Abstract
Orofacial clefts (OFCs) are common, complex birth defects with extremely heterogeneous phenotypic presentations. Two common subtypes – cleft lip alone (CL) and cleft lip plus cleft palate (CLP) - are typically grouped into a single phenotype for genetic analysis (i.e. cleft lip with or without cleft palate, CL/P). However, mounting evidence suggests there may be unique underlying pathophysiology and/or genetic modifiers influencing expression of these two phenotypes. To this end, we performed a genome-wide scan for genetic modifiers by directly comparing 450 CL cases with 1692 CLP cases from 18 recruitment sites across 13 countries from North America, Central or South America, Asia, Europe and Africa. We identified a region on 16q21 that is strongly associated with different cleft type (p=5.611×10−8). We also identified significant evidence of gene-gene interactions between this modifier locus and two recognized CL/P risk loci: 8q21 and 9q22 (FOXE1) (p=0.012 and p=0.023, respectively). SNPs in the 16q21 modifier locus demonstrated significant association with CL over CLP. The marker alleles on 16q21 that increased risk for CL were found at highest frequencies among individuals with a family history of CL (p=0.003). Our results demonstrate the existence of modifiers for which type of OFC develops and suggest plausible elements responsible for phenotypic heterogeneity, further elucidating the complex genetic architecture of OFCs.
Keywords: complex trait, gene-gene interaction, genetic modifier, orofacial cleft
Introduction
Orofacial clefts (OFCs) are common, complex birth defects with heterogeneous phenotypes. OFCs arise early in human development due to failure of one or more steps in a complicated, highly coordinated series of events governing craniofacial morphogenesis. As a result, there are numerous subtypes of OFCs, but the term most commonly refers to defects of the lip and/or palate. The three most common types of OFCs are cleft lip alone (CL), cleft lip plus cleft palate (CLP), and cleft palate alone (CP). CL and CLP are historically grouped into a single phenotype—cleft lip with or without cleft palate (CL/P)—for genetic studies, as they are thought to share common etiology through disruptions in the development of the lip (Marazita, 2012) which precedes development of CP alone in embryology and both CL and CLP show a higher risk to males whereas CP occurs more often in females.
Multiple genome-wide association studies and candidate gene studies have investigated genetic associations with CL/P (Beaty et al., 2010; Leslie et al., 2016). In such studies of CL/P, one underlying hypothesis was that the effect on risk of cleft is identical for both CL and CLP subgroups. However, investigations where CL/P was the sole phenotype have reduced power to detect variants for which the risk of OFC differs. Identification of genetic factors that act as modifiers of cleft subtypes is still critical for understanding the substantial variability of OFCs between individuals.
The possibility of cleft type modifiers is supported by mounting evidence suggesting there may be distinct underlying genetic causes for CL and CLP. These include studies from Norway citing epidemiologic differences between CL and CLP (Harville, Wilcox, Lie, Vindenes, & Abyholm, 2005), and from Denmark demonstrating subtype-specific recurrence risks (Grosen et al., 2010). Despite this population-based evidence, few studies have examined these differences in a genetic context. Previous studies have indicated that variants in IRF6 are more strongly associated with CL than CLP (Marazita, 2012; Rahimov et al., 2008), whereas SPRY2 has demonstrated some evidence of CLP-specific association (Jia et al., 2015; Ludwig et al., 2012). Similarly, GREM1 may be specifically associated with clefts in the lip and soft palate (Ludwig et al., 2016). However, beyond these few subtype-specific associations, relatively little is known about the biological mechanisms controlling these different phenotypic types of OFCs. Given the overall phenotypic heterogeneity of OFCs, combined with the success of detecting subtype-specific signals in recent studies, further investigation of subtype-specific variants and genetic modifiers is warranted.
As several lines of evidence—distinct embryological origins of the lip and palate, epidemiology, and genetic studies—already support somewhat distinct genetic architectures for CL/P and CP, genetic differences between these groups was not of interest (Dixon, Marazita, Beaty, & Murray, 2011; Leslie et al., 2017; Leslie et al., 2016). Instead, we hypothesized the presence of genetic modifiers for CL and CLP and tested this hypothesis by performing a case-case comparison, directly comparing allele frequencies at each SNP between the CL and CLP cases. This type of analysis has high power to find genetic risk factors that differ between the two groups, but it has no power to find factors that are important in both groups (Lee et al., 2011). Thus, this design is strictly a test for heterogeneity in the genotype/phenotype relationship, not an overall test for genetic effects on risk. Ideally, this test will reveal new loci for which there is an effect in only one subgroup; such loci may be masked in an overall scan when the two groups are combined. Therefore, the goals of this paper are to identify potential mechanisms through which CL and CLP arise by identifying genetic modifiers of OFC subtype, and also to lay the framework for investigating genetic modifiers of complex and heterogeneous diseases such as OFC.
Methods
GWAS Sample and SNP information
The cohort for this study was derived from a previously described worldwide sample recruited from 18 sites across 13 countries from North America, Central or South America, Asia, Europe and Africa (Leslie et al., 2016). Recruitment sites were part of ongoing genetic studies conducted by the University of Pittsburgh Center for Craniofacial and Dental Genetics and the University of Iowa. Informed consent was obtained for all participants, and all sites had both local IRB approvals and approvals at the University of Pittsburgh or the University of Iowa. A total of 1700 unaffected controls (from families with no known history of OFC or other craniofacial anomaly), 450 CL cases (44% female) and 1692 CLP cases (38% female) were extracted from all available participants. All individuals for this analysis were independent (i.e. unrelated) (Table I, Supplementary Table 1).
Table I.
Recruitment sites and sample sizes of cases
| Analysis | Site | Cleft Lip (CL) | Cleft Lip and Palate (CLP) | Controls* |
|---|---|---|---|---|
| GWAS | Argentina | 18 | 93 | 30 |
| China | 50 | 107 | 27 | |
| Colombia | 75 | 606 | 277 | |
| Denmark | 20 | 26 | 0 | |
| Ethiopia | 36 | 47 | 0 | |
| Guatemala | 21 | 81 | 208 | |
| Hungary | 33 | 72 | 253 | |
| India | 38 | 13 | 38 | |
| Nigeria | 17 | 33 | 68 | |
| Philippines | 33 | 126 | 96 | |
| Puerto Rico | 22 | 62 | 106 | |
| Spain | 5 | 29 | 0 | |
| Turkey | 10 | 162 | 171 | |
| United States | 72 | 235 | 415 | |
| Total | 450 | 1692 | 1700 | |
| Replication I | Brazil | 176 | 287 | - |
| Mongolia | 168 | 374 | - | |
| Philippines | 16 | 64 | - | |
| Total | 360 | 725 | - | |
| Replication II | European | 213 | 385 | - |
| Replication III | Asian | 224 | 703 | - |
controls used in gene-gene interaction analyses only
The methods for genotyping, quality control, imputation, and derivation of principal components (PCs) of ancestry have been described in detail by Leslie et al. (Leslie et al., 2016) and are also available online in the Quality Control Report issued by the University of Washington Genetics Coordinating Center (http://www.ccdg.pitt.edu/docs/Marazita_ofc_QC_report_feb2015.pdf). Briefly, samples were genotyped for 589,945 SNPs on the Illumina HumanCore + Exome panel at the Center for Inherited Disease Research. Genetic data were phased using SHAPEIT, and imputation was performed with IMPUTE2 software using the 1000 Genomes Phase 3 as a reference panel. At the time of imputation, chromosome X data were unavailable in IMPUTE2 format for the Phase 3 release; the X chromosome was imputed using 1000 Genomes Phase 1 (integrated variant set version 3, March 2012 release). A masked variant analysis indicated high-quality imputation, with a mean concordance of 0.995 for SNPs with MAF < 0.05 and 0.960 for SNPs with MAF ≥ 0.05. Genotypic probabilities were converted to most-likely genotype calls with the GTOOL software (http://www.well.ox.ac.uk/~cfreeman/software/gwas/gtool.html), using a genotype probability threshold of 0.9. Prior to statistical analysis, imputed SNPs with low info score (info<0.50) or with severe deviations (p<0.0001) from Hardy-Weinberg Equilibrium (HWE) in a set of independent, unaffected individuals of European ancestry (genotyped together with OFC cases from the current study) were excluded from analysis. Principal components (PCs) of ancestry were generated on the multi-ethnic cohort as described in Leslie et al. (Leslie et al., 2016). The PCs strongly tracked global recruitment site and self-reported race/ethnicity.
Genome-wide scan for genetic modifiers of cleft subtype
To identify potential genetic modifiers of cleft subtype, we analyzed the association between cleft subtype with 532,917 genotyped and 9,868,566 imputed SNPs where the minor allele frequency (MAF) was greater than 0.01 by directly comparing the two case subtypes using logistic regression (i.e., treating cleft subtype as the outcome) in PLINK (v1.9) assuming an additive genetic model and adjusting for 18 PCs of ancestry in order to protect against genomic inflation due to population structure. For the analysis of the X-chromosome, genotypes were coded 0, 1, and 2 under the additive genetic model for females, and coded 0, 2 for males to maintain the same scale between sexes. Genetic associations with p-values less than 5.0×10−8 were considered genome-wide significant based on a Bonferroni threshold for multiple testing of one million SNPs.
Additionally, rare variants (MAF<0.01) were evaluated for association with cleft subtype using the same framework; variants within exons of canonical transcripts for each gene were interrogated using gene-based versions of the Collapsed Multivariate and Combining (CMC) test (Li & Leal, 2008) and the Sequence Kernel Association Test (SKAT) (Wu et al., 2011). Statistical significance was determined using a Bonferroni threshold of 3.674×10−6, adjusting for 13,610 gene regions with at least two variants (on chromosomes 1–22).
Replication of 16q21
Replication of the top locus from the genome-wide scan was attempted using an independent sample of 360 CL cases and 725 CLP cases from Brazilian, Filipino, and Mongolian populations (Replication I, Table I). Three variants in high linkage disequilibrium (LD) with the lead SNP (rs7199325) were genotyped using Taqman SNP genotyping assays and read on an Applied Biosystems 7900HT instrument. SNPs were tested for association using a logistic regression model in R (version 3.3.3) including indicator variables for recruitment site. In silico replication for these SNPs was attempted in an independent sample of 437 CL cases and 1088 CLP cases of European and Asian ancestry (Replication II and III, respectively) using logistic regression while adjusting for 3 PCs of ancestry (Beaty et al., 2010).
Segregation of modifier variant of cleft types within families
The relationship between the 16q21 locus and family-level OFC patterns of the CL and CLP cases was examined to understand the genetic architecture and inheritance patterns of OFCs and the phenomenon that OFC subtypes often segregate within families. Each independent CL and CLP case was categorized based on the types of OFCs present within their reported pedigree (up to third-degree relatives of the affected proband). Because reported pedigrees can be incomplete across recruitment strategies, both multiplex and simplex pedigrees were included in this test. Three family OFC categories were considered in this analysis: CL cases from families where all affected individuals had CL (n=221, 25.8% multiplex), CLP cases from families where all affected individuals had CLP (n=1,017, 26.4% multiplex), and CL or CLP cases from families with a mixture of CL and CLP (n=287). Individual cases from the genome-wide scan that also belonged to one of these groups were included in this analysis (n=1,525). A linear regression model was used to assess the association between the average minor allele frequency at rs2848063, a variant selected from SNPs with the lowest p-values at the 16q21 locus, and family OFC pattern using linear regression, adjusting for 18 PCs of ancestry.
Interaction scans for modifying OFC risk
To investigate the potential modifying behavior of SNPs at the 16q21 locus identified in the genome-wide scan on the impact of known OFC risk loci, we conducted tests of gene-gene interaction. We tested the statistical interaction between a set of varaints previously associated with CL/P risk and a CL-risk variant from the 16q21 modifier locus. A total of 23 CL/P-risk SNPs demonstrating strong statistical evidence of association from 14 loci identified in two previous GWASs of CL/P (Leslie et al., 2017; Leslie et al., 2016), and those with MAF greater than 0.05 were selected for these interaction tests (Table III). The rs28480638 SNP was selected to represent the 16q21 locus identified in the genome-wide scan in the present study, because it had the highest MAF of all variants within the 16q21 region (MAF=0.30) while still yielding statistical evidence of association (p=5.303×10−6).
Table III.
Gene-gene interaction results with rs28480638 in chr.16q21
| CL/P GWAS Locus | SNP | Study | Minor Allele Frequency | CL | CLP | ||
|---|---|---|---|---|---|---|---|
| G×G P-value | Model | G×G P-value | Model | ||||
| PAX7 1p36.13 |
rs9439713 | A | 0.29 | 0.164 | DOM | 0.972 | ADD |
| ARHGAP29 1p22.1 |
rs66515264 | A | 0.2 | 0.198 | ADD | 0.762 | ADD |
| IRF6 1q32 |
rs75477785 | A | 0.14 | 0.159 | DOM | 0.571 | DOM |
| FAM49A 2p24.2 |
rs7566780 | A | 0.48 | 0.861 | ADD | 0.747 | ADD |
| 8q21 | rs12543318 | A | 0.45 | 0.018 | DOM | 0.399 | DOM |
| 8q24A | rs55658222 | A | 0.16 | 0.191 | ADD | 0.651 | ADD |
| 8q24B | rs7278734 | B | 0.16 | 0.122 | DOM | 0.834 | ADD |
| FOXE1 9q21.31 |
rs6559624 | B | 0.31 | 0.023 | DOM | 0.185 | ADD |
| VAX1 10q25 |
rs10886040 | A | 0.24 | 0.342 | ADD | 0.373 | ADD |
| SPRY2 13q31 |
rs11841646 | A | 0.39 | 0.759 | DOM | 0.452 | ADD |
| ARID3B 15q24 |
rs11072494 | A | 0.33 | 0.12 | DOM | 0.091 | DOM |
| NTN1 17p13.1 |
rs11273201 | B | 0.27 | 0.83 | ADD | 0.279 | ADD |
| NOG 17q22 |
rs227727 | B | 0.41 | 0.096 | ADD | 0.492 | ADD |
| RHPN2 19q13.11 |
rs73039428 | B | 0.08 | 0.209 | DOM | 0.523 | ADD |
| MAFB 20q12 |
rs6072081 | A | 0.47 | 0.776 | DOM | 0.991 | DOM |
A: (Leslie et al., 2017)
B: (Leslie et al., 2016)
Gene-gene interactions were tested via two logistic regression models in R (450 CL cases vs. 1700 unaffected controls and 1692 CLP cases vs. the same 1700 unaffected controls), each model containing terms for main effects of both genotypes and the potential interaction between them, while adjusting for 18 PCs of ancestry. Both additive and dominant genetic models were tested and compared using Akaike information criteria (AIC); the p-value of the interaction term from the best fitting model was used to assess the extent of interaction. The interaction effects were individually examined for evidence of association for each SNP. This approach has the potential to identify genetic risk factors for CL or CLP whose risk changes based on the genotype of the target 16q21 variant.
Results
Genome-wide scan for genetic modifiers of OFC subtype
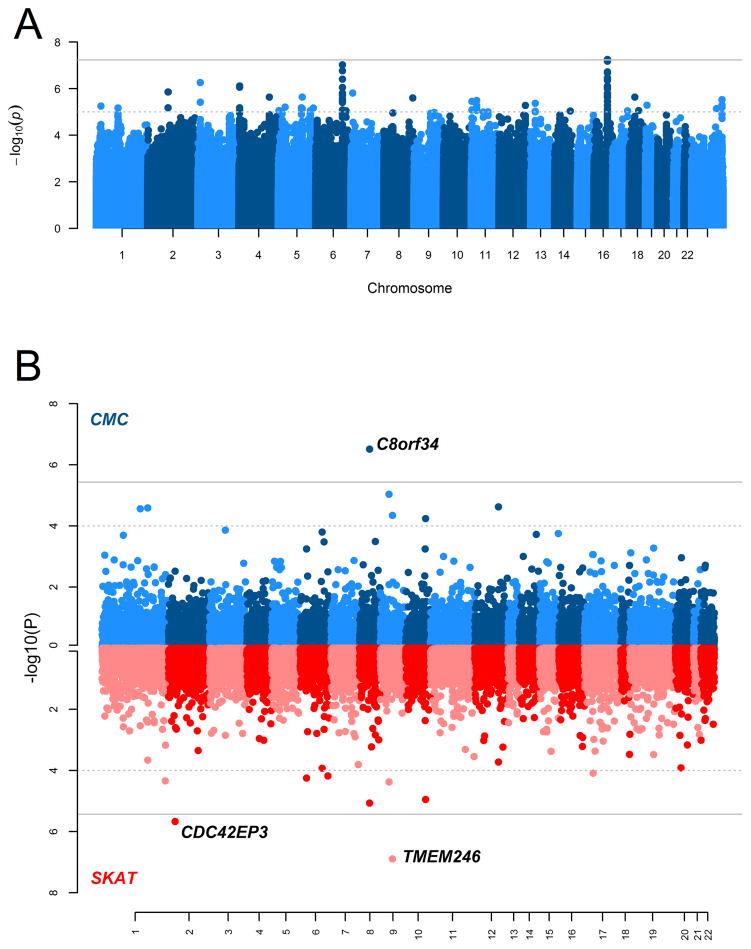
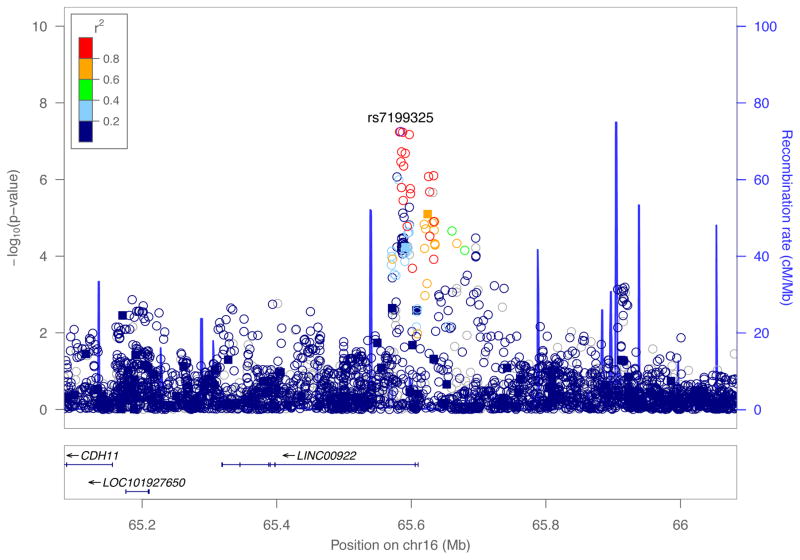
We performed a GWAS of 532,917 genotyped and 9.8 million imputed SNPs to detect modifiers of cleft type (CL vs. CLP) in a sample of 450 CL cases and 1692 CLP cases. Although no locus reached formal genome-wide statistical significance (i.e. p<5.0×10−8), we observed a suggestive association on chromosome 16 spanning LINC00922, a long non-protein-coding RNA (Figures 1 and 2). At this locus on 16q21, one genotyped SNP and 21 imputed SNPs showed at least suggestive evidence of association (p<1.0×10−5), with the lead SNP (rs7199325; p=5.611×10−8) demonstrating stronger evidence of association with CL compared with CLP (OR=0.406, 95% CI: [0.294, 0.562]). In fact, for all 22 genetic variants showing suggestive evidence of association, the minor allele was strongly associated with CL over CLP. There was no evidence of genomic inflation in this scan (λ=0.995). Table S2 contains results for all SNPs yielding p-values less than 1.0×10−5.
Figure 1.
Manhattan plots of the −log10(p-values) from the (A) common variant case-case analysis, (B) rare variant case-case analyses using the Collapsed Multivariate and Combining (CMC) test or the Sequence Kernel Association Test (SKAT). The solid grey lines denote the Bonferroni-threshold for statistical significance (A, 5.0 x 10−8; B, 3.647 x 10−6) and the dotted grey lines denote suggestive significance thresholds (A, 1.0 x 10−5; B, 1.0 x 10−4).
Figure 2.
Regional association plot for 16q21 showing −log10(p-values) for imputed (circle) and genotyped (square) SNPs from the common variant case-case analysis. Plots were generated using LocusZoom. The recombination overlay (blue line, right y-axis) indicates the boundaries of the LD-block. Points are color coded according to pairwise linkage disequilibrium (R2) with the index SNP.
In the rare variant analyses, rare variants within three genes demonstrated evidence of association with cleft subtype differences – C8orf34 (CMC scan p=3.095×10−7), TMEM246 (SKAT scan p=1.272×10−7), and CDC42EP3 (SKAT scan p=2.139×10−6). Rare variants in C8orf34 tended to have slightly higher frequencies in CL cases than CLP cases, whereas those within C9orf125 and CDC42EP3 did not show any consistent trends in frequency between CL and CLP cases. Results from all gene regions demonstrating at least suggestive evidence of association (p<5.0×10−4) are given in Table S4. The biologic relevance of these genes and their potential impact on risk to OFCs are unknown, and no replication data exist at this time.
Replication of genetic modifier locus
Three SNPs in modest to high LD with the top-associated variant (rs7199325) from the GWAS were tested in an independent sample of CL and CLP cases: rs6499007 (R2=0.83, D′=1.00 in the Admixed American (AMR) population from 1000 Genomes), rs16969175 (R2=0.41, D′=0.84), and rs16969137 (R2=0.86, D′=1.00). One variant, rs6499007, showed a significant association (p<0.05; Table 2) in the samples from Brazil, the Philippines, and Mongolia but not in the European population.
Table II.
Replication results for 16q21 locus
| rs6499007 | rs16969175 | rs16969137 | ||
|---|---|---|---|---|
| Discovery Sample | OR | 0.444 | 0.538 | 0.417 |
| 95% CI | 0.325,0.607 | 0.410, 0.707 | 0.303, 0.573 | |
| P | 3.463×10−7 | 7.997×10−6 | 6.732×10−8 | |
| Replication I Sample | OR | 0.678 | 0.897 | 1.258 |
| 95% CI | 0.465, 0.988 | 0.738, 1.090 | 0.770, 2.056 | |
| P | 0.043 | 0.273 | 0.36 | |
| Replication II Sample | OR | 0.812 | 0.948 | 0.804 |
| 95% CI | 0.533, 1.237 | 0.617, 1.458 | 0.527, 1.226 | |
| P | 0.332 | 0.808 | 0.313 | |
| Replication III Sample | OR | 1.023 | 0.815 | 1.120 |
| 95% CI | 0.454, 2.348 | 0.527, 1.261 | 0.493, 2.547 | |
| P | 0.957 | 0.357 | 0.786 |
Segregation of modifier variants within families
Twenty to thirty percent of OFC cases are considered multiplex with some family history of the disorder. Although any combination of OFC subtypes can occur within a family, recurrence risks are highest for the same type of cleft. As a result, it is not uncommon to find families where most, if not all, affected family members have the same type of OFC. We hypothesized that the OFC pattern within families may correlated with the genotype at 16q21. We assigned each independent CL or CLP case to a family cleft group based on the type of OFCs present within the reported pedigree. Within the 1,525 CL and CLP cases with known family OFC patterns (i.e. CL-only, CLP-only, or mixed CL+CLP families), the association between average frequency of the A allele at rs28480638 (associated with increased odds of CL vs. CLP, p=5.303×10−6) and family OFC pattern was examined using a linear regression model, adjusting for ancestry. We found differences in the average A-allele frequency by family OFC pattern (p=0.003), with the highest frequency of the A-allele found in cases from the CL-only families (Table S3). This result is consistent with the 16q21 locus modifying OFC subtype in favor of CL.
Interaction scan for modification of OFC risk
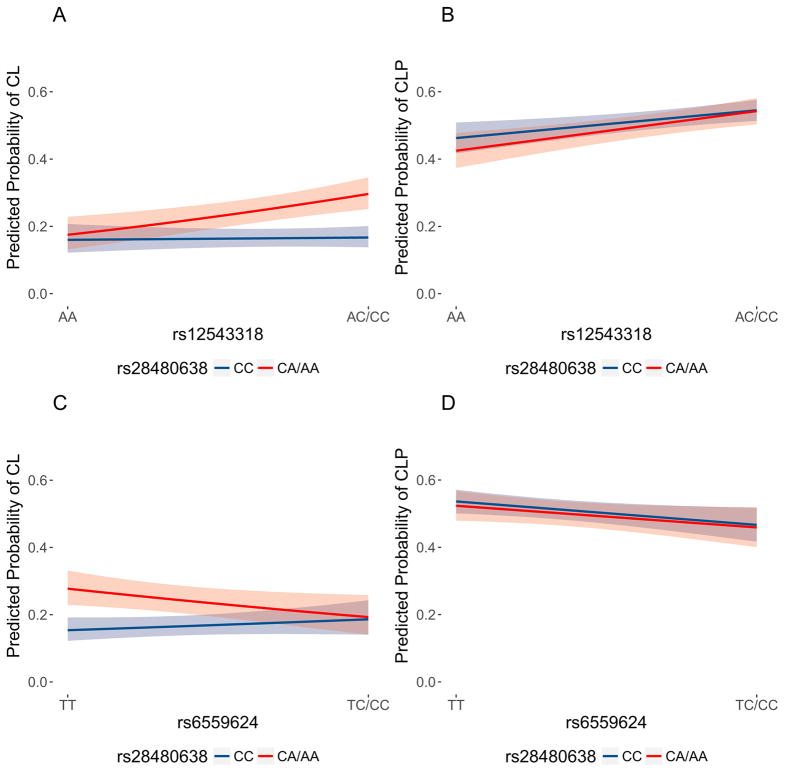
We next hypothesized wanted to explore the effect of the 16q21 locus when found in combination with other CL/P risk alleles. In simple two-locus model, an individual carrying the CL-associated 16q21 allele and a second allele for another CL/P risk locus would have an increased risk of CL over CLP. Although in practice, each individual carries multiple risk alleles, testing potential gene-gene interactions in a pairwise fashion can inform downstream analyses and identify biological mechanisms driving pathogenesis of OFCs. Therefore, we tested the hypothesis that the 16q21 modifier locus genetically interacts with previously-identified OFC risk loci to increase risk of CL. To this end, we tested for gene-gene interactions between markers in 16q21 and 14 known risk loci identified in previous GWASs of OFCs. Although no gene-gene interactions surpassed the Bonferroni threshold for statistical significance of 0.0017 (i.e. 0.05/30), two CL/P risk variants demonstrated evidence of nominal interaction with the most significant 16q21 variant (rs28480638): rs12543318 (8q21, p=0.012) and rs6559624 (FOXE1, p=0.023). This 8q21 locus was previously identified in a GWAS of CL/P (Ludwig et al., 2012), where the minor allele at rs12543318 was associated with increased risk of CL/P. This additive pattern held for CLP cases, however, risk of CL only increased in a similar manner for individuals carrying at least one copy of the minor allele at rs28480638 (CA or AA genotypes; Figure 3A). At the FOXE1 locus, the homozygous genotype (CC) at rs6559624 was associated with increased risk of CL/P. This pattern held for all CLP cases, however, the risk of CL only increased in for individuals with at least one copy of the rs28480638 minor allele (Figure 3B).
Figure 3.
(A) Predicted probabilities and 95% confidence bands of cleft lip for genotypes of the 8q21 variant, rs12543318, and the 16q21 modifier variant, rs28480638. (B) Predicted probabilities and 95% confidence bands of cleft lip and palate (CLP) for genotypes of the 8q21 variant, rs12543318, and the 16q21 modifier variant, rs28480638. (C) Predicted probabilities and 95% confidence bands of cleft lip for genotypes of the FOXE1 variant, rs6559624, and the 16q21 modifier variant, rs28480638. (D) Predicted probabilities and 95% confidence bands of cleft lip and palate (CLP) for genotypes of the FOXE1 variant, rs6559624, and the 16q21 modifier variant, rs28480638. Predicted probabilities were calculated using the gene-gene interaction models, holding the 18 principal components of ancestry constant at their average values. In each plot, the predicted probabilities for AC/CC genotypes at rs28480638 (which are associated with increased risk of cleft lip) are shaded in red, and those for AA genotypes at rs28480638 are shaded in blue.
Discussion
We performed a genome-wide scan for genetic modifiers of cleft type differences by comparing allele frequencies between CL and CLP cases in a case-case comparison using a large multi-site study of OFCs. We also performed gene-based tests of low frequency variants and identified three genes associated with such cleft type differences. In the scan of common variants (SNPs), the locus demonstrating the greatest statistical evidence of association, 16q21, also showed evidence of association in an independent sample of CL and CLP cases. In a subset of the discovery CL and CLP cases, the frequency of the CL-associated 16q21 allele was highest in cases without relatives with CLP, further supporting its role as a modifier of OFC subtype. We then tested for potential gene-gene interaction between a variant in this modifier region and variants in recognized CL/P risk loci. We found significant evidence of interaction between the most significant SNP at 16q21 with both a recognized genetic risk factor at 8q21 and a second recognized risk SNP at FOXE1. In both interactions, the risk of CL conferred by the known CL/P risk variant was increased for individuals containing the CL-associated allele at this putative modifier variant.
The loci identified in this study contain several candidate genes with pathophysiological relevance for CL vs. CLP differentiation. Identifying specific gene(s) responsible for the apparent modifying effect of these loci will require functional studies; however, the top regions found here do contain genes with biologic plausibility. The peak 16q21 signal is located proximal to LINC00922, a long non-coding RNA (lncRNA). LncRNAs belong to a class of RNA molecules with diverse functions, however, unlike protein-coding genes, their function is difficult to infer from sequence or structure alone. Increasing evidence supports the involvement of lncRNAs in gene regulation through chromatin modification, transcription, or post-transcriptional processing (Cech & Steitz, 2014). A role for LINC00922 is difficult to predict for craniofacial development as lncRNAs typically have low conservation between species, and relevant human tissue is not easily accessible for RNA-sequencing experiments. Nonetheless, lncRNAs represent reasonable candidate molecules to act as phenotypic modifiers and should be a focus of future studies.
The 16q21 locus also contains CDH11, encoding cadherin 11. Cadherins are proteins that mediate cell-cell adhesion and play important roles in proliferation, differentiation, and tissue morphogenesis, all critical processes for craniofacial development. CDH11 is expressed in the branchial arches during mouse embryonic development (Kimura et al., 1995), and in osteoblasts, mesenchymal cells, and epithelial cells undergoing epithelial-mesenchymal transition (Zeisberg & Neilson, 2009). In both human and mouse, CDH11 regulates extracellular matrix production via TGF-β and ROCK signaling pathways (Row, Liu, Alimperti, Agarwal, & Andreadis, 2016). This makes CDH11 a compelling candidate gene for palate morphogenesis, specifically shelf elevation, occurs through mesenchymal proliferation and changes to the extracellular matrix infrastructure.
A similar theme emerges from other top variants including the 6q22 locus, containing ARHGAP18, and the scan of low frequency variants, where variants in CDC42EP3 were associated with cleft type differentiation. CDC42EP3 is a Rho-GTPase effector protein involved in matrix remodeling (Calvo et al., 2015), and ARHGAP18 is a Rho-GTPase regulating RhoA which in turn controls cell shape, spreading, and migration (Maeda et al., 2011). However, specific roles in craniofacial development for these genes, C8orf34, and TMEM246 (identified in the low frequency variant scan) remain unknown. Further investigations into these loci and replication studies will be required to fully understand their contribution to cleft type differentiation.
There is some evidence in the literature of cleft type specific associations including SPRY2 and GREM1 with CLP (Ludwig et al., 2016; Ludwig et al., 2012); and IRF6 has been suggested to have a stronger effect in CL than CLP (Marazita, 2012; Rahimov et al., 2008). However, these loci were not among the top association signals in our analysis. Our genome-wide scan for modifiers of cleft type was well-powered to detect common variants with strong genetic effect (approximately 80% power for a variant with MAF of 30% and genotypic relative risk of 1.5). It is possible that weaker effects, especially with less frequent variants, would not be detected in our multiethnic cohort. This may be the case for IRF6 and SPRY2, where the reported association signals were strongest in some populations but not others. In a similar vein, we found evidence of replication in a sample that included an admixed Brazilian population, which more closely resembles our discovery sample than the European or Asian in silico replication sample. The 16q21 SNPs with the strongest evidence of association were most frequent among African and admixed populations with African ancestry; thus, further studies of multiethnic and diverse populations are needed.
We identified evidence of gene-gene interactions between the markers in 16q21 which seems to modify risk to CL and CLP (both forms of OFC) and two recognized genetic risk loci associated with OFCs. Although a mechanism for how these interactions contribute to specific cleft types is currently unknown, these results build upon our knowledge of the complex and heterogeneous genetic architecture of OFCs, and could inform future biological experiments. Substantial evidence now exists that the FOXE1 locus is associated with all subtypes of OFCs (Leslie et al., 2017; Ludwig et al., 2014; Marazita et al., 2009; Moreno et al., 2009). As the lip develops prior to the palate, it is tempting to speculate that the function of the novel genetic element in 16q21 is to promote proper palatogenesis; however, the genetic architecture of OFCs and, in general, craniofacial development is very complex and interactions with other genetic, environmental, or stochastic factors are likely to contribute overall risk to OFC and the specific type of cleft that results in the child. As gene-gene and higher-ordered interaction analyses require very large sample sizes, we view this study as the first step toward building comprehensive risk models for OFCs. Furthermore, these analyses do not consider maternal genetic effects, epigenetics, or environmental exposures that may also contribute to the cleft subtype observed in affected individuals.
Collectively, this study adds to our understanding of the genetic architecture of OFCs by identifying genetic markers differentially associated with CL and CLP cases. This approach may be applied to other aspects of OFC subtypes, including cleft laterality and subclinical phenotypes of OFCs (Marazita, 2012). As gene mapping studies move beyond GWAS and into whole-exome or whole-genome sequencing studies, this approach can be adapted for studies of rare variants (Carlson et al., 2017). This study demonstrates the power of detailed statistical analysis to generate novel hypotheses and motivate further study of potential biological mechanisms for craniofacial development. Applied to other complex traits or diseases with phenotypic heterogeneity, modifier GWASs such as this could create an opportunity to identify therapeutic targets and enhance individualized treatment, prognosis, or management.
Supplementary Material
Acknowledgments
The authors thank the dedicated field staff, collaborators, and participating families for their important contributions to this study. This work was supported by grants from the National Institutes of Health (NIH) including: R00-DE025060 [EJL], X01-HG007485 [MLM, EF], R01-DE016148 [MLM, SMW], U01-DE024425 [MLM], R37-DE008559 [JCM, MLM], R01-DE009886 [MLM], R21-DE016930 [MLM], R01-DE014667 [LMM], R01-DE012472 [MLM], R01-DE011931 [JTH], R01-DE011948 [KC], U01-DD000295 [GLW], R00-Grant DE022378 and Robert Wood Johnson Foundation Grant number 72429 [AB], K99-DE024571 [CJB], S21-MD001830 [CJB], R01-DE014581 [TB], U01-DE018993 [TB]. Other support for this work was provided by JSPS KAKENHI Grant-in-Aid for Scientific Research (C) Grant Number JP17K11863 [SS] and Grant-in-Aid for Scientific Research (A) Grant Number JP 24249092 [SS]. Genotyping and data cleaning were provided via an NIH contract to the Johns Hopkins Center for Inherited Disease Research: HHSN268201200008I.
References
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, … Scott AF. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature Genetics. 2010;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo F, Ranftl R, Hooper S, Farrugia AJ, Moeendarbary E, Bruckbauer A, … Sahai E. Cdc42EP3/BORG2 and Septin Network Enables Mechano-transduction and the Emergence of Cancer-Associated Fibroblasts. Cell Rep. 2015;13(12):2699–2714. doi: 10.1016/j.celrep.2015.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JC, Taub MA, Feingold E, Beaty TH, Murray JC, Marazita ML, Leslie EJ. Identifying Genetic Sources of Phenotypic Heterogeneity in Orofacial Clefts by Targeted Sequencing. Birth Defects Res. 2017 doi: 10.1002/bdra.23605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nature Reviews Genetics. 2011;12(3):167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosen D, Chevrier C, Skytthe A, Bille C, Molsted K, Sivertsen A, … Christensen K. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: support for the multifactorial threshold model of inheritance. Journal of Medical Genetics. 2010;47(3):162–168. doi: 10.1136/jmg.2009.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harville EW, Wilcox AJ, Lie RT, Vindenes H, Abyholm F. Cleft lip and palate versus cleft lip only: are they distinct defects? Am J Epidemiol. 2005;162(5):448–453. doi: 10.1093/aje/kwi214. [DOI] [PubMed] [Google Scholar]
- Jia Z, Leslie EJ, Cooper ME, Butali A, Standley J, Rigdon J, … Murray JC. Replication of 13q31.1 association in nonsyndromic cleft lip with cleft palate in Europeans. Am J Med Genet A. 2015;167(5):1054–1060. doi: 10.1002/ajmg.a.36912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Matsunami H, Inoue T, Shimamura K, Uchida N, Ueno T, … Takeichi M. Cadherin-11 expressed in association with mesenchymal morphogenesis in the head, somite, and limb bud of early mouse embryos. Dev Biol. 1995;169(1):347–358. doi: 10.1006/dbio.1995.1149. [DOI] [PubMed] [Google Scholar]
- Lee PH, Bergen SE, Perlis RH, Sullivan PF, Sklar P, Smoller JW, Purcell SM. Modifiers and subtype-specific analyses in whole-genome association studies: a likelihood framework. Hum Hered. 2011;72(1):10–20. doi: 10.1159/000327158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Butali A, Buxo CJ, Castilla EE, … Marazita ML. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum Genet. 2017;136(3):275–286. doi: 10.1007/s00439-016-1754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Liu H, Carlson JC, Shaffer JR, Feingold E, Wehby G, … Marazita ML. A Genome-wide Association Study of Nonsyndromic Cleft Palate Identifies an Etiologic Missense Variant in GRHL3. Am J Hum Genet. 2016;98(4):744–754. doi: 10.1016/j.ajhg.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Ahmed ST, Bohmer AC, Sangani NB, Varghese S, Klamt J, … Peters H. Meta-analysis Reveals Genome-Wide Significance at 15q13 for Nonsyndromic Clefting of Both the Lip and the Palate, and Functional Analyses Implicate GREM1 As a Plausible Causative Gene. PLoS Genetics. 2016;12(3):e1005914. doi: 10.1371/journal.pgen.1005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, … Nothen MM. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44(9):968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Wahle P, Reutter H, Paredes-Zenteno M, Munoz-Jimenez SG, Ortiz-Lopez R, … Mangold E. Evaluating eight newly identified susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a Mesoamerican population. Birth Defects Res A Clin Mol Teratol. 2014;100(1):43–47. doi: 10.1002/bdra.23209. [DOI] [PubMed] [Google Scholar]
- Maeda M, Hasegawa H, Hyodo T, Ito S, Asano E, Yuang H, … Senga T. ARHGAP18, a GTPase-activating protein for RhoA, controls cell shape, spreading, and motility. Molecular Biology of the Cell. 2011;22(20):3840–3852. doi: 10.1091/mbc.E11-04-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML. The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet. 2012;13:263–283. doi: 10.1146/annurev-genom-090711-163729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, Goldstein McHenry T, … Arcos-Burgos M. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Human Heredity. 2009;68(3):151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, … Lidral AC. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Human Molecular Genetics. 2009;18(24):4879–4896. doi: 10.1093/hmg/ddp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, … Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nature Genetics. 2008;40(11):1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row S, Liu Y, Alimperti S, Agarwal SK, Andreadis ST. Cadherin-11 is a novel regulator of extracellular matrix synthesis and tissue mechanics. J Cell Sci. 2016;129(15):2950–2961. doi: 10.1242/jcs.183772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119(6):1429–1437. doi: 10.1172/jci36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.