Light inhibits the COP1-directed degradation of ICE transcription factors, which act as activators of stomatal development in Arabidopsis.
Abstract
Stomata are epidermal openings that facilitate plant-atmosphere gas exchange during photosynthesis, respiration, and water evaporation. Stomatal differentiation and patterning are spatially and temporally regulated by the master regulators SPEECHLESS (SPCH), MUTE, and FAMA, which constitute a central gene regulatory network along with Inducer of CBF Expression (ICE) transcription factors for this developmental process. Stomatal development is also profoundly influenced by environmental conditions, such as light, temperature, and humidity. Light induces stomatal development, and various photoreceptors modulate this response. However, it is unknown how light is functionally linked with the master regulatory network. Here, we demonstrate that, under dark conditions, the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) degrades ICE proteins through ubiquitination pathways in leaf abaxial epidermal cells in Arabidopsis thaliana. Accordingly, the ICE proteins accumulate in the nuclei of leaf abaxial epidermal cells in COP1-defective mutants, which constitutively produce stomata. Notably, light in the blue, red, and far-red wavelength ranges suppresses the COP1-mediated degradation of the ICE proteins to induce stomatal development. These observations indicate that light is directly linked with the ICE-directed signaling module, via the COP1-mediated protein surveillance system, in the modulation of stomatal development.
INTRODUCTION
Stomata are epidermal openings that facilitate the shoot-atmosphere gas exchange in plants. Therefore, stomatal development is critical for photosynthesis, which ensures plant propagation and is a key factor in the global carbon cycle (Hetherington and Woodward, 2003; Driscoll et al., 2006). It is also important for respiration and water circulation in plants (Lawson and Blatt, 2014).
In Arabidopsis thaliana, stomatal development is initiated by an asymmetric cell division in stomatal progenitor cells, named meristemoid mother cells, thereby producing meristemoids (Bergmann and Sack, 2007; MacAlister et al., 2007; Pillitteri and Torii, 2012). The meristemoids subsequently differentiate into guard mother cells, which divide symmetrically and finally differentiate into guard cells (Ohashi-Ito and Bergmann, 2006; Pillitteri et al., 2007). A pair of guard cells form a stoma, which is more prevalent in the abaxial epidermis of leaves. Notably, mature stomata are never formed adjacent to each other; they are always separated by at least one epidermal cell. Asymmetric cell divisions occur only in those stomatal lineage ground cells that are located away from existing stomata (Geisler et al., 2000, 2003). It is now thought that the spatial regulation of stomatal development is important for proper stomatal physiology, such as modulation of stomatal aperture by osmosis, and water and ion supply to guard cells by stomatal lineage ground cells (Nadeau and Sack, 2002; Hetherington and Woodward, 2003; Shpak et al., 2005).
Stomatal formation and patterning is mediated by several developmental regulators, including mitogen-activated protein kinase (MAPK) signaling components and leucine-rich repeat receptor-like proteins (Bergmann et al., 2004; Shpak et al., 2005; Wang et al., 2007). In addition, a group of basic helix-loop-helix transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA, act sequentially during stomatal development by spatially and temporally triggering the initiation, proliferation, and termination of the stomatal lineage cells, respectively (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007). Interestingly, they interact with Inducer of CBF Expression (ICE) transcription factors (Kanaoka et al., 2008), also known as SCREAM (SCRM) proteins (Kanaoka et al., 2008), to induce stomatal development.
Stomatal development is also modulated by environmental signals, such as light, temperature, humidity, and atmospheric CO2 concentration (Gray et al., 2000; Crawford et al., 2012; Tricker et al., 2012; Casson and Hetherington, 2014), among which the effects of light have been most extensively studied. Light triggers the division of stomatal lineage cells (Kang et al., 2009). Consequently, the division of stomatal lineage cells is halted in the epidermis of etiolated seedlings (Kang et al., 2009; Balcerowicz et al., 2014).
The E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) is a photomorphogenic repressor that directs the degradation of light-regulated transcription factors (Osterlund et al., 2000; Jang et al., 2005). Notably, it also acts as a negative regulator of stomatal development (Kang et al., 2009). Photoactivation of the red and far-red light photoreceptors phytochromes (phyA to phyE) and the blue light photoreceptors cryptochromes (CRY1 and CRY2) suppresses the action of COP1, inducing stomatal development (Boccalandro et al., 2009; Kang et al., 2009; Lian et al., 2011; Casson and Hetherington, 2014). The phyA, phyB, and CRY photoreceptors inhibit the activity of COP1 by disrupting the formation of COP1-SUPPRESSOR OF PHYA-105 (SPA) protein complex during photomorphogenic development (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011; Lu et al., 2015; Sheerin et al., 2015). As a result, just like the COP1-defective mutants (Kang et al., 2009), transgenic plants overexpressing the COP1-interacting domain of CRY1 and SPA-defective mutants produce stomata even in darkness, supporting the suppressive role of COP1 in light-induced stomatal development.
Here, we demonstrate that COP1 degrades the ICE transcription factors through ubiquitin-proteasome pathways in darkness to inhibit stomatal development. The ICE proteins accumulated in the nuclei of leaf abaxial epidermal cells in COP1-defective mutants that constitutively produce stomata under both light and dark conditions. Notably, light suppresses the COP1-mediated degradation of ICE proteins, triggering stomatal development. Our observations indicate that, via the ICE-mediated signaling network, the COP1-mediated light signals are directly linked with the developmental programs that regulate stomatal development.
RESULTS
Light Induces ICE Accumulation in the Nuclei of Leaf Abaxial Epidermal Cells
Light triggers stomatal differentiation (Casson et al., 2009; Kang et al., 2009). ICE transcription factors are core components of the gene regulatory networks that specify stomatal differentiation through interactions with SPCH, MUTE, and FAMA (Kanaoka et al., 2008). Therefore, our first objective was to determine how the ICE-mediated signaling module is linked with light signaling.
We examined the effects of different light wavelengths on ICE transcription. Reverse transcription-mediated quantitative real-time RT-PCR (RT-qPCR) assays revealed that the transcription of ICE is slightly altered by different light wavelengths (Supplemental Figure 1A). Fluorescent examination of gene promoter activities in the abaxial epidermal cells of cotyledons showed that ICE transcription is only marginally affected by the light regimes examined (Supplemental Figures 1B to 1D). We therefore concluded that light does not play a prominent role in regulating ICE transcription.
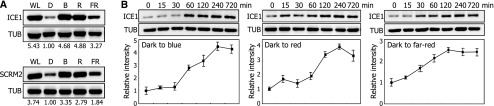
We next asked whether light influences the accumulation of ICE proteins. Seedlings expressing either a MYC-ICE1 or MYC-SCRM2/ICE2 gene fusion driven by their own promoters were exposed to various light wavelengths, and the levels of ICE proteins were examined. We found that the levels of ICE proteins were higher in seedlings exposed to red, far-red, and blue light, compared with the etiolated seedlings (Figure 1A; Supplemental File 1). Kinetic analysis of ICE1 accumulation revealed that its level gradually increases in the light and peaks at 4 h of light exposure (Figure 1B; Supplemental File 1).
Figure 1.
Light Induces ICE Accumulation.
(A) Light-induced accumulation of ICE proteins. Four-day-old ProICE1:MYC-ICE1 and ProSCRM2:MYC-SCRM2 transgenic seedlings grown on MS-agar plates in darkness were exposed to different light wavelengths for 12 h before extracting total proteins from whole seedlings. ICE proteins were detected immunologically using an anti-MYC antibody. Tubulin (TUB) was detected similarly as the loading control. WL, white light; D, darkness; B, blue light; R, red light; FR, far-red light. Blots on the membranes were quantitated using ImageJ software.
(B) Kinetics of ICE1 accumulation. Four-day-old 35S:MYC-ICE1 transgenic seedlings grown on MS-agar plates in darkness were exposed to different light wavelengths for up to 12 h before extracting total proteins from whole seedlings. Immunological detection of ICE1 was performed as described above. Blots on the membranes were quantitated as described above. Quantitations of three blots from independent biological samples were averaged. Bars indicate se of the mean.
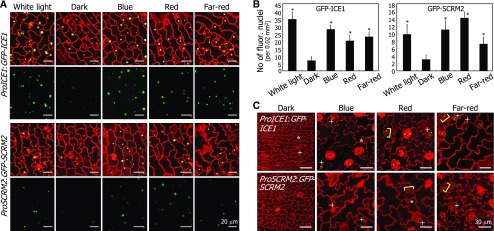
To verify the effect of light on ICE accumulation, dark-grown ProICE1:GFP-ICE1 and ProSCRM2:GFP-SCRM2 seedlings, in which GFP-ICE gene fusions were expressed with endogenous gene promoters, were exposed to different light wavelengths. Fluorescence microscopy showed that both ICE1 and SCRM2/ICE2 accumulate in the nuclei of abaxial epidermal cells of cotyledons following light illumination (Figures 2A and 2B). Accumulation of ICE1 was more prominent than SCRM2 accumulation. The light-induced ICE accumulation was also evident in guard cells and stomatal lineage cells (Figure 2C). These observations indicate that light stimulates ICE accumulation in leaf abaxial epidermal cells during stomatal development.
Figure 2.
Light-Induced ICE Accumulation Occurs in the Nuclei of Abaxial Epidermal Cells of Cotyledons.
(A) ICE accumulation in the nuclei of abaxial epidermal cells. Five-day-old ProICE1:GFP-ICE1 and ProSCRM2:GFP-SCRM2 seedlings grown on MS-agar plates in darkness were exposed to different light wavelengths for 4 h before obtaining confocal images.
(B) Counting of fluorescent nuclei in abaxial epidermal cells. Counting was performed using the cotyledon samples described in (A). Three independent counts, each consisting of 10 seedlings, were averaged and statistically analyzed (t test, *P < 0.01). Bars indicate se.
(C) Effects of light on ICE protein stability in stomata and stomatal lineage cells. The ProICE1:GFP-ICE1 and ProSCRM2:GFP-SCRM2 transgenic seedlings were grown on MS-agar plates for 5 d under different light wavelengths. Confocal images of abaxial epidermal cells of cotyledons were displayed. Asterisks indicate meristemoids. Brackets indicate guard mother cells. Plus signs indicate immature stomata.
We also examined the patterns of ICE1 accumulation in hypocotyl and root cells. Fluorescent detection of ICE1 proteins showed that, while light-induced ICE1 accumulation occurs in hypocotyl guard cells (Supplemental Figure 2A), ICE1 accumulation and transcription of its gene were not discernibly affected by light in the root cells (Supplemental Figures 2B to 2D and Supplemental File 1).
COP1 Interacts with ICE Proteins
COP1 acts as a repressor of light-induced stomatal development (Kang et al., 2009; Balcerowicz et al., 2014). We found that ICE accumulation diminishes in the dark, when a large proportion of COP1 proteins is nuclear localized (von Arnim and Deng, 1994; Stacey et al., 1999), raising the possibility that the reduction of ICE levels in the dark is associated with COP1 function.
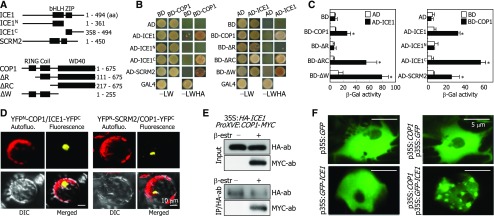
Yeast two-hybrid assays using genes encoding a set of ICE and COP1 proteins, including their full-size forms, showed that full-size ICE1 and SCRM2/ICE2 forms physically interact with COP1 (Figures 3A and 3B). Assays using truncated ICE1 forms revealed that the ICE1-COP1 interactions occur via the leucine zipper motif of ICE1 (Figures 3A to 3C). Meanwhile, assays using truncated forms of COP1 suggested that, for the ICE-COP1 interactions, the intact conformation of COP1 protein is more important than its specific protein domain(s), as discussed previously (Ang et al., 1998; Torii et al., 1998).
Figure 3.
ICE Proteins Interact with COP1.
(A) ICE and COP1 constructs. Numbers indicate amino acid (aa) positions. bHLH, basic helix-loop-helix; ZIP, leucine zipper; RING, ring domain; Coil, coiled-coil.
(B) ICE-COP1 interactions in yeast cells. –LW indicates Leu and Trp dropout plates. –LWHA indicates Leu, Trp, His, and Ade dropout plates.
(C) β-Gal activity assays. Interactions of ICE proteins with COP1 in yeast cells were examined by measuring β-Gal activity. Five measurements were averaged and statistically analyzed (t test, *P < 0.01). Bars indicate se.
(D) COP1-ICE interactions in Arabidopsis protoplasts. The YFPN-COP1 and ICE1-YFPC constructs and the YFPN-SCRM2 and COP1-YFPC constructs were coexpressed transiently in Arabidopsis protoplasts and visualized by differential interference contrast (DIC) and fluorescence microscopy. Bars = 10 μm.
(E) Coimmunoprecipitation. Five-day-old 35S:HA-ICE1 ProXVE:COP1-MYC seedlings were incubated in the presence of 25 μM β-estradiol and 50 μM MG132 for 1 d in darkness before extracting total proteins from whole seedlings. Immunoprecipitation (IP) was performed using an anti-HA antibody. ICE1 and COP1 were detected immunologically using anti-HA and anti-MYC antibodies, respectively.
(F) Localization of ICE1 into nuclear speckles. The COP1 and GFP-ICE1 constructs were coexpressed transiently in Arabidopsis protoplasts, and the nuclei were visualized by fluorescence microscopy. Bars = 5 μm.
Bimolecular fluorescence complementation (BiFC) assays using Arabidopsis protoplasts showed that the ICE-COP1 interactions occur in the nuclei (Figure 3D; Supplemental Figure 3). To confirm the ICE1-COP1 interactions in vivo, we performed coimmunoprecipitation assays using transgenic plants expressing a hemagglutinin (HA)-ICE1 fusion driven by the CaMV 35S promoter and a COP1-MYC fusion under the control of a β-estradiol-inducible promoter. The ICE1-COP1 interactions occurred when the expression of COP1-MYC was induced (Figure 3E; Supplemental File 1), whereas the protein-protein interactions were not detected in the absence of the inducer. These observations indicate that COP1 interacts with ICE proteins in plant cells.
To investigate the ICE-COP1 interactions further, we transiently expressed p35S:GFP-ICE1 and p35S:COP1 constructs separately or together in Arabidopsis protoplasts. Fluorescent assays showed that ICE1 is evenly distributed in the nuclei in the absence of COP1 (Figure 3F). When the two constructs were coexpressed, nuclear speckles emitting bright green fluorescence were observed in the nuclei, similar to the nuclear localization pattern of COP1 (Jang et al., 2005), supporting the interactions of ICE1 with COP1 in planta. By contrast, we found no discernible interactions of COP1 with SPCH, MUTE, and FAMA in yeast two-hybrid and BiFC assays (Supplemental Figures 4A and 4B, respectively).
COP1 Ubiquitinates ICE Proteins
COP1 is a well-characterized E3 ubiquitin ligase that targets a variety of proteins involved in light responses, including phytochrome photoreceptors (Osterlund et al., 2000; Jang et al., 2010). Based on the interactions of COP1 with ICE1 and SCRM2/ICE2, we postulated that ICE proteins would be subjected to COP1-mediated ubiquitination.
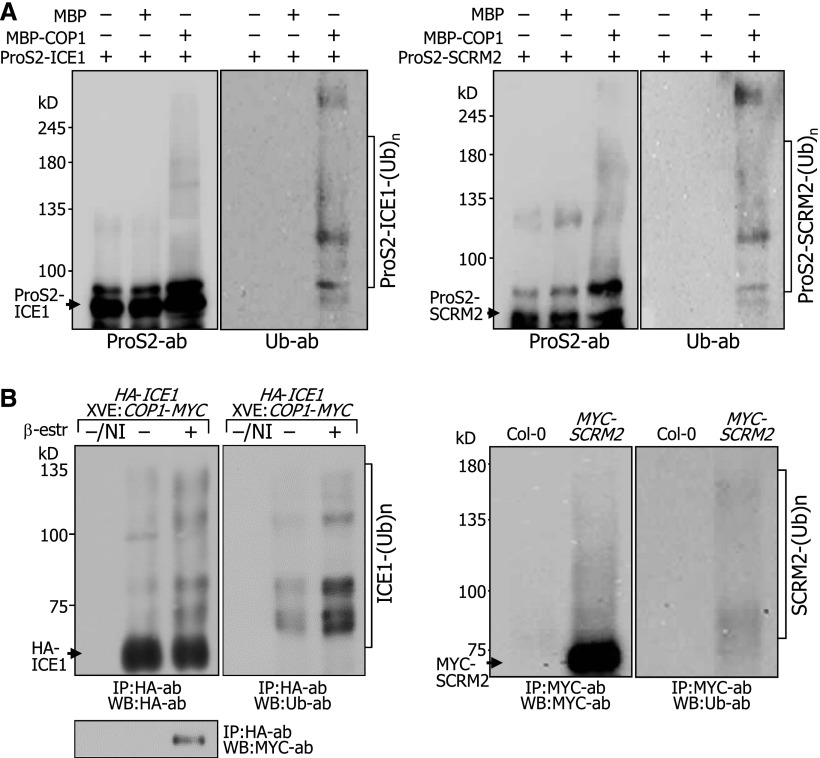
Using recombinant proteins, we examined in vitro whether COP1 ubiquitinates ICE proteins. Immunological assays using an antiubiquitin antibody showed that protein bands having higher molecular weights compared with those of native ICE1 and SCRM2 proteins were observed in the presence of COP1 but not detected in the absence of COP1 (Figure 4A; Supplemental Figure 5 and Supplemental File 1), indicating that COP1 ubiquitinates ICE proteins. To further examine the COP1-mediated ubiquitination of ICE proteins, we performed in vivo ubiquitination assays. The 35S:HA-ICE1 ProXVE:COP1-MYC plants expressing HA-ICE1 and COP1-MYC fusions were incubated in darkness in the presence of MG132, a potent 26S proteasome inhibitor (Lee and Goldberg, 1998). ICE1 was immunoprecipitated using an anti-HA antibody, and ubiquitinated proteins were detected immunologically using an antiubiquitin antibody. The results showed that a ladder of ubiquitinated ICE1 proteins was evident in the presence of β-estradiol, indicating that COP1-mediated ubiquitination of ICE1 occurs in planta (Figure 4B; Supplemental File 1). We also verified in a similar way that SCRM2 is ubiquitinated in vivo, but to a lesser extent than ICE1, suggesting that the primary target of the COP1-mediated ubiquitination is ICE1.
Figure 4.
COP1 Ubiquitinates ICE Proteins.
(A) ICE ubiquitination in vitro. Ubiquitination assays on ProS2 fusions in vitro were performed using human E1, E2, and recombinant MBP-COP1 that was pretreated with 20 μM ZnCl2 prior to the reactions. MBP protein was included as a negative control. Ubiquitinated ICE1 and SCRM2 proteins were detected immunologically using an antiubiquitin (Ub) antibody (left and right panels, respectively). kDa, kilodaltons.
(B) ICE ubiquitination in vivo. Ten-day-old 35S:HA-ICE1 ProXVE:COP1-MYC seedlings were incubated in the presence of 25 μM β-estradiol and 50 μM MG132 for 24 h in darkness before extracting total proteins from whole seedlings (left panel). IP was performed using an anti-HA antibody. Ubiquitinated ICE1 proteins were detected by immunoblot hybridization using an anti-Ub antibody. NI, no immunoprecipitation. Ten-day-old 35S:MYC-SCRM2 seedlings were incubated in the presence of 50 μM MG132 for 24 h in darkness before extracting total proteins from whole seedlings (right panel). IP and detection of ubiquitinated SCRM2 were performed as described above.
The ICE transcription factors form heterodimers with SPCH, MUTE, and FAMA (Kanaoka et al., 2008; Horst et al., 2015). Therefore, we examined whether SPCH, MUTE, and FAMA proteins are the substrates of the COP1-mediated ubiquitination. COP1 did not ubiquitinate SPCH, MUTE, and FAMA in vitro (Supplemental Figure 6 and Supplemental File 1).
COP1 Degrades ICE Proteins through Ubiquitin/Proteasome Pathways
We observed that COP1 ubiquitinates ICE proteins. Therefore, we examined whether COP1 directs the degradation of ICE proteins through ubiquitin/proteasome pathways.
Using 35S:MYC-ICE transgenic plants, we first investigated whether ubiquitinated ICE proteins undergo degradation. The plants were incubated in darkness with or without MG132, and the levels of ICE proteins were analyzed immunologically. We found that the levels of ICE1 and SCRM2/ICE2 in MG132-treated plants in darkness were comparable to those in light-grown plants (Figure 5A; Supplemental File 1). However, the protein levels were significantly lower in dark-grown plants without MG132 treatments, indicating that ICE degradation occurs through ubiquitin/proteasome-dependent pathways.
Figure 5.
COP1 Degrades ICE Proteins through Ubiquitin/Proteasome Pathways.
(A) Ubiquitin-mediated ICE degradation. Ten-day-old 35S:MYC-ICE1 and 35S:MYC-SCRM2 seedlings were incubated in the presence of 50 μM MG132 for 24 h in darkness before extracting total proteins from whole seedlings. An anti-MYC antibody was used to detect ICE proteins. Nontransformed Col-0seedlings were also included in the assays.
(B) Reduced ICE degradation in COP1-defective mutants. The 35S:MYC-ICE1 and 35S:MYC-SCRM2 plants were crossed with Col-0 plants and cop1 mutants (upper and lower panels, respectively). Seedling growth and detection of ICE proteins were performed as described in (A). Nontransformed Col-0seedlings were also included in the assays.
(C) COP1-mediated degradation of ICE1. Ten-day-old 35S:HA-ICE1 ProXVE:COP1-MYC seedlings were incubated in darkness for 24 h in the presence of 10 μM MG132 and 25 μM β-estradiol before extracting total proteins from whole seedlings. Anti-HA and anti-MYC antibodies were used to detect ICE1 and COP1 proteins, respectively. Nontransformed Col-0 seedlings were also included in the assays.
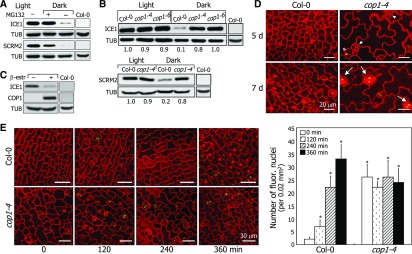
(D) ICE1 accumulation in cop1-4 stomatal lineage cells. The ProICE1:GFP-ICE1 and ProICE1:GFP-ICE1 cop1-4 seedlings were grown for 5 or 7 d in darkness. Confocal images of abaxial epidermal cells were obtained. Arrowheads indicate division-competent cells. Asterisk indicates meristemoid. Arrows indicate stomata.
(E) Kinetics of ICE1 accumulation. Three-day-old ProICE1:GFP-ICE1 and ProICE1:GFP-ICE1 cop1-4 seedlings grown in darkness were exposed to white light for the indicated time periods before obtaining confocal images (left panel). Bars = 30 μm. Three independent measurements, each consisting of 10 counts of fluorescent nuclei in abaxial epidermal cells, were averaged and statistically analyzed (t test, *P < 0.01) (right graph). Bars in the graph indicate se.
To test whether the ubiquitin-mediated degradation of ICE proteins is mediated by COP1, we overexpressed the MYC-ICE construct driven by the CaMV 35S promoter in COP1-defective mutants, and the levels of ICE proteins were investigated. We found that ICE degradation was significantly reduced in cop1-4 and cop1-6 mutants (Figure 5B; Supplemental File 1). We also examined the COP1-mediated degradation of ICE1 using the 35S:HA-ICE1 ProXVE:COP1-MYC plants. The ICE1 levels were considerably lower when COP1 expression was induced by β-estradiol (Figure 5C; Supplemental File 1), confirming that COP1 is responsible for the ubiquitin-mediated degradation of ICE proteins.
Our data showed that ICE proteins accumulate in cotyledon epidermal cells in the light but disappear in darkness. To examine whether the COP1-mediated degradation of ICE1 occurs in abaxial epidermal cells, we produced transgenic plants expressing a GFP-ICE1 fusion in the Col-0 and cop1-4 backgrounds. While GFP signals were evident in the stomata lineage cells of cop1-4 in darkness, the fluorescent signals were not detected in the Col-0 background under identical conditions (Figure 5D), indicating that the COP1-mediated degradation of ICE1 occurs in the nuclei of abaxial epidermal cells in darkness. We next examined the kinetics of ICE1 accumulation. The GFP signals appeared rapidly in the stomatal precursor cells following light illumination in the Col-0 background (Figure 5E). By contrast, GFP signals were evident in the stomatal precursor cells and immature stomata in the cop1-4 background even in darkness, confirming that COP1-mediated degradation of ICE1 occurs in the nuclei of abaxial epidermal cells in darkness.
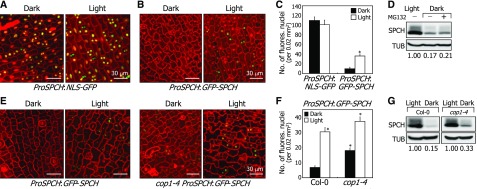
SPCH forms protein complexes with ICE proteins (Kanaoka et al., 2008; Horst et al., 2015), whose accumulation is enhanced by light during stomatal development. Promoter activity assays using ProSPCH:NLS-GFP and ProSPCH:GFP-SPCH constructs revealed that while SPCH transcription is not discernibly affected by light, its protein abundance was elevated in the light, as indicated by the numbers of green fluorescent nuclei (Figures 6A to 6C). Notably, the reduction in SPCH abundance in darkness could not be undone by MG132 (Figure 6D; Supplemental File 1). Similar assays showed that the number of green fluorescent nuclei was 2.2-fold higher in the cop1-4 background than in the Col-0 background under dark conditions (Figures 6E and 6F). In addition, light increased the SPCH abundance in both Col-0 plants and cop1-4 mutants (Figures 6E to 6G; Supplemental File 1). Therefore, we concluded that, while the protein abundance of SPCH is elevated by light, the dark-induced degradation of SPCH is not mediated by ubiquitin/proteasome pathways, unlike ICE proteins.
Figure 6.
SPCH Degradation Is Not Associated with Ubiquitin/Proteasome Pathways.
(A) Fluorescence-assisted detection of SPCH promoter activity. Three-day-old ProSPCH:NLS-GFP transgenic seedlings grown in darkness were exposed to white light for 6 h. Confocal images of abaxial epidermal cells of cotyledons were obtained. All scale bars represent the same value. Bars = 30 μm.
(B) and (C) SPCH accumulation in the nuclei of abaxial epidermal cells of cotyledons. Three-day-old ProSPCH:GFP-SPCH transgenic seedlings grown in darkness were exposed to white light for 6 h before obtaining confocal images (B). Bars = 30 μm. Fluorescent nuclei in abaxial epidermal cells were counted (C). Three independent measurements, each consisting of 10 counts, were averaged and statistically analyzed (t test, *P < 0.01). Bars indicate se.
(D) Effects of MG132 on SPCH degradation. Five-day-old transgenic seedlings overexpressing a MYC-SPCH fusion were incubated in darkness for 24 h in the presence of 50 μM MG132 before extracting total proteins from whole seedlings. Immunological detection of SPCH proteins was performed using an anti-MYC antibody. Blots on the membranes were quantitated using ImageJ software.
(E) and (F) Dark-induced degradation of SPCH in cop1-4 stomatal lineage cells. Three-day-old ProSPCH:GFP-SPCH and cop1-4 ProSPCH:GFP-SPCH seedlings grown in darkness were exposed to white light for 6 h before obtaining confocal images of abaxial epidermal cells of cotyledons (E). Bar = 30 μm. All scale bars represent the same value. Fluorescent nuclei in abaxial epidermal cells were counted (F). Three independent measurements, each consisting of 15 counts, were averaged and statistically analyzed (t test, *P < 0.01). Bars indicate se.
(G) SPCH accumulation in the cop1-4 background. Five-day-old 35S:MYC-SPCH seedlings were incubated in darkness for 24 h. Immunological detection of SPCH proteins was performed as described in (D). Blots on the membranes were quantitated as described above.
Light Inhibits COP1-Mediated Degradation of ICE proteins
Light inhibits the nuclear function of COP1 during photomorphogenic responses and stomatal development (Stacey et al., 1999; Kang et al., 2009; Sheerin et al., 2015). Therefore, we hypothesized that light would inhibit the COP1-mediated degradation of ICE proteins during stomatal development.
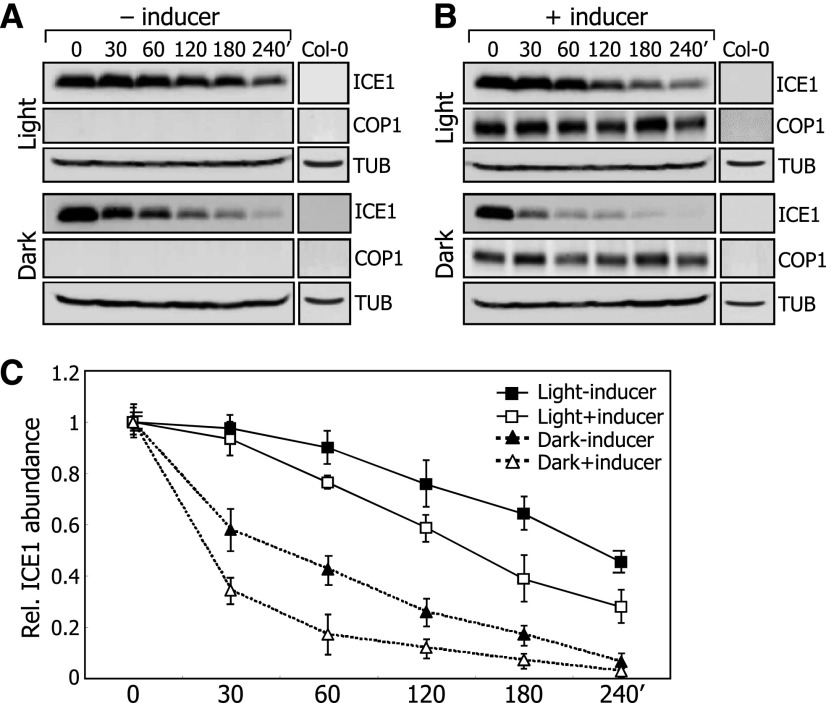
To investigate the effects of light on the COP1-mediated degradation of ICE1, the 35S:HA-ICE1 ProXVE:COP1-MYC transgenic plants were pretreated with MG132 in the presence or absence of β-estradiol, and the degradation kinetics of ICE1 were investigated in the presence of cycloheximide. In the absence of the inducer, ICE1 degradation was slower in light than in darkness (Figures 7A and 7C; Supplemental File 1), consistent with the degradation of ICE1 in darkness. In the inducer-pretreated plants, the degradation rate of ICE1 was discernibly faster than that in plants without the induction of COP1 production in the light (Figures 7B and 7C; Supplemental File 1). Similarly, in darkness, the rate of ICE1 degradation in the inducer-treated plants was faster than that in mock-treated plants, indicating that light inhibits the ability of COP1 to degrade ICE1. ICE1 degradation still occurred slowly in the light regardless of the induction of COP1 production, owing to the presence of endogenous COP1 produced in the 35S:HA-ICE1 ProXVE:COP1-MYC plants.
Figure 7.
Light Inhibits COP1-Mediated Degradation of ICE1.
Five-day-old 35S:HA-ICE1 ProXVE:COP1-MYC seedlings were pretreated with 50 μM MG132 in the absence (A) or presence (B) of 25 μM β-estradiol for 16 h and subsequently incubated in the presence of 0.5 mM cycloheximide either in the light or darkness. Whole seedlings were harvested at the indicated time points for total protein extraction. Anti-HA and anti-MYC antibodies were used to detect ICE1 and COP1 proteins, respectively. Nontransformed Col-0 seedlings were also included in the assays.
(C) Blots on the membranes were quantitated using ImageJ software. Quantitations of three blots of biological triplicates from independent plant samples incubated under identical conditions were averaged. Bars indicate se.
Light-Mediated Stabilization of ICE proteins Is Critical for Stomatal Development
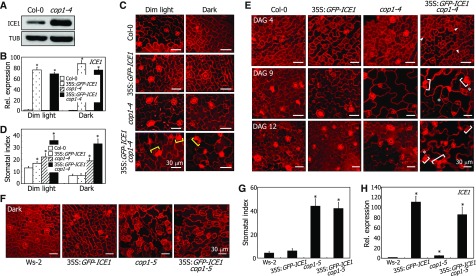
A critical question was whether the light-mediated stabilization of ICE proteins is essential for the induction of stomatal development. We measured the stomatal index (SI) for transgenic plants overexpressing a GFP-ICE1 fusion driven by the CaMV 35S promoter in Col-0 and cop1-4 backgrounds. It has been reported that Col-0 plants and cop1-4 mutants do not exhibit any differences in stomatal phenotypes under normal light conditions (Kang et al., 2009), which is due to the saturated accumulation of ICE proteins (Figure 5B). Therefore, plants were grown under dim light (1.5 μmol m−2 s−1), as ICE1 levels were different in Col-0 plants and cop1-4 mutants under these conditions (Figure 8A; Supplemental File 1).
Figure 8.
Light-Mediated Stabilization of ICE1 Is Important for Stomatal Development.
(A) ICE1 accumulation in the cop1-4 mutant. Col-0 and cop1-4 seedlings overexpressing a MYC-ICE1 fusion driven by the CaMV 35S promoter were grown in dim light (1.5 μmol m−2 s−1) for 5 d before extracting total proteins from whole seedlings. ICE1 was immunodetected using an anti-MYC antibody.
(B) Transcript levels of GFP-ICE1 transgene in the cop1-4 background. Seedlings were grown for 5 d on MS-agar plates under either dim light or dark conditions. Transcript levels were examined by RT-qPCR. Biological triplicates using independent seedling samples grown under identical conditions were statistically analyzed (t test, *P < 0.01). Bars indicate se.
(C) and (D) Stomatal phenotypes in dim light. Seedlings were grown under either dim light or dark conditions for 5 d. Confocal images of abaxial epidermal cells of cotyledons were obtained (C). Yellow brackets mark adjacent stomata. SI was measured (D). Three independent measurements, each consisting of 10 seedlings, were averaged and statistically analyzed (t test, *P < 0.01). Bars indicate se.
(E) Kinetics of stomatal differentiation in the cop1-4 background. Seedlings were grown on MS-agar plates in dim light. Confocal images of abaxial epidermal cells of cotyledons were obtained. DAG, days after germination. Arrowheads indicate compact division-competent cells. Asterisks indicate small cells, which readily differentiate into guard cells. Brackets indicate adjacent stomata.
(F) and (G) Stomatal phenotypes in darkness. The 35S:GFP-ICE1 cop1-5 transgenic seedlings were grown on MS-agar plates in darkness for 8 d. Confocal images of abaxial epidermal cells of cotyledons were obtained (F). Three independent measurements of SI, each consisting of 10 seedlings, were statistically analyzed (t test, *P < 0.01) (G). Bars indicate se.
(H) Transcript levels of GFP-ICE1 transgene in the cop1-5 background. Transcript levels were analyzed by RT-qPCR using the dark-grown seedlings described in (F). Whole seedlings were used for total RNA extraction. Biological triplicates using independent seedling samples grown under identical conditions were statistically analyzed (t test, *P < 0.01). Bars indicate se.
The 35S:GFP-ICE1 seedlings exhibited enhanced stomatal development compared with Col-0 plants under dim light conditions, as verified by SI measurements (Figures 8C and 8D). The cop1-4 mutant exhibited higher SI than Col-0 plants under both dim light and dark conditions, consistent with the negative regulatory role of COP1 in stomatal development (Kang et al., 2009). Notably, ICE1 overexpression further enhanced the cop1-4 stomatal phenotype in 35S:GFP-ICE1 cop1-4 seedlings (Figures 8C and 8D), producing adjacent stomata and increasing the number of small cells, which are known to readily differentiate into guard cells (Wang et al., 2007). A similar enhancement of the stomatal phenotype has also been observed for the severe COP1-defective allele cop1-5 (Kang et al., 2009). Meanwhile, we found no discernible differences in the levels of GFP-ICE1 transcripts between the cop1-4 mutants and Col-0 plants (Figure 8B).
Time sequence analysis of stomatal differentiation revealed that the 35S:GFP-ICE1 cop1-4 plants produced compact, division-competent cells during the early stages of stomatal differentiation, at 4 d after germination (Figure 8E). It was evident that the transgenic plants produced adjacent stomata and small cells at 9 d after germination, demonstrating that light-induced ICE1 accumulation is critical for stomatal development.
To further verify that the light-mediated inhibition of COP1 function in inducing ICE1 accumulation is important for stomatal development, a GFP-ICE1 fusion was overexpressed driven by the strong CaMV 35S promoter in the cop1-5 mutant in the Ws-2 background, which exhibits a severe stomatal phenotype, having more stomata than the cop1-4 mutant (Kang et al., 2009). The transgenic plants were grown in darkness, and the SI was measured. The results showed that ICE1 overexpression does not enhance the stomatal phenotype of the cop1-5 seedlings (Figures 8F to 8H), which would be because ICE1 accumulation is already higher in this phenotypically severe mutant compared with that in the cop1-4 mutant having mild phenotypes.
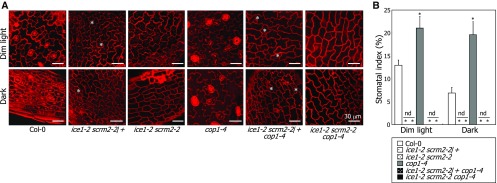
To further investigate the functional relationship between ICE transcription factors and COP1 in regulating stomatal development, the ice1-2 scrm2-2 double mutant was crossed with the cop1-4 and cop1-5 mutants, and the stomatal phenotypes of the resultant plants were analyzed. The ice1-2 scrm2-2 double mutant exhibited no symptoms of stomatal development in abaxial epidermal cells of cotyledons under both dim-light and dark conditions (Figures 9A and 9B). Notably, the ice1-2 scrm2-2 double mutation compromised the constitutive stomatal development of the dark-grown cop1-4 and cop1-5 mutants (Figures 9A and 9B; Supplemental Figure 7), confirming that suppression of COP1-mediated ICE degradation is required for light-induced stomatal development.
Figure 9.
ICE1 and SCRM2 Mediate Light-Mediated Induction of Stomatal Development.
Seedlings were grown on MS-agar plates under either dim light or dark conditions for 5 d. Confocal images of abaxial epidermal cells of cotyledons were obtained (A). Asterisks indicate aborted stomatal lineage cells. Three independent measurements of SI, each consisting of 10 seedlings, were averaged and statistically analyzed (t test, *P < 0.01) (B). Bars indicate se. nd, not detectable.
Light induces the division of stomatal lineage cells, but its effects are not prominent before entering the stomatal lineage (Kang et al., 2009). Therefore, we speculated that the suppression of the cop1-4 stomatal phenotype by the ice1-2 scrm2-2 double mutation occurs because the double mutant does not have stomatal lineage cells capable of responding to light. In support of this notion, it has been reported that spch mutations, which block the entry into the stomatal lineage, suppress the cop1-5 stomatal phenotype (Kang et al., 2009).
To resolve this uncertainty, we prepared ice1-2 scrm2-2/+ seedlings, in which the entry into stomatal lineage occurs properly but division of the stomatal lineage cells is aborted, unlike ice1-2 scrm2-2 seedlings (Kanaoka et al., 2008). We found that the ice1-2 scrm2-2/+ mutation still compromised the constitutive stomatal phenotype of the cop1-4 mutant (Figures 9A and 9B), further supporting the physiological significance of the light-induced suppression of COP1-mediated ICE degradation during stomatal development.
The MAPKK kinase YODA (YDA) acts downstream of COP1 in repressing stomatal development, and its defective mutants produce stomata even in darkness (Kang et al., 2009; Balcerowicz et al., 2014). A question was whether YDA is functionally associated with ICE proteins in stomatal development. We observed that the stomatal phenotype of the YDA-defective yda-10 mutant is still light-responsive, producing more stomata in the light than in darkness (Balcerowicz et al., 2014) (Supplemental Figure 8A). The yda-10 mutant harbors more cells that express ICE1 than Col-0plants under both light and dark conditions (Balcerowicz et al., 2014). To examine whether the stomatal phenotype of the mutant is associated with ICE1 accumulation, we produced transgenic plants expressing a GFP-ICE1 fusion in Col-0 and yda-10 backgrounds. While the number of green fluorescent nuclei was larger in the yda-10 background than in the Col-0 background, it further increased following light exposure in the yda-10 background (Supplemental Figures 8B and 8C), consistent with the stomatal phenotype of the mutant. Immunological assays also showed that ICE1 accumulation is still light-responsive in the yda-10 background but to a lesser degree compared with that in the Col-0background (Supplemental Figures 8D and 8E and Supplemental File 1). These observations suggest that YDA is functionally independent of ICE-mediated light signaling in regulating stomatal development. It is likely that YDA and ICE1 function in parallel during stomatal development.
ICE Proteins Integrate Light and Developmental Signals into Stomatal Development
Our data indicated that light triggers ICE accumulation to direct stomatal development. It is known that ICE and SPCH transcription factors cooperatively regulate genes that modulate stomatal development, such as EPIDERMAL PATTERNING FACTOR2 (EPF2) and TOO MANY MOUTH (TMM) (Horst et al., 2015). Therefore, we examined whether the ICE targets are involved in light-mediated stomatal development.
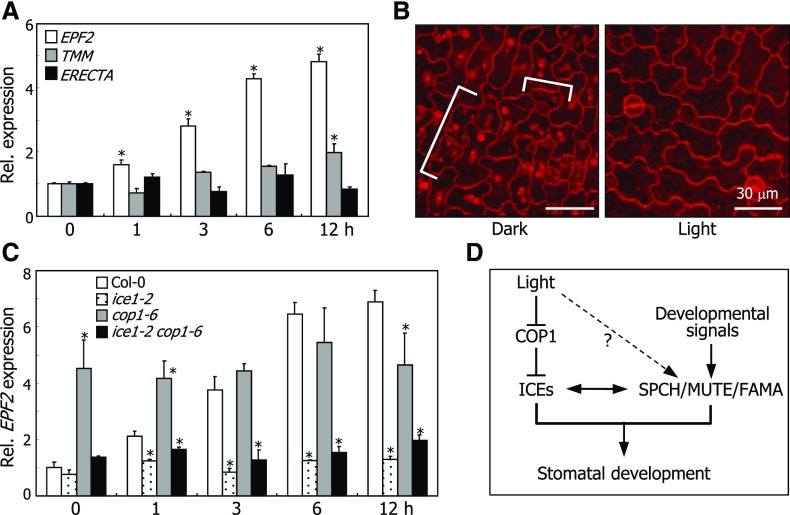
RT-qPCR analysis revealed that light promotes the transcription of EPF2. Its transcript levels gradually increased in the light, peaking 12 h following light illumination (Figure 10A). The transcription of TMM was also induced, albeit to a lesser degree, by light exposure.
Figure 10.
ICE Proteins Integrate Light Signals into Stomatal Development.
(A) Effects of light on the transcription of stomatal regulator genes. Col-0 plants were grown in darkness for 3 d and then exposed to white light for the indicated time periods before harvesting whole seedlings. Transcript levels were examined by RT-qPCR. Biological triplicates using independent seedling samples grown under identical conditions were statistically analyzed (t test, *P < 0.01). Bars indicate se. ERECTA was used as the negative control (Horst et al., 2015).
(B) Stomatal spacing and distribution. Col-0 plants were grown under either dark or light conditions for 3 d. Confocal images of abaxial epidermal surfaces of cotyledons were obtained. Brackets indicate small clustering cells that occur only in etiolated seedlings.
(C) Transcript levels of EPF2 in the ice1-2 and cop1-6 mutant. Seedling growth and RT-qPCR were performed as described in (A).
(D) ICE-mediated light signaling scheme during stomatal development. In this signaling scheme, ICE transcription factors integrate light signals into stomatal development. Light would also stabilize SPCH via a COP1-independent pathway.
Spatial regulation of initial stomatal development is conducted by ICE/SPCH transcription factors and its target EPF2 (Horst et al., 2015). Light-induced EPF2, which functions as a secreted signaling peptide, restricts entry into the stomatal lineage in neighboring cells (Hunt and Gray, 2009). Consequently, EPF2-defective mutants exhibit small-clustering-cell phenotypes under light conditions (Hunt and Gray, 2009; Yamamuro et al., 2014). Notably, dark-grown wild-type seedlings exhibited numerous small clustering cells (Figure 10B), similar to those observed in EPF2-defective mutants, supporting the notion that the ICE-mediated activation of EPF2 is associated with light regulation of stomatal development.
To examine whether the light-mediated induction of EPF2 is related to ICE transcription factors, we examined the levels of EPF2 transcripts in ice1-2 and cop1-6 mutants grown in the light. We found that while the transcript levels were elevated in Col-0plants, they were not elevated in the ice1-2 and ice1-2 cop1-6 mutants (Figure 10C). Meanwhile, high levels of EPF2 transcript were maintained in the cop1-6 mutant, in which ICE/SCRM proteins accumulate (Figure 5B). These observations demonstrate that ICE/SCRM transcription factors mediate the light-mediated induction of EPF2.
In conclusion, our data demonstrated that COP1 degrades the ICE transcription factors through ubiquitin/proteasome pathways in darkness, inhibiting stomatal development. Accordingly, ICE proteins accumulate in the nuclei of abaxial epidermal cells in COP1-defective mutants even in darkness. However, light suppresses the COP1-mediated degradation of ICE proteins, underlying the induction of stomatal development in light-grown plants. Our data provide a molecular mechanism by which light signals are integrated into the ICE-SPCH/MUTE/FAMA-mediated developmental programs during stomatal development (Figure 10D).
DISCUSSION
Photostabilization of ICE Proteins during Stomatal Development
Stomatal development progresses through a series of cell fate determination/specification steps from initial meristemoid mother cells to guard mother cells, which finally differentiate into guard cells. The initial stomatal development is specified by the coordinated actions of ICE and SPCH transcription factors, while it is restricted by the signaling peptide EPF2 and receptor-like proteins, such as TMM. The stomatal regulators function through a network of negative and positive feedback regulatory circuits (Horst et al., 2015). Meanwhile, it is known that light promotes stomatal development. Light inhibits the function of COP1, a well-known repressor of light responses, thus inducing stomatal development (Kang et al., 2009). While it has been generally accepted that the link between light signaling and stomatal development is mediated by COP1, the exact mechanism of how light regulates stomatal development at the molecular level is not well understood.
Here, we observed that COP1 degrades ICE proteins through ubiquitination pathways, thus suppressing stomatal development under dark conditions. As a result, the ICE proteins accumulate to high levels in the nuclei of abaxial epidermal cells of leaves in COP1-defective mutants, which constitutively produce stomata even in darkness. Notably, light suppresses the COP1-mediated degradation of the ICE proteins to sustain stomatal development.
ICE gene expression and their protein accumulation are largely unchanged during stomatal development, and the ICE transcription factors specify all rounds of stomatal division steps (Kanaoka et al., 2008). The ICE proteins form heterodimers with SPCH, MUTE, and FAMA, for promoting stomatal cell fate transitions during this developmental process. It has been reported that SPCH transcription is induced by red light (Klermund et al., 2016). We observed that, while SPCH transcription is only marginally affected by light, its protein accumulation is markedly promoted in the light (Figures 6A to 6C). In addition, the ICE and SPCH transcription factors are functionally interrelated through a feedback regulation pathway (Kanaoka et al., 2008; Horst et al., 2015). We propose that light-mediated stabilization of ICE and SPCH proteins contribute synergistically to the light-mediated induction of stomatal development.
COP1 is a central E3 ubiquitin ligase that directs the degradation of diverse target proteins involved in plant photomorphogenesis (Osterlund et al., 2000; Jang et al., 2005). Light suppresses the action of COP1, thereby stabilizing target proteins and commencing photomorphogenesis (Stacey et al., 1999; Liu et al., 2011; Lu et al., 2015; Sheerin et al., 2015). It is known that light-mediated suppression of COP1 function occurs through two distinct molecular mechanisms: nuclear depletion of COP1 proteins and dissociation of COP1-SPA protein complex. While the former mechanism is exerted around 24 h following light exposure (von Arnim and Deng, 1994; von Arnim et al., 1997), the latter mechanism is effective within 6 h after light exposure (Lu et al., 2015; Sheerin et al., 2015).
Our data indicate that light triggers the accumulation of ICE proteins by suppressing COP1-mediated degradation in the several hours following light exposure. Meanwhile, it is known that the effects of light on most plant cell divisions occur within 24 h under light conditions (Reichler et al., 2001). Therefore, we believe that light stabilizes the ICE proteins primarily by reorganizing the COP1-SPA complex during stomatal development. However, the light-mediated nucleocytoplasmic relocalization of COP1 would also contribute, at least in part, to the stabilization of ICE proteins (Pacín et al., 2014). We observed that ICE degradation still occurs in COP1-defective mutants (Figure 5B), suggesting that COP1 is not the sole E3 ubiquitin ligase that regulates ICE abundance during stomatal development. It will be worth investigating the additional E3 enzyme(s) responsible for ICE degradation during this developmental process. It is also possible that residual degradation of ICE proteins might still occur through COP1, considering that both the cop1-4 and cop1-6 mutants are not loss-of-function mutants.
Integration of Light Signals into Stomatal Development by ICE Proteins
The SPCH/MUTE/FAMA regulatory network constitutes a central developmental pathway for stomatal development and possibly functions downstream of the YDA-MAPK signaling cascade (Lampard et al., 2008; Dong and Bergmann, 2010). YDA acts as a negative regulator of stomatal development; thus, plants expressing a constitutively active YDA form fail to form stomata. Meanwhile, we found that the stomatal development of a YDA-defective mutant is still light-responsive, as has been reported previously (Balcerowicz et al., 2014). We believe that ICE-mediated light signaling is functionally distinct from the role of YDA in regulating stomatal development.
Our data illustrate that light stabilizes ICE proteins by suppressing COP1 function. ICE transcription factors are functionally interrelated with SPCH/MUTE/FAMA in regulating stomatal development (Kanaoka et al., 2008; Horst et al., 2015). Interestingly, COP1 does not interact with SPCH, MUTE, and FAMA, which interacts physically and functionally with the ICE proteins. Based on the previous reports and our own observations, we propose that the ICE transcription factors integrate light signals into the YDA-SPCH/MUTE/FAMA developmental regulatory network during light-regulated stomatal development.
EPF2 encodes a signaling peptide that restricts entry into the stomatal lineage (Hara et al., 2009; Hunt and Gray, 2009). We found that light induces ICE accumulation, which upregulates EPF2 activity at the transcriptional level, thereby conferring proper stomatal spacing and distribution. Interestingly, it has been recently shown that CO2 concentration affects the transcription of EPF2 (Engineer et al., 2014). Thus, it is likely that the COP1-ICE-EPF2 signaling module integrates environmental information, including light signals, into developmental programs that govern stomatal development.
Recent studies have shown that ICE transcription factors are conserved in a wide range of plant species and play versatile roles in diverse plant developmental processes (Lee et al., 2015; Raissig et al., 2016), raising the possibility that they serve as a molecular node that links environmental signals and plant developmental cues. Among the external signals mediated by ICE proteins, temperature responses have been studied extensively (Dong et al., 2006; Kim et al., 2015). It is known that stomatal development is also influenced by environmental temperatures (Thomashow, 1999; Crawford et al., 2012). It will be worthy of investigating whether and how temperature signals interact with light signals during stomatal development.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used were in the Columbia (Col-0) background except for the cop1-5 mutant, which was in the Wassilewskija (Ws-2) background (McNellis et al., 1994). The ice1-2 and scrm2-2 mutants have been described previously (Kanaoka et al., 2008; Kim et al., 2015). The cop1-4 and cop1-6 mutants have been described previously (Deng et al., 1991).
A MYC-coding sequence was fused in-frame to the 5′ ends of the ICE1 and SCRM2/ICE2 genes, and the fusions were expressed driven by endogenous ICE promoters in Col-0 plants, resulting in ProICE1:MYC-ICE1 and ProSCRM2:MYC-SCRM2 plants, respectively. To produce ProICE1:GFP-ICE1, ProSCRM2:GFP-SCRM2, and ProSPCH:GFP-SPCH plants, a GFP-coding sequence was fused in-frame to the 5′ ends of ICE and SPCH genes, and the fusions were subcloned under the control of endogenous promoters into the modified pBA002a vector (Jung et al., 2013). The 35S:MYC-ICE transgenic plants have been described previously (Kim et al., 2015; Lee et al., 2015). A full-size SPCH cDNA was fused in-frame to the 3′ end of a MYC-coding sequence, and the fusion construct was overexpressed driven by the CaMV 35S promoter in the Col-0 background to generate 35S:MYC-SPCH plants. To produce 35S:GFP-ICE1 plants, a full-size ICE1 cDNA was fused in-frame to the 3′ end of a GFP-coding sequence, and the fusion construct was subcloned under the control of the CaMV 35S promoter into the binary pBA002 vector (Kim et al., 2006). A full-size ICE1 cDNA was also fused in-frame to the 3′ end of a HA-coding sequence, and the fusion construct was overexpressed driven by the CaMV 35S promoter to generate 35S:HA-ICE1 plants. To produce transgenic plants expressing COP1-MYC under the control of a β-estradiol-inducible promoter, the full-length COP1-coding sequence was subcloned into the pER8 vector (Zuo et al., 2000). For generating ProICE:NLS-GFP and ProSPCH:NLS-GFP plants, the gene promoter sequences were transcriptionally fused to nuclear localization sequence (NLS) and GFP-coding sequence, and the fusion constructs were transformed into Col-0 plants.
Arabidopsis plants were grown in soil in a controlled culture room set at 23°C with relative humidity of 55% under long days (16 h light and 8 h dark) with white light illumination (120 μmol m−2 s−1) provided by fluorescent FLR40D/A tubes (Osram). For treatments with different light wavelengths, plants were grown on 0.5× Murashige and Skoog-agar (MS-agar) plates with white light (50 μmol m−2 s−1), blue (35 μmol m−2 s−1), red (40 μmol m−2 s−1), or far-red (15 μmol m−2 s−1) illumination. Arabidopsis transformation was performed by the floral dip method (Clough and Bent, 1998).
Gene Expression Analysis
Preparation of RNA samples and RT-qPCR were performed according to the rules proposed to ensure reproducible measurements of mRNA levels (Udvardi et al., 2008). RT-qPCR reactions were performed in 96-well blocks with the Applied Biosystems 7500 real-time PCR system using the SYBR Green I master mix in a volume of 20 μL. PCR primers used are listed in Supplemental Table 1. The two-step thermal cycling profile employed was 15 s at 95°C for denaturation and 1 min at 60°C for annealing and polymerization. An eIF4A gene was included as an internal control in the PCR reactions to normalize the variations in the amounts of cDNA used. All RT-qPCR reactions were performed in biological triplicates using RNA samples extracted from three independent plant materials grown under identical conditions. The threshold cycle (CT) was automatically determined for each reaction by the system set with default parameters. The reaction specificity was determined by the melt curve analysis of amplified products using the standard method installed in the system.
ICE Protein Stability Assay
Five-day-old 35S:HA-ICE1 ProXVE:COP1-MYC plants grown on MS-agar plates were incubated in MS liquid medium supplemented with 50 μM MG132 and/or 25 μM β-estradiol for 16 h in the light. Seedlings were washed three times with fresh MS liquid and transferred to MS liquid medium containing 0.5 mM cycloheximide under either light or dark conditions. Total proteins were extracted from whole seedlings. Immunoblot hybridization was conducted using specific antibodies (anti-HA antibody, Millipore, catalog no. 05-904; anti-MYC antibody, Millipore, catalog no. 05-724), as described previously (Lee et al., 2015).
Fluorescence Imaging and Microscopy Analysis
A Leica SP8 confocal laser scanning microscope was used to capture propidium iodide (PI) staining and GFP fluorescence images. Reconstitution of GFP fluorescence was observed with the following filter setup: excitation of 561 nm and emission of 570 to 650 nm for PI; excitation of 488 nm and emission of 490 to 540 nm for GFP using a HyD detector. Images were taken at the same laser intensity to compare intensities between genotypes. To counterstain epidermal cell shapes, plant tissues were stained with 50 μg/mL PI for 2 min and rinsed briefly with deionized water before visualization (Kang et al., 2009).
COP1-ICE Interaction
Yeast two-hybrid assays were conducted using the BD Matchmaker system (Clontech). The pGADT7 vector was used for GAL4 AD (activation domain), and the pGBKT7 vector was used for GAL4 BD (DNA binding domain). Yeast strain AH109 (leu−, trp−, ade−, his−), which harbors chromosomally integrated reporter genes lacZ and HIS driven by the GAL1 promoter, was used for transformation with the vector constructs. The COP1-ICE interactions were examined by both cell growth on selective media and measurements of β-galactosidase activity according to the system procedures.
BiFC assay was performed essentially as described previously (Lee et al., 2015). The YFPN-COP1 and ICE1-YFPC construct pair and the YFPN-SCRM2 and COP1-YFPC construct pair were coexpressed transiently in Arabidopsis mesophyll protoplasts.
For coimmunoprecipitation, the 35S:HA-ICE1 ProXVE:COP1-MYC plants grown on MS-agar plates for 5 d were incubated in the presence of 25 μM β-estradiol and 50 μM MG132 for 1 d in darkness before extracting total proteins from whole seedlings. Protein complexes were coimmunoprecipitated using an anti-HA antibody (Sigma-Aldrich), and the precipitates were analyzed immunologically using anti-HA and anti-MYC antibodies (Millipore).
Colocalization assays were performed by cotransfection of non-tagged COP1 and GFP-ICE1 constructs into Arabidopsis mesophyll protoplasts by a polyethylene glycol-calcium transfection method (Yoo et al., 2007). The subcellular distribution of GFP-ICE1 was visualized by fluorescence microscopy.
ICE Ubiquitination
Recombinant ProS2-ICE and MBP-COP1 fusion proteins were prepared in Escherichia coli cells. In vitro ubiquitination assays were performed using 400 ng ProS2-ICE and 200 ng MBP–COP1 proteins in a reaction buffer (50 mM Tris, pH 7.4, 5 mM MgCl2, 2 mM ATP, and 2 mM DTT) containing 100 ng human E1 (Boston Biochem), 100 ng human E2 UbcH5b (Boston Biochem), and 1 μg ubiquitin (Sigma-Aldrich). Recombinant MBP-COP1 protein was pretreated with 20 μM ZnCl2 before assays. Reactions were performed at 30°C for 2.5 h with gentle shaking and terminated by adding SDS-PAGE loading buffer. The reaction mixtures were separated on 6% SDS-PAGE gels, and ubiquitinated ProS2-ICE proteins were detected using an anti-ProS2 (Takara Bio) or an anti-ubiquitin (Sigma-Aldrich) antibody. Ubiquitination assays in vivo were performed as described previously (Kim et al., 2015).
Stomatal Index Measurement
SI is given as the percentage of the number of stomata compared with the total number of pavement cells per unit area of leaf (Kang et al., 2009). SI was measured using the central area of the abaxial epidermis of cotyledons. Only stomata having visible pores were counted.
Statistical Analysis
The statistical significance between two means of measurements were determined using Student’s t test with P values of <0.01.
Accession Numbers
Sequence data from this article can be obtained from the Arabidopsis Genome Initiative databases under the following accession numbers: eIF4A (At3g13920), ICE1 (At3g26744), SCRM2/ICE2 (At1g12860), COP1 (At2g32950), SPCH (At5g53210), MUTE (At3g06120), FAMA (At3g24140), YDA (At1g63700), EPF2 (At1g34245), TMM (At1g80080), and ERECTA (At2g26330).
Supplemental Data
Supplemental Figure 1. Effects of Light on ICE Transcription.
Supplemental Figure 2. Effects of Light on ICE1 Accumulation in Different Plant Organs.
Supplemental Figure 3. Control Reactions for Bimolecular Fluorescence Complementation.
Supplemental Figure 4. COP1 Does Not Interact with SPCH, MUTE, and FAMA.
Supplemental Figure 5. Autoubiquitination Activity of COP1 in Vitro.
Supplemental Figure 6. COP1 Does Not Ubiquitinate SPCH, MUTE, and FAMA.
Supplemental Figure 7. Stomatal Phenotypes of ice1-2 scrm2-2 Mutants Harboring the cop1-5 Mutation.
Supplemental Figure 8. Functional Relationship between YDA and ICE1 in Light-Induced Stomatal Development.
Supplemental Table 1. Primers Used.
Supplemental File 1. Original Images of Representative Immunoblots.
Acknowledgments
This work was supported by the National Research Foundation of Korea programs Leaping Research (NRF-2015R1A2A1A05001636) and Global Research Lab (NRF-2012K1A1A2055546) and by the Rural Development Administration of Korea Next-Generation BioGreen 21 Program (PJ0111532015).
AUTHOR CONTRIBUTIONS
C.-M.P. designed the experiments. C.-M.P. wrote the manuscript with contributions from J.-H.L. J.-H.L. performed most key experiments. C.-M.P. and J.-H.L. analyzed the data. J.-H.J. provided scientific discussions.
References
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Balcerowicz M., Ranjan A., Rupprecht L., Fiene G., Hoecker U. (2014). Auxin represses stomatal development in dark-grown seedlings via Aux/IAA proteins. Development 141: 3165–3176. [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497. [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Sack F.D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58: 163–181. [DOI] [PubMed] [Google Scholar]
- Boccalandro H.E., Rugnone M.L., Moreno J.E., Ploschuk E.L., Serna L., Yanovsky M.J., Casal J.J. (2009). Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiol. 150: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S.A., Franklin K.A., Gray J.E., Grierson C.S., Whitelam G.C., Hetherington A.M. (2009). phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr. Biol. 19: 229–234. [DOI] [PubMed] [Google Scholar]
- Casson S.A., Hetherington A.M. (2014). phytochrome B Is required for light-mediated systemic control of stomatal development. Curr. Biol. 24: 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crawford A.J., McLachlan D.H., Hetherington A.M., Franklin K.A. (2012). High temperature exposure increases plant cooling capacity. Curr. Biol. 22: R396–R397. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Bergmann D.C. (2010). Stomatal patterning and development. Curr. Top. Dev. Biol. 91: 267–297. [DOI] [PubMed] [Google Scholar]
- Driscoll S.P., Prins A., Olmos E., Kunert K.J., Foyer C.H. (2006). Specification of adaxial and abaxial stomata, epidermal structure and photosynthesis to CO2 enrichment in maize leaves. J. Exp. Bot. 57: 381–390. [DOI] [PubMed] [Google Scholar]
- Engineer C.B., Ghassemian M., Anderson J.C., Peck S.C., Hu H., Schroeder J.I. (2014). Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., Nadeau J., Sack F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M.J., Deppong D.O., Nadeau J.A., Sack F.D. (2003). Stomatal neighbor cell polarity and division in Arabidopsis. Planta 216: 571–579. [DOI] [PubMed] [Google Scholar]
- Gray J.E., Holroyd G.H., van der Lee F.M., Bahrami A.R., Sijmons P.C., Woodward F.I., Schuch W., Hetherington A.M. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716. [DOI] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., Torii K.U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Hetherington A.M., Woodward F.I. (2003). The role of stomata in sensing and driving environmental change. Nature 424: 901–908. [DOI] [PubMed] [Google Scholar]
- Horst R.J., Fujita H., Lee J.S., Rychel A.L., Garrick J.M., Kawaguchi M., Peterson K.M., Torii K.U. (2015). Molecular framework of a regulatory circuit initiating two-dimensional spatial patterning of stomatal lineage. PLoS Genet. 11: e1005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869. [DOI] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Park J.H., Lee S., To T.K., Kim J.M., Seki M., Park C.M. (2013). The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25: 4378–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.Y., Lian H.L., Wang F.F., Huang J.R., Yang H.Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Kim S.G., Park J.E., Park H.Y., Lim M.H., Chua N.H., Park C.M. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Lee M., Lee J.H., Lee H.J., Park C.M. (2015). The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 89: 187–201. [DOI] [PubMed] [Google Scholar]
- Klermund C., Ranftl Q.L., Diener J., Bastakis E., Richter R., Schwechheimer C. (2016). LLM-domain B-GATA transcription factors promote stomatal development downstream of light signaling pathways in Arabidopsis thaliana hypocotyls. Plant Cell 28: 646–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Lawson T., Blatt M.R. (2014). Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 164: 1556–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. (1998). Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8: 397–403. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Jung J.H., Park C.M. (2015). INDUCER OF CBF EXPRESSION 1 integrates cold signals into FLOWERING LOCUS C-mediated flowering pathways in Arabidopsis. Plant J. 84: 29–40. [DOI] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.D., Zhou C.M., Xu P.B., Luo Q., Lian H.L., Yang H.Q. (2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8: 467–478. [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540. [DOI] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Pacín M., Legris M., Casal J.J. (2014). Rapid decline in nuclear costitutive photomorphogenesis1 abundance anticipates the stabilization of its target elongated hypocotyl5 in the light. Plant Physiol. 164: 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505. [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63: 591–614. [DOI] [PubMed] [Google Scholar]
- Raissig M.T., Abrash E., Bettadapur A., Vogel J.P., Bergmann D.C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. USA 113: 8326–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichler S.A., Balk J., Brown M.E., Woodruff K., Clark G.B., Roux S.J. (2001). Light differentially regulates cell division and the mRNA abundance of pea nucleolin during de-etiolation. Plant Physiol. 125: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293. [DOI] [PubMed] [Google Scholar]
- Stacey M.G., Hicks S.N., von Arnim A.G. (1999). Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11: 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Torii K.U., McNellis T.W., Deng X.W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17: 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker P.J., Gibbings J.G., Rodríguez López C.M., Hadley P., Wilkinson M.J. (2012). Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J. Exp. Bot. 63: 3799–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M.K., Czechowski T., Scheible W.R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A.G., Deng X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045. [DOI] [PubMed] [Google Scholar]
- von Arnim A.G., Osterlund M.T., Kwok S.F., Deng X.W. (1997). Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 114: 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C., Miki D., Zheng Z., Ma J., Wang J., Yang Z., Dong J., Zhu J.K. (2014). Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat. Commun. 5: 4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]