Abstract
A phylogenetic analysis of seven different species (human, mouse, rat, worm, fly, yeast, and plant) utilizing all (541) basic helix-loop-helix (bHLH) genes identified, including expressed sequence tags (EST), was performed. A super-tree involving six clades and a structural categorization involving the entire coding sequence was established. A nomenclature was developed based on clade distribution to discuss the functional and ancestral relationships of all the genes. The position/location of specific genes on the phylogenetic tree in relation to known bHLH factors allows for predictions of the potential functions of uncharacterized bHLH factors, including EST’s. A genomic analysis using microarrays for four different mouse cell types (i.e. Sertoli, Schwann, thymic, and muscle) was performed and considered all known bHLH family members on the microarray for comparison. Cell-specific groups of bHLH genes helped clarify those bHLH genes potentially involved in cell specific differentiation. This phylogenetic and genomic analysis of the bHLH gene family has revealed unique aspects of the evolution and functional relationships of the different genes in the bHLH gene family.
Keywords: bHLH, testis, Sertoli cell, Schwann cell, muscle cell, thymic cell, microarray, phylogenetic human, rat, mouse, Drosophila, inset, C. elegans, Arabidopsis, plant, yeast
Introduction
Identification of the basic helix-loop-helix (bHLH) motif first occurred in 1989 (Murre et al., 1989) when E12 and E47 were discovered in the murine genome. Since this time, numerous bHLH proteins have been identified in animals, plants, and fungi. In 1997, the first large-scale phylogenetic analysis was performed (Atchley and Fitch, 1997) leading to a “natural” classification of different families of bHLH transcription factors. This classification was performed using only the bHLH motif because the flanking regions for proteins from independent clades are very divergent. This classification led to the postulation of four distinct groups based on amino-acid patterns and E-box-binding specificity (Atchley and Fitch, 1997). This classification segregated bHLH proteins under Class A, B, C, or D in an attempt to functionally segregate bHLH proteins. Unfortunately the majority of the bHLH genes do not have known functions or have multiple functions such that only a small sub-group of bHLH proteins can utilize this original classification. Class A includes several tissue-specific bHLH proteins (Hassan and Bellen, 2000) as well as several ubiquitously expressed bHLH proteins such as the E2A gene products E12 and E47, HEB, and E2-2 (Murre et al., 1989; Atchley and Fitch, 1997). Class B proteins represent a large group of functionally unrelated proteins that are involved in various cellular and developmental processes (Henriksson and Luscher, 1996; Facchini and Penn, 1998; Goding, 2000). Proteins in this group include Myod and myogenin, involved in muscle cell differentiation (Ishibashi et al., 2005; Tang et al., 2006), Ngn and Mash1 involved in neurogenesis (Nakada et al., 2004; Kageyama et al., 2005) and Hand involved in heart development (Thattaliyath et al., 2002). Several of the proteins in this group contain another functionally important motif known as the leucine zipper (Atchley and Fitch, 1997). The leucine zipper (Zip) motif is a protein interaction domain also present in the CREB family of transcription factors and can heterodimerize or homodimerize to bind DNA (Vinson et al., 2006). A subclass of the class B bHLH proteins that function as repressors (i.e. hairy and enhancer-of-split proteins) were first identified in Drosophila. Many vertebrate homologs have been subsequently identified including the Hes group of genes (Davis and Turner, 2001). These proteins contain another common structure known as the Orange domain located just C-terminal to the bHLH domain (Taelman et al., 2004). Members of the bHLH/Orange subclass of the class B proteins act as repressors that inhibit target gene expression by acting as direct or indirect DNA-binding-dependent transcriptional repressors or by sequestering positive bHLH factors or their common heterodimer partners (Chin et al., 2000; Giagtzoglou et al., 2003). The function of the Orange domain is not well understood, but may play a role in conferring specificity of binding to certain family members (Dawson et al., 1995) or have a role in transcriptional repression (Castella et al., 2000). An additional structural characteristic of the Hairy and E(spl) bHLH/Orange proteins is the presence of a C-terminal WRPW motif that binds the co-repressor Groucho and its mammalian homologs, the TLE proteins (Paroush et al., 1994; Fisher and Caudy, 1998; Chen and Courey, 2000). Class B bHLH proteins are postulated to not homodimerize, rather they are believed to heterodimerize with Class A bHLH proteins. Class C bHLH proteins also contain one or more PAS domains (Crews, 1998). This domain allows for dimerization between PAS proteins, non-PAS proteins and the binding of small molecules (e.g. dioxin) (Crews, 1998). Examples of class C bHLH proteins include HIF1 involved in regulation of hypoxia, and Sim proteins involved in food intake behavior (Yang et al., 2004). These proteins tend to be ubiquitous and are believed to bind a DNA sequence different from the common E-box (Crews, 1998; Crews and Fan, 1999; Taylor and Zhulin, 1999). Class D includes HLH proteins that lack a basic domain and are thus unable to bind DNA. These proteins are called inhibitors of differentiation (Id). In mammals, there are four known Id proteins that appear to have differential expression based on cell type (Chaudhary et al., 2001). Id1, Id2, and Id3 are thought to be ubiquitously expressed, while Id4 is primarily expressed in the testis (Chaudhary et al., 2001), brain and kidney (van Cruchten et al., 1998). A fifth group of bHLH proteins has been suggested (Crozatier et al., 1996), but phylogenetic analysis of this group is difficult as the HLH domain is highly divergent from the conserved bHLH motif. This group is known as the COE family and is characterized by the presence of an additional COE domain involved in dimerization and DNA binding. Owing to the increased size and diversity of the bHLH gene family, this original classification (i.e. Class A–D) has become inadequate and misleading. A classification that can incorporate the entire gene family and show relatedness is required.
Several large and small scale phylogenetic analysis of the bHLH transcription factor family have been performed for mammals (Atchley and Fitch, 1997; Ledent et al., 2002) and plants (Buck and Atchley, 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003). Most of these analyses have utilized only the bHLH domain while the remaining portion of the protein is considered to be too divergent. A recent analysis in plants has used the entire coding sequence (Li et al., 2006). Owing to a number of additional domains being associated with the bHLH proteins, utilizing the entire coding region in a large-scale phylogenetic analysis for classification is needed to allow better identification of the proper class structure of the bHLH genes. In addition, having a large-scale analysis of the known mammalian bHLH transcription factors will allow for the entire gene family to be used in concert with expression analysis in the investigation of cellular differentiation.
Terminal cellular differentiation occurs when a cell exits the cell cycle, becomes post mitotic, and develops specialized cellular functions associated with the differentiated gene expression profile. These terminally differentiated cells can often not be replaced if lost. Examples of terminally differentiated cells include myocytes (Tam et al., 1995; Wei and Paterson, 2001), neurons (Yoshikawa, 2000), and Sertoli cells (Skinner, 1991). While the role of the bHLH family of transcription factors has been partially identified in this terminal differentiated state for myocytes and neurons (Nakada et al., 2004; Ishibashi et al., 2005; Kageyama et al., 2005; Tang et al., 2006), factors responsible for Sertoli cells to undergo terminal differentiation remain to be elucidated.
Phylogenetic analysis of an entire gene family between species allows for the appropriate structural and functional distribution of genes, identifies gene duplications and species conservation, as well as reveals evolutionary considerations. The phylogenetic analysis of the bHLH gene family identifies new structural and functional relationships between bHLH genes and helps organize the gene family. A comparison of the phylogenetic bHLH gene family information with DNA microarray analyses from divergent cell types identifies the genes specific or unique to the different cells. This phylogenetic and genomic approach enhances the analysis of a transcription factor family associated with cellular differentiation and allows the investigation of the entire bHLH gene family.
Materials and methods
Identification of bHLH transcription factors
A search of the Ensembl database using know bHLH transcription factors was performed for human, rat, mouse, Saccharomyces cervacea, Drosophila, and Caenorhabditis elegans. Following identification of bHLH transcription factors in Ensembl, verification of these genes was performed using a NCBI protein search. A basic phylogenetic tree was generated in Vector NTI to discern clade distribution of the known proteins. One bHLH protein from each clade was then chosen; its bHLH domain selected and blasted in NCBI BLASTp to identify more genes not listed in the Ensembl database as well as possible EST’s. The Arabidopsis bHLH genes were taken from a previous analysis (Toledo-Ortiz et al., 2003). After generating the total list of bHLH genes for these seven species (i.e. 541 genes), a phylogenetic tree was generated in Vector NTI to determine if there were duplicate genes listed. Any EST with high homology to a known gene was then aligned (NCBI) with the known gene to determine similarity. If identical, the EST was deleted from the final analysis.
Generation of phylogenetic tree
An initial phylogenetic tree was generated using an amino acid alignment created using ClustalX 1.83 (Thompson et al., 1997) and analyzed with PHYML v.2.4.4 (Guindon and Gascuel, 2003), employing the JTT amino-acid substitution model (Jones et al., 1992). Because of the large numbers of genes (i.e. 541), this starting phylogenetic hypothesis was then divided into six major clades based on the preliminary tree structure for more rigorous analysis. After separating the matrix into separate matrices of 27, 28, 99, 107, 131, and 136 sequences, these amino-acid sequences were realigned with ClustalX to minimize the potential effects of having divergent sequences in the alignment. These final alignments were then analyzed using Bayesian inference analysis and was performed on the various matrices using MrBayes v.3.1 (Huelsenbeck and Ronquist, 2001). Ten million generations were run with four chains (Markov Chain Monte Carlo) and a tree was saved every 100 generations. In order to test for the occurrence of stationarity, convergence and mixing within ten million generations, multiple analyses were started from different random locations in tree space. The posterior probability distributions from these separate replicates were compared for convergence with the same posterior probabilities across branches. Majority rule consensus trees of those sampled in Bayesian inference analyses yielded probabilities that the clades are monophyletic (Lewis, 2001). The trees from the MrBayes analysis were loaded into PAUP*4.0 (Sinauer Associates Inc., Sunderland, MA), discarding the trees generated within the first 2,000,000 generations (those sampled during the “burnin” of the chain (Huelsenbeck and Ronquist, 2001), to only include trees after stationarity was established. Consensus trees were then created to display branches with posterior probabilities greater than 50%. A single tree was not created in this case because a point estimate might mislead interpretation of inferred relationships. The preference is to focus on those branches with strong support.
Affymetrix probe search for bHLH genes
The complete mouse bHLH gene list was taken from the phylogenetic analysis and used in a search of the Affymetrix (Affymetrix, Santa Clara, CA) database for MGU74v2 array chips in an effort to locate all bHLH genes and their associated Affymetrix ID’s on the chips. If the gene name did not yield an Affymetrix ID the actual sequence of the gene was blasted against the MGU74v2 probe set to ensure the gene was or was not on the array chips. Because there are many EST’s in the phylogenetic tree, these gene sequences were also used in an Affymetrix blast search for their associated Affymetrix ID. The MGU74v2 array chips contain three separate chips, the A chip contains 12,480 total genes of which the majority are fully annotated in NCBI. The B and C chips contain 12,477 and 11,934 genes respectively, many of these genes are EST’s or not fully annotated genes. Of the 107 identified mouse bHLH genes in the phylogenetic analysis, 81 were identified as existing on the MGU74v2 array chips. Owing to limitations in obtaining microarray analysis for other cell types that were also performed on all three MGU74v2 array chips, we limited our bHLH gene list to only those genes on the A chip. Therefore, only a subset of the potential bHLH genes expressed will be assessed.
Microarray analysis
High quality mouse RNA samples of at least 5 μg and with a minimum OD260/280 ratio of 1.8 were analyzed as previously described (McLean et al., 2002; Nef et al., 2005; Small et al., 2005). Briefly, RNA was transcribed into cDNA, which was transcribed into biotin labeled RNA. Biotin labeled RNA was then hybridized to either mouse MGU74v2a arrays containing approximately 37,000 total transcripts (Affymetrix) and visualized by labeling with phycoerythrin-coupled avidin. Hybridized chips were visualized on an Affymetrix Scanner 3000 (Affymetrix). Once raw data was obtained, data was processed using GCOS version 1.1 software (Affymetrix) and analyzed by Genespring version 7.2 (Silicon Genetics, Redwood City, CA) software.
Initial analysis of microarray data was performed as previously described (Small et al., 2005). Microarray hybridization data was examined for physical anomalies on the chip and background noise above a value of 3. Default GCOS statistical values were used for analysis. All probe sets were scaled to a mean of 125. An absolute analysis was performed with GCOS to assess the relative abundance of the transcripts on the chips based on signal and detection calls (present, absent, or marginal). This information was imported into GeneSpring (Silicon Genetics) and normalized using the recommended default normalization methods. This includes setting signal values below 0.01–0.01, total chip normalization to the 50th percentile, and normalization of each chip to the median which allows visualization of data based on relative abundance for a given sample rather than by comparison to a specific control value (Small et al., 2005). Transcripts with statistically significant presence calls (p<0.05) and raw signal values above 75 were selected for comparison to other microarray data. All the cell types used for the microarrays were freshly isolated cell preparations using fluorescent- activated cell sorting (FACS) and/or enzymatic digestion and gravity sedimentations. Gene expression data from mouse Schwann cells (Buchstaller et al., 2004) (GEO:GSE972), mouse thymic medullary epithelial cells (Anderson et al., 2002) (GEO: GSE85) and the mouse muscle cell (GEO:6487) (http://pepr.cnmcresearch.org/browse.do?action=list_prj_exp&projectId=151) were obtained from the Gene Expression Omnibus (GEO) available through NCBI. This data was obtained from MGU74v2a arrays. A minimum of two different microarray chips and experiments were used to obtain the mean data utilized. The raw data was pulled through GCOS and GeneSpring in the same manor as the mouse chips above and was used for comparison of the different cell types. Validation of gene expression from the microarray analysis used previous literature reports confirming (e.g. quantitative PCR) the expression of the selected genes. Over 16 different experiments from the various cell types and literature all confirmed the gene expression from the microarray data.
Results
bHLH phylogenetic analyses
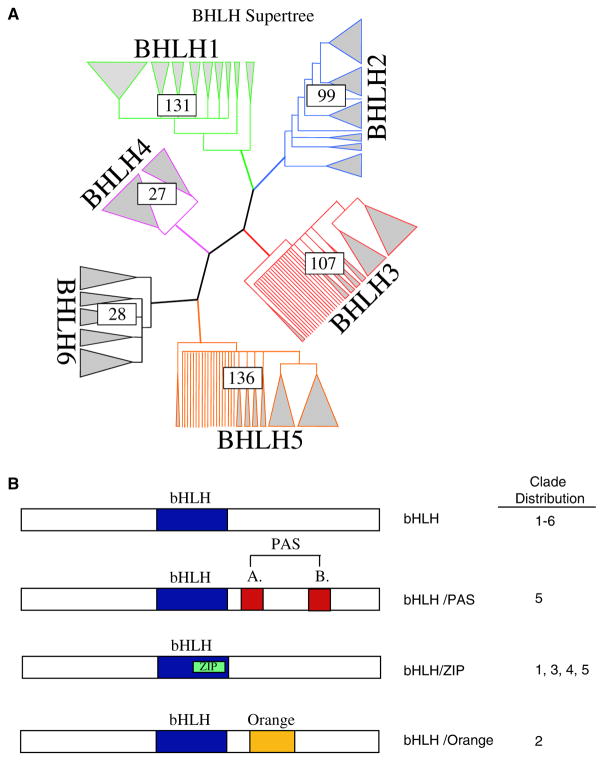
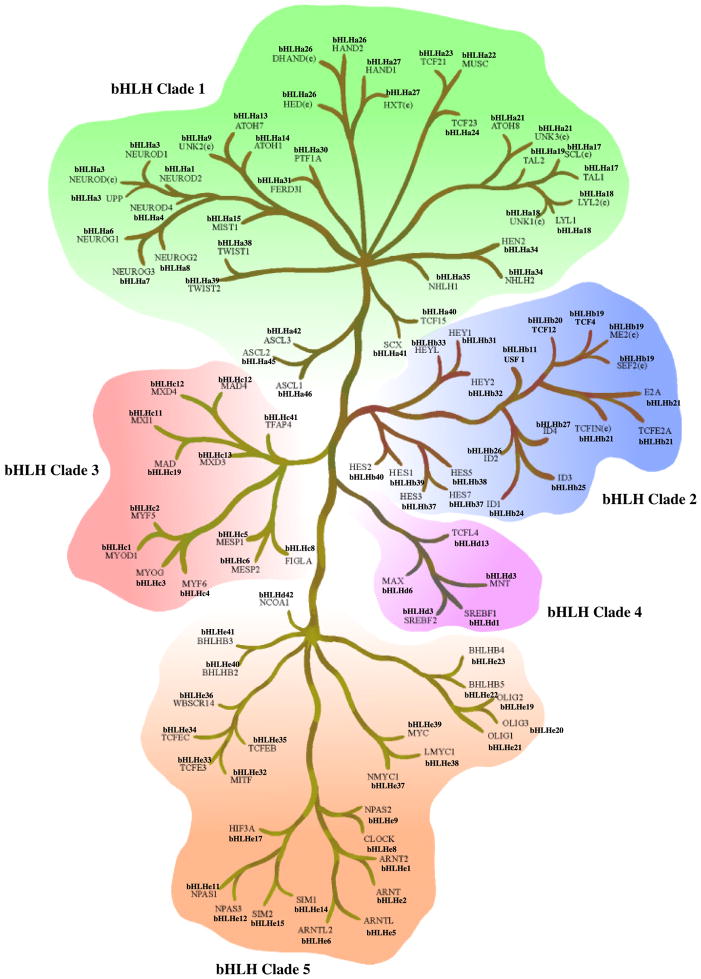
The preliminary maximum likelihood analysis of all the 541 bHLH genes found in the seven species (human, mouse, rat, worm, fly, yeast, plant) resulted in a single topology. Previous phylogenetic analyses of this gene family primarily used only the bHLH region of the protein, limiting the potential informative sequence. Previous studies also limited the number of genes in their total analysis, making a priori assumptions about gene copy relatedness before analysis. This has the potential to introduce misinterpretation of bHLH copy relatedness. Therefore, these a priori assumptions were minimized in the current study by including the entire coding region where available and all nonidentical gene copies from the seven species. Owing to the size of the matrix involved, it was necessary to break the tree into smaller sub-trees for more rigorous analysis. These divisions separated major clades of reasonable size from the initial maximum likelihood tree. It should be noted that these divisions were made subjectively and focused on creating related matrices of reasonable size for further analysis. Detailed analysis of these matrices demonstrated the divisions made were accurate. Six clades resulted from this division (Fig. 1A), with clade 1 containing only mammalian genes, clades 2–5 contained genes from a mixture of species, and clade 6 including the plant Arabidopsis bHLH genes only. All of the first five clades (Fig. 1A) contain bHLH genes that are typically thought of as Classes A and B factors showing that no unique division based on earlier phylogenetic analysis for these groups is present.
Fig. 1.
(A) bHLH supertree showing individual clade distribution, how many genes are in each clade, and representative grouping of individual genes within each clade. (B) bHLH protein categories with additional domains (i.e. Zip, PAS, and Orange) and the clade distribution of the categories.
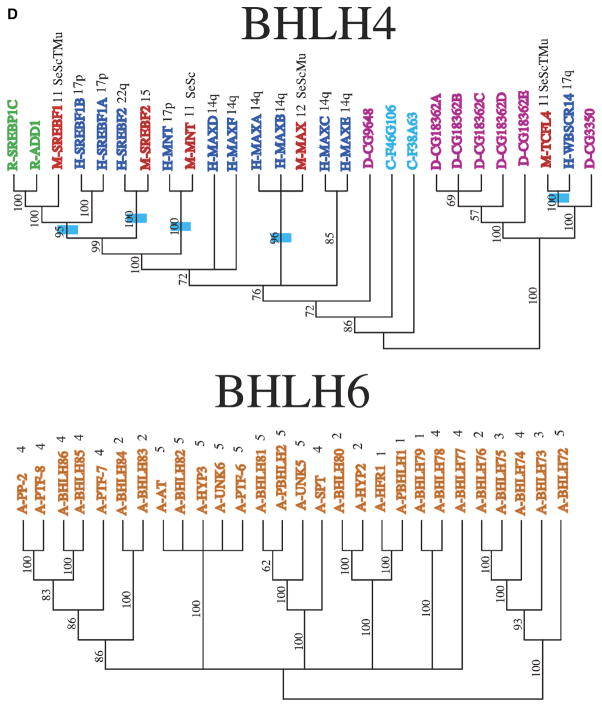
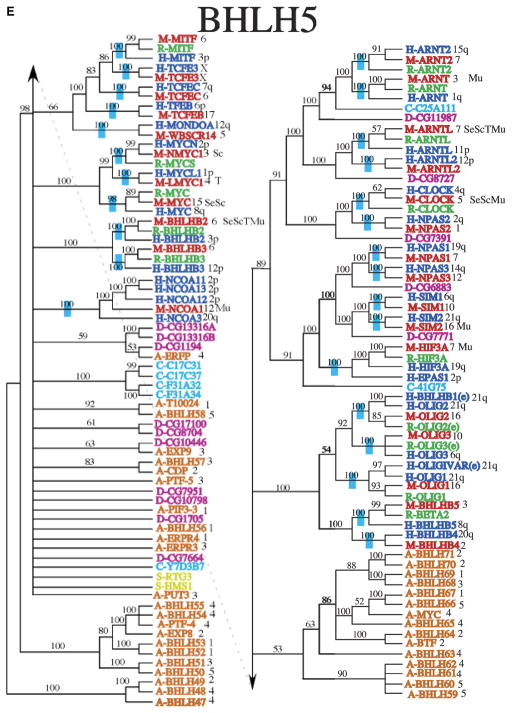
Analysis of each clade was performed to determine similarities among extra domains and allow comparative protein functional analysis (Fig. 1B). Minimally four different categories of bHLH proteins have been identified. All contain a bHLH domain and the additional domains include orange, PAS and Zip domains (Fig. 1B). Clade 1 is made up primarily of mammalian genes that were previously considered group B proteins (Fig. 2A). Many of the genes in this clade are also ESTs that have not been fully characterized. Clade 2 is made up of the Id genes that are involved in sequestering other bHLH proteins and inhibiting them from binding DNA (Fig. 2B), as well as the inhibitory Hey and Hes (bHLHb37-b39) genes. Similar to the Id proteins, these genes are involved in the negative regulation of differentiation by sequestering other bHLH proteins. The Hey and Hes genes contain a second domain, the Orange domain, the function of which is currently unknown. All the bHLH proteins containing an Orange domain localized to clade 2 (Fig. 1B). Clade 2 includes bHLH proteins previously classified in bHLH groups A, B, and D (Fig. 2B) suggesting no phylogenetic segregation of this previous bHLH family categorization. Clade 3 contains a large proportion of Arabidopsis genes, as well as mammalian genes that are involved in myogenesis and max (bHLHd4-d8) interaction proteins (Fig. 2C). The mammalian genes were all previously classified as group B proteins. Clade 4 contains mostly genes that contain both a bHLH region and a leucine zipper (Zip) region (Fig. 2D). The Zip region is involved in protein interactions and DNA binding, and was previously classified as group B proteins. Clade 4 is small and distantly related to clades 3 and 5 in the initial analysis, which is why this clade was not included within those larger clades. However, there are several genes in clade 1, 3, and 5 that also contain a leucine zipper region (Figs. 1B,2E), suggesting that the functional structure of this region needs to be explored further. In addition to genes containing a Zip region, clade 5 also contains genes that have one or more PAS domains (Figs. 1B,2E). These genes were previously included in the group C proteins. Clade 6 contains Arabidopsis plant genes only (Fig. 2D). The distribution of the different categories of bHLH proteins that contain other domains (Fig. 1B) demonstrates several of the bHLH categories are isolated to an individual clade. The phylogenetic bHLH family tree generated demonstrates an appropriate segregation of bHLH proteins with similar protein domains and functionally localized genes (Fig. 2). Those genes, previously shown to be related functionally, co-localized into appropriate clades and clusters. Observations suggest that the previous Classes, A, B, C, D categories, do not segregate. Based on the current phylogenetic analysis unknown or noncharacterized genes localized with functionally known bHLH genes can be predicted to potentially have a functional relationship that can now be investigated.
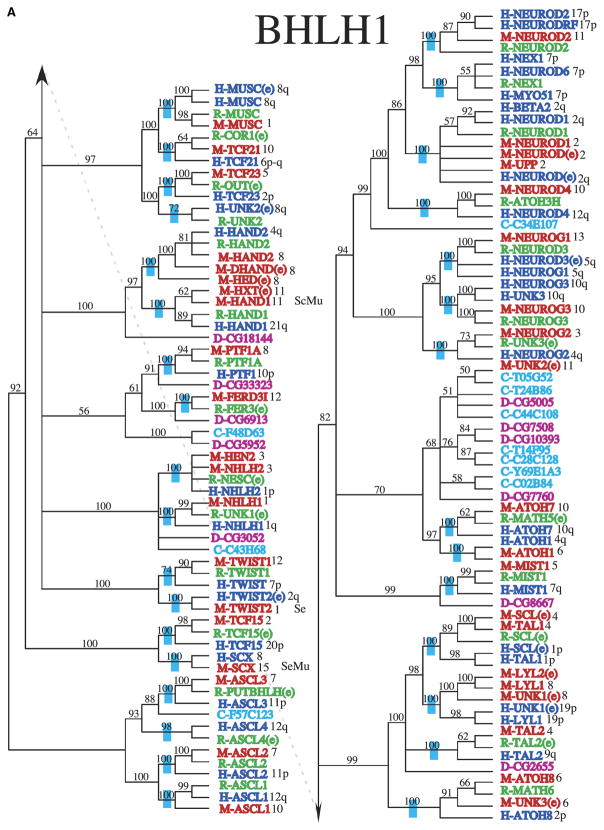
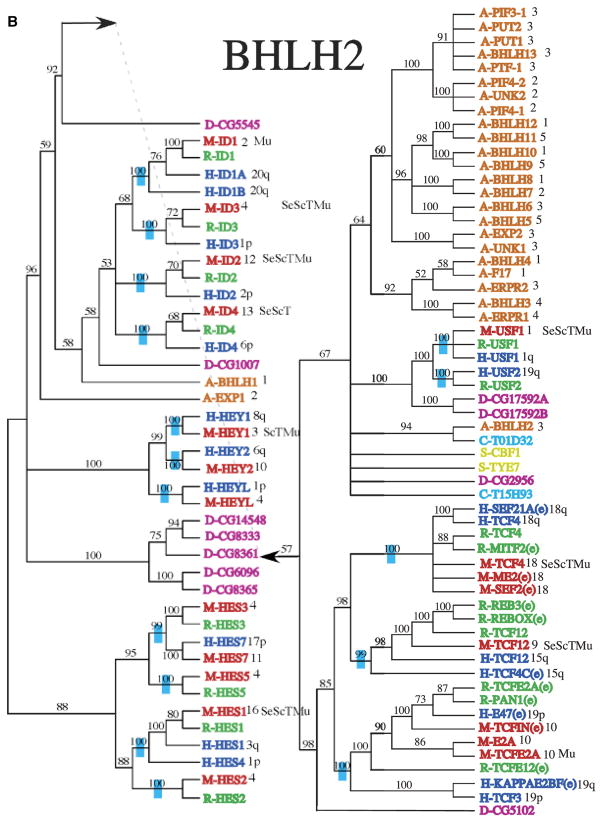
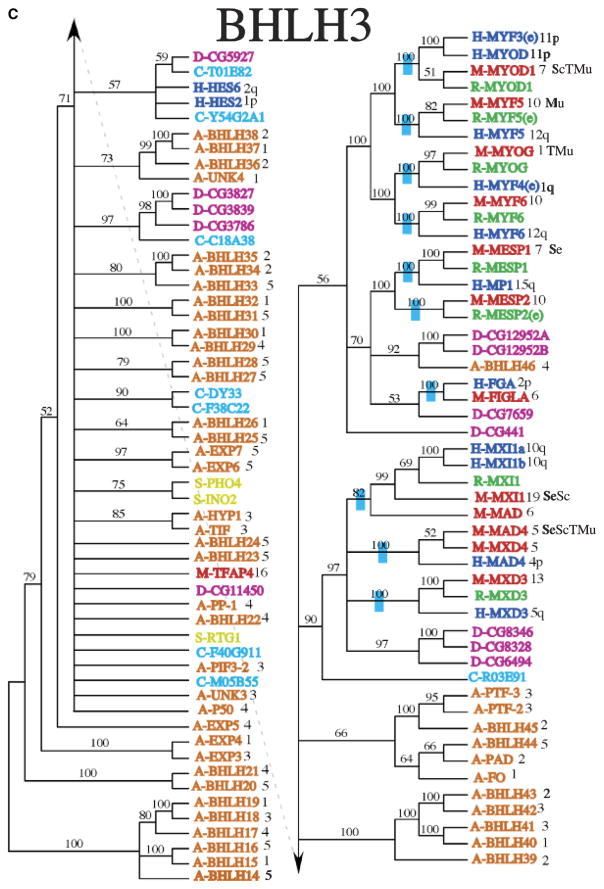
Fig. 2.
Phylogenetic relationship of individual genes in (A) clade 1, (B) clade 2, (C) clade 3, (D) clade 4 and 6, and (E) clade 5 with relatedness as determined by Bayesian posterior probability indicated by the blue hatch bar and number to the left of the genes. Chromosomal location for mouse and human genes is given to the right of the gene names. The species is indicated by blue (human), green (rat), red (mouse), magenta (Drosophila), pink (C. elegans), and orange (Arabidopsis), with the species letter in front of the name.
Nomenclature
Further exploration of the supertree and Bayesian consensus trees indicates a preponderance of genes with multiple gene names that have little phylogenetic information and the presence of a large number of EST’s without proper names. This has led to the need for a consistent nomenclature to allow a discussion of this gene family. During the blast searches for bHLH genes it became apparent that there are many different names for the same transcription factor. An example of this is TCF4 (bHLHb19), which has aliases of 5730422P05Rik, ASP-I2, E2-2, E2.2, Hnf-4, ITF-2b, ITF2, ME2, MITF-2A, MITF-2B, SEF2, SEF2-1, TFE, and Tcf-4. Owing to the confusion this causes and due to the number of bHLH ESTs identified in the current study, a nomenclature for the bHLH gene family that is consistent with the phylogenetic analysis was developed (Supporting Table S1). Genes with a high degree of homology as supported by Bayesian posterior probability values among human, rat, and mouse now have the same designation, since the probability that the gene has the same phylogenetic origin and potential functional role is high (Supporting Table S2). The nomenclature is based on clade distribution with a letter a-f indicating the clades 1–6 (e.g. bHLHa for clade 1) and numbering the genes within the clade with attention to gene relationships (e.g. 1–10), such that the first gene is bHLHa1 with the species designation, Supporting Table S1. The 541 genes were named and close homologs (e.g. splice variants) were also given letter designates when required [e.g. E2A products E12 (bHLHb21a) and E47 (bHLHb21b) splice variants]. The genomic information of specific genes can include sequencing errors to suggest multiple homologs when the different accession numbers are actually for the same gene. Therefore, some apparently closely related genes may be the same gene [e.g. Hxt and Hand1 (bHLHa27)], but further experimental information is needed for confirmation. This is a limitation of the current study, however; rather than arbitrarily infer relatedness, the current genomic information was used, and all comparative analyses that suggested sequence differences were used to assign separate genes when appropriate.
An example of the use of the nomenclature and the super-tree distribution for the mouse genes is shown in Fig. 3. The mouse has 107 bHLH genes that appear distinct with a clade distribution shown in Fig. 3. This mouse bHLH super-tree demonstrates the clade relationships and presents the related nomenclature. Each species bHLH gene family is listed in Supporting Table S2. Those genes conserved between species can also be observed. Many of the genes have multiple names and due to relatedness on the phylogenetic tree were given the same number. For example, the supertree in Fig. 3 has several gene clusters with different names, but have been given the same number (e.g. HAND1 and HXT(e), being bHLHa27) and are similar genes, while others (e.g. LYL1, LYL2(e), UNR1e, being bHLHa18) could be similar genes or splice variants. Future analysis of related genes is required to fine-tune this phylogenetic analysis. In the event they are splice variants a letter designation can be provided, versus the same gene when only one name/number should be used. The proposed nomenclature is suggested to allow a large number of EST to be assigned names and to clarify relatedness of clustered genes. Whether the nomenclature is generally used will require acceptance by the bHLH research community. The current nomenclature is not suggested to replace the current bHLH names, but instead to provide an approach to understand and discuss the functional, structural and species relationships of this large gene family.
Fig. 3.
Mouse bHLH super-tree for the five different clades and all 107 genes were relatedness presented. Clade 2 contains the inhibiting bHLH/Orange genes, as well as the Id genes. Clade 5 contains bHLH/PAS family genes. bHLH/bZip genes are spread throughout. Both the current and proposed new nomenclature are presented.
Epithelial cell distribution of bHLH transcription factors
To further elucidate the phylogenetic analysis and investigate bHLH gene expression differences that define differentiated cells, microarray analysis for four different mouse cell types were analyzed (Table 1). Freshly isolated mouse cell types were collected and used in the microarray analysis. Only a subset of the total bHLH genes was present on the microarray chip used (i.e. MGU74v2a), such that the genes listed do not reflect the total cohort of bHLH genes potentially expressed. All bHLH genes expressed above a signal of 75 with a statistically significant present call (p<0.05) and were present on the microarray chip are listed for each of the cell types. To verify the utility of such an approach the muscle cell expression data was compared with known information about muscle cell differentiation. Myogenic genes such as Myod1 (bHLHc1) and Myog (bHLHc3) are expressed at high levels in the differentiated muscle cells (Table 1). These genes are known to play a role in the differentiation of muscle cells (Nakada et al., 2004; Ishibashi et al., 2005) which validates the microarray data presented for these genes. The bHLH genes expressed in the muscle, thymic, Schwann, and Sertoli cells are shown in Table 1. Twist2 (bHLHa39) and Mesp1 (bHLHc5) are only expressed in Sertoli cells when compared with the other three cell types. Genes such as Mxi1 (bHLHc11) and Scx (bHLHa41) are differentially expressed among the different cell types. This differential expression pattern may be an indication that these four genes could be important in the differentiation of Sertoli cells. Previously scleraxis (bHLHa41), Id1-4 (bHLHb24-b27), and TCF12 (bHLHb20) were shown to be expressed by Sertoli cells (Chaudhary et al., 1999, 2001; Muir et al., 2005), which validates the microarray data for these genes. Genes that may be expressed at high levels during an earlier stage of differentiation that are then turned off during the maintenance of cellular differentiation are missed in this type of analysis. Harvesting Sertoli cells during different developmental stages for analysis would be one way to obtain this information. Comparing the clade distribution of bHLH genes expressed in the Sertoli cell versus other cell types does not indicate that any one clade is over- or under-represented in the Sertoli cell when compared with the other cell types, Figure S1.
Table 1.
Cell type differential bHLH gene expression
| Sertoli | Schwann | Thymic | Muscle | ||
|---|---|---|---|---|---|
| bHLHc11 | Mxi1 | 611.4 | 117.5 | ||
| bHLHc12 | Mad4 | 422.1 | 820.6 | 172.2 | 174.2 |
| bHLHa41 | Scx | 361.9 | 834.5 | ||
| bHLHb20 | Tcf12 | 324.8 | 590.1 | 379 | 696.6 |
| bHLHd1 | Srebf1 | 298.4 | 475.3 | 162.8 | 666.6 |
| bHLHb26 | Id2 | 267.6 | 2,362.6 | 457 | 113.8 |
| bHLHb12 | Usf2 | 210.7 | 345.7 | 241.2 | |
| bHLHd13 | Tcfl4 | 162.1 | 143.4 | 181.4 | 144 |
| bHLHd6 | Max | 152.1 | 221.2 | 94.9 | |
| bHLHe8 | Clock | 130.9 | 125.9 | 90.1 | |
| bHLHb39 | Hes1 | 121.8 | 341.1 | 93.7 | 244.3 |
| bHLHb11 | Usf1 | 116.3 | 94.5 | 82.6 | 112.8 |
| bHLHd3 | mnt | 112.5 | 116.9 | ||
| bHLHc5 | Mesp1 | 112.3 | |||
| bHLHe39 | Myc | 103.9 | 153.5 | ||
| bHLHde40 | BHLHb2 | 101.1 | 114.1 | 82.6 | 474.8 |
| bHLHe5 | Arntl | 99.5 | 98.5 | 89.2 | 85.6 |
| bHLHb25 | Id3 | 88.5 | 84.4 | 102.7 | 101.5 |
| bHLHb27 | Id4 | 86.1 | 159.2 | 124.1 | |
| bHLHb19 | Tcf4 | 82.9 | 139.1 | 133 | 239.3 |
| bHLHa39 | Twist2 | 79.4 | |||
| bHLHc1 | Myod1 | 76.6 | 78.1 | 646.1 | |
| bHLHa27 | Hand1 | 100.5 | 124 | ||
| BHLHb31 | Hey1 | 339.4 | 95.8 | 103.6 | |
| bHLHe37 | Nmyc1 | 108.9 | |||
| bHLHa22 | Msc | 279.7 | |||
| bHLHc3 | Myog | 213.6 | 2,660.3 | ||
| bHLHe38 | Lmyc1 | 107.4 | |||
| bHLHe2 | Arnt | 119.4 | |||
| bHLHb21 | Tcfe2a | 111.5 | |||
| bHLHe17 | Hif3a | 81.7 | |||
| bHLHe42 | Ncoa1 | 89.6 | |||
| bHLHb24 | Id1 | 132.3 | |||
| bHLHc2 | Myf5 | 139.2 | |||
| bLHe15 | Sim2 | 76.7 |
Values are mean raw microarray signals and the colors indicate which clade the gene is expressed in (red=clade 3, blue=clade 2, green=clade 1, orange=clade 5, and purple=clade 4). The lack of a number for a corresponding bHLH protein indicates an absence or no expression above 75 for that cell type.
The bHLH gene unique to Schwann cells was Nmyc1 (bHLHe37), with others of interest including Hand1 (bHLHa27), Mnt (bHLHd3), Id2 (bHLHb26), and Mad4 (bHLHc12), Table 1. Previously Schwann cells have been shown to express TCF12 (bHLHb20), Id2 (bHLHb26), Id3 (bHLHb25), and Id4 (bHLHb27) (Stewart et al., 1997; Thatikunta et al., 1999), confirming the microarray data for these genes. The bHLH genes unique to thymic cells are Lmyc1 (bHLHe38) and Msc (bHLHa22), Table 1. Previously thymic cells have been shown to express Id3 (bHLHb25), Id2 (bHLHb26), and TCF12 (bHLHb20) (Blom et al., 1999; Morrow et al., 1999; Bergqvist et al., 2000; Temchura et al., 2005), which validates the microarray data for these genes. As discussed above, the bHLH genes unique to muscle cells include Myog (bHLHc3), Myf5 (bHLHc2), Arnt (bHLHe2), Myod1 (bHLHc1), and Scx (bHLHa41), Table 1. Previously muscle cells have been shown to express Myod1 (bHLHc1), myogenin (bHLHa3), Myf5 (bHLHc2), Hand1 (bHLHa27), and scleraxis (bHLHa41) (Braun et al., 1992; Morikawa and Cserjesi, 2004; Ishibashi et al., 2005; Tang et al., 2006; Pryce et al., 2007), which validates the microarray data for these genes. As observed with the Sertoli cells, no clade distribution was unique to the different cells, Table 1 and Figure S1. Although some subsets of genes did cluster to specific clades (e.g. myogenic) most clades were presented in each cell type, Figure S1. Therefore, clade distribution and functional relationships of expressed bHLH genes was not a major factor in comparing the different cell types. This observation suggests a diversity of bHLH genes with various functions is likely required for cell differentiation. The set of bHLH genes for the different cell types is likely associated with cellular differentiation. In regards to validation of the microarray data, 16 different analyses in the various cell types confirmed the expression observed, validating the array data for these genes. Those genes listed in Table 1 not previously shown to be expressed now need to be further investigated. Not all the bHLH genes known are present on the microarray chip used, so this is a minimal subset of genes and further analysis of the entire gene family will likely reveal additional insights into the bHLH genes associated with the differentiation of these cell types. The bHLH genes identified are good initial candidates for further analysis of the cellular differentiation of these cell types.
Discussion
Phylogenetic analysis of the bHLH transcription factor family has been performed several times in the past with the first analysis in 1997 (Atchley and Fitch, 1997). In most cases only the bHLH domain was utilized in the analysis and a limited number of genes or species were represented. In the current study, Blast searches were performed of genomes to identify all potential bHLH genes for seven divergent species (i.e. mouse, rat, human, fly, worm, yeast, and plant). This analysis identified 541 total bHLH genes in all the species. Sequence alignments were performed to verify that genes were not identical in sequence before performing the phylogenetic analysis. Phylogenetic analysis was then performed utilizing the entire protein sequence rather than just the bHLH domain. This allowed the inclusion of extra domains associated with many of these proteins (e.g. zip, Orange, PAS), Fig. 1B. These additional domains play known roles in protein interactions (i.e. zip, Orange, and PAS) and may function in transcriptional repression (i.e. Orange). As expected, the majority of the bHLH proteins with similar domains clustered together in the different clades of the phylogenetic analysis, Fig. 1B. This suggests the gene groupings are appropriately related to structure and potentially similar in function.
The bHLH gene family has been previously divided into several “groups” (Atchley and Fitch, 1997; Ledent et al., 2002; Buck and Atchley, 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003), yet there has been a lack of consistency in how many groups are accepted and relevant. While group A, B, and D proteins are generally classified consistently, group E and C proteins may be classified with the group B proteins. Previous classification revolves around E-box-binding specificity and bHLH domain amino-acid patterns (Atchley and Fitch, 1997). However, amino-acid patterns in the current study would indicate that previous classification of this gene family is incorrect. For example, previously identified group B proteins are spread throughout all five clades containing mammalian bHLH genes. Similarly, the ability of the genes to homodimerize or heterodimerize does not correlate to a given region of the super-tree. Genes that can only heterodimerize are spread throughout all of the clades. E-box-binding specificity was not considered in the current study as this specificity may change with different dimerization partners. In addition, the majority of genes in the family have no known function or name. Therefore, the current study provides a system of classification that better fits the entire coding sequence and presents a phylogenetic basis of classification.
The Bayesian consensus trees suggest several interesting evolutionary patterns. First, it is clear that many of the duplication events predate vertebrate diversification (Figs. 2A–2F). Neither Drosophila nor C. elegans have as many bHLH gene family copies as mammalian species (Supporting Table S2). However, many bHLH clades are sister to Drosophila and/or C. elegans sequences suggesting these might be orthologs of the vertebrate copies and may provide candidates for studying functional diversification, Supporting Table S2. In some cases, the Drosophila or C. elegans sequence is sister to a single clade of vertebrate sequences, such as is found with the relationship among D-CG8667 (bHLHa16) and the vertebrate Mist1 (bHLHa15) sequences (Fig. 2), suggesting that this ortholog originated before the split of Drosophila and the vertebrates. In others, these non-vertebrate sequences are sister to a set of clades of vertebrate gene copies, as is found with D-CG5102 (bHLHb22) sister to the combination of the TCF4 (bHLHb19), TCF12 (bHLHb20), and TCF3 (bHLHb21) clades (Fig. 2B), which suggests that there were several duplication events before vertebrate diversification but after divergence from Drosophila. In all, it appears that approximately 89 paralogs of the bHLH gene family were present in the ancestor of human, mouse, and rat, as is noted by the blue bars on branches where these gene copies coalesce, Figs. 2A–2F.
While there are fewer bHLH gene copies in Drosophila and C. elegans with some paralogs that seem closely related to those found in vertebrates, there do seem to be some “insect specific” duplication events, particularly notable in the bHLH1, 2, and 4 clades (Figs. 2A–2C). This suggests that separate and different functions might be present for these paralogs as related copies are not found in vertebrates. Similarly, most of the Arabidopsis paralogs do not show close relationships to any of the animal sequences, and many of the duplicates form clades separate from other sequences (i.e. the bHLH clade 6; Fig. 2C). As this is the only plant included, the timing of these duplications is unclear, however, given the very few sequences with similarity to any of the animal sequences, it appears that few “orthologs” predate the divergence of plants and animals. The yeast sequences similarly do not group with sequences from any of the other lineages, not surprising given the phylogenetic distance between fungi and plants and animals. Comparatively, yeast has far fewer paralogs of the bHLH gene family than the other species, and most of them seem quite divergent from each other suggesting very old duplications. Sequences from additional fungi will be necessary to explore the origins of these duplication events.
As complete or nearly complete sequences are present for the human, mouse, and Arabidopsis genomes, chromosome location of the bHLH paralogs can be mapped to chromosomal location to explore the issue of possible origins/mechanisms for paralogous copies. These are mapped onto the phylogenies following the sequences names (Figs. 2A–2E). For instance, in the bHLH 4 clade (Fig. 2D), the paralogs MaxA-MaxF (bHLHd4-d8) are all found on chromosome arm 14q, suggesting these were either tandem duplications or duplications to near chromosomal locations. Conversely, the human paralogs Hey1 (bHLHb29), Hey2 (bHLHb32), and HeyL (bHLHb33), all closely related, are found on chromosome arms 8q, 6q, and 1p (Fig. 2B), suggesting a very different pattern of duplication.
Several expressed sequence tags (ESTs) have been included in these analyses due to the lack of a characterized copy identical to the EST copy. The placement of these ESTs follow a few basic patterns. In some cases, the EST fills in a gap where a gene copy is expected but not yet characterized. One example of this is the rat Tal2 (bHLHa19) EST (Fig. 2A). Characterized copies of Tal2 (bHLHa19) are present for mouse and human, and the uncharacterized rat EST is placed in the tree sister to the mouse copy. Because no other closely related rat copies exist, observations suggest this is the likely functional ortholog of the other vertebrate Tal2 (bHLHa19) genes. Alternatively, some ESTs are sister to functionally characterized genes from the same organism. One example is the human Scl (bHLHa17) EST sister to the human Tal1 (bHLHa17) gene (Fig. 2A). These sequences are not identical suggesting either duplication, modified splicing, or possibly mistakes in sequencing. Similarly there are fully characterized genes with several names that either contain errors in sequencing or have actual differences in their amino-acid sequence as suggested by sequence alignment. These putative paralogs need to be further analyzed in order to clarify their identity and relationships. There are a number of clades where there are not copies from human, mouse, and rat. In many cases this might be due to incomplete knowledge of the rat genome versus the human and mouse genomes. Alternatively, this may represent missed copies in mouse or human, or possibly more recent loss of those gene copies. An example of this is the presence of rat and human copies of the Nex1 (bHLHa2) (Fig. 2A), but no mouse copy.
Several functional domains other than the bHLH domain are associated with many of these proteins. Previous focus has been on the bHLH domain, but the other domains should not be ignored as they may play a vital role in protein dimerization and/or DNA binding. The first additional domain discussed is the leucine zipper coiled-coil dimerization domain (Zip) that is found in several families of transcription factors. Zip proteins can homodimerize or heterodimerize to bind DNA (Vinson et al., 2006). The presence of a bHLH domain along with the Zip domain may influence protein interactions and dimerization partners (Baxevanis and Vinson, 1993). In a recent study (Muir et al., 2008) it was found that bHLH and basic leucine zipper (bZip) proteins can directly interact leading to the complexity of the protein function through heterodimerization. Interestingly, the Zip domains in bHLH proteins is located in the C terminal helix domain (Fig. 1B) (Baxevanis and Vinson, 1993). The bHLH/Zip proteins did not localize to an individual clade suggesting a more general function for the Zip domain.
Similar to the bHLH/Zip proteins, the bHLH/PAS proteins have unique functions including ligand binding and protein–protein interactions. Unlike bHLH/Zip proteins, the PAS region is located a short distance C-terminal of the bHLH domain (Fig. 1B). In contrast to the bHLH/ZIP proteins, all of the bHLH/PAS proteins are located within one clade. The structure of the PAS portion of the bHLH/PAS proteins consist of a five- or six-stranded antiparallel β-sheet that is flanked by α-helices and loops (Crews and Fan, 1999). This structure forms a fold in which ligands can bind, potentially altering the conformation of the bHLH/PAS protein thereby allowing interactions with downstream signaling components (Crews and Fan, 1999). While many of the bHLH/PAS genes are conserved between mammals, Drosophila and C. elegans, the function of these genes differs. For example, the Aryl hydrocarbon receptor (Ahr) (i.e. dioxin receptor) and ARNT proteins form a bHLH protein DNA-binding complex that controls the physiological response to environmental compounds (e.g. dioxin) (Rowlands and Gustafsson, 1997) in mammals, but not in non-mammalian species (Hahn et al., 1997). The bHLH/PAS gene HIF (bHLH) has a role in cellular oxidative stress responses. The relationship of the different bHLH/PAS genes in clade 5, Fig. 3, suggests functional relationships between the different bHLH/PAS genes should be considered.
Similar to the bHLH/PAS proteins, the bHLH/Orange proteins are located within one clade and the Orange domain is located a short distance C-terminal of the bHLH domain (Davis and Turner, 2001), Fig. 1B. Interestingly, these proteins are located within clade 2 that contains the Id proteins and the bHLH/Orange proteins that function as transcriptional repressors. A direct interaction has been observed between bHLH/PAS and bHLH/Orange proteins (Chin et al., 2000). Further analysis indicates that transcriptional repression requires both the bHLH and Orange domains of these proteins. It has been speculated that the Orange domain function may be associated with subfamily specificity (Dawson et al., 1995). bHLH/Orange proteins bind to either an E-box or an N-box. Binding to an E-box raises the possibility that bHLH/Orange proteins can compete with bHLH activator proteins, thus functioning as a repressor. The WRPW motif is present in most bHLH/Orange proteins and is able to bind to the transcriptional co-repressor groucho and its mammalian homolog the TLE proteins (Paroush et al., 1994). Groucho/TLE proteins do not bind directly to DNA, rather they are recruited to target genes by a variety of DNA bound repressors (Fisher and Caudy, 1998) and appear to function in part by recruiting histone deacetylases to repress target genes (Chen and Courey, 2000). The bHLH/Orange proteins appear to be functionally related with other bHLH repressors localized to clade 2.
This phylogenetic analysis developed a classification of the bHLH gene family that can be used to elucidate cellular differentiation considering the gene family as a whole. The current study utilized a bioinformatics approach combining the phylogenetic information with microarray analysis to determine the association of various members of the bHLH gene family in cellular differentiation. The potential role of clade distribution in relation to expressed bHLH genes was examined. To verify the utility of this approach muscle cell microarray data was analyzed for bHLH transcription factors known to play a role in muscle cell differentiation. The myogenic proteins (Myog (bHLHc3), MyoD (bHLHc1), and myf5 (bHLHc2) are all expressed in the differentiated muscle cell as was expected. Scx (bHLHa41) that has a known role in chondrocyte development during embryogenesis is also expressed at a high level in the differentiated muscle cell. Making a direct comparison among four different cell types (i.e. Sertoli, Schwann, thymic, muscle) allows for differential expression of bHLH genes to be determined. All the cell types had unique bHLH genes expressed, as well as others expressed in common. These groups of bHLH genes expressed by the various cell types provide candidates for further analysis of the role of bHLH genes in cellular differentiation. For example, four genes that may play a role in pubertal differentiation of Sertoli cells were identified, Mxi1 (bHLHc11), Srebf1 (bHLHd1), Scx (bHLHa41), and Id4 (bHLHb27). Future studies involving knockdown and over-expression of these genes will help elucidate their respective roles in Sertoli cell differentiation. A limitation to this microarray analysis is that the chip used only provides a subset of bHLH genes within the total bHLH family. In addition, only a single adult developmental time-point was used in the analysis. Many bHLH genes expressed during development and involved in the induction of cellular differentiation may be absent from this bHLH gene set. An example is the role that Tcfe2a (bHLHb21) has in thymic cell differentiation (Jones and Zhuang, 2007), but was absent from the bHLH gene set, Table 1. Therefore, further developmental analysis using the entire bHLH family is needed in the future. In relation to the current study, the bHLH gene sets associated with the different cell types demonstrated the bHLH genes expressed represented multiple clades, Figure S1 and Table 1. Cellular differentiation appears to require a mixture of different functionally related bHLH genes present throughout the phylogenetic tree and clades.
The current study uses phylogenetic analysis in concert with microarray data to classify the bHLH gene family and identify genes that may be important in the process of cellular differentiation. Investigation of the entire gene family significantly enhances the ability to study the role of bHLH genes in cellular differentiation and development. Observations provided suggest a consideration that the group or network of bHLH genes present may be more physiologically relevant and important than the role of an individual gene. Owing to the differential functions and ability to heterodimerize, understanding the interplay of the network of bHLH genes is critical to investigating the role of the bHLH family in cellular differentiation. The phylogenetic bHLH super-tree presented and bHLH gene relationships identified will be critical for future investigations.
Supplementary Material
Acknowledgments
Grant Sponsorship: This work was supported by an NIH grant to Michael K. Skinner, 5R01 HD04381-04.
We acknowledge the technical assistance of Dr. Ingrid Sadler-Riggleman and Dr. Marina Savenkova. We thank John R. Clark for help with design and preparation of Fig. 3 and thank Ms. Jill Griffin for assistance in preparing the manuscript. None of the authors have any financial interests or disclosures. This work was supported by NIH grants to Michael K. Skinner.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Table S1. bHLH gene family nomenclature.
Table S2. Species distribution of bHLH homolog.
Fig. S1. Mouse bHLH super-tree for the 5 different clades and all 107 genes with the cellular expression of specific genes listed for Sertoli, Schwann, thymic and muscle cells. The expression is shown in the pie chart diagrams for each gene.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Contributor Information
Jeffrey D. Stevens, Center for Reproductive Biology, School of Molecular Biosciences, Pullman, WA 99164-4231, USA
Eric H. Roalson, School of Biological Sciences, Washington State University, Pullman, WA 99164-4231, USA
Michael K. Skinner, Center for Reproductive Biology, School of Molecular Biosciences, Pullman, WA 99164-4231, USA
References
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis AD, Vinson CR. Interactions of coiled coils in transcription factors: where is the specificity? Curr Opin Genet Dev. 1993;3:278–285. doi: 10.1016/0959-437x(93)90035-n. [DOI] [PubMed] [Google Scholar]
- Bergqvist I, Eriksson M, Saarikettu J, Eriksson B, Corneliussen B, Grundstrom T, Holmberg D. The basic helix- loop-helix transcription factor E2-2 is involved in T lymphocyte development. Eur J Immunol. 2000;30:2857–2863. doi: 10.1002/1521-4141(200010)30:10<2857::AID-IMMU2857>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Blom B, Heemskerk MH, Verschuren MC, van Dongen JJ, Stegmann AP, Bakker AQ, Couwenberg F, Res PC, Spits H. Disruption of alpha beta but not of gamma delta T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. EMBO J. 1999;18:2793–2802. doi: 10.1093/emboj/18.10.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Bober E, Arnold HH. Inhibition of muscle differentiation by the adenovirus E1a protein: repression of the transcriptional activating function of the HLH protein Myf-5. Genes Dev. 1992;6:888–902. doi: 10.1101/gad.6.5.888. [DOI] [PubMed] [Google Scholar]
- Buchstaller J, Sommer L, Bodmer M, Hoffmann R, Suter U, Mantei N. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24:2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Atchley WR. Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol. 2003;56:742–750. doi: 10.1007/s00239-002-2449-3. [DOI] [PubMed] [Google Scholar]
- Castella P, Sawai S, Nakao K, Wagner JA, Caudy M. HES-1 repression of differentiation and proliferation in PC12 cells: role for the helix 3-helix 4 domain in transcription repression. Mol Cell Biol. 2000;20:6170–6183. doi: 10.1128/mcb.20.16.6170-6183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary J, Johnson J, Kim G, Skinner MK. Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells. Endocrinology. 2001;142:1727–1736. doi: 10.1210/endo.142.5.8134. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Kim G, Skinner MK. Expression of the basic helix-loop-helix protein REBalpha in rat testicular Sertoli cells. Biol Reprod. 1999;60:1244–1250. doi: 10.1095/biolreprod60.5.1244. [DOI] [PubMed] [Google Scholar]
- Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- Chin MT, Maemura K, Fukumoto S, Jain MK, Layne MD, Watanabe M, Hsieh CM, Lee ME. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr Opin Genet Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr Biol. 1996;6:707–718. doi: 10.1016/s0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- Davis RL, Turner DL. Vertebrate hairy and enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- Dawson SR, Turner DL, Weintraub H, Parkhurst SM. Specificity for the hairy/enhancer of split basic helix-loophelix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- Fisher A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N, Alifragis P, Koumbanakis KA, Delidakis C. Two modes of recruitment of E(spl) repressors onto target genes. Development. 2003;130:259–270. doi: 10.1242/dev.00206. [DOI] [PubMed] [Google Scholar]
- Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–1728. [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Natl Acad Sci USA. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan BA, Bellen HJ. Doing the MATH: is the mouse a good model for fly development? Genes Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ishibashi J, Perry RL, Asakura A, Rudnicki MA. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J Cell Biol. 2005;171:471–482. doi: 10.1083/jcb.200502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3:RESEARCH0030. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PO. Phylogenetic systematics turns over a new leaf. Trends Ecol Evolut. 2001;16:30–37. doi: 10.1016/s0169-5347(00)02025-5. [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DJ, Friel PJ, Pouchnik D, Griswold MD. Oligonucleotide microarray analysis of gene expression in follicle- stimulating hormone-treated rat Sertoli cells. Mol Endocrinol. 2002;16:2780–2792. doi: 10.1210/me.2002-0059. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development. 2004;131:2195–2204. doi: 10.1242/dev.01091. [DOI] [PubMed] [Google Scholar]
- Morrow MA, Mayer EW, Perez CA, Adlam M, Siu G. Overexpression of the helix-loop-helix protein Id2 blocks T cell development at multiple stages. Mol Immunol. 1999;36:491–503. doi: 10.1016/s0161-5890(99)00071-1. [DOI] [PubMed] [Google Scholar]
- Muir T, Sadler-Riggleman I, Skinner MK. Role of the basic helix-loop-helix transcription factor, scleraxis, in the regulation of Sertoli cell function and differentiation. Mol Endocrinol. 2005;19:2164–2174. doi: 10.1210/me.2004-0473. [DOI] [PubMed] [Google Scholar]
- Muir T, Wilson-Rawls J, Stevens JD, Rawls A, Kang C, Skinner MK. Integration of CREB and bHLH transcriptional signaling pathways through direct heterodimerization of proteins. Mol Reprod Develop. 2008 doi: 10.1002/mrd.20802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, Parker KL, Vassalli JD. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- Rowlands JC, Gustafsson JA. Aryl hydrocarbon receptor- mediated signal transduction. Crit Rev Toxicol. 1997;27:109–134. doi: 10.3109/10408449709021615. [DOI] [PubMed] [Google Scholar]
- Skinner MK. Cell-cell interactions in the testis. Endocr Rev. 1991;12:45–77. doi: 10.1210/edrv-12-1-45. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart HJ, Zoidl G, Rossner M, Brennan A, Zoidl C, Nave KA, Mirsky R, Jessen KR. Helix-loop-helix proteins in Schwann cells: a study of regulation and subcellular localization of Ids, REB, and E12/47 during embryonic and postnatal development. J Neurosci Res. 1997;50:684–701. doi: 10.1002/(SICI)1097-4547(19971201)50:5<684::AID-JNR6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Taelman V, Van Wayenbergh R, Solter M, Pichon B, Pieler T, Christophe D, Bellefroid EJ. Sequences downstream of the bHLH domain of the Xenopus hairy-related transcription factor-1 act as an extended dimerization domain that contributes to the selection of the partners. Dev Biol. 2004;276:47–63. doi: 10.1016/j.ydbio.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Tam SK, Gu W, Mahdavi V, Nadal-Ginard B. Cardiac myocyte terminal differentiation. Potential for cardiac regeneration. Ann NY Acad Sci. 1995;752:72–79. doi: 10.1111/j.1749-6632.1995.tb17407.x. [DOI] [PubMed] [Google Scholar]
- Tang H, Veldman MB, Goldman D. Characterization of a muscle-specific enhancer in human MuSK promoter reveals the essential role of myogenin in controlling activity-dependent gene regulation. J Biol Chem. 2006;281:3943–3953. doi: 10.1074/jbc.M511317200. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temchura VV, Frericks M, Nacken W, Esser C. Role of the aryl hydrocarbon receptor in thymocyte emigration in vivo. Eur J Immunol. 2005;35:2738–2747. doi: 10.1002/eji.200425641. [DOI] [PubMed] [Google Scholar]
- Thatikunta P, Qin W, Christy BA, Tennekoon GI, Rutkowski JL. Reciprocal Id expression and myelin gene regulation in Schwann cells. Mol Cell Neurosci. 1999;14:519–528. doi: 10.1006/mcne.1999.0799. [DOI] [PubMed] [Google Scholar]
- Thattaliyath BD, Firulli BA, Firulli AB. The basichelix- loop-helix transcription factor HAND2 directly regulates transcription of the atrial naturetic peptide gene. J Mol Cell Cardiol. 2002;34:1335–1344. doi: 10.1006/jmcc.2002.2085. [DOI] [PubMed] [Google Scholar]
- Thompson JA, Gold PJ, Fefer A. Outpatient chemoimmunotherapy for the treatment of metastatic melanoma. Semin Oncol. 1997;24:S44–S48. [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Cruchten I, Cinato E, Fox M, King ER, Newton JS, Riechmann V, Sablitzky F. Structure, chromosomal localisation and expression of the murine dominant negative helix- loop-helix Id4 gene. Biochim Biophys Acta. 1998;1443:55–64. doi: 10.1016/s0167-4781(98)00196-1. [DOI] [PubMed] [Google Scholar]
- Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wei Q, Paterson BM. Regulation of MyoD function in the dividing myoblast. FEBS Lett. 2001;490:171–178. doi: 10.1016/s0014-5793(01)02120-2. [DOI] [PubMed] [Google Scholar]
- Yang C, Boucher F, Tremblay A, Michaud JL. Regulatory interaction between arylhydrocarbon receptor and SIM1, two basic helix-loop-helix PAS proteins involved in the control of food intake. J Biol Chem. 2004;279:9306–9312. doi: 10.1074/jbc.M307927200. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K. Cell cycle regulators in neural stem cells and postmitotic neurons. Neurosci Res. 2000;37:1–14. doi: 10.1016/s0168-0102(00)00101-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.