Summary
Prostate cancer exhibits a lineage-specific dependence on androgen signaling. Castration resistance involves reactivation of androgen signaling or activation of alternative lineage programs to bypass androgen requirement. We describe an aberrant gastrointestinal lineage transcriptome expressed in ~5% of primary prostate cancer that is characterized by abbreviated response to androgen deprivation therapy and in ~30% of castration-resistant prostate cancer. This program is governed by a transcriptional circuit consisting of HNF4G and HNF1A. Cistrome and chromatin analyses revealed that HNF4G is a pioneer factor that generates and maintains enhancer landscape at gastrointestinal lineage genes, independent of androgen receptor signaling. In HNF4G/HNF1A-double negative prostate cancer, exogenous expression of HNF4G at physiologic levels recapitulates the gastrointestinal transcriptome, chromatin landscape and leads to relative castration resistance.
Keywords: Prostate cancer, castration resistance, HNF4G, HNF1A, SPINK1, pioneer factor, ChIP-seq, androgen deprivation therapy, enzalutamide
Graphical Abstract
Shukla et al. identify an aberrantly expressed gastrointestinal-lineage transcriptome governed by HNF4G and HNF1A in ~30% of castration-resistant prostate cancer. HNF4G is a pioneer factor for this transcriptional program and its ectopic expression at physiologic levels reduces sensitivity to hormone deprivation.
Introduction
The prostate gland is an androgen-dependent male reproductive organ. Upon oncogenic transformation, prostate cancer retains a remarkable lineage-specific dependence on androgen-receptor (AR) signaling and is characterized by an almost universal initial response to androgen deprivation therapy (Huggins and Hodges, 1941). However, the depth and duration of response is highly variable with eventual progression to the lethal castration-resistant prostate cancer (CRPC). Multiple mechanisms that contribute to castration resistance have been elucidated. One class reactivate AR signaling in the castrate environment, such as by AR mutations, AR splice variants, AR amplification, or aberrant expression of the glucocorticoid receptor (GR) that activates AR target genes (Antonarakis et al., 2014; Arora et al., 2013; Chen et al., 2004; Taplin et al., 1995; Watson et al., 2015). Alternatively, increased cellular plasticity to bypass the prostate lineage-specific dependence on AR signaling, best exemplified by neuroendocrine transdifferentiation, is increasingly appreciated as a mechanism of castration resistance (Beltran et al., 2016). Nevertheless, even with next generation AR pathway inhibitors, the majority of CRPC remain histologically adenocarcinoma (Robinson et al., 2015).
Cancer outlier gene expression analysis identified SPINK1 overexpression in ~10% of all primary prostate cancers, representing a distinct subtype among those prostate cancers without ETS fusion (Tomlins et al., 2008). While prognostic role of SPINK1 overexpression in resected primary prostate cancer is controversial (Flavin et al., 2014; Tomlins et al., 2008), studies indicate that SPINK1 overexpression is associated with more rapid progression to castration resistance. In a Finish cohort of prostate cancer patients treated with primary hormone therapy, SPINK1 IHC positivity in prostate cancer was associated with a significantly more rapid progression to castration resistance (Leinonen et al., 2010). In a Chinese cohort diagnosed with metastatic prostate cancer to the bone, SPINK1 IHC positivity was associated with decreased progression free survival after hormone therapy (Pan et al., 2016). In a Hopkins cohort of intermediate and high risk prostate cancer patients treated with prostatectomy who subsequently recurred, SPINK1 expression was associated with more rapid progression to metastasis and to death (Johnson et al., 2016).
SPINK1, also known as pancreatic secretory trypsin inhibitor, protects the gastrointestinal (GI) tract from protease degradation and its expression is normally restricted to the GI organs. The mechanism by which this GI-restricted gene is expressed in prostate cancer is unknown and suggests that an alternative GI-lineage transcriptome is activated and may lead to decreased dependence on AR signaling. In this study we have explored the mechanism and significance of activation of this GI-lineage transcriptome in prostate cancer tumorigenesis and castration resistance
Results
HNF4G and HNF1A regulate a GI transcriptome in SPINK1-positive prostate cancer
Since normal SPINK1 expression is confined to GI tissues, we compared SPINK1 expression in normal prostate and prostate cancer with that in normal and malignant GI tissues using TCGA cohorts. SPINK1 is highly expressed in normal GI tissues and preserved in corresponding GI cancers. In prostate, SPINK1 demonstrates an outlier overexpression pattern, with a subset of cancers expressing high levels comparable to GI tissues while most prostate cancers and normal tissue express very low levels (Figure S1A). To determine whether SPINK1 overexpression in prostate cancer is part of an aberrant transcriptome, we examined three large high-quality prostate cancer gene expression datasets to identify genes whose expression is correlated with SPINK1 (Beltran et al., 2016; Cancer Genome Atlas Research, 2015; Taylor et al., 2010). We generated a transcriptome signature of SPINK1-correlated genes, consisting of 129 genes that were in the top 500 most correlated genes in two of three gene sets (Figure 1A, Table S1). Examination of normal tissue RNA-seq data from Genotype Tissue Expression GTEX (Consortium, 2015) demonstrated that the expression of SPINK1-correlated genes is not high in the normal prostate (Figure 1B). Instead, similar to SPINK1 itself, these genes are enriched in tissues of the GI tract including the liver and intestines. The signature included well-known GI genes such as albumin (ALB), complement factor 5 (C5), Coagulation Factor V (F5), vitamin D binding protein (GC), prealbumin (TTR), growth arrest-specific 2 (GAS2), and multiple UDP-glucuronosyltransferase and aldo-keto reductase genes involved in detoxification and steroid metabolism (Table S2). This analysis suggests that in prostate cancer, the outlier expression of SPINK1 is part of an aberrantly activated GI transcriptome and we thus name the SPINK1-correlated genes as the PCa-GI signature.
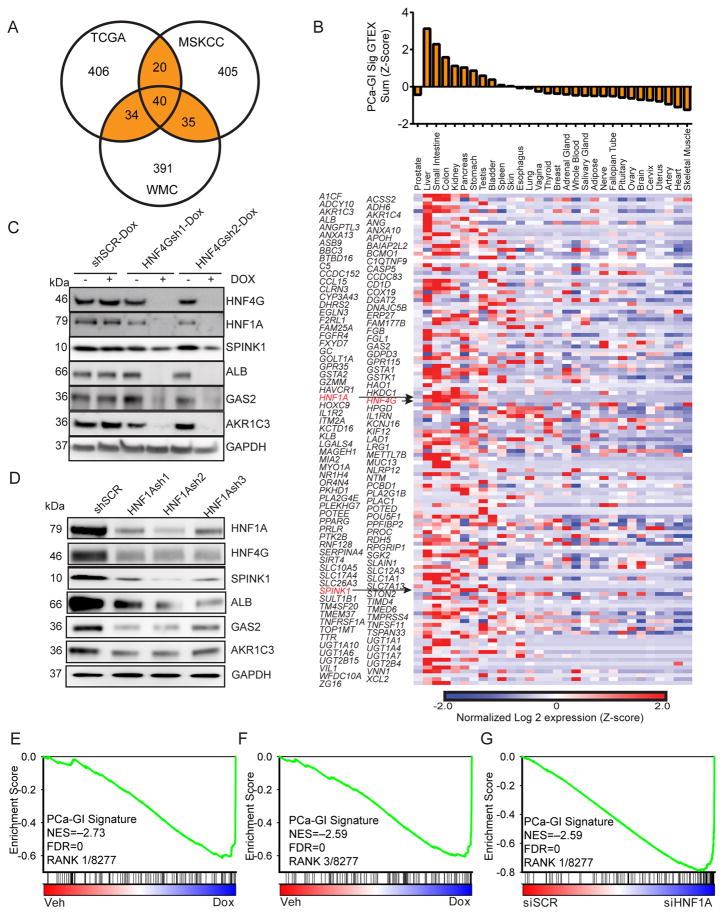
Figure 1. HNF4G and HNF1A regulate a GI gene signature in SPINK1-positive prostate cancer.
(A) Venn diagram generated using top 500 genes whose expressions most correlated with SPINK1 expression in three different gene expression datasets. Highlighted is the PCa-GI signature of 129 genes that are common in two of three datasets. The 40 genes that are in all three sets are called Core PCa-GI signature.
(B) Heat map of RNA-seq gene expression of the 129 individual SPINK1 correlated genes in normal tissues from GTEX, expressed as Z-score of log2 of read-per-kilobase mapped (RPKM). Top panel shows the sum expression of the 129 genes (Z-score).
(C) Immunoblot against indicated PCa-GI signature proteins of indicated derivatives of 22Rv1 cells treated with vehicle or doxycycline for 72 hours.
(D) Immunoblot against indicated PCa-GI signature proteins of 22Rv1 cells 72 hours after transduction with lentiviral shRNAs against HNF1A (HNF1Ash1, HNF1Ash2, HNF1Ash3) and a scrambled control (shSCR).
(E, F) GSEA plot of PCa-GI gene signature in 22Rv1-HNF4Gsh1-Dox cells (E) or 22Rv1-HNF4Gsh2-Dox cells (F) treated with doxycycline compared to vehicle. NES: Normalized enrichment score. FDR: False discovery rate.
(G) GSEA plot of PCa-GI gene signature in 22Rv1 cells transfected with HNF1A siRNA compared to scrambled siRNA.
To identify master regulator transcription factors that may be responsible for aberrant expression of the GI transcriptome in prostate cancer, we noted the presence of hepatocyte nuclear factor 4-gamma gene (HNF4G) and hepatocyte nuclear factor 1-alpha gene (HNF1A) in the PCa-GI signature (Figure 1B). HNF4G is highly homologous to the well-known master regulator hepatocyte nuclear factor 4-alpha (HNF4A) and can bind and transactivate a similar set of genes in hepatocytes (Daigo et al., 2011; Parviz et al., 2003). In liver and pancreas, HNF4A, HNF1A and FOXA (also known as HNF3) family transcription factors form a core GI transcriptome regulatory circuit where they reinforce each other’s expression to maintain lineage specification (Odom et al., 2006; Odom et al., 2004). Indeed, exogenous expression of HNF4A or HNF1A with a FOXA member can reprogram murine fibroblasts to gut endoderm that forms functional liver and colon depending on area of engraftment (Huang et al., 2011; Sekiya and Suzuki, 2011). Examination of HNF1A and HNF4G in TCGA cohorts of prostate and GI cancers revealed that their expression mirrored that of SPINK1, with high endogenous expression in GI tissues and aberrant overexpression in a subset of prostate cancers to levels comparable to GI tissues (Figure S1B, C).
To determine the role of HNF4G and HNF1A in regulation of the PCa-GI transcriptome, we employed 22Rv1 prostate cancer cells, previously characterized to express high levels of SPINK1, for our studies (Tomlins et al., 2008). 22Rv1 cells notably also expressed high-levels of HNF4G and HNF1A. To identify HNF4G-dependent genes, we generated two 22Rv1 derivatives with doxycycline-inducible HNF4G hairpins (HNF4Gsh1-Dox, HNF4Gsh2-Dox) and a control derivative with doxycycline-inducible scrambled hairpin (SCR-Dox). Knockdown of HNF4G with doxycycline treatment decreased protein and/or mRNA levels of HNF1A as well as several selected PCa-GI genes including albumin, SPINK1, GAS2, MUC13 and AKR1C3 (Figure 1C, S1D). Next, to identify HNF1A-dependent genes, we transduced 22Rv1 cells with three lentiviral hairpins against HNF1A. HNF1A-knockdown decreased protein and/or mRNA levels of HNF4G as well as the same selected PCa-GI genes (Figure 1D, S1E).
To define the global HNF4G regulated transcriptome, we performed gene expression profiling of doxycycline treatment in 22Rv1cells stably expressing doxycycline-inducible hairpins. PCa-GI signature genes are among the most downregulated genes, including ALB, SPINK1, MUC13, C5, GC, and TMED6 (Figure S1F). To study the perturbation induced by HNF4G knockdown in an unbiased fashion, we employed gene set enrichment analysis (GSEA), using >8,200 gene sets from the Molecular Signatures Database (MSigDB) (Subramanian et al., 2005) and the PCa-GI signature. For both HNF4Gsh1-Dox and HNF4Gsh2-Dox cells, the PCa-GI signature gene set was among the most significantly enriched gene set downregulated by doxycycline treatment (Figures 1E–F, Table S3). Next, to define the global HNF1A regulated transcriptome, we performed gene expression profiling of 22Rv1 cells with siRNA-mediated knockdown of HNF1A (siHNF1A) or scrambled control (siSCR). As with HNF4G knockdown, many PCa-GI signature genes were among the most downregulated following HNF1A knockdown (Figure S1F). GSEA showed that the PCa-GI signature gene set was the most significantly enriched gene set downregulated by HNF1A siRNA (Figure 1G, Table S4). The PCa-GI signature included another transcription factor Nuclear Receptor Subfamily 1 Group H Member 4 (NR1H4) that was expressed in 22Rv1 cells. NR1H4 knockdown using two different lentiviral hairpins did not show any decrease in select PCa-GI genes, instead we found it to be a downstream target of HNF4G and HNF1A (Figure S1G). These data indicate that HNF1A and HNF4G form a transcriptional regulatory circuitry to reinforce each other to regulate aberrant expression of a GI transcriptome in prostate cancer.
There are no other prostate cancer cell lines that express the PCa-GI transcriptome. We have generated a bank of patient-derived prostate cancer organoids from biopsy specimens of patients with castration-resistant metastatic prostate cancer (Gao et al., 2014). Among these, MSK-PCa10 expresses high levels of HNF4G and HNF1A. To determine if HNF1A and HNF4G regulate a similar PCa-GI transcriptional program in MSK-PCa10 organoids, we knocked down HNF4G and HNF1A using two different hairpins each. We found that HNF4G and HNF1A regulate the expression of these GI transcriptome genes in MSK-PCa10 prostate cancer cells (Figure S1H). This observation further confirms the role of both these factors in governing the aberrantly activated GI-transcriptional program in prostate cancer.
HNF4G/HNF1A axis is required for growth in PCa-GI positive prostate cancer
We examined the requirement of HNF4G and HNF1A for in vitro and in vivo growth of prostate cancers that express the PCa-GI transcriptome. Downregulation of either HNF1A or HNF4G, each using two independent shRNA sequences, in 22Rv1 cells caused significant growth suppression compared to scrambled shRNA (Figures 2A, B). Consistently, GSEA of transcriptomes from HNF4G or HNF1A knockdown revealed that multiple cell cycle gene sets were enriched among downregulated genes (Tables S3, S4).
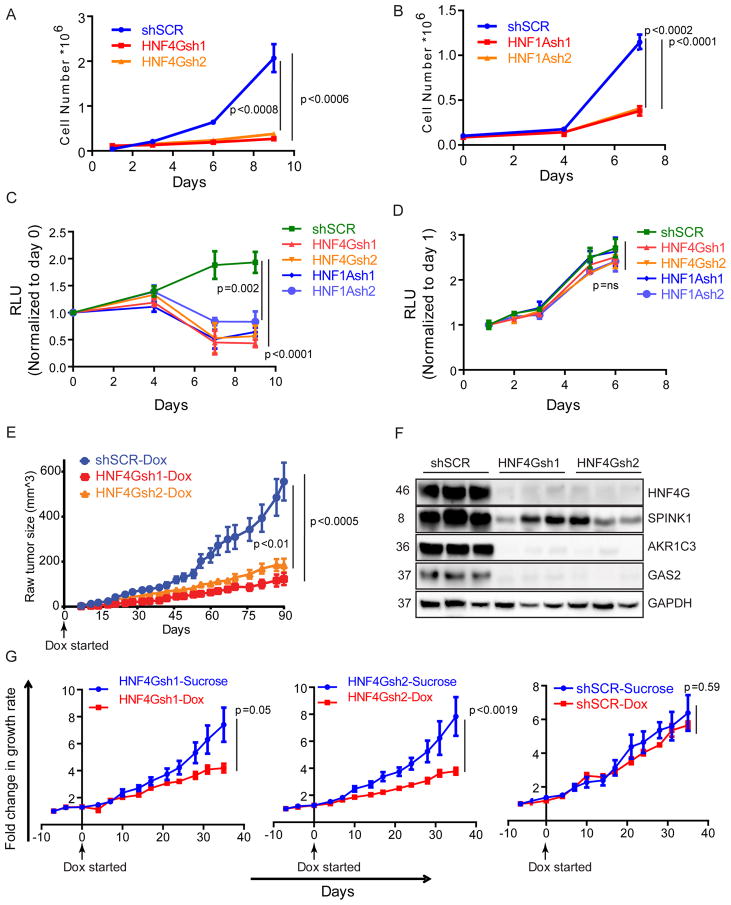
Figure 2. SPINK1 positive prostate cancer require HNF4G/HNF1A axis for growth.
(A, B) Cell growth curve of 22Rv1 following shRNA-mediated HNF4G (A) or HNF1A (B) knockdown and control. Mean ± SD. Two-tailed unpaired t-test, n=3.
(C, D) Cell growth curve of human patient derived CRPC cell lines MSK-PCa10 (C) and MSK-PCa1 (D) following shRNA-mediated HNF4G and HNF1A knockdown and control. Mean ± SD. Two-tailed unpaired t-test, n=3.
(E) Tumor formation and growth rate of indicated 22Rv1 cells when mice were fed with doxycycline drinking water beginning the same day as grafting. 2.0 × 106 cells were subcutaneously injected into 6–8 weeks old CB17-SCID mice; n=10 for all groups. Mean ± SEM. Two-tailed unpaired t-test.
(F) Immunoblots of three representative 22Rv1 explants obtained at the end of the experiment shown in (E).
(G) Response of indicated 22Rv1 xenograft tumors in SCID mice upon starting doxycycline water diet when tumors reached approximately 100 mm3; for shSCR-sucrose and shSCR-DOX n=6; For HNF4Gsh1-sucrose and HNF4Gsh1-DOX n=8; for HNF4Gsh2-sucrose and HNF4Gsh2-DOX n=6 and 8 respectively. Fold change in tumor volume over day 0 (start of doxycycline water) is plotted. Mean ± SEM. Two-tailed unpaired t-test.
See also Figure S2.
To investigate the effect of complete depletion of HNF4G and HNF1A, we performed CRISPR/Cas9-mediated knockout of HNF4G or HNF1A in 22Rv1. Due to outgrowth of cells that escape knockout in our preliminary experiments, we employed growth competitions assays. 22Rv1-Cas9 cells were then transduced with dual expression vector containing GFP and CRISPR guide RNAs. We verified Cas9-mediated genome editing by next-generation amplicon-sequencing (Figures S2A, S2B) and immunoblotting of HNF4G, HNF1A and downstream targets (Figure S2C, S2D). Next, we transduced 22Rv1-Cas9 cells at MOI~0.4. The relative growth of GFP-positive sgRNA-expressing cells compared to the GFP-negative cells were monitored over time by FACS analyses (Figure S2E). We observed depletion of GFP-positive sgHNF4G or sgHNF1A-expressing cells compared to GFP-negative control cells, but not of the sgNTC-expressing GFP-positive cells (Figure S2F), confirming that HNF4G or HNF1A loss leads to significant growth defect in prostate cancer cells.
To further examine the growth inhibitory effect of HNF4G and HNF1A on PCa-GI positive prostate cancer, we utilized two CRPC prostate cancer organoid lines: MSK-PCa10 (HNF4G+, HNF1A+) and MSK-PCa1 (HNF4G−, HNF1A−(Gao et al., 2014). We found that knockdown of HNF4G and HNF1A resulted in significant growth suppression in MSK-PCa10 but not in MSK-PCa1 (Figures 2C and D).
To determine the role of HNF4G in tumorigenesis in vivo, we employed 22Rv1 lines HNF4Gsh1-Dox, HNF4Gsh2-Dox and SCR-Dox (see Figure 1C). When mice were treated with doxycycline drinking water beginning the same day of grafting, we observed that HNF4Gsh1-Dox, HNF4Gsh2-Dox grafts grew significantly slower than SCR-Dox grafts (Figure 2E). Tumors explanted at the end of the experiment exhibited a decrease in HNF4G and its target proteins AKR1C3, GAS2 and SPINK1 in HNF4Gsh1-Dox and HNF4Gsh2-Dox xenografts compared to SCR-Dox xenografts (Figure 2F). To determine the requirement of HNF4G on tumor growth and maintenance in established tumors, we allowed HNF4Gsh1-Dox, HNF4Gsh2-Dox and SCR-Dox xenografts to reach a size of 100 mm3 and then started doxycycline or vehicle treatment in drinking water. Tumors explanted 2 days after doxycycline or vehicle administration show that the xenografts retained doxycycline induced knockdown of HNF4G (Figure S2G). In SCR-Dox xenografts, doxycycline had no significant effect on tumor growth. However, in HNF4Gsh1-Dox and HNF4Gsh2-Dox grafts, doxycycline treatment caused a significant reduction in tumor growth rate (Figure 2G). These data indicate that in the subset of prostate cancers with aberrantly expressed HNF4G/HNF1A transcriptional circuitry, maintaining the circuitry is required for prostate cancer cell growth and tumorigenesis.
HNF4G has a distinct cistrome from AR and maintains enhancer chromatin features at its binding sites in prostate cancer
The cellular landscape of enhancers, hubs of permissive chromatin where multiple transcription factors bind, is highly lineage-specific and reflects the cell-type specific gene expression patterns (Heintzman et al., 2009). While many transcription factors bind to existing enhancers to modulate transcription, some master regulators commonly referred to as “pioneer factors” can generate and maintain the enhancer sites de novo (Zaret and Carroll, 2011). In the prostate lineage, FOXA1 is a well-established pioneer factor that shapes the enhancer landscape and AR cistrome (Iwafuchi-Doi et al., 2016; Lupien et al., 2008; Wang et al., 2011). To determine how HNF4G regulates the GI transcriptome and the interplay of HNF4G, FOXA1 and AR signaling in prostate cancer pathogenesis, we mapped HNF4G, FOXA1, and AR genome-wide binding sites using ChIP-seq and performed cistrome analyses with and without doxycycline-mediated HNF4G knockdown in 22Rv1-HNF4Gsh2-Dox cells. Further, to determine the role of HNF4G chromatin binding on the enhancer landscape and chromatin accessibility, we performed ChIP-seq of H3K4me1 and H3K27ac, two histone marks of all enhancers and active enhancers respectively as well as Assay for Transposase Accessible Chromatin (ATAC-seq) (Buenrostro et al., 2013).
At baseline (vehicle treated HNF4Gsh2-Dox cells), we identified ~9,500 high confidence (q<10−5) HNF4G peaks, with 90% in enhancer (non-promoter) regions and 10% in promoter regions, consistent with previous observations for the homolog HNF4A (Wallerman et al., 2009) (Figure S3A). Approximately 17% of HNF4G peaks overlapped with FOXA1 peaks, and ~13% overlapped with AR peaks (Figure S3B). In contrast, ~50% of ~24,000 AR peaks overlapped with FOXA1 (Figure S3B), consistent with prior observation and the prominent role of FOXA1 in defining the AR cistrome (Lupien et al., 2008; Wang et al., 2011). The de novo motif of top HNF4G peaks identified the HNF4 motif, centered at the peak summit, consistent with specific HNF4G chromatin immunoprecipitation (ChIP) (Figure S3C). Similarly, de novo motif of top AR peaks identified the AR motif, centered at the peak summit. While the FOXA1 motif was the second most enriched motif around both HNF4G and AR peaks, it was substantially more significant in the AR cistrome than the HNF4G cistrome (Figure S3C).
We next examined the effect of HNF4G depletion. At baseline, top HNF4G (blue) and top AR (green) binding sites are largely distinct with little overlap (Figure 3A), with a higher percentage of AR binding sites compared to HNF4G binding sites also bound by FOXA1. Both top HNF4G binding sites and top AR binding sites exhibit enrichment for H3K4me1 that mark enhancers and the majority of these sites also exhibit enrichment of H3K27ac suggesting they are active. ATAC-seq reveals that both top HNF4G and AR binding sites are at assessable chromatin. Doxycycline-mediated HNF4G knockdown decreased the mean tag densities of both H3K27ac and H3K4me1 at HNF4G binding sites, with a shift from a bimodal profile to a more unimodal profile. This change in profile suggests loss of the central nucleosome depleted regions with HNF4G downregulation (Figure 3A, blue) (He et al., 2010). Consistently, HNF4G knockdown also decreased ATAC signal at HNF4G binding sites, indicating decreased chromatin accessibility. In contrast, HNF4G knockdown did not affect the H3K27ac and H3K4me1 ChIP profiles or the ATAC-seq profile at top AR binding peaks (Figure 3A, green).
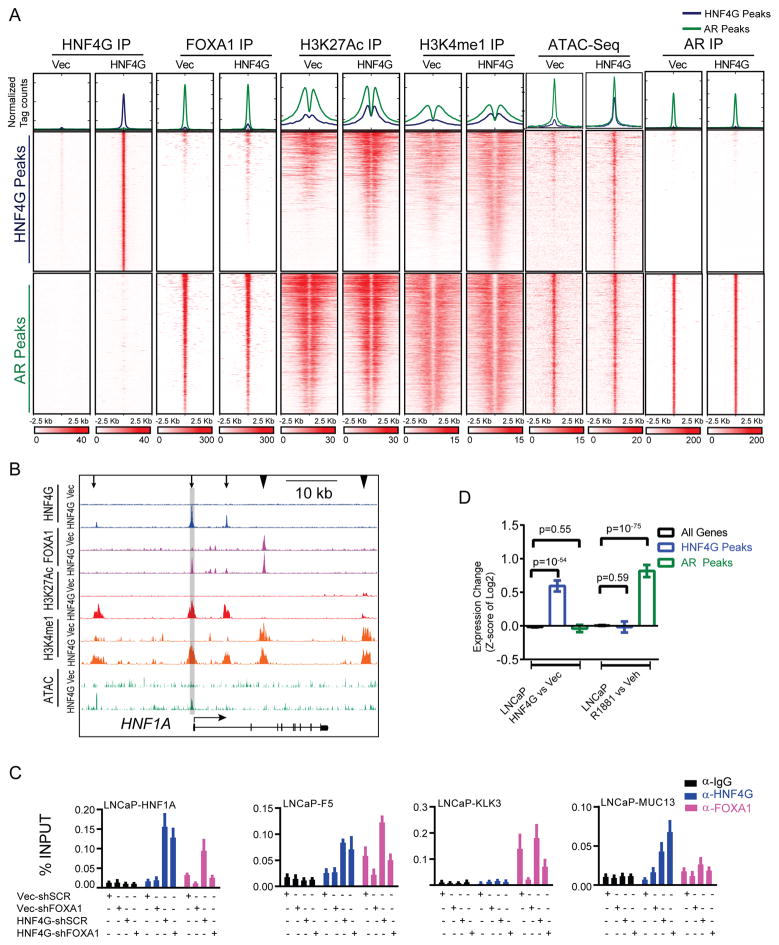
Figure 3. HNF4G binding maintains enhancer chromatin at binding sites.
(A) Histograms (top) show the average normalized tag counts of HNF4G, FOXA1, H3K27Ac, H3K4me1 and AR ChIP-seq as well as ATAC-Seq in vehicle and doxycycline treated 22Rv1-HNF4Gsh2-Dox cells at HNF4G (blue) and AR (green) binding sites. Heatmap shows the tag densities of HNF4G, FOXA1, H3K27Ac, H3K4me1, AR and ATAC-signal at the top 1,000 HNF4G (middle) or AR (bottom, as internal control) binding sites upon vehicle or doxycycline treatment in 22Rv1-HNF4Gsh2-Dox cells.
(B) ChIP-seq and ATAC-Seq profiles of HNF4G, FOXA1, H3K27Ac, and H3K4me1 at the HNF1A locus with or without HNF4G knockdown. Arrows indicate enhancers with HNF4G peaks and arrowheads indicate control enhancers without HNF4G peaks. Locus used for ChIP-qPCR is highlighted.
(C) ChIP-re-ChIP showing co-binding of HNF4G and FOXA1 at select HNF4G/FOXA1 co-binding loci (HNF1A and RNASE4) as well as HNF4G non-occupied locus (KLK3) and a HNF4G and FOXA1 non-occupied locus (GPR20) as controls. First ChIP was performed in 22Rv1 cells with HNF4G and FOXA1antibodies and no antibody as a control. Sequential HNF4G and FOXA1 ChIP were then performed with eluates from 1st ChIP of FOXA1. Input is 0.1% for 1st ChIP, FOXA1 1st ChIP is 10% for subsequent 2nd ChIPs.
(D) ChIP-qPCR of HNF4G and FOXA1 at selected HNF4G and FOXA1 co-binding loci (HNF1A and F5), HNF4G alone locus (MUC13) and FOXA1 alone locus (KLK3) in 22Rv1 cells. For each bar graph: left axis is fold enrichment over input for IgG, HNF4G ChIP and right axis is fold enrichment over input for FOXA1 ChIP Mean ± SD, n=3.
(E) Bar graph of gene expression change by HNF4G knockdown (dox treatment of 22Rv1-HNF4Gsh2-Dox) or AR activation (DHT treatment of 22Rv1) of all genes (black), genes mapped to top 1,000 HNF4G peaks (blue) and top 1,000 AR peaks (green). Mean ± 95% confidence. Two-tailed unpaired t-test.
See also Figure S3.
Examination of ChIP-seq profiles at representative HNF4G target genes HNF1A, F5, CLRN3, GAS2, and MUC13 illustrates a general diminishment of H3K4me1, H3K27ac, ATAC, and FOXA1 binding at many HNF4G binding sites with doxycycline treatment (Figures 3B, S3D, arrows, S3E), whereas non-HNF4G binding sites showed little change (Figures 3B, S3D, arrowheads). Among the spectrum of HNF4G binding sites, some are characterized by loss of H3K4me1 and ATAC signal (HNF1A, CLRN3) suggesting HNF4G is required for enhancer maintenance and some are characterized by loss of H3K27ac (GAS2, RNASE4) with preservation of H3K4me1 suggesting that HNF4G is required for enhancer activation.
At HNF4G and FOXA1 co-bound sites, FOXA1 binding is decreased in some (HNF1A, GAS2, F5) and preserved in others (RNASE4) (Figure 3B, S3D, arrows). Given the well-established role of FOXA1 as a pioneer factor in the prostate lineage, we were surprised by the requirement of HNF4G for FOXA1 binding, suggesting that the presence of FOXA1 alone is insufficient for maintaining open chromatin at these sites. Using ChIP-reChIP, we confirmed that genomic sites with both HNF4G peaks and FOXA1 peaks were indeed co-bound in the same cell by HNF4G and FOXA1 (Figure 3C). We performed combinatorial depletion of HNF4G (using doxycycline vs vehicle) and FOXA1 (using shSCR vs shFOXA1 lentiviral hairpins) to determine their effect on HNF4G and FOXA1 DNA binding using ChIP-qPCR. HNF4G and FOXA1 did not regulate each other and knockdown efficiency of one gene was unaffected by the other (Figure S3F). At the KLK3 site bound only by FOXA1 and the MUC13 site bound only by HNF4G, depletion of FOXA1 and HNF4G depleted FOXA1 and HNF4G binding respectively (Figure 3D). At the HNF1A and F5 co-bound sites (Figures 3B, S3D), depletion of HNF4G significantly decreased FOXA1 binding whereas depletion of FOXA1 did not decrease HNF4G binding (Figure 3D). These data indicate that HNF4G is required to maintain enhancer chromatin at certain HNF4G binding sites, as well as recruitment of other transcription factors, including FOXA1. Co-immunoprecipitation experiments failed to detect stable interaction between HNF4G and FOXA1, suggesting that the two transcription factors do not form stable complexes prior to DNA binding (Figure S3G).
We next performed integrative analysis of the HNF4G and AR cistrome with the HNF4G- and AR-regulated transcriptome. Compared to all genes, genes mapped to HNF4G peaks were significantly downregulated after HNF4G knockdown (p<10−17) but unaffected by DHT treatment (p=0.75), while genes mapped to AR peaks were significantly upregulated by DHT treatment (p<10−108) but unaffected by HNF4G knockdown (p=0.23) (Figure 3E). These data indicate that HNF4G is required to maintain enhancer chromatin context and allow binding of other transcription factors including FOXA1 at certain HNF4G binding sites and regulate transcriptional targets. Further, HNF4G binds to and maintains a cistrome that regulates a PCa-GI transcriptome distinct from AR signaling and contributes to prostate cancer oncogenesis.
Exogenous HNF4G or HNF1A expression activates the PCa-GI gene signature and chromatin signature, independent of AR signaling
The SPINK1-positive subset of prostate cancers have been shown to progress more rapidly to castration-resistance in multiple studies (Johnson et al., 2016; Leinonen et al., 2010; Pan et al., 2016) and several other PCa-GI signature genes including AKR1C3 and UGT2B15 are clinical biomarkers of castration-resistance (Stanbrough et al., 2006). We therefore asked whether activation of the PCa-GI-signature by HNF4G/HNF1A regulatory circuitry can alter AR dependence and lead to castration-resistance.
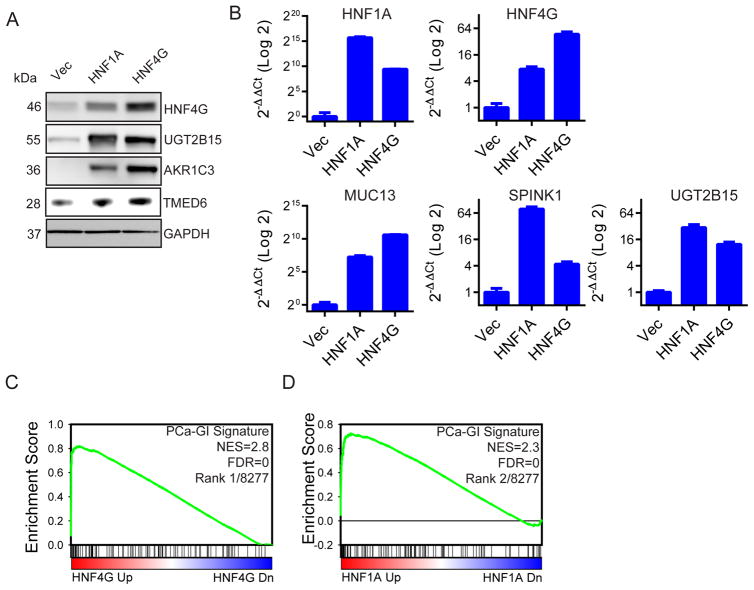
First, to determine if exogenous HNF4G or HNF1A expression can recapitulate the GI lineage transcriptome in prostate cancer, we stably expressed HNF4G or HNF1A in the LNCaP prostate cancer cell line that is SPINK1 negative and harbor an ETV1 translocation (Chen et al., 2013; Tomlins et al., 2007). We observed that exogenous expression of HNF4G resulted in expression of the endogenous HNF1A and vice versa. Further, exogenous expression of either transcription factor resulted in upregulation of PCa-GI signature genes including AKR1C3, MUC13, TMED6, SPINK1, UGT2B15 (Figures 4A, B). To characterize the global transcriptome in response to HNF4G or HNF1A expression, we performed gene expression profiling. GSEA revealed that for each transcriptome, the PCa-GI gene signature was among the most enriched gene sets (Figures 4C, 4D, S4, Table S5). Other enriched gene sets include steroid metabolism genes, liver and pancreas specific genes, and known HNF4A and HNF1A dependent genes. These data indicate that expression of either HNF1A or HNF4G in prostate cancer cells can activate the HNF1A/HNF4G circuitry to express the PCa-GI gene signature.
Figure 4. Exogenous HNF4G or HNF1A expression recapitulates the PCa-GI signature.
(A) Immunoblots of indicated proteins in LNCaP cells transduced for stable expression of HNF4G, HNF1A or empty vector control against the indicated proteins.
(B) qRT-PCR showing the expression of selected PCa-GI signature genes after exogenous expression of HNF4G and HNF1A in LNCaP cells. Data is presented as mean ± SD.
(C, D) GSEA plot of PCa-GI signature in LNCaP cells exogenously expressing HNF4G (C) or HNF1A (D) compared to empty vector control. NES: normalized enrichment score. FDR: false discovery rate.
To determine the effect of activation of HNF1A/HNF4G circuitry on the chromatin enhancer landscape, chromatin accessibility and FOXA1 and AR cistrome, we mapped the global localization of HNF4G, AR, FOXA1, H3K4me1, and H3K27ac by ChIP-seq and ATAC-Seq in LNCaP cells stably expressing HNF4G vs. vector control. In LNCaP cells, there were a large number of FOXA1 peaks (>150,000) and the majority of AR co-localized with FOXA1 (Figure S5A), consistent with prior reports (Jin et al., 2014; Wang et al., 2011). With exogenous HNF4G expression, 34% of HNF4G peaks co-localized with FOXA1 peaks, and 6.5% of HNF4G peaks with AR peaks (Figure S5A). We compared the HNF4G, AR, and FOXA1 peaks of LNCaP-HNF4G cells with 22Rv1-HNF4Gsh2-Dox vehicle treated cells. We found the ~70% of induced HNF4G peaks in LNCaP cells overlapped with endogenous peaks in 22Rv1 cells, suggesting that we have faithfully recapitulated HNF4G cistrome (Figure S5B). De novo motif analysis of the top 1,000 HNF4G peaks and AR peaks found that the most enriched motifs were HNF4 and AR respectively, centered at peak center. The FOXA1 motif was the next most enriched motif for both HNF4G and AR binding sites, but is much more prevalent and significant for AR than that of HNF4G binding sites (Figure S5C).
We next examined the effects of HNF4G exogenous expression on FOXA1 and AR cistrome, as well as on DNA accessibility and enhancer chromatin marks (e.g. H3K4me1, H3K27ac) at top HNF4G and AR binding sites in LNCaP cells. HNF4G exogenous expression did not affect the AR cistrome, but enhanced FOXA1 binding at a subset of HNF4G sites (Figure 5A). In addition, the mean ATAC signal, H3K4me1 and H3K27ac profile exhibited increased tag density and a shift in peak profile toward a bimodal distribution upon HNF4G exogenous expression in LNCaP cells at top HNF4G binding sites (Figure 5A, blue). In contrast, exogenous HNF4G expression did not affect the ATAC signal, H3K4me1 and H3K27ac distribution at top AR binding sites (Figure 5A, green). The ChIP-Seq and ATAC-Seq profiles at representative HNF4G target genes (HNF1A, CLRN3, F5, MUC13) illustrates gain of enhancer marks and increased chromatin accessibility specifically at sites of HNF4G binding (Figures 5B, S5D). At some sites (HNF1A, CLRN3), LNCAP cells were completely devoid of H3K4me1, H3K27ac marks and HNF4G expression generated them de novo. Other sites (F5, MUC13, RNASE4) were pre-marked by H3K4me1signfiying poised enhancer and HNF4G increased H3K27ac and ATAC signal suggesting enhancer activation. Upon HNF4G expression, FOXA1 binding was induced (HNF1A, CLRN3), enhanced (F5) or unaffected (RNASE4). These patterns of different effects of exogenous HNF4G expression in LNCaP cells were remarkable similar to HNF4G depletion in 22Rv1 cells. We performed combinatorial exogenous HNF4G expression and FOXA1 knockdown and assayed FOXA1 and HNF4G binding at specific sites by ChIP-qPCR. At control FOXA1-only KLK3 and HNF4G-only MUC13 sites, FOXA1 depletion and HNF4G expression resulted in expected changed in binding. At the co-bound sites at HNF1A and F5, HNF4G expression caused increased FOXA1 binding whereas FOXA1 knockdown did not significantly affect HNF4G binding (Figure 5C, S5E).
Figure 5. Exogenous HNF4G expression recapitulates chromatin landscape of endogenous HNF4G expression in 22Rv1.
(A) Histograms (top) show the average normalized tag counts of HNF4G, FOXA1, H3K27Ac, H3K4me1, AR ChIP-seq and ATAC-signal in LNCaP cells with exogenous expression of HNF4G or vector control at top 1,000 HNF4G and AR binding sites. Heatmap shows the tag densities of HNF4G, FOXA1, H3K27Ac, H3K4me1, AR and ATAC-signal at the top 1,000 HNF4G (middle) or AR (bottom) binding sites.
(B) Representative ChIP-seq and ATAC-Seq profiles of HNF4G, FOXA1, H3K27Ac, and H3K4me1 at HNF1A locus in LNCaP cells with or without exogenous HNF4G expression. Arrows indicate enhancers with HNF4G peaks and arrowheads indicate control enhancers without HNF4G peaks. Locus used for ChIP-qPCR is highlighted.
(C) ChIP-qPCR of HNF4G and FOXA1 at select HNF4G and FOXA1 co-binding loci (HNF1A and F5) as well as HNF4G alone (MUC13) and FOXA1 alone (KLK3) loci in LNCaP cells. Mean ± SD, n=3.
(D) Bar graph of gene expression change by HNF4G expression or AR activation (R1881 treatment) of all genes (black), genes mapped to top 1,000 HNF4G peaks (blue) and top 1,000 AR peaks (green) in LNCaP cells. Mean ± 95% confidence. Two-tailed unpaired t-test
See also Figure S5.
There was a notable absence of HNF4G binding at the SPINK1 locus, which contains two canonical HNF1 sites in intron 1 and ~7.5 kb upstream of the promoter. The upstream site is notable for decreased H3K27ac and ATAC signal in 22Rv1 cells after HNF4G knockdown and increase in H3K27Ac and ATAC signal in LnCaP cells after HNF4G expression (Figure S5F). SPINK1 expression is induced to a higher extent by HNF1A overexpression compared to HNF4G overexpression (Figures S4, S5G). These data suggest that SPINK1 is a direct HNF1A target gene.
Integrating with transcriptome, we found that exogenous HNF4G expression significantly increased expression of genes mapped to HNF4G peaks but not AR peaks, whereas androgen treatment significantly upregulated genes mapped to AR peaks but not HNF4G peaks (Figure 5D). These data indicate that HNF4G can establish enhancers de novo and transcriptionally activate the PCa-GI transcriptome independent of AR signaling in prostate cancer.
HNF4G expression imparts resistance to androgen ablation and enzalutamide treatment in LNCaP cells
While the prognostic role of SPINK1 expression in surgically resected early disease is controversial, three independent studies have shown that SPINK1 overexpression predicts for decreased response to androgen deprivation therapy and more rapid progression to castration resistance and cancer-related death in advanced prostate cancer (Johnson et al., 2016; Leinonen et al., 2010; Pan et al., 2016). We further noted that the SPINK1 correlated genes (i.e. the PCa-GI signature) is enriched for steroid metabolism genes including AKR1C3 and UGT2B15 that are associated with castration resistance (Table S2) (Stanbrough et al., 2006). We therefore examined the role of HNF4G expression in castration resistance, using the established LNCaP/AR prostate cancer cell line (Arora et al., 2013; Tran et al., 2009). This LNCaP derivative has exogenous AR overexpression to mimic clinically observed AR overexpression seen in CRPC. It readily forms tumor in castrate mice but is sensitive to the second generation antiandrogen enzalutamide treatment in vitro and in vivo.
We performed in vitro colony formation assays in full serum (FS) and charcoal-stripped serum (CSS) that is depleted of steroid hormones (e.g. androgen). In FS, there was a slight increase in number of colonies formed. In CSS, LNCaP/AR cells expressing HNF4G were able to form significantly more colonies than those expressing empty vector (Figures 6A, S6A).
Figure 6. HNF4G expression imparts resistance to androgen ablation and enzalutamide treatment in vitro and in vivo.
(A) Number of colonies formed by LNCaP/AR cells with exogenous expression of HNF4G or vector control in media with full serum or stripped-serum. n=3, Mean ± SD. Two-tailed unpaired t-test.
(B) Growth curve of LNCaP/AR cells exogenously expressing HNF4G or vector control cultured in media with stripped-serum. Arrows shows the time points at which cells were harvested for RNA. n=2, Mean ± SD.yhu
(C) The sum Z-score of individual PCa-GI genes (mean ± SD) in LNCaP/AR cells expressing vector control at 9 days of growth in CSS media and HNF4G at day 9 and day 32 of growth in CSS media (top) and heatmap shows the expression of individual SPINK1 signature genes (bottom). n=2.
(D) Treatment response of LNCaP/AR cells xenografts exogenously expressing HNF4G or vector control in SCID mice when treated with enzalutamide (10 mg/kg) or vehicle (1% carboxymethyl cellulose) once a day. For Vec-vehicle and Vec-enzalutamide n=4 and 13 respectively; for HNF4G-Veh and HNF4G-enzalutamide, n=4 and 18 respectively. Treatment was started when tumors reached a volume of approximately 400 mm3. Fold change in growth rate over day 0 (start of treatment) is shown. Mean ± SEM. Two-tailed unpaired t-test.
(E) Box plot showing HNF4G mRNA levels of explanted xenografts at the end of the experiment. Box plots show median, quartiles, min and max, with each sample dot plotted. Statistical analysis was performed using two-tailed unpaired t-test.
While exogenous HNF4G expression increased colony formation in CSS, it did not fully restore colony formation to the level of full serum. This suggested that HNF4G primed a subset of cells to become castration resistant. We observed that when grown long-term in CSS, both LNCaP/AR-Vec and LNCaP/AR-HNF4G cells grew slowly for 10 days. Afterwards, multiple LNCaP/AR-HNF4G cell clones grew to take over the culture while LNCaP/AR-Vec cells arrested (Figure 6B). To study the underlying mechanism of castration-resistant growth we performed transcriptome analysis of LNCaP/AR-Vec and LNCaP/AR-HNF4G at day 9 (D9) of growth in CSS and LNCaP/AR-HNF4G at 32 days (D32) of growth in CSS to identify the HNF4G transcriptome, as well as determinants of castration-resistant growth among bulk cells with exogenous expression of HNF4G (Figure 6B). As expected, expression of PCa-GI signature genes was increased in LNCaP/AR-HNF4G cells compared to LNCaP/AR-Vec cells (Figures 6C, S6B). Notably, at day 32 (D32) in CSS when LNCaP/AR-HNF4G cells have started to grow rapidly, there was a further increase in PCa-GI signature gene expression (Figure 6C, S6B). GSEA revealed that the PCa-GI signature is significantly and highly enriched not only among genes upregulated by HNF4G expression compared to vector control after 9 days in CSS (D9 HNF4G vs.Vec CSS), but also among genes upregulated by HNF4G expression after 32 days of CSS compared to 9 days of CSS in HNF4G-expressing cells (D32 vs. D9 HNF4G CSS) (Figure S6C, Table S6). These data suggest that further upregulation of the PCa-GI gene signature by HNF4G correlates with progression to castration-resistant growth in prostate cancer cells.
To examine the role of HNF4G expression on AR directed therapy resistance in vivo, we utilized the enzalutamide treatment of LNCaP/AR cells grafted into castrate mice, a well-established in vivo system in the laboratory (Arora et al., 2013; Balbas et al., 2013; Tran et al., 2009). We grafted LNCaP/AR-Vec and LNCaP/AR-HNF4G cells and assessed for response to enzalutamide. LNCaP/AR-Vec tumor grafts exhibited an average tumor size reduction of ~50% and slow progression after ~60 days of treatment similar to previously reported (Arora et al., 2013). In contrast, LNCaP/AR-HNF4G tumor grafts exhibited decreased depth and duration of response, reaching pre-treatment size by 45 days of therapy (Figure 6D). As expected, the HNF4G transcript level of explanted tumors at the end of experiment was higher in LNCaP/AR-HNF4G compared to LNCaP/AR-Vec tumor grafts (Figure 6E). Notably, in both LNCaP/AR-Vec and LNCaP/AR-HNF4G grafts, enzalutamide treatment resulted in higher HNF4G transcript levels compared to vehicle controls. This suggests that expression of HNF4G can be induced in response to AR inhibition, which provides an effective alternative pathway for growth and survival in CRPCs
Aberrant PCa-GI activation is prevalent in human CRPC
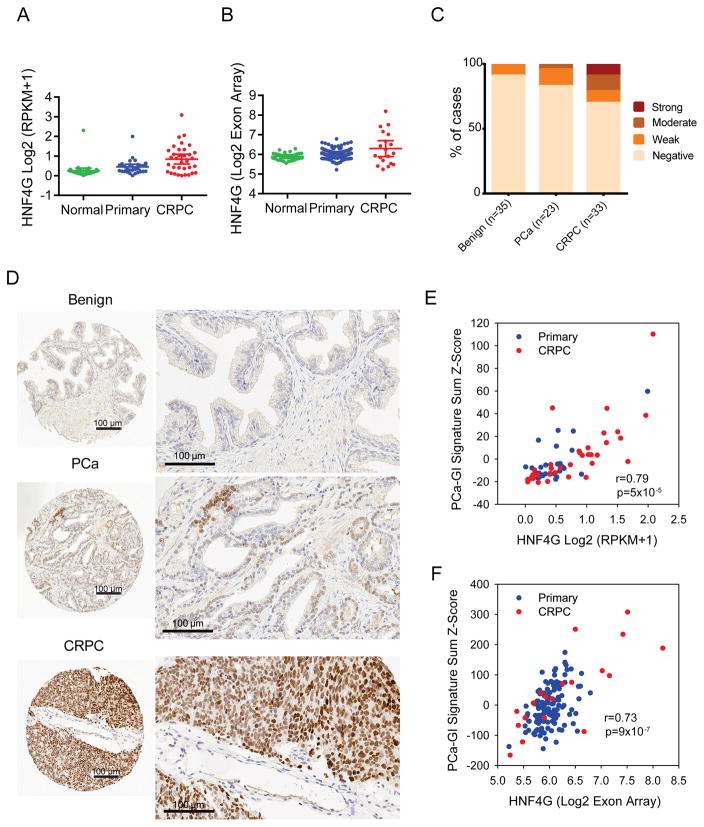
Since HNF4G expression can lead to activation of PCa-GI expression signature and castration resistance in prostate cancer, we asked if HNF4G and the PCa-signature are activated in human CRPC. In three transcriptome datasets with normal prostate, primary prostate cancer and CRPC clinical samples (Beltran et al., 2016; Grasso et al., 2012; Taylor et al., 2010), there were both an increase in percentage of CRPC tumors with HNF4G aberrant expression, as well as an increase in expression level of HNF4G in positive cases, with ~30% of CRPC showing outlier expression (Figures 7A, B, S7A). Similarly, HNF1A is progressively overexpressed from benign prostate, to localized cancer, to CRPC (Figure S7B). To validate this observation, we performed immunohistochemistry staining of HNF4G on a set of tissue microarray consisting of benign, primary and CRPC cases. We found that in a small percentage of “benign” prostate samples, there were some cells with low HNF4G expression, with the important caveat that the samples are from patients with prostate cancer. As the disease progress to primary cancer and CRPC, HNF4G increased in both intensity as well as fraction of cases, with ~30 % of CRPC cases positive (Figure 7C, D).
Figure 7. HNF4G overexpression is prevalent in CRPC.
(A, B) HNF4G expression in normal prostate, primary prostate cancer, and CRPC from the WCMC (A) and the MSKCC (B) datasets. Mean ± 95% CI.
(C) Quantification of HNF4G nuclear staining by immunohistochemistry (IHC) analysis on tissue microarrays (TMAs) of benign, primary prostate cancer (PCa) and castration-resistant prostate cancer (CRPC) from the WCMC cohort.
(D) Representative images of HNF4G nuclear staining in benign, primary prostate cancer and CRPC tissue to show negative, weak and strong HNF4G staining respectively. Each scale bar is 100 μm.
(E, F) Scatter plot showing correlation between HNF4G expression and PCa-GI signature sum (Z-score) in primary and CRPC cases from WCMC dataset (E) and MSKCC dataset (F).
See also Figure S7.
To examine if HNF4G is transcriptionally active in CRPC, we calculated the PCa-GI score for each sample and found that HNF4G expression level and PCa-GI sum expression score was highly correlated (Figure 7E, F). This data suggests that HNF4G is not only aberrantly highly expressed and transcriptionally active in a subset of primary prostate cancers that contribute to pathogenesis, but also in a substantial subset of CRPC during clinical progression with AR signaling-targeted therapies.
Discussion
During development, cell identity is specified by activation of lineage-specific master regulatory transcription factors, which in concert with chromatin modifying enzymes define the enhancer chromatin landscape (Stergachis et al., 2013). Upon tumorigenic transformation, many cancer types retain gene expression patterns and dependence on master regulators of the cell-of-origin lineage (Bass et al., 2009; Chi et al., 2010; Garraway et al., 2005). Prostate cancer is characterized by dependence on lineage specific master regulators FOXA1, HOXB13, and AR (Huggins and Hodges, 1941; Pomerantz et al., 2015), with AR dependence forming the scientific basis and clinical success for androgen deprivation therapy (ADT).
Here, we have uncovered a mechanism involved in prostate cancer pathogenesis and castration resistance by a HNF4G/HNF1A transcriptional circuitry. HNF4G/HNF1A expression activates PCa-GI signature characteristic of the GI lineage genes, distinct from the AR-dependent transcriptome, in ~5% of primary prostate cancer and ~30% of castration-resistant prostate cancer. Prostate cancers that activate this circuitry are dependent on it for growth and survival. Further, exogenous expression of HNF4G to activate this circuit results in more rapid progression to androgen independent growth in vitro and enzalutamide resistance in vivo. While exogenous expression of HNF4G does not convey immediate androgen independence to all cells, the cells that eventually propagate in androgen-deplete conditions express higher levels of HNF4G and GI signature genes. These experimental observations correlate with clinical data that SPINK1-positive prostate cancers that have an activated HNF4G/HNF1A circuit progress more rapidly to castration resistance (Johnson et al., 2016; Leinonen et al., 2010; Pan et al., 2016). Interestingly, enzalutamide selection pressure in vivo increases HNF4G expression. These data are consistent with a prominent role of HNF4G in resistance to AR-targeted therapy,
Previous studies of key transcription factors involved in prostate pathogenesis and clinical progression to CRPC, e.g. FOXA1, AR, ETV1/ERG, GR, RORγ, etc all involve the activation of AR signaling in primary prostate cancer and reactivation of AR signaling in presence of ADT. In contrast, HNF4G/HNF1A expression does not alter the AR cistrome or AR signaling. Instead, HNF4G can establish de novo enhancers and maintain as well as augment pre-existing enhancers at HNF4G binding sites, which can facilitate the binding of other transcription factors and mediate the GI-lineage transcriptome in prostate cancer pathogenesis. Importantly, our data indicate that HNF4G-mediated castration-resistant mechanisms will not only evade the current available ADT and anti-androgen therapies, but also will predict therapeutic resistance to newer generations of therapeutics targeting AR-signaling reactivation in CRPCs.
In prostate cancer, ETS translocations serves as a paradigm that aberrant expression of transcription factors at levels endogenous in other tissue types (e.g., ERG in endothelial cells and ETV1 in interstitial cells of Cajal) can be tumorigenic (Chi et al., 2010; Miettinen et al., 2011). An important question raised by our work is how HNF4G is aberrantly expressed in prostate cancer and whether it is induced in CRPC. IHC studies have identified low HNF4G expression in a small percentage of cells in some benign prostate specimens (with caveat that they are adjacent to cancerous prostate), raising the possibility that they may be the cell of origin of this subtype of prostate cancer. We further observed that HNF4G and PCa-GI signature genes are induced by androgen deprivation. During development, FOXA1/2 expression specifies the definitive endoderm and establishes competence for subsequent lineage specific regulators including AR in prostate, HNF4A in liver, and PDX1 in pancreas (Jozwik and Carroll, 2012; Wang et al., 2015). The prostate is derived from the hindgut endoderm and shares expression of master regulators HOXB13 and FOXA1 (Iwafuchi-Doi and Zaret, 2014; Zaret and Carroll, 2011). The suppression of AR activity may therefore activate a latent development program.
HNF4G is a nuclear receptor homologous to retinoid and peroxisome proliferator-activated receptor with fatty acids as its endogenous ligand (Wisely et al., 2002; Yuan et al., 2009). While HNF4A is essential, Hnf4g knockout mice exhibit minimal defects (Baraille et al., 2015). These data suggest that HNF4G may be a clinically practical therapeutic target in prostate cancer, especially in CRPCs.
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Please direct all requests for reagents and resource sharing to the Lead Contact, Yu Chen (cheny1@mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and cell culture
The LNCaP and 22Rv1 cell lines were obtained from the American Type Culture Collection (ATCC). The LNCaP/AR line was a kind gift from Dr. Charles Sawyers (Memorial Sloan-Kettering Cancer Center). All these cell lines were maintained in RPMI supplemented with 10% Fetal bovine serum (Omega), L-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml). MSK-PCa1 and MSK-PCa10 were generated from patient samples by organoid culture technique and cultured as described previously (Gao et. al., 2014). Cell lines were authenticated by RNA-seq analysis of SNP’s compared to exome data from Cancer Cell Line Encyclopedia. Cell lines were confirmed mycoplasma free by PCR testing.
Clinical Specimen
For Prostate tissue microarray staining, archival Formalin-fixed paraffin-embedded (FFPE) material was used under an IRB-approved protocol (Weill Cornell Medicine IRB 1007011157A015). For benign prostate tissue, TMA cores were obtained for areas localized at a distance from prostate cancer. For hormone naïve, organ-confined prostate adenocarcinomas (23 patients in total). Tumor tissue was obtained from radical prostatectomy specimens. The Gleason scores ranged from 6 (3+3) to 9 (4+5). For castration-resistant prostate cancer (33 patients in total), castration resistance was defined as disease progression in spite of an androgen-deprivation treatment. Seven samples were obtained from metastatic sites, the remaining ones represented locally advanced tumor. Each case was represented at least in duplicate (two cores per case) on the TMA. Most cases were represented in triplicate (three cores per case).
Animal Studies
All mice procedures were performed under MSKCC approved IACUC protocol #11-12-027. 6–8 weeks old CB17-SCID mice were purchased from Taconic and maintained under standard pathogen free conditions. For Enzalutamide treatment, we calculated that 7 xenografts are required to give 90% power to detect a 50% difference in growth with α=0.05. We used an intragroup variance of 50% and a treatment effect of 2-fold.
METHOD DETAILS
Antibodies
Antibodies to the following were used for Western blotting, ChIP, IHC and IF: rabbit anti-HNF4G (Sigma; HPA005438; 1:50 for IHC, 10 μg for ChIP-seq), rabbit anti-HNF4G (Proteintech; 25801-1-AP; 10 μg for ChIP-Seq) mouse anti-HNF4G (Clone B6502A; a kind gift from Dr. Takao Hamakubo, University of Tokyo (Daigo et al., 2011); 1:2000 for Western blotting), goat anti-HNF1A (Santa Cruj Biotechnology, sc6547X; 1:2000 for Western blotting), rabbit anti-H3K4me1 (for ChIP; Abcam; ab8895), rabbit anti-H3K27ac (for ChIP; Abcam; ab4729), rabbit anti-AR (Abcam, ab108341; 1:1000 for Western blotting, 5 μg for ChIP-seq), goat anti-FOXA1 (for ChIP; Abcam; ab5089), mouse anti-SPINK1(R&D Systems; MAB7496; 2 μg/ml for Western Blotting), rabbit anti-GAS2 (Abcam; ab109762; 1:200 for Western blotting), mouse anti-AKR1C3 (Sigma; A6229; 1:1000 for Western blotting), rabbit anti-Albumin (Cell signaling; 4929S; 1:1000 for Western blotting), mouse anti-GAPDH (abmgood; G041; 1:5000 for Western blotting), rabbit anti-UGT2B15 (Abcam; ab154864; 1:1000 for Western blotting), rabbit anti-TMED6 (Abcam; ab182489; 1:500 for Western blotting).
Lentiviral knockdown, siRNA knockdown and CRISPR/Cas9-Mediated Knockout
The following hairpins were used to knockdown HNF4G:
HNF4Gsh1: TRCN0000019243, HNF4Gsh2: TRCN0000420190 and shSCR (Addgene plasmid # 1864) (Sarbassov et al., 2005). See Table S7 for hairpins sequences. These were sub-cloned into Tet-pLKO-puro, a gift from Dmitri Wiederschain (Addgene plasmid # 21915) (Wiederschain et al., 2009). Lentiviral particles were generated by co-transfecting the lentiviral constructs with psPax2 and pVSV-G into 293FT cells using XtremeGene 9 (Roche) or Fugene 6 (Promega; E269A). Viral supernatants were collected 48–72 hours post transfection. Stable cell lines were generated after puromycin selection. Doxycycline at a concentration of 100 ng/ml was used to achieve HNF4G knockdown.
The following hairpins were used to knockdown HNF1A:
HNF1Ash1: TRCN0000017193, HNF1Ash2: TRCN0000017196, HNF1Ash3: TRCN0000017195 and shSCR (as above). See Table S7 for hairpins sequences.
Cell lysates and RNA were isolated 72 hours after transduction for immunoblot and qRT-PCR.
For siRNA knockdown, 22Rv1 cells were transfected with pooled siRNA (20 nM) against HNF1A (Dharmacon #L-008215-00-0005) or scrambled control (Dharmacon #D-001810-10-05) using Lipofectamine RNAiMAX transfection reagent (Invitrogen #13778100). RNA was harvested 72 hours after transfection.
For CRISPR/Cas9 knock-out of HNF4G and HNF1A in 22Rv1 cell line, single guide RNA (sgRNA) sequences were designed using CRISPR design tool from Feng Zhang lab (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). See Table S7 for sgRNA sequences. We first generated 22Rv1 cells stably expressing Cas9 using Lenti-Cas9-blast, a gift from Feng Zhang (Addgene plasmid # 52962) (Sanjana et al., 2014). We next transduced the 22Rv1-Cas9 cells with sgRNAs and GFP co-expression. The sgRNAs were cloned into the pLKO5.sgRNA.EFS.GFP a gift from Benjamin Ebert (Addgene plasmid # 57822) (Heckl et al., 2014).
CRISPR knockout Validation by Sequencing
For HNF4G and HNF1A knockout validation, 22Rv1-Cas9 cells were transduced with either sgNTC, sgHNF4G or sgHNF1A expressing viruses at a high MOI. After two days of puromycin selection and at seven days of infection genomic DNA was isolated from each population. Region specific primers were designed to amplify sgHNF4G and sgHNF1A target regions using genomic DNA extracted from each of the three cell population. The amplicons were subjected to 125 bp paired-end deep sequencing. The reads obtained were aligned to Refseq sequence using software CRISPResso (Pinello et al., 2016) and were analyzed for indels at or around the sgRNA target regions.
Stable Gene Expression
cDNA for HNF4G in pDONR201 vector was obtained from Harvard medical school Plasmid database (ID:HsCD00022314) and cloned into an murine stem cell virus (MSCV)-based retroviral vector with puromycin selection marker (Addgene) using Gateway technology. The HNF1A overexpression plasmid was purchased from Origene (RC211201L1). The ORF was subsequently sub-cloned into pMSCVhygro (Addgene). Retroviruses were produced in 293FT cells by standard methods using amphoteric packaging vector. Virus-containing supernatant was harvested at 48 and 72 h after transfection, pooled and filtered through a 0.45 μm PVDF membrane, and used for transduction in the presence of polybrene (8 μg ml−1). LNCaP cells were selected with 2 μg/ml puromycin or 400 μg/ml of hygromycin for 4 days at 48 hours after infection. The lines generated were used for subsequent studies.
RNA isolation and qRT-PCR
To isolate RNA from cell lines, E.Z.N.A total RNA kit (Omega) was used. To isolate RNA from xenograft tumor explants, the tumor samples were ground in 1 ml Trizol (Invitogen) using a PowerGen homogenizer (Fisher Scientific), followed by the addition of 200 μL chloroform. The samples were then centrifuged at 10,000 g for 15 minutes. The upper phase was mixed with an equal volume of 70% ethanol, and the RNA was further purified using the E.Z.N.A total RNA kit (Omega). For qRT-PCR, RNA was reverse transcribed using the High-Capacity CDNA Reverse Transcription Lit (ABI). Power SYBR Master Mix (ABI) was used to run PCR on a ViiA7 Real Time PCR System (Life Technologies).
Immunoblot
Cell lysates were prepared in RIPA buffer supplemented with proteinase/phosphatase inhibitor. Proteins were resolved on NuPAGE Novex 4–12% Bis-Tris Protein Gels (Life Technologies) or Tris -glycine 4–20% acrylamide gels (Invitrogen) and transferred electrophoretically onto a PVDF 0.45 μm membrane (BioRad). Membranes were blocked for 1 hour at room temperature in blocking buffer consisting of 5% milk or 1 % BSA diluted in Tris buffer saline plus 0.1% Tween 20 (TBST) and were incubated overnight at 4 °C with the primary antibodies diluted in the same buffer. After 3 washes of 10 min in TBST, membranes were incubated with secondary antibodies diluted in blocking buffer for 1 hour at room temperature. After 3 washes of 10 minutes in TBST, Enhanced Chemiluminiscence (ECL) was performed using ECL kit (Thermo Scientific, 80196).
Analysis of public human gene expression datasets
To identify SPINK1 correlated genes, we obtained normalized Affymetrix Human Exon 1.0 ST array gene expression data from MSKCC (GEO GSE21034), normalized RNA-seq data from prostate cancer TCGA (www.firebrowse.org) and RNA-seq prostate cancer data from Weill Cornell (dbGap phs000909.v.p1). We identified top 1,000 SPINK1 correlated genes by Pearson analysis and overlapped them. We defined the PCa-GI signature as the 129 genes in two of three datasets and the core PCa-GI signature as the 40 genes in all three datasets. To determine the expression of PCa-GI signature genes in normal tissue, we downloaded the GTEx v1.18 RNA-seq gene expression data in RPKM (http://www.gtexportal.org/home/datasets). We used the mean expression of each tissue type. Z-score was calculated as the standard deviations away from mean. To determine the expression of SPINK1, HNF1A, HNF4G in normal and cancerous tissue in TCGA datasets or prostate, colon, liver, pancreas, rectum and stomach cancers, we used Tukey box-and-whisker plots from http://firebrowse.org/.
Gene expression analysis
For gene expression profiling of 22Rv1 cells after HNF4G knockdown, doxycycline inducible 22Rv1-HNF4Gsh1-dox, 22Rv1-HNF4Gsh2-Dox, and 22Rv1-shSCR-dox lines were treated with vehicle or doxycycline (100 ng/ml) for 72 hours in triplicate prior to RNA isolation. Microarray was performed on an Illumina HumanHT-12 platform.
For gene expression profiling of 22Rv1 cells after HNF1A knockdown, 22Rv1 cells were transfected with pooled siSCR and siHNF1A. At 72 hours after transfection, RNA was isolated and gene expression profiling was performed using RNA-seq.
For gene expression profiling of 22Rv1 cells after DHT treatment, 22Rv1 cells treated with vehicle (DMSO) or 1 nM DHT for 8 hours. RNA was isolated and gene expression profiling was performed using RNA-seq. For gene expression profiling of LNCaP cells after R1881 treatment, LNCaP cells treated with vehicle (DMSO) or 1 nM R1881 for 24 hours in triplicate. RNA was isolated and gene expression profiling was performed using Illumina HumanHT-12.
For gene expression profiling of LNCaP cells expressing exogenous HNF4G, LNCaP cells were transduced with viruses containing MSCV-HNF4G or empty vector control in triplicate. After 48 hours, cells were selected with puromycin (2 μg/ml) for 4 days. RNA was isolated 2 days later for gene expression analysis using Illumina HumanHT-12 platform.
For gene expression profiling of LNCaP cells expressing exogenous HNF1A, LNCaP cells were transduced with viruses containing MSCV-HNF1A or empty vector control in duplicate. After 48 hours, cells were selected with hygromycin (400 μg/ml) for 4 days. RNA was isolated 2 days later for gene expression analysis using RNA-seq.
To generate the transcriptome of LNCaP/AR-HNF4G and LNCaP/AR-Vec cells after 9 days of growth in charcoal-stripped serum and of LNCaP/AR-HNF4G cells after 32 days of growth in charcoal-stripped serum, we performed the entire 32-day experiment in duplicate. Cell growth analysis was performed using cell counting and gene expression was analyzed using RNA-seq.
RNA-seq was performed by MSKCC genomics core facility using poly-A capture. The libraries were sequenced on an Illumina HiSeq-2500 platform with 50 bp single reads to obtain a minimum yield of 40 million reads per sample. The sequence data were processed and mapped to the human reference genome (hg19) using STAR v2.3 (Dobin et al., 2013). Gene expression was quantified to reads-per-kilobase mapped (RPKM) using the Cufflinks (Trapnell et al., 2010) and Log 2 transformed. GSEA was performed using JAVA GSEA 2.0 program (Subramanian et al., 2005), using difference of mean between replicates and gene permutation. The gene sets used were the Broad Molecular Signatures Database gene sets v5, c2 (curated geen sets), c5 (gene ontology gene sets), c6 (oncogenic signatures), c7 (immunologic signatures) as well as PCa-GI custom gene set generated by us.
Heatmaps were generated using GENE-E software (http://www.broadinstitute.org/cancer/software/GENE-E).
Chromatin Immunoprecipitation and Sequencing
Chromatin isolation from cell lines and immunoprecipitation was performed following the protocol previously described (Chi et al., 2010). For HNF4G knockdown experiments, chromatin was isolated from 22Rv1-HNF4Gsh2-Dox cell lines expressing doxycycline inducible HNF4Gsh2 hairpin 72 hours post doxycycline or mock treatment. For HNF4G overexpression, chromatin was isolated from LNCaP cells expressing HNF4G or a vector control. HNF4G, AR, FOXA1, H3K4me1 and H3K27ac ChIP were performed using the antibodies described in reagents section. Input DNA was also sequenced. For AR ChIP, cells were treated with 10 nM DHT for 8 hours to induce maximal AR binding sites. Next generation sequencing was performed on an Illumina HiSeq2000 platform with 50-bp or 100 bp single reads.
Reads were aligned to the human genome (hg 19) using the Bowtie alignment software (Langmead et al., 2009). Duplicate reads were eliminated for subsequent analysis. For FOXA1 and AR in both 22Rv1 and LNCaP cells and for HNF4G in LNCaP cells, peak calling was performed using MACS 2.1 callpeak function comparing immunoprecipitated chromatin with input chromatin, using a standard parameters and a q-value cutoff of 10−2 (Zhang et al., 2008). For HNF4G in 22Rv1-HNF4Gsh2-Dox cells, we used MACS2.1 bdgdiff function using profiles of vehicle and doxycycline treated HNF4G ChIP and input DNA to identify differential peaks in vehicle treated cells compared to doxycycline treated cells. After running MACS2.1, we further filtered the peaks file for q-value < 10−5 for downstream application of high-confidence peaks.
To determine overlap of AR, FOXA1, and HNF4G peaks we used Homer mergePeaks algorithm (Heinz et al., 2010) and consider two peaks overlapping if the summits are within 200 bp of each other. The ChIP-seq profiles presented were generated using Integrated Genome Browser (IGB) software of bigWig format files, generated using the bamCoverage tool from deepTool2 (Ramirez et al., 2016). We identified top 1,000 AR and top 1,000 HNF4G peaks, based on lowest q-values and plotted the ChIP-seq profiles around the sites using deepTools2. For motif analysis, we employed the MEME-ChIP suite (Machanick and Bailey, 2011), 400 bp sequences (200 bp upstream and downstream of the peak summit) of the top 1,000 peaks. To annotate peaks as promoter, gene body, and intergenic, we used Homer annotatePeaks program.
For integrative analysis of gene expression and ChIP-seq in 22Rv1 cells, we determined the gene expression change of HNF4G knockdown (22Rv1-HNF4Gsh2-Dox treated with doxycycline or vehicle) and AR activation (22Rv1 cells treated with vehicle or 10 nM DHT for 8 hours) at the closest genes which mapped to top HNF4G and AR peaks, using Homer annotatePeaks program. Similarly, for LNCaP cells, we determined the gene expression change of HNF4G overexpression (LNCaP-HNF4G vs LNCaP-Vec) and AR activation (LNCaP cells treated with vehicle or 1nM R1881 for 24 hours) at the closest genes that mapped to top HNF4G and AR peaks.
ChIP-re-ChIP
The ChIP-re-ChIP assay was performed with Re-ChIP-IT kit according to manufacturer’s instructions (Active Motif, 53016). Anti-HNF4G (Proteintech; 25801-1-AP) and anti-FOXA1 (Abcam; ab5089) were used for the first ChIP; eluates from FOXA1 ChIP were used to perform 2nd ChIP with same anti-HNF4G, anti-FOXA1 or no antibody control.
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) and analysis
ATAC-seq was performed as previously described (Buenrostro et al., 2013) with exception that digitonin was used instead of NP-40 for nuclei isolation. For each sample, cell nuclei were prepared from 50,000 cells, and incubated with 2.5 μl of transposase (Illumina) in a 50 μl reaction for 30 min at 37 °C. After purification of transposase-fragmented DNA, the library was amplified by PCR and subjected to high-throughput sequencing on an Illumina HiSeq2500 platform.
ATAC-seq reads were quality and adapter trimmed using ‘trim_galore’ before aligning to human genome assembly hg19 with bowtie2 using the default parameters. Aligned reads with the same start position and orientation were collapsed to a single read before subsequent analysis. Density profiles were created by extending each read to the average library fragment size and then computing density using the BEDTools suite, with subsequent normalization to a sequencing depth of ten million reads for each library. Subsequent data analysis and display is as described in the ChIP-seq analysis section.
Colony formation assay
LNCaP/AR cells overexpressing HNF4G or vector control were pre-grown in media containing charcoal-stripped serum for about 7 days. After 7 days, 2500 cells from both group were plated on a well of a six-well tissue culture dish and allowed to grow for an additional 10–12 days. The colonies obtained were then washed in PBS buffer, fixed in 70% ethanol for 15 minutes and then stained with a 0.5% crystal violet solution in 25% ethanol. The dishes were washed in a gentle stream of water through the side of the wells and were then air dried. The colonies obtained were then counted by an automated colony counter.
Cell Proliferation assay
For HNF4G and HNF1A knockdown in 22Rv1 cells, cells were plated at 0.1× 106 cells per well in a 12-well plate on day 0 and infected with pLKO.1 hairpin viruses at MOI ~2.0 on day 1. Cells were not further puromycin selected. Cells were counted in triplicate using Vicell XR cell viability analyzer at the indicated days.
For LNCaP/AR growth curve in charcoal-stripped serum with exogenous HNF4G or Vector expression, cells were plated at 0.1 million in a six well plate in duplicates. The cells were counted and replated at a density of 0.1 million at indicated days. The difference in split ratios to plate the same number of cells was noted and was used in growth curve plotting to extrapolate the total number of cells obtained every week.
For competition growth assay, 22Rv1 cells were stably transduced for Cas9 expression. 22Rv1-Cas9 cells were then transduced at ~ 0.4 MOI for dual expression of GFP and CRISPR guide RNAs against HNF4G, HNF1A or non target control (NTC). FACS analysis was performed at regular intervals to determine any changes in percentage of GFP-positive cells over the course of the experiment.
For HNF4G and HNF1A knockdown in patient derived organoid cell lines, MSK-PCa1 and MS-PCa10, about 2 million cells were plated per well of a six-well plate and were transduced next day with desired pLKO.1 hairpin viruses at MOI ~2.0. After a day of infection, cells were split and plated at a density of 20,000 cells per well of a 96 well plate. At indicated time points, the number of cells was determined using a CelltiterGlo assay (Promega)
Mouse Xenograft procedures
For LNCaP/AR xenograft studies, 2.0 × 106 cells resuspended in 100 μL of 1:1 mix of growth media and Matrigel (BD Biosciences) were subcutaneously injected into 6–8 weeks old CB17-SCID castrated male mice (Taconic). Tumor sizes were measured weekly with callipers starting 10 weeks after xenografting and were calculated using the following formula: tumor volume = (D2 × d2× h2)/6, whereby D, d and h refers to long diameter, short diameter and height of the tumor, respectively. Treatment with enzalutamide (10 mg/kg) or vehicle was begun at a tumor size of 400 mm3. Mice were treated once daily until the end of the experiments.
For 22Rv1 xenografts, 2.0 × 106 cells resuspended in 100 μL of 1:1 mix of growth media and Matrigel (BD Biosciences) were subcutaneously injected into 6–8 weeks old CB17-SCID mice (Taconic). The mice were fed with doxycycline water (200 mg/L in 0.5% sucrose) from the beginning of the grafting or when the tumors reached a size of 100 mm3. Tumors were measured twice a week after 1 week of grafting.
Analysis of HNF4G IHC in prostate cancer tissue microarrays
Immunohistochemistry (IHC) was performed using the anti-HNF4G antibody HPA005438 (MilliporeSigma, Billerica, MA, USA) at a 1:50 dilution, on a Bond III automated immunostainer (Leica Microsystems, IL, USA). Formalin-fixed paraffin-embedded (FFPE) tissue sections were de-paraffinized and endogenous peroxidase was inactivated. Antigen retrieval was performed using the Bond Epitope Retrieval Solution 1 (ER1) at 99–100°C for 60 minutes (Leica Microsystems). Sections were then incubated sequentially with the primary antibody overnight, post-primary for 15 minutes and polymer for 25 minutes, followed by a 10 minute colorimetric development with diaminobenzidine (DAB) (Bond Polymer Refine Detection; Leica Microsystems). FFPE material from 22Rv1 prostate cancer cell line xenografts with known levels of HNF4G expression were used as controls.
IHC was performed on tissue microarrays composed of representative cases of benign human prostate tissue, hormone naïve prostatic carcinoma (HNPCa), castration-resistant prostatic adenocarcinoma (CRPC). Staining was evaluated by a pathologist with experience in genitourinary pathology (JC). Nuclear HNF4G expression was considered positive if at least 10% of nuclei in a given tissue type showed brown staining. Nuclear staining intensity was scored following a three-tiered system (negative=0, weak=1, medium=2 and strong=3).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical comparisons between two groups were performed by GraphPad Prism software 6.0 using a two-tailed unpaired t-test. The variance between the statistically compared groups was similar.
DATA AND SOFTWARE AVAILABILITY
Gene Expression Omnibus (GEO) Accession numbers of datasets generated
GSE85242: Illumina HumanHT-12 bead array expression profile of doxycycline mediate HNF4G knockdown in 22Rv1 cells.
GSE85244: Illumina HumanHT-12 bead array expression profile of HNF4G expression in LNCaP cells.
GSE85556: RNA-seq expression profile of HNF1A knockdown in 22Rv1 cells and HNF1A overexpression in LNCaP cells respectively.
GSE85557: RNA-seq expression profile of LNCaP/AR cells with or without HNF4G exogenous expression grown in charcoal-stripped serum.
GSE85558: HNF4G, AR, FOXA1, H3K4me1, H3K27acetyl ChIP-seq and ATAC-seq in 22Rv1 and LNCaP cells with knockdown and overexpression of HNF4G respectively.
Supplementary Material
Table S1, related to Figure 1: PCa-GI Signature.
Table S2, related to Figure 1: Pathways enriched for PCa-GI signature genes.
Table S3, related to Figure 1: GSEA downregulated gene sets in HNF4G knockdown 22Rv1 cells.
Table S4, related to Figure 1: GSEA downregulated gene sets in siHNF1A vs siSCR in 22Rv1 cells.
Table S5, related to Figure 4: GSEA upregulated gene sets in LNCaP cells transduced with HNF4G or HNF1A expression.
Table S6, related to Figure 6: GSEA upregulated gene sets in LNCaP/AR/HNF4G cells upon growth in Charcoal stripped-serum.
Table S7, related to STAR Methods: List of oligonucleotides used in the study.
Significance.
Lineage-directed therapy using androgen deprivation has been the mainstay of prostate cancer treatment for 70 years and can be circumvented by activation of survival programs of alternative lineages. We found that 5% of primary untreated prostate cancers and 30% of castration-resistant prostate cancers aberrantly express a gastrointestinal-lineage (PCa-GI) transcriptome. This PCa-GI transcriptome is regulated by master regulators HNF1A and HNF4G. Integrative cistrome, transcriptome, and chromatin analysis shows that HNF4G is a pioneer factor that reprograms the enhancer chromatin landscape and mediates AR therapy resistance.
Highlights.
A GI lineage transcriptome is prevalent in castration- resistant prostate cancer
HNF4G and HNF1A regulate each other and the GI lineage transcriptome
HNF4G is a pioneer factor that generates and maintains enhancers at GI lineage genes
Exogenous HNF4G expression in prostate cancer leads to castration resistance
Acknowledgments
We like to thank Dr. Ling Cai and Daniel Neil Weinberg for reagent development, and Dr. Takao Hamakubo for HNF4G antibody. Next generation sequencing and gene expression arrays were done at the Memorial Sloan Kettering Cancer Center (MSKCC) Genomics Core Facility (for ChIP-Seq and RNA-Seq) and New York Genome Center (for ATAC-Seq). ATAC-Seq was performed by the Epigenomics Core at MSKCC. Immunohistochemistry was performed at the Translational Research Program facility, Department of Pathology and Laboratory Medicine at Weill Cornell Medicine. We like to thank support and feedback from the Prostate Cancer Foundation. This work was supported by grants from the NCI (K08CA140946 YC; R01 CA208100, YC; R01CA193837, YC; P50CA092629, YC, HIS, AG; P30CA008748, PC, YC, HIS; P50CA140146, PC; K08CA151660, PC; DP2 CA174499, PC; U01 CA111275-09, MAR and JMM), US DOD (W81XWH-10-1-0197), Starr Cancer Consortium (YC, PC, GS, MAR), Geoffrey Beene Cancer Research Center (YC, PC), Gerstner Family Foundation (YC), Bressler Scholars Fund (YC), GIST Cancer Research Fund (PC), Shuman Fund (PC) and GIST Cancer Awareness Fund (PC). JC was supported by the Nuovo-Soldati foundation
Footnotes
Author Contribution
Experime ntal design: S.S., P.C. and Y.C.; Western blots and qRT-PCRs: S.S.; ChIP-seq, Microarray and RNA-seq: S.S.; ChIP-re-ChIP: L.R.; Generation of the expression vectors: S.S., P.A., Y.X., Z.C.; Xenografts and drug treatments: D.M., E.G.W., S.S., S.W., J.S., Y.C. and J.W.; Growth curves: S.S., Y.Z.; Colony formation assays, Growth Competition Assay and FACS: S.S.; Immunohistochemistry: J.C., J.M.M.; Bioinformatic Analysis: A.S., H.B. R.P.K. and Y.C. Clinical samples acquisition: M.A.R, J.M.M. Manuscript writing: S.S., P.C. and Y.C. All authors reviewed and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. The New England journal of medicine. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, Shah N, Cai L, Efstathiou E, Logothetis C, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155:1309–1322. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbas MD, Evans MJ, Hosfield DJ, Wongvipat J, Arora VK, Watson PA, Chen Y, Greene GL, Shen Y, Sawyers CL. Overcoming mutation-based resistance to antiandrogens with rational drug design. eLife. 2013;2:e00499. doi: 10.7554/eLife.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraille F, Ayari S, Carriere V, Osinski C, Garbin K, Blondeau B, Guillemain G, Serradas P, Rousset M, Lacasa M, et al. Glucose Tolerance Is Improved in Mice Invalidated for the Nuclear Receptor HNF-4gamma: A Critical Role for Enteroendocrine Cell Lineage. Diabetes. 2015;64:2744–2756. doi: 10.2337/db14-0993. [DOI] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature medicine. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nature medicine. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nature medicine. 2013;19:1023–1029. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, Fletcher JA, Dewell S, Maki RG, Zheng D, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–853. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigo K, Kawamura T, Ohta Y, Ohashi R, Katayose S, Tanaka T, Aburatani H, Naito M, Kodama T, Ihara S, et al. Proteomic analysis of native hepatocyte nuclear factor-4alpha (HNF4alpha) isoforms, phosphorylation status, and interactive cofactors. The Journal of biological chemistry. 2011;286:674–686. doi: 10.1074/jbc.M110.154732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin R, Pettersson A, Hendrickson WK, Fiorentino M, Finn S, Kunz L, Judson GL, Lis R, Bailey D, Fiore C, et al. SPINK1 protein expression and prostate cancer progression. Clin Cancer Res. 2014;20:4904–4911. doi: 10.1158/1078-0432.CCR-13-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nature genetics. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nature biotechnology. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Huggins C, Hodges CV. Studies on Prostatic Cancer. I. The Effect of Castration, of Estrogen and of Androgen Injection on Serum Phosphatases in Metastatic Carcinoma of the Prostate. Cancer research. 1941;1:293–297. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Molecular cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes & development. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HJ, Zhao JC, Wu L, Kim J, Yu J. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nature communications. 2014;5:3972. doi: 10.1038/ncomms4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH, Ross AE, Alshalalfa M, Erho N, Yousefi K, Glavaris S, Fedor H, Han M, Faraj SF, Bezerra SM, et al. SPINK1 defines a molecular subtype of prostate cancer in men with more rapid progression in an at-risk, natural history radical prostatectomy cohort. J Urol. 2016;196:1436–1444. doi: 10.1016/j.juro.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nature reviews Cancer. 2012;12:381–385. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen KA, Tolonen TT, Bracken H, Stenman UH, Tammela TL, Saramaki OR, Visakorpi T. Association of SPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine-treated prostate cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2845–2851. doi: 10.1158/1078-0432.CCR-09-2505. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen M, Wang ZF, Paetau A, Tan SH, Dobi A, Srivastava S, Sesterhenn I. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am J Surg Pathol. 2011;35:432–441. doi: 10.1097/PAS.0b013e318206b67b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2:20060017. doi: 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Zhang X, Gong J, Tan J, Yin X, Tang Q, Shu K, Shen P, Zeng H, Chen N. The expression profile and prognostic value of SPINK1 in initially diagnosed bone metastatic prostate cancer. Prostate. 2016;76:823–833. doi: 10.1002/pros.23173. [DOI] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nature genetics. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, Yuan GC. Analyzing CRISPR genome-editing experiments with CRISPResso. Nature biotechnology. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, Cejas P, Vazquez F, Cook J, Shivdasani RA, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nature genetics. 2015;47:1346–1351. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic acids research. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, Febbo PG, Balk SP. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer research. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. The New England journal of medicine. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, Demichelis F, Helgeson BE, Laxman B, Morris DS, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer cell. 2008;13:519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallerman O, Motallebipour M, Enroth S, Patra K, Bysani MS, Komorowski J, Wadelius C. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic acids research. 2009;37:7498–7508. doi: 10.1093/nar/gkp823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nature reviews Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, Chen Y, Caponigro G, Yao YM, Lengauer C, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Willson TM, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PloS one. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 1: PCa-GI Signature.
Table S2, related to Figure 1: Pathways enriched for PCa-GI signature genes.
Table S3, related to Figure 1: GSEA downregulated gene sets in HNF4G knockdown 22Rv1 cells.
Table S4, related to Figure 1: GSEA downregulated gene sets in siHNF1A vs siSCR in 22Rv1 cells.
Table S5, related to Figure 4: GSEA upregulated gene sets in LNCaP cells transduced with HNF4G or HNF1A expression.
Table S6, related to Figure 6: GSEA upregulated gene sets in LNCaP/AR/HNF4G cells upon growth in Charcoal stripped-serum.
Table S7, related to STAR Methods: List of oligonucleotides used in the study.