A leaf senescence accelerator involving the transcription factor WRKY75, salicylic acid, and H2O2 is driven by three interlinking positive feedback loops.
Abstract
Leaf senescence is a highly coordinated, complicated process involving the integration of numerous internal and environmental signals. Salicylic acid (SA) and reactive oxygen species (ROS) are two well-defined inducers of leaf senescence whose contents progressively and interdependently increase during leaf senescence via an unknown mechanism. Here, we characterized the transcription factor WRKY75 as a positive regulator of leaf senescence in Arabidopsis thaliana. Knockdown or knockout of WRKY75 delayed age-dependent leaf senescence, while overexpression of WRKY75 accelerated this process. WRKY75 transcription is induced by age, SA, H2O2, and multiple plant hormones. Meanwhile, WRKY75 promotes SA production by inducing the transcription of SA INDUCTION-DEFICIENT2 (SID2) and suppresses H2O2 scavenging, partly by repressing the transcription of CATALASE2 (CAT2). Genetic analysis revealed that the mutation of SID2 or an increase in catalase activity rescued the precocious leaf senescence phenotype evoked by WRKY75 overexpression. Based on these results, we propose a tripartite amplification loop model in which WRKY75, SA, and ROS undergo a gradual but self-sustained rise driven by three interlinking positive feedback loops. This tripartite amplification loop provides a molecular framework connecting upstream signals, such as age and plant hormones, to the downstream regulatory network executed by SA- and H2O2-responsive transcription factors during leaf senescence.
INTRODUCTION
Leaf senescence is a common developmental phenomenon observed in nature that helps create amazing scenery in autumn. This last stage of leaf development ultimately leads to leaf death (Gan and Amasino, 1997; Lim et al., 2007). During the senescence of plant organs, nutrients released from the senescent leaves are recycled to actively growing young leaves, developing fruits, and seeds (Lim et al., 2007; Zhou et al., 2009). This crucial and complicated process is influenced by both environmental factors such as nutrient deficiency, drought or salt stress, extreme temperatures, pathogen attack, and internal factors including age, phytohormones, reactive oxygen species (ROS), and reproduction (Gan and Amasino, 1995; Beers and McDowell, 2001; Lim et al., 2007; Woo et al., 2013). Among phytohormones, cytokinins and auxins delay leaf senescence, while ethylene, salicylic acid (SA), abscisic acid (ABA), and jasmonates (JA) accelerate leaf aging (Lim et al., 2007; Jibran et al., 2013).
SA has long been known to play a promoting role in natural leaf senescence (Buchanan-Wollaston et al., 2005; Lim et al., 2007; Rivas-San Vicente and Plasencia, 2011). The endogenous SA concentration increases gradually as a leaf ages, which induces the expression of several senescence-associated genes (SAGs) during leaf senescence (Morris et al., 2000; Lim et al., 2007). Arabidopsis thaliana plants with defects in the SA response or biosynthetic pathway, such as npr1 (NONEXPRESSER OF PR GENES1) and pad4 (PHYTOALEXIN DEFICIENT4) mutants and NahG transgenic plants, exhibit considerably reduced expression of SAGs and delayed senescence phenotypes (Morris et al., 2000; Lim et al., 2007). Additionally, autophagy-defective mutants (atg3 and atg5), mutants of pat14 (PROTEIN S-ACYL TRANSFERASE14) or s3h (SA3-HYDROXYLASE), all of which accumulate high levels of free SA, display precocious leaf senescence phenotypes (Yoshimoto et al., 2009; Zhao et al., 2016; Zhang et al., 2013). Another interesting observation is that SA treatment and age-dependent leaf senescence showed a high degree of overlap in genome-wide transcriptome analysis (Lim et al., 2007), further supporting the stimulating effect of SA on leaf senescence.
High levels of endogenous H2O2, which are generated by a variety of environmental stresses such as excess light irradiation, drought, or high salinity, and extreme temperatures, as well as some critical developmental changes, are also responsible for accelerated leaf senescence (Apel and Hirt, 2004; Mittler et al., 2004; Khanna-Chopra, 2012). This elevated H2O2 content might lead to increases in protein and lipid oxidation and dysfunction as leaf aging proceeds, which represents a component of senescence-related syndrome (Vanacker et al., 2006). Arabidopsis mutants that generate excessive amounts of H2O2, such as cpr5 (CONSTITUTIVE EXPRESSION OF PR GENES5) and jub1 (JUNGBRUNNEN1), exhibit precocious leaf senescence (Jing et al., 2008; Wu et al., 2012). Conversely, mutants with reduced H2O2 accumulation, such as ntl4 (NAC WITH TRANSMEMBRANE MOTIF 1-LIKE4) and aaf-KO (ARABIDOPSIS A-FIFTEEN knock-out), show delayed drought-induced and age-dependent leaf senescence, respectively (Lee et al., 2012; Chen et al., 2012). Interestingly, H2O2 induces the accumulation of SA, and vice versa. For instance, exogenous application of H2O2 induces SA biosynthesis in tobacco (Nicotiana tabacum) leaves via a yet unknown mechanism (Leon et al., 1995). Meanwhile, SA directly inactivates catalases (CATs) and ascorbate peroxidases (APXs), two types of H2O2 scavengers, thus leading to H2O2 accumulation (Durner and Klessig, 1995, 1996; Rao et al., 1997).
Genomic studies have revealed that dozens of transcription factors (TFs) are highly upregulated as leaf senescence progresses (Woo et al., 2016). The roles of many of these so-called sen-TFs in the regulation of leaf senescence have recently been functionally characterized. For instance, several WRKY TFs, such as Arabidopsis WRKY6, WRKY22, WRKY53, WRKY54, WRKY57, and WRKY70, play either positive or negative roles in leaf senescence (Robatzek and Somssich, 2001; Miao et al., 2004; Miao and Zentgraf, 2007; Zentgraf et al., 2010; Zhou et al., 2011; Besseau et al., 2012; Jiang et al., 2014). We previously developed a leaf senescence database, wherein 5357 SAGs and 324 senescence-altered mutants were collected from 44 species (Liu et al., 2011; Li et al., 2014). Using data mining combined with phenotypic screening, WRKY75 was identified as another sen-TF that positively regulates leaf senescence (Li et al., 2012). In this study, we investigated the molecular mechanism underlying WRKY75-regulated leaf senescence in Arabidopsis. We found that, on one hand, SA and H2O2 increased the expression of WRKY75; on the other hand, WRKY75 increased SA production by inducing SA INDUCTION-DEFICIENT2 (SID2) transcription and repressing ROS scavenging by suppressing CAT2 transcription. Genetic studies revealed that concomitant WRKY75-induced SA biosynthesis and H2O2 accumulation is critical for accelerating the progression of leaf senescence. Thus, our study establishes a tripartite amplification loop involving WRKY75, SA, and ROS, which undergo mutual promotion via distinct mechanisms, providing insights into the molecular regulatory network of leaf senescence.
RESULTS
WRKY75 Transcription Is Induced by Age, ROS, and Multiple Phytohormones
We previously showed that WRKY75 promotes the natural leaf senescence process (Li et al., 2012), but the underlying molecular mechanism has been unclear. To this end, we first investigated how the expression of WRKY75 is regulated. Arabidopsis leaves at four different developmental stages, including young, mature, early stage of senescence, and late stage of senescence, were used to analyze WRKY75 transcript levels. Similar to WRKY53 and SAG12, two well-known senescence-induced genes (Noh and Amasino, 1999; Hinderhofer and Zentgraf, 2001; Miao et al., 2004) (Supplemental Figures 1A and 1B), WRKY75 expression gradually increased during the progression of leaf senescence (Figure 1A). To further verify the age-dependent regulation of WRKY75, we generated transgenic plants expressing a GUS (β-glucuronidase) gene driven by the WRKY75 promoter containing a 1.5-kb genomic sequence upstream of the start codon (ProWRKY75:GUS/Col-0). GUS staining analysis of 30-d-old ProWRKY75:GUS/Col-0 rosette leaves showed that yellowing leaves displayed higher GUS activity than green leaves (Figure 1B), supporting the finding that WRKY75 transcription is upregulated during leaf aging.
Figure 1.
WRKY75 Is a Senescence-Associated Gene Induced by SA and H2O2.
(A) Transcript levels of WRKY75 increase in an age-dependent manner. Y, young leaves of 10-d-old seedlings; M, fully expanded mature leaves; ES, early senescence leaves, with <25% leaf area yellowing; LS, late senescence leaves, with >50% leaf area yellowing. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(B) GUS staining of rosette leaves of 30-d-old plant harboring the GUS transgene driven by the WRKY75 promoter (ProWRKY75:GUS/Col-0). Arrows show senescent leaf areas with higher GUS activity.
(C) GUS staining of 10-d-old ProWRKY75:GUS/Col-0 seedlings treated with SA or H2O2. Seedlings were treated with water (Mock), 500 µM SA, or 10 mM H2O2 for 2 h before GUS staining.
(D) and (E) qRT-PCR analysis of WRKY75 expression in 10-d-old Col-0 seedlings treated with 500 µM SA (D) or 10 mM H2O2 (E) for the indicated time. Data are represented as means ± sd, n = 3. The experiments were performed three times with similar results.
We also investigated the effects of other senescence-regulating signals such as plant hormones and ROS on WRKY75 transcription. We found that treatment with SA and H2O2 greatly enhanced WRKY75 transcription in ProWRKY75:GUS/Col-0 transgenic plants (Figure 1C) compared with mock-treated plants, which was further confirmed by quantitative analysis of WRKY75 transcript levels in wild type (Col-0) plants (Figures 1D and 1E). The induction of PR1 (PATHOGENESIS-RELATED GENE1; an SA-responsive gene) (Blanco et al., 2009) and DEFL (DEFENSIN-LIKE; an H2O2-responsive gene) (Gadjev et al., 2006) ensured that the treatments were effective (Supplemental Figures 1C and 1D). Other hormones, such as ethylene, JA, and ABA, induced, whereas cytokinins repressed, WRKY75 expression (Supplemental Figure 2A). Furthermore, we found that ethylene-induced WRKY75 expression was largely diminished in ein3 eil1, an ethylene-insensitive mutant, similar to that of ERF1, a well-known target gene of EIN3/EIL1 (Solano et al., 1998) (Supplemental Figures 2B and 2C). Consistently, six putative EIN3 binding elements (Itzhaki et al., 1994; An et al., 2012; Li et al., 2013) (TTCAAA, E1 to E6) were found in the promoter of WRKY75 (Supplemental Figure 2D). Chromatin immunoprecipitation (ChIP) experiments using inducible EIN3-FLAG/ein3 eil1 ebf1 ebf2 (iE/qm) plants (An et al., 2012) revealed that EIN3 was able to bind to the promoter of WRKY75, particularly in the E1 and E4 regions (Supplemental Figure 2E). Collectively, these results suggest that WRKY75 is a direct target gene of EIN3.
WRKY75 Positively Regulates Leaf Senescence in an Age-Dependent Manner
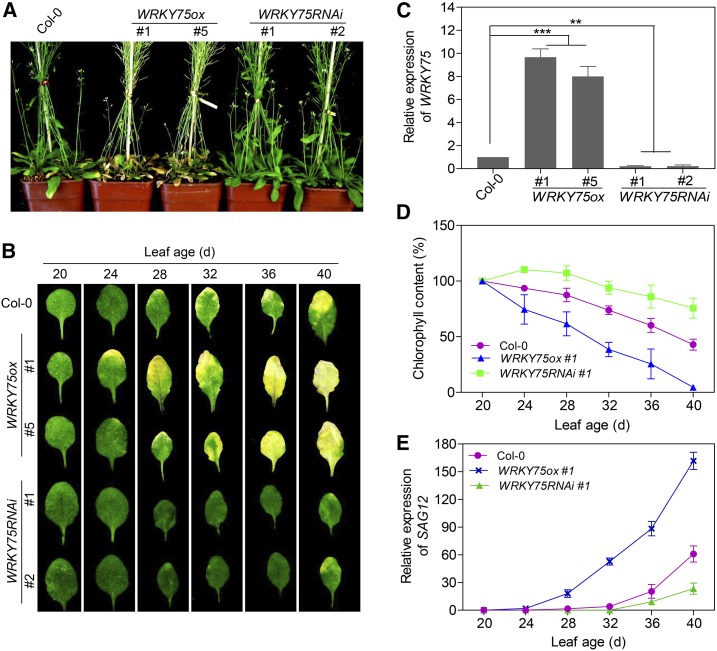
We previously found that the reduction of WRKY75 expression led to a delayed leaf senescence phenotype in WRKY75RNAi plants (Li et al., 2012). In this study, we generated WRKY75-overexpressing transgenic plants (WRKY75ox) harboring the WRKY75 coding sequence driven by the 35S promoter. Quantitative RT-PCR analysis showed that WRKY75 transcript levels were 8- to 10-fold that of the wild type (Col-0) in WRKY75ox but only ∼10% that of Col-0 in WRKY75RNAi plants (Figure 2C). In agreement with their respective expression levels, WRKY75ox plants displayed precocious leaf senescence symptoms, while WRKY75RNAi plants exhibited a delayed senescence phenotype (Figures 2A and 2B). WRKY75ox rosette leaves started to turn yellow from the tip at 24 d old, whereas Col-0 and WRKY75RNAi leaves began to turn yellow at 32 and 40 d old, respectively (Figure 2B). Consistently, the decline in chlorophyll contents and the expression of SAG12 upon leaf aging showed a similar pattern to the senescence phenotypes among the three genotypes (Figures 2D and 2E). Together, these results suggest that WRKY75 positively regulates leaf senescence in an age-dependent manner.
Figure 2.
WRKY75 Positively Regulates Leaf Senescence in an Age-Dependent Manner.
(A) The senescence phenotype of 40-d-old Col-0, WRKY75-knockdown plants obtained via RNAi (WRKY75RNAi), and WRKY75 overexpressing plants (WRKY75ox). Two independent lines of each transgenic plant were examined.
(B) The fourth rosette leaves detached from whole plants at the indicated ages.
(C) qRT-PCR analysis of WRKY75 expression in 7-d-old Col-0, WRKY75RNAi, and WRKY75ox plants. Student’s t test, **P < 0.01 and ***P < 0.001. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(D) and (E) Chlorophyll contents (D) and SAG12 expression (E) in the leaves shown in (B). WRKY75RNAi #1 and WRKY75ox #1 were used for all experiments involving the two types of transgenic plants in subsequent analyses. Data are represented as means ± sd, n = 3. The experiments were performed three times with similar results.
We recently obtained the previously reported T-DNA insertion line N121525 (wrky75-25) (Rishmawi et al., 2014), which is likely a null allele, as WRKY75 transcript was not detected in the mutant line (Supplemental Figures 3A and 3B). As expected, the wrky75-25 mutants exhibited a delayed leaf senescence phenotype (Supplemental Figures 3C and 3D). Meanwhile, we sought to generate additional wrky75 mutant alleles using the CRISPR-Cas9 system (Feng et al., 2013), in which the target sequence is located in the second exon that encodes part of the WRKY75 DNA binding domain (Supplemental Figure 4A). Two wrky75 mutant alleles were obtained, designated wrky75-c1 (with a four-base deletion) and wrky75-c2 (with a single-base insertion leading to a premature stop codon) (Supplemental Figures 4B and 4C). Phenotype analysis revealed that WRKY75RNAi, wrky75-25, and wrky75-c1 showed comparable delayed senescence phenotypes (Supplemental Figures 5A and 5B), providing clear genetic evidence supporting a positive role for WRKY75 in regulating leaf senescence. Additionally, we observed a weak delayed-flowering phenotype in WRKY75 knockout or knockdown mutants (Supplemental Figure 5C), suggesting that WRKY75 promotes the reproductive transition coupled with leaf aging, which in turn shortens a plant’s lifespan.
To further investigate the regulatory role of WRKY75 in leaf senescence, we generated transgenic plants expressing estradiol-inducible WRKY75-HA (iWRKY75-HA) (Supplemental Figures 6A and 6C). While no obvious effect was observed in young iWRKY75-HA plants treated with estradiol, the tips of some leaves started to turn yellow in mature iWRKY75-HA plants after 1 week of estradiol treatment (Supplemental Figure 6B). Consistently, a decline in chlorophyll contents was observed only in mature estradiol-treated leaves (Supplemental Figure 6D). Together, these results suggest that temporary induction of WRKY75 is sufficient to promote leaf senescence in an age-dependent manner.
WRKY75 Positively Regulates SA Production and Disease Resistance
Next, we sought to explore how WRKY75 promotes leaf senescence. As expected, WRKY75 is a nuclear protein, as evidenced by the subcellular localization of GFP in transgenic plants expressing WRKY75-GFP fusion protein (Supplemental Figure 7). We then performed transcriptome profile analysis of 28-d-old Col-0 and WRKY75RNAi leaves by RNA-seq. Compared with the wild type, 998 genes were downregulated (log2 less than or equal to −1, P < 0.01), while 1360 genes were upregulated (log2 greater than or equal to 1, P < 0.01) in WRKY75RNAi leaves (Supplemental Data Set 1). Gene Ontology enrichment analysis of these differentially expressed genes showed that the downregulated genes are highly enriched in functional categories, including those related to biotic stress, SA-related biological processes, oxidative stress, and hydrogen peroxide-mediated programmed cell death (Supplemental Figure 8).
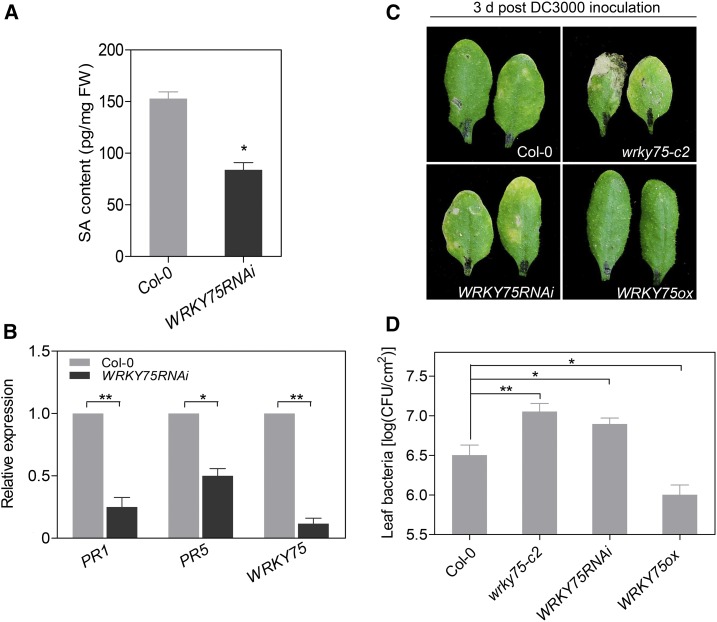
Next, we measured the SA contents in WRKY75RNAi and Col-0 plants. The free SA levels in WRKY75RNAi leaves were approximately half that in Col-0 (Figure 3A). Consistently, the expression of PR1 and PR5, two widely used SA-responsive genes (Blanco et al., 2009), was significantly downregulated in WRKY75RNAi (Figure 3B). Given that SA biosynthesis is strongly induced upon pathogen infection and reduced SA production enhances susceptibility to bacterial pathogens (Nawrath and Métraux, 1999; Chen et al., 2009), we performed a disease resistance experiment against pathogenic bacteria Pseudomonas syringae pv tomato DC3000 (Pst DC3000) (Xin and He, 2013). Upon inoculation with DC3000 for 3 d, the yellowish areas around the inoculation sites were larger in 3-week-old WRKY75RNAi and wrky75-c2 leaves and smaller in WRKY75ox leaves compared with Col-0 (Figure 3C). We counted the bacterial populations in the inoculated leaves, finding much greater bacterial growth in WRKY75RNAi and wrky75-c2 and less bacterial growth in WRKY75ox plants (Figure 3D) compared with the wild type, indicating that WRKY75 positively regulates resistance to Pst DC3000. Taken together, these results suggest that WRKY75 positively regulates SA production and bacterial resistance in Arabidopsis.
Figure 3.
WRKY75 Is Involved in SA Biosynthesis and Plant Defense Responses.
(A) Measurement of free SA levels in the third and fourth rosette leaves from 32-d-old plants. Student’s t test, *P < 0.05. Data are represented as means ± sd, n = 3.
(B) qRT-PCR analysis of transcript levels of PR1 and PR5 in leaves (A). Student’s t test, *P < 0.05 and **P < 0.01. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(C) Phenotypes of 3-week-old Col-0, wrky75-c2, WRKY75RNAi, and WRKY75ox leaves inoculated with DC3000 bacteria for 3 d. The experiment was performed twice, with similar results.
(D) The bacterial populations in inoculated leaves shown in (C). Student’s t test, *P < 0.05 and **P < 0.01. Data are represented as means ± sd, n = 8.
WRKY75 Promotes SA Biosynthesis by Directly Activating SID2 Transcription
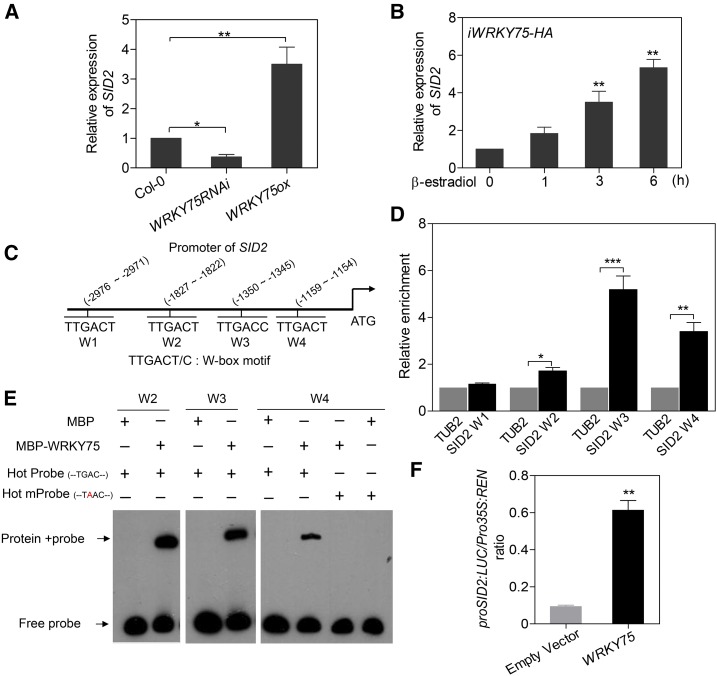
Based on the above data, we hypothesized that WRKY75 might promote SA biosynthesis or metabolism. Two distinct SA biosynthesis pathways have been proposed in plants, the isochorismate pathway, which is the major SA biosynthesis pathway in Arabidopsis, and the phenylalanine ammonia-lyase pathway (Dempsey et al., 2011). Two genes, ISOCHORISMATE SYNTHASE1 (Wildermuth et al., 2001), also called SID2 (Nawrath and Métraux, 1999), and PHE AMMONIA LYASE1 (PAL1) (Cochrane et al., 2004), encode key enzymes of the two pathways, respectively. We thus examined the expression of the two genes and found that SID2 transcript levels markedly declined in WRKY75RNAi leaves (Figure 4A), whereas PAL1 transcript levels did not (Supplemental Figure 9A), which is consistent with the RNA-seq data. Conversely, SID2 transcript levels increased in WRKY75ox plants compared with Col-0 (Figure 4A). A time-course analysis revealed that SID2 expression was rapidly induced in iWRKY75-HA plants upon treatment with β-estradiol (Figure 4B). We also examined other previously reported regulators of SID2 expression, such as WRKY28 (van Verk et al., 2011), CAM BINDING PROTEIN 60-LIKE G (CBP60G), and SAR DEFICIENT1 (SARD1) (Zhang et al., 2010; Wang et al., 2011) and found that the expression of CBP60G and SARD1 but not WRKY28 significantly differed in WRKY75RNAi compared with Col-0 (Supplemental Figure 9A), suggesting that WRKY75 might directly or indirectly induce SID2 expression.
Figure 4.
WRKY75 Activates the SID2 Transcription.
(A) qRT-PCR analysis of SID2 expression in the third and fourth rosette leaves of 32-d-old plants. Student’s t test, *P < 0.05 and **P < 0.01. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(B) qRT-PCR analysis of SID2 expression in iWRKY75-HA plants treated with 20 µM β-estradiol for the indicated time. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(C) Schematic diagram indicating the locations of four putative WRKY75 binding sites (W1 to W4) in the promoter of SID2.
(D) ChIP-qPCR analysis of relative WRKY75 binding to the promoter of SID2. An anti-GFP monoclonal antibody was used for DNA immunoprecipitation from 32-d-old Pro35S:WRKY75-GFP/Col-0 transgenic plants. Black bars indicate the enrichment fold changes normalized to TUB2. Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(E) EMSA analysis of the binding of recombinant WRKY75 protein to the promoter of SID2 (W2 to W4). Hot Probe is biotin-labeled, while mProbe contains a single nucleic acid mutation from TGAC to TAAC.
(F) Transient dual-luciferase reporter assay. Construct pGreenII-0800 LUC containing the SID2 promoter and construct p62-SK with or without the WRKY75 coding region were transiently cotransformed into Col-0 protoplasts. Firefly luciferase (LUC) and Renilla luciferase (REN) activity were measured after culturing the protoplasts under low-light conditions for 16 h. The ProSID2:LUC/Pro35S:REN ratio represents the relative activity of SID2 transcription. Student’s t test, **P < 0.01. Data are represented as means ± sd, n = 6. The experiment was performed three times with similar results.
To assess whether WRKY75 directly binds to the SID2 promoter to regulate its transcription, we performed ChIP experiments using Pro35S:WRKY75-GFP/Col-0 plants. As WRKY TFs specifically bind to the W-box (TTGACT) sequence, we identified four putative W-box sequences in the SID2 promoter, which were designated as SID2 W1 to W4, respectively (Figure 4C). The ChIP-PCR results showed significant enrichment of WRKY75 in three putative W-box regions, particularly the W3 and W4 sites (Figure 4D), indicating that WRKY75 binds to these regions in vivo. Additionally, we performed electrophoretic mobility shift assays (EMSAs) to determine the in vitro binding of WRKY75 to these regions. Full-length WRKY75 protein tagged with maltose binding protein (MBP) (MBP-WRKY75) was capable of binding to probes containing W2, W3, or W4, while MBP alone was not (Figure 4E). When the W4 probe harboring a single nucleic acid mutation (TTAACT) was used for EMSA, the binding was abolished (Figure 4E). Collectively, these results demonstrate that WRKY75 directly binds to the promoter of SID2.
We further investigated the positive regulation of WRKY75 on SID2 transcription using a dual-luciferase (LUC) reporter assay. The SID2 promoter-driven firefly luciferase reporter (ProSID2:LUC) and 35S promoter-driven Renilla luciferase (Pro35S:REN; as an internal control) were introduced into the same plasmid and transiently expressed in Arabidopsis protoplasts. Another plasmid with or without the WRKY75 coding region was coexpressed into the protoplasts, followed by monitoring of the LUC:REN ratio, which reflects the transcriptional activity of the SID2 promoter in vivo. We found that coexpression of WRKY75 and ProSID2:LUC/Pro35S:REN markedly increased the LUC:REN ratio (Figure 4F), indicating that WRKY75 is able to activate the SID2 promoter-driven transcription.
Additionally, we also examined the expression of key genes involved in SA modification or metabolism, such as BSMT1, SAGT1, and PBS3 (Dempsey et al., 2011), in WRKY75RNAi leaves. No significant changes were observed, except for elevated expression of BSMT1, encoding an enzyme that catalyzes the formation of methyl salicylate (Chen et al., 2003) (Supplemental Figure 9B). Taken together, these results suggest that WRKY75 promotes SA production, mainly by activating SID2 transcription.
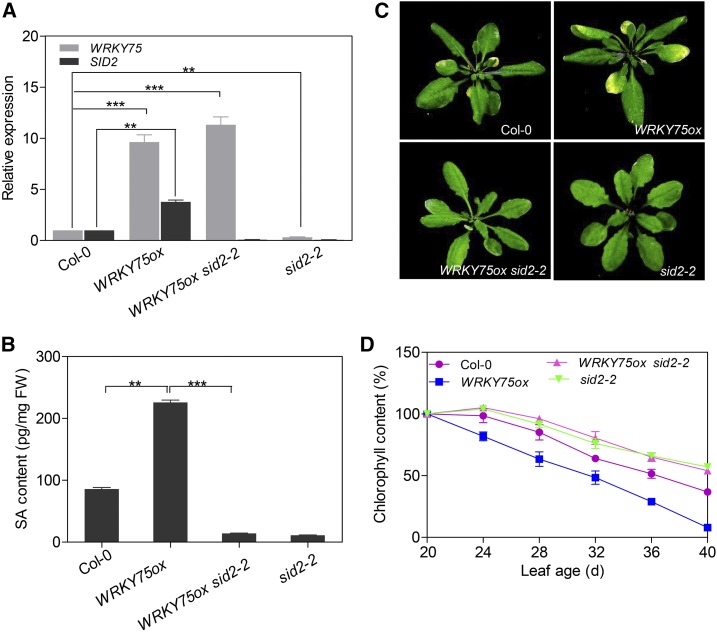
SID2 Mutation Suppresses the Elevated SA Content and Accelerated Senescence Phenotype of WRKY75ox
To explore the genetic relationship between WRKY75 and SID2 in regulating leaf senescence, we generated WRKY75ox sid2-2 plants by crossing the sid2-2 mutant with the WRKY75ox transgenic line (Figure 5A). In line with the finding that SID2 is a downstream target gene of WRKY75, the elevated SA production in WRKY75ox plants was abolished by sid2 mutation (Figure 5B). The free SA levels in WRKY75ox sid2-2 plants were as low as that in sid2-2 (Figure 5B), suggesting that the higher SA accumulation in WRKY75ox is mainly attributed to the induction of SID2 expression. Likewise, the accelerated leaf senescence phenotype of WRKY75ox was also suppressed by sid2 mutation (Figure 5C). Measurement of chlorophyll contents further confirmed this genetic interaction (Figure 5D). Taken together, we conclude that the loss of SID2 function suppresses the elevated SA production and accelerated leaf senescence phenotype of WRKY75ox.
Figure 5.
Mutation of SID2 Suppresses the Early Leaf Senescence Phenotype of WRKY75ox Plants.
(A) qRT-PCR analysis of WRKY75 and SID2 expression in the third and fourth rosette leaves from 28-d-old plants. Student’s t test, **P < 0.01 and ***P < 0.001. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(B) Measurement of free SA levels in the leaves shown in (A). Student’s t test, **P < 0.01 and ***P < 0.001. Data are represented as means ± sd, n = 3.
(C) The senescence phenotypes of 28-d-old plants with the indicated genotypes.
(D) Chlorophyll contents in Col-0, WRKY75ox, WRKY75ox/sid2-2, and sid2-2 leaves. The third and fourth rosette leaves were used. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
We found that SID2 is also a senescence-associated gene whose transcription increases with leaf aging (Supplemental Figure 10A). As expected, overexpression of SID2 (SID2ox) led to elevated SA production and an accelerated leaf senescence phenotype (Supplemental Figures 10B and 10C). Interestingly, we also detected an increase in WRKY75 expression in SID2ox (Supplemental Figure 10B), which is consistent with the finding that WRKY75 expression is induced by SA (Figure 1D). Accordingly, a decrease in WRKY75 transcript levels was observed in the sid2-2 mutant (Figure 5A), supporting the existence of mutual promotion between SID2-mediated SA production and WRKY75 expression.
WRKY75RNAi Plants Exhibit Enhanced Tolerance to Oxidative Stress
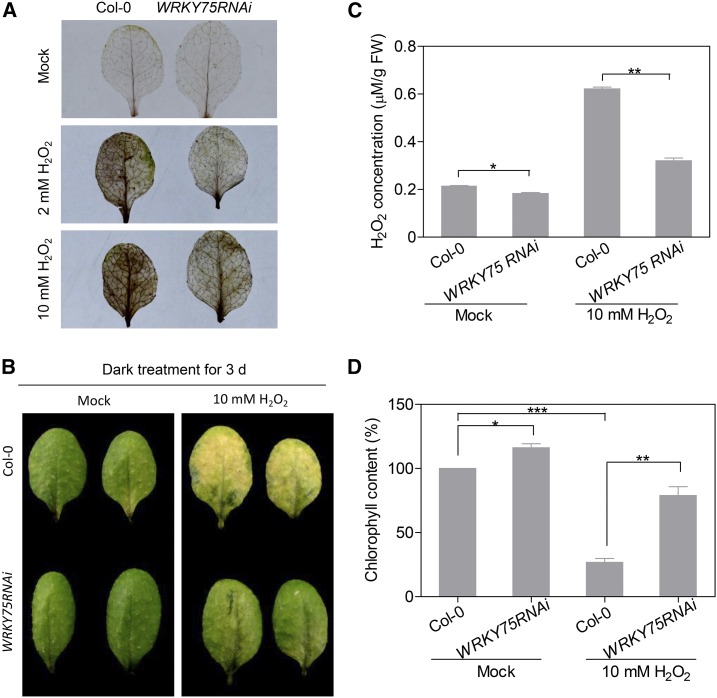
During leaf senescence, endogenous H2O2 levels gradually rise; the accumulated H2O2 plays an important role in the development of senescence symptoms (Mittler et al., 2004; Khanna-Chopra, 2012). Based on our findings that WRKY75 expression is induced by H2O2 and that oxidative stress pathways are downregulated in WRKY75RNAi plants (Supplemental Figure 8), we next investigated whether WRKY75 participates in oxidative stress-induced senescence. Interestingly, the 3,3′-diaminobenzidine (DAB) staining showed that upon exogenous H2O2 treatment, WRKY75RNAi leaves displayed less H2O2 accumulation than Col-0 (Figure 6A), implying that WRKY75RNAi leaves had higher H2O2 scavenging capacity.
Figure 6.
WRKY75RNAi Displays Enhanced Resistance to Oxidative Stress.
(A) DAB staining of detached leaves treated with H2O2. The third and fourth rosette leaves of 3-week-old plants were detached and incubated in MES buffer (2 mM MES, pH 5.8) with the indicated H2O2 concentration for 3 d, followed by DAB staining. The brown color represents H2O2 accumulation.
(B) The senescence phenotypes of detached leaves treated with H2O2 and incubated in darkness for 3 d. The third and fourth rosette leaves of 3-week-old plants were detached and incubated in MES buffer (2 mM MES, pH 5.8) treated with or without H2O2 under dark conditions for 3 d.
(C) and (D) Measurement of H2O2 contents (C) and chlorophyll contents (D) in the leaves shown in (B). Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001. Data are represented as means ± sd, n = 3. The experiments were performed three times with similar results.
We also examined the H2O2-induced senescence phenotype of Col-0 and WRKY75RNAi leaves under dark conditions. When subjected to H2O2 treatment for 3 d, all detached Col-0 leaves turned yellow, while WRKY75RNAi leaves largely remained green (Figure 6B), indicating that WRKY75RNAi plants displayed enhanced tolerance to H2O2-induced leaf senescence. Quantitative measurement also revealed that WRKY75RNAi leaves accumulated less endogenous H2O2 than Col-0 upon exogenous H2O2 application (Figure 6C). Measurement of chlorophyll contents in dark/H2O2-treated Col-0 and WRKY75RNAi leaves reinforced the observed phenotype (Figure 6D). Furthermore, similar results were obtained from comparisons between Col-0 and wrky75-25 (Supplemental Figures 11A and 11B). Conversely, WRKY75ox leaves showed reduced resistance to H2O2 treatment compared with Col-0 (Supplemental Figures 11C and 11D). Taken together, knockdown or knockout of WRKY75 enhances tolerance to oxidative stress (H2O2) and delays leaf senescence.
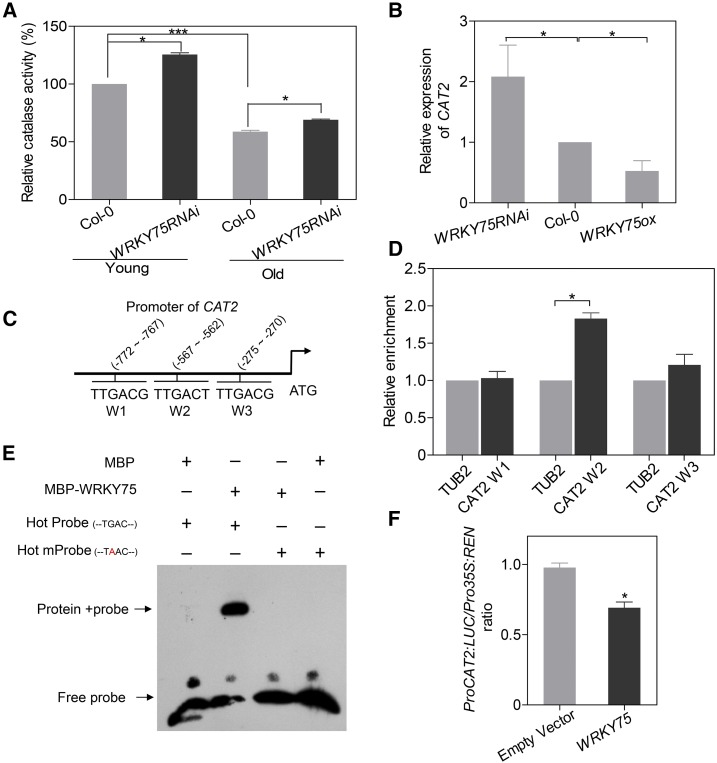
WRKY75 Suppresses Catalase Activity by Repressing CAT2 Transcription
Given that WRKY75RNAi plants showed reduced H2O2 accumulation and enhanced tolerance to exogenous H2O2 treatment (Figure 6), we investigated the relationship between WRKY75 and H2O2 scavengers. Plants contain several antioxidant enzymes or proteins, such as superoxide dismutases, CATs, APXs, thiodoxins (Trxs), and guaiacol peroxidases; CATs and APXs are primarily responsible for H2O2 scavenging (Willekens et al., 1997; Finkel and Holbrook, 2000; Blokhina et al., 2003; Mittler et al., 2004; Khanna-Chopra, 2012). We first measured catalase activity and found that WRKY75RNAi plants exhibited significantly higher catalase activity in both young and old leaves compared with Col-0 (Figure 7A), indicating that WRKY75 suppresses catalase activity. Three catalase genes (CAT1, CAT2, and CAT3) have been annotated in the Arabidopsis genome (Frugoli et al., 1996). We found that the expression of CAT2 and CAT3, but not CAT1, was induced in WRKY75RNAi plants (Figure 7B; Supplemental Figure 12A). Given the finding that CAT2 is the major catalase isoform, accounting for 90% of catalase activity in vivo (Queval et al., 2007; Mhamdi et al., 2010), we focused our analysis on CAT2. As expected, CAT2 expression declined in WRKY75ox transgenic plant but increased in WRKY75RNAi, wrky75-25, and wrky75-c1 plants compared with the wild type (Figure 7B; Supplemental Figure 12B). Interestingly, three putative W-box sequences (designated as CAT2 W1-W3) were found in the promoter of CAT2 (Figure 7C). ChIP experiments revealed significant enrichment of WRKY75 merely in the CAT2 W2 regions (Figure 7D). EMSA results confirmed that WRKY75 was able to directly and specifically bind to the CAT2 W2 sequence (Figure 7E). Moreover, dual-LUC reporter assays revealed that WRKY75 repressed CAT2 transcription in Arabidopsis protoplasts (Figure 7F). Together, these results demonstrate that WRKY75 represses catalase activity by inhibiting CAT2 transcription.
Figure 7.
WRKY75 Suppresses Catalase Activity by Repressing CAT2 Transcription.
(A) Measurement of catalase activity in Col-0 and WRKY75RNAi plants. Ten-day-old (Young) green seedlings and the third and fourth rosette leaves from 32-d-old (Old) plants were used, respectively. Student’s t test, *P < 0.05 and ***P < 0.001. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(B) qRT-PCR analysis of CAT2 expression in the third and fourth rosette leaves from 32-d-old plants. Student’s t test, *P < 0.05. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(C) Schematic diagram indicating the locations of three putative WRKY75 binding sites (W1 to W3) in the promoter of CAT2.
(D) ChIP-qPCR analysis of the relative binding of WRKY75 to the promoter of CAT2. An anti-GFP monoclonal antibody was used for DNA immunoprecipitation from 32-d-old Pro35S:WRKY75-GFP/Col-0 transgenic plants. Black bars indicate the enrichment fold changes normalized to TUB2. Student’s t test, *P < 0.05. Data are represented as means ± sd, n = 3. The experiment was performed three times with similar results.
(E) EMSA analysis of the binding of recombinant WRKY75 protein to the promoter of CAT2 (CAT2 W2). Hot mProbe is the biotin-labeled probe with a single nucleic acid mutation from TGAC to TAAC.
(F) Transient dual-luciferase reporter assay. Construct pGreenII-0800 LUC containing the CAT2 promoter and construct p62-SK with or without the WRKY75 coding region were transiently cotransformed into Col-0 protoplasts. Firefly luciferase (LUC) and Renilla luciferase (REN) activity were measured after culturing the protoplasts under low-light conditions for 16 h. The ProCAT2:LUC/Pro35S:REN ratio represents the relative activity of CAT2 transcription. Student’s t test, *P < 0.05. Data are represented as means ± sd, n = 6. The experiment was performed three times with similar results.
Besides catalases, we also determined the expression of other genes encoding major H2O2 scavenging enzymes such as APXs (Khanna-Chopra, 2012). The relatively high levels of several APX transcripts in WRKY75RNAi leaves indicated that WRKY75 also repressed their expression (Supplemental Figure 12A), but whether WRKY75 directly acts on their transcription needs to be further explored.
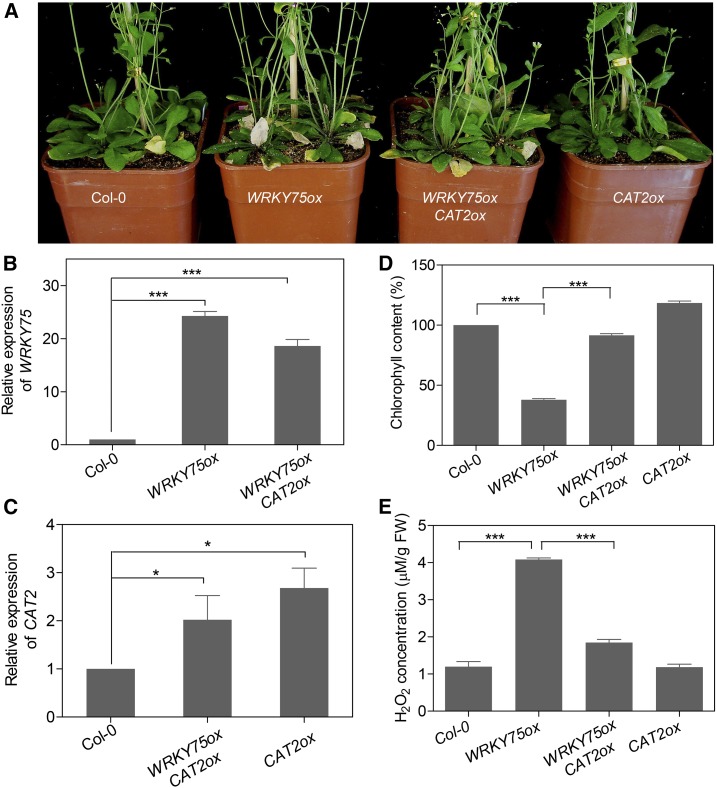
Increased Catalase Activity Suppresses Early Senescence Phenotype of WRKY75ox
Given our findings that WRKY75 represses CAT2 transcription and catalase activity, we investigated whether an increase in catalase activity in WRKY75ox plants could suppress their early senescence phenotype. To this end, we generated WRKY75ox/CAT2ox plants by crossing CAT2ox (Pro35S:CAT2-GFP/Col-0) into WRKY75ox (Figures 8B and 8C). We found that overexpression of CAT2 partly rescued the early senescence phenotype of WRKY75ox, including the reduced numbers of yellow leaves and increased chlorophyll content (Figures 8A and 8D). Consistent with the visible phenotype, the increase in H2O2 contents observed in WRKY75ox plants was also suppressed by overexpression of CAT2 (Figure 8E). Interestingly, while the cat2-1 mutant showed an early senescence phenotype (Supplemental Figure 13), CAT2ox plants in the Col-0 background displayed only a marginal delay in leaf senescence and virtually no difference in H2O2 content compared with Col-0 (Figures 8A and 8D), implying that catalase activity is probably at an adequate level to scavenge oxidative signals during normal development. Taken together, these results suggest that the elevated H2O2 content in WRKY75ox serves as another mechanism underlying its accelerated leaf senescence phenotype.
Figure 8.
Overexpression of CAT2 Represses the Early Senescence Phenotype of WRKY75ox Plants.
(A) The senescence phenotypes of 40-d-old Col-0, WRKY75ox, WRKY75ox/CAT2ox, and CAT2ox plants.
(B) and (C) qRT-PCR analysis of the expression of WRKY75 (B) and CAT2 (C) in the leaves shown in (A). The third and fourth rosette leaves were used. Student’s t test, *P < 0.05 and ***P < 0.001. Data are represented as means ± sd, n = 3. The experiments were performed three times with similar results.
(D) and (E) Measurement of chlorophyll contents (D) and H2O2 contents (E) in the leaves shown in (A). The third and fourth rosette leaves were used. Student’s t test, ***P < 0.001. Data are represented as means ± sd, n = 3. The experiments were performed three times with similar results.
DISCUSSION
Leaf senescence is a highly coordinated and complicated process requiring the integration of a wide variety of internal and environmental signals (Lim et al., 2007; Woo et al., 2013). Genome-wide studies have revealed that thousands of genes (SAGs) are differentially expressed in senescing leaves, suggesting that leaf senescence is under sophisticated transcriptional control. We previously identified a WRKY transcription factor, WRKY75, which acts as a positive regulator of leaf senescence (Li et al., 2012). In this study, we demonstrate that WRKY75 is a functional SAG whose transcription is induced by numerous signals, such as age, phytohormones including SA, ethylene, JA, and ABA, as well as H2O2. Knockdown or knockout of WRKY75 delayed leaf senescence, whereas overexpression of WRKY75 led to premature senescence. Genome-wide transcriptome profiling analysis revealed the enrichment of SA-related biological processes and biotic stress response that were substantially downregulated in WRKY75 knockdown transgenic plants. Accordingly, we observed reduced SA levels and a compromised defense response against a bacterial pathogen in WRKY75RNAi plants. Further analysis demonstrated that WRKY75 induces the transcription of SID2, the key gene responsible for SA biosynthesis. Loss of SID2 function suppressed the elevated SA production and precocious senescence phenotype of WRKY75ox, providing genetic evidence supporting the importance of SID2 induction by WRKY75 to leaf senescence and SA accumulation.
In addition to promoting SA biosynthesis, we discovered that WRKY75 also functions in plant tolerance to oxidative signals such as H2O2. Exogenous application of H2O2 can dramatically accelerate the leaf senescence process, particularly in the dark (Lin and Kao, 1998; Weaver and Amasino, 2001). Knockdown or knockout of WRKY75 led to enhanced tolerance to exogenous H2O2 application, as less endogenous H2O2 accumulated in plant leaves. One reason behind this effect is the enhanced catalase activity in WRKY75RNAi plants, which is mainly due to the upregulation of CAT2. Like SID2, CAT2 is also a direct target gene of WRKY75, although the regulatory mechanisms of these two genes are distinct. We identified three W-box motifs in the promoter of SID2 that mediate its induction by WRKY75, whereas at least one W-box motif in the promoter of CAT2 mediates its repression by WRKY75. Currently, it is not known how these seemingly identical W-box sequences play opposite roles in mediating WRKY75-directed transcription.
Collectively, our study provides mechanistic insights into how WRKY75 positively regulates leaf senescence, which is to activate at least two parallel pathways: SA production and H2O2 accumulation. It is conceivable that, upon leaf senescence, age-dependent WRKY75 expression progressively leads to the concomitant increase in SA and H2O2 levels, two well-defined inducers of leaf senescence. In fact, WRKY75 has been reported to modulate diverse biological processes, particularly stress responses such as phosphate deficiency (Devaiah et al., 2007), root hair development (Rishmawi et al., 2014), oxalic acid stress resistance (Chen et al., 2013), and the unfolded protein response (Hossain et al., 2016). Given that the downstream regulatory networks dictated by WRKY75 in these processes are still unclear, our findings about the regulatory role of WRKY75 in SA biosynthesis and ROS scavenging in leaf senescence offers a potential mechanism for these processes as well.
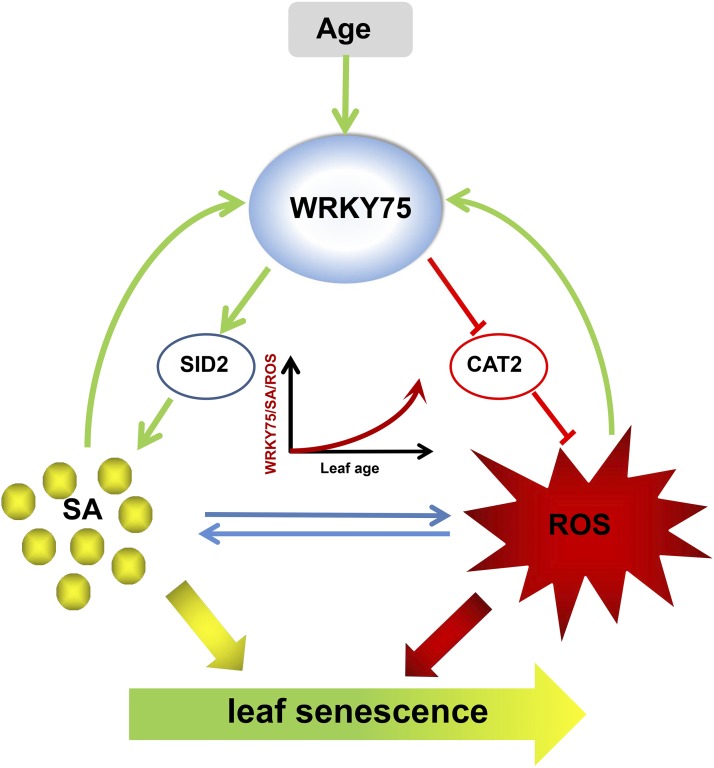
On the other hand, while WRKY75 promotes SA biosynthesis and H2O2 accumulation, we found that WRKY75 expression was greatly induced by SA and H2O2 treatment. Interestingly, SA and ROS levels are highly correlated, and these compounds mutually induce each other’s accumulation during many biological processes, such as plant defense responses (Leon et al., 1995; Rao et al., 1997). Based on previous and current findings, we propose the “tripartite amplification loop” model describing how WRKY75, SA, and ROS are responsible for accelerating leaf senescence (Figure 9). In this model, each of the three players promotes the accumulation of the other two via distinctive mechanisms. Thus, the levels of WRKY75, SA, and ROS undergo a gradual but self-sustaining rise driven by interlinking positive feedback loops along with leaf aging. It is conceivable that suppressing any of the three players would weaken the amplification loops, therefore leading to a slowdown of senescence progression.
Figure 9.
A Proposed Model Illustrating the Tripartite Amplification Loop Involving WRKY75, SA, and ROS That Accelerates Leaf Senescence.
WRKY75 is a senescence-associated gene that is induced by SA and ROS, two well-documented leaf senescence inducers. Meanwhile, WRKY75 promotes SA biosynthesis by activating SID2 transcription and enhances the H2O2 accumulation by repressing CAT2 transcription. Given that SA and ROS also mutually promote each other’s accumulation, WRKY75, SA, and ROS form a tripartite amplification loop to accelerate leaf senescence. As a result, the levels of WRKY75, SA, and ROS undergo a gradual but irreversible rise driven by the interlinking positive feedback loops during leaf aging.
This “tripartite amplification loop” model provides possible explanations for several well-known features of leaf senescence, for instance, irreversibility at its late phase and age dependency for environmental signals such as ethylene. In essence, leaf senescence is an unstoppable degenerative process, as any regulator can shorten or extend the time of its development but cannot prevent it from ultimately happening. Our model proposes that as leaves get older, the contents of WRKY75, SA, and ROS reach a critical threshold level, and such high levels are self-sustained and adequate to induce leaf aging even if the amplification loops are abrogated by other negative stimuli. At this point, this would make it difficult to pause the leaf senescence program, let alone reverse it. Meanwhile, ethylene is a well-documented senescence inducer, and such induction normally requires leaves to reach a defined age (Jing et al., 2005). It has been hypothesized that ethylene relies on some age-related factors to effectively induce leaf senescence (Jibran et al., 2013), although these factors remain to be identified. Our results demonstrate that WRKY75 is a direct target gene of EIN3, suggesting that the WRKY75-SA-ROS amplification loop acts as one such age-related factor in ethylene-induced leaf senescence. It would be interesting to examine this hypothesis in the future.
Although our study establishes an intertwined signaling network involving WRKY75, SA, and ROS, many other components could also be involved in (or associated with) this tripartite positive feedback loop. WRKY28, another known positive regulator of SID2, plays a similar role to WRKY75 in plant resistance (van Verk et al., 2011; Chen et al., 2013) and likely works in conjunction with WRKY75 in the regulation of leaf senescence. Another positive regulator of the amplification loop might be the bZIP transcription factor GBF1, which induces H2O2 accumulation by downregulating CAT2 expression during leaf senescence (Smykowski et al., 2015). A recent report indicates that the mitochondrial protease FtSH4 promotes leaf senescence by inducing the expression of a number of WRKY transcription factor genes (including WRKY75) as well as SA accumulation (Zhang et al., 2017). By contrast, the transcription of WRKY70 and WRKY54 is induced by SA, but their double mutants exhibit increased SA levels and an early senescence phenotype (Besseau et al., 2012), suggesting that they act as a brake for the amplification loop and are thus negative regulators of leaf senescence. Other sen-TFs, including SA-responsive WRKYs such as WRKY6 and WRKY53 and H2O2-responsive NACs such as ORS1 (Balazadeh et al., 2011), JUB1 (Wu et al., 2012), and AtNAP (Guo and Gan, 2006), may function downstream of the tripartite loop. Thus, the “tripartite amplification loop” model provides a molecular framework connecting upstream signals, such as age, ethylene, JA, ABA, and other environmental stresses, with the downstream regulatory network executed by these SA-responsive and H2O2-responsive TFs during leaf senescence.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Col-0 is the parental strain for all mutants and transgenic lines used in this study. The WRKY75RNAi line was previously described (Devaiah et al., 2007). The wrky75-25 mutant was described previously (Rishmawi et al., 2014). The sid2-2 (Chen et al., 2009) and cat2-1 (Li et al., 2015) mutants were ordered from the ABRC.
Seeds were surface-sterilized and plated on Murashige and Skoog (MS) medium (4.3 g/L MS salts, 1% [w/v] sucrose, pH 5.7 to 5.8, and 8 g/L agar). After stratification at 4°C for 3 d, the plates were exposed to white light (PAR of 100 to 150 µE m−2 s−1) for 5 d. The light-grown seedlings were transferred to soil and grown at 22°C under a 16-h-light/8-h-dark cycle.
Construction of Plasmids and Generation of Transgenic Plants
To construct ProWRKY75:GUS/Col-0, a 2522-bp genomic promoter sequence upstream of the coding region of WRKY75 was amplified, digested with SalI and BamHI, and inserted into the pBI101 vector (Jefferson et al., 1987). To create Pro35S:WRKY75, Pro35S:SID2, and Pro35S:CAT2 constructs, the corresponding cDNA sequences were amplified, digested with XbaI and KpnI, and inserted into the pQG110 vector (Qin et al., 2005). To generate Pro35S:WRKY75-GFP construct, the WRKY75 cDNA sequence without a stop codon was amplified, digested with KpnI and XbaI, and inserted into the pCHF3-GFP vector (Yin et al., 2002). To generate the iWRKY75-HA construct, the WRKY75 cDNA sequence without stop codon was amplified, digested with SpeI and XhoI, and linked with HA tag sequence and then introduced into the pER8 vector (Zuo et al., 2000). All DNA constructs were verified by DNA sequencing analysis and were electroporated into Agrobacterium tumefaciens GV3101, which was used to transform Col-0 plants by the floral dip method (Clough and Bent, 1998).
To generate WRKY75 knockout plants by CRISPR-Cas9 (Feng et al., 2013), the target site (TGGTCGTGACAACCTACGA) preceding an AGG was introduced into single guide RNA using WRKY75-CRISPR target primers. The plasmid was introduced into plants by the floral dip method. Successfully transformed T1 plants were selected on MS medium with 50 μg/mL hygromycin. Total DNA was isolated from individual T2 or T3 plants and used as a template to PCR amplify the 200-bp DNA region containing the single guide RNA target site, followed by sequencing. All primer sequences used here are listed in Supplemental Data Set 2.
Chemical Solution Preparation
SA, H2O2 (30%), 1-aminocyclopropane-1-carboxylic acid, methyl jasmonate, ABA, gibberellin (GA3), 6-benzylaminopurine, and β-estradiol were purchased from Sigma-Aldrich and prepared as stock solutions. SA (1 mM) was dissolved in 75% ethanol, 1-aminocyclopropane-1-carboxylic acid (500 µM) was dissolved in double distilled water, and methyl jasmonate (500 µM), ABA (500 µM), 6-benzylaminopurine (500 µM), and β-estradiol (500 µM) were dissolved in DMSO. The working solution was diluted from the stock solution.
Assay for Natural and H2O2-Induced Leaf Senescence
Natural leaf senescence analysis was performed as described previously (Li et al., 2013). The third and fourth rosette leaves from plants at different developmental ages were used for chlorophyll content measurement and for detection of SAG12 expression. For H2O2-induced leaf senescence, the third and fourth rosette leaves were detached and incubated in MES buffer (2 mM MES, pH 5.8) with or without H2O2 under dark conditions for 3 d.
Measurement of Chlorophyll Contents
Chlorophyll contents were measured in the third and fourth leaves using a SPAD Chlorophyll Meter (SPAD-502 Plus; Konica Minolta). Each leaf was evenly divided into 10 spots, and one measurement was taken per spot. The average value of the 10 measurements (SPAD Unit) represents a single data point and one biological replicate. At least three individual leaves of each genotype are measured, and three biological replicates were performed.
RNA Extraction and qRT-PCR
Total RNA was extracted from green seedlings or leaves of different ages using Trizol reagent (Invitrogen). Reverse transcription was performed using M-MLV reverse transcriptase (Promega), followed by PCR on a Light Cycler 480 system (Roche) with SYBR Premix ExTaq reagents (TaKaRa). All oligonucleotide sequences used here are listed in Supplemental Data Set 2.
RNA-Seq
The third and fourth leaves of Col-0 and WRKY75RNAi plants were collected and ground into a powder in liquid nitrogen. Total RNA was extracted using an RNeasy Plant Mini kit (Qiagen). RNA-seq and differential gene expression analysis were performed by the BIOPIC institution at Peking University. In brief, RNA quality was evaluated on a Bioanalyzer 2100 instrument (Agilent). Sequencing libraries were prepared following the standard protocol of the TruSeq RNA Library Prep Kit (Illumina). The 100-nucleotide, single-end, high-throughput sequencing was performed on an Illumina HiSeq 2000 (Illumina). Low-quality sequencing reads were removed and adaptor trimmed using Fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The TAIR10 genome was used as the Arabidopsis genome reference (www.arabidopsis.org). Reads were mapped to the genome with TopHat2 (https://ccb.jhu.edu/software/tophat/index.shtml) software, and differential expression analysis was conducted using Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/). Gene Ontology enrichment analysis was performed using the BiNGO app (https://www.psb.ugent.be/cbd/papers/BiNGO/Home.html) in the Cytoscape software package (http://cytoscape.org). Default parameters were used for all bioinformatics software.
Microscopy
WRKY75-GFP fluorescence was detected using a Zeiss Axio Imager M2 microscope (Carl Zeiss) with 20× objectives. Roots from Pro35S:WRKY75-GFP/Col-0 transgenic plants were mounted on standard microscope slides with water. An excitation wavelength of 488 nm and emission at 509 nm were used for GFP detection.
Protein Expression and Purification
To generate the MBP-WRKY75 fusion protein, the coding sequence of WRKY75 was amplified, digested with BamHI and SalI, and cloned into pMAL-p2X (GE Healthcare), and the plasmid was transformed into Escherichia coli BL21 (DE3) competent cells. Isopropyl-β-d-thiogalactopyranoside (0.5 mM) was used to induce the expression of MBP-WRKY75 protein. After expression at low temperature (16°C) for 12 h, bacterial cells were collected for protein purification. Purification of MBP-WRKY75 protein was performed using the ÄKTA pure chromatography system (GE Healthcare) following the manufacturer’s instructions.
EMSA
Oligonucleotide probes (SID2 W2, W3, and W4 as well as CAT2 W2) were synthesized and labeled with biotin at the 3′ hydroxyl end of the sense strand. For the mutated probes, the single mutation site is located at the core sequence of the W-box (change from TGAC to TAAC). EMSA was performed using a Light Shift Chemiluminescent EMSA Kit (Pierce). Briefly, 40 fmol of labeled wild-type or mutated probes was incubated with proteins in binding buffer (2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA) at room temperature for 30 min. The reaction products were transferred to a 6% polyacrylamide gel, followed by electrophoresis in 0.5× Tris-borate-EDTA buffer for 40 min. All oligonucleotide sequences used here are listed in Supplemental Data Set 2.
ChIP-qPCR
ChIP was performed as described previously (Saleh et al., 2008). Briefly, ?2.5 g of 4-week-old WRKY75-GFP leaf tissue was collected and fixed in 1% formaldehyde for 15 min under a vacuum, followed by the neutralization using 0.125 M glycine for an additional 5 min. The leaves were then washed twice with water, frozen in liquid nitrogen, and ground into a fine powder. Next, chromatin DNA was isolated from the ground tissue and sonicated. The sonicated chromatin solution (300 µL) was diluted with ChIP dilution buffer (10% Triton, 1 µM EDTA, 16.7 µM Tris-Cl, pH 8.0, and 167 µM NaCl) to 3 mL and divided equally into two new tubes; 40 μL of protein G-agarose beads was added to each tube, and the solution was precleared by incubating at 4°C for 1 h. The solutions were transferred into two fresh tubes, and 10 μL anti-GFP antibody with a dilution of 1:150 (v/v) (Abmart; catalog no. M20004) was added to one tube but not the other (as a negative control). The two tubes were incubated at 4°C overnight with gentle rotation, followed by the addition of 50 μL of protein G-agarose beads for immunoprecipitation. Elution of the immunoprecipitated complex was performed with TE buffer (10 µM Tris-Cl, pH 8.0, and 1 µM EDTA), followed by Proteinase K (10 mg/mL; Sigma-Aldrich) treatment and reverse cross-linking with 5 M NaCl. DNA was extracted with phenol/chloroform (1:1, v/v) and resuspended in 20 μL of sterile distilled water. The DNA samples were used as templates for PCR. All oligonucleotide sequences used here are listed in Supplemental Data Set 2.
Transient Dual-Luciferase Reporter System
A 1.8-kb promoter sequence of SID2 and a 2.0-kb promoter sequence of CAT2 were amplified from Arabidopsis genomic DNA, digested with KpnI and BamHI, KpnI and SalI, respectively, inserted into the pGreen II 0800-LUC vector, and used as reporter plasmids (Hellens et al., 2005). The coding sequence of WRKY75 was amplified by PCR, digested with XbaI and KpnI, inserted into pGreen II 62-SK, and used as an effector plasmid. Arabidopsis protoplasts were prepared as described previously (Li et al., 2013) and cotransfected with the constructs following the instructions for the Dual-Luciferase reporter assay system (Promega). The ratio of LUC to REN was determined for the dual-luciferase reporter system (Promega) on a GLO-MAX 20/20 luminometer (Promega) after culturing the protoplasts under low-light conditions for 16 h. The LUN/REN ratio indicates transcriptional activity.
GUS Staining
GUS staining was performed as described previously (Jefferson et al., 1987). Briefly, tissues were washed three times with PBS buffer (100 mM Na3PO4, pH 7.0), incubated with GUS staining solution (100 mM Na3PO4, pH 7.0, 1 mM EDTA, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 1% Triton X-100, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) for 8 to 12 h or longer at 37°C in the dark, and washed three times with PBS buffer, followed by decolorization using 95% ethanol.
SA Measurements
Tissues were fully ground into a powder in liquid nitrogen, weighed, and delivered to the plant hormone platform at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, for quantitative free-SA measurements by HPLC-tandem mass spectrometry (Fu et al., 2012).
Bacterial Growth Assay
The bacterial growth assay was performed as described (Chen et al., 2009). Briefly, mature rosette levels from 3-week-old Arabidopsis plants were infiltrated with DC3000 bacterial cells for 3 d, followed by photography and bacterial growth (colony-forming units/cm2) determination.
DAB Staining and H2O2 Measurements
For DAB staining, tissues were washed three times with PBS buffer, incubated in DAB staining solution (1 mg/mL DAB, 10 mM Na2HPO4, and 0.05% Tween 20, pH 3.8) in the dark for 4 to 8 h, and decolorized in 95% ethanol. The intensity of brown coloration indicates H2O2 contents.
Quantitative H2O2 measurement was performed using an Amplex Red hydrogen peroxide/peroxidase assay kit (Molecular Probes). Briefly, the samples were frozen in liquid nitrogen and ground into a fine powder; 30 mg of each sample was fully suspended in 200 μL H2O2 extraction buffer (25 mM sodium phosphate buffer, pH 6.5). The extract was centrifuged at 12,000 rpm for 15 min at 4°C, and the supernatant was prepared for the quantitative assay. Measurement of the absorbance was performed using a Tecan infinite F200/M200 at 560 nm. H2O2 concentration is indicated in µM/g fresh weight.
Catalase Activity Assay
The catalase activity assay was performed using a catalase assay kit (Beyotime Biotechnology). Briefly, samples were frozen in liquid nitrogen and fully ground into a powder; each sample was suspended in 100 μL extraction buffer (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 1% Nonidet P-40) and centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was used to analyze catalase activity. Protein concentration was measured using a BCA protein assay kit (Beyotime Biotechnology). Catalase activity is indicated as units/mg. One unit of catalase activity represents the amount of enzyme that catalyzes the decomposition of 1 µM H2O2 per minute at 25°C.
Flowering Time Measurement
Flowering time was measured as described (Putterill et al., 1995). The number of rosette leaves was counted when the flower bud appeared in the center of the rosette leaves under long-day conditions. The means ± sd of the number of rosette leaves represent flowering time.
Statistical Analysis
Two-tailed Student’s t tests were used to determine significance between populations. Graphs throughout the article show the mean value and error bars sd.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: WRKY75 (At5g13080), WRKY53 (At4g23810), SID2 (At1g74710), PR1 (At2g14610), PR5 (At1g75040), PAL1 (At2g37040), WRKY28 (At4g18170), CBP60G (At5g26920), SARD1 (At1g73805), BSMT1 (At3g11480), SAGT1 (At2g43820), PBS3 (At5g13320), SAG12 (At3g20770), CAT1 (At1g20630), CAT2 (At4g35090), CAT3 (At1g20620), DEFL (At2g43510), APX1 (At1g07890), APX2 (At3g09640), APX3 (At4g35000), APX4 (At4g09010), APX5 (At4g35970), APX6 (At4g32320), EIN3 (At3g20770), ERF1 (At3g23240), ACTIN2 (At3g18780), and TUBULIN2 (TUB2; At5g62690). RNA-seq raw data are available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE102302.
Supplemental Data
Supplemental Figure 1. Gene Expression in Response to Senescence, SA, and ROS.
Supplemental Figure 2. Expression Patterns of WRKY75 upon Treatment with Various Hormones.
Supplemental Figure 3. The wrky75-25 Mutant Shows a Delayed Leaf Senescence Phenotype.
Supplemental Figure 4. Generation of Two wrky75 Mutants Using CRISPR-Cas9.
Supplemental Figure 5. Senescence and Flowering Phenotypes of Col-0, WRKY75RNAi, wrky75-25, and wrky75-c1 Plants.
Supplemental Figure 6. Inducible Overexpression of WRKY75 Promotes Leaf Senescence.
Supplemental Figure 7. WRKY75 Is a Nuclear Protein.
Supplemental Figure 8. Enrichment of Selected Gene Ontology Categories of Downregulated Genes Identified by Transcriptome Sequencing.
Supplemental Figure 9. Expression of Genes Involved in SA Biosynthesis and Metabolism.
Supplemental Figure 10. SID2 Is a Senescence-Associated Gene That Promotes Leaf Senescence.
Supplemental Figure 11. Oxidative Stress Resistance Is Enhanced in the wrky75-25 Mutant and Reduced in WRKY75ox Plants.
Supplemental Figure 12. Expression Patterns of CATs and APXs.
Supplemental Figure 13. The cat2-1 Mutant Exhibits an Early Senescence Phenotype.
Supplemental Data Set 1. Differentially Expressed Genes in WRKY75RNAi Compared with Col-0.
Supplemental Data Set 2. Primer and Oligonucleotide Sequences Used in This Study.
Acknowledgments
We thank members of the Guo Lab for helpful discussions and critical reading of the manuscript. We also thank Kashchandra Raghothama (Purdue University) for kindly providing the WRKY75RNAi seeds, Martin Hülskamp (University of Cologne, Germany) for kindly providing the wrky75-25 seeds, Jian-Kang Zhu (Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences) for providing the CRISPR-Cas9 system plasmids, and Jian-Min Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for assistance with the bacterial growth assay. The T-DNA mutant lines were purchased from the ABRC. This work was funded by grants from the National Natural Science Foundation of China (Grant 31570286 to H.G.), start-up funding from Southern University of Science and Technology to H.G., the China Postdoctoral Science Foundation (2014M560015 and 2015T80013 to Z.L.), and grants from Peking-Tsinghua Center for Life Sciences to H.G. and Z.L.
AUTHOR CONTRIBUTIONS
H.G., P.G., and Z.L. conceived the project and designed the experiments. P.G. and Z.L. performed and analyzed the phenotypes, performed GUS staining, and generated transgenic Arabidopsis plants. P.G. performed the ChIP, EMSA, H2O2 measurements, CAT activity assay, bacterial growth assay, and CRISPR-Cas9 system. P.G. and P.H. performed qRT-PCR and transient dual-luciferase assay. P.G. and B.L. performed and analyzed the RNA-seq data. J.C. and F.S. performed the quantitative free-SA measurement. P.G., Z.L., and H.G. wrote the article. All authors analyzed and discussed the data in the article.
References
- An F., Zhang X., Zhu Z., Ji Y., He W., Jiang Z., Li M., Guo H. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Kwasniewski M., Caldana C., Mehrnia M., Zanor M.I., Xue G.P., Mueller-Roeber B. (2011). ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4: 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers E.P., McDowell J.M. (2001). Regulation and execution of programmed cell death in response to pathogens, stress and developmental cues. Curr. Opin. Plant Biol. 4: 561–567. [DOI] [PubMed] [Google Scholar]
- Besseau S., Li J., Palva E.T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63: 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F., Salinas P., Cecchini N.M., Jordana X., Van Hummelen P., Alvarez M.E., Holuigue L. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70: 79–102. [DOI] [PubMed] [Google Scholar]
- Blokhina O., Virolainen E., Fagerstedt K.V. (2003). Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 91: 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585. [DOI] [PubMed] [Google Scholar]
- Chen F., D’Auria J.C., Tholl D., Ross J.R., Gershenzon J., Noel J.P., Pichersky E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36: 577–588. [DOI] [PubMed] [Google Scholar]
- Chen G.H., Liu C.P., Chen S.C., Wang L.C. (2012). Role of ARABIDOPSIS A-FIFTEEN in regulating leaf senescence involves response to reactive oxygen species and is dependent on ETHYLENE INSENSITIVE2. J. Exp. Bot. 63: 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xue L., Chintamanani S., Germain H., Lin H., Cui H., Cai R., Zuo J., Tang X., Li X., Guo H., Zhou J.M. (2009). ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu J., Lin G., Wang A., Wang Z., Lu G. (2013). Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 32: 1589–1599. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cochrane F.C., Davin L.B., Lewis N.G. (2004). The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65: 1557–1564. [DOI] [PubMed] [Google Scholar]
- Dempsey D.A., Vlot A.C., Wildermuth M.C., Klessig D.F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B.N., Karthikeyan A.S., Raghothama K.G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143: 1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J., Klessig D.F. (1995). Inhibition of ascorbate peroxidase by salicylic acid and 2,6-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Natl. Acad. Sci. USA 92: 11312–11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J., Klessig D.F. (1996). Salicylic acid is a modulator of tobacco and mammalian catalases. J. Biol. Chem. 271: 28492–28501. [DOI] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D.L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., Zhu J.K. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T., Holbrook N.J. (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247. [DOI] [PubMed] [Google Scholar]
- Frugoli J.A., Zhong H.H., Nuccio M.L., McCourt P., McPeek M.A., Thomas T.L., McClung C.R. (1996). Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 112: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Chu J., Sun X., Wang J., Yan C. (2012). Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28: 1081–1087. [DOI] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S., Amasino R.M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988. [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R.M. (1997). Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46: 601–612. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderhofer K., Zentgraf U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469–473. [DOI] [PubMed] [Google Scholar]
- Hossain M.A., Henríquez-Valencia C., Gómez-Páez M., Medina J., Orellana A., Vicente-Carbajosa J., Zouhar J. (2016). Identification of novel components of the unfolded protein response in Arabidopsis. Front. Plant Sci. 7: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki H., Maxson J.M., Woodson W.R. (1994). An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. USA 91: 8925–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibran R., A Hunter D., P Dijkwel P. (2013). Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 82: 547–561. [DOI] [PubMed] [Google Scholar]
- Jing H.C., Hebeler R., Oeljeklaus S., Sitek B., Stühler K., Meyer H.E., Sturre M.J., Hille J., Warscheid B., Dijkwel P.P. (2008). Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol (Stuttg) 10 (Suppl 1): 85–98. [DOI] [PubMed] [Google Scholar]
- Jing H.C., Schippers J.H., Hille J., Dijkwel P.P. (2005). Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J. Exp. Bot. 56: 2915–2923. [DOI] [PubMed] [Google Scholar]
- Khanna-Chopra R. (2012). Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249: 469–481. [DOI] [PubMed] [Google Scholar]
- Lee S., Seo P.J., Lee H.J., Park C.M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70: 831–844. [DOI] [PubMed] [Google Scholar]
- Leon J., Lawton M.A., Raskin I. (1995). Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol. 108: 1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., et al. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27: 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2012). Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant Biol. 54: 526–539. [DOI] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao Y., Liu X., Peng J., Guo H., Luo J. (2014). LSD 2.0: an update of the leaf senescence database. Nucleic Acids Res. 42: D1200–D1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136. [DOI] [PubMed] [Google Scholar]
- Lin J.N., Kao C.H. (1998). Effect of oxidative stress caused by hydrogen peroxide on senescence of rice leaves. Bot. Bull. Acad. Sin. 39: 161–165. [Google Scholar]
- Liu X., Li Z., Jiang Z., Zhao Y., Peng J., Jin J., Guo H., Luo J. (2011). LSD: a leaf senescence database. Nucleic Acids Res. 39: D1103–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61: 4197–4220. [DOI] [PubMed] [Google Scholar]
- Miao Y., Zentgraf U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55: 853–867. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498. [DOI] [PubMed] [Google Scholar]
- Morris K., MacKerness S.A., Page T., John C.F., Murphy A.M., Carr J.P., Buchanan-Wollaston V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23: 677–685. [DOI] [PubMed] [Google Scholar]
- Nawrath C., Métraux J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y.S., Amasino R.M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41: 181–194. [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L.J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52: 640–657. [DOI] [PubMed] [Google Scholar]
- Rao M.V., Paliyath G., Ormrod D.P., Murr D.P., Watkins C.B. (1997). Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol. 115: 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L., Pesch M., Juengst C., Schauss A.C., Schrader A., Hülskamp M. (2014). Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol. 165: 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62: 3321–3338. [DOI] [PubMed] [Google Scholar]
- Robatzek S., Somssich I.E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28: 123–133. [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Smykowski A., Fischer S.M., Zentgraf U. (2015). Phosphorylation affects DNA-binding of the senescence-regulating bZIP transcription factor GBF1. Plants (Basel) 4: 691–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H., Sandalio L., Jiménez A., Palma J.M., Corpas F.J., Meseguer V., Gómez M., Sevilla F., Leterrier M., Foyer C.H., del Río L.A. (2006). Roles for redox regulation in leaf senescence of pea plants grown on different sources of nitrogen nutrition. J. Exp. Bot. 57: 1735–1745. [DOI] [PubMed] [Google Scholar]
- van Verk M.C., Bol J.F., Linthorst H.J. (2011). WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tsuda K., Truman W., Sato M., Nguyen V., Katagiri F., Glazebrook J. (2011). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67: 1029–1041. [DOI] [PubMed] [Google Scholar]
- Weaver L.M., Amasino R.M. (2001). Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127: 876–886. [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Willekens H., Chamnongpol S., Davey M., Schraudner M., Langebartels C., Van Montagu M., Inzé D., Van Camp W. (1997). Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 16: 4806–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Kim H.J., Nam H.G., Lim P.O. (2013). Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 126: 4823–4833. [DOI] [PubMed] [Google Scholar]
- Woo H.R., et al. (2016). Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 171: 452–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X.F., He S.Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf U., Laun T., Miao Y. (2010). The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 89: 133–137. [DOI] [PubMed] [Google Scholar]
- Zhang K., Halitschke R., Yin C., Liu C.J., Gan S.S. (2013). Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 110: 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Li C., Wang R., Chen Y., Shu S., Huang R., Zhang D., Li J., Xiao S., Yao N., Yang C. (2017). The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol. 173: 2294–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu S., Ding P., Wang D., Cheng Y.T., He J., Gao M., Xu F., Li Y., Zhu Z., Li X., Zhang Y. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 107: 18220–18225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.Y., Wang J.G., Song S.J., Wang Q., Kang H., Zhang Y., Li S. (2016). Precocious leaf senescence by functional loss of PROTEIN S-ACYL TRANSFERASE14 involves the NPR1-dependent salicylic acid signaling. Sci. Rep. 6: 20309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Cai Z., Guo Y., Gan S. (2009). An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 150: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jiang Y., Yu D. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]