Linear tetrapyrroles (bilins) are needed for light-dependent greening and for assembly and maintenance of a functional photosynthetic apparatus in the green alga Chlamydomonas.
Abstract
In land plants, linear tetrapyrrole (bilin)-based phytochrome photosensors optimize photosynthetic light capture by mediating massive reprogramming of gene expression. But, surprisingly, many green algal genomes lack phytochrome genes. Studies of the heme oxygenase mutant (hmox1) of the green alga Chlamydomonas reinhardtii suggest that bilin biosynthesis in plastids is essential for proper regulation of a nuclear gene network implicated in oxygen detoxification during dark-to-light transitions. hmox1 cannot grow photoautotrophically and photoacclimates poorly to increased illumination. We show that these phenotypes are due to reduced accumulation of photosystem I (PSI) reaction centers, the PSI electron acceptors 5′-monohydroxyphylloquinone and phylloquinone, and the loss of PSI and photosystem II antennae complexes during photoacclimation. The hmox1 mutant resembles chlorophyll biosynthesis mutants phenotypically, but can be rescued by exogenous biliverdin IXα, the bilin produced by HMOX1. This rescue is independent of photosynthesis and is strongly dependent on blue light. RNA-seq comparisons of hmox1, genetically complemented hmox1, and chemically rescued hmox1 reveal that tetrapyrrole biosynthesis and known photoreceptor and photosynthesis-related genes are not impacted in the hmox1 mutant at the transcript level. We propose that a bilin-based, blue-light-sensing system within plastids evolved together with a bilin-based retrograde signaling pathway to ensure that a robust photosynthetic apparatus is sustained in light-grown Chlamydomonas.
INTRODUCTION
Biogenesis of the eukaryotic photosynthetic apparatus requires coordinated synthesis of nuclear- and chloroplast-encoded polypeptide subunits of photosynthetic complexes, targeting of these nascent polypeptides to their correct sites within chloroplasts, and supramolecular assembly of proteins, pigments, and cofactors into the various complexes that modulate necessary functions in the thylakoid membranes (Rochaix, 2004). All of these processes are linked to diurnal light-dark cycles, which entrain them to the cellular energy status, influence the turnover of photosynthetic complexes, and inform expression of both plastid-encoded and photosynthesis-associated nuclear genes (PhANGs) (reviewed in Eberhard et al., 2008).
Proper regulation of chlorophyll synthesis and integration into functional protein complexes are especially critical in the light because free pigments can readily generate reactive oxygen species (ROS) within chloroplasts (reviewed in Schmidt and Schippers, 2015). ROS can also be generated by the two photosystems themselves, when electron acceptors, particularly those of photosystem I (PSI), become overreduced. ROS-derived retrograde signals can then be relayed to the nucleus to regulate both PhANGs and genes involved in ROS detoxification (reviewed in Chi et al., 2013). In plants and green algae, the light-harvesting complexes (LHCs) of PSI and photosystem II (PSII) contain the bulk of cellular chlorophylls (reviewed in Merchant and Sawaya, 2005; Rochaix, 2014). Thus, when PSI and PSII reaction centers (RCs) become saturated or inactivated by excess light, LHCs become the major source for light-dependent ROS generation (Tripathy and Oelmüller, 2012). Multiple short-term (acute) acclimation and long-term adaptation responses have evolved to minimize ROS production and damage in both plants and green algae (reviewed in Li et al., 2009).
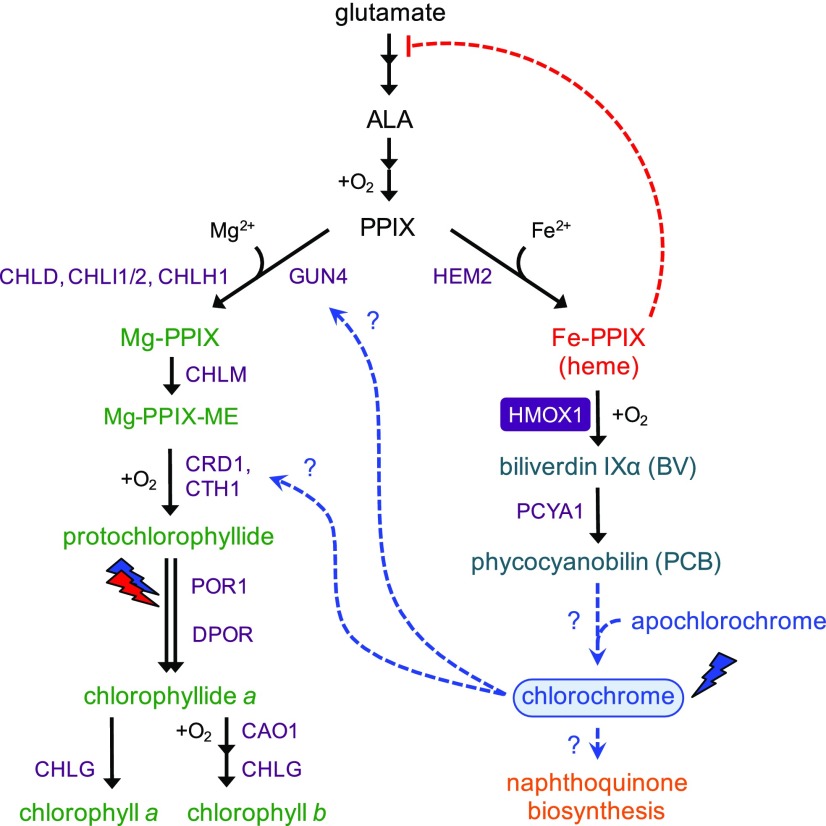
One key regulatory mechanism associated with photoacclimation, the process whereby the photosynthetic apparatus is remodeled in response to changes in light quality and fluence rate, is the restriction or activation of chlorophyll biosynthesis (Brzezowski et al., 2015). The synthesis of tetrapyrroles is highly regulated in all eukaryotes, and this control is especially important for photosynthetic organisms. Faulty regulation is potentially lethal due to release of photosensitizing intermediates such as protoporphyrin IX (PPIX) or Mg-protoporphyrin IX (Mg-PPIX) in the presence of oxygen and light (Rebeiz et al., 1990; Mochizuki et al., 2010; Busch and Montgomery, 2015). Heme and chlorophyll biosynthesis in plants and algae share a trunk pathway in which PPIX is derived from the amino acid glutamate (reviewed in Grossman et al., 2004; Tanaka and Tanaka, 2007; Kobayashi and Masuda, 2016). PPIX is the penultimate intermediate in both pathways, which diverge upon addition of either Fe2+ for heme or Mg2+ for chlorophyll (Figure 1). Both Mg-PPIX and Fe-PPIX (heme) are pro-oxidants, which mediate deleterious oxidative chemistry in the presence of oxygen and/or light, so sequestration and limiting their ability to react with oxygen is critical to ensure plant and algal survival. All eukaryotes possess feedback pathways for regulating the accumulation of 5-aminolevulinic acid (ALA), the first common intermediate of chlorophyll and heme synthesis (Figure 1). Furthermore, most of the later steps of the pathway are strongly inhibited by enzymatic products, ensuring that pro-oxidative intermediates are not generated at high levels. In plants and algae, chlorophyll synthesis is confined to plastids, shielding the rest of the cell from direct contact with these damaging molecules.
Figure 1.
Crosstalk among Chlorophyll, Bilin, and Naphthoquinone Biosynthesis in Chlamydomonas.
The biosynthesis of chlorophyll (Chl), heme, and linear tetrapyrroles (bilins) in Chlamydomonas share a common pathway from glutamate to PPIX via the intermediate ALA. The heme and chlorophyll pathways diverge with the insertion of iron or magnesium, respectively, into PPIX. The bilins biliverdin IXα (BV) and PCB are derived from heme via the successive action of HMOX1 (purple rectangle) and PCYA1. PCB is hypothesized to be the chromophore cofactor of chlorochrome, which may target key enzymes involved in chlorophyll biosynthesis (represented by dashed blue lines). Feedback inhibition of ALA synthesis by heme is depicted with a dashed red line. Light-regulated steps of chlorophyll biosynthesis are indicated with red and blue lightning bolts, which represent red- and blue-light-dependent activity of the light-dependent protochlorophyllide oxidoreductase. Intermediates committed to chlorophyll, heme, and bilin pathways are colored in green, red, and teal, respectively. Enzymes, cofactors, and cosubstrates are shown next to arrows. Gene names of key enzymes include Mg-chelatase (CHLD, CHLH1, CHLI1/2, and GUN4), Mg-protoporphyrin O-methyltransferase (CHLM), copper response defect 1 (CRD1), copper target 1 (CTH1), light-dependent protochlorophyllide oxidoreductase (POR1), light-independent (dark) protochlorophyllide oxidoreductase (DPOR; encoded by plastid genes chlB, chlL, and chlN), chlorophyllide a oxygenase (CAO1), chlorophyll synthase (CHLG), ferrochelatase (HEM2), heme oxygenase (HMOX1), and phycocyanobilin:ferredoxin oxidoreductase (PCYA1). Based on this work, hypothetical targets of blue-light-activated chlorochrome are shown with blue dashed arrows.
Despite many mechanisms to control tetrapyrrole synthesis, it is inevitable that some heme and chlorophyll will be lost from chromoprotein complexes. Free heme can be detoxified by heme oxygenases (HMOXs), oxygen-dependent enzymes that convert heme to biliverdin (BV), carbon monoxide, and iron. HMOXs also perform critical roles in iron recycling, gas signaling, and linear tetrapyrrole (bilin) pigment biosynthesis in many organisms in the absence of ROS production (reviewed in Wilks, 2002). Owing to the rate-limiting release of BV product, high-throughput HMOX turnover requires a coupled reaction for efficient heme detoxification. This is generally accomplished by bilirubin-generating NADPH-dependent biliverdin reductases in animals and by phytobilin-generating ferredoxin-dependent bilin reductases (FDBRs) in cyanobacteria and photosynthetic eukaryotes (Frankenberg and Lagarias, 2003; Rockwell et al., 2014). FDBRs and HMOXs are localized in plastids, consistent with their roles in synthesizing phycobiliprotein antennae and phytochrome chromophores (Frankenberg-Dinkel and Terry, 2009). This subcellular compartmentalization ensures efficient detoxification of heme that is released from the photosynthetic apparatus during hemoprotein turnover.
The reference green alga Chlamydomonas reinhardtii possesses two canonical heme oxygenases (HMOX1 and HMOX2) and one FDBR phycocyanobilin:ferredoxin oxidoreductase (PCYA1), and yet it lacks phytochromes and phycobiliproteins (Merchant et al., 2007). Recent studies show that HMOX1 and PCYA1 comprise a plastid-localized pathway for the synthesis of the phytobilin phycocyanobilin (PCB) and that the cytosol-localized HMOX2 participates in iron recycling and probably also heme detoxification (Duanmu et al., 2013). These studies also showed that an hmox1 null mutant accumulates chlorophyll poorly during photoautotrophic growth, whereas hmox1 cultures accumulate wild-type levels of chlorophyll when grown heterotrophically in the dark (Duanmu et al., 2013). Comparative transcriptome analyses of dark-acclimated and 0.5 h light-treated hmox1 cultures with the 4A+ WT (wild type) parent and two genetically HMOX1-complemented lines suggest that plastid bilins serve as retrograde signals that regulate a small network of nuclear genes implicated in ROS detoxification during dark-to-light transitions (Duanmu et al., 2013).
This investigation was undertaken to determine the mechanistic basis of defects in photoautotrophic growth and light-dependent chlorophyll accumulation in the light-grown Chlamydomonas hmox1 mutant. Our studies show that the hmox1 mutant is profoundly deficient in PSI activity, reflecting decreased light-dependent accumulation of PSI core subunits, quinone cofactors, and associated LHCs. Supplementing with BV, the product of the HMOX1 reaction, rescues most of the biosynthetic deficiencies observed in the hmox1 mutant, through a blue light-mediated process that is independent of photosynthesis. RNA-seq analyses of cultures acclimated to either 0.5 or 4 h light reveal that the hmox1 mutant does not impact tetrapyrrole biosynthesis, known photoreceptors, and photosynthesis-associated genes at the transcript level. These analyses provide compelling evidence that bilins, in conjunction with blue light photoperception, play an essential role in the biogenesis of the photosynthetic apparatus in Chlamydomonas. Our results suggest the existence of a bilin-based photoreceptor that impacts the assembly/stability of photosynthetic complexes in the light.
RESULTS
Light-Dependent Growth, Greening, and Photosynthesis Are Compromised in the hmox1 Mutant
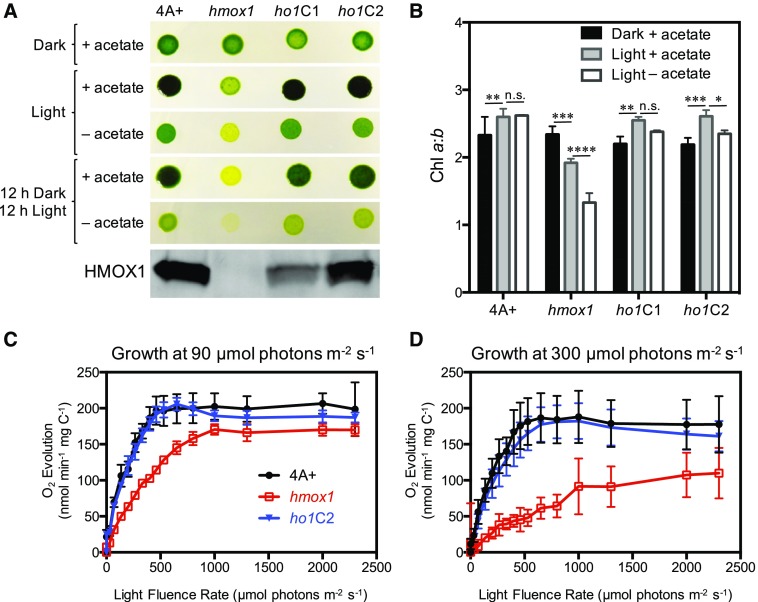
The Chlamydomonas hmox1 mutant is defective in photoautotrophic growth and light-dependent chlorophyll accumulation (Duanmu et al., 2013). These phenotypes are even more exaggerated in hmox1 cultures grown under diurnal 12-h-dark/12-h-light cycles either mixotrophically (with acetate) or photoautotrophically (without acetate) (Figure 2A), suggesting an impairment of diurnal photoacclimation. To probe the molecular basis of these phenotypes, we compared the chlorophyll a/b ratios of fully acclimated cultures of hmox1, two complemented lines (ho1C1 and ho1C2), and the parental 4A+ WT strain that were grown in darkness heterotrophically or under continuous white light (120 µmol photons m−2 s−1) either mixotrophically or photoautotrophically. All except the hmox1 mutant had a constant or slightly increased chlorophyll a/b ratio under all light regimes compared with dark-acclimated cultures (Figure 2B). Although the chlorophyll a/b ratio of the hmox1 mutant grown in darkness was similar to that of other lines, this ratio decreased significantly for light-grown hmox1 cultures and dropped most for those grown photoautotrophically (P < 0.001, two-way ANOVA and Fisher’s least significant difference test). This reduced chlorophyll a/b ratio in light-grown hmox1 is consistent with a reduced level of RCs relative to LHC complexes.
Figure 2.
Growth Characteristics of hmox1 under Different Trophic Conditions.
(A) Comparative growth of 4A+, hmox1, and two cDNA complemented lines (ho1C1 and ho1C2) either in the dark, under constant light (∼120 μmol photons m−2 s−1) or under a 12-h-light/12-h-dark cycle. Thirty micrograms of total protein from photoautotrophically growing exponential phase cell cultures were used for immunoblot analysis of HMOX1 expression.
(B) Chlorophyll a/b ratios under different growth conditions. Data show means of three biological replicates ± sd. Two-way ANOVA and Fisher’s least significant difference test was applied for multiple pairwise comparisons (see Supplemental File 1). Asterisks above pairs of bars indicate that the corresponding values are significantly different (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; n.s., not statistically significant).
(C) and (D) Light saturation curves of O2 evolution rates. Cells were grown mixotrophically in TAP medium under constant light at either 90 (C) or 300 (D) μmol photons m−2 s−1, and O2 evolution was measured as a function of light fluence rate. Photosynthetic O2 evolution rates were normalized to total organic carbon. Biological culture triplicates were analyzed and data show means ± sd. Maximal photosynthesis rates were calculated by fitting the curves with the Box-Lucas method.
We next compared photosynthetic electron transfer (PET) activity in exponentially growing mixotrophic cultures of the wild-type, hmox1, and ho1C2 strains. All cultures were acclimated to continuous white light at fluence rates of 90 or 300 µmol photons m−2 s−1. Rates of photosynthetic O2 evolution were measured as a function of increasing light fluence rate. Wild-type and ho1C2 cultures acclimated to 90 µmol photons m−2 s−1 exhibited saturation of O2 evolution at ∼500 µmol photons m−2 s−1 white light (Figure 2C). In contrast, hmox1 cultures displayed reduced O2 evolution compared with the wild type and ho1C2 and higher fluence rates were required to saturate O2 evolution in hmox1. However, the maximum level achieved for hmox1 was similar to levels in the wild type and ho1C2 when normalized to total organic carbon. These results suggest that linear electron flow was not severely impacted in the mutant strain when acclimated to 90 µmol photons m−2 s−1 white light. When grown at a fluence rate of 300 µmol photons m−2 s−1, the wild type and ho1C2 exhibited saturated O2 evolution at ∼500 µmol photons m−2 s−1, whereas hmox1 exhibited a smaller light-dependent increase in O2 evolution and appeared to saturate at ∼1000 µmol photons m−2 s−1 (Figure 2D). Moreover, the maximum capacity of hmox1 cells for O2 evolution (131 ± 13 nmol O2 min−1 mg C−1) was markedly lower than that of the wild type and ho1C2 (174∼186 nmol O2 min−1 mg C−1, P < 0.01 by two-tailed Student’s t test). These results underscore a critical role for HMOX1 in maintaining photosynthetic activity and chlorophyll levels in the light.
The hmox1 Mutant Has Reduced Activity of Both Photosystems in the Light
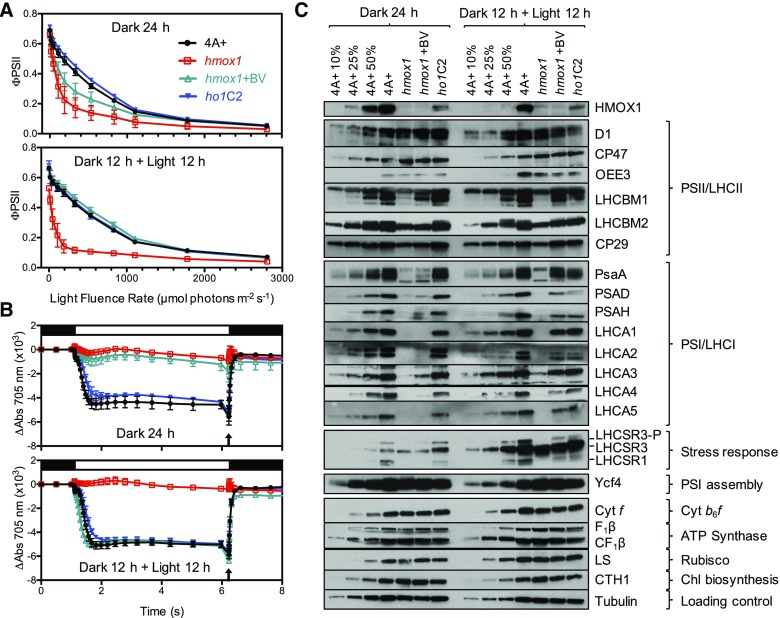
To compare linear PET in hmox1, wild type, ho1C2, and hmox1 + BV supplementation, we used a chlorophyll fluorescence-based assay. All strains were grown mixotrophically (light + acetate) under low white light (∼30 µmol photons m−2 s−1) for these experiments. After dilution into fresh medium, half of each culture was transferred to complete darkness for 24 h (Figures 3A and 3B, top), while the other half was transferred to darkness for 12 h followed by illumination with ∼100 µmol photons m−2 s−1 white light for 12 h (Figures 3A and 3B, bottom) prior to fluorescence or spectroscopic measurements. ΦPSII (quantum efficiency of PSII) values of 24 h dark-acclimated hmox1 cultures were similar to those of wild-type cells and the ho1C2 complemented strain at very low light fluence rates, but decreased to a much greater extent at fluence rates above 50 µmol photons m−2 s−1 (Figure 3A, top). This reduction in ΦPSII was only slightly rescued when BV was present during growth in the dark (e.g., compare hmox1+BV with ho1C2). A severe reduction in ΦPSII was also observed after 12 h of light acclimation of the hmox1 mutant (Figure 3A, bottom). Intriguingly, ΦPSII of hmox1 cultures was fully rescued under these conditions when BV (0.1 mM) was included in the growth medium at the onset of the 24 h period prior to photosynthesis measurements.
Figure 3.
PSII and PSI Activities and Protein Abundance of Dark-Maintained and Light-Acclimated Cultures.
(A) and (B) Cells grown in moderate light (∼30 μmol photons m−2 s−1) were diluted to ∼1 × 106 cells/mL and either acclimated to darkness for 24 h (top panels) or acclimated to darkness for 12 h and then transferred to light (∼100 μmol photons m−2 s−1) for another 12 h (bottom panels). All measurements were from three biological replicates with error bars representing sd.
(A) PSII efficiency (ΦPSII) measurements were performed with a DUAL-PAM 100 on the light curve setting. Each light fluence rate step lasted 60 s followed by a saturating pulse. Cells were adjusted to 10 µg/mL chlorophyll in fresh TAP. Sodium bicarbonate (1 mM) was added as an electron acceptor.
(B) PSI oxidation-reduction kinetics were measured by absorption changes at 705 nm with a JTS10 spectrophotometer. Cells were exposed to ∼500 μmol photons m−2 s−1 light for 5 s (open bar) followed by a saturating pulse to achieve full oxidation of P700 (indicated by arrow). Cells were adjusted to 30 µg/mL chlorophyll in 20 mM HEPES-KOH, pH 7.4, and 10% Ficoll; 20 µM DCMU and 1 mM HA were added as PSII inhibitors.
(C) Immunoblot analyses using monospecific antibodies against photosynthetic proteins. Lanes were loaded based on equal amounts of total cellular protein, the concentration of which was assessed using tubulin as a loading control. A dilution series (10, 25, and 50%) of the 4A+ WT strain is provided for both growth conditions.
These analyses show that the hmox1 mutant has impaired electron flow downstream of PSII because the maximum quantum yield of PSII of the mutant is similar to that of the wild type and ho1C2 (Fv/Fm, initial ΦPSII value following the dark adaptation period; Figure 3A, bottom). The partial rescue by BV of the dark-acclimated hmox1 mutant could reflect light attenuation by the added BV and fluence rate-dependent instability of PSI. However, complete rescue of ΦPSII in 12-h-dark/12-h-light hmox1 cultures supplemented with BV cannot be explained by light attenuation and instead suggests a bilin-based rescue of the mutant phenotype that is also light dependent.
To more specifically localize the photosynthetic defect in hmox1, we measured PSI activity for both dark- and light-acclimated cultures by assaying light-dependent absorption changes at 705 nm. Such measurements are used to determine the amount of light-driven oxidation of P700 to P700+, and the re-reduction of P700+ back to P700 in the dark. The amount of photooxidizable P700 (i.e., functional PSI) was normalized to the chlorophyll levels. In comparison to the wild-type strain, dark-acclimated hmox1 had low levels of photooxidizable P700 and there was only a minor effect on this activity when the mutant cells were supplemented with BV (Figure 3B, top). A similar loss of active PSI in the mutant was observed following the 12-h-dark/12-h-light treatment (Figure 3B, bottom). However, in contrast to the results of dark-acclimated hmox1 cultures, exogenous BV restored P700 photooxidation to the same levels observed in light-acclimated wild-type and ho1C2 cultures (Figure 3B, bottom). These experiments establish that BV supplementation can fully rescue both PSI and ΦPSII deficiencies during the 12-h-dark/12-h-light acclimation period.
The hmox1 Mutant Is Deficient in Light-Dependent PSI and LHC Polypeptide Accumulation
The observations described above prompted us to determine whether the PSI-deficient phenotype was accompanied by changes in photosynthesis-related polypeptide accumulation. Therefore, we analyzed total cellular protein from dark- and light-acclimated cultures of the wild type, hmox1, hmox1 + BV, and ho1C2 from the same cultures that were used to measure photosynthetic activities (ΦPSII and P700 redox changes). Whereas many photosynthetic proteins accumulated to similar levels in the four lines under both light regimes, e.g., subunits of the PSII core (D1, CP47), oxygen-evolving complex (OEE3), the stress response proteins LHCSR1/3, the cytochrome b6f complex (Cyt f), the chloroplast (and mitochondrial) ATP synthase (β subunits of each, respectively), and Rubisco (large subunit [LS]), we observed striking differences in levels of polypeptides associated with PSI and LHCs (Figure 3C).
After acclimation for 24 h in darkness, the hmox1 mutant (+/−BV) possessed very little PSI (representative subunits: PsaA, PSAD, and PSAH). Based on a dilution series with extracts from the wild type, dark-acclimated hmox1 cells (−BV) contained <10% of wild-type levels of PSI polypeptides when normalized to tubulin levels. Such hmox1 cultures were also notably deficient in LHCI polypeptides (representative subunits: LHCA1, LHCA2, LHCA3, LHCA4, and LHCA5). Addition of BV prior to 24 h of dark acclimation failed to appreciably increase the levels of these LHCA polypeptides. Those LHCAs that did increase during this 24-h dark incubation period in the presence of BV never accumulated to more than 25% of the levels seen in wild-type cells and the genetically complemented ho1C2 control lines. Dark-acclimated hmox1 cultures were also deficient in PSII-associated antennae polypeptides, LHCBM1, LHCBM2, and CP29. BV treatment did partially restore levels of some of these polypeptides, i.e., notably LHCBM1. However, their levels never reached those of wild-type or ho1C2 lines. Overall, these results suggest that the reduced PSI activity of dark-acclimated hmox1 cultures (Figure 3B) mainly reflects lower abundances of PSI-associated polypeptides and partial loss of LHCII polypeptides.
Similar to dark-acclimated hmox1 cultures, hmox1 cultures grown for 12 h in darkness followed by 12 h in the light were severely deficient in PSI-associated polypeptides (Figure 3C). All of the PSI and LHCA polypeptides analyzed were either not detectable (PSAD, PSAH, LHCA1, LHCA2, LHCA4, and LHCA5) or were present at very low (<10%) levels (PsaA and LHCA3). When BV was added to hmox1 cells at the onset of the 12-h-dark/12-h-light period, most of the PSI and LHCA polypeptides reappeared, accumulating in many cases to similar levels seen in wild-type and ho1C2 control lines. This reappearance of PSI polypeptides was also accompanied by a significant light-dependent increase in 5′-monohydroxyphylloquinone and phylloquinone, which are one-electron carriers within PSI of Chlamydomonas (Figure 4A; P < 0.05, two-way ANOVA and Fisher’s least significant difference test) (Ozawa et al., 2012). BV also rescued reduced levels of PSII-associated antennae polypeptides to levels comparable to those seen in the wild-type and ho1C2 lines (Figure 3C). These results indicate that BV can compensate for the loss of PSI-associated proteins/cofactors and LHCIIs in the hmox1 mutant and that such compensation is light dependent.
Figure 4.
Effect of BV and Light Quality on Accumulation of Naphthoquinone Cofactors, Pigment-Protein Complexes, and Photosynthetic Proteins.
(A) to (C) Cells grown in moderate light (∼30 μmol photons m−2 s−1) were diluted to ∼1 × 106 cells/mL and either acclimated to darkness for 24 h (black bars) or acclimated to darkness for 12 h and then transferred to light (∼100 μmol photons m−2 s−1) for another 12 h (gray bars).
(A) 5′-Monohydroxyphylloquinone + phylloquinone content is plotted on a per cell basis. Data show means of three biological replicates ± sd. Asterisks above the bars indicate that the corresponding values are significantly different as determined by Fisher’s least significant difference test with ANOVA (*P < 0.05 and **P < 0.01; n.s., not statistically significant; see Supplemental File 1).
(B) Separation of solubilized thylakoid membranes by BN-PAGE. Purified thylakoids were solubilized in 1% β-DM; 2.5 µg chlorophyll was loaded in each lane. Assignments of photosynthetic complexes to specific bands were based on immunoblot analyses.
(C) Rescue of PSI/LHCI accumulation at different light quantities and qualities. Strains were grown in darkness for at least 24 h to ensure loss of PSI/LHCI in hmox1 before transfer either to white light at ∼100 μmol photons m−2 s−1 or to white, blue, or red light at 10 μmol photons m−2 s−1, respectively. The contribution of blue light in 100 µmol photons m−2 s−1 white light was ∼10 µmol photons m−2 s−1. DCMU (20 µM) was added as a photosynthesis inhibitor.
Given the impact on PSI, we next examined how the loss of HMOX1 affected accumulation of the major photosynthetic pigment-protein complexes using blue native (BN)-PAGE analysis. Thylakoid membranes were isolated from cells grown under similar conditions as described above. These membranes were solubilized with the mild, nonionic detergent n-dodecyl β-d-maltoside (β-DM), and the major photosynthetic complexes were resolved by BN-PAGE (Figure 4B). Dark-acclimated hmox1 (+/−BV) displayed reduced accumulation of the large photosystem-antenna supercomplexes (PSII-LHCII and PSI-LHCI), and this was not surprising given the loss or reduction of major PSI and LHC components. The accumulation of PSII-LHCII and PSI-LHCI supercomplexes was highly reduced in light-acclimated hmox1 cultures grown in the absence of BV. However, BV supplementation fully restored the levels of both PSII-LHCII and PSI-LHCI in light-acclimated hmox1 cultures. These results confirm a critical role of HMOX1 in maintaining photosynthetic pigment-protein supercomplexes, and they also demonstrate that the rescue of supercomplex accumulation requires both BV and light.
BV Rescue Is Blue Light Dependent
Many photosynthesis-associated processes, including protein complex assembly, stability, and functionality, are regulated by diurnal dark/light cues. These processes are also responsive to changes in both light quantity and quality. To determine whether the rescue of PSI-LHCI accumulation in the hmox1 mutant was dependent on light quality and fluence rate, we grew the same strains (including hmox1 with and without BV) in darkness and then transferred them to different light conditions for 24 h prior to analyzing representative PSI-associated proteins. We grew cells under two fluence rates of white light (100 and 10 µmol photons m−2 s−1) as well as under blue or red light (10 µmol photons m−2 s−1 of each). While BV was able to fully rescue accumulation of PSAD and LHCA1 under 100 µmol photons m−2 s−1 white light, this rescue was incomplete under 10 µmol photons m−2 s−1 white light (Figure 4C). Surprisingly, we found that rescue of PSAD and LHCA1 levels in hmox1 was specifically enhanced by blue light. Indeed, BV-dependent accumulation of PSAD and LHCA1 in 10 µmol photons m−2 s−1 blue light was only slightly less effective than under 100 µmol photons m−2 s−1 white light (the contribution of blue wavelengths in this white light is ∼10 µmol photons m−2 s−1). In contrast, 10 µmol photons m−2 s−1 red light resulted in no detectable recovery of PSAD and LHCA1 in hmox1. These results suggest that BV rescue of PSI and LHCI in hmox1 is not a consequence of the level of photosynthetic activity, but is instead dependent on a blue-light-responsive signaling pathway. This hypothesis is supported by the lack of complete DCMU suppression of the blue light- and BV-dependent recovery of PSAD and LHCA1 in hmox1 (Figure 4C).
HMOX1 Is Dispensable for Light-Dependent Transcript Accumulation of Tetrapyrrole, Known Photoreceptors, and Photosynthesis-Associated Pathway Genes
To identify the mechanism by which HMOX1 affects accumulation of photosynthetic proteins in the light, an earlier study of the transcriptome in dark-grown hmox1 (Duanmu et al., 2013) was extended to study the dark-to-light transition in chemically and genetically complemented strains. These results suggest that an as yet undiscovered transcriptional or posttranscriptional response to light is BV-dependent. We therefore analyzed gene expression in duplicate, in the wild-type parental strain 4A+, the hmox1 mutant, and two complemented strains (ho1C1 and ho1C2) in the dark, at 0.5 and 4 h after transition into the light. In addition, expression was analyzed in 4A+ and hmox1 cells supplemented with exogenous BV, at the same time points before and after transition into the light. All reads were mapped against the most recent assembly of the Chlamydomonas genome (www.phytozome.net/chlamy), utilizing version 5.5 gene models to calculate gene expression estimates. The complete set of expression estimates for all genes can be found in Supplemental Data Set 1.
We first analyzed the differences of gene expression in the different strains in the dark and found a surprisingly high number of genes (1362 genes) in the parental strain 4A+ that is consistently different from those of the mutant and two independent complemented strains. A similar analysis of the other three lines resulted in only 8 to 44 consistently differentially expressed genes (Figure 5A; Supplemental Data Set 1). In addition, pairwise comparisons revealed a larger overlap of the dark-to-light transition hmox1 mutant transcriptome with those of both complemented strains (Figure 5B; Supplemental Data Set 1). The two complemented lines therefore represent better controls for identification of HMOX1-dependent genes in this experiment, similar to what was observed in other Chlamydomonas transcriptome analyses (Hemschemeier et al., 2013; Zones et al., 2015). Consequently, we focused our analysis on the differences between hmox1 and chemically (hmox1+BV) or genetically (ho1C1 and ho1C2) rescued lines at 0.5 and 4 h light acclimation.
Figure 5.
Transcriptome Profiling during Dark-to-Light Transition.
(A) Differences of gene expression levels among various strains in the dark. The bar graph shows the number of consistently differentially expressed genes in 4A+ (versus hmox1, ho1C1, and ho1C2), hmox1 (versus 4A+, ho1C1, and ho1C2), ho1C1 (versus hmox1, 4A+, and ho1C2), and ho1C2 (versus hmox1, ho1C1, and 4A+) at the beginning of the experiment in the dark.
(B) Responses of the transcriptome in hmox1 and control strains during the transition from dark to light. Venn diagrams show the overlap of genes changing during the transition from darkness to 0.5 h in the light (top) and from darkness to 4 h in the light (bottom). The number of genes changing similarly in hmox1, hmox1+BV, ho1C1, and ho1C2 is shown in the central overlap regions in yellow and orange. The number of genes that were consistently changing only in the genetically (ho1C1 or ho1C2) and chemically (hmox1+BV) complemented (comp) strains are shown on the right in pink and red. The numbers of genes found to be consistently different in hmox1 compared with complemented strains are shown on the left in cyan and green.
(C) HMOX1-dependent gene expression. The bar graph shows the number of consistently differentially expressed genes in hmox1 compared with chemically (hmox1+BV) and genetically (ho1C1 or ho1C2) complemented strains in the dark and 0.5 or 4 h after transition into the light. The mRNAs found to be changed at 0.5 and 4 h in the light in hmox1 specifically are found in the central intersection of the Venn diagram underneath the graph, defining the number of differentially accumulating mRNAs common to both 0.5 and 4 h transcriptomes.
(D) Transcript abundance changes of 50 genes consistently different between hmox1 and both genetically and chemically complemented strains, at both 0.5 and 4 h after transition from dark to light. Box color (blue, increase; red, decrease) indicates log2-transformed fold change of transcript abundances between hmox1 and hmox1+BV, hmox1 and ho1C1, and hmox1 and ho1C2 at 0.5 or 4 h in the light, and transcript abundances between dark acclimated cells and 4 h upon transition into the light (0 → 4 h). Red outlines indicate nonsignificant changes (1% FDR).
In order to identify genes whose expression was affected by the absence of HMOX1, we required the transcript abundances in the hmox1 mutant not only to be significantly different (false discovery rate [FDR] < 1%) from the chemically rescued (hmox1+BV) and at least one of the two genetically complemented (ho1C1 or ho1C2) lines but also to be consistently different (either consistently higher or consistently lower) compared with all the rescued lines. Differences in expression were predominantly found in the light: Only a single gene was found to be consistently different in the dark (Cre09.g398400, encoding a putative membrane protein), whereas 546 and 358 genes were consistently differentially expressed in hmox1 at 0.5 and 4 h in the light, respectively (Figure 5C; Supplemental Data Set 1). In contrast, the vast majority of light-regulated transcripts were expressed similarly in the mutant and complemented strains, i.e., 4753 transcripts at 0.5 h and 2464 at 4 h (Figure 5B; Supplemental Data Set 1). Moreover, among the genes whose expression change was dependent on HMOX1 at 0.5 and 4 h in the light, only 50 overlapped (Figures 5C and 5D; Supplemental Data Set 1), with 45 (90%) accumulating more mRNA in hmox1 than in the complemented lines. This gene set included heat shock proteins (HSP22C, HSP22E, and CLPB3), putative heme binding proteins (SOUL2 and SOUL4), and many poorly annotated genes (Figure 5D; Supplemental Data Set 1). Of the 50 overlapping genes, 34 were induced in all strains and at both time points during the transition from dark to light, whereas no transcript of the 50 genes was consistently reduced.
In a more targeted analysis, we focused on the expression of nuclear genes involved in tetrapyrrole metabolism and light sensing and those involved in photosynthetic electron transfer (Supplemental Data Set 2). As expected, tetrapyrrole and photosynthesis-related genes were mostly induced upon transition into the light, whereas among the genes involved in light sensing, transcripts from the cryptochrome photoreceptor family showed differential expression after 4 h light illumination. For tetrapyrrole pathway genes, most of the transcripts had significantly different abundances at the 4-h time point, whereas 10 out of the 30 inducible transcripts were already elevated after 0.5 h in the light. Six early-induced genes encode enzymes involved in heme and siroheme metabolism, including all of the committed enzymes in siroheme (precorrin-2 synthase UPM1, sirohydrochlorin ferrochelatase SIRB, and UROS1), heme (ferrochelatase HEM2), and linear tetrapyrrole (heme oxygenase HMOX1 and the ferredoxin-dependent bilin reductase PCYA1) biosynthetic pathways. Our analyses also show that HMOX2 expression was repressed by light, a result consistent with the observed lack of a light-dependent phenotype of the hmox2 mutant (Duanmu et al., 2013). In addition to these heme, siroheme, and bilin pathway genes, expression of 4 out of the 12 induced genes in the chlorophyll-specific branch from PPIX to chlorophyllide a also increased early. Among these are genes encoding one of the Mg-chelatase subunits (CHLH1), Mg-PPIX methyltransferase (CHLM), Mg-PPIX monomethyl ester cyclase (CTH1), and divinyl (proto) chlorophyllide vinyl reductase (DVR1). With one exception, i.e., precorrin-2 synthase (UPM1), all of these tetrapyrrole pathway genes were regulated similarly in the hmox1 mutant and both chemically and genetically complemented lines. Overall, these results indicate that HMOX1 is dispensable for normal light-dependent regulation of tetrapyrrole gene expression at the transcript level.
Among the genes associated with light sensing in Chlamydomonas, cryptochromes and phototropin appear to be obvious candidates to play roles in blue-light-dependent regulatory mechanisms (Huang et al., 2002; Zorin et al., 2009; Ahmad, 2016; Petroutsos et al., 2016; Duanmu et al., 2017, Müller et al., 2017; Zou et al., 2017). Among the five genes encoding cryptochrome (CRY) photoreceptors (for a recent review on algal CRYs, see Duanmu et al., 2017; Kottke et al., 2017), expression levels for the gene encoding plant CRY (pCRY) are highest, peak in the dark, and are rapidly repressed upon the onset of light. This is similar to diurnal conditions where transcripts of pCRY were found to be highest during the night and rapidly declined during the day (Zones et al., 2015). Animal cryptochrome (aCRY) and the Cry-DASH (Drosophila melanogaster, Arabidopsis thaliana, Synechocystis sp PCC6803, and human) cryptochrome instead are both induced upon the transition from dark to light. Despite the changes in transcript levels upon onset of light, expression levels for cryptochromes are very similar in hmox1 and both chemically and genetically complemented lines. Phototropin (PHOT1) is slightly induced upon transition to the light, but no significant difference in expression of PHOT1 was seen in hmox1. Other candidate photoreceptors include UVR8 (Tilbrook et al., 2016; Allorent et al., 2016), channel rhodopsins, histidine kinase rhodopsins (HKRs), and GAF (cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA) domain-containing proteins. Although most of these genes show light-dependent changes of transcript levels, especially in the first 30 min after transition from the dark into the light, none of them appears to be affected by the absence of HMOX1.
With respect to genes encoding proteins of the photosynthetic electron transfer chain, only PSB28, PETC, PETN, and LHCB7 (a subset of PhANGs) were induced early, while increased transcript levels for most other PhANGs were not evident until 4 h after transfer to the light (Supplemental Data Set 2). Among the late-induced PhANGs, several PSI and LHCI mRNAs appeared to accumulate to a lesser extent in hmox1 by comparison with the complemented lines, i.e., PSAD, PSAE, PSAG, PSAH, PSAI, PSAK, PSAL, PSAO1, LHCA1, LHCA2, and LHCA3. However, these differences were not found to be significant. Overall, these results suggest that hmox1 is not impaired in light-dependent changes in PSI and LHCI transcript accumulation. Therefore, the observed marked reduction in PSI and LHCI protein abundance in the hmox1 mutant reflects the loss of a bilin-dependent blue light regulatory system that targets processes downstream of transcription and transcript homeostasis.
DISCUSSION
BV Rescues Defective Light-Dependent Greening in hmox1
These studies identify a critical role for BV, the bilin product of Chlamydomonas HMOX1, in assembly and maintenance of a functional photosynthetic apparatus in the light. We show that BV is sufficient to rescue the lack of light-dependent greening seen in the hmox1 mutant, implicating either BV itself or a bilin metabolite derived from BV (e.g., PCB) to be responsible. Because BV rescue of the hmox1 mutant is blue light specific, we favor the hypothesis that plastid-derived bilins are used as chromophore precursors of an as yet unknown blue-light-sensing biliprotein that we name chlorochrome (Figure 1). Cells lacking this bilin-dependent photosensory system acclimate poorly to changes in light fluence rate. Our studies establish that this inability to acclimate is not due to global differences in transcript abundance of tetrapyrrole biosynthesis, known light sensor or photosynthesis-associated genes. In light of previous studies (Duanmu et al., 2013), bilins thus appear to perform dual regulatory roles in Chlamydomonas, i.e., as retrograde signals to anticipate the dark-to-light transition and as potential chromophore precursors of a blue light sensor critical for maintaining a functional photosynthetic apparatus in plastids.
Light-Grown hmox1 Mutants Have Reduced Chlorophyll a/b Ratios
We have shown that transfer of dark-adapted wild-type cultures to the light leads to a rapid and pronounced increase in chlorophyll levels accompanied by an increase in the chlorophyll a/b ratio. Since hmox1 mutants are impaired in this response, light-adapted hmox1 mutants exhibit reduced chlorophyll a/b ratios relative to the wild type. These studies establish that hmox1 mutants are primarily impaired in the light-dependent accumulation of PSI and its associated LHCI antennae (Figure 3C). This defect leads to altered thylakoid membrane architecture, particularly regarding the accumulation of photosynthetic pigment-protein supercomplexes (Figure 4B). Although a number of Chlamydomonas mutants have been identified with lesions that affect accumulation of LHCs (Olive et al., 1981; de Vitry and Wollman, 1988), none of them fully phenocopy the hmox1 mutant. Among these are tla2 and tla3 mutants, which have reduced antennae systems (Kirst et al., 2012a, 2012b). Compared with wild-type cells, both of these tla mutants have elevated chlorophyll a/b ratios due to the loss of chlorophyll b-rich LHCs relative to RCs. By contrast, the reduced chlorophyll a/b ratio we observed in hmox1 was not due to a specific increase in antennae relative to the photosystems. Instead, the decreased chlorophyll a/b ratio in hmox1 is more consistent with a marked decline in PSI complexes relative to LHCII antennae because PSI is highly enriched in chlorophyll a (chlorophyll a/b > 20) (Bassi et al., 1992). Therefore, the reduced levels of PSI RCs and LHC components in the hmox1 mutant therefore must arise from aberrant translation, assembly, and/or stability of the complexes because the levels of mRNAs encoding these components are essentially identical to those in the complemented cells at the analyzed time points.
It is also possible that aberrant chlorophyll a and chlorophyll b association with the LHC/RC proteins or with complexes not directly associated with PSI or PSII function is responsible for the decreased accumulation of PSI and LHCs observed in hmox1. Alternatively, this decrease may be due to the altered synthesis or integration of specific xanthophylls. This latter possibility, though not addressed in this manuscript, has been demonstrated for the Arabidopsis nox mutant that lacks xanthophylls and accumulates almost no PSI (Dall’Osto et al., 2013). The nox mutant lacks LHCII polypeptides and accumulates very little LHCI polypeptides, but unlike the Chlamydomonas hmox1 mutant, it does accumulate an appreciable amount of LHCA2 (Dall’Osto et al., 2013). Regardless of the distribution of chlorophyll a and chlorophyll b in the mutant, the failure of hmox1 cells to green reflects their inability to accumulate new PSI complexes in the light that apparently arises from an aberrant bilin-dependent posttranscriptional process that is light mediated. Our studies also demonstrate that hmox1 failed to upregulate the production of 5′-monohydroxyphylloquinone and phylloquinone upon transfer from darkness to light (Figure 4A). However, this may be a secondary effect of the reduced PSI levels in this mutant because loss of phylloquinone does not cause complete loss of PSI in Chlamydomonas (Emonds-Alt et al., 2017).
HMOX1 Deficiency Affects Accumulation of a Distinct Subset of Photosynthetic Complexes
In Chlamydomonas, PSI is comprised of a single core complex surrounded by nine LHCI antenna proteins (Drop et al., 2011). Several proteins implicated in the assembly of PSI and LHCI have been identified in plants and algae (reviewed in Wittkopp et al., 2016). PSI assembly factors include chloroplast-encoded Ycf3 (Naver et al., 2001) and Ycf4 (Boudreau et al., 1997; Krech et al., 2012) as well as nucleus-encoded Y3IP1 (Albus et al., 2010), PPD1 (Liu et al., 2012), Ycf37/PYG7/CGL71 (Wilde et al., 2001; Stöckel et al., 2006; Heinnickel et al., 2016), and PSA2 (Fristedt et al., 2014). Deficiencies of these factors impact only PSI core subunits, with no effect on the levels of LHCI and LHCII. We observed that at least one of these assembly factors (Ycf4) accumulated to wild-type levels in the hmox1 mutant (Figure 3C). In Arabidopsis, the nucleus-encoded, chloroplast-localized ALBINO3 (Alb3) was shown to have a critical role in thylakoid membrane biogenesis (Sundberg et al., 1997) and was needed for integration of LHC proteins into thylakoid membranes (Moore et al., 2000). The Chlamydomonas genome contains two Alb3 homologs (Alb3.1 and Alb3.2) (Bellafiore et al., 2002), and a loss of Alb3.1 in the ac29 strain resulted in a >10-fold reduction in LHCI and LHCII content and a 2-fold reduction in PSII content (Bellafiore et al., 2002). However, ac29 accumulates normal levels of PSI, the cytochrome b6f complex, and chloroplast ATP synthase (Bellafiore et al., 2002). Depletion of Chlamydomonas Alb3.2 by RNA interference also caused a major decrease in PSI, PSII, and LHCII content (LHCI abundance was not assessed) (Göhre et al., 2006). A recently characterized LHC-like transmembrane protein, Msf1 (Maintenance Factor for PSI), interacts with CTH1, and the loss of Msf1 causes reduced accumulation of several PSI RC and peripheral subunits in Chlamydomonas (Zhao et al., 2017). However, levels of most LHCA proteins were unchanged in the msf1 mutant (Zhao et al., 2017). Thus, HMOX1 appears to affect a different pathway for the assembly and maintenance of photosynthetic complexes than those already identified.
BV-Dependent Rescue of the hmox1 Photosynthetic Apparatus Reveals Both Blue-Light-Dependent and Light-Independent Pathways
Our studies establish that both genetic and BV complementation restore PSI activities and abundance of associated LHC proteins that are reduced in hmox1. We show that this BV-dependent rescue of the hmox1 photosynthetic apparatus is light dependent (Figure 3C), being mediated by a blue light photosensory system that is independent of linear PET (Figure 4C). These results thus highlight a photosensory role for the tetrapyrrole product(s) of HMOX1 catalysis, BV and/or PCB, rather than the HMOX1 protein itself. HMOX1 enzymatic activity is thus dispensable for chemical rescue of the photosynthetic apparatus of hmox1 in the light. In the absence of light, BV supplementation of hmox1 cultures does partially restore wild-type PSI and PSII activities (Figures 3A to 3C). Taken together, these results highlight a dual role for bilins in the proper maintenance and functioning of the photosynthetic apparatus, and especially PSI, both in the light and dark.
Phenotypic Similarities of Chlorophyll Biosynthesis Mutants with the hmox1 Mutant
The hmox1 mutant shares phenotypic similarities with mutants disrupted in committed nonessential genes of the chlorophyll biosynthetic pathway. The phenotype of the Chlamydomonas gun4 mutant, which lacks the GUN4 regulatory subunit of the Mg-chelatase complex, is notably similar to hmox1. Like hmox1, gun4 mutants are chlorophyll-deficient and accumulate reduced amounts of PSI, LHCI, and LHCII polypeptides despite normal accumulation of transcripts encoding these proteins (Formighieri et al., 2012; Brzezowski et al., 2014). This suggests that chlorophyll insufficiency can produce phenotypes similar to hmox1. The phenotype of a mutant in CRD1, one of the two Mg-PPIX monomethyl ester cyclase genes (the other being CTH1), is also similar to hmox1. However, whereas the crd1 mutant lacks LHCI and displays reduced PSI activity like hmox1, these phenotypes require copper-deficient medium (Moseley et al., 2000). Under these conditions, expression of the paralogous CTH1 is inhibited and therefore cannot compensate for loss of CRD1 (Moseley et al., 2002). Because our immunoblot measurements also reveal that CTH1 levels are unaffected by the absence of HMOX1 (Figure 3C), reduced cyclase protein level does not account for the chlorophyll deficiency of the hmox1 mutant. This suggests that BV-dependent rescue of PSI activity in hmox1 is mechanistically independent of CRD1/CTH1 levels. However, we cannot rule out the possibility that BV or a BV-derived metabolite modulates the activity of CTH1 (and/or CRD1).
In contrast with the gun4 and crd1 mutants, phenotypes of other mutants in the chlorophyll biosynthetic pathway are more severe. These include mutants of both the light- and the dark-dependent protochlorophyllide reductases (POR1 and DPOR) of the Mg-PPIX methyltransferase (CHLM) and of the three subunits of Mg chelatase (CHLH1, CHLI1/2, and CHLD). These mutants are photosensitive, severely chlorotic, and/or yellow in the dark (Li and Timko, 1996; Chekounova et al., 2001; Falciatore et al., 2005; von Gromoff et al., 2008; Meinecke et al., 2010; Grovenstein et al., 2013). It was recently demonstrated that the specific loss of chlorophyll b in Chlamydomonas does not disrupt accumulation of LHCI and LHCII polypeptides as long as chlorophyll a biosynthesis remains intact (Bujaldon et al., 2017). Despite the phenotypic discrepancies among these mutants, our transcriptomic data suggest that aberrant accumulation of PSI, LHCI, and LHCII polypeptides in the hmox1 mutant is not due to inhibition of chlorophyll biosynthesis-related gene expression.
Heme Feedback Regulation Is Not Responsible for the Chlorophyll-Deficient Phenotype of the hmox1 Mutant
Heme is a feedback regulator of ALA synthesis in most eukaryotes, including plants (Figure 1; Terry and Smith, 2013). Hence, the hmox1 mutant should be chlorophyll deficient on regulatory grounds alone. However, rescue of the hmox1 photosynthesis deficiency by exogenous BV and blue light strongly argues that heme feedback is not responsible. Alternatively, bilins themselves could regulate tetrapyrrole biosynthesis at the protein level or they could mitigate damage by ROS generated from intermediates in the tetrapyrrole biosynthetic pathway. However, this would require bilins to be located at/or near the sites of chlorophyll biosynthesis or ROS generation in membranes, and yet there is no evidence that this is the case. In addition, x-ray crystallographic studies have not identified linear tetrapyrroles as integral components of either photosystems or light-harvesting complexes in the Viridiplantae (Qin et al., 2015), which includes plants and green algae. We also show that the expression patterns of light-regulated genes in hmox1 are mostly indistinguishable from those seen in both genetically and chemically rescued lines (Figure 5B). This indicates that the bilin-dependent transcriptional response to light is independent of the presence of HMOX1 protein. Besides HMOX1, the only other known bilin binding protein in Chlamydomonas is PCYA1, the plastid-localized enzyme that converts BV to PCB. Whereas it is unlikely that PCYA1 itself is a blue light sensor, PCYA1 is partially membrane associated (Duanmu et al., 2013) and could therefore be a candidate to function as a bilin binding protein that could regulate the activity and/or stability of chlorophyll biosynthetic enzymes and/or factors involved in PSI assembly/stability. Unfortunately, our attempts to isolate a pcya1 mutant have so far been unsuccessful. Because the Chlamydomonas photoacclimation response is both bilin and blue light dependent, the loss of a bilin-based blue light sensor that uses PCB as a prosthetic group is the simplest explanation for the phenotypes of the hmox1 mutant.
The HMOX1-Dependent Transcriptome May Represent a Heme-Derived Retrograde Pathway That Operates during Photoacclimation
Our transcriptome measurements do not show dramatic differences between the hmox1 mutant and genetically and chemically complemented lines. Only a small group of 50 genes is significantly affected by the absence of HMOX1 and the majority of these genes are more significantly upregulated in the hmox1 mutant compared with the control lines at one or both time points. Many of these genes encode proteins of unknown function, but those that can be recognized include members of the SOUL heme binding protein family and stress-activated genes, i.e., HSP22CandE, LCI7, and GPX3 (Figure 5D). The enhanced expression of these genes is consistent with an increased need for heme-derived ROS detoxification and possibly heme sequestration in the light when damage to the photosynthetic apparatus occurs. We speculate that this network is activated by sustained export of heme from the plastid to the cytosol in the hmox1 mutant, although other ROS-derived signaling molecules could serve this function. The ability of exogenous BV to mitigate this response is consistent with a decreased efflux of heme during the BV-enhanced, light-dependent biogenesis of the photosynthetic apparatus.
A Model for Bilin-Dependent Regulation of Greening in Chlamydomonas
Chlorophyte algae diverged from the streptophyte lineage, which includes land plants and streptophyte algae, >700 million years ago (Leliaert et al., 2012). All lineages that diverged prior to this time possessed bilin-based photosynthetic antennae, so retention of bilin biosynthesis during the ascendance of the Viridiplantae is biologically relevant. One important reason for retention of bilin biosynthesis in the Viridiplantae is the need for the bilin chromophore precursors of phytochromes, which are critical regulators of PhANG expression in the streptophyte lineage (Rockwell et al., 2006). Despite repeated loss of phytochromes within various photosynthetic eukaryote lineages, including chlorophyte algae such as Chlamydomonas, all phototrophs derived from primary and secondary endosymbiosis retain plastid-localized FDBRs (Rockwell et al., 2014; Duanmu et al., 2017). FDBRs are critical for driving HMOX turnover, which is highly product inhibited. Our studies suggest that FDBR retention may also reflect an essential role for their bilin products, i.e., PCB, to coordinate synthesis of functional PSI and LHC complexes with changing light environments.
The observed blue light responses mediated by chlorochrome are distinct from those of the flavin-containing phototropins and cryptochromes, which regulate expression of many PhANGs at the transcript level (Im et al., 2006; Beel et al., 2012). The lack of HMOX1 dependence on light-regulated accumulation of LHSCR3 (Figure 3C) is also inconsistent with a role for PHOT1 in the bilin-dependent blue light response (Petroutsos et al., 2016). Blue-light-absorbing phytochromes and cyanobacteriochromes that use PCB as chromophores are well known (Rockwell et al., 2011, 2014). Phycobiliproteins that absorb blue light are also known, although these proteins require different bilin precursors (Ting et al., 2002). A bilin-based blue light sensor might be more important for chlorophyte algae, in which the composition and activities of the light-harvesting antennae systems differ from those of plants (Rochaix, 2014; Iwai et al., 2015; Wobbe et al., 2016). It is therefore reasonable that this chlorochrome photoreceptor would have evolved among the phytochrome-deficient chlorophyte lineage. Chlorochrome is probably targeted to the plastid, where it may influence the activity of chlorophyll biosynthetic enzymes as depicted in Figure 1. However, we cannot rule out existence of a cytosolic chlorochrome that functions like a plant phytochrome to regulate expression of nuclear-encoded, chloroplast-targeted factors that stimulate/derepress the activity of these key enzymes.
Taken together with previous studies, we conclude that bilins play a dual regulatory role in the biosynthesis and maintenance of the photosynthetic apparatus in Chlamydomonas. On one hand, bilins are retrograde signaling molecules that can induce expression of genes involved in oxygen consumption or detoxification during transitions from hypoxia in darkness to normoxia at daybreak (Duanmu et al., 2013). This pathway does not require light, yet it is an unavoidable consequence of light exposure that drives photosynthetic oxygen evolution, HMOX turnover, and a burst in bilin production/release. Consequently, plastid-derived bilins function as inhibitory signals to suppress the expression of photoprotective genes such as LHCSR1, PSBS1, and ELI3 (Duanmu et al., 2013), without major impacts on transcript abundances of PhANGs (Supplemental Data Set 2). A light-independent (dark-operative), bilin-dependent pathway may also be needed for maintenance of PSI and LHCs, but not for cofactor biosynthesis in prolonged darkness. Observations that BV failed to rescue the hmox1 mutant in the dark could reflect the inability of exogenous BV to be properly modified or assembled with the chlorochrome apoprotein in the absence of light. Alternatively, HMOX1 itself may be required for regulation in the dark. In this regard, our previous transcriptome analysis showed that BV feeding does not cause a robust response in the hmox1 mutant compared with the wild-type strain, i.e., 6 and 76 genes were found to be responsive to BV in hmox1 and the wild type, respectively (Duanmu et al., 2013).
The second signaling pathway is mediated by a bilin-dependent photoreceptor whose regulatory activity is critical for light-dependent PSI biogenesis. Although C. reinhardtii lacks phytochromes, genomic data reveal a proliferation of a family of Ser/Thr kinases in this alga, many of which possess GAF domains distantly related to bilin binding domains of phytochromes (Merchant et al., 2007). At least three members of this family (Cre12.g552150, Cre14.g628200, and Cre17.g739250) possess putative chloroplast targeting sequences, making them attractive candidates for a chlorochrome sensor that regulates key steps in biosynthetic pathways for chlorophyll and possibly other cofactors (e.g., naphthoquinone) (Figure 1). Such a photoreceptor could preserve energy resources of the algal cell by both minimizing accumulation of photosynthetic complexes during periods of prolonged darkness and stimulating their reaccumulation when light becomes available. In this regard, this pathway is analogous to the etiolation/deetiolation cycle of plants that leverages phytochromes to coordinate expression of PhANGs with the light-dependent stimulation of chlorophyll synthesis, except that this pathway would directly impact enzymatic activities rather than transcript abundance. Therefore, chlorophyte algae may have lost phytochromes, while retaining other photoreceptors for global regulation of PhANG expression. Bilin biosynthesis may have been retained to report the state of tetrapyrrole metabolism in the plastid and coordinate photoreception, photosynthesis, and photoprotection, the combined action of which is vital for acclimation of microalgae to changing light conditions (Allorent and Petroutsos, 2017).
METHODS
Chlamydomonas Strains, Growth Conditions, and Light Treatments
The Chlamydomonas reinhardtii hmox1 mutant (CC-4601, mt+) was isolated in a classical genetic screen following insertional mutagenesis of the wild-type parent 4A+ (CC-4051, mt+, 137C) (Dent et al., 2005). The ho1C1 and ho1C2 strains were generated by complementation of the hmox1 mutant with HMOX1 cDNA (Duanmu et al., 2013). Strains were maintained on Tris acetate phosphate (TAP) solid medium, pH 7.0 (Harris, 1989), at 20°C under very low white light (<10 µmol photons m−2 s−1) conditions. For experiments on solid medium, similar amounts of cells were spotted onto TAP or TP (TAP with no acetate) agar with special K elements (Kropat et al., 2011) and grown either for ∼1 week in the dark under continuous broad spectrum fluorescent light (GE Daylight 6500K F40T12/DX/ECO lamps, ∼120 µmol photons m−2 s−1) or on a diurnal cycle (12 h light/12 h dark). For experiments in liquid medium, cells were grown in continuous cool white fluorescent light at 30 to ∼40 µmol photons m−2 s−1 unless otherwise indicated. To maintain darkness, flasks were wrapped in aluminum foil. Specific light treatments were performed using Lumibar LED strip lights (Lumigrow) containing blue (452 nm) and red (655 nm) LEDs at PAR ∼10 µmol photons m−2 s−1. The quanta of blue wavelengths (400–480 nm) in white light was determined spectroradiometrically to be ∼10% of the total. Determination of cell densities was performed using a Countess II FL automated cell counter (Thermo Fisher Scientific). BV hydrochloride (VWR) was dissolved in methanol and used at a final concentration of 0.1 mM.
Chlorophyll and Prenylated Naphthoquinone Analyses
For chlorophyll analysis, liquid cultures in mid-log growth phase were collected and pigments were extracted with N,N-dimethylformamide (Sigma-Aldrich). Chlorophyll was quantified according to the following equations: chlorophyll a (µg/mL) = 12.70 A664.5 – 2.79 A647; chlorophyll b (µg/mL) = 20.70 A647 – 4.62 A664.5 (Inskeep and Bloom, 1985). For naphthoquinone quantification, strains were grown to mid-log phase in TAP under continuous cool white fluorescent light (GE 80994 6500 K F40DX; 30 to ∼40 µmol photons m−2 s−1). Cells were collected by centrifugation (1000g, 4 min, 20°C) and resuspended to 1 × 106 cells mL−1 in TAP (4A+, hmox1, and ho1C2; triplicate cultures starting from three separate colonies) or TAP + 0.1 mM BV (hmox1; n = 3 biological replicates). Cultures were grown either in darkness for 24 h or for 12 h in the dark and then transferred to the light (∼100 µmol photons m−2 s−1) for 12 h. After 24 h, cell densities and chlorophyll concentration were determined as described above. From each biological replicate, 2 mL of cells were collected by centrifugation (16,100g, 2 min, 20°C), and the cell pellet was immediately flash frozen in liquid N2 and stored at −80°C. Thawed cell pellets were resuspended in 300 μL of 95% (v/v) ethanol, spiked with 385 pmol of menaquinone-4 as an internal standard, and homogenized in 5 mL Pyrex tissue grinders. The grinders were then washed twice with 300 μL of 95% (v/v) ethanol, and the washes were combined with the initial extract in a Pyrex screw-cap tube containing 600 μL of water. The samples were partitioned twice with 5 mL hexane and by vigorous shaking. The hexane phases were combined and evaporated to dryness with gaseous N2. The residues were resuspended in 300 μL of methanol/dichloromethane (10:1), and the resulting extracts were cleared by centrifugation (18,000g, 5 min, 20°C). HPLC separation with fluorescence detection was performed as previously described (Widhalm et al., 2012). Retention times were 5.5 min for 5′-monohydroxyphylloquinone, 8.3 min for menaquinone-4, and 13 min for phylloquinone. Phylloquinone and 5′-monohydroxyphylloquinone levels were quantified based on the amount of external standard of phylloquinone added and then corrected based on the recovery of the internal standard of menaquinone-4.
Photosynthetic O2 Evolution
Strains were grown in TAP liquid medium (with special K trace elements) with constant agitation (200 rpm) at light intensities of either ∼90 or ∼300 µmol photons m−2 s−1 provided by a Sylvania M1000/U/BT37 metal halide bulb at 3800K. Cultures were diluted once before making the measurements in order to achieve a final concentration of ∼2 × 106 cells/mL. Chlorophyll was measured spectrophotometrically as described above, and total organic carbon was analyzed on a TOC-L/TNM-L analyzer (Shimadzu). Photosynthetic O2 evolution rates were measured using the OxyLab oxygen electrode control unit. Maximum photosynthesis rates were calculated by fitting the curves with the Box-Lucas method in OriginPro 9.1 (OriginLab). All strains were analyzed as triplicate cultures starting from three separate colonies.
Chlorophyll Fluorescence
Cells were collected by centrifugation (3200g, 10 min, 20°C), resuspended in fresh TAP medium and then dark-adapted with vigorous shaking for at least 20 min prior to measurements. Sodium bicarbonate (1 mM) was added as a terminal electron acceptor. Chlorophyll fluorescence was measured using a DUAL-PAM 100 (Walz) on the light curve setting, with 60-s steps at each light fluence rate (Heinnickel et al., 2013). Saturating pulses were given immediately following each intensity step to determine Fv/Fm and ΦPSII values. Fv/Fm = (Fm − F0)/Fm and ΦPSII = (Fm’ − Fs)/Fm’, where Fv is variable fluorescence Fm is dark-adapted maximum fluorescence, F0 is dark-adapted minimum fluorescence, Fm’ is light-adapted maximum fluorescence, and Fs is steady state fluorescence (Maxwell and Johnson, 2000). All strains were analyzed as triplicate cultures starting from three separate colonies.
PSI Activity Measurements
Cells were collected by centrifugation (3200g, 10 min, 20°C) and resuspended in a solution containing 20 mM HEPES-KOH, pH 7.5, and 10% Ficoll (w/v). For all samples, the final chlorophyll concentration was adjusted to 30 µg/mL. The oxidation and reduction kinetics of P700 were measured as absorption changes at 705 nm using a JTS10 spectrophotometer (BioLogic). All strains were analyzed as triplicate cultures starting from three separate colonies.
Protein Extraction and Immunoblot Analysis
Total cellular protein was extracted by pelleting cells (16,100g, 5 min, 20°C), resuspending in a protein extraction buffer (5 mM HEPES-KOH, pH 7.5, 100 mM DTT, 100 mM Na2CO3, 2% SDS, and 12% sucrose), and boiling the suspension for 50 s. Extracted proteins were flash frozen in liquid N2 and stored at −80°C prior to resolving them by SDS-PAGE (10% monomer). Resolved proteins were transferred to PVDF membranes (Bio-Rad) using a semidry transfer apparatus (Bio-Rad) according to the manufacturer’s recommendations (15 V, 30 min). For immunoblot analysis, membranes were blocked in TBS + 0.1% Tween (TBST) containing 5% nonfat dry milk (Molecular Products). Incubations with the primary antibodies were performed in TBST containing 3% milk. Primary antibodies from Agrisera include ATPB (AS05 085), CP29 (AS04 045), CP47 (AS04 038), D1 (AS10 704), LHCA1 (AS01 005), LHCA2 (AS01 006), LHCBM1 (AS01 004), LHCBM2 (AS01 003), LHCSR1/3 (AS14 2766), OEE3 (AS06 142-16), PsaA (AS06 172-100), PSAD (AS09 461), PSAH (AS06 143), Rubisco LS (AS03 037), and Ycf4 (AS07 274). Antitubulin antibody was purchased from Sigma-Aldrich (T6074). The anti-Cyt f antibody was a generous gift from Francis-André Wollman, and Jean-David Rochaix provided anti-LHCA3/4/5 antibodies. Immunoreactive proteins were detected using a standard enhanced chemiluminescence procedure (GE Healthcare).
Thylakoid Membrane Purification, Solubilization, and Separation of Complexes by BN-PAGE
Thylakoid membranes were purified according to Takahashi et al. (2006). Purified thylakoids were pelleted and resuspended in BN solubilization buffer (50 mM Bis-Tris-HCl, pH 7.0, 750 mM ε-amino-α-caproic acid, and 20% [v/v] glycerol) plus β-DM at a final concentration of 1% (w/v). Solubilization was performed on ice and in the dark for 40 min. Following this incubation, insoluble membranes were pelleted by centrifugation (1000g, 10 min, 4°C) and the supernatant was transferred to a new tube. Coomassie Brilliant Blue G 250 stain (Serva) was added so that the ratio of detergent/stain was 4:1 (w/w). Chlorophyll (2.5 µg) from the solubilized preparation were loaded onto NativePAGE Novex 4-16% Bis-Tris gels (Life Technologies) and electrophoresed according to the manufacturer’s instructions. Electrophoresis was initiated with dark blue cathode buffer (0.02% Coomassie G 250), and after the proteins had migrated ∼60% down the gel, light-blue cathode buffer (0.002% Coomassie G 250) was used. Following electrophoresis, the gel was destained with 50:40:10 (v/v/v) water:methanol:acetic acid.
RNA-Seq Analysis
Chlamydomonas cells were harvested and total RNA was extracted using a Trizol and Qiagen RNeasy mini kit. Library preparation and sequencing were described previously (Duanmu et al., 2013), with one additional time point incorporated into the analysis presented here (4 h after dark-to-light transition), and all of the data were reanalyzed for HMOX1-dependent gene expression using the most recent version (v5.5) of the Chlamydomonas transcriptome. Raw and processed sequence files are available at the NCBI Gene Expression Omnibus (accession number GSE40031). Short reads were aligned using STAR (Dobin et al., 2013) with a maximum tolerance of four mismatches every 100 bases. V5.5 transcript sequences, based on the 5th assembly of the Chlamydomonas genome (www.phytozome.net/chlamy), were used as a reference for assembly using Cufflinks v2.21 (Trapnell et al., 2013).
For gene expression estimates, the counts-per-gene matrix was normalized by the total number of aligned reads and the transcript mappable length (Trapnell et al., 2010). Independent filtering was applied to the normalized expression matrix to filter out poorly expressed genes (below 1 FPKM accumulated expression across the entire data set). The counts matrix for the remaining 15,535 genes was analyzed for differential expression at 1% FDR using pairwise comparisons with the DESeq2 package in R (Love et al., 2014). The obtained set of differentially expressed genes (DEG) was additionally filtered by comparison to DEG sets obtained with either CuffDiff (Trapnell et al., 2013) or DESeq (Anders and Huber, 2010), using either the same count matrix as described above or a second count matrix containing multireads (obtained with Cufflinks v2.02), requiring a significant change (1% FDR) to be observed in every DEG set in order to retain only the most robust significantly changing genes that were consistently identified with multiple methods. To identify genes affected by HMOX1, the overlap of the filtered set of DEG between the hmox1 mutant and the chemically complemented mutant (hmox1+BV) and at least one of the two genetically complemented strains (ho1C1 and ho1C2) was analyzed in the dark, at 0.5 and 4 h in the light, respectively. The overlap was additionally filtered for genes responding uniformly to either complementation. For dark-acclimated strains, conserved and significantly differentially expressed genes (Supplemental Data Set 1) were identified by comparing 4A+ to (hmox1, ho1C1, ho1C2) or comparing hmox1 to (hmox1+BV, ho1C1, ho1C2), respectively. Light-regulated genes at 0.5 or 4 h were derived by comparing dark versus 0.5 or 4 h in hmox1, hmox1+BV, ho1C1, and ho1C2. Conserved, HMOX1-dependent genes were identified by comparing hmox1 at 0.5 or 4 h to chemically and genetically complemented lines (hmox1+BV, ho1C1, and ho1C2).
Statistical Analysis
ANOVA and Fisher’s least significant difference test was applied for multiple pairwise comparisons (GraphPad Prism 7). Means were compared using Student’s t test. Asterisks in the figures denote significant differences as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Accession Numbers
Sequence data from this article can be found in the Chlamydomonas reinhardtii v5.5 database under the following accession numbers: CF1 β (cp_atpB), CP29 (Cre17.g720250), CP47 (cp_psbB), CTH1 (Cre12.g510050), Cyt f (cp_petA), D1 (chloroplastic, cp_psbA), F1 β (Cre17.g698000), HMOX1 (Cre10.g423500), LHCA1 (Cre06.g283050), LHCA2 (Cre12.g508750), LHCA3 (Cre11.g467573), LHCA4 (Cre10.g452050), LHCA5 (Cre10.g425900), LHCBM1 (Cre01.g066917), LHCBM2 (Cre12.g548400), LHCSR1 (Cre08.g365900), LHCSR3 (Cre08.g367400, Cre08.g367500), OEE3 (Cre08.g372450), PCYA1 (Cre13.g587100), PsaA (cp_psaA), PSAD (Cre05.g238332), PSAH (Cre07.g330250), Rubisco LS (cp_rbcL), Tubulin (Cre03.g190950, Cre04.g216850), and Ycf4 (cp_ycf4). RNA-seq data analyzed in this study are available in the Gene Expression Omnibus database under accession number GSE40031. Chlamydomonas strains used are wild-type parental strain 4A+ (CC-4051, mt+, 137C), hmox1 mutant (CC-4601, mt+), and ho1C1 and ho1C2 (Duanmu et al., 2013).
Supplemental Data
Supplemental File 1. ANOVA tables.
Supplemental Data Set 1A. Summary of expression estimates (FPKM) of all 17,741 genes during the transition from dark to light in 4A+, 4A+ +BV, hmox1, hmox1+BV, ho1C1, and ho1C2.
Supplemental Data Set 1B. Conserved and significantly differentially expressed genes in 4A+ in the dark compared with other genotypes (hmox1, ho1C1, and ho1C2).
Supplemental Data Set 1C. Conserved and significantly light-regulated genes (0.5 h in the light versus dark) in hmox1, hmox1 +BV, ho1C1, and ho1C2.
Supplemental Data Set 1D. Conserved and significantly light-regulated genes (4 h in the light versus dark) in hmox1, hmox1 +BV, ho1C1, and ho1C2.
Supplemental Data Set 1E. Conserved and significantly differentially expressed genes in hmox1 in the dark compared with chemically and genetically complemented lines (hmox1 +BV, ho1C1, and ho1C2).
Supplemental Data Set 1F. Conserved and significantly differentially expressed genes in hmox1 at 0.5 h in the light compared with chemically and genetically complemented lines (hmox1 +BV, ho1C1, and ho1C2).
Supplemental Data Set 1G. Conserved and significantly differentially expressed genes in hmox1 at 4 h in the light compared with chemically and genetically complemented lines (hmox1 +BV, ho1C1, and ho1C2).
Supplemental Data Set 1H. Conserved and significantly differentially expressed genes in hmox1 at both 0.5 and 4 h in the light compared with chemically and genetically complemented lines (hmox1 +BV, ho1C1, and ho1C2).
Supplemental Data Set 2A. Transcript abundance changes of photosynthesis-related genes during the dark-to-light transition in Chlamydomonas strains hmox1, hmox1+BV, ho1C1, and ho1C2.
Supplemental Data Set 2B. Transcript abundance changes of tetrapyrrole biosynthesis genes during the dark-to-light transition in Chlamydomonas strains hmox1, hmox1+BV, ho1C1, and ho1C2.
Supplemental Data Set 2C. Transcript abundance changes of photoreceptor genes during the dark-to-light transition in Chlamydomonas strains hmox1, hmox1+BV, ho1C1, and ho1C2.
Acknowledgments
We thank Francis-André Wollman for the anti-Cyt f antibody and Jean-David Rochaix for the anti-LHCA3/4/5 antibodies. We thank Ian K. Blaby, David Casero, and Matteo Pellegrini for assistance with the initial RNA-seq data analysis. This work was supported by the National Natural Science Foundation of China (project no. 31570233) awarded to D.D., by NIH National Institute of General Medical Sciences Grant 2RO1 GM068552 awarded to J.C.L., by NSF Division of Molecular and Cellular Biosciences Grant MCB-0951094 awarded to A.R.G., by NSF CAREER Grant MCB-1148968 awarded to G.J.B., and by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S Department of Energy (Grant DE-FD02-04ER15529 awarded to S.S.M.). Research in D.D.’s lab was also supported by the Junior Thousand Talents Program of China for young researchers and Huazhong Agricultural University Scientific & Technological Self-Innovation Foundation (Program No. 2014RC018). T.M.W. was supported in part by the NIH National Institute of General Medical Sciences under Award T32GM007276.
AUTHOR CONTRIBUTIONS
T.M.W., St.S., A.R.G., D.D., and J.C.L. conceived the study and designed the experiments. T.M.W., St.S., Sh.S., W.H., Q.F., W.Z., S.D.G., E.S., and D.D. performed the experiments. T.M.W., St.S., M.T.L., G.J.B., A.R.G., D.D., and J.C.L. analyzed the data. T.M.W., St.S., D.D., and J.C.L. wrote the article. All authors discussed the results and commented on the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ahmad M. (2016). Photocycle and signaling mechanisms of plant cryptochromes. Curr. Opin. Plant Biol. 33: 108–115. [DOI] [PubMed] [Google Scholar]
- Albus C.A., Ruf S., Schöttler M.A., Lein W., Kehr J., Bock R. (2010). Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22: 2838–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allorent G., Lefebvre-Legendre L., Chappuis R., Kuntz M., Truong T.B., Niyogi K.K., Ulm R., Goldschmidt-Clermont M. (2016). UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 113: 14864–14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allorent G., Petroutsos D. (2017). Photoreceptor-dependent regulation of photoprotection. Curr. Opin. Plant Biol. 37: 102–108. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R., Soen S.Y., Frank G., Zuber H., Rochaix J.D. (1992). Characterization of chlorophyll a/b proteins of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 267: 25714–25721. [PubMed] [Google Scholar]
- Beel B., Prager K., Spexard M., Sasso S., Weiss D., Müller N., Heinnickel M., Dewez D., Ikoma D., Grossman A.R., Kottke T., Mittag M. (2012). A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24: 2992–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellafiore S., Ferris P., Naver H., Göhre V., Rochaix J.D. (2002). Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14: 2303–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J.D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16: 6095–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezowski P., Schlicke H., Richter A., Dent R.M., Niyogi K.K., Grimm B. (2014). The GUN4 protein plays a regulatory role in tetrapyrrole biosynthesis and chloroplast-to-nucleus signalling in Chlamydomonas reinhardtii. Plant J. 79: 285–298. [DOI] [PubMed] [Google Scholar]
- Brzezowski P., Richter A.S., Grimm B. (2015). Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 1847: 968–985. [DOI] [PubMed] [Google Scholar]
- Bujaldon S., Kodama N., Rappaport F., Subramanyam R., de Vitry C., Takahashi Y., Wollman F.A. (2017). The functional accumulation of antenna proteins chlorophyll b-less mutants of Chlamydomonas reinhardtii. Mol. Plant 10: 115–130. [DOI] [PubMed] [Google Scholar]
- Busch A.W.U., Montgomery B.L. (2015). Interdependence of tetrapyrrole metabolism, the generation of oxidative stress and the mitigative oxidative stress response. Redox Biol. 4: 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekounova E., Voronetskaya V., Papenbrock J., Grimm B., Beck C.F. (2001). Characterization of Chlamydomonas mutants defective in the H subunit of Mg-chelatase. Mol. Genet. Genomics 266: 363–373. [DOI] [PubMed] [Google Scholar]
- Chi W., Sun X., Zhang L. (2013). Intracellular signaling from plastid to nucleus. Annu. Rev. Plant Biol. 64: 559–582. [DOI] [PubMed] [Google Scholar]
- Dall’Osto L., Piques M., Ronzani M., Molesini B., Alboresi A., Cazzaniga S., Bassi R. (2013). The Arabidopsis nox mutant lacking carotene hydroxylase activity reveals a critical role for xanthophylls in photosystem I biogenesis. Plant Cell 25: 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R.M., Haglund C.M., Chin B.L., Kobayashi M.C., Niyogi K.K. (2005). Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry C., Wollman F.A. (1988). Changes in phosphorylation of thylakoid membrane proteins in light-harvesting complex mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 933: 444–449. [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drop B., Webber-Birungi M., Fusetti F., Kouřil R., Redding K.E., Boekema E.J., Croce R. (2011). Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J. Biol. Chem. 286: 44878–44887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D., Casero D., Dent R.M., Gallaher S., Yang W., Rockwell N.C., Martin S.S., Pellegrini M., Niyogi K.K., Merchant S.S., Grossman A.R., Lagarias J.C. (2013). Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc. Natl. Acad. Sci. USA 110: 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D., Rockwell N.C., Lagarias J.C. (2017). Algal light sensing and photoacclimation in aquatic environments. Plant Cell Environ. 40: 2558–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Finazzi G., Wollman F.A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42: 463–515. [DOI] [PubMed] [Google Scholar]
- Emonds-Alt B., Coosemans N., Gerards T., Remacle C., Cardol P. (2017). Isolation and characterization of mutants corresponding to the MENA, MENB, MENC and MENE enzymatic steps of 5′-monohydroxyphylloquinone biosynthesis in Chlamydomonas reinhardtii. Plant J. 89: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A., Merendino L., Barneche F., Ceol M., Meskauskiene R., Apel K., Rochaix J.D. (2005). The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes Dev. 19: 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formighieri C., Ceol M., Bonente G., Rochaix J.D., Bassi R. (2012). Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol. Plant 5: 1242–1262. [DOI] [PubMed] [Google Scholar]
- Frankenberg N., Lagarias J.C. (2003). Biosynthesis and biological functions of bilins. In Porphyrin Handbook, Vol. 13, Kadish K.M., Smith K.M., Guilard R., eds (San Diego, CA: Academic Press; ), pp. 211–235. [Google Scholar]
- Frankenberg-Dinkel N., Terry M.J. (2009). Synthesis and role of bilins in photosynthetic organisms. In Tetrapyrroles: Birth, Life, and Death, Warren M.J., Smith A.G., eds (New York: Springer; ), pp. 208–220. [Google Scholar]
- Fristedt R., Williams-Carrier R., Merchant S.S., Barkan A. (2014). A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants. J. Biol. Chem. 289: 30657–30667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Ossenbühl F., Crèvecoeur M., Eichacker L.A., Rochaix J.D. (2006). One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18: 1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Lohr M., Im C.S. (2004). Chlamydomonas reinhardtii in the landscape of pigments. Annu. Rev. Genet. 38: 119–173. [DOI] [PubMed] [Google Scholar]
- Grovenstein P.B., Wilson D.A., Lennox C.G., Smith K.P., Contractor A.A., Mincey J.L., Lankford K.D., Smith J.M., Haye T.C., Mitra M. (2013). Identification and molecular characterization of a novel Chlamydomonas reinhardtii mutant defective in chlorophyll biosynthesis. F1000 Res. 2: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook. (San Diego, CA: Academic Press; ). [Google Scholar]
- Heinnickel M.L., Alric J., Wittkopp T., Yang W., Catalanotti C., Dent R., Niyogi K.K., Wollman F.A., Grossman A.R. (2013). Novel thylakoid membrane GreenCut protein CPLD38 impacts accumulation of the cytochrome b6f complex and associated regulatory processes. J. Biol. Chem. 288: 7024–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinnickel M., Kim R.G., Wittkopp T.M., Yang W., Walters K.A., Herbert S.K., Grossman A.R. (2016). Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proc. Natl. Acad. Sci. USA 113: 2774–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A., Casero D., Liu B., Benning C., Pellegrini M., Happe T., Merchant S.S. (2013). Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell 25: 3186–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Merkle T., Beck C.F. (2002). Isolation and characterization of a Chlamydomonas gene that encodes a putative blue-light photoreceptor of the phototropin family. Physiol. Plant. 115: 613–622. [DOI] [PubMed] [Google Scholar]
- Im C.S., Eberhard S., Huang K., Beck C.F., Grossman A.R. (2006). Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 48: 1–16. [DOI] [PubMed] [Google Scholar]
- Inskeep W.P., Bloom P.R. (1985). Extinction coefficients of chlorophyll a and B in n,n-dimethylformamide and 80% acetone. Plant Physiol. 77: 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M., Yokono M., Kono M., Noguchi K., Akimoto S., Nakano A. (2015). Light-harvesting complex Lhcb9 confers a green alga-type photosystem I supercomplex to the moss Physcomitrella patens. Nat. Plants 1: 14008. [DOI] [PubMed] [Google Scholar]
- Kirst H., García-Cerdán J.G., Zurbriggen A., Melis A. (2012a). Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol. 158: 930–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H., Garcia-Cerdan J.G., Zurbriggen A., Ruehle T., Melis A. (2012b). Truncated photosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiol. 160: 2251–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Masuda T. (2016). Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 7: 1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke T., Oldemeyer S., Wenzel S., Zou Y., Mittag M. (2017). Cryptochrome photoreceptors in green algae: Unexpected versatility of mechanisms and functions. J. Plant Physiol. 217: 4–14. [DOI] [PubMed] [Google Scholar]
- Krech K., Ruf S., Masduki F.F., Thiele W., Bednarczyk D., Albus C.A., Tiller N., Hasse C., Schöttler M.A., Bock R. (2012). The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D. (2011). A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66: 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F., Smith D.R., Moreau H., Herron M.D., Verbruggen H., Delwiche C.F., De Clerck O. (2012). Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 31: 1–46. [Google Scholar]
- Li J., Timko M.P. (1996). The pc-1 phenotype of Chlamydomonas reinhardtii results from a deletion mutation in the nuclear gene for NADPH:protochlorophyllide oxidoreductase. Plant Mol. Biol. 30: 15–37. [DOI] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260. [DOI] [PubMed] [Google Scholar]
- Liu J., Yang H., Lu Q., Wen X., Chen F., Peng L., Zhang L., Lu C. (2012). PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24: 4992–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence--a practical guide. J. Exp. Bot. 51: 659–668. [DOI] [PubMed] [Google Scholar]
- Meinecke L., Alawady A., Schroda M., Willows R., Kobayashi M.C., Niyogi K.K., Grimm B., Beck C.F. (2010). Chlorophyll-deficient mutants of Chlamydomonas reinhardtii that accumulate magnesium protoporphyrin IX. Plant Mol. Biol. 72: 643–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S., Sawaya M.R. (2005). The light reactions: a guide to recent acquisitions for the picture gallery. Plant Cell 17: 648–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A.G., Tanaka A., Terry M.J. (2010). The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15: 488–498. [DOI] [PubMed] [Google Scholar]
- Moore M., Harrison M.S., Peterson E.C., Henry R. (2000). Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275: 1529–1532. [DOI] [PubMed] [Google Scholar]
- Moseley J., Quinn J., Eriksson M., Merchant S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19: 2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F., Theg S.M., Hippler M., Merchant S. (2002). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N., Wenzel S., Zou Y., Künzel S., Sasso S., Weiß D., Prager K., Grossman A., Kottke T., Mittag M. (2017). A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiol. 174: 185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naver H., Boudreau E., Rochaix J.D. (2001). Functional studies of Ycf3: its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13: 2731–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive J., Wollman F.A., Bennoun P., Recouvreur M. (1981). Ultrastructure of thylakoid membranes in C. reinhardtii: evidence for variations in the partition coefficient of the light-harvesting complex-containing particles upon membrane fracture. Arch. Biochem. Biophys. 208: 456–467. [DOI] [PubMed] [Google Scholar]
- Ozawa S., Kosugi M., Kashino Y., Sugimura T., Takahashi Y. (2012). 5′-monohydroxyphylloquinone is the dominant naphthoquinone of PSI in the green alga Chlamydomonas reinhardtii. Plant Cell Physiol. 53: 237–243. [DOI] [PubMed] [Google Scholar]
- Petroutsos D., Tokutsu R., Maruyama S., Flori S., Greiner A., Magneschi L., Cusant L., Kottke T., Mittag M., Hegemann P., Finazzi G., Minagawa J. (2016). A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537: 563–566. [DOI] [PubMed] [Google Scholar]
- Qin X., Suga M., Kuang T., Shen J.R. (2015). Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348: 989–995. [DOI] [PubMed] [Google Scholar]
- Rebeiz C.A., Reddy K.N., Nandihalli U.B., Velu J. (1990). Tetrapyrrole-dependent photodynamic herbicides. Photochem. Photobiol. 52: 1099–1117. [Google Scholar]
- Rochaix J.D. (2004). Genetics of the biogenesis and dynamics of the photosynthetic machinery in eukaryotes. Plant Cell 16: 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.D. (2014). Regulation and dynamics of the light-harvesting system. Annu. Rev. Plant Biol. 65: 287–309. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.S., Lagarias J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Feoktistova K., Lagarias J.C. (2011). Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. USA 108: 11854–11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Lagarias J.C., Bhattacharya D. (2014). Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes. Front. Ecol. Evol. 2: 10.3389/fevo.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Schippers J.H. (2015). ROS-mediated redox signaling during cell differentiation in plants. Biochim. Biophys. Acta 1850: 1497–1508. [DOI] [PubMed] [Google Scholar]
- Stöckel J., Bennewitz S., Hein P., Oelmüller R. (2006). The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141: 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg E., Slagter J.G., Fridborg I., Cleary S.P., Robinson C., Coupland G. (1997). ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Iwai M., Takahashi Y., Minagawa J. (2006). Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103: 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58: 321–346. [DOI] [PubMed] [Google Scholar]
- Terry M.J., Smith A.G. (2013). A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K., Dubois M., Crocco C.D., Yin R., Chappuis R., Allorent G., Schmid-Siegert E., Goldschmidt-Clermont M., Ulm R. (2016). UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 28: 966–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C.S., Rocap G., King J., Chisholm S.W. (2002). Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10: 134–142. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy B.C., Oelmüller R. (2012). Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 7: 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff E.D., Alawady A., Meinecke L., Grimm B., Beck C.F. (2008). Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell 20: 552–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widhalm J.R., Ducluzeau A.L., Buller N.E., Elowsky C.G., Olsen L.J., Basset G.J. (2012). Phylloquinone (vitamin K(1) ) biosynthesis in plants: two peroxisomal thioesterases of Lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-CoA. Plant J. 71: 205–215. [DOI] [PubMed] [Google Scholar]
- Wilde A., Lünser K., Ossenbühl F., Nickelsen J., Börner T. (2001). Characterization of the cyanobacterial ycf37: mutation decreases the photosystem I content. Biochem. J. 357: 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks A. (2002). Heme oxygenase: evolution, structure, and mechanism. Antioxid. Redox Signal. 4: 603–614. [DOI] [PubMed] [Google Scholar]
- Wittkopp T.M., Saroussi S., Yang W., Grossman A.R. (2016). The GreenCut: functions and relationships of proteins conserved in green lineage organisms. In Chloroplasts: Current Research and Future Trends, Kirchhoff H., ed (Norfolk, UK: Horizon), : pp. 241–278. [Google Scholar]
- Wobbe L., Bassi R., Kruse O. (2016). Multi-level light capture control in plants and green algae. Trends Plant Sci. 21: 55–68. [DOI] [PubMed] [Google Scholar]
- Zhao L., et al. (2017). A light harvesting complex-like protein in maintenance of photosynthetic components in Chlamydomonas. Plant Physiol. 174: 2419–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones J.M., Blaby I.K., Merchant S.S., Umen J.G. (2015). High-Resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorin B., Lu Y., Sizova I., Hegemann P. (2009). Nuclear gene targeting in Chlamydomonas as exemplified by disruption of the PHOT gene. Gene 432: 91–96. [DOI] [PubMed] [Google Scholar]
- Zou Y., Wenzel S., Müller N., Prager K., Jung E.M., Kothe E., Kottke T., Mittag M. (2017). An animal-like cryptochrome controls the Chlamydomonas sexual cycle. Plant Physiol. 174: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]