Abstract
Equilibrative nucleoside transporters (ENTs) are polytopic integral membrane proteins that mediate the transport of nucleosides, nucleobases, and therapeutic analogs. The best-characterized ENTs are the human transporters hENT1 and hENT2. However, non-mammalian eukaryotic ENTs have also been studied (e.g., yeast, parasitic protozoa). ENTs are major pharmaceutical targets responsible for modulating the efficacy of more than 30 approved drugs. However, the molecular mechanisms and chemical determinants of ENT-mediated substrate recognition, binding, inhibition, and transport are poorly understood. This review highlights findings on the characterization of ENTs by surveying studies on genetics, permeant and inhibitor interactions, mutagenesis, and structural models of ENT function.
Nucleosides are important biologically active molecules formed by the amalgamation of a pyrimidine or purine nitrogenous base with either a ribose or 2′-deoxyribose pentose sugar. Pyrimidine and purine nucleosides, and their derivatives, play critical roles in the physiology of prokaryotic and eukaryotic organisms by serving as metabolic precursors in the synthesis of nucleic acids, as major elements of energy metabolism (ATP and GTP), and as ligands for purinergic receptors (adenosine, and inosine) (1, 2). Nucleoside analogs also represent important classes of antineoplastic and antiviral therapeutics (3). Since the activity of many of these hydrophilic compounds relies upon their entry into intracellular metabolic pathways to exert their effectiveness, crossing the cellular membranes is a prerequisite to downstream function.
Two classes of nucleoside transporters mediate physiologic nucleoside transport across cellular membranes: equilibrative nucleoside transporters (ENTs, SLC29 family) and concentrative nucleoside transporters (CNTs, SLC28 family) (4, 5). The CNT and ENT families are structurally unrelated nucleoside transporters with overlapping substrate specificities. CNTs are evolutionarily conserved symporters that require an inwardly directed sodium-dependent, or proton-dependent, coupling (reviewed elsewhere (3, 5, 6)). In contrast, ENTs are sodium-independent uniporters with no definitive prokaryotic orthologs. While passive transport is a hallmark of the ENT family, active, proton-linked, equilibrative transporters have been identified in protozoa (7) and activity of the human ENT3 and ENT4 transporters have been shown to be stimulated at lower pH (8). Mammalian ENTs were initially classified into two main groups: the es transporters were sensitive to nM concentrations of the inhibitor NBMPR (nitrobenzylthioinosine, NBTI), while the ei transporters were either unaffected by NBMPR or inhibited at higher concentrations (μM or higher) (3). Later studies identified 3 archetypical human isoforms (hENT1-3), which display the customary broad substrate selectivity (3). In addition, an evolutionarily divergent transporter (hENT4) was later shown to mediate adenosine transport in a pH-dependent manner with optimal transport occurring at approximately pH 6.0 (9). In spite of this, hENT4 is more commonly known as the plasma membrane monoamine transporter (PMAT) due to its ability to transport organic cations including biogenic amines, cationic therapeutics, and neurotoxins (9, 10). PMAT has substantial substrate overlap and inhibitor specificity with the organic cation transporters OCT1 - 3 in the SLC22 gene family (11–15). Another coinciding feature with OCTs is that PMAT-mediated transport is sensitive to membrane potential and sodium independent (16). Additionally, PMAT-mediated adenosine transport is likely insignificant under normal physiological conditions due to the low affinity and low activity of PMAT towards adenosine and the presence of other adenosine transporters (e.g., ENT1) (10). While PMAT may play a role in adenosine transport in times of ischemia or hypoxia where ENT1 activity is repressed (hypoxia) (17), PMAT will be excluded from this review because functionally it is viewed as a polyspecific organic cation transporter rather than the prototypical ENT.
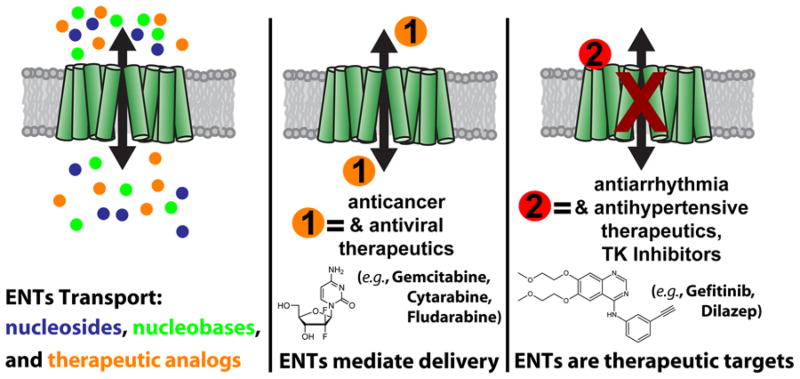
The ability of ENTs to regulate the flux of nucleosides, nucleobases, and nucleoside-derived therapeutics (Figure 1) has far reaching implications. Adenosine is of particular interest because of its wide-ranging effects on multiple organ systems by interacting with adenosine receptors Adora1, Adora2a, Adora2b, and Adora3 (1, 18, 19) which govern cellular functions via regulation of downstream heterotrimeric G-proteins (20, 21). ENTs also modulate efficacy for a chemically diverse range of therapeutics (>30 FDA/EMA approved drugs) including anticancer (e.g., gemcitabine, cytarabine) (22), antiarrhythmia (e.g., dilazep, dipyridamole) (23, 24), antiviral (e.g., ribavirin, azidothymidine) (25–27), and antihypertensive (e.g., nifedipine) (28, 29) medications (Figure 1). However, medications that exert their effects in the cardiovasuclar (e.g., dilazep, dipyridamole, nifedipine) system are known to have overlapping functions and can affect vasodilation (33–36) as well as platelet activity (37–40). It should be noted that not all studies assessing therapeutic interactions with ENTs have been performed using clinically relevant drug concentrations and this is likely due to the nature of the systems being studied, where external manipulations (e.g., overexpression of an ENT, knockdown of an ENT, expression of a human ENT in a non-human cell line, etc.) and methods utilized require concentrations beyond clinical dosing schemes in vivo. In addition, one should consider the complex milieu that exists in cells and tissues. ENTs may have protein or small molecule functional modulators yet to be characterized. hENT1 expression levels have been linked to increased patient survival for pancreatic cancer patients receiving gemcitabine treatment (41–49). Despite the pharmacological significance of ENT transport and activity, a detailed understanding of how ENTs function at the molecular level remains unknown due to hurdles associated with studying purified transporters (50–53) and native ENT recycling and expression in cells.
FIGURE 1. ENTs are diverse transporters and serve as major pharmaceutical targets.
ENTs regulate the flux of pyrimidine and purine nucleosides, nucleobases, and therapeutic analogs. ENTs also modulate therapeutic efficacy by mediating the transport of medications across cellular membranes to their ultimate site of action (orange “1”) or serve as the direct target for drug binding (red “2”) resulting in transporter inhibition.
Functional Characterization of Mammalian ENT Proteins
To date, there are three archetypical human ENTs (hENT1-3) (3), and one putative ENT (ENT4), and each transporter has a varied but overlapping substrate transport profile (Table 1). Human ENT1-3 are broadly expressed, however, they are localized with more abundance in certain tissues. Early characterization of ENTs was done in erythrocytes (54, 55), followed by mammalian tumor cell lines (56, 57), then Xenopus laevis oocytes (3), and this eventually lead to attempts of characterizing purified transporters (50–53, 58) – but the ENTs proved to be resistant to characterization in detergent solubilized, purified form. Only recently have reports been made that ENTs are amenable to heterologous expression, purification, and functional characterization (59, 60).
Table 1.
Permeant Selectivities of Nucleoside Transporters
| Substratea | Human (Km (mM)b)* | Yeast | Parasitic | Plant |
|---|---|---|---|---|
| Nucleosides and Nucleobases | ||||
| 2-Deoxyuridine | ScENT1 | |||
| Adenine | hENT1 (0.12 – 3.2), hENT2 (1.1 – 1.8), hENT3 | ScENT1 | LmaNT3, TbNT8.1 | |
| Adenosine | hENT1 (0.011 – 0.040), hENT2 (0.1 – 0.14), hENT3 (1.86), hENT4 (0.78 – 7.8) | ScENT1 | LdNT1.1, TbNT2, TbNT5, TbNT6, TbNT7, TbNT9, TbNT10 | AtENT1, AtENT3, AtENT6, AtENT7, StENT1, HvENT1, OsENT2 |
| Cytidine | hENT1 (0.21 – 0.58), hENT2 (5.6), hENT3 | ScENT1 | LdNT1.1 | AtENT1, AtENT3, AtENT4, AtENT6, AtENT7, StENT3, HvENT1 |
| Cytosine | hENT2 | ScENT1 | ||
| Guanine | hENT1, hENT2 | LmaNT3, TbNT8.1 | ||
| Guanosine | hENT1 (0.048 – 0.14), hENT2, hENT3 | ScENT1 | LdNT2, TbNT2, TbNT5, TbNT6, TbNT7, TbNT9, TbNT10 | AtENT3, AtENT4, AtENT6, AtENT7, StENT1, |
| Hypoxanthine | hENT1 (0.096 – 6.0), hENT2 (0.70 – 1.5) | ScENT1 | LmaNT3, TbNT5, TbNT6, TbNT7, TbNT8.1, TbNT9 | |
| Inosine | hENT1 (0.029 – 0.17), hENT2 (0.05 – 0.18), hENT3 | ScENT1 | LdNT2, TbNT2, TbNT5, TbNT6, TbNT7, TbNT9, TbNT10 | |
| Thymidine | hENT1 (0.30), hENT2 (0.71), hENT3 | ScENT1 | LdNT1.1 | AtENT3 |
| Thymine | hENT1 (6.3), hENT2 (6.0), hENT3 | |||
| Uracil | hENT1, hENT2 (2.6) | ScENT1 | ||
| Uridine | hENT1 (0.043 – 0.40), hENT2 (0.2 – 0.73), hENT3 (2.0) | ScENT1 | LdNT1.1 | AtENT1, AtENT3, AtENT4, AtENT6, AtENT7, StENT1, StENT3, HvENT1, OsENT2 |
| Xanthine | LmaNT3, TbNT8.1 | |||
| Nucleotides or Triphosphorylated Compounds | ||||
| ATP | hENT3 | |||
| UTP | ScENT1 | |||
| Therapeutic Analogs | ||||
| 5-Fluorouracil | hENT1 (2.3), hENT2 (2.6) | |||
| 6-Mercaptopurine | hENT1 (1.2), hENT2 (1.1) | |||
| Cladribine | hENT1 (0.023), hENT2, hENT3 | |||
| Clofarabine | hENT1 (0.11 – 0.15), hENT2 (0.33), hENT3 | |||
| Cytarabine | hENT1, hENT2 | |||
| Didanosine | hENT1 (2.3 – 3.0), hENT2, hENT3 | |||
| Fludarabine | hENT1 (0.11), hENT2, hENT3 | |||
| Gemcitabine | hENT1 (0.16), hENT2 (0.74), hENT3 | |||
| Ribavirin | hENT1 (0.16 – 3.5), hENT2 (0.33 – 3.8) | |||
| Zalcitabine | hENT1, hENT2 (>7.5), hENT3 | |||
| Zebularine | hENT3 | |||
| Zidovudine | hENT2, hENT3 | |||
| 5′-Deoxy-5-fluroyridine | hENT1 | |||
| Pentostatin | hENT1, hENT2 | |||
| Azacutidine | hENT1, hENT2, hENT3 | |||
| Decitabine | hENT1, hENT2 | |||
| Imaging Agent | ||||
| 3′-Deoxy-3′-fluorothymidine | hENT1 (3.4), hENT2 (2.6) | |||
Substrate selectivities compiled using the works of Young, et al.( 3), Landdear, et al.( 7), Girke, et al.( 128), Molina-Arcas, et al.( 90), Damaraju, et al. (22), Pastor-Anglada, et al.( 176), Boswell-Casteel, et al. (59), and unpublished data from Boswell-Casteel, et al. Empty cells signify either untested substrates or substrates that were deemed untransportable.
Km values in different expression systems vary, however, relative affinities are generally consistent
Km values for human ENTs are obtained from the works of Young, et al. (3), Parkinson, et al.( 104), Bone, et al. (177), Molina-Arcas, et al. (90), SenGupta, et al. (141), Ward, et al., (99), Visser, et al. (135), Visser, et al. (94), and Yao, et al. (89) and are meant to demonstrate the differences in subtype preference. A range of published Km values are shown for substrates that have multiple reported values.
Tissue Localization
While all ENTs are considered to be ubiquitously expressed across most tissue types, they each have specific tissues that have an overall higher abundance of expressed protein or mRNA. According to the Human Protein Atlas (http://www.proteinatlas.org (61)), hENT1 protein expression is highest in the adrenal gland, ovary, stomach, duodenum, small intestine, and colon while mRNA was most abundant in the adrenal gland. hENT2 protein expression was highest across a broader range of tissue types including various neurological tissues, segments of the gastrointestinal tract, skin, placenta, parathyroid gland, appendix, testis, urinary bladder, heart muscle, nasopharynx, pancreas, and gallbladder with mRNA being most abundant in skeletal muscle. Protein expression of hENT3 predominates in cerebral cortex, lateral ventricle, ovary, adrenal gland, and testis with higher levels of mRNA expression in placenta, urinary bladder, and ovary. ENT1 and ENT2 are primarily found in the plasma membrane while ENT3 contains an N-terminal dileucine motif (DE)XXXL(LI) (65) (characteristic motif for endosomal/lysosomal targeting) in the hydrophilic region of the sequence that precedes the first transmembrane domain (TMD) leading to an enrichment of ENT3 in the intracellular membranes of the endosome/lysosome and mitochondria (66). It should be noted that mutation of the dileucine motif causes the protein to be targeted to the plasma membrane (65).
SNPs, splice variants, and knockout models
Unlike other transporter families, the SLC29 genes show infrequent genetic variation (67–69) suggesting that the SLC29A1-2 genes are under substantial selective pressure, with nonsynonymous mutations being selected against. While few variations have been identified in hENT1, hENT1 polymorphisms have been associated with patients that are non-responsive to gemcitabine treatment, a nucleoside analog chemotherapeutic (70). There are currently no reported splice variants of hENT1, however, a variant of a mouse homolog (mENT1) has been reported as a product of alternative splicing at the end of exon 7 (mENT1.2) and is widely distributed (71). Choi et al. (72) developed the first reported ENT1-null mouse and demonstrated that it maintained normal reproductive behavior, had no gross anatomical abnormalities, and survival rates were similar to wild-type mice, although, bodyweight was significantly less than wild-type littermates (72–74). Several studies have utilized ENT1 null mice (20, 72–77) and found that these mice possess elevated circulating adenosine and thymidine levels in the plasma, reduced cellular uptake of adenosine (20, 73, 75), aberrant bone density (73, 74), dysregulation of the calcification of soft tissues associated with the enthesis regions of the vertebral column and sternum (73), increased resistance to oxidative stress (76), deficit in locomotor activity and motor coordination (74, 77), increased voluntary ethanol self-seeking behaviors associated with increased resistance to acute ethanol intoxication and reduced aversive effects of ethanol (72, 77), and that the absence of ENT1 is associated with reduced anxiety-like behavior in mice (78).
Two deletion variants have been found in the coding region of hENT2 (67, 69). The first is a splice variant that results in a frameshift encoding a 326-residue protein (nHNP36) that lacks the first three TMDs of hENT2 (64, 67, 79). HNP36 in both humans and mice has been shown to be nonfunctional as a transporter (64, 67), and HNP36 is associated with growth factor-induced delayed early response genes, but the function of HNP36 remains elusive (64, 79). The second variant results in a two-amino acid deletion and a nonsynonymous substitution of a third residue (67, 69). ENT2 knockout mice show increased levels of adenosine in bronchoalveolar fluid and alveolar space (80, 81), and that knockdown of ENT2 provided protection to acute lung injury (80, 81).
In marked contrast to hENT1-2, mutations in hENT3 have been linked to multiple disease states (82–85). SLC29A3 (hENT3) germline mutations are generally associated with autosomal recessive disorders such as H syndrome (83), pigmented hypertrichosis with insulin-dependent diabetes (PHID) syndrome, (82, 83), Faisalabad histiocytosis (83), and sinus histiocytosis with massive lymphadenopathy (SHML, Rosai Dorfman disease) (83). Mutations in hENT3 have also been associated with depression (85), osteoporosis (84), and increased survival of non-small-cell lung cancer patients treated with gemcitabine (86). Several of these disease causing variants were studied (G247S, G437R, M116R, and T449R) and all were shown to alter nucleoside transport with the C-terminal mutations resulting in a partial reduction of transport (8). Residue G427 was also shown to be critical for transporter function (8). In addition to nonsynomyous mutations, two deletion mutations resulting in frameshifts with early C-terminal truncations at residues 404 and 444 significantly reduced transport due to changes in protein stability (8). The nonsense mutation, E444X, leads to truncation at residue 444 and retains a higher level of function (8). ENT3 null mice developed spontaneous and progressive macrophage-dominated histiocytosis, altered macrophage function, lysosomal nucleoside buildup, and elevated intralysosomal pH (87). Mice lacking ENT3 also developed spontaneous splenomegaly and lymphadenopathy by eight weeks of age, and had a significanlty shorter life span relative to wild-type littermates (87). These studies suggest that defects in ENT3 have strong involvement in lysosomal storage disorders associated with histiocytosis.
Permeant Interactions
ENTs regulate the plasmalemmal flux of purine and pyrimidine nucleosides and nucleobases. A summary of currently known substrate selectivities for various human, yeast, parasitic protozoa, and plants can be found in Table 1. In addition to endogenous ligands, ENTs modulate efficacy for a variety of FDA/EMA approved therapeutics, and ENTs are known biomarkers for drug efficacy in the treatment of certain human cancers (41, 88). hENTs 1–3 have demonstrated varying levels of nucleoside, nucleobase, and nucleoside-analog transport (3, 65, 89). However, unlike hENT1-2, hENT3 transport appears to be proton coupled (8). Generally, ENTs have Km values for the transport of nucleosides in the high micromolar range (~ 100 – 800 μM) (3, 90). While ENT1 and ENT3 are capable of transporting nucleobases, hENT2 has a slightly higher affinity for nucleobases compared to hENT1 (3.2 mM – 6.3 mM vs. 1.5 mM – 6.0 mM ) for transporters assessed in Xenopus oocytes (3). ENT3 has a greater sensitivity to antivirals, and is stimulated at acidic pH (3, 8, 66). ENTs are known for their inability to transport nucleotides, however it has been reported that ENT3 is capable of transporting ATP and other therapeutics with triphosphate modifications (66). Therefore, understanding the chemical basis of permeant interactions is pivotal to engineering the next generation of antiviral and antineoplastic therapeutics. Understanding the chemical and structural requirements of the permeants on an atomic level has remained elusive, as no atomic resolution structure of an ENT from any species has been obtained.
Chemically, nucleosides are the consolidation of a nitrogenous base (purine or pyrimidine) with a pentose sugar. The pentose sugar has been repeatedly shown to be the primary determinant for ENT transport (91–96). ENT1 is selective for ribose or arabinose moieties (92), and sensitive to modifications at the C(2′) and C(5′) positions with the C(3′) hydroxyl being essential for substrate binding (93–95, 97, 98). The C(3′) position is also essential for permeants of ENT2 (94, 95), and ENT2 is also sensitive to modification at the C(5′) position (94, 95). Furthermore, chemical based studies have revealed the following: 1) ENTs have a weak preference for permeants adopting the C(2′)-endo/C(3′)-exo (South) sugar pucker conformation (96), 2) modifications at the C(3′)-position, a lack of conformational flexibility, and loss of a portion of the sugar ring are factors capable of decreasing the ability of some nucleosides to function as transportable substrates (93), 3) the pyrimidine moiety is the essential base component (97), and 4) there is a nitrogen to carbon bond specificity between the nitrogenous base and the sugar (97). It is currently unclear if ENT-mediated transport is: 1) affected by regions of electronegativity, 2) sensitive to the orientation of the purine/pyrimidine ring about the glycosidic linkage (anti vs. syn), 3) facilitated by hydrophobic interactions, and 4) what role hydrogen bonding plays in the transport mechanism. Therefore, a detailed chemometric understanding of ENT transportable ligands remains unresolved.
Functional characterization of transporter-permeant interactions is preferable using purified protein in a defined environment to exclude any overlap from the presence of other endogenous transporters, pumps, and/or metabolic activity. Given the difficulties associated with obtaining purified, active ENTs (50–53, 58), the use of other informative flux assays have been used including: 1) recombinant proteins produced in NT-deficient cells such as S. cerevisiae cells (knockout strains that lack nucleoside transporter activity), X. laevis oocytes (no endogenous nucleoside transport activity) (3), human or porcine cell lines (mutated to have null nucleoside transport activity (e.g., CCRF-CM, PK15-NTD) (99, 100), or 4) using cells that produce a single transporter type (e.g., studying hENT1 in S. cerevisiae) (54, 55, 100). Generally, the use of radioisotopes is used to determine ligand flux for all the assays mentioned above, and for transporters that generate rapid bidirectional substrate flux the use of an inhibitor may need to be added prior to collection to prevent substrate efflux and subsequent loss of substrate from the luminal volume of proteoliposomes or the cytoplasm of cells.
Inhibition
The distinguishing characteristic of mammalian ENTs is that hENT1 is inhibited by nM concentrations of NBMPR, while hENT2 and 3 are less sensitive to NBMPR (hENT2 ≫ hENT3) (3, 65). Although NBMPR potently inhibits hENT1, it is not an effective combinatorial chemotherapeutic (101) agent due to significant off target effects in the cardiovascular system (102, 103). Dipyridamole and dilazep have also been shown to affect hENT1-3 (3, 65). In addition to these inhibitors, hENT1-2 have also been shown to be affected to a lesser degree by tyrosine- and serine/threonine-kinase inhibitors and benzodiazepines (104). Another potent inhibitor of ENT1 is CBD (cannabidiol) with a reported Ki of < 250 nM (105). It should be noted that this is not an exhaustive list of ENT inhibitors, but highlights the broad classifications of therapeutics that have been shown to interact with ENTs.
Interestingly, the transport of permeants by ENT1 is also inhibited in the presence of ethanol (106–108). Specifically, acute ethanol exposures has been show to inhibit ENT1-mediated transport in human lymphocytes (106), primary cultures of hepatocytes isolated from rat (109), human placental cells (110), human bronchial epithelial cells (108), HL-1 cardiomyocytes (111), S49 mouse lymphoma cells and a hybrid rodent neuronal cell line (NG108-15) (107, 112–114). Ethanol has also been observed to attenuate transport mediated by purified and reconstituted yeast ENT, FUN26 (Boswell-Casteel, unpublished data). Moreover, ENT1 sensitivity to ethanol has been shown to be regulated in a kinase-dependent manner by PKA and PKC (107, 111, 112), which have been previously shown to phosphorylate mouse ENT1 in the intercellular loop region between TMDs 6 and 7 (115). The means by which ethanol may modulate ENT function is unknown and the effect may be indirect of ENT-mediated binding. Furthermore, ENT1 contributes to the behavioral effects of ethanol (72), alcohol consumption and preference (72), and genetic polymorphisms of hENT1 are associated with alcoholism and an increased risk of alcohol withdrawal seizures (116). Additionally, the acute inhibition of ENT1 contributes to the regulation of glutamatergic neurotransmission by controlling adenosine flux (117). Studies have shown that adenosine inhibits neuronal activity by suppressing synchronous discharges associated with the Adora1 receptor (118–122). Specifically, adenosine acts via the Adora1 receptor presynaptically to inhibit glutamatergic synaptic transmission within the hippocampus (123, 124). Reduced adenosine signaling has also been implicated in decreased sensitivity to the intoxicating effect of ethanol and increased ethanol consumption in mice (125). Inhibition of the Adroa1 receptor increases glutamate-evoked postsynaptic transmission in the nucleus accumbens (126), and ENT1 null mice have reduced Adora1-mediated inhibition of glutamate excitatory postsynaptic currents within the nucleus accumbens (72). It has been shown that short term inhibition of ENT1 reduces seizure load (117), however we postulate that chronic inhibition of ENT1 may contribute to seizure activity associated with substance withdrawal (e.g., alcohol, benzodiazepines) due to the reestablishment of adenosine flux eliciting changes in glutamate signaling. Given that chronic ethanol exposure/consumption evokes an adaptive response where increases in extracellular adenosine levels are no longer observed due to downregulation of ENT1 gene expression (113).
Functional Characterization of Non-mammalian ENT Proteins
Given the high pharmacological value of ENTs, multiple studies of non-mammalian ENTs have been conducted. A number of studies have been performed on ENTs from parasitic protozoa and plants, [reviewed in (7, 127, 128)]. Table 1 contains a representative list of transportable substrates for non-mammalian ENTs. ENTs are invaluable in the lifecycle of parasitic protozoa, because they lack the ability to synthesize purines de novo and are therefore reliant on salvage pathways mediated my plasma membrane nucleoside and nucleobase transporters to provide substrate-specific permeation routes (7). This reliance make ENTs of parasitic protozoa prime therapeutic targets for the delivery of subversive substrates (129), but development of inhibitors directed at the ENTs of parasitic protozoa have been limited due to the multiplicity of expressed purine nucleoside and nucleobase transporters (7). Parasitic protozoa transporters LdNT1.1, LdNT1.2, and LdNT2 (Leishmania donovani) display 20 to 100-fold increase in substrate affinity compared to mammalian ENTs, they are electrogenic proton symporters, and are less effected by the mammalian inhibitors NMBPR, dipyridamole, and dilazep (7). The transporters from Trypanosoma brucei also exhibit a higher affinity for substrates than mammalian ENTs (7), while transporters from Plasmodium falciparum have affinities more comparable to mammalian ENTs. Another frequently studied family of ENT proteins comes from Arabidopsis [reviewed in (128)]. Transporters from A. thaliana display broad substrate selectivity and affinity (3 to 100 μM), function as substrate-proton symporters (with exception to AtENT7), and are insensitive to the inhibitors NMBPR, dilazep, and dipryidamol (128). In addition to these transporters, FUN26 (Function Unknown Now 26) from Saccharomyces cerevisiae was recently identified as a broadly selective, high-affinity, nucleoside and nucleobase transporter (59). FUN26 is not stimulated by a pH differential nor is it sensitive to NBMPR (59, 130). Like hENT3, FUN26 has also demonstrated limited ability to transport nucleotides, albeit with lower affinity than nucleosides or nucleobases (Boswell-Casteel, unpublished data). Importantly, FUN26 and AtENT7 are the first ENTs to be functionally characterized in purified form (59, 60), and this marks a major advancement in efforts to obtain a molecular structure of an ENT protein.
Mutagenesis Studies of ENTs
Site specific mutagenesis has been utilized to identify critical residues conferring function and structure of ENTs in the absence of an atomic resolution structure. The architecture of ENTs consists of 11 TMDs, a cytoplasmic N-terminus and an extracellular C-terminus that was confirmed by glycosylation scanning mutagenesis and through the use of antipeptide antibodies as topological probes (131, 132). ENT1 was found to be N-glycosylated at N48 (133), ENT2 also contains glycosylation sites at residues N48 and N57 (134). ENT3 is expected to be glycosylated, but this is yet to be proved (65). Multiple studies have identified residues that affect substrate transport or inhibitor binding (8, 25, 36, 67, 82, 135–153), mitochondria targeting (144), and targeting to the plasma membrane (59, 142, 149, 150). A list of mutated residues with their extrapolated function is available in Table 2 for human ENTs and Table 3 for the non-mammalian ENTs. A recent study focusing on deletion mutants (deletion of intra- and extra-cellular loops as well as TMDs 9–11 and TMD11) demonstrated that the C-terminal TMDs were essential for proper trafficking and protein folding, while the loop regions appeared to be dispensable (154). Studies of ENT chimeras have also revealed important functional information surrounding the putative translocation pore and inhibitor binding sites in TMDs 3–6 (137, 140). Studies in S. cerevisiae have shown that SNPs conserved in ENT3 and FUN26 have functional overlap (59) (Boswell-Casteel, unpublished data). Collectively, these studies suggest that highly conserved residues throughout the ENT family will have overlapping functional duties, and be critical in unraveling ENT function. However, additional mutagenesis data is needed to fully understand the breadth of ENT-mediated substrate transport.
Table 2.
Mutagenesis of Human ENTs
| Residue | Location | Studied Mutation(s) | Functional Role | Reference(s) |
|---|---|---|---|---|
| hENT1 | ||||
| G24 | TMD1 | A/R/E | Nucleoside recognition and uptake | (147) |
| W29 | TMD1 | C/G/T | inhibitor sensitivity, substrate selectivity, nucleoside transport efficacy | (36) |
| M33 | TMD1 | inhibitor sensitivity | (36, 136) | |
| N48 | loop between TMD 1 & 2 | Q | glycosylation | (131) |
| P71 | loop between TMD 1 & 2 | L | mitochondrial targeting | (144) |
| E72 | loop between TMD 1 & 2 | G | mitochondrial targeting | (144) |
| N74 | loop between TMD 1 & 2 | P | mitochondrial targeting | (144) |
| C87 | TMD2 | S | inhibitor sensitivity | (178) |
| M89 | TMD2 | C/L/V/T/Q | inhibitor sensitivity, nucleoside transport efficacy | (143) |
| L92 | TMD2 | P/Q | inhibitor sensitivity, nucleoside transport efficacy | (143) |
| G154 | TMD4 | S | inhibitor sensitivity | (141) |
| S160 | TMD4 | C/N | inhibitor sensitivity | (143) |
| G179 | TMD5 | A/L/V/C/S | inhibitor sensitivity, nucleoside transport efficacy | (142) |
| G184 | TMD5 | A/L/V/C/S | Targeting protein to plasma membrane | (142) |
| C193 | TMD5 | S | inhibitor sensitivity | (178) |
| C213 | TMD6 | S | nucleoside transport efficacy | (178) |
| I216 | TMD6 | T | ethanol sensitivity, nucleoside transport efficacy | (68) |
| C222 | TMD6 | S | inhibitor sensitivity | (178) |
| F334 | TMD8 | Y | catalytic turnover | (145) |
| N338 | TMD8 | Q/C | protein folding | (145) |
| C387 | loop between TMD 9 & 10 | S | inhibitor sensitivity, protein folding | (179) |
| C414 | loop between TMD 10 & 11 | S | nucleobase transport | (89, 179) |
| C416 | loop between TMD 10 & 11 | S/A | inhibitor sensitivity, substrate selectivity, nucleoside transport efficacy | (179) |
| C439 | TMD11 | A | inhibitor sensitivity, nucleoside transport efficacy | (179) |
| L442 | TMD11 | I | substrate selectivity, nucleoside transport efficacy | (36, 135) |
| hENT2 | ||||
| D5 | N-terminus | Y | nucleoside transport efficacy | (67) |
| I33 | TMD1 | M/S/A/C | inhibitor sensitivity | (94, 135) |
| N48 | loop between TMD 1 & 2 | D | glycosylation | (134) |
| N57 | loop between TMD 1 & 2 | D | glycosylation | (134) |
| hENT3 | ||||
| M116 | TMD2 | R | retention in endoplasmic reticulum, nucleoside transport selectivity, nucleoside transport efficacy | (8, 82) |
| R363 | TMD8 | Q | (180) | |
| G427 | TMD10 | S/R/A/F/Y | nucleoside transport selectivity, nucleoside transport efficacy | (8) |
| G437 | loop between TMD 10 & 11 | R | catalytic turnover, nucleoside transport efficacy | (8, 82) |
| T449 | loop between TMD 10 & 11 | R | accumulation in late endosomes | (82) |
Table 3.
Mutagenesis of Non-Mammalian ENTs
| Residue | Location | Studied Mutation(s) | Functional Role | Reference(s) |
|---|---|---|---|---|
| LdNT1.1 | ||||
| M40 | TMD1 | D | nucleoside transport efficacy | (148) |
| F48 | TMD1 | A/W | nucleoside transport efficacy | (148) |
| W75 | TMD2 | A | nucleoside transport efficacy | (148) |
| E94 | TMD2 | Q/D | nucleoside transport efficacy | (148, 149) |
| E121 | TMD2 | Q/D | nucleoside transport efficacy | (149) |
| K153 | TMD4 | R/A | nucleoside transport efficacy, substrate selectivity | (149) |
| E157 | TMD4 | Q/D | substrate selectivity | (148) |
| Y161 | TMD4 | A | nucleoside transport efficacy | (150) |
| G163 | TMD4 | A | nucleoside transport efficacy | (150) |
| M175 | TMD5 | A | nucleoside transport efficacy, substrate selectivity | (150) |
| G183 | TMD5 | D/A/N | nucleoside transport efficacy | (151) |
| D215 | TMD6 | N/E | nucleoside transport efficacy | (149) |
| C337 | TMD7 | Y/F/S | nucleoside transport efficacy | (151) |
| F341 | TMD7 | A | substrate selectivity | (148) |
| R404 | TMD9 | A/K | targeting to plasma membrane | (149) |
| E429 | TMD9 | Q/D | (149) | |
| V445 | TMD10 | A | nucleoside transport efficacy | (150) |
| G467 | TMD11 | A | nucleoside transport efficacy | (150) |
| I468 | TMD11 | A | targeting to plasma membrane | (150) |
| S469 | TMD11 | F | nucleoside transport efficacy | (148) |
| I470 | TMD11 | A | nucleoside transport efficacy | (150) |
| T478 | TMD11 | F | nucleoside transport efficacy | (148) |
| LdNT2 | ||||
| R393 | L/E/N/K | nucleoside transport efficacy, substrate selectivity | (152) | |
| D389 | N//E | targeting to plasma membrane | (152) | |
| CeNT1 | ||||
| I49 | TMD1 | M/A/L/T | nucleoside transport efficacy | (153) |
| I429 | TMD11 | L/T | inhibitor sensitivity | (153) |
| ScENT1 | ||||
| G216 | TMD5 | A | loss of protein expression | (59) |
| F249 | TMD6 | I | nucleoside transport efficacy, substrate selectivity | (59) |
| L390 | TMD8 | A | nucleoside transport efficacy, substrate selectivity | (59) |
| G463 | TMD10 | A | nucleoside transport efficacy, substrate selectivity | (59) |
| AtENT3 | ||||
| G281 | 7 | R | fluorouridine sensitivity, pyrimidine transport | (181, 182) |
Structural Modeling and Mechanism of ENT Transport
ENTs are members of the Major Facilitator Superfamily (MFS), (155) due to observations of structural commonality between other members of the MFS (e.g., LacY, GlpT), putative structural models (homology and ab initio) have been constructed (148, 150, 156, 157). In the absence of an atomic resolution structure, computational models provide a platform for future studies aimed at probing structure-function relationships. Based on a canonical MFS fold (158), the computational ENT models predict an inner bundle of transmembrane domains (TMDs) 1, 2, 4, 5, 7, 8, 10, and 11 surrounding a central hydrophobic cavity, while TMDs 3, 6, and 9 are peripheral to the central pore and face the surrounding membrane (148, 150, 156, 157). The use of these models in combination with mutagenesis or cysteine-crosslinking data has shown that TMDs 5, 7, and 8 are important to transporter function (148, 157). Aromatic residues at the distal ends of TMDs 1, 2, 7, and 6 are expected to form the extracellular gate (or cytoplasmic gate for intracellular ENTs) in the inward-open configuration. Additionally, the intracellular gate (luminal gate) is expected to contain residues from TMDs 4, 5, 10, and 11 (150). Finally, TMDs 1, 8, 10, and 11 are expected to participate in the permeation pathway (148). Collectively, these studies point to an alternating access mechanism, which is common among MFS transporters (155, 158). In this model, substrate binds to a central cavity in the open configuration, followed by a series of intermediate, occluded, states (159) leading to conformational switching which, ultimately, leads to the release of bound substrate on the opposite side of the membrane bilayer (158). This model predicts that each substrate will be able to reciprocally inhibit the uptake of other permeants. However, there is evidence that some parasitic ENTs exhibit nonreciprocal inhibition, which implies that the simple model of competitive inhibition by structurally similar substrates binding to overlapping sites may not always hold true for ENTs (160, 161). Additionally, biphasic uptake was observed for the fluorescent probe FuPmR by ENT1, but is believed to be the result initial uptake combined with intracellular metabolism of FuPmR (162). Another transport mechanism was recently suggested for members of the MFS and it is termed the clamp-and-switch model (163). In this model transitions between the inward-open, outward-open, and multiple occluded states involves rigid-body rotation between the N- and C- domains, but also includes changes in the individual TMDs – particularly the A helices (TMDs 1, 4, 7, and 10) that bend to form the occluded states (clamp) (163). Once the occluded state (clamp) has been formed, the N- and C- domains rotate to expose a binding pocket to the opposite face of the membrane (switching) (163). While structural modeling can serve as a platform for directing future structure-function studies, until multiple atomic resolution structures of various conformations have been ascertained the alternating access or clamp-and-switch model of transport will remain ambiguous.
Physiological Roles of ENT Function
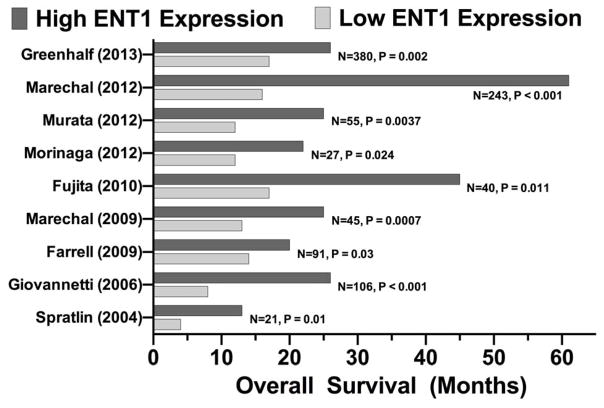
The ability of ENTs to transport nucleosides and nucleobases contributes to maintaining cellular nucleoside homeostasis. Nucleoside recycling is essential for nucleotide, DNA, and RNA synthesis, intracellular signaling pathways (e.g., cAMP, cGMP) and phospholipid synthesis (e.g., CDP) (1, 2). ENTs also have essential roles in delivering nucleoside analogs to intracellular targets for the treatment of numerous hematological and solid tumors, and viral infections (e.g., HIV, hepatitis C) (164, 165). Specifically, hENT1 expression has been associated with increased patient survival for pancreatic cancer patients receiving gemcitabine treatment (Figure 2) (41–49). Nucleoside analogs are routinely used in conjunction with platinum-based chemotherapeutics, such as cisplatin, and have been shown to have an enhanced effect when compared to individual treatments for a wide range of cancers (e.g., pancreatic, breast, non-small-cell lung cancers) (166–169). ENT inhibitors are also used to treat epilepsy and various cardiac conditions that require use of antiplatelet agents, calcium channel blockers, and vasodilators (31, 32, 81, 104).
FIGURE 2. Human ENT1 expression levels correlate to increased patient survival.
Multiple studies have shown that higher levels of hENT1 expression results in increased patient survival for pancreatic cancer patients receiving gemcitabine treatment. Data was compiled from multiple previously published works (41–49), red bars indicate high hENT1 expression levels, while blue bars represent low hENT1 expression levels.
By extension, ENTs contribute to the regulation of a plethora of metabolic functions and cell signaling cascades by controlling intra- and extracellular adenosine concentrations. Dihydropyridine-type calcium channel blockers (e.g., nimodipine, nitrendipine, nifedipine, nicardipine) are a class of vasodilators that also inhibit the cellular uptake of adenosine by targeting hENT1 and hENT2 (2). Nimodipine inhibits hNET1 at nM concentrations and nifedipine, nitrendipine, and nimodipine inhibit hENT2 at μM concentrations (2). Acting through distinct GPCRs, adenosine signaling mediates a variety of physiological responses such as vasodilation, coronary blood flow, myocardial oxygen supply-demand balance, inflammation, neurotransmission, hypoxia, trauma, and ischemia (32, 81, 94, 170–174). In fact, adenosine analogs have been exploited in clinical settings for their antiarrhythmic and cardioprotective effects. Inhibitors of ENTs confer a protective advantage in ischemia, trauma, hypoxia, and certain types of seizure disorders by blocking adenosine uptake (32, 81, 94, 117, 172, 173, 175). Also, as described above, hENT3 is associated with a number of autosomal recessive disorders. Given the far-reaching effects of ENTs on human physiology, it is imperative that a continued focused effort be maintained, especially in the area of drug delivery, cardiology, neurology, and functional/structural characterization.
Perspective
Major advances have been made in the ability to heterologously express, purify, and functionally characterize ENTs. The use of computational structural models has served as an insightful guide for designing mutagenesis strategies and aiding in understanding substrate/inhibitor interactions. However, despite progress unraveling the individual roles of ENTs, understanding the molecular architecture of the ENT family, the transport mechanism, and the significance of the ENT family for whole organism physiology remains at its infancy. Accumulating a greater understanding of ENT structure and function will require the use of transgenic animal models, studies using purified protein, and ultimately the acquisition of multiple atomic resolution structures. Headway in these areas will be essential to developing novel therapeutics and exploiting the remedial potential of ENTs.
Acknowledgments
We thank Jennifer M. Johnson and Dr. Yuko Tsutsui at the University of Oklahoma Health Sciences Center for helpful comments and suggestions.
FUNDING SOURCES
Authors of this review would like to acknowledge support from an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103639, Oklahoma Center for the Advancement of Science grant HR11-046 (to F.A.H.), OUHSC College of Medicine Alumni Association seed grant (to F.A.H.), and American Heart Association predoctoral fellowship 13PRE17040024 (to R.C.B-C.).
Footnotes
NOTES
The authors declare no competing financial interest.
References
- 1.Jacobson KA. Introduction to adenosine receptors as therapeutic targets. Handbook of experimental pharmacology. 2009;(193):1–24. doi: 10.1007/978-3-540-89615-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li RW, et al. Physiological and pharmacological roles of vascular nucleoside transporters. Journal of cardiovascular pharmacology. 2012;59(1):10–15. doi: 10.1097/FJC.0b013e31820eb788. [DOI] [PubMed] [Google Scholar]
- 3.Young JD, Yao SY, Baldwin JM, Cass CE, Baldwin SA. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol Aspects Med. 2013;34(2–3):529–547. doi: 10.1016/j.mam.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin SA, et al. The equilibrative nucleoside transporter family, SLC29. Pflugers Archiv : European journal of physiology. 2004;447(5):735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 5.Gray JH, Owen RP, Giacomini KM. The concentrative nucleoside transporter family, SLC28. Pflugers Archiv : European journal of physiology. 2004;447(5):728–734. doi: 10.1007/s00424-003-1107-y. [DOI] [PubMed] [Google Scholar]
- 6.Cano-Soldado P, Pastor-Anglada M. Transporters that translocate nucleosides and structural similar drugs: structural requirements for substrate recognition. Med Res Rev. 2012;32(2):428–457. doi: 10.1002/med.20221. [DOI] [PubMed] [Google Scholar]
- 7.Landfear SM, Ullman B, Carter NS, Sanchez MA. Nucleoside and nucleobase transporters in parasitic protozoa. Eukaryot Cell. 2004;3(2):245–254. doi: 10.1128/EC.3.2.245-254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang N, et al. Human equilibrative nucleoside transporter-3 (hENT3) spectrum disorder mutations impair nucleoside transport, protein localization, and stability. The Journal of biological chemistry. 2010;285(36):28343–28352. doi: 10.1074/jbc.M110.109199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes K, et al. Distribution and functional characterization of equilibrative nucleoside transporter-4, a novel cardiac adenosine transporter activated at acidic pH. Circulation research. 2006;99(5):510–519. doi: 10.1161/01.RES.0000238359.18495.42. [DOI] [PubMed] [Google Scholar]
- 10.Zhou M, Duan H, Engel K, Xia L, Wang J. Adenosine transport by plasma membrane monoamine transporter: reinvestigation and comparison with organic cations. Drug metabolism and disposition: the biological fate of chemicals. 2010;38(10):1798–1805. doi: 10.1124/dmd.110.032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engel K, Zhou M, Wang J. Identification and characterization of a novel monoamine transporter in the human brain. The Journal of biological chemistry. 2004;279(48):50042–50049. doi: 10.1074/jbc.M407913200. [DOI] [PubMed] [Google Scholar]
- 12.Engel K, Wang J. Interaction of organic cations with a newly identified plasma membrane monoamine transporter. Molecular pharmacology. 2005;68(5):1397–1407. doi: 10.1124/mol.105.016832. [DOI] [PubMed] [Google Scholar]
- 13.Zhou M, Engel K, Wang J. Evidence for significant contribution of a newly identified monoamine transporter (PMAT) to serotonin uptake in the human brain. Biochemical pharmacology. 2007;73(1):147–154. doi: 10.1016/j.bcp.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M, Xia L, Wang J. Metformin transport by a newly cloned proton-stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug metabolism and disposition: the biological fate of chemicals. 2007;35(10):1956–1962. doi: 10.1124/dmd.107.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho HT, et al. Molecular analysis and structure-activity relationship modeling of the substrate/inhibitor interaction site of plasma membrane monoamine transporter. The Journal of pharmacology and experimental therapeutics. 2011;339(2):376–385. doi: 10.1124/jpet.111.184036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itagaki S, et al. Electrophysiological characterization of the polyspecific organic cation transporter plasma membrane monoamine transporter. Drug metabolism and disposition: the biological fate of chemicals. 2012;40(6):1138–1143. doi: 10.1124/dmd.111.042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eltzschig HK, et al. HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J Exp Med. 2005;202(11):1493–1505. doi: 10.1084/jem.20050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koeppen M, Eckle T, Eltzschig HK. Interplay of hypoxia and A2B adenosine receptors in tissue protection. Adv Pharmacol. 2011;61:145–186. doi: 10.1016/B978-0-12-385526-8.00006-0. [DOI] [PubMed] [Google Scholar]
- 19.Eltzschig HK, Sitkovsky MV, Robson SC. Purinergic signaling during inflammation. N Engl J Med. 2012;367(24):2322–2333. doi: 10.1056/NEJMra1205750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose JB, et al. Absence of equilibrative nucleoside transporter 1 in ENT1 knockout mice leads to altered nucleoside levels following hypoxic challenge. Life Sci. 2011;89(17–18):621–630. doi: 10.1016/j.lfs.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Ribeiro JA, Sebastiao AM. Modulation and metamodulation of synapses by adenosine. Acta Physiol (Oxf) 2010;199(2):161–169. doi: 10.1111/j.1748-1716.2010.02115.x. [DOI] [PubMed] [Google Scholar]
- 22.Damaraju VL, et al. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene. 2003;22(47):7524–7536. doi: 10.1038/sj.onc.1206952. [DOI] [PubMed] [Google Scholar]
- 23.Sono K, Akimoto Y, Kurahashi K, Fujiwara M. Effects of antiarrhythmic drugs on the extraneuronal accumulation of isoprenaline in perfused rat hearts. Naunyn Schmiedebergs Arch Pharmacol. 1986;334(2):145–148. doi: 10.1007/BF00505814. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida Y, et al. Antiarrhythmic efficacy of dipyridamole in treatment of reperfusion arrhythmias : evidence for cAMP-mediated triggered activity as a mechanism responsible for reperfusion arrhythmias. Circulation. 2000;101(6):624–630. doi: 10.1161/01.cir.101.6.624. [DOI] [PubMed] [Google Scholar]
- 25.Yao SY, et al. Transport of antiviral 3′-deoxy-nucleoside drugs by recombinant human and rat equilibrative, nitrobenzylthioinosine (NBMPR)-insensitive (ENT2) nucleoside transporter proteins produced in Xenopus oocytes. Molecular membrane biology. 2001;18(2):161–167. doi: 10.1080/09687680110048318. [DOI] [PubMed] [Google Scholar]
- 26.Endres CJ, Moss AM, Govindarajan R, Choi DS, Unadkat JD. The role of nucleoside transporters in the erythrocyte disposition and oral absorption of ribavirin in the wild-type and equilibrative nucleoside transporter 1−/− mice. The Journal of pharmacology and experimental therapeutics. 2009;331(1):287–296. doi: 10.1124/jpet.109.153130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endres CJ, et al. The role of the equilibrative nucleoside transporter 1 (ENT1) in transport and metabolism of ribavirin by human and wild-type or Ent1−/− mouse erythrocytes. The Journal of pharmacology and experimental therapeutics. 2009;329(1):387–398. doi: 10.1124/jpet.108.145854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masotti G, Morettini A, Galanti G, Paoli G, Poggesi L. Antihypertensive action of nifedipine: effects on arteries and veins. J Clin Pharmacol. 1985;25(1):27–35. doi: 10.1002/j.1552-4604.1985.tb02797.x. [DOI] [PubMed] [Google Scholar]
- 29.Snider ME, Nuzum DS, Veverka A. Long-acting nifedipine in the management of the hypertensive patient. Vasc Health Risk Manag. 2008;4(6):1249–1257. doi: 10.2147/vhrm.s3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Sadee W. Membrane transporters and channels in chemoresistance and -sensitivity of tumor cells. Cancer letters. 2006;239(2):168–182. doi: 10.1016/j.canlet.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 31.Marvi M, et al. Nucleoside transporter expression profiles in human cardiac tissue show striking individual variability with overall predominance of hENT1. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2010;41(5):685–691. doi: 10.1016/j.ejps.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Rose JB, et al. Equilibrative nucleoside transporter 1 plays an essential role in cardioprotection. American journal of physiology. Heart and circulatory physiology. 2010;298(3):H771–777. doi: 10.1152/ajpheart.00711.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitakaze M, et al. Nifedipine-induced coronary vasodilation in ischemic hearts is attributable to bradykinin- and NO-dependent mechanisms in dogs. Circulation. 2000;101(3):311–317. doi: 10.1161/01.cir.101.3.311. [DOI] [PubMed] [Google Scholar]
- 34.Weis M, Pehlivanli S, von Scheidt W. Vasodilator response to nifedipine in human coronary arteries with endothelial dysfunction. Journal of cardiovascular pharmacology. 2002;39(2):172–180. doi: 10.1097/00005344-200202000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Mustafa SJ. Effects of coronary vasodilator drugs on the uptake and release of adenosine in cardiac cells. Biochemical pharmacology. 1979;28(17):2617–2624. doi: 10.1016/0006-2952(79)90037-6. [DOI] [PubMed] [Google Scholar]
- 36.Paproski RJ, et al. Mutation of Trp29 of human equilibrative nucleoside transporter 1 alters affinity for coronary vasodilator drugs and nucleoside selectivity. The Biochemical journal. 2008;414(2):291–300. doi: 10.1042/BJ20080074. [DOI] [PubMed] [Google Scholar]
- 37.Deguchi H, et al. Dilazep, an antiplatelet agent, inhibits tissue factor expression in endothelial cells and monocytes. Blood. 1997;90(6):2345–2356. [PubMed] [Google Scholar]
- 38.Nakamura T, et al. Effect of the antiplatelet drug dilazep dihydrochloride on urinary podocytes in patients in the early stage of diabetic nephropathy. Diabetes Care. 2000;23(8):1168–1171. doi: 10.2337/diacare.23.8.1168. [DOI] [PubMed] [Google Scholar]
- 39.Chakrabarti S, Freedman JE. Dipyridamole, cerebrovascular disease, and the vasculature. Vascul Pharmacol. 2008;48(4–6):143–149. doi: 10.1016/j.vph.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 40.Chou TC. New mechanisms of antiplatelet activity of nifedipine, an L-type calcium channel blocker. Biomedicine (Taipei) 2014;4:24. doi: 10.7603/s40681-014-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenhalf W, et al. Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. Journal of the National Cancer Institute. 2014;106(1):djt347. doi: 10.1093/jnci/djt347. [DOI] [PubMed] [Google Scholar]
- 42.Marechal R, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology. 2012;143(3):664–674. e661–666. doi: 10.1053/j.gastro.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Murata Y, et al. Human equilibrative nucleoside transporter 1 expression is a strong independent prognostic factor in UICC T3-T4 pancreatic cancer patients treated with preoperative gemcitabine-based chemoradiotherapy. J Hepatobiliary Pancreat Sci. 2012;19(4):413–425. doi: 10.1007/s00534-011-0440-3. [DOI] [PubMed] [Google Scholar]
- 44.Morinaga S, et al. Immunohistochemical analysis of human equilibrative nucleoside transporter-1 (hENT1) predicts survival in resected pancreatic cancer patients treated with adjuvant gemcitabine monotherapy. Annals of surgical oncology. 2012;19(Suppl 3):S558–564. doi: 10.1245/s10434-011-2054-z. [DOI] [PubMed] [Google Scholar]
- 45.Fujita H, et al. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12(10):807–817. doi: 10.1593/neo.10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marechal R, et al. Human equilibrative nucleoside transporter 1 and human concentrative nucleoside transporter 3 predict survival after adjuvant gemcitabine therapy in resected pancreatic adenocarcinoma. Clin Cancer Res. 2009;15(8):2913–2919. doi: 10.1158/1078-0432.CCR-08-2080. [DOI] [PubMed] [Google Scholar]
- 47.Farrell JJ, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136(1):187–195. doi: 10.1053/j.gastro.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 48.Giovannetti E, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66(7):3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 49.Spratlin J, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. 2004;10(20):6956–6961. doi: 10.1158/1078-0432.CCR-04-0224. [DOI] [PubMed] [Google Scholar]
- 50.Hammond JR. Enhancement of the functional stability of solubilized nucleoside transporters by substrates and inhibitors. Biochemical pharmacology. 1997;53(5):623–629. doi: 10.1016/s0006-2952(96)00857-x. [DOI] [PubMed] [Google Scholar]
- 51.Hammond JR. Functional reconstitution of pharmacologically distinct subtypes of nucleoside transporters in liposomal membranes. The Journal of pharmacology and experimental therapeutics. 1994;271(2):906–917. [PubMed] [Google Scholar]
- 52.Tse CM, et al. Reconstitution studies of the human erythrocyte nucleoside transporter. The Journal of biological chemistry. 1985;260(6):3506–3511. [PubMed] [Google Scholar]
- 53.Hammond JR, Zarenda M. Effect of detergents on ligand binding and translocation activities of solubilized/reconstituted nucleoside transporters. Archives of biochemistry and biophysics. 1996;332(2):313–322. doi: 10.1006/abbi.1996.0347. [DOI] [PubMed] [Google Scholar]
- 54.Cass CE, Paterson AR. Nitrobenzylthionionosine binding sites in the erythrocyte membrane. Biochimica et biophysica acta. 1976;419(2):285–294. doi: 10.1016/0005-2736(76)90354-0. [DOI] [PubMed] [Google Scholar]
- 55.Jarvis SM, Hammond JR, Paterson AR, Clanachan AS. Species differences in nucleoside transport. A study of uridine transport and nitrobenzylthioinosine binding by mammalian erythrocytes. The Biochemical journal. 1982;208(1):83–88. doi: 10.1042/bj2080083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch TP, Lauzon GJ, Naik SR, Cass CE, Paterson AR. Inhibition of nucleoside uptake in HeLa cells by nitrobenzylthioinosinate. Biochemical pharmacology. 1978;27(8):1303–1304. doi: 10.1016/0006-2952(78)90473-2. [DOI] [PubMed] [Google Scholar]
- 57.Hammond JR. Comparative pharmacology of the nitrobenzylthioguanosine-sensitive and -resistant nucleoside transport mechanisms of Ehrlich ascites tumor cells. The Journal of pharmacology and experimental therapeutics. 1991;259(2):799–807. [PubMed] [Google Scholar]
- 58.Hammond JR. Effect of membrane lipid composition on the functional activity of a reconstituted nucleoside transporter derived from Ehrlich ascites cells. Adv Exp Med Biol. 1991;309A:423–426. doi: 10.1007/978-1-4899-2638-8_97. [DOI] [PubMed] [Google Scholar]
- 59.Boswell-Casteel RC, et al. FUN26 (function unknown now 26) protein from saccharomyces cerevisiae is a broad selectivity, high affinity, nucleoside and nucleobase transporter. The Journal of biological chemistry. 2014;289(35):24440–24451. doi: 10.1074/jbc.M114.553503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Girke C, et al. High yield expression and purification of equilibrative nucleoside transporter 7 (ENT7) from Arabidopsis thaliana. Biochimica et biophysica acta. 2015;1850(9):1921–1929. doi: 10.1016/j.bbagen.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 61.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 62.Pennycooke M, Chaudary N, Shuralyova I, Zhang Y, Coe IR. Differential expression of human nucleoside transporters in normal and tumor tissue. Biochem Biophys Res Commun. 2001;280(3):951–959. doi: 10.1006/bbrc.2000.4205. [DOI] [PubMed] [Google Scholar]
- 63.Thompson CG, et al. Penetrating eye injuries in rural New South Wales. Aust N Z J Ophthalmol. 1997;25(1):37–41. doi: 10.1111/j.1442-9071.1997.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 64.Crawford CR, Patel DH, Naeve C, Belt JA. Cloning of the human equilibrative, nitrobenzylmercaptopurine riboside (NBMPR)-insensitive nucleoside transporter ei by functional expression in a transport-deficient cell line. The Journal of biological chemistry. 1998;273(9):5288–5293. doi: 10.1074/jbc.273.9.5288. [DOI] [PubMed] [Google Scholar]
- 65.Baldwin SA, et al. Functional characterization of novel human and mouse equilibrative nucleoside transporters (hENT3 and mENT3) located in intracellular membranes. The Journal of biological chemistry. 2005;280(16):15880–15887. doi: 10.1074/jbc.M414337200. [DOI] [PubMed] [Google Scholar]
- 66.Govindarajan R, et al. Facilitated mitochondrial import of antiviral and anticancer nucleoside drugs by human equilibrative nucleoside transporter-3. Am J Physiol Gastrointest Liver Physiol. 2009;296(4):G910–922. doi: 10.1152/ajpgi.90672.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Owen RP, et al. Functional characterization and haplotype analysis of polymorphisms in the human equilibrative nucleoside transporter, ENT2. Drug metabolism and disposition: the biological fate of chemicals. 2006;34(1):12–15. doi: 10.1124/dmd.105.006270. [DOI] [PubMed] [Google Scholar]
- 68.Osato DH, et al. Functional characterization in yeast of genetic variants in the human equilibrative nucleoside transporter, ENT1. Pharmacogenetics. 2003;13(5):297–301. doi: 10.1097/00008571-200305000-00010. [DOI] [PubMed] [Google Scholar]
- 69.Leabman MK, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu F, et al. Effect of hENT1 polymorphism G-706C on clinical outcomes of gemcitabine-containing chemotherapy for Chinese non-small-cell lung cancer patients. Cancer Epidemiol. 2014;38(6):728–732. doi: 10.1016/j.canep.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 71.Handa M, et al. Cloning of a novel isoform of the mouse NBMPR-sensitive equilibrative nucleoside transporter (ENT1) lacking a putative phosphorylation site. Gene. 2001;262(1–2):301–307. doi: 10.1016/s0378-1119(00)00555-2. [DOI] [PubMed] [Google Scholar]
- 72.Choi DS, et al. The type 1 equilibrative nucleoside transporter regulates ethanol intoxication and preference. Nat Neurosci. 2004;7(8):855–861. doi: 10.1038/nn1288. [DOI] [PubMed] [Google Scholar]
- 73.Warraich S, et al. Loss of equilibrative nucleoside transporter 1 in mice leads to progressive ectopic mineralization of spinal tissues resembling diffuse idiopathic skeletal hyperostosis in humans. J Bone Miner Res. 2013;28(5):1135–1149. doi: 10.1002/jbmr.1826. [DOI] [PubMed] [Google Scholar]
- 74.Hinton DJ, et al. Aberrant bone density in aging mice lacking the adenosine transporter ENT1. PloS one. 2014;9(2):e88818. doi: 10.1371/journal.pone.0088818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paproski RJ, et al. Biodistribution and uptake of 3′-deoxy-3′-fluorothymidine in ENT1-knockout mice and in an ENT1-knockdown tumor model. J Nucl Med. 2010;51(9):1447–1455. doi: 10.2967/jnumed.110.076356. [DOI] [PubMed] [Google Scholar]
- 76.Bone DB, Antic M, Quinonez D, Hammond JR. Hypoxanthine uptake by skeletal muscle microvascular endothelial cells from equilibrative nucleoside transporter 1 (ENT1)-null mice: effect of oxidative stress. Microvasc Res. 2015;98:16–22. doi: 10.1016/j.mvr.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 77.Chen J, et al. Altered glutamatergic neurotransmission in the striatum regulates ethanol sensitivity and intake in mice lacking ENT1. Behav Brain Res. 2010;208(2):636–642. doi: 10.1016/j.bbr.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen J, et al. The type 1 equilibrative nucleoside transporter regulates anxiety-like behavior in mice. Genes Brain Behav. 2007;6(8):776–783. doi: 10.1111/j.1601-183X.2007.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams JB, Lanahan AA. A mammalian delayed-early response gene encodes HNP36, a novel, conserved nucleolar protein. Biochem Biophys Res Commun. 1995;213(1):325–333. doi: 10.1006/bbrc.1995.2133. [DOI] [PubMed] [Google Scholar]
- 80.Morote-Garcia JC, et al. Repression of the equilibrative nucleoside transporters dampens inflammatory lung injury. Am J Respir Cell Mol Biol. 2013;49(2):296–305. doi: 10.1165/rcmb.2012-0457OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eckle T, et al. Crosstalk between the equilibrative nucleoside transporter ENT2 and alveolar Adora2b adenosine receptors dampens acute lung injury. FASEB J. 2013;27(8):3078–3089. doi: 10.1096/fj.13-228551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cliffe ST, et al. SLC29A3 gene is mutated in pigmented hypertrichosis with insulin-dependent diabetes mellitus syndrome and interacts with the insulin signaling pathway. Hum Mol Genet. 2009;18(12):2257–2265. doi: 10.1093/hmg/ddp161. [DOI] [PubMed] [Google Scholar]
- 83.Morgan NV, et al. Mutations in SLC29A3, encoding an equilibrative nucleoside transporter ENT3, cause a familial histiocytosis syndrome (Faisalabad histiocytosis) and familial Rosai-Dorfman disease. PLoS Genet. 2010;6(2):e1000833. doi: 10.1371/journal.pgen.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campeau PM, et al. Whole-exome sequencing identifies mutations in the nucleoside transporter gene SLC29A3 in dysosteosclerosis, a form of osteopetrosis. Hum Mol Genet. 2012;21(22):4904–4909. doi: 10.1093/hmg/dds326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gass N, et al. Contribution of adenosine related genes to the risk of depression with disturbed sleep. J Affect Disord. 2010;126(1–2):134–139. doi: 10.1016/j.jad.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 86.Chen X, et al. Genomic polymorphisms of SLC29A3 associated with overall survival in advanced non-small-cell lung cancer treated with gemcitabine. Med Oncol. 2014;31(4):865. doi: 10.1007/s12032-014-0865-z. [DOI] [PubMed] [Google Scholar]
- 87.Hsu CL, et al. Equilibrative nucleoside transporter 3 deficiency perturbs lysosome function and macrophage homeostasis. Science. 2012;335(6064):89–92. doi: 10.1126/science.1213682. [DOI] [PubMed] [Google Scholar]
- 88.Jordheim LP, Durantel D, Zoulim F, Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov. 2013;12(6):447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- 89.Yao SY, Ng AM, Cass CE, Baldwin SA, Young JD. Nucleobase transport by human equilibrative nucleoside transporter 1 (hENT1) The Journal of biological chemistry. 2011;286(37):32552–32562. doi: 10.1074/jbc.M111.236117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Molina-Arcas M, Casado FJ, Pastor-Anglada M. Nucleoside transporter proteins. Curr Vasc Pharmacol. 2009;7(4):426–434. doi: 10.2174/157016109789043892. [DOI] [PubMed] [Google Scholar]
- 91.Cass CE, Paterson AR. Mediated transport of nucleosides in human erythrocytes. Accelerative exchange diffusion of uridine and thymidine and specificity toward pyrimidine nucleosides as permeants. The Journal of biological chemistry. 1972;247(10):3314–3320. [PubMed] [Google Scholar]
- 92.Cass CE, Paterson AR. Mediated transport of nucleosides by human erythrocytes. Specificity toward purine nucleosides as permeants. Biochimica et biophysica acta. 1973;291(3):734–746. doi: 10.1016/0005-2736(73)90477-x. [DOI] [PubMed] [Google Scholar]
- 93.Gati WP, Misra HK, Knaus EE, Wiebe LI. Structural modifications at the 2′- and 3′-positions of some pyrimidine nucleosides as determinants of their interaction with the mouse erythrocyte nucleoside transporter. Biochemical pharmacology. 1984;33(21):3325–3331. doi: 10.1016/0006-2952(84)90101-1. [DOI] [PubMed] [Google Scholar]
- 94.Vickers MF, et al. Comparison of the interaction of uridine, cytidine, and other pyrimidine nucleoside analogues with recombinant human equilibrative nucleoside transporter 2 (hENT2) produced in Saccharomyces cerevisiae. Biochem Cell Biol. 2002;80(5):639–644. doi: 10.1139/o02-148. [DOI] [PubMed] [Google Scholar]
- 95.Vickers MF, et al. Uridine recognition motifs of human equilibrative nucleoside transporters 1 and 2 produced in Saccharomyces cerevisiae. Nucleosides Nucleotides Nucleic Acids. 2004;23(1–2):361–373. doi: 10.1081/ncn-120028333. [DOI] [PubMed] [Google Scholar]
- 96.Damaraju VL, et al. Influence of sugar ring conformation on the transportability of nucleosides by human nucleoside transporters. Chembiochem : a European journal of chemical biology. 2011;12(18):2774–2778. doi: 10.1002/cbic.201100567. [DOI] [PubMed] [Google Scholar]
- 97.Taube RA, Berlin RD. Membrane transport of nucleosides in rabbit polymorphonuclear leukocytes. Biochimica et biophysica acta. 1972;255(1):6–18. doi: 10.1016/0005-2736(72)90003-x. [DOI] [PubMed] [Google Scholar]
- 98.Turnheim K, Plank B, Kolassa N. Inhibition of adenosine uptake in human erythrocytes by adenosine-5′-carboxamides, xylosyladenine, dipyridamole, hexobendine, and p-nitrobenzylthioguanosine. Biochemical pharmacology. 1978;27(18):2191–2197. doi: 10.1016/0006-2952(78)90076-x. [DOI] [PubMed] [Google Scholar]
- 99.Ward JL, Sherali A, Mo ZP, Tse CM. Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. Ent2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. The Journal of biological chemistry. 2000;275(12):8375–8381. doi: 10.1074/jbc.275.12.8375. [DOI] [PubMed] [Google Scholar]
- 100.King KM, et al. A comparison of the transportability, and its role in cytotoxicity, of clofarabine, cladribine, and fludarabine by recombinant human nucleoside transporters produced in three model expression systems. Molecular pharmacology. 2006;69(1):346–353. doi: 10.1124/mol.105.015768. [DOI] [PubMed] [Google Scholar]
- 101.Adjei AA, Dagnino L, Wong MM, Paterson AR. Protection against fludarabine neurotoxicity in leukemic mice by the nucleoside transport inhibitor nitrobenzylthioinosine. Cancer Chemother Pharmacol. 1992;31(1):71–75. doi: 10.1007/BF00695997. [DOI] [PubMed] [Google Scholar]
- 102.Choi JS, Berdis AJ. Nucleoside transporters: biological insights and therapeutic applications. Future Med Chem. 2012;4(11):1461–1478. doi: 10.4155/fmc.12.79. [DOI] [PubMed] [Google Scholar]
- 103.Tromp RA, Spanjersberg RF, von Frijtag Drabbe Kunzel JK, APIJ Inhibition of nucleoside transport proteins by C8-alkylamine-substituted purines. Journal of medicinal chemistry. 2005;48(1):321–329. doi: 10.1021/jm049303k. [DOI] [PubMed] [Google Scholar]
- 104.Parkinson FE, et al. Molecular biology of nucleoside transporters and their distributions and functions in the brain. Current topics in medicinal chemistry. 2011;11(8):948–972. doi: 10.2174/156802611795347582. [DOI] [PubMed] [Google Scholar]
- 105.Carrier EJ, Auchampach JA, Hillard CJ. Inhibition of an equilibrative nucleoside transporter by cannabidiol: a mechanism of cannabinoid immunosuppression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(20):7895–7900. doi: 10.1073/pnas.0511232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Krauss SW, Ghirnikar RB, Diamond I, Gordon AS. Inhibition of adenosine uptake by ethanol is specific for one class of nucleoside transporters. Molecular pharmacology. 1993;44(5):1021–1026. [PubMed] [Google Scholar]
- 107.Coe IR, Dohrman DP, Constantinescu A, Diamond I, Gordon AS. Activation of cyclic AMP-dependent protein kinase reverses tolerance of a nucleoside transporter to ethanol. The Journal of pharmacology and experimental therapeutics. 1996;276(2):365–369. [PubMed] [Google Scholar]
- 108.Allen-Gipson DS, et al. Ethanol blocks adenosine uptake via inhibiting the nucleoside transport system in bronchial epithelial cells. Alcohol Clin Exp Res. 2009;33(5):791–798. doi: 10.1111/j.1530-0277.2009.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nagy LE. Ethanol metabolism and inhibition of nucleoside uptake lead to increased extracellular adenosine in hepatocytes. Am J Physiol. 1992;262(5 Pt 1):C1175–1180. doi: 10.1152/ajpcell.1992.262.5.C1175. [DOI] [PubMed] [Google Scholar]
- 110.Acevedo CG, et al. Effect of ethanol on human placental transport and metabolism of adenosine. Placenta. 1997;18(5–6):387–392. doi: 10.1016/s0143-4004(97)80038-0. [DOI] [PubMed] [Google Scholar]
- 111.Ramadan A, Naydenova Z, Stevanovic K, Rose JB, Coe IR. The adenosine transporter, ENT1, in cardiomyocytes is sensitive to inhibition by ethanol in a kinase-dependent manner: implications for ethanol-dependent cardioprotection and nucleoside analog drug cytotoxicity. Purinergic signalling. 2014;10(2):305–312. doi: 10.1007/s11302-013-9391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coe IR, Yao L, Diamond I, Gordon AS. The role of protein kinase C in cellular tolerance to ethanol. The Journal of biological chemistry. 1996;271(46):29468–29472. doi: 10.1074/jbc.271.46.29468. [DOI] [PubMed] [Google Scholar]
- 113.Nagy LE, Diamond I, Casso DJ, Franklin C, Gordon AS. Ethanol increases extracellular adenosine by inhibiting adenosine uptake via the nucleoside transporter. The Journal of biological chemistry. 1990;265(4):1946–1951. [PubMed] [Google Scholar]
- 114.Nagy LE, et al. Adenosine is required for ethanol-induced heterologous desensitization. Molecular pharmacology. 1989;36(5):744–748. [PubMed] [Google Scholar]
- 115.Reyes G, et al. The Equilibrative Nucleoside Transporter (ENT1) can be phosphorylated at multiple sites by PKC and PKA. Molecular membrane biology. 2011;28(6):412–426. doi: 10.3109/09687688.2011.604861. [DOI] [PubMed] [Google Scholar]
- 116.Kim JH, et al. Functional role of the polymorphic 647 T/C variant of ENT1 (SLC29A1) and its association with alcohol withdrawal seizures. PloS one. 2011;6(1):e16331. doi: 10.1371/journal.pone.0016331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu Z, et al. ENT1 inhibition attenuates epileptic seizure severity via regulation of glutamatergic neurotransmission. Neuromolecular Med. 2015;17(1):1–11. doi: 10.1007/s12017-014-8338-2. [DOI] [PubMed] [Google Scholar]
- 118.Huber A, et al. Grafts of adenosine-releasing cells suppress seizures in kindling epilepsy. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13):7611–7616. doi: 10.1073/pnas.131102898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dale N, Frenguelli BG. Release of adenosine and ATP during ischemia and epilepsy. Curr Neuropharmacol. 2009;7(3):160–179. doi: 10.2174/157015909789152146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Etherington LA, et al. Astrocytic adenosine kinase regulates basal synaptic adenosine levels and seizure activity but not activity-dependent adenosine release in the hippocampus. Neuropharmacology. 2009;56(2):429–437. doi: 10.1016/j.neuropharm.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sebastiao AM. Adenosine and epilepsy-thinking beyond A(1) receptors. Purinergic signalling. 2010;6(1):1–2. doi: 10.1007/s11302-010-9179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fredholm BB, Chen JF, Cunha RA, Svenningsson P, Vaugeois JM. Adenosine and brain function. Int Rev Neurobiol. 2005;63:191–270. doi: 10.1016/S0074-7742(05)63007-3. [DOI] [PubMed] [Google Scholar]
- 123.Lupica CR, Proctor WR, Dunwiddie TV. Presynaptic inhibition of excitatory synaptic transmission by adenosine in rat hippocampus: analysis of unitary EPSP variance measured by whole-cell recording. J Neurosci. 1992;12(10):3753–3764. doi: 10.1523/JNEUROSCI.12-10-03753.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dunwiddie TV, Masino SA. The role and regulation of adenosine in the central nervous system. Annu Rev Neurosci. 2001;24:31–55. doi: 10.1146/annurev.neuro.24.1.31. [DOI] [PubMed] [Google Scholar]
- 125.Ruby CL, Adams CA, Knight EJ, Nam HW, Choi DS. An essential role for adenosine signaling in alcohol abuse. Curr Drug Abuse Rev. 2010;3(3):163–174. doi: 10.2174/1874473711003030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Harvey J, Lacey MG. A postsynaptic interaction between dopamine D1 and NMDA receptors promotes presynaptic inhibition in the rat nucleus accumbens via adenosine release. J Neurosci. 1997;17(14):5271–5280. doi: 10.1523/JNEUROSCI.17-14-05271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dean P, Major P, Nakjang S, Hirt RP, Embley TM. Transport proteins of parasitic protists and their role in nutrient salvage. Front Plant Sci. 2014;5:153. doi: 10.3389/fpls.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Girke C, Daumann M, Niopek-Witz S, Mohlmann T. Nucleobase and nucleoside transport and integration into plant metabolism. Front Plant Sci. 2014;5:443. doi: 10.3389/fpls.2014.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marr JJ. Purine analogs as chemotherapeutic agents in leishmaniasis and American trypanosomiasis. J Lab Clin Med. 1991;118(2):111–119. [PubMed] [Google Scholar]
- 130.Vickers MF, Yao SY, Baldwin SA, Young JD, Cass CE. Nucleoside transporter proteins of Saccharomyces cerevisiae. Demonstration of a transporter (FUI1) with high uridine selectivity in plasma membranes and a transporter (FUN26) with broad nucleoside selectivity in intracellular membranes. The Journal of biological chemistry. 2000;275(34):25931–25938. doi: 10.1074/jbc.M000239200. [DOI] [PubMed] [Google Scholar]
- 131.Sundaram M, et al. Topology of a human equilibrative, nitrobenzylthioinosine (NBMPR)-sensitive nucleoside transporter (hENT1) implicated in the cellular uptake of adenosine and anti-cancer drugs. The Journal of biological chemistry. 2001;276(48):45270–45275. doi: 10.1074/jbc.M107169200. [DOI] [PubMed] [Google Scholar]
- 132.Griffiths M, et al. Cloning of a human nucleoside transporter implicated in the cellular uptake of adenosine and chemotherapeutic drugs. Nature medicine. 1997;3(1):89–93. doi: 10.1038/nm0197-89. [DOI] [PubMed] [Google Scholar]
- 133.Vickers MF, et al. Functional production and reconstitution of the human equilibrative nucleoside transporter (hENT1) in Saccharomyces cerevisiae. Interaction of inhibitors of nucleoside transport with recombinant hENT1 and a glycosylation-defective derivative (hENT1/N48Q) The Biochemical journal. 1999;339( Pt 1):21–32. [PMC free article] [PubMed] [Google Scholar]
- 134.Ward JL, Leung GP, Toan SV, Tse CM. Functional analysis of site-directed glycosylation mutants of the human equilibrative nucleoside transporter-2. Archives of biochemistry and biophysics. 2003;411(1):19–26. doi: 10.1016/s0003-9861(02)00718-x. [DOI] [PubMed] [Google Scholar]
- 135.Visser F, et al. Residue 33 of human equilibrative nucleoside transporter 2 is a functionally important component of both the dipyridamole and nucleoside binding sites. Molecular pharmacology. 2005;67(4):1291–1298. doi: 10.1124/mol.104.005884. [DOI] [PubMed] [Google Scholar]
- 136.Visser F, et al. Mutation of residue 33 of human equilibrative nucleoside transporters 1 and 2 alters sensitivity to inhibition of transport by dilazep and dipyridamole. The Journal of biological chemistry. 2002;277(1):395–401. doi: 10.1074/jbc.M105324200. [DOI] [PubMed] [Google Scholar]
- 137.Sundaram M, et al. Chimeric constructs between human and rat equilibrative nucleoside transporters (hENT1 and rENT1) reveal hENT1 structural domains interacting with coronary vasoactive drugs. The Journal of biological chemistry. 1998;273(34):21519–21525. doi: 10.1074/jbc.273.34.21519. [DOI] [PubMed] [Google Scholar]
- 138.Sundaram M, et al. Equilibrative nucleoside transporters: mapping regions of interaction for the substrate analogue nitrobenzylthioinosine (NBMPR) using rat chimeric proteins. Biochemistry. 2001;40(27):8146–8151. doi: 10.1021/bi0101805. [DOI] [PubMed] [Google Scholar]
- 139.Yao SY, et al. Identification of Cys140 in helix 4 as an exofacial cysteine residue within the substrate-translocation channel of rat equilibrative nitrobenzylthioinosine (NBMPR)-insensitive nucleoside transporter rENT2. The Biochemical journal. 2001;353(Pt 2):387–393. doi: 10.1042/0264-6021:3530387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yao SY, et al. Functional and molecular characterization of nucleobase transport by recombinant human and rat equilibrative nucleoside transporters 1 and 2. Chimeric constructs reveal a role for the ENT2 helix 5–6 region in nucleobase translocation. The Journal of biological chemistry. 2002;277(28):24938–24948. doi: 10.1074/jbc.M200966200. [DOI] [PubMed] [Google Scholar]
- 141.SenGupta DJ, Unadkat JD. Glycine 154 of the equilibrative nucleoside transporter, hENT1, is important for nucleoside transport and for conferring sensitivity to the inhibitors nitrobenzylthioinosine, dipyridamole, and dilazep. Biochemical pharmacology. 2004;67(3):453–458. doi: 10.1016/j.bcp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 142.SenGupta DJ, et al. A single glycine mutation in the equilibrative nucleoside transporter gene, hENT1, alters nucleoside transport activity and sensitivity to nitrobenzylthioinosine. Biochemistry. 2002;41(5):1512–1519. doi: 10.1021/bi015833w. [DOI] [PubMed] [Google Scholar]
- 143.Endres CJ, Sengupta DJ, Unadkat JD. Mutation of leucine-92 selectively reduces the apparent affinity of inosine, guanosine, NBMPR [S6-(4-nitrobenzyl)-mercaptopurine riboside] and dilazep for the human equilibrative nucleoside transporter, hENT1. The Biochemical journal. 2004;380(Pt 1):131–137. doi: 10.1042/BJ20031880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee EW, Lai Y, Zhang H, Unadkat JD. Identification of the mitochondrial targeting signal of the human equilibrative nucleoside transporter 1 (hENT1): implications for interspecies differences in mitochondrial toxicity of fialuridine. The Journal of biological chemistry. 2006;281(24):16700–16706. doi: 10.1074/jbc.M513825200. [DOI] [PubMed] [Google Scholar]
- 145.Visser F, et al. Residues 334 and 338 in transmembrane segment 8 of human equilibrative nucleoside transporter 1 are important determinants of inhibitor sensitivity, protein folding, and catalytic turnover. The Journal of biological chemistry. 2007;282(19):14148–14157. doi: 10.1074/jbc.M701735200. [DOI] [PubMed] [Google Scholar]
- 146.Endres CJ, Unadkat JD. Residues Met89 and Ser160 in the human equilibrative nucleoside transporter 1 affect its affinity for adenosine, guanosine, S6-(4-nitrobenzyl)-mercaptopurine riboside, and dipyridamole. Molecular pharmacology. 2005;67(3):837–844. doi: 10.1124/mol.104.008102. [DOI] [PubMed] [Google Scholar]
- 147.Zimmerman EI, et al. Identification of a novel point mutation in ENT1 that confers resistance to Ara-C in human T cell leukemia CCRF-CEM cells. FEBS letters. 2009;583(2):425–429. doi: 10.1016/j.febslet.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Valdes R, Arastu-Kapur S, Landfear SM, Shinde U. An ab Initio structural model of a nucleoside permease predicts functionally important residues. The Journal of biological chemistry. 2009;284(28):19067–19076. doi: 10.1074/jbc.M109.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Valdes R, Liu W, Ullman B, Landfear SM. Comprehensive examination of charged intramembrane residues in a nucleoside transporter. The Journal of biological chemistry. 2006;281(32):22647–22655. doi: 10.1074/jbc.M602366200. [DOI] [PubMed] [Google Scholar]
- 150.Valdes R, Elferich J, Shinde U, Landfear SM. Identification of the intracellular gate for a member of the equilibrative nucleoside transporter (ENT) family. The Journal of biological chemistry. 2014;289(13):8799–8809. doi: 10.1074/jbc.M113.546960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vasudevan G, Ullman B, Landfear SM. Point mutations in a nucleoside transporter gene from Leishmania donovani confer drug resistance and alter substrate selectivity. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(11):6092–6097. doi: 10.1073/pnas.101537298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Arastu-Kapur S, Ford E, Ullman B, Carter NS. Functional analysis of an inosine-guanosine transporter from Leishmania donovani. The role of conserved residues, aspartate 389 and arginine 393. The Journal of biological chemistry. 2003;278(35):33327–33333. doi: 10.1074/jbc.M305141200. [DOI] [PubMed] [Google Scholar]
- 153.Visser F, Baldwin SA, Isaac RE, Young JD, Cass CE. Identification and mutational analysis of amino acid residues involved in dipyridamole interactions with human and Caenorhabditis elegans equilibrative nucleoside transporters. The Journal of biological chemistry. 2005;280(12):11025–11034. doi: 10.1074/jbc.M410348200. [DOI] [PubMed] [Google Scholar]
- 154.Aseervatham J, Tran L, Machaca K, Boudker O. The Role of Flexible Loops in Folding, Trafficking and Activity of Equilibrative Nucleoside Transporters. PloS one. 2015;10(9):e0136779. doi: 10.1371/journal.pone.0136779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yan N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu Rev Biophys. 2015;44:257–283. doi: 10.1146/annurev-biophys-060414-033901. [DOI] [PubMed] [Google Scholar]
- 156.Arastu-Kapur S, Arendt CS, Purnat T, Carter NS, Ullman B. Second-site suppression of a nonfunctional mutation within the Leishmania donovani inosine-guanosine transporter. The Journal of biological chemistry. 2005;280(3):2213–2219. doi: 10.1074/jbc.M408224200. [DOI] [PubMed] [Google Scholar]
- 157.Papageorgiou I, De Koning HP, Soteriadou K, Diallinas G. Kinetic and mutational analysis of the Trypanosoma brucei NBT1 nucleobase transporter expressed in Saccharomyces cerevisiae reveals structural similarities between ENT and MFS transporters. Int J Parasitol. 2008;38(6):641–653. doi: 10.1016/j.ijpara.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 158.Yan N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci. 2013;38(3):151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 159.Kaback HR. A chemiosmotic mechanism of symport. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(5):1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Henriques C, Sanchez MA, Tryon R, Landfear SM. Molecular and functional characterization of the first nucleobase transporter gene from African trypanosomes. Mol Biochem Parasitol. 2003;130(2):101–110. doi: 10.1016/s0166-6851(03)00167-1. [DOI] [PubMed] [Google Scholar]
- 161.Sanchez MA, Tryon R, Pierce S, Vasudevan G, Landfear SM. Functional expression and characterization of a purine nucleobase transporter gene from Leishmania major. Molecular membrane biology. 2004;21(1):11–18. doi: 10.1080/0968768031000140845. [DOI] [PubMed] [Google Scholar]
- 162.Zhang J, et al. Studies of nucleoside transporters using novel autofluorescent nucleoside probes. Biochemistry. 2006;45(4):1087–1098. doi: 10.1021/bi0520535. [DOI] [PubMed] [Google Scholar]
- 163.Quistgaard EM, Low C, Guettou F, Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nature reviews. Molecular cell biology. 2016;17(2):123–132. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 164.King AE, Ackley MA, Cass CE, Young JD, Baldwin SA. Nucleoside transporters: from scavengers to novel therapeutic targets. Trends in pharmacological sciences. 2006;27(8):416–425. doi: 10.1016/j.tips.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 165.Hu XH, et al. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics. 2007;175(3):1479–1487. doi: 10.1534/genetics.106.065292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Le Chevalier T, et al. Efficacy of gemcitabine plus platinum chemotherapy compared with other platinum containing regimens in advanced non-small-cell lung cancer: a meta-analysis of survival outcomes. Lung Cancer. 2005;47(1):69–80. doi: 10.1016/j.lungcan.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 167.Scagliotti GV, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(21):3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 168.Heinemann V, et al. Randomized phase III trial of gemcitabine plus cisplatin compared with gemcitabine alone in advanced pancreatic cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24(24):3946–3952. doi: 10.1200/JCO.2005.05.1490. [DOI] [PubMed] [Google Scholar]
- 169.Heinemann V, et al. High efficacy of gemcitabine and cisplatin in patients with predominantly anthracycline- and taxane-pretreated metastatic breast cancer. Cancer Chemother Pharmacol. 2006;57(5):640–646. doi: 10.1007/s00280-005-0093-5. [DOI] [PubMed] [Google Scholar]
- 170.Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature. 2014;509(7500):310–317. doi: 10.1038/nature13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5(3):247–264. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Parkinson FE, Ferguson J, Zamzow CR, Xiong W. Gene expression for enzymes and transporters involved in regulating adenosine and inosine levels in rat forebrain neurons, astrocytes and C6 glioma cells. J Neurosci Res. 2006;84(4):801–808. doi: 10.1002/jnr.20988. [DOI] [PubMed] [Google Scholar]
- 173.Parkinson FE, et al. Effects of nitrobenzylthioinosine on neuronal injury, adenosine levels, and adenosine receptor activity in rat forebrain ischemia. J Neurochem. 2000;75(2):795–802. doi: 10.1046/j.1471-4159.2000.0750795.x. [DOI] [PubMed] [Google Scholar]
- 174.Idzko M, Ferrari D, Riegel AK, Eltzschig HK. Extracellular nucleotide and nucleoside signaling in vascular and blood disease. Blood. 2014;124(7):1029–1037. doi: 10.1182/blood-2013-09-402560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang G, Franklin PH, Murray TF. Manipulation of endogenous adenosine in the rat prepiriform cortex modulates seizure susceptibility. The Journal of pharmacology and experimental therapeutics. 1993;264(3):1415–1424. [PubMed] [Google Scholar]
- 176.Pastor-Anglada M, Perez-Torras S. Nucleoside transporter proteins as biomarkers of drug responsiveness and drug targets. Front Pharmacol. 2015;6:13. doi: 10.3389/fphar.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bone DB, Hammond JR. Nucleoside and nucleobase transporters of primary human cardiac microvascular endothelial cells: characterization of a novel nucleobase transporter. American journal of physiology. Heart and circulatory physiology. 2007;293(6):H3325–3332. doi: 10.1152/ajpheart.01006.2007. [DOI] [PubMed] [Google Scholar]
- 178.Park JS, Hughes SJ, Cunningham FK, Hammond JR. Identification of cysteines involved in the effects of methanethiosulfonate reagents on human equilibrative nucleoside transporter 1. Molecular pharmacology. 2011;80(4):735–746. doi: 10.1124/mol.111.072587. [DOI] [PubMed] [Google Scholar]
- 179.Park JS, Hammond JR. Cysteine residues in the transmembrane (TM) 9 to TM11 region of the human equilibrative nucleoside transporter subtype 1 play an important role in inhibitor binding and translocation function. Molecular pharmacology. 2012;82(5):784–794. doi: 10.1124/mol.112.079616. [DOI] [PubMed] [Google Scholar]
- 180.Melki I, et al. Mutation in the SLC29A3 gene: a new cause of a monogenic, autoinflammatory condition. Pediatrics. 2013;131(4):e1308–1313. doi: 10.1542/peds.2012-2255. [DOI] [PubMed] [Google Scholar]
- 181.Traub M, et al. The fluorouridine insensitive 1 (fur1) mutant is defective in equilibrative nucleoside transporter 3 (ENT3), and thus represents an important pyrimidine nucleoside uptake system in Arabidopsis thaliana. Plant J. 2007;49(5):855–864. doi: 10.1111/j.1365-313X.2006.02998.x. [DOI] [PubMed] [Google Scholar]
- 182.Wu K, King J. Biochemical and genetic characterization of 5-fluoro-2′-deoxyuridine-resistant mutants of Arabidopsis thaliana. Planta. 1994;194(1):117–122. [Google Scholar]