Abstract
In this study, confocal Raman microspectroscopy was used to detect lipids in microalgae rapidly and non-destructively. Microalgae cells were cultured under nitrogen deficiency. The accumulation of lipids in Scenedesmus obliquus was observed by Nile red staining, and the total amount of lipids accumulated in the cells was measured by gravimetric method. The signals from different microalgae cells were collected by confocal Raman microspectroscopy to establish a prediction model of intracellular lipid content, and surface scanning signals for drawing pseudo color images of lipids distribution. The images can show the location of pyrenoid and lipid accumulation in cells. Analyze Raman spectrum data and build PCA-LDA model using four different bands (full bands, pigments, lipids, and mixed features). Models of full bands or pigment characteristic bands were capable of identifying S. obliquus cells under different nitrogen stress culture time. The prediction accuracy of model of lipid characteristic bands is relatively low. The correlation between the fatty acid content measured by the gravimetric method and the integral Raman intensity of the oil characteristic peak (1445 cm−1) measured by Raman spectroscopy was analyzed. There was significant correlation (R 2 = 0.83), which means that Raman spectroscopy is applicable to semi-quantitative detection of microalgal lipid content.
Keywords: Raman microspectroscopy, Microalgae, Lipids
Background
The world energy crisis is intensified continuously nowadays, which raises interest in looking for renewable energy resources. Among the existing renewable energy sources, biodiesel is the most widely used variety which also have the fastest speed of development. Microalgae have good potential for fuel production because of their promising biomass feedstock and ability to synthesize large amounts of certain chemical compounds from sunlight and carbon dioxide [1, 2]. Under the environment of nitrogen stress, some kind of microalgae (e.g., Chlorella pyrenoidosa, Scenedesmus obliquus…) significantly synthesize and accumulate lipids, mainly in the form of triacylglycerol (TAG) [3, 4].
Several methodologies used for quantitative analysis of microalgae lipids, such as gas chromatography–mass spectrometry (GC–MS), are not just time-consuming and ruinous, but also requires sophisticated sample preparation process which produces hazardous chemical waste. Although GC–MS provide precise analysis of lipid composition, it cannot provide the detailed information of lipid metabolism. Vital staining methods using Nile Red or BODIPY 505/515 can show lipid distribution in a single cell but cannot provide some desired lipid characteristics such as chain length and degree of unsaturation [5, 6]. Studies showed that the lipid content of one cell was significantly correlated with the fluorescence intensity of Nile Red combined with the lipid inside the cell [7, 8].
Confocal Raman microscopy is a powerful tool for physicochemical characterization of biological samples, which directly detects vibrations of biochemical bonds through the inelastic scattering by a laser light [9]. It enables single-cell, in vivo monitoring of various cellular components in a rapid, non-destructive, label-free and quantitative manner [10–13]. With the assistance of chemometric methods, Raman spectroscopy can be applied to wide research area like microalgae species identification [14], water pollution identification, and nutritional status identification [15–17]. A study showed that the water bodies under different nitrogen nutrition conditions can be effectively identified by the LDA classification model established based on lipid-related Raman shift [18]. Heraud described a in vivo method for predicting the nutrient status of individual algae cells using Raman microscopy and partial least squares discriminant analysis [19]. Samek employed the characteristic peaks in the Raman scattering spectra at 1656 and 1445 cm−1 as the markers defining the ratio of unsaturated-to-saturated carbon–carbon bonds of the fatty acids in the algal lipids [20]. In situ and in vivo chemical compounds distribution and concentration can be shown by Raman spectrum data processing [21–24].
At the present time, the research of microalgae lipid production in China is mostly focused on promoting the ability of lipid production of microalgae through cultivation environment modifying or genetic engineering as well as optimization of lipid conversion technology. Development of rapid and non-destructive Raman spectrum testing process toward microalgae lipid will accelerate the research on biodiesel production. Problems still remain in the direct quantitative and qualitative detection research of microalgae lipid using Raman spectrum. In our preliminary experiments, we found that the characteristic peaks of lipids are not obvious due to the concealment effect of pigments (e.g., chlorophyll, carotenoids). The object of the preliminary experiment was Chlorella sp. Although this kind of microalgae can accumulate large amounts of lipids, its cellular volume is so small that obstructs the acquisition of spectrum and the process of data. The object of this study was a different kind of microalgae, S. obliquus. Scenedesmus obliquus is a Chlorophyta that the dry weight of its cellular lipid content can be accounted for more than 50% of the weight of cells. The large cell individual is suitable for the detection research of Raman point and surface scan. Under normal growth conditions, the cells of S. obliquus are fusiform. Combined living form of four cells is common, which can be changed under the stress environment.
This study, using S. obliquus as the experimental subject, verified the ability of Raman spectroscopy to classify cells collected from different days cultivated under nitrogen stress, and discussed the optimization of detection process according to the Raman mapping measurement imaging results of single cells.
We first list the materials and methods of the experiment, including the cultivation of microalgae, Nile Red staining and observing process, gravimetric analysis of total lipid content, GC–MS analysis of lipid composition and Raman spectral acquisition. We next build a PCA-LDA model to classify microalgae cells collected from different days, and its correct classification rate reached to 100%. We then make visual analysis of cellular lipids and pigments and cell growth pattern based on the Raman mapping data. The paper also attempts to discuss the correlations between the lipid content measured by gravimetric analysis and Raman mapping measurement.
Methods
Algae species and culture conditions
Scenedesmus obliquus, FACHB-276, was purchased in Freshwater Algae Culture Collection at the Institute of Hydrobiology, FACHB-collection. The SE basal medium was configured according to the standards provided by the Institute of Hydrobiology. Scenedesmus obliquus was expanded in the SE medium for 30 days and cultured to a stable growth stage. 2 l BG11 nitrogen-deficient (BG11-N) medium was configured with NaNO3 concentration of 0.5 g/l. 1 l uniform algae fluid was centrifugated and then liquid supernatant was discarded. The washed algal mud was quickly added to the BG11-N medium, placed at a constant temperature (25–27 °C), under uniform illumination (2000–3000 lx), and under periodic illumination conditions (12 l:12 days) cultured continuously for 9 days. Before each experiment, the microalgae cell concentration was estimated by the cell count plate method to ensure that the microalgae were in stable growth during the whole experiment.
Intracellular lipid accumulation observation
The intracellular lipid of S. obliquus was observed by Nile red staining. 1.5 ml uniform algae fluid and 0.5 ml DMSO were mixed, and oscillated 10 min for breaking cytoderm. 20 μl NR liquor (0.1 g/l) was then added, and oscillated 10 min for dyeing evenly. A 2 μl stained algae fluid was taken for the observation by fluorescence microscope (Nikon Eclipse 90i, 20× objective lens). The excitation wavelength range is 505–566 nm. Pictures of the stained algae cells were taken using corresponding image processing software.
Determination of total lipid content
The total fat content was determined by gravimetric method. 50-ml centrifuge tube (M0) was washed and weighed after drying. 30 ml uniform algae fluid was added in the centrifuge tube, and liquid supernatant was discarded after centrifugation. The algae mod was dried in the centrifuge tube in the drying oven under 60 °C then weighed (M1). Some 1 mol/l HCL was added and then heated the mixture in 80 °C water bath for 30 min: liquid supernatant was discarded after centrifugation. The extraction methods were performed as per the Bligh–Dyer method [25]. 6 ml methanol:chloroform = 2:1 mixed solution was added into the residual, and oscillated by the vortex oscillator until all the algal residue turned white. After 4-h stewing, the chloroform layer was transferred to a culture dish that had been washed, dried and weighed (M2). It was dried to constant weighing in the draught cupboard then weighed (M3). Lipid content (LC) was calculated according to the following formula:
| 1 |
Determination of lipid composition
The oil composition of algae cells was directly treated with methyl esterification. Fatty acid methyl ester (FAMEs) was analyzed by gas chromatograph-mass spectrometer (GC–MS). At ninth day, 240 ml uniform algae fluid was centrifugated and the supernatant was discarded. The algae mud was transferred to a 50-ml round-bottomed flask and crashed it using ultrasonication for 10 min. 10 ml reactant mixture (methanol:chloroform: HCL = 10:1:1) was added into the algae mud, mixed, and then refluxed in 90 °C water baths for 4 h. After the reaction is complete, the flask was removed and cooled it to room temperature. 3 ml hexane was added into the mixture for extracting the FAMEs, and repeated three times. The extract was dried with nitrogen and then added with 1 ml hexane. 37 FAMEs mixture standard heating process was used for GC–MS detection.
Raman spectrum acquisition
Sample preparation
1.5 g powdered agar and 50 ml distilled water were mixed, boiled for 2 min, and then cooled to 40 °C. 2 ml algae fluid was transferred to a clean centrifuge tube, and 2 ml prepared agar solution was added. The mixture was cooled and solidified The solidified sample was cut into thin slices and was placed it on the slide.
Data acquisition
The Renishaw laser confocal micro Raman spectrometer (Renishaw PLC, United Kingdom/InVia–Reflex 532/XYZ) was the main instruments for this study. Data acquisition software WiRE3.3 was used to adjust the acquisition parameters. Specific parameters were as follows: the excitation wavelength is 532 nm; the spectral collection range is 633–1813 Raman shift/cm−1; the laser intensity is 1.5 W; and the exposure time is 2 s. 15 single data were acquired . S. obliquus cells were selected with different growth forms and mapping data were acquired, after the target area was selected; acquisition step is 0.5 μm. 1–2 sets of mapping data were acquired for each growth morphology of the cells. The experiment was performed every other day; 75 sets of single data and 20 sets of mapping data were acquired altogether.
Spectral data processing
Single data processing and modeling
In this paper, we use Baseline correction, Savitzky–Golay smoothing (SG) and data normalization to preprocess single data. The purpose of the baseline correction is to eliminate the baseline drift caused by the instrument or other interference and reduce the data error through polynomial fitting and other mathematical algorithms [26]. Through the least square fitting of spectral data, SG can reduce data noise level and retain the characteristics of spectral distribution, such as characteristic peak height, relative maximum, minimum and spectral peak width to a great extent [27]. Different spectral data processings apply different data normalization formulas. In this paper, the relative content of oil and chlorophyll is used to describe the accumulation of oil over time.
In order to reduce modeling variables and computation, principal components analysis (PCA) was used to reduce the dimension of spectral data after pretreatment [28]. Linear discriminant analysis (LDA) was used for modeling. LDA is a classical algorithm for pattern recognition. The purpose is to obtain the best separability of the model in the space by vector projection [29]. Data processing softwares were MATLAB R2015b and The Unscrambler X®.
Mapping data processing
Baseline correction and SG smoothing were used for preprocessing the mapping data. After selecting the peak of target characteristic, the pseudo color image of lipids distribution is drawn by interpolation method.
Results and discussion
Content and composition of lipids in S. obliquus
Under nitrogen stress, S. obliquus showed significant accumulation and accumulation of lipids. The total lipid content of microalgae was 20.54% before stress culture; this value increased to 30.77% at the ninth day of stress culture (Fig. 1). Rate of increase was 49.80%.
Fig. 1.
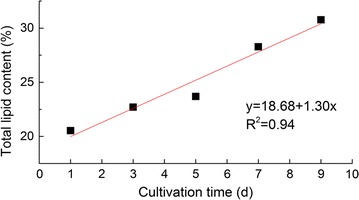
Fitting curve of total lipid content of S. obliquus
The GC/MS result (Table 1) shows that saturated fatty acids (SFA) and unsaturated fatty acids (UFA) synthesized by S. obliquus cells accounted for 55.66 and 44.34%, respectively. Monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA) accounted for 14.46 and 29.89%, respectively. In addition, small amount of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) were detected. As for the number of fatty acids, 47.83% of the accumulated oil in this algae species under nitrogen deficiency environment is odd carbon fatty acid (OCFA). Microorganisms tend to synthesize high ratio of OCFA, because of its unique metabolic process different from higher animals and plants [30]. The above analysis results show that, with the capability of synthesizing large number of long-chain fatty acids, S. obliquus can be used as potential algae for biodiesel production. UFA synthesized by these algae has nutrient value; OCFA has industrial and health care value [31].
Table 1.
Fatty acid composition of S. obliquus
| Fatty acids | Content/% |
|---|---|
| C14:0 | 1.42 |
| C15:0 | 35.32 |
| C16:0 | 4.77 |
| C16:1 | 4.20 |
| C16:3 | 1.94 |
| C17:0 | 10.30 |
| C18:0 | 3.84 |
| C18:1 | 8.05 |
| C18:2 | 1.71 |
| C18:3 | 25.12 |
| C19:1 | 2.21 |
| C20:5 (EPA) | 0.40 |
| C22:6 (DHA) | 0.72 |
| SFA | 55.66 |
| UFA | 44.34 |
| MUFA | 14.46 |
| PUFA | 29.89 |
| OCFA | 47.83 |
NR microscopic examination of S. obliquus
The results of NR staining microscopy (Fig. 2) showed that the lipid content of S. obliquus cells accumulated obviously during the period of nitrogen stress. Most of the cells were spindle shaped, and the pyrenoid in the cells was clearly visible. During the cultivation period, size and intracellular chlorophyll content of S. obliquus cells had not changed. Because of the polar lipids on the surface of the algal cells, the color of cells in the fluorescent images was basically orange, and the region of pyrenoid was also orange. Bright yellow spots appeared over time, which were the lipid particles that were accumulated in the algae cells.
Fig. 2.
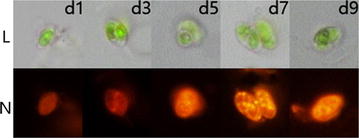
Light-field images (L) and NR fluorescent images (N) of S. obliquus at different days after lipid accumulation induction
Visual analysis of intracellular constituents in S. obliquus
In order to assess the ability of Raman spectroscopy to detect the intracellular substances of S. obliquus, visualization processing of Raman mapping data was carried out. In Raman spectrum of S. obliquus cells, the peaks of 1266 and 1656 cm−1 corresponded to the cis = C–H in-plane deformation and the cis C = C stretch, respectively. These two peaks mainly represented the degree of unsaturation of the fat chain. The peaks of 1302 and 1445 cm−1 corresponded to the CH2 twist and the CH2 bend, respectively. These two peaks were assigned to saturated carbon chain [32]. Raman images of saturated fatty acid (1445 cm−1) and unsaturated fatty acid (1266 cm−1) were constructed at the ninth day of nitrogen stress (Fig. 3b–d). A typical Raman spectrum of S. obliquus cell is shown in Fig. 3a. There was no obvious Raman response in a circular region in the Raman images of algal cells, indicating that there was no accumulation of neutral lipids in these areas, which should be the region of pyrenoid. This result was consistent with the results of NR microscopy. In single algae cells, the Raman response high value is dotted, corresponding to the accumulation of lipid particles in the cells. The distribution of saturated fatty acids is slightly different from the distribution of unsaturated fatty acids. Scenedesmus obliquus often grow in four-cell combination form. Figure 3d displays a Raman image of four-cell combination form of S. obliquus, of which four pyrenoids are obvious. The overall Raman response of four-cell combination was significantly lower than that of single cell, indicating that lipid accumulation level was relatively low, probably due to the combined growth form which is not conducive to lipid accumulation.
Fig. 3.
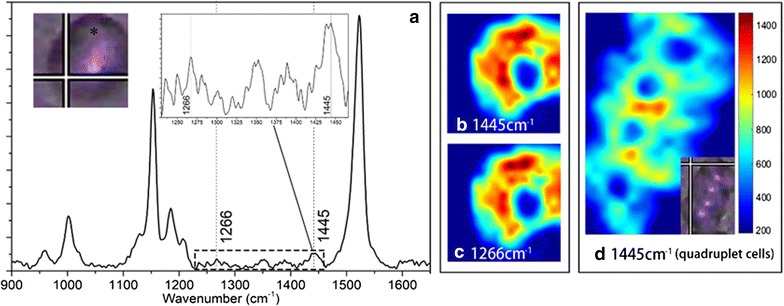
Raman image obtain of S. obliquus. a Raman spectra after pretreatment locate at the black asterisk in the inset image. The left inset image shows the bright-field image of a single cell of S. obliquus, and the other inset image shows partial enlarged detail of lipid characteristic peaks. b, c Raman images of the same cell at b 1445 cm−1, c 1266 cm−1. d Raman image and bright-field image of quadruplet cells of S. obliquus at 1445 cm−1
PCA-LDA modeling based on Raman spectroscopy
Raman single data were divided into modeling set and predicting set with a ratio of 2:1. Using PCA-LDA algorithm to model the data of modeling set to discriminate different days of nitrogen stress. The Raman spectra of the cells in different culture days showed obvious clustering in PC-1 (56%) and PC-2 (26%) directions (Fig. 4a). Figure 4b shows the loading plot of PC-1, and shows the contribution of each spectral band of PC-1. The first two PCs described most of chemical information of the Raman spectra of S. obliquus. In the PC-1 loading plot, the characteristic peaks with greater contribution rate were basically attributed to β-carotene (1001, 1152, 1161, 1521 cm−1) and chlorophyll a (1182, 1534 cm−1), which means that these two substances play a major role in the identification of the data in this experiment. During the process of nitrogen stress culture, the contents of β-carotene and chlorophyll a in the microalgae cells in the stable stage were changed greatly.
Fig. 4.
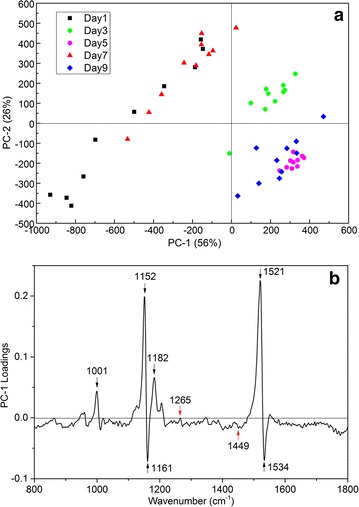
a Score plot of PC-1 and PC-2. b Corresponding loading plot of PC-1
PCA-LDA discriminant models were established based on the Raman characteristic band of typical chemical substances of S. obliquus cells. Four different modeling schemes were used: full band modeling (F), characteristic band of pigments (β-carotene, chlorophyll a) modeling (P—965, 1007, 1156, 1189, 1526 cm−1), characteristic band of lipids modeling (L—1266, 1302, 1445, 1656, 1742 cm−1), and mixed characteristic band modeling (PL—1007, 1156, 1266, 1445, 1526 cm−1). The prediction results (Table 2) of these four models were analyzed. The prediction accuracy of model F, P, and PL can reach 100%, and model L was relatively low, from high to low, was F > P > PL > L. The accuracy of predicting set was slightly lower than of modeling set. These results indicated that the model constructed by Raman spectrum of full band or pigment characteristic band can discriminate S. obliquus cells under different nitrogen stress culture times effectively, but accuracy of the model based on lipid characteristic band was relatively low. On this basis, we combined partial Raman bands of pigment and lipid to model (modeling scheme PL) and obtained better results, which means that this prediction method was effective.
Table 2.
The prediction accuracy of PCA-LDA models
| Time | Modeling schemesa | |||||||
|---|---|---|---|---|---|---|---|---|
| F | P | L | PL | |||||
| Modeling (%) | Prediction (%) | Modeling (%) | Prediction (%) | Modeling (%) | Prediction (%) | Modeling (%) | Prediction (%) | |
| d1 | 100 | 100 | 100 | 100 | 80 | 40 | 100 | 100 |
| d3 | 100 | 100 | 100 | 100 | 100 | 40 | 100 | 100 |
| d5 | 100 | 100 | 100 | 80 | 90 | 100 | 100 | 100 |
| d7 | 100 | 100 | 100 | 100 | 80 | 60 | 100 | 80 |
| d9 | 100 | 80 | 100 | 80 | 60 | 60 | 100 | 60 |
| Total | 100 | 96 | 100 | 92 | 82 | 60 | 100 | 88 |
aThe modeling scheme is defined based on the Raman characteristic bands used to build the model: F—full band modeling; P—characteristic band of pigment (β-carotene, chlorophyll a) modeling (965, 1007, 1156, 1189, 1526 cm−1); L—characteristic band of lipid modeling (1266, 1302, 1445, 1656, 1742 cm−1); PL—partial characteristic band of pigment (1007, 1156, 1526 cm−1) and partial characteristic band of lipid (1266, 1445 cm−1) modeling
The response of lipid characteristic peak was significantly lower than that of pigment characteristic peak, which directly resulted in the low discriminant accuracy of modeling scheme L. The prediction accuracy of modeling scheme P can reach 100%, which indicated that the nitrogen deficiency caused the significant change of pigments in S. obliquus cells of stable stage. Although the main purpose of nitrogen stress in this experiment was to induce the accumulation of intracellular lipids, this growth condition also affected the pigments in cells. The effect of pigment change on the model establishment was significant, and the discriminant accuracy of modeling using pigment characteristic bands was better than that of lipid characteristic bands.
Correlation of Raman mapping and gravimetric method for lipid determination
The above results show that Raman spectroscopy is effective for the detection of intracellular lipid in S. obliquus. Correlation analysis of total lipid contents measured by gravimetric method and Raman intensity of characteristic peak of lipid (1445 cm−1) was carried out (Fig. 5). There was significant correlation (R 2 = 0.83), indicating that Raman spectroscopy can be applied to the semi-quantitative detection of microalgal lipid content.
Fig. 5.
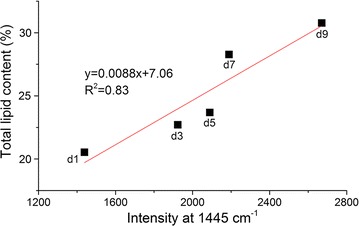
Relationships between total lipid contents and Raman intensity at 1445 cm−1 (lipid)
Conclusions
Under the nitrogen stress, significant accumulation of lipid was observed in S. obliquus cells. This microalgae species has the ability to synthesize large quantities of long-chain fatty acids and can be used as potential algae for biodiesel production. UFA synthesized by these algae has nutrient value; OCFA has industrial and health care value. The accumulation of lipid in S. obliquus was observed by Nile red staining: light yellow spots appeared in cells over time, which were the lipid particles produced by S. obliquus.
Raman mapping data of algae cells were visualized. The pyrenoid and intracellular lipid could be located in the pseudo color graph. Raman response high value was dotted in single algae cells, corresponding to the accumulation of lipid particles in the cells. The distribution of SFA and UFA was slightly different. Lipid characteristic peaks were obvious in Raman spectrum, but pigment peaks were more significant.
The Raman spectra of the cells in different culture days showed obvious clustering in PC-1 (56%) and PC-2 (26%) directions. In the PC-1 loading plot, the characteristic peaks with greater contribution rate were basically attributed to β-carotene and chlorophyll a. During the process of nitrogen stress culture, the contents of β-carotene and chlorophyll a in the microalgae cells in the stable stage were changed greatly. Four different models (F—full band, P—characteristic band of pigments, L—characteristic band of lipids, PL—mixed characteristic band) were build and analyzed: the model constructed by Raman spectrum of full band or pigment characteristic band can discriminate S. obliquus cells under different nitrogen stress culture times effectively, but accuracy of the model based on lipid characteristic band was relatively low. On this basis, we combined partial Raman bands of pigment and lipid to model (modeling scheme PL) and obtained better results, which means that this prediction method was effective.
Result of correlation analysis of total lipid contents measured by gravimetric method and Raman intensity of characteristic peak of lipid (1445 cm−1) showed that there was significant correlation (R 2 = 0.83), indicating that Raman spectroscopy can be applied to the semi-quantitative detection of microalgal lipid content.
From the above experimental results, it is possible to apply Raman spectroscopy to identify different algal cells with different nitrogen stress days and analyze the intracellular lipid content of algae qualitatively and quantitatively.
Authors’ contributions
The work presented here was carried out in collaboration between all authors. YS and HF conceived the idea. HF, HZ, QW co-worked on associated data collection and carried out the experimental work. HF drafted the manuscript, YS helped to renew the original paper. YZ have provided experience on paper revision. YH, in which institute, supported the instrument and technology. All authors have contributed, reviewed and improved the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The research presented in this paper was partially supported by The National Natural Science Foundation of China (31,402,318, 61,605,173), the Public welfare project in Zhejiang Province (2016C32055), and the Ph.D. Programs Foundation of Ministry of Education of China (20133120110007).
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongni Shao, Email: ynshao@zju.edu.cn.
Hui Fang, Email: eviefang@zju.edu.cn.
Hong Zhou, Email: cutezh@foxmail.com.
Qi Wang, Email: 3120100288@zju.edu.cn.
Yiming Zhu, Email: ymzhu@usst.edu.cn.
Yong He, Email: yhe@zju.edu.cn.
References
- 1.Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008;26(3):126. doi: 10.1016/j.tibtech.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Harwood JL, Guschina IA. The versatility of algae and their lipid metabolism. Biochimie. 2009;91(6):679. doi: 10.1016/j.biochi.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Tang H, Ma H, et al. Effect of nutrients on growth and lipid accumulation in the green algae Dunaliella tertiolecta. Bioresour sTechnol. 2011;102(2):1649–1655. doi: 10.1016/j.biortech.2010.09.062. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y, Pei ZJ, Yuan WQ, et al. Effect of nitrogen and extraction method on algae lipid yield. Int J Agric Biol Eng. 2009;2(1):51–57. [Google Scholar]
- 5.Feng GD, Zhang F, Cheng LH, et al. Evaluation of FT-IR and Nile Red methods for microalgal lipid characterization and biomass composition determination. Bioresour Technol. 2013;128(1):107–112. doi: 10.1016/j.biortech.2012.09.123. [DOI] [PubMed] [Google Scholar]
- 6.Satpati GG, Mallick SK, Pal R. An alternative high-throughput staining method for detection of neutral lipids in green microalgae for biodiesel applications. Biotechnol Bioprocess Eng. 2015;20(6):1044–1055. doi: 10.1007/s12257-015-0281-z. [DOI] [Google Scholar]
- 7.Zhang JJ, Lv XJ, Li AF, Wan LL, Wu H, Yin SJ, Zhang CW. Rapid estimation of lipids in microalgae cells. China Biotechnol. 2012;32(1):64–72. [Google Scholar]
- 8.Chen W, Zhang C, Song L, et al. A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods. 2009;77(1):41–47. doi: 10.1016/j.mimet.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Huang WE, Li M, Jarvis RM, et al. Chapter 5—Shining light on the microbial world: the application of raman microspectroscopy. In: Allen IL, Sima S, Geoffrey MG, et al., editors. Advances in applied microbiology. Cambridge: Academic Press; 2010. pp. 153–186. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Ji Y, Wang Y, et al. Quantitative dynamics of triacylglycerol accumulation in microalgae populations at single-cell resolution revealed by Raman microspectroscopy. Biotechnol Biofuels. 2014;7(1):58. doi: 10.1186/1754-6834-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samek O, Zemánek P, Jonáš A, et al. Characterization of oil-producing microalgae using Raman spectroscopy. Laser Phys Lett. 2011;8(10):701–709. doi: 10.1002/lapl.201110060. [DOI] [Google Scholar]
- 12.Wu H, Volponi JV, Oliver AE, et al. In vivo lipidomics using single-cell Raman spectroscopy. Proc Natl Acad Sci USA. 2011;108(9):3809. doi: 10.1073/pnas.1009043108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma SK, Nelson DR, Abdrabu R, et al. An integrative Raman microscopy-based workflow for rapid in situ analysis of microalgal lipid bodies. Biotechnol Biofuels. 2015;8(1):164. doi: 10.1186/s13068-015-0349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao YN, Pan J, He Y. Algal variety identification study with Raman microspectroscopy technology. Spectrosc Spectr Anal. 2015;7:1908–1911. [PubMed] [Google Scholar]
- 15.Shao YN, Li Y, Jiang LJ, Pan J, He Y, Dou XM. Identification of pesticide varieties by detecting characteristics of Chlorella pyrenoidosa using visible/near infrared hyperspectral imaging and Raman microspectroscopy technology. Water Res. 2016;104:432–440. doi: 10.1016/j.watres.2016.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Shao YN, Jiang LJ, Zhou H, Pan J, He Y. Identification of pesticide varieties by testing microalgae using visible/near infrared hyperspectral imaging technology. Sci Rep. 2016;6:24221. doi: 10.1038/srep24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heraud P, Beardall J, Mcnaughton D, et al. In vivo prediction of the nutrient status of individual microalgal cells using Raman microspectroscopy. FEMS Microbiol Lett. 2007;275(1):24–30. doi: 10.1111/j.1574-6968.2007.00861.x. [DOI] [PubMed] [Google Scholar]
- 18.Fang H, Jiang LJ, Pan J, He Y, Gong AP, Shao YN. Research on microalgae lipid change under nitrogen-based stress by Raman microspectroscopy. Spectrosc Spectr Anal. 2017;37(10):3108–3111. [Google Scholar]
- 19.Heraud P, Beardall J, Mcnaughton D, et al. In vivo, prediction of the nutrient status of individual microalgal cells using Raman microspectroscopy. FEMS Microbiol Lett. 2007;275(1):24–30. doi: 10.1111/j.1574-6968.2007.00861.x. [DOI] [PubMed] [Google Scholar]
- 20.Samek O, Jonáš A, Pilát Z, et al. Raman microspectroscopy of individual algal cells: sensing unsaturation of storage lipids in vivo. Sensors. 2010;10(9):8635–8651. doi: 10.3390/s100908635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaczor A, Turnau K, Baranska M. In situ Raman imaging of astaxanthin in a single microalgal cell. Analyst. 2011;136(136):1109–1112. doi: 10.1039/c0an00553c. [DOI] [PubMed] [Google Scholar]
- 22.He XN, Allen J, Black PN, et al. Coherent anti-Stokes Raman scattering and spontaneous Raman spectroscopy and microscopy of microalgae with nitrogen depletion. Biomed Opt Express. 2012;3(11):2896. doi: 10.1364/BOE.3.002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YY, Beal CM, Cai WW, et al. Micro-Raman spectroscopy of algae: composition analysis and fluorescence background behavior. Biotechnol Bioeng. 2010;105(5):889. doi: 10.1002/bit.22617. [DOI] [PubMed] [Google Scholar]
- 24.Cavonius L, Fink H, Kiskis J, et al. Imaging of lipids in microalgae with coherent anti-stokes Raman scattering microscopy. Plant Physiol. 2015;167(3):603–616. doi: 10.1104/pp.114.252197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- 26.Bonizzoni L, Bruni S, Fanti G, Tiberio P, Zaffino C. Ageing of flax textiles: fingerprints in micro-Raman spectra of single fibres. Microchem J. 2016;125:69–74. doi: 10.1016/j.microc.2015.11.011. [DOI] [Google Scholar]
- 27.Zhai C, Xu TF, Peng YK, Li YY. Detection of chlorpyrifos on spinach based on surface enhanced Raman spectroscopy with silver colloids. Spectrosc Spectr Anal. 2016;36:2835–2840. [PubMed] [Google Scholar]
- 28.Rousson V, Gasser T. Simple component analysis. J R Stat Soc Ser C Appl Stat. 2004;53:539–555. doi: 10.1111/j.1467-9876.2004.05359.x. [DOI] [Google Scholar]
- 29.Shu X, Lu HT. Linear discriminant analysis with spectral regularization. Appl Intell. 2014;40:724–731. doi: 10.1007/s10489-013-0485-x. [DOI] [Google Scholar]
- 30.Tagliamonte B, Tomassi G. Fatty acids with an odd number of carbon atoms: metabolic and nutritional aspects. S & TA & NU. Rivista di scienza e tecnologia degli alimenti e di nutrizione umana. 1976;6(1):7. [PubMed] [Google Scholar]
- 31.Roe C R. Five and fifteen carbon fatty acids for treating metabolic disorders and as nutritional supplements: US, US8399515[P]. 2013.
- 32.Hosokawa M, et al. In vivo live cell imaging for the quantitative monitoring of lipids by using Raman microspectroscopy. Anal Chem. 2014;86:8224–8230. doi: 10.1021/ac501591d. [DOI] [PubMed] [Google Scholar]


