inhibition of JAK kinase activity. (b) The protein inhibitor of activated STAT (PIAS) family consists of five major proteins, each containing four conserved motifs. The amino-terminal scaffold attachment factor (SAF)-A/B, Acinus and PIAS (SAP) motif facilitates PIAS-DNA interactions. The RING-finger-like zinc-binding (RLD) motif is required for the SUMO-E3-ligase activity of PIAS proteins. The SUMO interaction motif (SIM) and the Pro-Ile-Asn-Ile-Thr (PINIT) motif have been proposed to regulate PIAS substrate specify and protein localization, respectively. Scale bars correspond to number of amino acids.
In accordance with these functional domains, SOCS proteins have been shown to inhibit JAK/STAT signaling in three distinct ways (Figure 1, Key Figure). First, all eight SOCS family members have an SH2 domain that enables them to bind phosphotyrosyl sites on the intracellular region of receptors and potentially compete with STATs for binding. However, direct evidence for this competition-based mechanism is lacking. Although CIS and SOCS2 interact with known STAT3 and STAT5 binding sites on the leptin receptor, direct competition between SOCS and STATs at these phosphotyrosyl sites has yet to be validated [33,34]. Additionally, Ram and Waxman have proposed that CIS inhibits growth hormone receptor (GHR) signaling in part by blocking STAT5b from binding to phosphotyrosyl sites on GHR. This conclusion was largely based on the indirect observation that CIS inhibition is more complete at lower STAT5b levels [35]. However, a more recent study using the mammalian protein-protein interaction trap (MAPPIT) technique has shown that neither CIS nor SOCS2 bind the same phosphotyrosyl sites as STAT5 on GHR [36].
Likewise, Endo et al. have demonstrated that CIS suppresses PRL receptor-mediated STAT5 activation by binding to a phosphotyrosyl site distinct from that of STAT5, raising the possibility that CIS inhibits STAT5 binding by steric hindrance or conformational changes rather than direct competition [37]. Therefore, more evidence is needed to determine whether SOCS proteins inhibit STAT signaling through direct competition at phosphotyrosyl sites, or whether other SH2-dependent mechanisms are at play, such as steric hindrance, SOCS-induced receptor modifications, or even direct SOCS-STAT interactions. An understanding of these mechanisms may inform the development of next-generation SOCS-SH2 mimetics that demonstrate increased selectivity.
The second mechanism of SOCS-mediated STAT inhibition is the ubiquitination and degradation of SH2 and amino-terminal bound SOCS substrates via the SOCS box domain [32]. It has been proposed that SOCS proteins facilitate the ubiquitination and proteasomal degradation of various components of the STAT signaling pathway, including ligands, receptors, tyrosine kinases, and other SOCS family members [38]. The primary SOCS E3 ligase substrates appear to be kinases. Both SOCS1 and SOCS3 have been implicated in the degradation of kinases, including JAK1, JAK2, TEL-JAK2, breast tumor kinase (BRK), and focal adhesion kinase (FAK) [31,38]. However, these studies have largely focused on substrate protein level changes following exogenous expression of either wild type or SOCS box mutant constructs or treatment with proteasome inhibitors. More direct experiments to identify endogenous substrates have been difficult due to the low abundance and transient nature of ubiquitinated complexes. Perhaps modified mass spectrometry techniques, such as proximity-based biotinylation (BioID), can be used to both validate known substrates and reveal novel substrates [39–41].
With regards to SOCS-dependent receptor degradation, relatively few receptors have been identified as SOCS E3 ligase substrates, and the few that have appear to undergo distinct degradation pathways in response to ubiquitination [42]. For example, ubiquitination of GHR by SOCS2 triggers proteasome-dependent receptor degradation [43], whereas ubiquitination of granulocyte CSF receptor (GCSFR) by SOCS3 facilitates receptor endocytosis and lysosomal degradation [42]. These divergent degradation pathways suggest that SOCS E3 ligases can differentially modify receptor substrates (monoubiquitination versus polyubiquitination). However, the precise mechanisms underlying these processes remain unclear, and many questions remain regarding the specificity, composition, and function of the SOCS E3 ubiquitin ligase complexes. In general, an understanding of these mechanistic details may inform the development of SOCS-specific drugs that enhance SOCS substrate recognition much like phthalimide-based derivatives.
The final mechanism of SOCS-dependent STAT inhibition is direct inhibition of JAK kinase activity via the KIR domain of SOCS1 and SOCS3. The strongest evidence for this mechanism comes from the recently elucidated crystal structure of SOCS3 bound to the kinase domain of JAK2 and gp130 [44]. In brief, SOCS3 forms a ternary complex with gp130 and JAK2. The SOCS3-gp130 interaction is a canonical SH2-phosphotyrosyl interaction, whereas the SOCS3-JAK2 interaction is phosphotyrosyl-independent, primarily hydrophobic, and centered on the JAK2 GQM motif. Unlike most tyrosine kinase inhibitors, which compete with ATP, the SOCS3 KIR domain blocks the substrate binding pocket on JAK2 and prevents JAK2-substrate interactions. Moreover, the affinity of the SOCS3-JAK2 interaction is much higher in the presence of a receptor containing a SOCS3-interaction motif, providing additional specificity to SOCS3-mediated JAK inhibition.
Since SOCS1 also contains a KIR domain, it might be conjectured that these SOCS3 structural-mechanistic findings also apply to SOCS1. However, previous studies have shown that SOCS1 can bind JAKs in the absence of a receptor with much higher affinity than SOCS3, raising the possibility that SOCS1-JAK interactions are distinct from gp130-SOCS3-JAK interactions [45]. Thus, it will be important to specifically determine the crystal structure of SOCS1 bound to JAK to more fully understand the mechanism of SOCS1-mediated kinase inhibition. Moreover, it will be interesting to determine how SOCS3 binds and inhibits other kinases, such as BRK and FAK, especially since the GQM motif is specific to JAK1, JAK2, and TYK2, and its absence seems to explain the inability of SOCS3 to bind and inhibit JAK3. A more complete mechanistic understanding of KIR-mediated kinase inhibition may inform the development of peptide mimetics and small molecules that function as a new class of kinase inhibitors [46].
Due to the critical role SOCS proteins play in inhibiting JAK/STAT signaling, loss of SOCS expression or function has been reported in many human cancers. Such mechanisms of SOCS loss include promoter hypermethylation, receptor truncations, and mutations (reviewed in [38,47]). Interestingly, several recent studies have demonstrated that microRNAs (miRNAs) can promote or suppress tumorigenesis by directly modulating SOCS expression. Zhang et al. reported that miR-572, a tumor promoting miRNA commonly upregulated in human ovarian cancer tissues and associated with poor overall survival, directly targets the 3’-untranslated region (UTR) of SOCS1 [48]. Additionally, they showed that knockdown of SOCS1 abrogated miR-572-mediated proliferation in ovarian cancer cell lines.
Similarly, Das and colleagues have shown that miR-194 drives prostate cancer metastasis by directly targeting the 3’-UTR of SOCS2. miR-194-mediated downregulation of SOCS2 resulted in de-repression of FLT3 and JAK2, leading to activation of ERK and STAT3 [49]. In contrast, Patel and associates have shown that miRNA let-7 acts as a tumor suppressor in pancreatic ductal adenocarcinoma (PDAC) by enhancing SOCS3 expression and suppressing STAT3 phosphorylation [50]. However, the mechanism by which let-7 stabilizes or increases SOCS3 levels remains unknown. Together, these findings suggest that re-expression of functional SOCS proteins may be an effective strategy for inhibiting oncogenic STATs. Such therapeutic strategies may include the use of epigenetic drugs, such as DNA methyltransferase inhibitors or histone deacetylase (HDAC) inhibitors, to restore SOCS expression, or miRNA-based techniques to inhibit or restore SOCS-specific miRNAs [51].
Protein Inhibitor of Activated STAT (PIAS) Proteins
The PIAS family consists of five major proteins, namely PIAS1, PIASxα, PIASxβ, PIAS3, and PIAS4 (PIASγ), and was originally named for the ability of these proteins to inhibit the transcriptional activity of STATs. PIAS1 and PIAS4 primarily interact with STAT1, whereas PIAS3 and PIASx interact with STAT3 and STAT4, respectively [52]. However, PIAS proteins have also been shown to regulate the activity of other transcription factors, including NF-κB and p53, and mediate various cellular processes by interacting with more than 60 proteins [53]. Structurally, PIAS family members are characterized by four conserved motifs: an amino-terminal scaffold attachment factor (SAF)-A/B, Acinus and PIAS (SAP) motif, a Pro-Ile-Asn-Ile-Thr (PINIT) motif, a RING-finger-like zinc-binding (RLD) motif, and a SUMO interaction motif (SIM; Figure 2B). In brief, the SAP motif is commonly found in chromatin-binding proteins and is thought to facilitate either sequence- or structure-specific DNA binding. The PINIT motif has been proposed to regulate protein localization and nuclear retention. The RLD is required for PIAS SUMO-E3-ligase activity, whereas the SIM motif has been proposed to contribute to substrate specificity (reviewed in [53,54]).
PIAS proteins have been shown to inhibit STAT transcriptional activity via three primary mechanisms [52,55] (Figure 1, Key Figure). First, PIAS proteins can interact with STATs and prevent STAT-DNA interactions. The Shuai laboratory used co-immunoprecipitation analyses in conjunction with in vitro electrophoretic mobility shift assays (EMSAs) to demonstrate that PIAS3 interacts with STAT3 in a phosphorylation-dependent manner and prevents both STAT3 homodimers and STAT1-STAT3 heterodimers from binding to DNA probes containing STAT3-specific binding sites. Interestingly, PIAS3 had no effect on the DNA binding ability of STAT1 homodimers [56]. They showed equivalent results for PIAS1-mediated inhibition of STAT1 [57]. Although these data are convincing, the mechanistic details describing how PIAS proteins prevent STAT-DNA interactions remain poorly understood. Additional studies are necessary to determine whether PIAS proteins mask STAT DNA-binding domains or disrupt STAT dimers. The second mechanism of PIAS-mediated STAT inhibition is recruitment of transcriptional co-repressors to STAT target genes. Pan et al. have recently demonstrated that PIAS3 and PIAS4 work together in lung adenocarcinoma to suppress STAT-dependent expression of SLUG, a major regulator of metastasis [58]. They showed that stimulation with OSM induces the phosphorylation and activation of both STAT1 and STAT3. Once in the nucleus, active STAT1 binds to the SLUG promoter and interacts with PIAS4, which recruits histone deacetylase 1 (HDAC1). HDAC1 removes gene-activating histone 3 lysine 9 acetylation (H3K9Ac) marks and further silences SLUG expression. In parallel, PIAS3 interacts with active STAT3 in the nucleus and prevents STAT3-binding at the SLUG promoter. Interestingly, these findings suggest that unlike PIAS3 binding to STAT3, PIAS4 binding to STAT1 does not alter STAT1-DNA interactions. Therefore, much remains to be understood about how PIAS family members physically interact with STATs.
The final mechanism of PIAS inhibition is PIAS-directed protein sumoylation. Although analogous to ubiquitination in terms of reaction scheme, sumoylation requires distinct enzymes that attach small ubiquitin-like modifiers (SUMOs), namely SUMO1, SUMO2, SUMO3, and SUMO4, to protein substrates. Depending on the SUMO isoform, the substrate, and the modification site, sumoylation has been reported to regulate sub-nuclear protein localization, protein-protein interactions, protein stability, and transcription factor activity [52,59]. Recent high-resolution mass spectrometry efforts have identified several STAT sumoylation sites, including five STAT1 sites (K114, K193, K652, K679, and K703) and one STAT3 site (K451) [60–62]. However, only two of these sites have been functionally characterized.
Due to the close proximity of the PIASxα-mediated STAT1 sumoylation site, K703, and the activating STAT1 tyrosine phosphorylation site, K701, the Niedenthal group hypothesized that STAT1 sumoylation inhibits STAT1 transcriptional activity by disrupting STAT1 phosphorylation and dimerization. To test this hypothesis, they fused a SUMO-conjugating enzyme to STAT1 and co-expressed it with or without EGFP-SUMO1. They found that sumoylation at K703 prevents tyrosine phosphorylation at Y701 [63]. Additionally, they found that tyrosine phosphorylation at Y701 prevents sumoylation at K703 [64]. They proposed that this mutually exclusive relationship between STAT1 sumoylation and tyrosine phosphorylation plays an important role in creating differentially modified pools of STAT1 and facilitating STAT1 transport back to the nucleus. This hypothesis remains to be formally tested.
More recently, Grönholm et al. used molecular modeling followed by mutational analyses to more fully characterize the functional significance of sumoylation at STAT1 K703. Using published structures of tyrosine-phosphorylated STAT1 homodimer bound to DNA and thymine-DNA glycosylase (TDG)-SUMO-1 as templates, they were able to model the location of the SUMO moiety at K703. Based on this model, they hypothesized that SUMO points towards the DNA and sterically hinders STAT1-DNA interactions. To test this hypothesis, they used oligonucleotide-mediated precipitation experiments to show that sumoylation deficient STAT1 has a higher affinity for DNA than wild type STAT1 [65]. It is important to note that although these findings demonstrate that PIAS-mediated sumoylation inhibits DNA binding via steric hindrance, PIAS proteins can also block STAT DNA-binding activity independently of sumoylation [59].
The second STAT sumoylation site that has been functionally characterized is STAT3 K451. Zhou and colleagues have demonstrated that sumoylation, specifically conjugation of SUMO2/3 with K451, inhibits STAT3 function by recruiting TC45, a SIM-containing nuclear phosphatase known to dephosphorylate STAT3 [66]. Moreover, they identified SENP3 as the SUMO2/3-specific protease responsible for STAT3 K451 desumoylation and hyperphosphorylation of STAT3 in head and neck cancer. In addition to elucidating the mechanism by which STAT3 sumoylation affects STAT3 transcriptional activity, these findings highlight a critical link between two classes of endogenous STAT inhibitors, namely PIAS proteins and phosphatases.
Although many mechanisms by which PIAS proteins inhibit STAT transcriptional activity have been elucidated, several questions remain. First, the source of PIAS substrate specificity remains poorly understood. Although PIAS1 and PIAS3 are unable to bind and inhibit STAT3 and STAT1, respectively, in vivo, this specificity is lost when PIAS1 and PIAS3 are overexpressed or analyzed in vitro, suggesting that PIAS-STAT specificity depends on posttranslational modifications, subcellular localization, or protein-protein interactions only present in vivo [57]. An understanding of this specificity may inform the development of STAT3- or STAT5-specific inhibitors that do not inhibit the primarily tumor suppressive role of STAT1.
Interestingly, PIAS3 has recently been shown to interact with and inhibit STAT5, in addition to STAT3, and this PIAS3-STAT5 inhibitory interaction depends on the amino-terminal domain of STAT5, which is missing from the oncogenic STAT5 truncation mutant found in prostate cancer cells [67]. Thus, loss of inhibition by PIAS3 promotes prostate cancer tumorigenesis. In contrast, overexpression of PIAS1 in local and metastatic prostate cancer appears to promote tumorigenesis in part by inhibiting STAT1 and indirectly promoting STAT3 [68]. These findings suggest that an understanding of the PIAS-STAT landscape within a particular tumor environment will be critical for the development of effective STAT inhibitors. PIAS mimetics may be particularly beneficial in some cancer types, while PIAS inhibitors may be effective in others.
Another therapeutically intriguing aspect of PIAS-mediated STAT inhibition is that PIAS deletion only affects a subset of STAT target genes as determined by the strength of the STAT binding site [52]. Thus, PIAS-sensitive genes contain weaker STAT binding sites than PIAS-insensitive genes. A better understanding of this selectivity may enable the development of STAT inhibitors that preferentially target STAT target genes involved in oncogenesis, potentially enhancing specificity and limiting toxicity.
Therapeutic Implications of Endogenous STAT Inhibitors
Although many questions remain regarding the mechanisms by which SOCS and PIAS proteins inhibit STAT activity, several groups have begun using known mechanistic details to develop novel therapeutic strategies. So far, the two most prominent strategies are to restore the expression of SOCS and PIAS family members and to mimic the kinase inhibitory activity of SOCS1.
As previously mentioned, SOCS expression is commonly suppressed in cancer via epigenetic gene silencing. SOCS promoter hypermethylation has been detected in 90% of head and neck cancers, as well as in nearly 50% of pancreatic cancers, hepatoblastoma, hepatocellular carcinoma, melanoma, AML, and multiple myeloma (reviewed in [47]). In many of these cases, as demonstrated by Niwa and colleagues for hepatocellular carcinoma, SOCS hypermethylation correlates with decreased SOCS expression and increased JAK/STAT activity [69]. Additionally, several groups have demonstrated that HDAC inhibitors can restore SOCS3 expression in hepatocellular carcinoma and myeloproliferative neoplasms, suggesting that hypoacetylation also plays a role in SOCS silencing [70–72]. Together, these findings support the therapeutic use of DNA methyltransferase inhibitors, such as azacytidine, and HDAC inhibitors, such as suberanilohydroxamic acid, in SOCS-low cancers to restore SOCS expression and inhibit oncogenic STAT activity.
Interestingly, loss of PIAS expression has also been reported in cancer, but the mechanism of PIAS loss appears to be distinct from that of SOCS loss. Brantley et al. reported that although PIAS3 mRNA levels are similar between control and human glioblastoma multiforme (GBM) tissues, PIAS3 protein levels are significantly lower in the GBM tissues. Accordingly, they showed that STAT3 activation, as measured by tyrosine and serine phosphorylation, is elevated in the GBM tissues [73]. Based on these findings, they hypothesized that PIAS3 loss in glioblastoma may be caused by rapid protein degradation. To test this hypothesis, they treated human glioblastoma cells with MG-132, a proteasome inhibitor, and looked at PIAS3 protein levels. Indeed, they found that treatment with MG-132 led to an increase in PIAS3 protein levels over time, suggesting that ubiquitin-mediated degradation may promote PIAS loss in vivo. These findings make a strong argument for the use of proteasomal inhibitors, such as bortezomib or marizomib, to increase PIAS3 protein levels and inhibit STAT3 activity in GBM, as well as other PIAS3-low cancers [74].
The second strategic approach has been to develop kinase inhibitors that mimic the unique kinase inhibitory activity of SOCS1 rather than functioning as general ATP analogs, which are somewhat limited by high intracellular levels of ATP and lack of specificity [32]. Flowers et al. provided the first proof-of-concept by developing a short 12-mer peptide mimetic of SOCS1 called JAK2 tyrosine kinase inhibitor peptide (Tkip) that binds to the autophosphorylation site of JAK2 and inhibits JAK2 tyrosine kinase activity [75]. Additionally, they demonstrated that Tkip inhibits proliferation, as well as constitutive and IL-6 induced STAT3 phosphorylation, in two prostate cancer cell lines [76]. Several other groups have since developed SOCS1 KIR peptide mimetics for use in STAT1-dependent immune-mediated diseases [77,78]. Although these peptide mimetics are intriguing from a proof-of-concept perspective, small molecule mimetics will likely be more effective for translational applications.
Concluding Remarks
Although this review focuses on only two of the three major families of endogenous STAT inhibitors, namely SOCS and PIAS proteins, an understanding of STAT phosphatases will also be important. Since each of these three families of negative regulators converge on STATs from a slightly different direction, a mechanistic investigation of each family has the potential to reveal novel therapeutic strategies that can be used in combination for the treatment of STAT-dependent cancers (see Outstanding Questions). In general, the ability to inhibit single STAT family members, and possibly even specific subsets of STAT target genes, will be key for developing therapeutically effective STAT inhibitors.
Outstanding Questions.
How does crosstalk between STAT family members functionally mediate STAT-dependent oncogenesis?
How do SOCS and PIAS proteins mechanistically modulate STAT function? For example: How and to what extent do SOCS proteins inhibit JAK-independent kinases? How do SOCS proteins inhibit STAT-receptor interactions? How does PIAS-mediated STAT sumoylation affect STAT sub-nuclear localization?
What determines the selectivity and specificity of endogenous STAT inhibitors?
What role do endogenous STAT inhibitors play in regulating mitochondrial STAT function?
Is it possible to develop therapeutically effective STAT inhibitors that modulate or mimic endogenous STAT inhibitors?
Trends Box.
Constitutively active signal transducer and activator of transcription (STAT) proteins drive tumor initiation and progression in many human cancers.
Therapeutic STAT inhibitors have been limited by insufficient specificity, potency, bioavailability, and delivery, as well as unknown mechanisms of action.
Recent chemical biology approaches have identified novel STAT inhibitors that may function by activating endogenous STAT inhibitors.
The mechanisms of action of endogenous STAT inhibitors, namely suppressor of cytokine signaling (SOCS) proteins, phosphatases, and protein inhibitor of activated STAT (PIAS) proteins, are complex and remain incompletely understood.
Text Box 1.
The STAT Family of Transcription Factors
The STAT family consists of seven members, namely STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6. Although STAT5a and STAT5b are encoded by separate genes, they share many overlapping functions and are often referred to collectively as STAT5 [21]. STATs are characterized by six functional domains, namely the amino terminal domain (NH2), the coiled-coil domain (CCD), the DNA-binding domain (DBD), the linker domain, the Src homology 2 domain (SH2), and the carboxyl-terminal transcriptional activation domain (TAD) [79]. Each of these domains has been shown to play an important role in STAT function by directly or indirectly modulating STAT phosphorylation, nuclear translocation, and DNA-binding.
STATs regulate gene expression in response to various cytokines and growth factors, including colony-stimulating factors (CSFs), interferons (IFNs), some interleukins (ILs), erythropoietin (EPO), thrombopoietin (TPO), growth hormone (GH), prolactin (PRL), and leptin [80]. These factors bind to cognate receptors and induce receptor oligomerization, which brings receptor-associated Janus kinases (JAKs; see Glossary) into close proximity with one another and facilitates JAK transphosphorylation. Active JAKs then phosphorylate nearby tyrosine residues on the intracellular region of the receptor, creating docking sites for cytoplasmic SH2-containing STATs. Following receptor docking, STATs are phosphorylated and activated by receptor-associated JAKs. Active STAT dimers then translocate to the nucleus where they bind DNA and regulate the transcription of target genes.
Glossary
- Signal transducer and activator of transcription (STAT)
a seven-member family of intracellular transcription factors that mediates various aspects of differentiation, immunity, inflammation, proliferation, and survival in response to cytokines and growth factors. Following activation by upstream kinases, STATs translocate from the cytoplasm to the nucleus where they bind to a consensus DNA-recognition motif (TTCNmGAA) and regulate transcription. STAT misregulation has been widely implicated in the pathogenesis of cancer
- Janus kinase (JAK)
a four-member family of non-receptor tyrosine kinases that associates with the intracellular region of membrane-bound receptors and plays a key role in JAK/STAT signal transduction. Following receptor-ligand engagement, receptor oligomerization facilitates JAK transphosphorylation and activation. Activated JAKs then phosphorylate and activate downstream STATs, which translocate to the nucleus and regulate the transcription of specific target genes. As upstream regulators of STAT activity, hyperactive JAK mutants tend to be oncogenic
- Suppressor of cytokine signaling (SOCS)
an eight-member family of JAK/STAT negative feedback regulators that limits JAK/STAT signaling to a relatively short pulse following cytokine stimulation. SOCS proteins have been shown to function via three mechanisms, namely inhibiting JAK kinase activity, preventing STAT-receptor docking, and facilitating the degradation of JAK/STAT signaling machinery
- Protein inhibitor of activated STAT (PIAS)
a family of transcriptional regulators that was originally named for the ability to inhibit STAT proteins. Four PIAS genes, PIAS1, PIAS2 (PIASx), PIAS3, and PIAS4 (PIASγ), encode five primary protein products, including PIAS1, PIASxα, PIASxβ, PIAS3, and PIASγ. PIAS proteins have been shown to inhibit STAT transcriptional activity via three distinct mechanisms, namely preventing STAT-DNA interactions, recruiting transcriptional co-repressors, and mediating STAT sumoylation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhagwat AS, Vakoc CR. Targeting Transcription Factors in Cancer. Trends in Cancer. 2015;1:53–65. doi: 10.1016/j.trecan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeh JE, et al. Targeting transcription factors: promising new strategies for cancer therapy. Curr. Opin. Oncol. 2013;25:652–658. doi: 10.1097/01.cco.0000432528.88101.1a. [DOI] [PubMed] [Google Scholar]
- 4.Miklossy G, et al. Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov. 2013;12:611–629. doi: 10.1038/nrd4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buettner R, et al. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 6.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32:2601–2613. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 7.Quintás-Cardama A, Verstovsek S. Molecular pathways: JAK/STAT pathway: Mutations, inhibitors, and resistance. Clin. Cancer Res. 2013;19:1933–1940. doi: 10.1158/1078-0432.CCR-12-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villarino AV, et al. Mechanisms of Jak/STAT Signaling in Immunity and Disease. J. Immunol. 2015;194:21–27. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, et al. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaler P, et al. The Role of STAT1 for Crosstalk between Fibroblasts and Colon Cancer Cells. Front. Oncol. 2014;4:88. doi: 10.3389/fonc.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wee ZN, et al. EZH2-mediated inactivation of IFN-γ-JAK-STAT1 signaling is an effective therapeutic target in MYC-driven prostate cancer. Cell Rep. 2014;8:204–216. doi: 10.1016/j.celrep.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 12.Kharma B, et al. STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Res. 2014;74:6519–6530. doi: 10.1158/0008-5472.CAN-14-0847. [DOI] [PubMed] [Google Scholar]
- 13.Kortylewski M, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 14.Yu H, et al. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 15.Avalle L, et al. STAT1 and STAT3 in tumorigenesis: A matter of balance. Jak-Stat. 2012;1:65–72. doi: 10.4161/jkst.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nivarthi H, et al. The ratio of STAT1 to STAT3 expression is a determinant of colorectal cancer growth. Oncotarget. 2016;5:51096–51106. doi: 10.18632/oncotarget.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoelbl A, et al. Stat5 is indispensable for the maintenance of Bcr/Abl-positive leukaemia. EMBO Mol. Med. 2010;2:98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warsch W, et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood. 2011;117:3409–3420. doi: 10.1182/blood-2009-10-248211. [DOI] [PubMed] [Google Scholar]
- 19.Warsch W, et al. STAT5 triggers BCR-ABL1 mutation by mediating ROS production in chronic myeloid leukaemia. Oncotarget. 2012;3:1669–1687. doi: 10.18632/oncotarget.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leong PL, et al. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene. 2002;11:2846–2853. doi: 10.1038/sj.onc.1205385. [DOI] [PubMed] [Google Scholar]
- 21.Walker SR, et al. Distinct roles of STAT3 and STAT5 in the pathogenesis and targeted therapy of breast cancer. Mol. Cell. Endocrinol. 2014;382:616–621. doi: 10.1016/j.mce.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H, Jove R. The STATs of cancer — new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 23.Darnell JE. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 24.Furtek SL, et al. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS Chem. Biol. 2016;11:308–318. doi: 10.1021/acschembio.5b00945. [DOI] [PubMed] [Google Scholar]
- 25.Furqan M, et al. STAT inhibitors for cancer therapy. J. Hematol. Oncol. 2013;6:90. doi: 10.1186/1756-8722-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorritie KA, et al. STAT transcription factors in hematopoiesis and leukemogenesis: opportunities for therapeutic intervention. Leukemia. 2014;28:248–257. doi: 10.1038/leu.2013.192. [DOI] [PubMed] [Google Scholar]
- 27.Nelson EA, et al. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112:5095–5102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takakura A, et al. Pyrimethamine inhibits adult polycystic kidney disease by modulating STAT signaling pathways. Hum. Mol. Genet. 2011;20:4143–4154. doi: 10.1093/hmg/ddr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson EA, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117:3421–3429. doi: 10.1182/blood-2009-11-255232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson EA, et al. The STAT5 Inhibitor Pimozide Displays Efficacy in Models of Acute Myelogenous Leukemia Driven by FLT3 Mutations. Genes Cancer. 2012;3:503–511. doi: 10.1177/1947601912466555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahony R, et al. SOCS3 revisited: a broad regulator of disease, now ready for therapeutic use? Cell. Mol. Life Sci. 2016;73:3323–3336. doi: 10.1007/s00018-016-2234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linossi EM, et al. Suppression of cytokine signaling: The SOCS perspective. Cytokine Growth Factor Rev. 2013;24:241–248. doi: 10.1016/j.cytogfr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavens D, et al. Review: Negative regulation of leptin receptor signalling. Eur. Cytokine Netw. 2006;17:211–219. [PubMed] [Google Scholar]
- 34.Lavens D, et al. A complex interaction pattern of CIS and SOCS2 with the leptin receptor. J. Cell Sci. 2006;119:2214–2224. doi: 10.1242/jcs.02947. [DOI] [PubMed] [Google Scholar]
- 35.Ram PA, Waxman DJ. SOCS/CIS protein inhibition of growth hormone-stimulated STAT5 signaling by multiple mechanisms. J. Biol. Chem. 1999;274:35553–35561. doi: 10.1074/jbc.274.50.35553. [DOI] [PubMed] [Google Scholar]
- 36.Uyttendaele I, et al. Mammalian protein-protein interaction trap (MAPPIT) analysis of STAT5, CIS, and SOCS2 interactions with the growth hormone receptor. Mol. Endocrinol. 2007;21:2821–2831. doi: 10.1210/me.2006-0541. [DOI] [PubMed] [Google Scholar]
- 37.Endo T, et al. CIS1 Interacts with the Y532 of the Prolactin Receptor and Suppresses Prolactin-Dependent STAT5 Activation. J. Biol. Chem. 2003;133:109–113. doi: 10.1093/jb/mvg004. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad A, Ahmad R. Understanding the mechanism of hepatic fibrosis and potential therapeutic approaches. Saudi J. Gastroenterol. 2012;18:155–167. doi: 10.4103/1319-3767.96445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyaud E, et al. BioID-based Identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 Ligase Substrates. Mol. Cell. Proteomics. 2015;14:1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haglund K, et al. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 41.Irandoust MI, et al. Suppressor of cytokine signaling 3 controls lysosomal routing of G-CSF receptor. EMBO J. 2007;26:1782–1793. doi: 10.1038/sj.emboj.7601640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croker BA, et al. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 2008;19:414–422. doi: 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vesterlund M, et al. The SOCS2 ubiquitin ligase complex regulates growth hormone receptor levels. PLoS One. 2011;6:1–10. doi: 10.1371/journal.pone.0025358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kershaw NJ, et al. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat. Struct. Mol. Biol. 2013;20:469–476. doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshimura A, Yasukawa H. JAK’s SOCS: A Mechanism of Inhibition. Immunity. 2012;36:157–159. doi: 10.1016/j.immuni.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Babon JJ, et al. Suppression of Cytokine Signaling by SOCS3: Characterization of the Mode of Inhibition and the Basis of Its Specificity. Immunity. 2012;36:239–250. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inagaki-Ohara K, et al. SOCS, inflammation, and cancer. Jak-Stat. 2013;2:e24053. doi: 10.4161/jkst.24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, et al. Upregulation of miR-572 transcriptionally suppresses SOCS1 and p21 and contributes to human ovarian cancer progression. Oncotarget. 2015;6:15180–93. doi: 10.18632/oncotarget.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das R, et al. MicroRNA-194 promotes prostate cancer metastasis by inhibiting SOCS2. Cancer Res. 2017;77:1021–1034. doi: 10.1158/0008-5472.CAN-16-2529. [DOI] [PubMed] [Google Scholar]
- 50.Patel K, et al. MicroRNA let-7 downregulates STAT3 phosphorylation in pancreatic cancer cells by increasing SOCS3 expression. Cancer Lett. 2014;347:54–64. doi: 10.1016/j.canlet.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah MY, et al. microRNA Therapeutics in Cancer — An Emerging Concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16:196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- 53.Rabellino A, et al. The Role of PIAS SUMO E3-Ligases in Cancer. Cancer Res. 2017;77:1542–1547. doi: 10.1158/0008-5472.CAN-16-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 55.Yuan J, et al. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci. Rep. 2016;5:1–10. doi: 10.1038/srep17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung CD, et al. Specific Inhibition of Stat3 Signal Transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 57.Liu B, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan C, et al. Oncostatin M suppresses metastasis of lung adenocarcinoma by inhibiting SLUG expression through coordination of STATs and PIASs signalings. Oncotarget. 2016;7:60395–60406. doi: 10.18632/oncotarget.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 2007;35:1405–1408. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- 60.Hendriks IA, et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014;21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matic I, et al. Site-Specific Identification of SUMO-2 Targets in Cells Reveals an Inverted SUMOylation Motif and a Hydrophobic Cluster SUMOylation Motif. Mol. Cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 62.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jakobs A, et al. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat. Methods. 2007;4:245–250. doi: 10.1038/nmeth1006. [DOI] [PubMed] [Google Scholar]
- 64.Zimnik S, et al. Mutually exclusive STAT1 modifications identified by Ubc9/substrate dimerization-dependent SUMOylation. Nucleic Acids Res. 2009;37:1–7. doi: 10.1093/nar/gkp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grönholm J, et al. Structure-function analysis indicates that sumoylation modulates DNA-binding activity of STAT1. BMC Biochem. 2012;13:20. doi: 10.1186/1471-2091-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Z, et al. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene. 2016;35:5826–5838. doi: 10.1038/onc.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dagvadorj A, et al. N-terminal truncation of Stat5a/b circumvents PIAS3-mediated transcriptional inhibition of Stat5 in prostate cancer cells. Int. J. Biochem. Cell Biol. 2010;42:2037–2046. doi: 10.1016/j.biocel.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puhr M, et al. PIAS1 is a crucial factor for prostate cancer cell survival and a valid target in docetaxel resistant cells. Oncotarget. 2014;5:12043–12056. doi: 10.18632/oncotarget.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niwa Y, et al. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 2005;24:6406–6417. doi: 10.1038/sj.onc.1208788. [DOI] [PubMed] [Google Scholar]
- 70.Xiong H, et al. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol. Carcinog. 2012;51:174–184. doi: 10.1002/mc.20777. [DOI] [PubMed] [Google Scholar]
- 71.Chen C, et al. Pure curcumin increases the expression of SOCS1 and SOCS3 in myeloproliferative neoplasms through suppressing class I histone deacetylases. Carcinogenesis. 2013;34:1442–1449. doi: 10.1093/carcin/bgt070. [DOI] [PubMed] [Google Scholar]
- 72.Gao S, et al. Histone deacetylases inhibitor sodium butyrate inhibits JAK2/STAT signaling through upregulation of SOCS1 and SOCS3 mediated by HDAC8 inhibition in myeloproliferative neoplasms. Exp. Hematol. 2013;41:261–270. doi: 10.1016/j.exphem.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 73.Brantley EC, et al. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: Implications for STAT-3 activation and gene expression. Clin. Cancer Res. 2008;14:4694–4704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manton CA, et al. Induction of cell death by the novel proteasome inhibitor marizomib in glioblastoma in vitro and in vivo. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flowers LO, et al. Characterization of a Peptide Inhibitor of Janus Kinase 2 That Mimics Suppressor of Cytokine Signaling 1 Function. J. Immunol. 2004;172:7510–7518. doi: 10.4049/jimmunol.172.12.7510. [DOI] [PubMed] [Google Scholar]
- 76.Flowers LO, et al. A SOCS-1 peptide mimetic inhibits both constitutive and IL-6 induced activation of STAT3 in prostate cancer cells. Oncogene. 2005;24:2114–2120. doi: 10.1038/sj.onc.1208437. [DOI] [PubMed] [Google Scholar]
- 77.Madonna S, et al. Therapeutical potential of a peptide mimicking the SOCS1 kinase inhibitory region in skin immune responses. Eur. J. Immunol. 2013;43:1883–1895. doi: 10.1002/eji.201343370. [DOI] [PubMed] [Google Scholar]
- 78.Doti N, et al. New mimetic peptides of the kinase-inhibitory region (KIR) of SOCS1 through focused peptide libraries. Biochem. J. 2012;443:231–240. doi: 10.1042/BJ20111647. [DOI] [PubMed] [Google Scholar]
- 79.Levy DE, Darnell JE. Signalling: STATs: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 80.O’Shea JJ, et al. JAKs and STATs in Immunity, Immunodeficiency, and Cancer. N. Engl. J. Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]