Abstract
Eukaryotic translation is tightly regulated to ensure that protein production occurs at the right time and place. Recent studies on abnormal repeat proteins, especially in age-dependent neurodegenerative diseases caused by nucleotide repeat expansion, have highlighted or identified two forms of unconventional translation initiation: usage of AUG-like sites (near cognates) or repeat-associated non-AUG (RAN) translation. We discuss how repeat proteins may differ due to not just unconventional initiation, but also ribosomal frameshifting and/or imperfect repeat DNA replication, expansion and repair, and highlight how research on translation of repeats may uncover insights into the biology of translation and its contribution to disease.
Keywords: ALS, C9ORF72, dipeptide repeat proteins, frontotemporal dementia, near-cognate start codon, repeat expansion, upstream open reading frame, translation
In 1961, Marshall W. Nirenberg and his postdoctoral fellow Johann H. Matthaei at the National Institutes of Health found that mixing polyuridylic acid (poly-U) with cell-free extracts from E. coli resulted in the production of proteins made entirely of phenylalanine (Nirenberg and Matthaei, 1961). Using additional synthetic RNAs, including those with repeating triplets, Nirenberg and H. Gobind Khorana produced different mono- and dipeptide proteins. Together, they deciphered the genetic code (Khorana, 1968), and for this work, shared the 1968 Nobel Prize in Physiology or Medicine, along with Robert W. Holley who first isolated tRNA.
Today, translation in eukaryote cells is recognized to be a highly regulated process. Initiation is the rate-limiting step and thus is the phase of protein synthesis most subject to regulation (Richter and Sonenberg, 2005). Although initiation itself is a multi-step process, it culminates in 5′ to 3′ “scanning” of a complex containing the 40S ribosome subunit—often referred to as a 43S preinitiation complex that includes a ternary complex of eIF2, GTP, and initiator methionine tRNA. Usually upon encountering the first AUG codon, which more often than not resides in a favorable context (the 5′ A/GCCAUGG 3′ Kozak sequence that promotes initiation (Kozak, 1986; Hinnebusch et al., 2016), the preinitiation complex stops, recruits the 60S subunit and polypeptide elongation begins (Figure 1A).
Figure 1. Mechanisms of translation initiation.
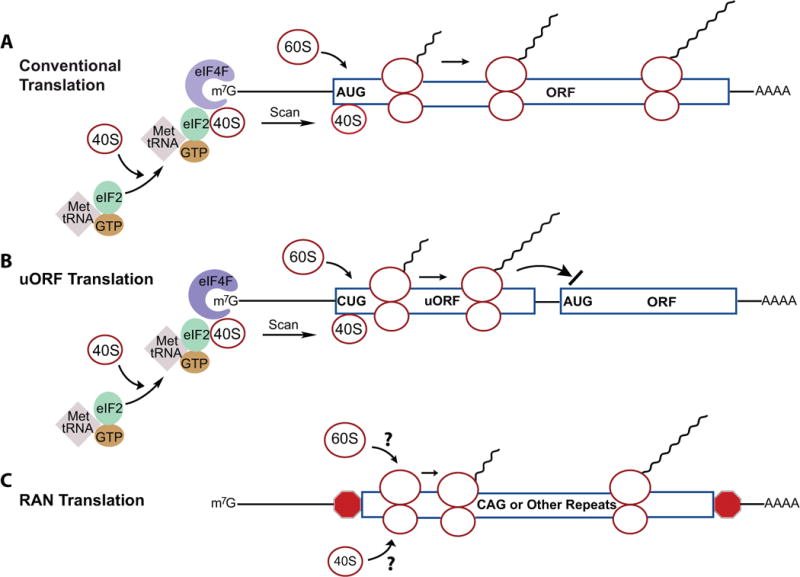
(A) During “conventional” translation, the ternary complex (TC) composed of eIF2, GTP, and Met-tRNA is joined by the 40S ribosomal subunit to form a 43S pre-initiation complex (PIC). The PIC interacts with eIF4F (composed of three factors: the cap binding protein eIF4E, the scaffold protein eIF4G, and the RNA helicase eIF4A) of the mRNA 5′ cap. The 40S then scans until if stalls at usually the first AUG, which often resides within a Kozak sequence. The 60S is then recruited and the full ribosome commences polypeptide synthesis.
(B) One type of uORF translation is illustrated, in which a non-canonical CUG in the uORF is the initiating codon and translation of the uORF inhibits translation of the downstream ORF.
(C) RAN translation proposes that the 40S and 60S ribosomal subunits directly bind the expanded repeats in mRNA through an unknown mechanism.
5′ to the classic AUG initiation codon, however, it is now clear that many RNAs have upstream open reading frames (uORFs), which can serve several different roles in translation. Some uORFs lack a stop codon and begin with a canonical AUG initiator codon in-frame with the downstream AUG. In these cases, “leaky” scanning may occur where peptide synthesis occurs at both the uORF and downstream ORF resulting in two peptides distinct only at the amino terminus (Hinnebusch et al., 2016). Other uORFs can suppress or prevent translation at the downstream ORF when a stop codon is present between them because ribosome re-initiation is not robust (Figure 1B). Moreover, scanning 40S ribosomal subunits can sometimes bypass the uORF altogether and initiate translation at the downstream AUG codon, which can be particularly robust when the uORF acts as an enhancer of downstream initiation (Ivanov et al., 2010).
With the advent of next-generation sequencing and ribosome profiling, which together can map the positions and relative amounts of ribosomes on mRNA (Ingolia et al 2009), it has become clear that 1) most mRNAs have uORFs and 2) these uORFs frequently use alternative (non-AUG) start codons. For example, Ingolia et al. (2011) and Lee et al. (2012) performed ribosome profiling in cultured cells in the presence of the translation inhibitor cycloheximide, which binds both initiating and elongating ribosomes, or other drugs such as harringtonine and lactimidomycin, which binds initiating ribosomes with an exit (E) site that is devoid of tRNA. These drugs allow elongating ribosomes to run-off the mRNA while maintaining the initiating ribosome in place, thereby enabling start codon identification on a global basis following sequencing of the RNAs protected by this initiating ribosome (an approach called global translation initiation sequencing - or GTI-seq). These and related analyses (e.g. Bazzini et al., 2014; Fields et al., 2015; Ji et al., 2015) have revealed that >50% of mRNAs contain uORFs that are occupied by ribosomes, in close agreement with a similar proportion estimated by an earlier RNA-seq effort (Calvo et al 2009). Use of GTI-seq has also demonstrated remarkable variation in translation initiation sites (uORF and ORF), with only 51% beginning with AUG, 16% with CUG, and 24% with other sequences (UUG, CUG, AGG, ACG, AAG, AUC, AUA, AUU). For uORFs in particular, 75% begin with non-AUG codons, with CUG (encoding leucine) being the most prevalent.
The preponderance of uORFs implies widespread regulation of downstream ORF translation (Morris and Geballe, 2000), and it is the ternary complex component eIF2α that is the key factor controlling uORF utilization. Phosphorylation of eIF2α serine 51 inhibits the activity of the guanine nucleotide exchange factor eIF2B, which in turn impedes formation of the 43S pre-initiation complex and consequently reduces general translation. However, some mRNAs escape this inhibition and are translated even though they have uORFs. This is caused by stress-induced eIF2α phosphorylation and resulting ternary complex limitation, which renders uORF initiation unfavorable; it is thus bypassed and initiation occurs at the downstream AUG. Examples of mRNAs under this type of regulation encode such proteins as the transcription factors CHOP (CCAAT-enhancer-binding protein homologous protein) and ATF4 and ATF5, the growth arrest and DNA damage-inducible protein GADD34, among many others (Baird et al., 2014; Young and Wek, 2016). Added to this, a peptide encoded by the uORF in CHOP mRNA regulates translation of the downstream ORF (Jousse et al., 2001).
uORF regulation of translation and eIF2α phosphorylation are central components of an integrated stress response (Young and Wek, 2016). Four kinases respond to different environmental stresses and phosphorylate eIF2α serine 51: PERK responds to ER stress, HRI responds to oxidative stress and heat shock, PKR responds to viral infection, and GCN2 is a nutrient sensor. Correspondingly, cellular stress, through any one of these kinases, can modulate uORF and downstream ORF translation, which may result in unique biological responses depending on the cell type.
Abnormal translation products and potential translation mechanisms in repeat expansion disorders
One highly specialized cell type is the neuron, which can live for decades without undergoing cell division. Although these cells can endure even when encountering various stresses, in some neurological diseases they undergo a remarkable molecular reorganization. Repeat expansion disorders are a set of more than 30 genetic diseases, mostly of the nervous system, that are caused by expansion of short repeat sequences of 3–6 nucleotides in a host gene’s coding sequences, introns, or 5′ and 3′ untranslated regions (UTRs) (Zhang and Ashizawa, 2017). During the last two decades, unconventional translation products from various expanded repeat sequences have been repeatedly detected in the nervous system of affected individuals. Three mechanisms have been proposed to account for the generation of these disease-specific proteins: 1) ribosomal frameshifting (Gaspar et al., 2000), 2) repeat-associated non-AUG (RAN) translation (Zu et al., 2011), and 3) near-cognate start codon initiated translation (Sellier et al., 2017).
Ribosomal frameshifting
Beginning with the work of Jacks and Varmus (Jacks and Varmus, 1985; Jacks et al., 1988), it has become clear that most retroviruses, including HIV, utilize ribosomal frameshifting as an obligatory component of their life cycle. Frameshifting can be highly efficient and driven by combination of a 7 base “slippery” RNA sequence and an RNA pseudoknot 3′ to the site of frameshifting (Chamorro et al., 1992). Beyond viruses, frameshifting is also found frequently in bacteria and known for some eukaryotic mRNAs (Caliskan et al., 2015). Similar frameshifting appears to be a component in some polyglutamine diseases in which polyglutamine tracts are synthesized from expanded CAG repeats in frame with the coding regions of respective host genes (Orr and Zoghbi, 2007). For example, in spinocerebellar ataxia type 3 (SCA3), ataxin 3 contains CAG repeats whose in-frame translation produces an expanded polyglutamine track; however, polyalanine-containing proteins are also generated in patient neurons (Gaspar et al., 2000). Using reporter constructs in which different tags were engineered into separate reading frames, polyalanine synthesis was initially proposed to be caused by ribosomal slippage into the GCA frame in a repeat length–dependent manner, although the slippage site was not identified (Toulouse et al., 2005). In both Drosophila and mammalian neurons, production of polyalanine may be a key component of CAG repeat toxicity (Stochmanski et al., 2012).
In Huntington’s disease (HD), the most common CAG repeat disease, the repeats are in-frame with the huntingtin ORF, producing polyglutamine after codon number 17 in huntingtin (HTT). Nevertheless, both polyserine and polyalanine, encoded by the +1 and −1 frames of expanded CAG repeats, respectively, have been detected in autopsy samples of HD patients and in a transgenic mouse model of HD (Davies and Rubinsztein, 2006; Bañez-Coronel et al., 2015). Frameshifting has been established to account for production of polyserine or polyalanine linked to the amino terminal portion of huntingtin. This frameshifting can occur within the expanded CAG repeats and is enhanced by depletion of the charged glutaminyl-tRNA that pairs to the CAG codon (Girstmair et al, 2013). Detailed mass spectrometry and molecular analysis revealed that a UUCC sequence at the 5′ end of CAG repeats in the Huntingtin coding region initiates the +1 frameshifting, and the effect of the UUCC sequence is enhanced by increased formation of stem-loop structures by expanded CAG repeats, providing a specific molecular mechanism for potential ribosomal frameshifting in HD patient cells (Saffert et al., 2016).
RAN translation
Polyalanine has also been detected in tissues of patients with another CAG repeat disease (SCA8) and in mouse models of the disease (Zu et al., 2011). Here, CAG repeats are in-frame with the coding sequence of ataxin 8 (ATXN8). Rather than frameshifting, it has been proposed that polyalanine is produced by a different unconventional translation mechanism, repeat associated non-AUG (RAN) translation, in which initiation in any of the three reading frames within expanded CAG (or other) repeats is thought to occur internally (Zu et al., 2011; Figure 1C), seemingly consistent with the internal ribosome binding of mRNA observed by Nirenberg and Khorana decades ago using cell-free extracts primed with synthetic polyribonucleotides (Nirenberg and Matthaei, 1961; Nishimura et al., 1964). Expression of engineered repeat-containing ATXN8 or HTT mRNA with multiple stop codons 5′ to the CAG repeats in the +1 and +2 frames has shown that RAN translation certainly occurs under certain experimental conditions (Zu et al., 2011; Bañez-Coronel et al., 2015). Abnormal disease-specific repeat proteins proposed to be synthesized from both sense and antisense transcripts through RAN translation have also been detected in brain tissues of patients with CTG repeat expansion in the 3′UTR of DMPK in myotonic dystrophy type 1 (DM1) (Zu et al., 2011), GGGGCC repeat expansion in the first intron of C9ORF72 that causes both amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Ash et al., 2013; Mori et al., 2013; Zu et al., 2013), and CCTG repeat expansion in the first intron of ZNF9 in myotonic dystrophy type 2 (DM2) (Zu et al., 2017). Thus, RAN translation is posited to be a general phenomenon in various repeat expansion disorders (Cleary and Ranum, 2017).
Near-cognate start codons
Near-cognate start codon–initiated translation is a third mechanism that can yield mono- or dipeptide-containing proteins from translation of nucleotide repeats. The founding example of this came from fragile X-associated tremor/ataxia syndrome (FXTAS), which is caused by CGG expansion (55–200 repeats) in the 5′UTR of the fragile X mental retardation 1 (FMR1) gene (Hagerman and Hagerman, 2016). The GGC frame of these repeats encodes polyglycine (FMRpolyG), which is indeed detected in FXTAS patient brains (Todd et al., 2013; Sellier et al., 2017). The production of polyglycine does not seem to be mediated by RAN translation; rather, it requires a cap-dependent scanning mechanism for translation initiation (Kearse et al., 2016) and depends on an upstream, in-frame ACG near-cognate start codon embedded in a Kozak consensus sequence (Sellier et al., 2017). The expanded CGG repeats create a large uORF 5′ to the FMR1 open reading frame. Cap-dependent ribosome scanning that initiates at the ACG codon produces a protein with an N-terminal methionine and a short polypeptide, followed by polyglycine, and then a 42–amino acid C-terminus, all encoded by the sequences in what is typically annotated as the FMR1 5′UTR (Kearse et al., 2016; Sellier et al., 2017). Both polyglycine and the 42–amino acid C-terminus that follows it appear to be key components driving disease, with deletion of the ACG near-cognate start codon abolishing polyglycine production and neurotoxicity (Sellier et al., 2017).
The translational complexity of expanded G4C2 repeats in C9ORF72-ALS/FTD
The complexity of atypical translation in repeat expansion diseases is most apparent in ALS, a fatal paralytic disease, and, FTD, the second most common form of pre-senile dementia. The most frequent genetic cause of these diseases is GGGGCC repeat expansion in the first intron of C9ORF72 with up to thousands of copies in the affected areas of the human nervous system (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Figure 2A). Since its discovery in 2011, rapid progress has been made in identifying dysregulated molecular pathways in C9ORF72-ALS/FTD (Gao et al., 2017). Although repeat RNAs themselves may be partially responsible for neurotoxicity, accumulating evidence suggests that dipeptide repeat (DPR) proteins are pathogenic (Gitler and Tsuiji, 2016). At least five DPR proteins [poly(GA), poly(GR), poly(GP), poly(PR), and poly(PA)] encoded in the multiple frames of sense and antisense repeat-containing RNAs, have been detected in C9ORF72-ALS/FTD patients (Ash et al., 2013; Mori et al., 2013; Zu et al., 2013), albeit different investigators have reported highly divergent levels and number of neurons affected. Although poly(GP) proteins are synthesized predominantly from the sense repeats (based using antisense oligonucleotides to trigger RNase H-dependent destruction of the C9ORF72 sense strand RNAs) (Gendron et al., 2017), they can also be produced from the antisense strand with a different C-terminus (Zu et al., 2013). Thus, they probably should be considered as two different proteins. In fact, because we do not know the precise translation initiation and termination sites on repeat-containing RNA in human cells, the diversity of DPR proteins could be much larger than detected thus far.
Figure 2. Different translation mechanisms that may operate in C9ORF72 ALS/FTD.
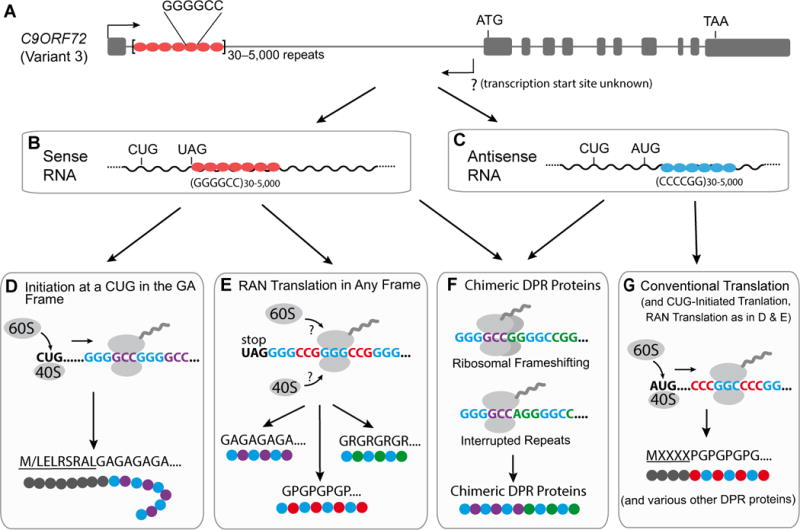
(A) Genomic organization of the C9ORF72 gene (variant 3) containing expanded G4C2 repeats.
(B) G4C2-contianing RNA that could be spliced intron or intron 1-retained mRNA.
(C) C4G2-containing RNA whose 5′ and 3′ ends are unknown.
(D) It is speculated that poly(GA) may be synthesized through CUG-initiated translation.
(E) Different DPR proteins are thought to be produced through RAN translation.
(F) The proposed mechanisms that may lead to the synthesis of chimeric DPR proteins from both sense and antisense repeat transcripts.
(G) DPR proteins may be also synthesized through conventional translation from expanded C4G2 repeats-containing RNA. Three AUG start codons embedded in Kozak sequences are in-frame with poly(PG) coding sequence. All three DPR proteins, poly(PA), poly(PG) and poly(PR), may contain a N-terminus synthesized from nucleotide sequences 5′ to C4G2 repeats.
Because the intron containing expanded G4C2 repeats appears to be spliced out efficiently in human cells (Tran et al., 2015), some DPR proteins might be produced from RNAs without other regulatory elements typically present in mRNAs, including the 5′ cap and the poly(A) tail. However, experimental evidence is still lacking to definitively demonstrate the nature of the RNA that are translated for DPR protein production. This is an important unresolved issue because the template for translation, spliced intron or unspliced pre-mRNA, may determine which translation mechanism(s) is/are at play. Nonetheless, here we speculate that ALS/FTD-linked DPR proteins can be synthesized by any of the three mechanisms introduced above: RAN translation of either the repeat-containing excised intron or the pre-mRNA, ribosomal scanning for a near-cognate start codon, or translational frameshifting producing chimeric DPR products after initiation by either RAN translation or a near-cognate start codon (Figure 2).
We would like to emphasize that it is not established yet which mechanism(s) is primarily responsible for production of endogenous DPR proteins in human cells where expanded G4C2 repeats are expressed in their native molecular context and at a low level. Consider, for example, that in the sense strand RNA 5′ to expanded G4C2 repeats, a near-cognate CUG start codon is embedded in a good Kozak sequence and in-frame with poly(GA), the most abundant DPR protein in C9ORF72-ALS/FTD patient brains (Mackenzie et al., 2015). It is thus possible that poly(GA) might be synthesized through a conventional ribosomal scanning mechanism (Figure 2D). Poly(GP), on the other hand, is mostly synthesized from the sense transcripts (Gendron et al., 2017) and encoded by the second reading frame that has an in-frame UAG translation terminator juxtaposed just 5′ to the G4C2 repeats. Thus, it may be produced either through RAN translation (Figure 2E) and/or in combination with other mechanisms. It is not obvious to predict where poly(GR) synthesis begins. RAN translation is plausible, although 26 codons are in-frame with poly(GR) in the sequence 5′ to the repeats but whose role in poly(GR) synthesis is unknown. Similarly, although the transcription start and termination sites for C9ORF72 repeats-containing antisense transcripts are unknown, long stretches of amino acids are potentially encoded upstream and in-frame with poly(PG) and poly(PR) in antisense transcripts that contain several AUG, CUG and putative Kozak sequences, raising the possibility that some or all of these DPR proteins are synthesized by usage of AUG or near-cognate start codons upstream of the expanded repeats (Figure 2G). These possibilities have not yet been tested experimentally.
In principle, ribosomal frameshifting can also occur in C9ORF72-ALS/FTD (Figure 2F), as this process is enhanced by downstream RNA pseudoknots (Chamorro et al., 1992) or G-quadruplexes (Yu et al., 2014), the latter a secondary structure readily formed by expanded G4C2 repeat RNA (Reddy et al., 2013). Moreover, although widely assumed to be pure repeats, we caution that this may not be the case in C9ORF72-ALS/FTD. It is plausible that the somatic expansion that produces up to thousands of G4C2 repeats is accompanied by loss or gain of one or more nucleotides, thereby driving production of chimeric dipeptides. Interrupted G4C2 repeats can potentially be generated by DNA replication stalling and/or template slippage at expanded repeats (Viterbo et al., 2016), resulting in the activation of post-replication repair pathways that can introduce errors in the sequence (Lee and Myung, 2008). Moreover, DPR proteins either directly or indirectly increase DNA damage in human C9ORF72 cells (Lopez-Gonalez et al., 2016; Farg et al., 2017; Walker et al., 2017). Loss of TDP-43, a key pathological protein in more than 95% of ALS cases (Ling et al., 2013), compromises transcription-coupled DNA repair (Hill et al., 2016). Thus, an improper repair process in postmitotic neurons may also result in interrupted repeat sequences. Recognizing this, we speculate that a wide variety of DPR proteins may be produced in C9ORF72-ALS/FTD patients: pure DPR proteins of different lengths, DPR proteins with unique N- and/or C-terminal amino acid sequences, chimeric DPR proteins, and potentially DPR proteins containing other amino acids encoded by unknown nucleotide sequences embedded within G4C2 repeats, each of which can be initiated either by near-cognate start codons or RAN translation.
Concluding remarks
The discovery of abnormal mono- and di-peptide proteins in repeat expansion diseases, especially in C9ORF72-ALS/FTD, has uncovered unexpected aspects of protein translation and generated enormous interest in how these proteins are synthesized. It remains to be determined the extent to which each of the proposed mechanisms – frameshifting, RAN translation, or uORF translation are used in human cells where expanded nucleotide repeats are expressed in their native molecular contexts. Of course, these mechanisms are not mutually exclusive. Possible therapeutic interventions to alleviate disease protein toxicity may be based on specific translational mechanisms, underscoring the importance of understanding these basic molecular processes. We speculate for example, that the composition of DPR proteins in C9ORF72-ALS/FTD may be much more complex than initially thought, depending on the translational mechanisms operating in a specific cell type, which may also partially explain selective neuronal vulnerability in these neurodegenerative disorders. Because aging is the major risk factor, it is possible that increased unconventional translation in disease may be one of the downstream responses to pathogenic stresses. It will be critical to determine how various chronic stresses influence the efficiency of each unconventional translation mode in human neurons. Finally, our prediction is that research on repeats and their translation is likely to continue to uncover novel insights into the biology of translation, as well as contributing to uncovering pathogenic mechanisms of disease.
Acknowledgments
We thank Sandra Almeida and Botao Liu for discussions. This work is supported by the NIH (NS101986 and NS093097 to F.-B.G., NS079415 and HD082013 to J.D.R., NS027036 to D.W.C), and the Target ALS Foundation (to F.-B.G. and J.D.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Palam LR, Fusakio ME, Willy JA, Davis CM, McCintock JN, Anthony TG, Wek RC. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKa. Mol Cell Biol. 2014;25:1686–1697. doi: 10.1091/mbc.E14-02-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, et al. RAN Translation in Huntington disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan N, Peske F, Rodnina MV. Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting. Trends Biochem Sci. 2015;40:265–274. doi: 10.1016/j.tibs.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro M, Parkin N, Varmus HE. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. New developments in RAN translation: insights from multiple diseases. Curr Opin Genet Dev. 2017;44:125–134. doi: 10.1016/j.gde.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JE, Rubinsztein DC. Polyalanine and polyserine frameshift products in Huntington’s disease. J Med Genet. 2006;43:893–896. doi: 10.1136/jmg.2006.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Konopka A, Ying Soo K, Ito D, Atkin JD. The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in Amyotrophic Lateral Sclerosis. Hum Mol Genet. 2017 doi: 10.1093/hmg/ddx170. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. A Regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell. 2015;60:816–827. doi: 10.1016/j.molcel.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Almeida S, Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorders. EMBO J. 2017 doi: 10.15252/embj.201797568. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar C, Jannatipour M, Dion P, Laganiere J, Sequeiros J, Brais B, Rouleau GA. CAG tract of MJD-1 may be prone to frameshifts causing polyalanine accumulation. Hum Mol Genet. 2000;9:1957–1966. doi: 10.1093/hmg/9.13.1957. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, Prudencio M, Carlomagno Y, Daughrity LM, Jansen-West K, Perkerson EA, et al. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci Transl Med. 2017;9:383. doi: 10.1126/scitranslmed.aai7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girstmair H, Saffert P, Rode S, Czech A, Holland G, Bannert N, Ignatova Z. Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Rep. 2013;3:148–159. doi: 10.1016/j.celrep.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Gitler AD, Tsuiji H. There has been an awakening: Emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res. 2016;1647:19–29. doi: 10.1016/j.brainres.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman P. Fragile X-associated tremor/ataxia syndrome -features, mechanisms and management. Nat Rev Neurol. 2016;12:403–412. doi: 10.1038/nrneurol.2016.82. [DOI] [PubMed] [Google Scholar]
- Hill SJ, Mordes DA, Cameron LA, Neuberg DS, Landini S, Eggan K, Livingston DM. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci U S A. 2016;113:E7701–E7709. doi: 10.1073/pnas.1611673113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resulotion using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucl Acids Res. 2010;38:353–359. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Jacks T, Varmus HE. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, Struhl K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015;4:e08890. doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C, Bruhat A, Carraro B, Urano F, Ferrara M, Ron D, Fafoumoux P. Inhibition of CHOP translation by a peptide encoded by an open reading from localized in the chop 5′ UTR. Nucl Acid Res. 2001;29:4341–4351. doi: 10.1093/nar/29.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, Todd PK. CGG repeat-associated non-ATG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol Cell. 2016;62:314–322. doi: 10.1016/j.molcel.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana HG. Synthetic nucleic acids and the genetic code. JAMA. 1968;206:1978–1982. [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang S-X, She B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA. 2012;109:E2424–2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, Petrucelli L, Miller BL, Almeida S, Gao FB. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D, et al. Quantitative analysis and clinicopathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015;130:845–861. doi: 10.1007/s00401-015-1476-2. [DOI] [PubMed] [Google Scholar]
- Mardon G, Varmus HE. Frameshift and intragenic suppressor mutations in a Rous sarcoma provirus suggest src encodes two proteins. Cell. 1983;32:871–879. doi: 10.1016/0092-8674(83)90072-7. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013a;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AR. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci U S A. 1961;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Jacob TM, Khorana HG. Synthetic Deoxyribopolynucleotides as Templates for Ribonucleic Acid Polymerase: The Formation and characterization of a ribopolynucleotide with a repeating trinucleotide sequence. Proc Natl Acad Sci U S A. 1964;52:1494–1501. doi: 10.1073/pnas.52.6.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Saffert P, Adamla F, Schieweck R, Atkins JF, Ignatova Z. An expanded CAG repeat in Huntingtin causes +1 frameshifting. J Biol Chem. 2016;291:18505–18513. doi: 10.1074/jbc.M116.744326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RA, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, et al. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to Fragile X Tremor Ataxia Syndrome. Neuron. 2017;93:331–347. doi: 10.1016/j.neuron.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stochmanski SJ, Therrien M, Laganiere J, Rochefort D, Laurent S, Karemera L, Gaudet R, Vyboh K, Van Meyel DJ, Di Cristo G, et al. Expanded ATXN3 frameshifting events are toxic in Drosophila and mammalian neuron models. Hum Mol Genet. 2012;21:2211–2218. doi: 10.1093/hmg/dds036. [DOI] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z, Gao FB. Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron. 2015;87:1207–1214. doi: 10.1016/j.neuron.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulouse A, Au-Yeung F, Gaspar C, Roussel J, Dion P, Rouleau GA. Ribosomal frameshifting on MJD-1 transcripts with long CAG tracts. Hum Mol Genet. 2005;14:2649–2660. doi: 10.1093/hmg/ddi299. [DOI] [PubMed] [Google Scholar]
- Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, Lukashchuk V, Chiang SC, Ray S, Mulcahy PJ, et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat Neurosci. 2017 doi: 10.1038/nn.4604. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viterbo D, Michoud G, Mosbach V, Dujon B, Richard GF. Replication stalling and heteroduplex formation within CAG/CTG trinucleotide repeats by mismatch repair. DNA Repair (Amst) 2016;42:94–106. doi: 10.1016/j.dnarep.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Young SK, Wek RC. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem. 2016;291:169927–16935. doi: 10.1074/jbc.R116.733899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CH, Teulade-Fichou MP, Olsthoorn RC. Stimulation of ribosomal frameshifting by RNA G-quadruplex structures. Nucleic Acids Res. 2014;42:1887–1892. doi: 10.1093/nar/gkt1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Ashizawa T. RNA toxicity and foci formation in microsatellite expansion diseases. Curr Opin Genet Dev. 2017;44:17–29. doi: 10.1016/j.gde.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Cleary JD, Liu Y, Bañez-Coronel M, Bubenik JL, Ayhan F, Ashizawa T, Xia G, Clark HB, Yachnis AT, et al. RAN translation regulated by Muscleblind proteins in myotonic dystrophy type 2. Neuron. 2017;95:1292–1305. doi: 10.1016/j.neuron.2017.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]


