Rice flowering depends on formation of transcriptional complexes, some of which act at the shoot apical meristem, whereas others promote or repress the floral transition by acting from the leaves.
Abstract
Plants measure day or night lengths to coordinate specific developmental changes with a favorable season. In rice (Oryza sativa), the reproductive phase is initiated by exposure to short days when expression of HEADING DATE 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1) is induced in leaves. The cognate proteins are components of the florigenic signal and move systemically through the phloem to reach the shoot apical meristem (SAM). In the SAM, they form a transcriptional activation complex with the bZIP transcription factor OsFD1 to start panicle development. Here, we show that Hd3a and RFT1 can form transcriptional activation or repression complexes also in leaves and feed back to regulate their own transcription. Activation complexes depend on OsFD1 to promote flowering. However, additional bZIPs, including Hd3a BINDING REPRESSOR FACTOR1 (HBF1) and HBF2, form repressor complexes that reduce Hd3a and RFT1 expression to delay flowering. We propose that Hd3a and RFT1 are also active locally in leaves to fine-tune photoperiodic flowering responses.
INTRODUCTION
The floral transition sets the beginning of the reproductive phase and is completed upon switching of the shoot apical meristem (SAM) from indeterminate vegetative to determinate reproductive growth. In many plant species, these changes are triggered by daylength (or photoperiod), which is measured in leaves to synchronize inflorescence development with the most favorable seasons. This signaling mechanism requires systemic communication signals that integrate environmental inputs and connect distant tissues of the plant.
Rice (Oryza sativa) preferentially flowers under short days (SDs). When daylength falls under a critical threshold, proteins encoded by the HEADING DATE 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1) loci are produced in leaves and delivered through the phloem to the SAM, where they induce developmental reprogramming (Tamaki et al., 2007, 2015; Komiya et al., 2009). Both proteins share homology with FLOWERING LOCUS T (FT) of Arabidopsis thaliana and belong to the phosphatidylethanolamine binding protein (PEBP) family of regulators, which includes also TERMINAL FLOWER1 (TFL1) homologs (Kojima et al., 2002; Ho and Weigel, 2014). However, whereas FT-like proteins are strong activators of flowering, TFL1-like proteins are flowering inhibitors (Wickland and Hanzawa, 2015).
Under inductive photoperiods, both Hd3a and RFT1 are transcribed, and their protein products are essential for flowering to the extent that artificial reduction of their mRNA expression results in never-flowering plants (Komiya et al., 2008; Tamaki et al., 2015). However, transcription of RFT1 can be induced also under long days (LDs), and its floral promotive activity under these conditions contributes to the facultative nature of the photoperiodic flowering response of rice (Gómez-Ariza et al., 2015; Komiya et al., 2009).
Induction of Hd3a and RFT1 expression in leaves results from the integration of photoperiodic information with diurnal timing set by the circadian clock. Environmental signals ultimately converge on the transcriptional activation of Early heading date 1 (Ehd1), encoding a B-type response regulator unique to rice (Brambilla and Fornara, 2013; Doi et al., 2004; Cho et al., 2016). Transcription of Ehd1, Hd3a, and RFT1 thus correlates under SD in leaves, showing a transient induction that persists only for the time required to irreversibly commit flowering at the SAM (Galbiati et al., 2016; Doi et al., 2004; Cho et al., 2016; Komiya et al., 2008). Once a sufficient amount of Hd3a and/or RFT1 proteins reaches the SAM, expression of target genes that promote inflorescence formation is induced (Taoka et al., 2011; Tamaki et al., 2015).
FT-like proteins have no DNA binding property. Therefore, upon reaching the cytoplasm of cells at the SAM, they bind to transcription factors of the bZIP family, including FD in Arabidopsis and OsFD1 in rice (Wigge et al., 2005; Taoka et al., 2011). The complex, originally found to be dimeric based on studies in Arabidopsis, was later demonstrated to contain also a 14-3-3 protein of the Gf14 family (G-box factor 14-3-3) that bridges the interaction between OsFD1 and Hd3a. The resulting ternary complex, named florigen activation complex (FAC), is targeted to the nucleus where it further dimerizes, forming a heterohexameric complex tethered by OsFD1 on target DNA sequences (Zhao et al., 2015; Taoka et al., 2011). Similar interactions take place in many plant species, including tomato (Solanum lycopersicum; Park et al., 2014), potato (Solanum tuberosum; Teo et al., 2017), wheat (Triticum aestivum) and barley (Hordeum vulgare; Li et al., 2015), maize (Zea mays; Danilevskaya et al., 2008), and hybrid aspen (Populus tremula × tremuloides; Tylewicz et al., 2015), suggesting that this molecular module is widely conserved among angiosperms. This conservation is further corroborated by interspecific interactions demonstrated to occur between Hd3a/RFT1 and FD (Jang et al., 2017). In many such examples, FD-like genes can provide DNA binding specificity by recognizing ACGT-containing consensus sequences on the DNA of target promoters (Izawa et al., 1993; Li and Dubcovsky, 2008; Taoka et al., 2011; Wigge et al., 2005). Competition between FT-like and TFL1-like proteins for interaction with FD and 14-3-3 proteins partly explains their opposite function on flowering and shoot architecture. Again, such competitive behavior is widespread among angiosperms (Pnueli et al., 2001; Randoux et al., 2014; Hanano and Goto, 2011; Park et al., 2014).
The rice genome encodes seven Gf14 proteins, four of which (the b, c, d, and e) can assemble into a FAC (Taoka et al., 2011). The Gf14c protein was the first to be functionally characterized as an Hd3a interactor (Purwestri et al., 2009; Taoka et al., 2011). Because of their redundancy and pleiotropic effects, it has not been possible to study gf14 mutants, but transgenic rice overexpressing Gf14c had delayed flowering (Purwestri et al., 2009). Despite the apparent contrast with the nature of a FAC, this result might indicate that a tightly regulated balance between FAC components needs to be achieved at the SAM to promote flowering. Alternatively, floral repressor complexes containing Gf14c might exist and become predominant upon overexpression of this specific 14-3-3 protein.
Besides FD-like transcription factors and 14-3-3 proteins, FT-like genes can interact with members of the TEOSINTE BRANCHED1, CYCLOIDEA, PCF (TCP) transcription factor family. The ability to bind distinct members of this group of regulators partly discriminates between FT- and TFL1-like proteins and indicates that TCPs are preferential interactors of FT-like proteins (Mimida et al., 2011; Niwa et al., 2013; Ho and Weigel, 2014). Finally, apple (Malus domestica) Vascular Plant One Zinc finger (MdVOZ1a) was isolated as an interactor of apple FT and shown to alter inflorescence architecture when expressed in Arabidopsis (Mimida et al., 2011). Whether interactions between FT-like and VOZ-like proteins are conserved among flowering plants is yet to be assessed.
Downstream targets of the FAC at the SAM include members of the MADS box transcription factor family that are necessary to switch the meristem to reproductive growth. In Arabidopsis, induction of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1, FRUITFULL (FUL), and APETALA1 takes place shortly after arrival of FT at the SAM (Andrés and Coupland, 2012). Similarly, OsMADS14, OsMADS15, and OsMADS18, genes belonging the FUL clade, and OSMADS34/PAP2, a SEPALLATA (SEP)-like gene, are progressively activated upon floral transition in rice (Kobayashi et al., 2012; Litt and Irish, 2003). Mutants in which all four genes are silenced develop inflorescence stems where flowers are replaced by vegetative shoots (Kobayashi et al., 2012). This general mode of action of the florigens at the SAM has been observed in several plant species (Jang et al., 2015; Jaudal et al., 2015; Li and Dubcovsky, 2008). However, FACs can be deployed also in tissues different from the SAM to control a broad spectrum of developmental processes different from inflorescence formation. For example, components of FACs governing leaf development have been reported in both Arabidopsis and rice (Teper-Bamnolker and Samach, 2005; Tsuji et al., 2013). Potato tuber formation depends on FACs forming at the stolon meristem in response to FT export from the leaves (Navarro et al., 2011; Teo et al., 2017). Seasonal growth cessation in trees is induced by FACs assembled in vegetative apical meristems that stop elongation and leaf production before the onset of winter (Tylewicz et al., 2015). These findings illustrate the plasticity and robustness of FACs as integrators of photoperiodic signals into distinct developmental networks.
Given the high number of OsbZIP-coding genes in rice, the combinatorial interactions possibly leading to different florigen-containing complexes are very high (Tylewicz et al., 2015; Park et al., 2014; Tsuji et al., 2013; Li et al., 2015). Additionally, the floral transition in rice is associated with both induction and repression of gene expression at the SAM, and different complexes could operate by promoting or repressing expression of specific targets (Tamaki et al., 2015). Here, we demonstrate that canonical FACs can also form in leaves where Hd3a and RFT1 interact through Gf14c with OsFD1. These complexes are required to activate a positive feedback loop on Ehd1, Hd3a, and RFT1 expression. This function is counterbalanced by two OsbZIP transcription factors closely related to OsFD1 that directly bind Hd3a and function as negative regulators of the Ehd1 florigens module in leaves. Finally, we provide evidence for a meristematic function of one such OsbZIP to repress the floral transition by reducing the expression of inflorescence identity genes. We propose that dynamic formation of distinct complexes fine tunes flowering in leaves and at the SAM of rice.
RESULTS
An Active Florigen Activation Complex Can Form in Leaves
The rice FAC is a transcriptional activation complex assembled in cells of the SAM by Hd3a or RFT1, a Gf14 protein and OsFD1, and its primary targets include members of the OsMADS transcription factor family (Kojima et al., 2002; Taoka et al., 2011; Tsuji et al., 2013; Tamaki et al., 2015; Kobayashi et al., 2012). It has been proposed that FAC complexes control a wide range of developmental processes in distinct tissues of several plant species, but to which extent a FAC might function outside of the SAM and in rice leaf tissues is unclear. The diurnal mRNA expression of components of the FAC was quantified under inductive and noninductive photoperiods, including SD (10 h light) and LD (16 h light) in the leaves (Supplemental Figures 1A to 1D). The expression of Gf14c did not depend upon the photoperiod and showed a peak at Zeitgeber (ZT) 15 (Supplemental Figure 1B). Expression of OsFD1 was detected under both photoperiods; however, its expression under LD was constant during the time course, whereas it oscillated under SD with a peak in the middle of the night (Supplemental Figure 1C). Similarly, expression of Hd3a and RFT1 was induced during the night and peaked toward the end of it (Supplemental Figure 1A).
Since all FAC components were coexpressed in leaves under SD, the expression of OsMADS14 was used as readout for the activity of the FAC. OsMADS14 mRNA showed a peak during the night only in leaves of plants grown under SDs, similarly to OsFD1 (Supplemental Figure 1D). Additionally, expression of both OsMADS14 and OsMADS15 was induced in leaves upon shifting plants from LD (16 h light) to SD (10 h light), as more Hd3a and RFT1 became available for FAC formation (Supplemental Figures 1E and 1F). Expression of OsMADS TFs is therefore sensitive to expression of FAC components in both leaves and meristem (Taoka et al., 2011; Kobayashi et al., 2012).
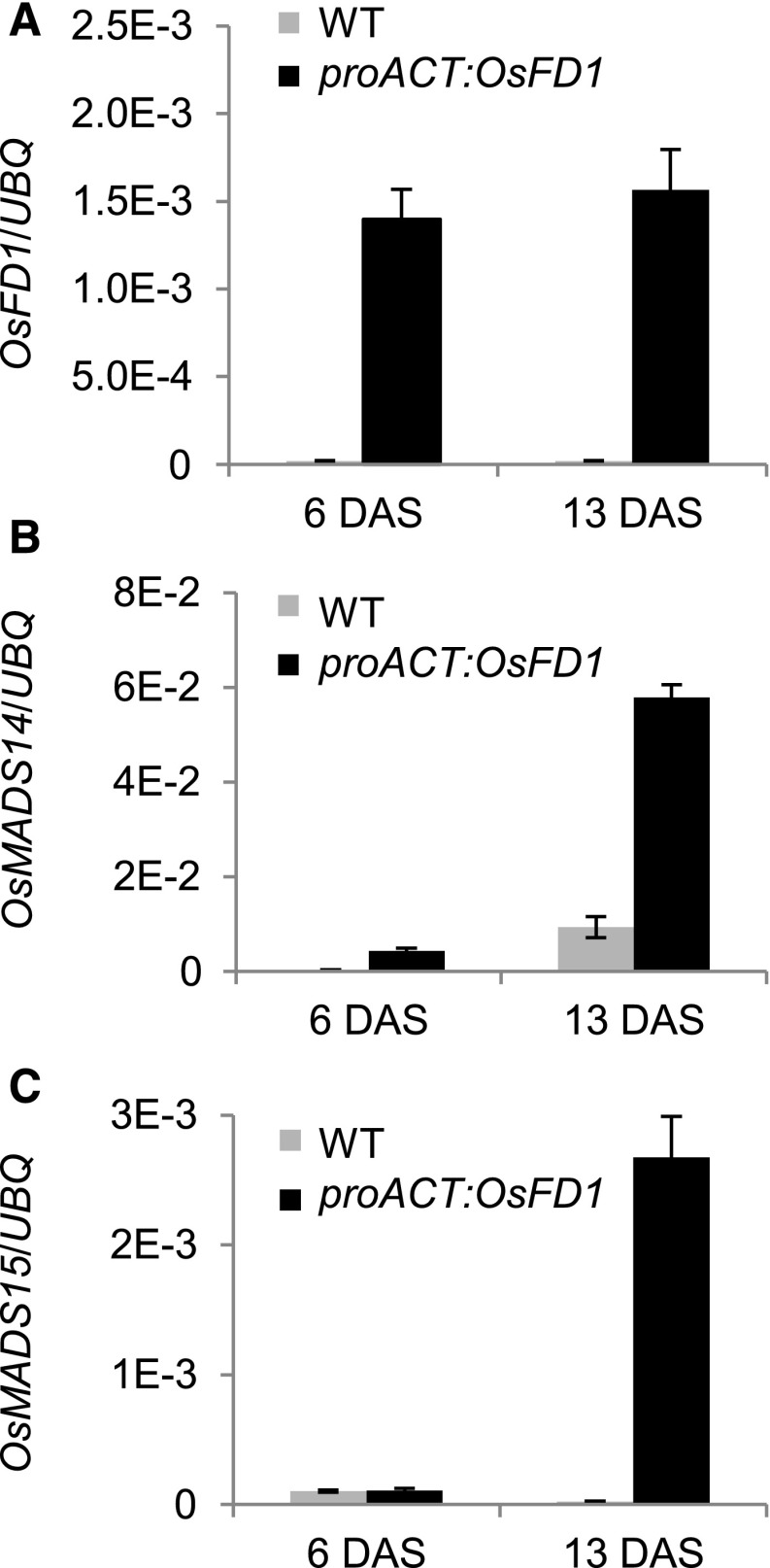
Based on relative transcript quantifications, OsFD1 maximum expression was 5 times lower relative to Hd3a or RFT1 and ∼50 times lower than Gf14c (compared with y axis scales in Supplemental Figures 1A to 1C). Although relative mRNA amounts cannot be accurately compared between genes, these data suggested that OsFD1 might be a limiting factor to FAC formation in leaves. To test this hypothesis, the coding sequence of OsFD1 was expressed under the constitutive rice ACTIN2 promoter (proACT:OsFD1), and expression of OsMADS14 and OsMADS15 was quantified at 6 and 13 d after shifting plants from LD to SD (Figures 1A to 1C). In proACT:OsFD1 plants, OsMADS14 and OsMADS15 expression was strongly upregulated in leaves at the indicated time points, compared with wild-type plants grown under the same conditions, indicating that increasing OsFD1 abundance results in higher induction of FAC target genes (Figures 1B and 1C).
Figure 1.
Overexpression of OsFD1 in Leaves Induces Transcription of Targets of the FAC.
Expression of OsFD1 (A), OsMADS14 (B), and OsMADS15 (C) in leaves of transgenic proACT:OsFD1 plants. Plants were grown under LD (14.5 h light) for 6 weeks and then shifted to SD (10 h light). Leaves were collected at ZT0 at 6 and 13 d after shift to SD (DAS). UBIQUITIN (UBQ) was used as standard for quantification of gene expression. Data are represented as mean ± sd. E-n = × 10−n. ANOVA tests for graphs in (A) to (C) are shown in Supplemental File 1.
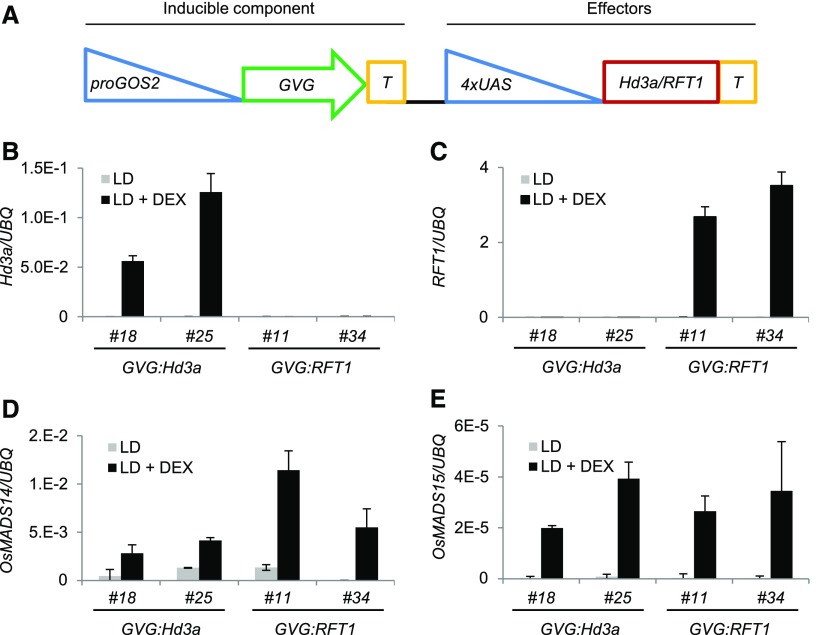
Following the same rationale, we conditionally overexpressed Hd3a or RFT1 in leaves under LD, when Gf14c and OsFD1, but not Hd3a or RFT1, are expressed. To control overexpression, dexamethasone-inducible (DEX) Hd3a- or RFT1-overexpressing plants were produced (proGOS2:GVG 4xUAS:Hd3a and proGOS2:GVG 4xUAS:RFT1; hereafter referred to as GVG:Hd3a and GVG:RFT1; Figure 2A). We used a previously validated system for inducible gene expression, composed of a DEX-inducible component that drives expression of the genes of interest (Ouwerkerk et al., 2001). Using this system, we avoided the need for a chimeric florigen-glucocorticoid receptor protein, whose size might impinge on Hd3a or RFT1 protein movement or activity.
Figure 2.
Expression of OsMADS14 and OsMADS15 in Leaves Is Dependent on Expression of Hd3a and RFT1.
(A) Schematics of the inducible system used in this study. The GVG chimeric protein is expressed under the GOS2 promoter to produce the inducible part of the vector. The Hd3a or RFT1 coding sequences are cloned under the control of the 4x UPSTREAM ACTIVATION SEQUENCE (UAS) to produce the effector component of the vector. T indicates the terminator.
(B) to (E) Expression of Hd3a (B), RFT1 (C), OsMADS14 (D), and OsMADS15 (E) in leaves of DEX-inducible transgenic plants grown under LD. Leaves were harvested at ZT0. GVG:Hd3a and GVG:RFT1 indicate DEX-inducible Hd3a- and RFT1-overexpressing lines, respectively. Two independent transgenic lines are shown for each construct. Plants were either DEX- or mock-treated, and transcripts were quantified using primers designed on the coding sequences. UBQ was used as standard for quantification of gene expression. Data are represented as mean ± sd. xE-n = × 10−n. ANOVA tests for graphs in (B) to (E) are shown in Supplemental File 1.
Transgenic plants containing GVG:Hd3a or GVG:RFT1 could overexpress transgenic Hd3a or RFT1 only upon DEX treatments (Figures 2B and 2C). While a negligible basal expression of OsMADS14 and OsMADS15 was observed in leaves of untreated plants under LD, expression of OsMADS14 and OsMADS15 was strongly activated 16 h after DEX treatment, concomitantly to Hd3a or RFT1 induction (Figures 2D and 2E).
Taken together, these experiments indicate that OsMADS14 and OsMADS15 transcription in leaves is activated upon coexpression of all FAC components that are likely to form an active complex, as in the SAM.
A Negative Feedback Loop Independent of OsFD1 Limits Florigen Expression in Leaves
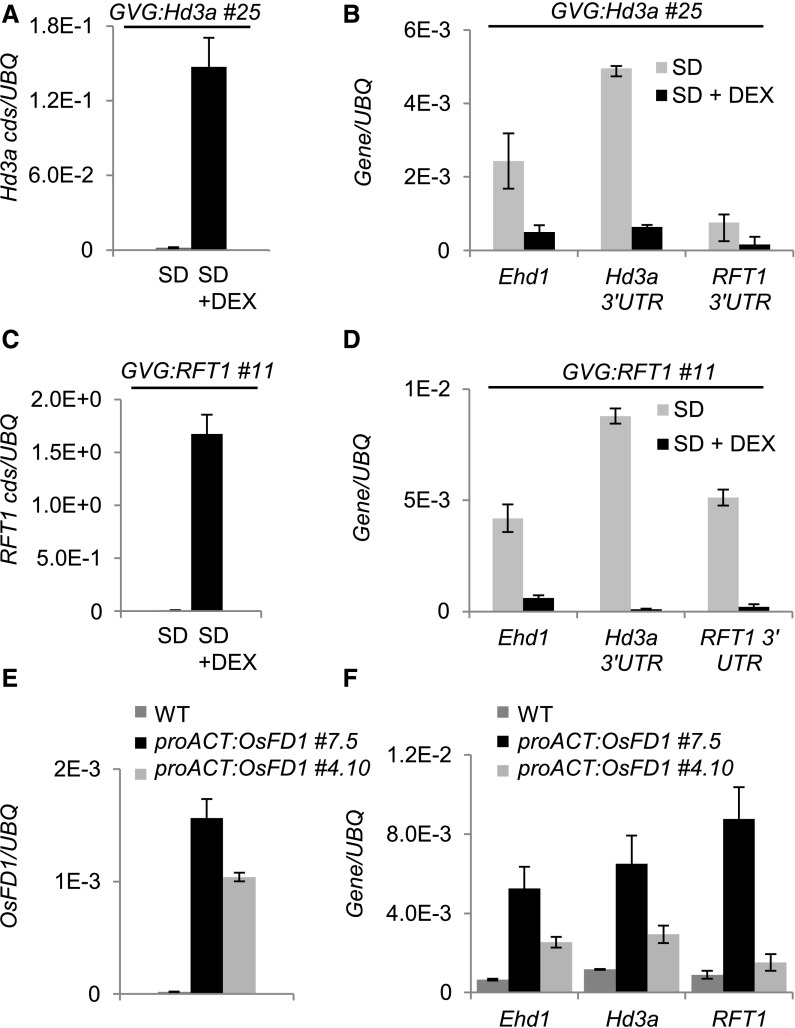
The expression of Hd3a and RFT1 is transiently activated in leaves of plants grown under natural field or artificial conditions. This observation suggests the existence of a mechanism that downregulates their expression upon commitment to flowering and that could possibly depend on Ehd1, encoding a common upstream promoter of Hd3a and RFT1 expression (Goretti et al., 2017; Ogiso-Tanaka et al., 2013; Gómez-Ariza et al., 2015). Under our growing conditions, expression of the florigens reached a peak ∼12 to 15 d after shifting plants from LD to SD (Galbiati et al., 2016). We tested whether Hd3a and RFT1 are causal to their own downregulation in leaves after the floral transition. The GVG:Hd3a or GVG:RFT1 transgenic plants were grown under LD (16 h light) and then shifted to SD (10 h light) to induce expression of the endogenous Hd3a and RFT1 transcripts in leaves. After 13 SD, half of the plants were DEX treated to overexpress transgenic Hd3a or RFT1 (Figures 3A and 3C). Leaf samples were harvested 16 h after DEX treatment at ZT0, when endogenous Ehd1, Hd3a, and RFT1 were highly expressed. Quantification of transcripts indicated that the endogenous Ehd1, Hd3a, and RFT1 transcripts were strongly downregulated in DEX-treated plants compared with mock-treated controls (Figures 3B and 3D). A similar reduction of transcripts abundance was observed when either of the two florigens was induced (Figures 3A to 3D). We tested several independent lines of both GVG:Hd3a and GVG:RFT1 for DEX-dependent control of Ehd1, Hd3a, and RFT1 transcripts. Despite a varying degree of inducibility among independent transgenic lines, as quantified by the increase in Hd3a and RFT1 expression in response to DEX, we consistently observed reduction of endogenous Ehd1, Hd3a, and RFT1 transcripts (Supplemental Figures 2A and 2B). Therefore, both Hd3a and RFT1 can mediate a negative feedback loop on Ehd1 and, indirectly, on their own expression. The negative loop is activated also at low levels of expression of transgenic Hd3a or RFT1, suggesting that it finely adjusts expression of the florigens during floral induction.
Figure 3.
A Negative Feedback Loop Independent of OsFD1 Reduces Ehd1, Hd3a, and RFT1 Expression during Floral Induction in Leaves.
(A) to (D) DEX-induced overexpression of Hd3a ([A] and [B]) or RFT1 ([C] and [D]) causes strong increase of Hd3a (A) or RFT1 (C) transcript accumulation from transgenic sequences, but downregulation of Ehd1, Hd3a, and RFT1 endogenous transcripts, compared with mock-treated controls ([B] and [D]).
(E) and (F) Two independent transgenic proACT:OsFD1 lines show increased expression of OsFD1 (E) and of Ehd1, Hd3a, and RFT1 in leaves compared with the wild type (F). DEX was applied at 13 DAS, and leaf samples were collected at ZT0, 16 h later. proACT:OsFD1 plants were collected at ZT0 and 12 DAS. Leaves from 10 plants per treatment were sampled. UBQ was used as standard for quantification of gene expression. Data are represented as mean ± sd. Primers on Hd3a or RFT1 coding sequences or on the 3′ untranslated regions were used to distinguish transgenic+endogenous ([A] and [C]) from endogenous transcripts, respectively ([B] and [D]). ANOVA tests for graphs in (A) to (F) are shown in Supplemental File 1.
A canonical OsFD-containing FAC could be required for negative regulation of Hd3a and RFT1 expression. Since OsFD1 is limiting to FAC formation in leaves at 12 d after shift (DAS), expression of the florigens was analyzed in proACT:OsFD1 plants at this time point. Compared with wild-type plants, constitutive expression of OsFD1 induced the upregulation of Hd3a, RFT1, and Ehd1 expression (Figures 3E and 3F). These data suggest that OsFD1 can promote expression of Ehd1, Hd3a, and RFT1 in leaves and is not part of the mechanism that self-limits expression of the florigens.
Identification of FAC Components Expressed in Leaves
In rice and other plant species, many bZIP TFs have been already described that form alternative FACs with the florigens and control different developmental processes (Tylewicz et al., 2015; Tsuji et al., 2013; Li et al., 2015). We evaluated whether other TFs abundant in leaves might form alternative FACs with a flowering repressive function. We performed untargeted and targeted yeast two-hybrid screens using Hd3a and RFT1 as baits. Only the results of targeted screens will be presented in this study. We selected members of the bZIP family of transcription factors based on sequence similarity with OsFD1, wheat TaFDL2 (Li et al., 2015; Li and Dubcovsky, 2008) and maize DLF1 (Muszynski et al., 2006) (Supplemental Figure 3A and Supplemental Data Set 1), and we tested their interaction with Hd3a and RFT1. Since it has been shown that bZIP TFs bind DNA by forming homo- and heterodimers, we also tested their ability to homo- and heterodimerize. OsFD1 interaction with Gf14c was used as positive control (Taoka et al., 2011). A summary of all interactions is reported in Table 1. We excluded from this analysis OsbZIP29 as we could not amplify it from cDNA of LD- or SD-grown plants, bZIP54/OsFD6 as it is inferred to be a pseudogene (Tsuji et al., 2013), and finally genes whose interaction patterns have already been determined (Tsuji et al., 2013). The OsbZIP24/OsFD3 and OsbZIP69/OsFD4 proteins could not interact in our yeast assay with Hd3a or RFT1, although a recent report indicates weak interaction with RFT1 (Jang et al., 2017). OsbZIP24/OsFD3 could interact with Gf14c, while OsbZIP69/OsFD4 could not. Conversely, OsbZIP62, OsbZIP42, and OsbZIP9 could interact with Hd3a but not with RFT1, indicating some binding preference for one of the florigens. However, they also interacted with Gf14c, which could possibly bridge the interaction with both florigens.
Table 1. Targeted Yeast Two-Hybrid Analysis between Hd3a, RFT1, Gf14c, and Selected OsbZIPs.
| AD Clones | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hd3a | RFT1 | Gf14c | OsFD1 | OsbZIP69/OsFD4 | OsbZIP24/OsFD3 | OsbZIP62 | OsbZIP9/HBF2 | OsbZIP42/HBF1 | Empty AD | ||
| BD Clones | Hd3a | – | – | 20 | - | – | – | 15 | 20 | 20 | – |
| RFT1 | – | – | 20 | - | – | – | – | – | – | – | |
| Gf14c | – | – | 20 | 20 | – | 15 | 10 | 20 | 20 | – | |
| OsFD1 | – | – | 10 | – | – | – | – | – | n.t. | – | |
| OsbZIP69/OsFD4 | – | – | – | – | 20 | 20 | – | – | – | – | |
| OsbZIP24/OsFD3 | – | – | 15 | – | – | 20 | – | – | – | – | |
| OsbZIP62 | – | – | 20 | – | – | – | – | n.t. | – | – | |
| OsbZIP9/HBF2 | – | – | 10 | – | – | – | – | n.t. | – | – | |
| OsbZIP42/HBF1 | 10 | – | 15 | – | – | – | – | – | n.t. | – | |
| Empty BD | – | – | – | – | – | – | – | – | – | – | |
Interaction strength is shown as the highest 3-aminotriazole concentration on which diploid colonies could grow when plated on selective medium. A dash indicates no interaction. n.t., not tested. BD fusions were expressed in yeast strain Y187 (matα), and AD fusions were expressed in yeast AH109 (matA). Diploid yeast was produced by mating. Growth was observed after 6 d at 30°C.
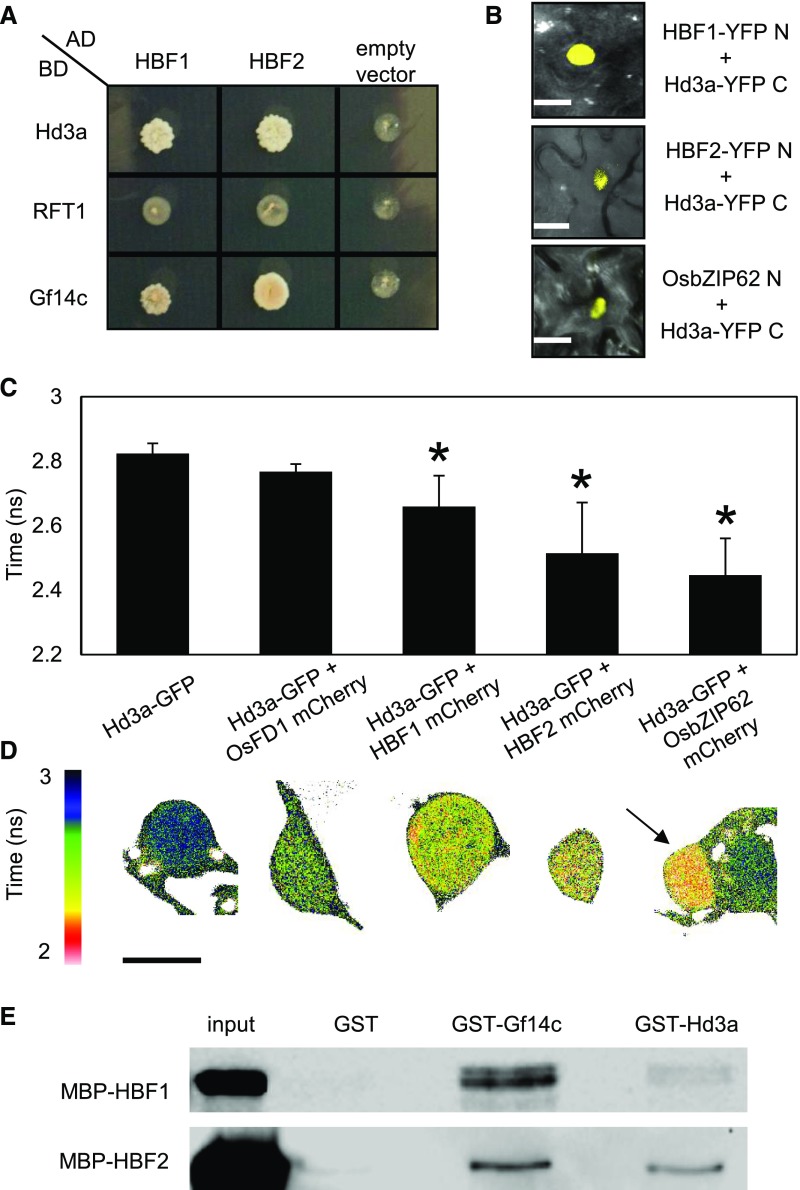
Among the bZIP TFs tested, we identified OsbZIP62, OsbZIP42, and OsbZIP9 as interactors of Hd3a and Gf14c (Table 1, Figure 4A). Based on their functional characterization, we renamed OsbZIP42 and OsbZIP9 as Hd3a BINDING REPRESSOR FACTOR1 (HBF1) and HBF2, respectively. The HBF1 and HBF2 proteins share 19.13% and 20.75% amino acid identity with OsFD1 and cluster in the same branch of the bZIP phylogenetic tree (Supplemental Figure 3A). They share 68% identity with each other when the full-length proteins are considered.
Figure 4.
HBF1 and HBF2 Interact with GF14c and Directly with Hd3a.
(A) Yeast two-hybrid assays between Hd3a, RFT1, and Gf14c fused to the binding domain (BD) and HBF1 or HBF2 fused to the activation domain (AD) of Gal4. Colonies were grown on selective -L-W-H medium supplemented with 10 mM 3-aminotriazole.
(B) BiFC assays showing restored YFP fluorescence in nuclei upon coexpression of Hd3a-YFP C with HBF1-YFP N, HBF2-YFP N, or OsbZIP62-YFP N. Bar = 10 μm.
(C) FRET-FLIM measurements of the Hd3a-GFP donor lifetime in the presence of the acceptors OsFD1-mCherry (no FRET), HBF1-mCherry, HBF2-mCherry, or OsbZIP62-mCherry. The average lifetime of 10 transformed nuclei per measurement is shown ± sd. An asterisk indicates significance for P < 0.0003 (Student’s t test). ANOVA test for the graph is shown in Supplemental File 1.
(D) Color code indicating the lifetime of GFP at each pixel in one representative nucleus for the interactions shown in (C). For the interaction between Hd3a and OsbZIP62 two adjacent cells are shown, where only the left nucleus (arrow) coexpresses both constructs, while the right one expresses only Hd3a-GFP. Accordingly, shortened lifetime is observed only in the left nucleus.
(E) GST pull-down assay showing interactions between MBP-HBF1 and MBP-HBF2 with GST-Gf14c and GST-Hd3a, but not with GST alone. An immunoblot using an anti-MBP antibody is shown. Protein sizes are MBP-HBF1, 79.5 kD, and MBP-HBF2, 79.5 kD. Resin loading control is shown in Supplemental Figure 3E.
To further validate the direct interactions of HBF1, HBF2, and OsbZIP62 with Hd3a, bimolecular fluorescent complementation (BiFC) experiments were performed. The YFP N terminus was fused to each bZIP transcription factor creating HBF1-YFP N, HBF2-YFP N, and bZIP62-YFP N chimeric proteins, whereas the YFP C terminus was fused to Hd3a (Hd3a-YFP C) (Figure 4B). Leaves of Nicotiana benthamiana were infiltrated with Hd3a-YFP C and each of the bZIP chimeric fusions, and nuclei of the epidermis showed strong YFP fluorescence, indicating physical interactions between Hd3a and HBF1, HBF2, or bZIP62 as well as nuclear localization of the heterodimers. No fluorescence was observed in nuclei coexpressing OsFD1-YFP N and Hd3a-YFP C, confirming the indirect interaction between OsFD1 and Hd3a (Taoka et al., 2011).
Interactions were also assessed by Förster resonance energy transfer (FRET) fluorescence lifetime imaging microscopy (FLIM) (Berezin and Achilefu, 2010). In FRET-FLIM measurements, the readout for FRET is a reduced lifetime of the donor molecule in the FRET sample, compared with the donor-only sample. FRET occurs when two molecules interact directly. A decrease in the Hd3a-GFP donor lifetime was observed in the presence of HBF1-mCherry, HBF2-mCherry, and OsbZIP62-mCherry, confirming direct interactions in N. benthamiana epidermal nuclei (Figures 4C and 4D). No significant reduction of donor lifetime was observed when coexpressing Hd3a-GFP and OsFD1-mCherry (Figures 4C and 4D).
Direct interactions between HBF1, HBF2, and Hd3a were conclusively assessed in vitro by GST pull-down assays. We fused HBF1 and HBF2 to the maltose binding protein (MBP) and incubated them with either Gf14c-GST or Hd3a-GST immobilized on a glutathione resin. Both bZIPs bound Gf14c-GST and Hd3a-GST, but not GST alone (Figure 4E; Supplemental Figure 3E). These data confirm that interactions between HBF1, HBF2, and Hd3a occur in nuclei and do not require an intermediate 14-3-3 protein.
Finally, since bZIP TFs bind the DNA as dimers (Schütze et al., 2008; Reinke et al., 2013), we also tested the possibility that HBF1 and HBF2 could heterodimerize with each other or with OsFD1. We did not observe heterodimerization between these proteins in yeast (Table 1) or using the FRET-FLIM system (data not shown), indicating that HBF1, HBF2, and OsFD1 are likely part of distinct transcriptional complexes.
Diurnal time courses were used to determine the spatiotemporal expression of OsbZIP62, HBF1, and HBF2 (Supplemental Figures 3B to 3D). The mRNA expression of OsbZIP62 was most abundant in the SAM under SD and showed no strong oscillation during the 24-h cycle, despite a slight decline during the night. Transcript abundance was negligible in leaves, indicating that OsbZIP62 is likely not part of a complex limiting Hd3a expression in leaves but is possibly part of an Hd3a-containing complex in cells of the SAM (Supplemental Figure 3D). Transcripts of HBF1 and HBF2 were highly expressed in the SAM and showed expression also in leaves. HBF1 transcription in leaves reached a peak during the night, when Hd3a transcripts are also abundant (Supplemental Figures 3B and 3C). Taken together, these data indicate that HBFs can potentially form distinct complexes both in the SAM and leaves.
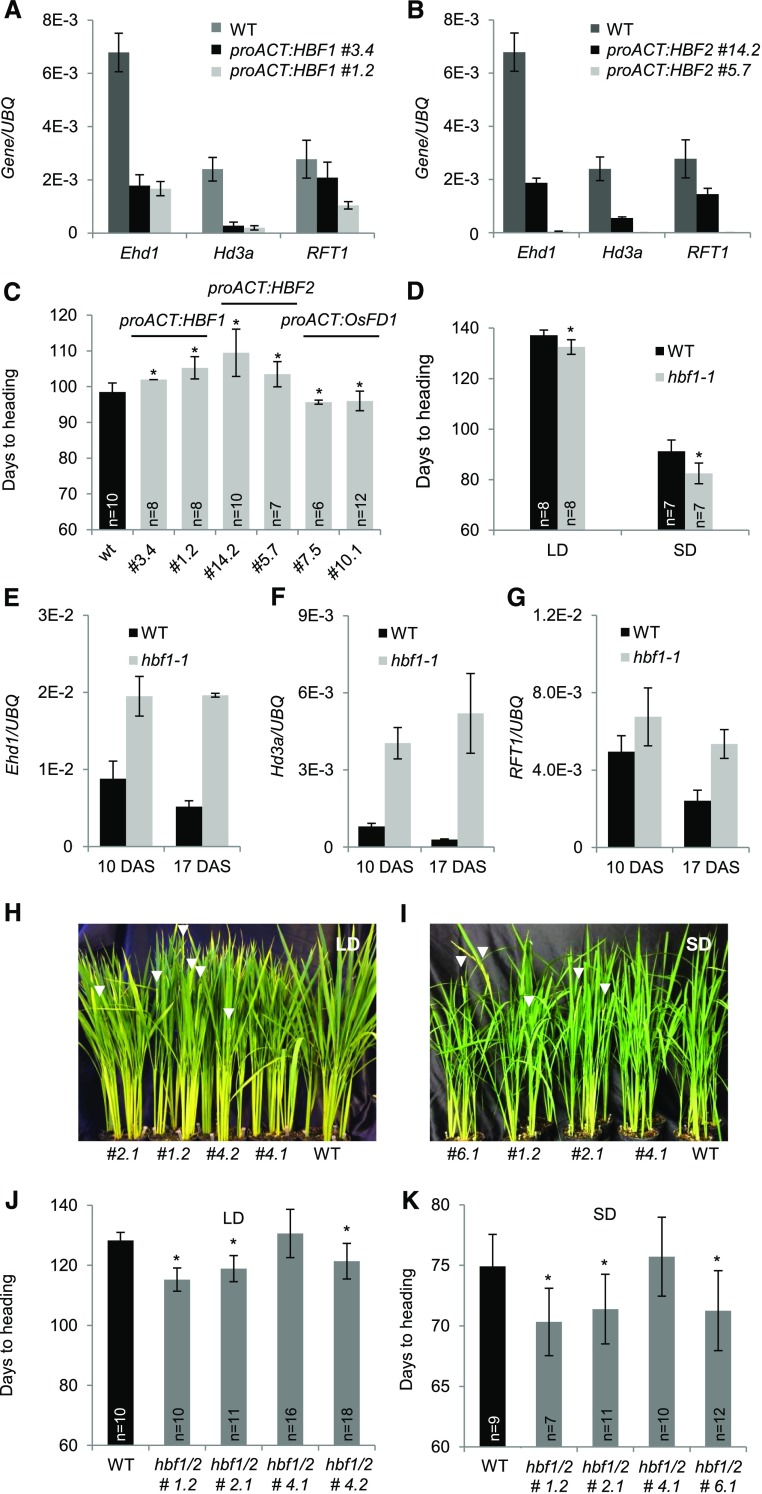
HBF1 and HBF2 Encode Floral Repressors That Reduce Ehd1, Hd3a, and RFT1 Expression in Leaves
Whether HBF1 and HBF2 could influence flowering or expression of the florigens in leaves was assessed by overexpressing them under the constitutive ACT promoter (Supplemental Figures 3F and 3G). Expression of Ehd1, Hd3a, and RFT1 was monitored during photoperiodic induction of plants shifted from LD (16 h light) to SD (10 h light). Leaves of the proACT:HBF1 and proACT:HBF2 plants showed a marked downregulation of Ehd1, Hd3a, and RFT1 expression compared with the wild type, unlike what observed in proACT:OsFD1 transgenic plants (Figures 5A and 5B). In agreement with the overall downregulation of the Ehd1-florigen module, proACT:HBF1 and proACT:HBF2 plants flowered late when grown for 2 months under LD and then shifted to SD (Figure 5C).
Figure 5.
HBF1 and HBF2 Encode Floral Repressors Reducing Ehd1 Expression.
(A) and (B) Quantification of mRNA levels of Ehd1, Hd3a, and RFT1 in leaves of proACT:HBF1 (A) and proACT:HBF2 (B) overexpression plants grown for 8 weeks under LD (16 h light) and then shifted to SD (10 h light). UBQ was used as standard for quantification of gene expression. Data are represented by mean ± sd.
(C) Days to heading of wild type, proACT:HBF1, proACT:HBF2, and proACT:OsFD1 overexpressors grown for 8 weeks under LD (16 h light) and then shifted to SD (10 h light).
(D) Heading dates of wild type (Dongjin) and hbf1-1 mutants grown under continuous LD (14.5 h light) or continuous SD (10 h light).
(E) to (G) Expression of Ehd1 (E), Hd3a (F), and RFT1 (G) in hbf1-1 mutant plants compared with the wild type.
(H) to (K) mRNA levels are shown at 10 and 17 d after shifting plants from LD to SD.
(H) and (I) Nipponbare wild type and T2 hbf1 hbf2 CRISPR mutants grown under continuous LD (14.5 h light) (H) or shifted from LD (16 h light) to SD (10 h light) 8 weeks after sowing (I). Arrowheads indicate the emerging panicles.
(J) and (K) Quantification of heading dates in the same plants as in (H) and (I), respectively (n indicates the number of plants scored). Asterisks indicate P < 0.05 in an unpaired two tailed Student’s t test. E-n = × 10−n. The detailed genotypes of the mutants are reported in Supplemental Figure 5C.
ANOVA tests for graphs in (A) to (G), (J), and (K) are shown in Supplemental File 1.
We obtained the hbf1-1 mutant from the PFG T-DNA collection in the cultivar Dongjin (Jeon et al., 2000). Quantification of transcripts in the mutant showed that expression of HBF1 was strongly reduced because of insertion of the T-DNA in the promoter (Supplemental Figures 4A and 4B). We analyzed the flowering behavior of the hbf1-1 mutant and observed that it headed earlier by ∼5 d compared with segregating wild-type siblings under continuous LD (14.5 h light) and by ∼9 d under SD (10 h light) (Figure 5D). To link the mutant phenotype with photoperiodic regulation of the Ehd1-florigen module, transcript abundance of Ehd1, Hd3a, and RFT1 was determined at two time points after shifting plants from LD to SD (10 and 17 DAS). The mRNA accumulation of all genes was higher in the hbf1-1 mutant compared with the wild type at both time points, indicating derepression of the module (Figures 5E to 5G). To exclude an indirect effect of HBF1 on Ehd1 expression, the expression of six genes upstream of Ehd1 was also measured (Supplemental Figures 4C and 4D). None of them showed a difference in gene expression between the wild type and the hbf1-1 mutant. The only exception was Ghd7, which was slightly downregulated in the mutant compared with the wild type (Supplemental Figure 4D).
To confirm that loss of HBF1 function promotes flowering and also to assess a possible functional redundancy between HBF1 and HBF2, we generated a series of double hbf1 hbf2 mutants in the cultivar Nipponbare, using the CRISPR/Cas9 technology (Miao et al., 2013). We designed a single-guide RNA (sgRNA) on a region highly conserved between HBF1 and HBF2 on their first exon, to simultaneously target both loci (Supplemental Figure 5A). Upon regeneration of transgenic plants, we obtained six independent lines harboring different combinations of biallelic or homozygous indels (Supplemental Figure 5B). We selected five T2 lines (#1.2, #2.1, #4.1, #4.2, and #6.1) from four independent T1s (#1, #2, #4, and #6), all of which were homozygous for hbf1 mutations and homozygous or biallelic for hbf2 (Supplemental Figure 5C). All lines were double hbf1 hbf2 loss-of-function mutants, except line #4.1, which contained a homozygous −27 bp in-frame deletion at the HBF1 locus, likely not causing loss of gene function (Supplemental Figure 5C). We measured their flowering time under LD (14.5 h light) and after growth for 8 weeks under LD followed by SD (10 h light). Under both conditions, all hbf1 hbf2 double loss-of-function mutants flowered earlier compared with the wild type (Figures 5H to 5K), but flowering was not accelerated in line #4.1. These data indicate that loss of nine amino acids (EDFLVKAGV before the bZIP domain) in the HBF1 protein likely does not affect its function. They further indicate that the hbf2 mutation does not additively contribute to the phenotype caused by single hbf1 mutations. As opposed to the effect of the hbf1-1 allele in Dongjin, the Nipponbare hbf1 hbf2 CRISPR mutants showed predominantly accelerated flowering under LD (∼13 d was the largest difference observed between line #1.2 and the wild type), rather than under SD (the same line #1.2 flowered ∼5 d earlier than the wild type). We attribute these differences to the different sensitivity of Dongjin and Nipponbare to loss of HBF1 function.
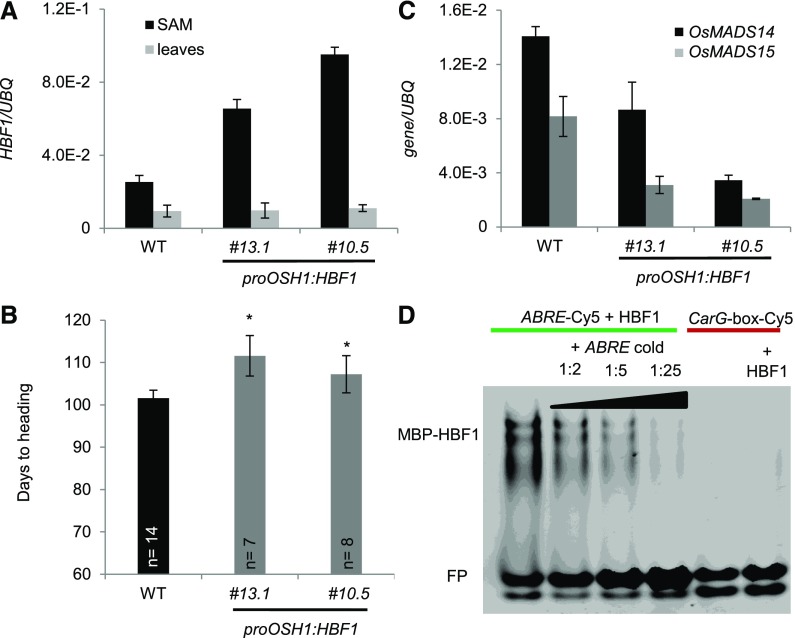
HBF1 Can Bind the Ehd1 Promoter
Expression of Ehd1 is dependent upon HBF1 activity. The Ehd1 promoter region was scanned in search of conserved motifs recognized by bZIP TFs, and we found three CACGTC motifs that are characteristic of abscisic acid response elements (ABREs) and G-boxes (Li and Dubcovsky, 2008) (Supplemental Figure 5D). As expected by the central position of Ehd1 in flowering regulatory networks, many other motifs were identified in its promoter region spanning 1.5 kb upstream of the ATG (Supplemental Figure 5D). The possibility of a direct interaction between HBF1 and the Ehd1 promoter was assessed using electrophoretic mobility shift assay. The HBF1 protein was purified and incubated with a Cy5-labeled oligonucleotide identical to the region of the Ehd1 promoter containing the ABRE, located at −482 bp (Supplemental Figure 5D). HBF1 binding to this oligonucleotide resulted in a band shift (Figure 6D). Addition of an excess of unlabeled oligonucleotide reversed the shift of the fluorescent probe. However, no band shift could be detected when HBF1 was incubated with a promoter fragment containing a CArG-box, demonstrating that HBF1 binding to the ABRE-containing region was specific (Figure 6D). No ABREs or G-boxes were identified by scanning the Hd3a or RFT1 promoters, although indirect binding of HBF1 to these genes cannot be completely excluded.
Figure 6.
HBF1 Represses Flowering at the SAM.
(A) Quantification of HBF1 expression in SAMs and leaves of plants misexpressing HBF1 from the OSH1 promoter. Two independent transgenic lines are shown.
(B) Heading dates of proOSH1:HBF1 transgenic plants grown for 8 weeks under LD (16 h light) and then shifted to SD (10 h light) (n indicates the number of plants scored). Asterisks indicate P < 0.05 in an unpaired two-tailed Student’s t test.
(C) Quantification of OsMADS14 and OsMADS15 expression in SAMs of transgenic proOSH1:HBF1 plants. Samples in (A) and (C) were collected from apical meristems grown under LD and then exposed to 12 inductive SD. UBQ was used as standard for quantification of gene expression. All data are represented by mean ± sd. E-n = × 10−n.
(D) Electrophoretic mobility shift assay between MBP-HBF1 and ABRE-Cy5 (lanes 1–4) and HBF1 and CArG-box-Cy5 (lane 6). The specificity of interaction between HBF1 and ABRE-Cy5 was tested by incubation with increasing amounts of unlabeled oligonucleotides (labeled/unlabeled oligonucleotide ratios 1:2, 1:5, and 1:25). HBF1 was incubated with an oligonucleotide containing a CArG-box-Cy5 (lanes 5 and 6) as a negative control. FP, free probe.
ANOVA tests for graphs in (A) to (C) are shown in Supplemental File 1.
HBF1 Represses Transcription of OsMADS14 and OsMADS15 in the Shoot Apical Meristem
The HBF1 and HBF2 transcripts could be identified in both leaves and SAMs, suggesting that they are expressed in both florigen-producing and -receiving tissues. Their overexpression delayed flowering, and in leaves it reduced mRNA expression of Hd3a and RFT1. Whether these proteins also had a role in the SAM to control flowering or gene expression was tested by misexpression studies. To this end, the promoter of ORYZA SATIVA HOMEOBOX1 (proOSH1) was cloned and used to drive expression of HBF1. OSH1 is expressed in undifferentiated cells of the SAM but not in organ primordia arising from it (Itoh et al., 2000; Sentoku et al., 1999). Transgenic proOSH1:HBF1 rice plants that overexpressed HBF1 were produced. Transcriptional analysis of leaves and SAMs of T2 lines indicated that expression driven by the OSH1 promoter was effective at increasing expression of HBF1 at the SAM but not in leaves (Figure 6A). The same plants had delayed flowering by a few days compared with nontransgenic segregating controls (Figure 6B). Our dissection of SAMs included also some of the youngest leaf primordia arising from the meristem; however, the OSH1 promoter is not active in this tissue (Tsuda et al., 2011). Thus, we conclude that the flowering delay is caused by increased expression of HBF1 in meristematic cells. Transcripts of Hd3a and RFT1 were not expressed at the meristem; therefore, although we cannot fully exclude the expression of other FT-like genes, feedback regulation of these florigens is likely not occurring at the apex.
Finally, the expression of OsMADS14 and OsMADS15 was found to be significantly reduced in SAMs (Figure 6C). These data indicate that HBF1 can repress flowering and expression of inflorescence identity genes at the SAM and therefore has a dual transcriptional repressive function in distinct plant compartments.
DISCUSSION
Dexamethasone treatment of plants expressing inducible versions of Hd3a and RFT1 indicated the existence of transcriptional repression of the florigens mediated by a feedback negative loop.
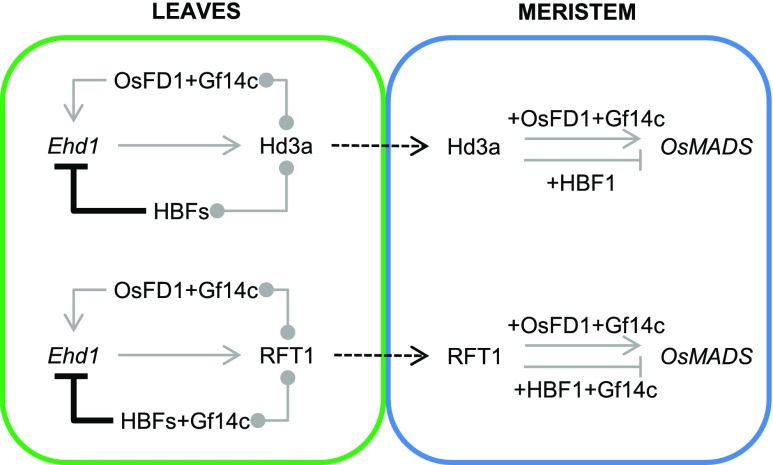
Thus, we propose a modification of the rice floral induction model to include an autoregulatory loop centered on Hd3a and RFT1. The florigens regulate their own expression in leaves by forming distinct FACs with several OsbZIP proteins (Figure 7). These complexes can either promote or repress Ehd1, Hd3a, and RFT1 depending on the interacting bZIP. In particular, OsFD1 acts as transcriptional activator in leaves, whereas the closely related HBFs repress expression of the florigens in the same tissue. Thus, Hd3a and RFT1 proteins can engage in both florigen activation and repression complexes. Binding of HBF1 to the promoter of Ehd1 further provides molecular evidence for feedback regulation of the florigens. The preference of RFT1 and Hd3a to interact with OsFD1 or the HBFs can be driven by relative expression patterns or modifications of OsFD1 and the HBFs under different growing conditions. Both the HBF1 and HBF2 transcripts are expressed in the SAM as well, and tissue-specific overexpression of HBF1 at least, could reduce the expression of targets of the FAC at the apex.
Figure 7.
Combinatorial Circuitry Controlling Production of and Response to Florigenic Proteins in Rice.
In leaves, Hd3a and RFT1 can promote expression of Ehd1 by forming a canonical FAC with OsFD1 and Gf14c, and they can repress it by interacting with HBFs. Hd3a can interact directly with HBFs, whereas RFT1 might interact indirectly with HBFs through GF14c. Binding of HBF1 to the Ehd1 promoter is direct. Upon translocation to the meristem, Hd3a and RFT1 proteins can promote transcription of OsMADS target genes by forming a canonical FAC. HBF1 at least can repress transcription of the same targets by forming a repressive FAC. Gray arrows and flat-end arrows indicate transcriptional activation and repression, respectively. Connectors indicate protein-protein interactions. Thick, black flat-end arrows indicate direct repression by protein-DNA binding. Dashed arrows indicate protein movement.
These data identify a previously unknown function for the rice florigens in leaves and suggest the existence of a regulatory layer limiting Hd3a and RFT1 signaling to fine-tune production of the florigens in leaves and their effect on gene regulatory networks at the apical meristem.
The Rice Florigens Act in Leaves to Regulate Their Own Expression
A growing number of studies demonstrate that FT-like proteins are involved in a wide range of developmental processes, including tuberization (Navarro et al., 2011), bulbing (Lee et al., 2013), stomatal opening (Kinoshita et al., 2011), leaf curling (Teper-Bamnolker and Samach, 2005), vegetative growth in trees (Hsu et al., 2011), plant architecture in tomato (Park et al., 2014), and tillering in rice (Tsuji et al., 2015). In many such instances, they function in tissues different from the SAM. However, FT-like proteins have been most prominently described in the context of flowering time control in response to environmental cues. During this process, they act as long distance flowering promoters produced in leaves and translocated to the SAM, inducing developmental switches upon the formation of a FAC (Lifschitz et al., 2006; Corbesier et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007). The data presented in this study suggest that a FAC can form also in rice leaves to activate expression of the same targets normally transcribed in the SAM. That a FAC is active also in leaves was initially suggested by experiments in Arabidopsis (Teper-Bamnolker and Samach, 2005). Expression of FT or Tomato FT (TFT) in transgenic Arabidopsis plants from the viral 35S promoter caused leaf curling that could be suppressed by mutating FD, SEP3, or FUL. These data indicated that a FAC formed in leaves under specific conditions could perturb leaf development by promoting transcription of targets usually expressed at the SAM (Teper-Bamnolker and Samach, 2005).
Whether a FAC has any biologically relevant function in leaves of Arabidopsis remains to be clarified. However, the identification of Ehd1, Hd3a, and RFT1 as targets of florigen-containing complexes in leaves of rice suggests that one function of these complexes is feedback tuning of the expression of some of its own components. In particular, by reducing transcription of Ehd1, florigen repressor complexes can indirectly limit expression of Hd3a and RFT1, downstream targets of Ehd1 (Doi et al., 2004; Zhao et al., 2015). Since seasonal expression of the rice florigens is transient and is strongly reduced upon completion of the floral transition, a plausible biological role for this autoregulatory loop could be to switch off transcription of the florigens upon floral commitment. Alternatively (or in parallel), it could fine-tune the production of Hd3a and RFT1 during photoperiodic induction (Gómez-Ariza et al., 2015; Ogiso-Tanaka et al., 2013). More data will be required to distinguish between these possibilities and validate them, but it is clear that reproductive commitment requires a tight balance between flowering promoting and repressive complexes, whose equilibrium could be controlled by modulating the expression levels of distinct bZIPs by developmental or environmental factors (Tang et al., 2016; Wu et al., 2014; Zhang et al., 2016) or by controlling their activity through phosphorylation (Kagaya et al., 2002; Choi et al., 2005; Furihata et al., 2006). Indeed, phosphorylation of OsFD transcription factors is required for binding to 14-3-3 proteins and is limiting to FAC function (Taoka et al., 2011).
Autoregulatory motifs are likely very common in gene regulatory networks but can be identified and studied only by quantifying endogenous transcripts in plants expressing transgenic copies of the same gene or its closely related homologs. Such approach has led to the identification of a loop regulating StSP6A expression, encoding a tuberigen, the mobile protein causing tuber formation at the apical meristem of potato stolons, and sharing high sequence similarity with Hd3a (Navarro et al., 2011). A similar autoregulatory loop in the expression of an endogenous florigen has been recently reported in chrysanthemum (Chrysanthemum seticuspe), where transcriptional induction of CsFTL3 required a complex formed by CsFTL3 and CsFDL1 proteins (Higuchi et al., 2013). It is noteworthy that regulatory loops involving two FT-like proteins are also very common among Angiosperms. The FT-like SP5G proteins of potato and tomato inhibit expression of the SINGLE FLOWER TRUSS (SFT) florigen and of StSP6A, respectively (Abelenda et al., 2016; Soyk et al., 2017). Similar modules in which an FT-like protein inhibits developmental transitions by repressing a second FT-like gene have been reported also for flowering in sugar beet (Beta vulgaris; Pin et al., 2010; Higuchi et al., 2013) and bulbing in onion (Allium cepa; Lee et al., 2013). In rice, both autoregulatory and relay mechanisms between Hd3a and RFT1 are possible under inductive conditions, when both proteins are expressed. Their differential ability to directly bind to HBFs might underlie differences in their capacity to take part in positive or negative relay mechanisms, but this type of cross-regulation is difficult to dissect genetically because of the redundancy between these factors. However, in general, autoregulatory and relay mechanisms among florigen-like proteins are emerging as very common modules controlling developmental switches.
Florigen-Containing Complexes Exhibit Combinatorial Properties
Florigen activation complexes from several species have a modular structure where distinct bZIP proteins can interact with different FT-like proteins in a combinatorial fashion (Sussmilch et al., 2015; Tsuji et al., 2013). Temporal and spatial dynamics of complex formation highly expand the regulatory possibilities of such complexes to control plant development. In rice leaves, Hd3a and RFT1 can form complexes displaying transcriptional promoting or repressive activity depending on the interacting bZIP. Since HBF1, HBF2, and OsFD1 do not heterodimerize, they cannot be part of the same complex, in agreement with their opposite functions. Additionally, since HBF1 and HBF2 do not interact with each other, they are possibly part of independent complexes.
Different examples in plants suggest that the functional specificity of these regulatory complexes can be provided by the bZIP as well as the FT-like protein. In rice, branching of shoots and altered panicle architecture are induced upon overexpression of OsFD2 (Tsuji et al., 2013). This bZIP can interact with Hd3a, and the interaction is bridged by the Gf14b protein. Given that OsFD2 controls patterns of vegetative growth, it could be speculated that FACs are active during distinct phases of the plant life cycle and not only during reproduction. Additionally, it raises the interesting possibility that complexes dynamically changing the Gf14 protein component might take on different roles. However, functional studies with Gf14 mutants are complicated by their pleiotropy and essential nature (Purwestri et al., 2009).
In hybrid aspen, overexpression of FDL1 but not FDL2 delays bud set and growth cessation, indicating FDL1 specificity for these developmental processes. However, both FDLs could interact with FT1 and FT2 to activate downstream targets in transient heterologous systems (Tylewicz et al., 2015). In these examples, specificity is likely contributed by the FD-like transcription factor.
Conversely, distinct PEBP components binding to the same bZIP protein can switch its function. Arabidopsis FD can interact with FT but also with TFL1, to form a flowering repressive complex (Hanano and Goto, 2011; Ho and Weigel, 2014). Similar interaction patterns are also possible in tomato between SP3G/SPP, an FD homolog, and the TFL1-like protein SELF PRUNING or the SFT florigen, where the balance between complexes regulates shoot architecture and, ultimately, yield (Pnueli et al., 2001; Park et al., 2014). Finally, the floral transition in Arabidopsis axillary meristems is controlled by the TCP transcription factor BRANCHED1, directly interacting with the PEBPs FT and TWIN SISTER OF FT but not with TFL1 (Niwa et al., 2013). Overall, these patterns indicate that a basal conserved module can be repurposed in distantly related species to control several developmental programs and that plasticity in complex assembly determines the balance between developmental programs.
METHODS
Plant Materials
The hbf1-1 mutant corresponds to the Salk line PFG_2D-00885 in the cultivar Donjing. Homozygous T-DNA insertional mutants were selected using primers listed in Supplemental Table 1. The cultivar Nipponbare was used in all other experiments.
Growth Conditions, Sampling, and Quantification of Gene Expression
Plants (rice [Oryza sativa]) were grown under LD (14.5 h light/9.5 h dark or 16 h light/8 h dark) or SD conditions (10 h light/14 h dark) in Conviron PGR15 growth chambers. Light was provided by T8 fluorescent and halogen incandescent lamps. Light intensity was adjusted to level 3 for both sets of lamps, resulting in ∼450 µmol/m2/s. Plant material was collected from the distal part of mature leaves, from at least three plants/time point, at ZT0. Only for the experiments described in Figures 5E to 5G and in Supplemental Figures 4C and 4D, plants were sampled at ZT20 under SD, as this time point corresponds to peak expression of Ehd1. Only for the data described in Figures 5A and 5B, all samples were quantified in the same experiments and then split into separate graphs for clarity of presentation. For SAM sampling, at least five apices/sample were manually dissected under a stereomicroscope using scalpels. Sample included the meristem, the two younger leaf primordia arising from it, as well as part of the rib meristem. RNA was extracted from leaves using the TRIzol reagent (Thermo Fisher Scientific) and from SAMs using the NucleoSpin RNA Plant kit (Macherey-Nagel). To prepare and quantify cDNAs, the RNA was retro-transcribed using the ImProm-II reverse transcriptase (Promega), and the Maxima SYBR qPCR master mix (Thermo Fisher Scientific) was used to measure gene expression in a Mastercycler Real Plex2 (Eppendorf). All primers used in RT-qPCR experiments have an annealing temperature of 60°C. For quantification of transcripts of Hd3a and RFT1 endogenous mRNAs, Ehd1, OsMADS14, OsMADS15, and UBQ, we used primers described by Galbiati et al. (2016) and Gómez-Ariza et al. (2015). All other primers used in this study are listed in Supplemental Table 1.
Construction of Transgenic Plants and DEX Treatments
The OsbZIP coding sequences were amplified from leaf or SAM cDNAs using primers listed in Supplemental Table 1 and subsequently cloned in pDONR207 (Invitrogen). Plant expression vectors were obtained by Gateway cloning, recombining the coding sequence after the ACTIN promoter in the pH2GW7 plasmid. The Hd3a and RFT1 coding sequences were amplified from leaves of Nipponbare with primers Os1-Os2 and Os3-Os2, respectively. The pINDEX2 vector was used for DEX-inducible expression of Hd3a and RFT1 (Ouwerkerk et al., 2001), but it was first turned into a Gateway-compatible (Invitrogen) destination vector by blunt cutting with PmlI and insertion of an EcoRV-digested Gateway RFC cassette. A proOSH1:Gateway destination construct was generated cloning a 1.5-kb promoter fragment using primers Os_6 and Os_7 (Supplemental Table 1). The pINDEX4 vector and proOSH1 were then cut using MunI and MluI and ligated to create pINDEX4 proOSH1. The RFA Gateway cassette was inserted into the proOSH1 pINDEX4 vector after blunt cutting using EcoRV and StuI. Subsequently, the DEX-inducible cassette was removed by blunt cutting using SwaI and BbrPI and self-ligation of the vector. The proOSH1:HBF1 vector was generated by LR recombination (Invitrogen).
For rice transformation, embryogenic calli were produced from Nipponbare seeds, prepared and transformed according to the protocol of Sahoo et al. (2011) using the EHA105 strain of Agrobacterium tumefaciens. Transgenic plants were selected on 50 mg/L and 100 mg/L hygromycin during selections I and II, respectively. Gene expression of Hd3a and RFT1 was induced by leaf spray with 10 μm DEX solution + 0.2% Tween, in transgenic homozygous T3 plants. DEX treatments were performed at ZT8 and sampling was done 16 h later at ZT0. Induction efficiency was assessed by RT-qPCR on leaves using primers specific for the Hd3a or RFT1 coding sequences.
Protein-Protein Interaction Studies
For yeast two-hybrid studies, the coding sequences were cloned into the vectors pGADT7 and pGBKT7 (Clontech) Gateway (Invitrogen) and transformed into AH109 and Y187 yeast strains, respectively. Interactions were tested by mating and growth of diploid yeast on selective -L-W-H medium supplemented with 3-aminotriazole. BiFC experiments were performed in Nicotiana benthamiana epidermal cells with the vectors pBAT TL-B sYFP-N and pBAT TL-B sYFP-C. FRET-FLIM experiments were performed in N. benthamiana epidermal cells transformed with the β-estradiol-inducible vectors pABIND-GFP and pABIND-mCherry (Bleckmann et al., 2010; Somssich et al., 2015). β-Estradiol induction of the transgenes was performed with 20 µM β-estradiol and 0.1% Tween 20 4 to 6 h before measurements. FRET-FLIM measurements were performed on 10 cotransformed nuclei at least, and mean, sd, and P value (Student’s t test) of the donor lifetime for the various sets of experiments were calculated, as described by Stahl et al. (2013).
GST Pull-Down
The GST-Hd3a and GTS-GF14c fusion proteins were obtained by recombining the coding sequence into pDEST15 (Invitrogen), expressing them using BL21 (DE3) cells (Invitrogen) and purifying them with Glutathione Sepharose 4b (Sigma-Aldrich). The concentration of each fusion protein was determined using Bradford assays. Equal amounts of GST-fusion proteins and GST were incubated in TIF buffer (150 mM NaCl, 20 mM Tris, pH 8.0, 1 mM MgCl2, 0.1% Nonidet P-40, and 10% glycerol) and added to 2 mL of clarified bacterial lysate of BL21 (DE3) cells expressing HBF1 and HBF2 proteins fused to MBP (pMAL vector adapted to Gateway system). The bacterial lysate was obtained by sonication of a bacterial pellet resuspended in TIF buffer supplemented with cOmplete Protease Inhibitor Cocktail (Roche). The reaction mixture was incubated for 2 h at 4°C under gentle rotation. After three washes with TIF buffer and two washes with PBS buffer, the resins were resuspended with SDS-PAGE loading buffer and eluted at 99°C for 5 min. The eluted proteins were resolved in 10% SDS-PAGE, and immunoblot analysis was performed using a monoclonal anti-MBP HRP-conjugated antibody (BioLabs).
Phylogenetic Analysis
Sequences of bZIP proteins were retrieved from public databases and aligned using the CLC Genomics Workbench program with the following parameters: gap open cost = 20.0; gap extension cost = 10.0; end gap cost = as any other; alignment mode = very accurate. An unrooted phylogenetic tree was created on the alignment using the neighbor-joining algorithm. Distances were measured using the Jukes-Cantor model. Bootstrap values are indicated at each node based on 1000 replicates. Sequence alignments are reported in Supplemental Data Set 1.
CRISPR-Cas9 Editing
The CRISPR-Cas9 vector was previously described (Miao et al., 2013). The single-guide RNA oligo (Os_934) targeting both HBF1 and HBF2 was designed based on the first exon of both genes, upstream of the region encoding the bZIP domain and expressed in transgenic Nipponbare. Transformation was performed as described above. The HBF1 and HBF2 loci in the regenerating plants were amplified and sequenced using primers Os_551-Os_338 and Os_976-Os_553, respectively, to identify the mutations introduced by nonhomologous end joining. The same primers were used to genotype the subsequent plant generations.
Electrophoretic Mobility Shift Assays
Consensus sequences in the Ehd1 promoter (1.5 kb upstream of the ATG) were identified using the Nsite software (Shahmuradov and Solovyev, 2015). The sequences of the ABRE and CArG-box containing primers are shown in Supplemental Table 1. The HBF1 protein fused to MBP was expressed in the Escherichia coli Rosetta strain and purified to homogeneity by passing it through a maltose column followed by an ion exchange step (MonoQ). Binding of HBF1 to the Ehd1 promoter was tested using 25 pmol of Cy5-labeled DNA duplexes (either ABRE or CArG-box sequences; Supplemental Table 1) mixed with 150 pmol of the purified protein in 20 mM Tris-HCl, pH 8.0, and 200 mM NaCl. In the competition studies, the mixture was supplemented with increasing amounts (1:2 to 1:25 molar ratio) of unlabeled DNA. Precast Novex TBE gels (Thermo Fisher Scientific) were used for the electrophoretic run.
Accession Numbers
Sequence data from this article can be found in the Rice MSU Genome Annotation Release 7 under the following accession numbers: LOC_Os06g06320.1 (Hd3a), LOC_Os06g06300 (RFT1), LOC_Os08g33370 (Gf14c), LOC_Os09g36910 (OsFD1), LOC_05g41070 (HBF1), LOC_Os01g59760 (HBF2), LOC_07g48660 (bZIP62), LOC_Os06g16370.1 (Hd1), LOC_Os10g32600.1 (Ehd1), LOC_Os07g15770.1 (Ghd7), LOC_Os07g49460.1 (PRR37), LOC_Os03g54160.1 (OsMADS14), and LOC_Os07g01820.1 (OsMADS15).
Supplemental Data
Supplemental Figure 1. Expression of FAC components and FAC targets in leaves.
Supplemental Figure 2. Independent Hd3a or RFT1 DEX-inducible transgenic lines show a range of Hd3a or RFT1 DEX-dependent induction and downregulation of Ehd1, Hd3a, and RFT1 endogenous expression.
Supplemental Figure 3. Selection of bZIP transcription factors putatively forming a transcriptional complex with the florigens.
Supplemental Figure 4. Analysis of the hbf1-1 mutant.
Supplemental Figure 5. Analysis of hbf1 hbf2 CRISPR mutants and of the HBF1 promoter.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis shown in Supplemental Figure 3A.
Supplemental File 1. ANOVA tables.
Acknowledgments
We thank Ludovico Dreni for providing the proACTIN overexpression vector, and Jin Miao and Li-Jia Qu for providing the rice CRISPR-Cas9 vectors. This work was supported by European Research Council Starting Grant 260963 to F.F.
AUTHOR CONTRIBUTIONS
V.B., Rü.S., and F.F. designed the research. V.B., D.M., D.G., M.S., M.d.R., M.C., F.G., R.S., and F.L. performed research. V.B. and F.F. wrote the article.
References
- Abelenda J.A., Cruz-Oró E., Franco-Zorrilla J.M., Prat S. (2016). Potato StCONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 26: 872–881. [DOI] [PubMed] [Google Scholar]
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Berezin M.Y., Achilefu S. (2010). Fluorescence lifetime measurements and biological imaging. Chem. Rev. 110: 2641–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A., Simon R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V., Fornara F. (2013). Molecular control of flowering in response to day length in rice. J. Integr. Plant Biol. 55: 410–418. [DOI] [PubMed] [Google Scholar]
- Cho L.-H., Yoon, J., Pasriga R., An G. (2016). Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 170: 2159–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.I., Park H.-J., Park J.H., Kim S., Im M.-Y., Seo H.-H., Kim Y.-W., Hwang I., Kim S.Y. (2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139: 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Hou Z., Ananiev E.V., Simmons C.R. (2008). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146: 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18: 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F., Chiozzotto R., Locatelli F., Spada A., Genga A., Fornara F. (2016). Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ. 39: 1982–1993. [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J., Galbiati F., Goretti D., Brambilla V., Shrestha R., Pappolla A., Courtois B., Fornara F. (2015). Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 66: 2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretti D., Martignago D., Landini M., Brambilla V., Gómez-Ariza J., Gnesutta N., Galbiati F., Collani S., Takagi H., Terauchi R., Mantovani R., Fornara F. (2017). Transcriptional and post-transcriptional mechanisms limit Heading Date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13: e1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Goto K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Narumi T., Oda A., Nakano Y., Sumitomo K., Fukai S., Hisamatsu T. (2013). The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 110: 17137–17142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W.W.H., Weigel D. (2014). Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26: 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.-Y., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 108: 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J.I., Kitano H., Matsuoka M., Nagato Y. (2000). Shoot organization genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12: 2161–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Foster R., Chua N.H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230: 1131–1144. [DOI] [PubMed] [Google Scholar]
- Jang S., Choi S.C., Li H.Y., An G., Schmelzer E. (2015). Functional characterization of phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS One 10: e0134987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Li H.-Y., Kuo M.-L. (2017). Ectopic expression of Arabidopsis FD and FD PARALOGUE in rice results in dwarfism with size reduction of spikelets. Sci. Rep. 7: 44477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaudal M., Zhang L., Che C., Putterill J. (2015). Three Medicago MtFUL genes have distinct and overlapping expression patterns during vegetative and reproductive development and 35S:MtFULb accelerates flowering and causes a terminal flower phenotype in Arabidopsis. Front. Genet. 6: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22: 561–570. [DOI] [PubMed] [Google Scholar]
- Kagaya Y., Hobo T., Murata M., Ban A., Hattori T. (2002). Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14: 3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Ono N., Hayashi Y., Morimoto S., Nakamura S., Soda M., Kato Y., Ohnishi M., Nakano T., Inoue S., Shimazaki K. (2011). FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 21: 1232–1238. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., Yoshida H., Nagamura Y., Kyozuka J. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Takahashi Y., Kobayashi Y., Monna L., Sasaki T., Araki T., Yano M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774. [DOI] [PubMed] [Google Scholar]
- Komiya R., Yokoi S., Shimamoto K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450. [DOI] [PubMed] [Google Scholar]
- Lee R., Baldwin S., Kenel F., McCallum J., Macknight R. (2013). FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat. Commun. 4: 2884. [DOI] [PubMed] [Google Scholar]
- Li C., Dubcovsky J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55: 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lin H., Dubcovsky J. (2015). Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 84: 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A., Irish V.F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165: 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.-J. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23: 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimida N., Kidou S., Iwanami H., Moriya S., Abe K., Voogd C., Varkonyi-Gasic E., Kotoda N. (2011). Apple FLOWERING LOCUS T proteins interact with transcription factors implicated in cell growth and organ development. Tree Physiol. 31: 555–566. [DOI] [PubMed] [Google Scholar]
- Muszynski M.G., Dam T., Li B., Shirbroun D.M., Hou Z., Bruggemann E., Archibald R., Ananiev E.V., Danilevskaya O.N. (2006). delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142: 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Abelenda J.A., Cruz-Oró E., Cuéllar C.A., Tamaki S., Silva J., Shimamoto K., Prat S. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122. [DOI] [PubMed] [Google Scholar]
- Niwa M., Daimon Y., Kurotani K., Higo A., Pruneda-Paz J.L., Breton G., Mitsuda N., Kay S.A., Ohme-Takagi M., Endo M., Araki T. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25: 1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso-Tanaka E., Matsubara K., Yamamoto S., Nonoue Y., Wu J., Fujisawa H., Ishikubo H., Tanaka T., Ando T., Matsumoto T., Yano M. (2013). Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS One 8: e75959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk P.B., de Kam R.J., Hoge J.H., Meijer A.H. (2001). Glucocorticoid-inducible gene expression in rice. Planta 213: 370–378. [DOI] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Pin P.A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J.J.L., Nilsson O. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330: 1397–1400. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Gutfinger T., Hareven D., Ben-Naim O., Ron N., Adir N., Lifschitz E. (2001). Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13: 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwestri Y.A., Ogaki Y., Tamaki S., Tsuji H., Shimamoto K. (2009). The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 50: 429–438. [DOI] [PubMed] [Google Scholar]
- Randoux M., Davière J.-M., Jeauffre J., Thouroude T., Pierre S., Toualbia Y., Perrotte J., Reynoird J.-P., Jammes M.-J., Hibrand-Saint Oyant L., Foucher F. (2014). RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytol. 202: 161–173. [DOI] [PubMed] [Google Scholar]
- Reinke A.W., Baek J., Ashenberg O., Keating A.E. (2013). Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 340: 730–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo K.K., Tripathi A.K., Pareek A., Sopory S.K., Singla-Pareek S.L. (2011). An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods 7: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K., Harter K., Chaban C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13: 247–255. [DOI] [PubMed] [Google Scholar]
- Sentoku N., Sato Y., Kurata N., Ito Y., Kitano H., Matsuoka M. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmuradov I.A., Solovyev V.V. (2015). Nsite, NsiteH and NsiteM computer tools for studying transcription regulatory elements. Bioinformatics 31: 3544–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M., Ma Q., Weidtkamp-Peters S., Stahl Y., Felekyan S., Bleckmann A., Seidel C.A.M., Simon R. (2015). Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci. Signal. 8: ra76. [DOI] [PubMed] [Google Scholar]
- Soyk S., Müller N.A., Park S.J., Schmalenbach I., Jiang K., Hayama R., Zhang L., Van Eck J., Jiménez-Gómez J.M., Lippman Z.B. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49: 162–168. [DOI] [PubMed] [Google Scholar]
- Stahl Y., et al. (2013). Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23: 362–371. [DOI] [PubMed] [Google Scholar]
- Sussmilch F.C., Berbel A., Hecht V., Vander Schoor J.K., Ferrándiz C., Madueño F., Weller J.L. (2015). Pea VEGETATIVE2 is an FD homolog that is essential for flowering and compound inflorescence development. Plant Cell 27: 1046–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- Tamaki S., Tsuji H., Matsumoto A., Fujita A., Shimatani Z., Terada R., Sakamoto T., Kurata T., Shimamoto K. (2015). FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc. Natl. Acad. Sci. USA 112: E901–E910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N. et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28: 2161–2177. [DOI] [PMC free article] [PubMed]
- Taoka K., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335. [DOI] [PubMed] [Google Scholar]
- Teo C.-J., Takahashi K., Shimizu K., Shimamoto K., Taoka K.I. (2017). Potato tuber induction is regulated by interactions between components of a tuberigen complex. Plant Cell Physiol. 58: 365–374. [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P., Samach A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Ito Y., Sato Y., Kurata N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Nakamura H., Taoka K., Shimamoto K. (2013). Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 54: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Tachibana C., Tamaki S., Taoka K., Kyozuka J., Shimamoto K. (2015). Hd3a promotes lateral branching in rice. Plant J. 82: 256–266. [DOI] [PubMed] [Google Scholar]
- Tylewicz S., Tsuji H., Miskolczi P., Petterle A., Azeez A., Jonsson K., Shimamoto K., Bhalerao R.P. (2015). Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways. Proc. Natl. Acad. Sci. USA 112: 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickland D.P., Hanzawa Y. (2015). The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant 8: 983–997. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wu J., Zhu C., Pang J., Zhang X., Yang C., Xia G., Tian Y., He C. (2014). OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberellin biosynthesis in Oryza sativa. Plant J. 80: 1118–1130. [DOI] [PubMed] [Google Scholar]
- Zhang C., Liu J., Zhao T., Gomez A., Li C., Yu C., Li H., Lin J., Yang Y., Liu B., Lin C. (2016). A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol. 171: 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Chen H., Ren D., Tang H., Qiu R., Feng J., Long Y., Niu B., Chen D., Zhong T., Liu Y.-G., Guo J. (2015). Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 208: 936–948. [DOI] [PubMed] [Google Scholar]