Key Points
H/F-LVs permit very-high-level transduction of human HSCs, even in the complete absence of cytokine stimulation.
H/F-LVs outperform VSV-G-LVs for efficient correction of Fanconi anemia patient cells in unfractionated BM.
Abstract
Hematopoietic stem cell (HSC)–based gene therapy trials are now moving toward the use of lentiviral vectors (LVs) with success. However, one challenge in the field remains: efficient transduction of HSCs without compromising their stem cell potential. Here we showed that measles virus glycoprotein–displaying LVs (hemagglutinin and fusion protein LVs [H/F-LVs]) were capable of transducing 100% of early-acting cytokine-stimulated human CD34+ (hCD34+) progenitor cells upon a single application. Strikingly, these H/F-LVs also allowed transduction of up to 70% of nonstimulated quiescent hCD34+ cells, whereas conventional vesicular stomatitis virus G (VSV-G)–LVs reached 5% at the most with H/F-LV entry occurring exclusively through the CD46 complement receptor. Importantly, reconstitution of NOD/SCIDγc−/− (NSG) mice with H/F-LV transduced prestimulated or resting hCD34+ cells confirmed these high transduction levels in all myeloid and lymphoid lineages. Remarkably, for resting CD34+ cells, secondary recipients exhibited increasing transduction levels of up to 100%, emphasizing that H/F-LVs efficiently gene-marked HSCs in the resting state. Because H/F-LVs promoted ex vivo gene modification of minimally manipulated CD34+ progenitors that maintained stemness, we assessed their applicability in Fanconi anemia, a bone marrow (BM) failure with chromosomal fragility. Notably, only H/F-LVs efficiently gene-corrected minimally stimulated hCD34+ cells in unfractionated BM from these patients. These H/F-LVs improved HSC gene delivery in the absence of cytokine stimulation while maintaining their stem cell potential. Thus, H/F-LVs will facilitate future clinical applications requiring HSC gene modification, including BM failure syndromes, for which treatment has been very challenging up to now.
Visual Abstract
Introduction
Hematopoietic stem cell (HSC)–based gene therapy holds promise for the cure of many inherited and acquired diseases. The field is now moving toward the use of lentiviral vectors (LVs) as evidenced by recent successful clinical trials for adrenoleukodystrophy,1 β-thalassemia,2 metachromatic leukodystrophy3 and Wiskott-Aldrich syndrome.4 Moreover, LV trials for X-SCID, Fanconi anemia (FA), and other monogenetic defects were initiated.5-7
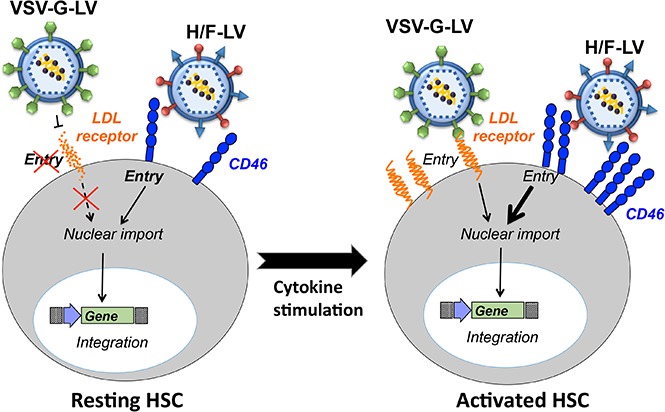
Genetically modified HSCs should retain engraftment and functional properties similar to those of unmodified HSCs. They should be able to self-renew and to differentiate into all hematopoietic lineages. HSCs are poorly permissive for classical vesicular stomatitis virus G (VSV-G)-LV transduction because 75% of them reside in the G0 phase of the cell cycle, which was proposed to restrict VSV-G-LV transduction.8 Indeed, in resting lymphocytes, LV postentry steps such as completion of reverse transcription, nuclear import, and chromosomal integration of the transgene do not readily occur.8-10
Moreover, we confirmed low expression levels of the VSV receptor (ie, low-density lipoprotein receptor11) at the surface of unstimulated CD34+ cells, coinciding with poor VSV-G-LV–mediated transduction.12 Only early-acting cytokine stimulation of human CD34+ (hCD34+) cells that upregulated the LDL-R permitted high-level VSV-G-LV transduction.12 However, intense exposure of HSCs to cytokines affects their homing and trafficking ability and might promote differentiation rather than expansion of the HSC pool.13,14 It was also shown that exit from dormancy provokes DNA damage–induced attrition in HSCs.15 High hCD34+-cell transduction was achieved only by combining high VSV-G-LV doses with strong cytokine stimulation that increased the risk for multicopy integration. Insertional mutagenesis under these conditions cannot be excluded,16,17 although current trials using LVs do not report adverse events. Therefore, mild cytokine stimulation that allows efficient HSC transduction is an important objective. Clearly, to achieve this objective, alternatives to classical VSV-G pseudotyped LVs are needed.
Previously, we engineered LVs pseudotyped with modified measles virus (MV) envelope glycoproteins (gp’s), hemagglutinin (H), and fusion protein (F). They represented the first tool that allowed efficient transduction of quiescent human T and B cells without inducing entry into the cell cycle or changes in phenotype.18-20 These vector particles (H/F-LVs) are able to mediate cell entry via the native MV receptors CD46 and signaling lymphocyte activation molecule (SLAM).19,21 CD46 is a complement regulatory molecule expressed on all human nucleated cells,22 whereas SLAM (CD150) is constitutively expressed at the surface of some T and B subsets and upregulated upon proliferation of T and B lymphocytes and mature dendritic cells.23,24 The third MV receptor, nectin-4, is not expressed on lymphocytes.25
Interestingly, the SLAM receptors, which are cell-cell interaction and signaling receptors, are differentially expressed on distinct type of leukocytes.26,27 CD150 receptors are selectively expressed among primitive mouse progenitors which allows us to highly purify murine bone marrow (BM) and fetal liver HSCs by staining these SLAM receptors (CD150+, CD244–, and CD48–).28,29 This led to the term “SLAM code” whose applicability to human HSCs (hHSCs) and progenitors is still controversial.28-32 Indeed, several reports already showed the absence of CD150 on the hCD34+CD38– hematopoietic early progenitors.31,33
Here, we evaluated H/F-LVs for their potential to transduce cytokine-stimulated as well as unstimulated HSCs. We achieved transduction efficiencies of up to 100% in prestimulated HSCs by using a low dose of H/F-LVs. Even more remarkable, unstimulated hCD34+ cells were transduced by H/F-LVs at levels reaching 70%. Even upon transplantation in primary and secondary NSG recipients, this high level of gene-modified cells persisted in all myeloid and lymphoid lineages, emphasizing that our system promoted a highly efficient transduction of quiescent HSCs, which maintained their stemness. In addition, we demonstrated the efficacy of therapeutic H/F-LVs to efficiently correct the phenotype of unfractionated BM progenitor cells from Fanconi anemia (FA) patients.
Materials and methods
Plasmids
The Edmonston measles Hgp- and Fgp-encoding plasmids were described previously.19 The SLAM-restricted vectors (H-D4/F-LVs), SLAM-blind Hgp (Ha533), and H-D4-YG were described previously21,34
LV production
Self-inactivating HIV-1–derived vectors were generated by transient transfection of 293T cells as described previously.35 Viral vectors were produced either pseudotyped with VSV-G or with MV gp’s. Transfection was performed by using the classical calcium phosphate method (solution A: 280 mM NaCl, 10 mM KCl, 1.5 mM Na2HPO4, 12 mM d-glucose, and 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) adjusted to pH 7.2 with NaOH; solution B: 2 mM CaCl2). For co-display of the different Hgp’s and Fgp’s, 3 µg of each envelope plasmid was transfected together with a Gag-Pol packaging plasmid (8.91) and a plasmid encoding a self-inactivating LV-expressing green fluorescent protein (GFP) under the control of the spleen focus foamy virus promoter ([SIN]-HIVSFFVGFP). For VSV-G-LV preparation, 3 µg of envelope plasmid was co-transfected. The HIV-FANCA and GFP corresponding control expressed the FANCA or GFP gene under the control of the pgk promoter. These vectors are based on the pCCL backbone: pCCL1-PGK GFP and pCCL1-PGK FANCA, both described in González-Murillo et al.36 Eighteen hours after transfection, the medium was replaced by Opti-MEM supplemented with HEPES (Invitrogen). Viral supernatants were harvested 48 hours after transfection and filtered. The vectors were concentrated at low speed by overnight centrifugation of the viral supernatants at 3000g at 4°C (details are provided in supplemental Methods).
Titration of the LVs
Details are provided in supplemental Methods.
Cells
Chinese hamster ovary cells expressing SLAM (CHO-SLAM) cells were grown in RPMI medium (Gibco Invitrogen, Auckland, New Zealand) supplemented with 10% fetal calf serumand 50 μg/mL of penicillin/streptomycin; human kidney epithelial cells (293T) were grown in Dulbecco’s modified Eagle medium (Gibco Invitrogen) medium supplemented as for RPMI.
Sample collection and isolation of CD34+ cells
Cord blood (CB) samples and mobilized blood were collected in sterile tubes containing citrate-dextrose (ACD; Sigma, Saint Quentin Fallavier, France). CB, mobilized blood, and unfractionated BM from FA-A patients were obtained after informed consent of donors or patients (or their parents), and approval was obtained by the ethics committees of the hospitals according to the Helsinki Declaration. Human and macaque CD34+ cells were isolated as described.37,38 Unfractionated BM was obtained by BM aspiration during clinical follow-up of the patients, and erythrocytes were depleted by hydroxyethyl-starch density gradient.39
Transduction of human and macaque CD34+ cells
Human and macaque CD34+ cells were incubated for 14 or 18 to 24 hours (as indicated) in 24-well plates in serum-free medium (CellGro; CellGenix, Freiburg, Germany) supplemented with human recombinant cytokines (as indicated): stem cell factor (SCF; 100 ng/mL), thyroid peroxidase (TPO; 20 ng/mL), Flt3-ligand (Flt3-L; 100 ng/mL), interleukin-6 (IL-6; 20 ng/mL), and IL-3 (20 ng/mL) (Preprotech, Neuilly-Sur-Seine, France). Then, 5 × 104 prestimulated CD34+ cells were transduced in 48-well plates with concentrated LVs at indicated multiplicities of infection (MOIs) in serum-free medium. Cells were replenished with cytokines every 3 days. Three, 6, and 14 days after transduction, the percent of GFP+ cells was determined by fluorescence-activated cell sorting (FACS). Alternatively, CD34+ cells were freshly isolated and transduced immediately in the absence of cytokines and kept for 3 days in the absence of cytokines before FACS analysis. Where indicated, transductions were performed on retronectin-coated plates according to manufacturer’s instructions (Takara, Kusatsu, Shiga, Japan). After 3 days, transduced unstimulated CD34+ cells were continued in a cytokine cocktail in the presence or absence of a retrotranscriptase inhibitor (10 µM lamivudine).
For myeloid differentiation of CD34+ cells, 3 days after transduction, cells were washed, counted, and seeded in methylcellulose medium (STEMCELL Technologies, Grenoble, France) in the presence of reverse-transcriptase (RT) inhibitor (10 µM lamivudine) to perform colony-forming cell (CFC) assay. CFCs were analyzed on day 14 of culture. Note that all transduction experiments were performed with at least 3 different vector preparations and CD34+ cells from 3 different donors.
For transduction of CB CD34+ cells under low oxygen, cells were left unstimulated or prestimulated overnight with SCF (100 ng/mL), Flt3-L (100 ng/mL), and TPO (20 ng/mL) under low oxygen (2% O2) or atmospheric oxygen (20% O2) in cellgro medium. Cells were transduced with the H/F-LVs at an MOI of 5 on retronectin (20 µg/cm2)–coated plates in 2% O2 or 20% O2; at 3 days after transduction, cells were analyzed for GFP+CD34+ cells.
For transduction of FA-A patient cells, all samples were prestimulated for 18 hours in X-VIVO 20 medium (Lonza; Basel, Switzerland) supplemented with 1% penicillin/streptomycin (Gibco/Thermofisher, Villebon-sur-Yvette, France) and human recombinant cytokines: SCF (100 ng/mL), Flt3-L (100 ng/mL), TPO (20 ng/mL; EuroBiosciences), anti-TNF-α (10 µg/mL; Enbrel-Etanercept; Pfizer, New York, NY) and N-acetylcysteine (NAC; 1 mM; Pharmazam, Spain). Anti-TNF-α and NAC are compounds commonly used when maintaining FA hematopoietic stem and progenitors cells in vitro. They share the common role of avoiding oxidative damage when the cells are manipulated ex vivo. Anti-TNF-α performs the antibody-mediated inhibition of TNF-α, a cytokine that has been described to be overproduced in FA cells. This feature increases the cell predisposition to enter into apoptosis. NAC is an antioxidant used to protect FA cells from the oxidative stress. Cells were maintained in hypoxic conditions (5% O2), 37°C, 5% CO2, and 95% relative humidity. Then, 2 × 106 FA total BM cells were transduced with H/F-LVs at an MOI of 5 or with VSV-G-LVs at an MOI of 300 on plates coated with retronectin (20 µg/cm2) (Takara). The cells were exposed to a single transduction hit to avoid the maintenance of FA cells in culture for long periods of time. At 20 hours after transduction, cells were washed, counted, and seeded in methylcellulose medium (STEMCELL Technologies) to perform CFC assays. CFCs were analyzed at day 14 of culture. As necessary, mitomycin C (10 nM; Sigma) was added to the medium to select the corrected cells.
Receptor blocking assay
CD34+ cells were seeded in 96-well plates and incubated for 2 hours with anti-CD46 (clone 13/42; gift from R. Buckland, International Center for Infectiology Research, Lyon, France), anti-SLAM (IPO-3; eBioscience/Thermofisher, Villebon-sur-Yvette, France), or anti-nectin-4 (R&D Systems, Minneapolis, MN) antibodies before transduction. Concentrated viral supernatants were added to the cells at an MOI of 10, and 3 days later, transduction efficiency was analyzed by FACS.
Lentiviral barcoding experiment
The GFP-BC32 lentiviral vector encoding for a barcode library described previously in Thielecke et al40 was used for H/F-LV production. These H/F-LVs were used to transduce unstimulated human CD34+ cells before transplantation into recipient NSG mice. Four months after transplantation, the mice were euthanized, and BM, spleen, and thymus were harvested. To investigate the barcode content, the following cell populations were sorted from BM: CD13+, CD34+CD19+, and CD34+CD19– cells; from spleen: CD3+, CD14+, CD19+ cells; from the thymus: CD3+ cells. DNA was extracted from all populations, and barcode retrieval, sequencing, and barcode analyses, including error correction, were performed as described.40 Venn diagrams were produced by using an in-house R script with the VennDiagram package.
Conditioning and reconstitution of NSG mice
NSG mice used in our study were housed in our animal facility (Plateau de Biologie Expérimentale de la Souris, Lyon, France). Experiments were performed in accordance with the European Union guidelines upon approval of the protocols by the local ethical committee (Authorization Agreement No. C2EA-15; local Animal Ethics Evaluation Committee [CECCAPP], Lyon, France).
Two- to 3-day-old newborn NSG mice were subjected to 1 Gy irradiation, and 2 × 105 transduced prestimulated CB CD34+ cells were injected intrahepatically. Unstimulated freshly isolated CD34+ cells were transduced for 24 hours, and 3 × 105 transduced CD34+ cells were injected into NSG mice as above. After 12 weeks of reconstitution, extensive cell phenotyping in the different hematopoietic tissues by flow cytometry (BM, spleen, thymus, blood) of these mice was performed (see supplemental Methods).
For secondary engraftments, the CD34+ cells were isolated from the BM of 1 primary engrafted mouse as above, and 4 × 105 to 5 × 105 isolated CD34+ cells were injected into 1 newborn NSG mouse following the same procedure and phenotypic analysis as above. The CD34+ cells isolated from different primary recipients were thus not pooled for injection into secondary recipient mice.
DNA extraction, quantitative polymerase chain reaction, integration site, and FACS analysis
Details are provided in supplemental Methods.
Results
H/F-LVs outperform VSV-G-LVs for the transduction of immature hCD34+ cells upon mild cytokine prestimulation
Because the H/F-LVs developed in our laboratory were successful in transducing multiple other hematopoietic cells, we tested them for early progenitor hCD34+ cell transduction. We prestimulated hCD34+ cells with a cytokine cocktail and transduced them with VSV-G-LV and H/F-LV–encoding GFP at the indicated MOI in the presence or absence of the transduction-facilitating agent retronectin (Figure 1A). The H/F-LVs outperformed the VSV-G-LV pseudotypes for the transduction of hCD34+ cells in the presence of retronectin (Figure 1A). Importantly, under these conditions, up to 100% of the hCD34+ cells were transduced by a single exposure to the H/F-LVs. Transduction of stimulated CD34+ cells with equivalent amounts of physical particles (HIV-p24) confirmed superior transduction by H/F-LVs (supplemental Figure 1).
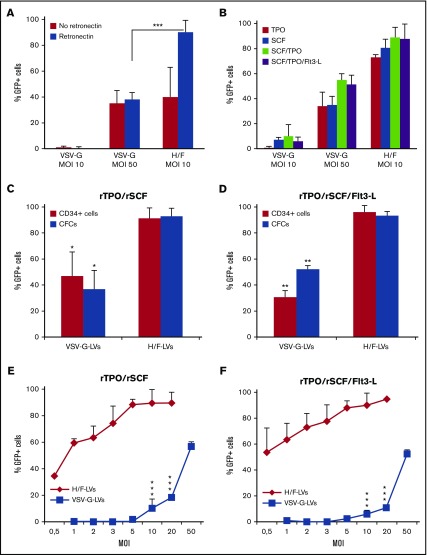
Figure 1.
H/F-LVs allow close to 100% transduction of prestimulated hCD34+cells. (A) hCD34+ cells were prestimulated with a cytokine cocktail (TPO, SCF, Flt3-L [Flt3-L]) and transduced with H/F-LVs or VSV-G-LVs at indicated MOIs in the presence or absence of retronectin. Six days after transduction, the cells were analyzed for the percentage of GFP+CD34+ cells by FACS (means ± standard deviation [SD]; n = 6). (B) hCD34+ cells were prestimulated for 18 hours as follows: SCF or TPO alone, SCF + TPO, or SCF + TPO + Flt3-L. Subsequently, they were transduced with H/F-LVs or VSV-G-LVs at indicated MOIs in the presence of retronectin. Six days after transduction, the cells were evaluated for the percentage of hCD34+GFP+ cells by FACS (mean ± SD; n = 5). (C-D) Comparison between the transduction level of CD34+ cells and the myeloid colonies derived from these cells (GFP+ CFCs) for CD34+ cells prestimulated with (C) SCF + TPO or (D) SCF + TPO + Flt3-L (mean ± SD; n = 3). (E-F) An MOI of 10 for H/F-LVs and an MOI of 50 for VSV-G-LVs was used. Subsequently, they were transduced with H/F-LVs or VSV-G-LVs at increasing MOIs in the presence of retronectin. Six days after transduction, the cells were analyzed for the percentage of hCD34+GFP+ cells by FACS (means ± SD; n = 3). Statistical analysis for comparison of VSV-G-LV vs H/F-LVs was performed by using the paired Student t test. *P < .05; **P < .01; ***P < .001.
Strong stimulation with a cytokine cocktail can induce HSC differentiation, loss of self-renewal, and homing capacity. Thus, the less the cells are prestimulated, the more they keep their stem cell characteristics.13,14,37,38,41 Therefore, we performed LV transductions of hCD34+ cells by using different cytokine prestimulation conditions (Figure 1B). Remarkably, stimulation with TPO or SCF was sufficient to transduce hCD34+ cells up to 75% (TPO) and 90% (SCF) using H/F-LVs, whereas VSV-G-LVs reached maximum transduction levels of 10% at an MOI of 10. TPO + SCF or TPO + SCF + Flt3-L prestimulation and a single hit with H/F-LVs resulted in up to 100% hCD34+ cell transduction, whereas VSV-G-LVs at same MOI achieved 10% transduction on average. Even when high doses of VSV-G-LVs were used, H/F-LV gene transfer was still significantly higher (Figure 1B).
For both recombinant (r) SCF + rTPO and rSCF + rTPO + rFlt3-L stimulations, stable transduction was demonstrated by differentiation of the transduced hCD34+ cells into myeloid lineages (Figure 1C-D). H/F-LVs transduced significantly higher levels of hCD34+ cells at lower vector doses than the VSV-G-LVs. They reached 90% transduction for both cytokine stimulation conditions at an MOI of 5, whereas VSV-G-LVs did not even reach 5% transduction (Figure 1E-F). Only at MOI 50 would the VSV-G-LVs allow efficient hCD34+ cell transduction rates (50% to 60%), equivalent to the performance of H/F-LVs at an MOI of 1 (Figure 1E-F). As expected, increasing vector doses coincided with increasing vector copy number per cell (supplemental Figure 2).
Mobilized peripheral blood (mPB) hCD34+ cells are readily accessible target cells for gene therapy. Importantly, H/F-LVs allowed >95% stable transduction of mPB hCD34+ cells (supplemental Figure 3).
Because MV can infect hematopoietic cells of nonhuman primates (an important preclinical animal model), we confirmed that macaque CD34+ cells were efficiently transduced by simian immunodeficiency virus–derived H/F-LVs (supplemental Figure 4). To summarize, H/F-LVs outperformed VSV-G-LVs by far for gene transfer in hCD34+ cells, even when using low stimulation combined with low vector doses.
H/F-LV pseudotypes permit high-level transduction of resting hCD34+ cells
We then hypothesized that H/F-LV binding and signaling through the MV receptors might be sufficient to transduce unstimulated hCD34+ cells.18,19 Moreover, omitting cytokine stimulation might avoid differentiation and loss of the HSCs. Importantly, the H/F-LVs were able to efficiently transduce unstimulated hCD34+ cells at levels not reported before (40% to 70%) after a single exposure at low vector doses (MOI, 10; Figure 2A). As we previously described,12,42,43 the VSV-G-LVs were unable to transduce efficiently resting hCD34+ cells (Figure 2A). In contrast to cytokine-prestimulated hCD34+ cells, their resting counterparts require at least an MOI of 10 for efficient H/F-LV transduction (Figure 1E-F vs Figure 2B). We continued these liquid cultures after 3 days of transduction for 10 days in the presence of cytokine with and without RT inhibitor to confirm that there was no rescue of transduction as a result of virus bound at the surface (supplemental Figure 5).
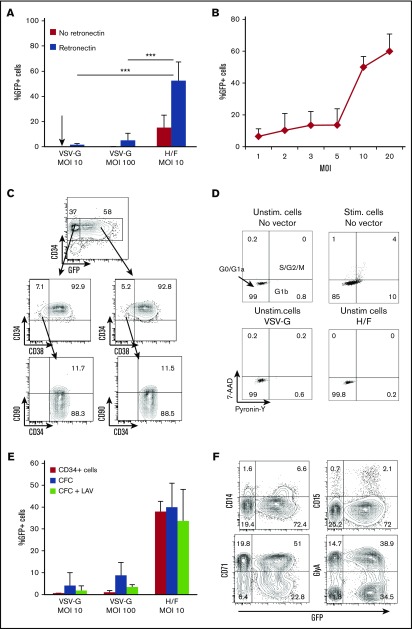
Figure 2.
H/F-LVs allow high-level transduction of unstimulated CD34+cells without altering their immature phenotype. (A) Freshly isolated hCD34+ cells were transduced with H/F-LVs or VSV-G-LVs at indicated MOIs in the presence or absence of retronectin. No cytokines were added. Three days after transduction, the cells were evaluated for the percentage of GFP+hCD34+ cells by FACS (mean ± SD; n = 6). (B) Unstimulated hCD34+ cells were transduced with H/F-LVs at increasing MOIs in the presence of retronectin. Three days after transduction, the cells were analyzed for the percentage of GFP+hCD34+ cells by FACS (mean ± SD; n = 6). (C) Surface marking of very immature progenitors (CD34+CD38–CD90+) for the transduced (GFP+) and untransduced (GFP–) cells. The arrows indicate the gated areas. (D) Cell cycle progression was monitored by simultaneously visualizing the RNA (Pyronine-Y) and DNA (7-AAD) content of the hCD34+ cells 3 days after transduction. The percentages of cells in the G0/G1a, G1b, and S/G2/M phase of the cell cycle are indicated in the dot blots. (E) Comparison between the percentage of GFP+ unstimulated (Unstim) CD34+ cells and the percentage of GFP+ myeloid colonies (CFCs) derived from these transduced CD34+ cells. In parallel, the CFC assay was performed in the presence of the RT inhibitor lamivudine (LAV) to exclude rescue of transduction resulting from H/F-LV bound to the cell surface upon cytokine stimulation (mean ± SD; n = 4). (F) Surface staining for the different transduced myeloid colonies derived from unstimulated transduced hCD34+ cells (GlyA for erythrocytes; CD15 and CD14 for granulocytes, monocytes, and macrophages; CD71 for immature erythrocyte progenitors). Data in panels C, D, and F are representative of 3 independent experiments. Statistical analysis for comparison of VSV-G-LVs vs H/F-LVs was performed by using a paired Student t test. ***P < .001. Stim, stimulated.
Upon H/F-LV transduction of resting hCD34+ cells, the most immature population (CD34+CD38–CD90+) enriched in HSCs was efficiently transduced, and no skewing of this subpopulation was induced (Figure 2C).
Because efficient transduction of hCD34+ cells with classical VSV-G-LVs requires cytokine prestimulation12 leading to cell cycle entry (Figure 2D) and probably differentiation, we wanted to exclude that H/F-LV binding to the MV receptors induced hCD34+ cell cycle entry. Although H/F-LVs resulted in high-level hCD34+ cell transduction (Figure 2A), these cells remained in their resting state (Figure 2D). This is essential because the majority of the hCD34+ cells with SCID repopulating potential reside in the G0 phase of the cell cycle.44,45 The vector copy number determined in G0 CD34+ cells transduced with H/F-LVs (MOI, 10) was as high as 5 copies per cell (supplemental Figure 2).
Notably, MV infections are characterized by a generalized immunosuppression. Transient lymphopenia is a hallmark of MV-induced immunosuppression. Reports on the effect of MV on BM cells in vivo and its consequence in hematopoiesis leading to immunosuppression are still contradictory.31,33 Since Boussaad et al33 reported a suppressing effect of MV on the development of hCD34+-derived short-term progenitors, we demonstrated that the H/F-LV hCD34+ gene transfer levels persisted in their derived myeloid colonies (CFCs) (Figure 2E). To exclude transduction rescue resulting from cytokine stimulation, the CFC assay was performed in the presence of an RT inhibitor (Figure 2E). Equivalent CFC numbers for H/F-LV transduced (152 ± 20; n = 4) and untransduced (160 ± 26; n = 4) cells were obtained, and the different myeloid lineages were all transduced to the same extent (Figure 2F). Thus, H/F-LVs allowed, for the first time, very high-level transduction of resting hCD34+ cells without skewing this cell population.
High-level gene transfer in CD34+ cells is the result of H/F-LV binding to the CD46 receptor
It was important to elucidate the roles of the MV receptors (CD46, SLAM, and nectin-4) in the H/F-LV transduction process of these primary gene therapy target cells. Therefore, the expression levels of the MV receptors were quantified by FACS analysis on hCD34+ cell surface (Figure 3A). CD46 was expressed on the vast majority of cytokine-stimulated and resting hCD34+ cells (98% and 99%, respectively). Of note, CD46 expression levels increased on cytokine-stimulated as compared with resting hCD34+ cells, marked by a twofold increase in mean fluorescent intensity (Figure 3A). In contrast, SLAM was almost undetectable at the surface of quiescent and stimulated hCD34+ cells as reported,31,33 which confirmed that SLAM, a valid surface marker for murine HSCs is not applicable for hHSCs (Figure 3A and Larochelle et al30). Nectin-4 was also not detected on HSCs.25 A gradual upregulation of CD46 density was detected at the hCD34+ cell surface at different time points of early-acting cytokine stimulation (Figure 3B). To establish a link between CD46 expression and hCD34+ cell transduction, resting and stimulated hCD34+ cells were sorted for high and low CD46 surface density and subsequently transduced with H/F-LVs. The lowest transduction levels perfectly coincided with the lowest CD46 surface density on these cells (Figure 3C). Antibody-mediated blocking of CD46 on stimulated and quiescent hCD34+ cells completely abrogated H/F-LV transduction, whereas blocking the SLAM or nectin-4 receptors had no significant effect (Figure 3D-E).
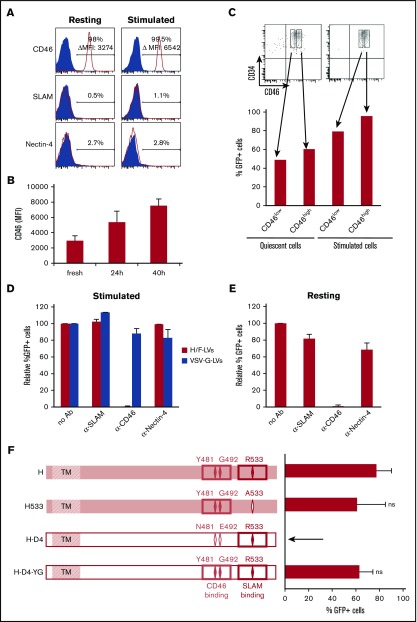
Figure 3.
H/F-LVs exclusively use the CD46 receptor for high-level transduction of stimulated and quiescent hCD34+cells. (A) Surface staining for the measles receptors (SLAM, CD46, and nectin-4) on resting or cytokine (TPO + SCF + Flt3-L) prestimulated hCD34+ cells. (B) Quantification of CD46 surface expression mean fluorescence intensity (MFI) of hCD34+ cells at different time points of cytokine stimulation (TPO + SCF + Flt3-L). (C) Prestimulated and resting CD34+ cells were stained for CD46, and each was sorted for low and high CD46 expression. Obtained subpopulations were transduced with H/F-LVs in the presence of retronectin, and transduction efficiency was analyzed by FACS 3 days later. (D) Prestimulated and (E) resting hCD34+ cells were transduced with H/F-LVs (MOI, 10) in the absence or presence of anti-CD46, anti-SLAM, or anti-nectin-4 blocking antibodies (Ab’s). Three days after transduction, GFP expression in these cells was determined by FACS. Transduction levels are presented as GFP expression relative to the H/F-LV transduction in the absence of antibody, set to 100% (mean ± SD; n = 3). (F) Schematic representation of the different MV hemagglutinin gp’s (Hgp’s). (H) The MV vaccinal Edmonston strain Hgp contains binding residues for both SLAM and CD46. CD46-tropic mutant Hgp was engineered by mutating 1 residue of Edmonston Hgp responsible for binding/fusion through SLAM (R533A; H533). SLAM-tropic Hgp was derived from the clinical strain D4 (H-D4). H-D4-YG was obtained by introducing CD46 binding residues (481Y and 492G) into the H-D4 envelope. Prestimulated hCD34+ cells were transduced with H/F-LVs, H-D4/F-LVs, H533/F-LVs, or H-D4-YG/F-LVs, and GFP expression was analyzed by FACS 6 days later (mean ± SD; n = 4). ns, not significantly different for H533 and HD4-YG compared with H. Statistical analysis was performed by using paired Student t test.
To further study the roles of SLAM and CD46 in H/F-LV transduction of hCD34+ cells, we used a series of LVs pseudotyped with (1) Hgp that contained both binding residues to SLAM and CD46 (H)19,35; (2) CD46-tropic H mutant in which 1 residue (R533A) of Edmonston-Hgp responsible for binding/fusion through SLAM was ablated (H533)34; (3) SLAM-tropic Hgp’s derived from clinical strains (H-D4)46-48; and (4) SLAM/CD46 binding mutant engineered by adding the mutation N481Y (responsible for CD46 binding) into the clinical strain H-D4. The mutation E492G was also included because it has been shown to stabilize CD46 binding (H-D4-YG49,50; Figure 3F).
H533/F-LVs, which do not recognize SLAM receptor, were capable of transducing cytokine-prestimulated hCD34+ cells, whereas H-D4/F-LVs, which do not recognize the CD46 receptor, were refractory for transduction of these cells. Introducing the CD46-binding sites in HD4-YG/F-LVs restored transduction. These results showed that MV receptor CD46 was essential in H/F-LV entry into hCD34+ cells and that the level of transduction was highly correlated with CD46 surface density.
Comparison of H/F-LV integration site distribution in activated and resting CD34+ cells
One important issue was to verify whether the genomic accessibility for H/F-LV proviral integration differed between resting and stimulated hCD34+ cells. To dissect the potential differences in integration site (IS) distribution between cytokine-stimulated and resting CD34+ cells, we performed a comparative LV IS study by using linear-amplification mediated polymerase chain reaction followed by high-throughput sequencing (supplemental Table 1). LV integrome analysis in H/F-LV transduction of unstimulated (G0) and stimulated (Stim) CD34+ cells retrieved 6631 and 2930 unique ISs, respectively (supplemental Table 1). Integration frequencies were analyzed for a set of genomic features such as transcriptional start sites, CpG islands, intragenic regions, and DNAse I cleavage-sensitive sites and compared them with a set of matched random integration sites (Figure 4; supplemental Table 1).
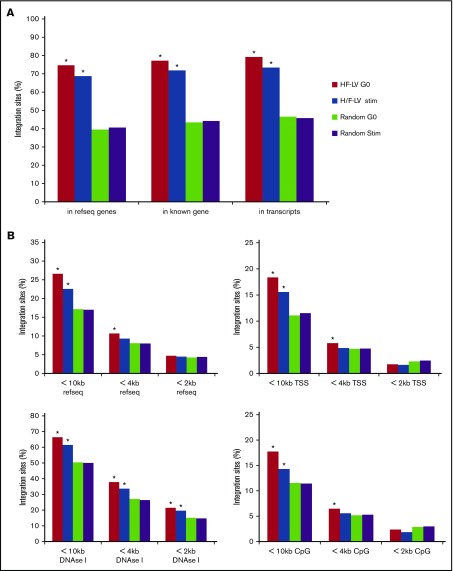
Figure 4.
H/F-LV IS distribution in hCD34+cells. Unstimulated (G0) or cytokine-stimulated CD34 cells (Stim) were transduced with H/F-LVs for 24 hours, washed, and kept in culture for 10 days and were processed for LV genomic IS analysis. (A) The percentage of lentiviral ISs located in RefSeq genes, in known genes, and in transcribed genes. Statistical tests have been performed by comparing the experimental data to the in silico–generated random IS data set (in gray). (B) The percentage of lentiviral ISs located in RefSeq genes, transcriptional start sites, DNAse I–sensitive sites, or CpG-dense regions, including the surrounding 2-kb, 4-kb, or 10-kb surrounding regions are shown. Statistical tests were performed by comparing the experimental data with the in silico–generated random IS data set (in gray). The asterisks indicate the statistically significant differences between the unstimulated (G0) or cytokine-stimulated CD34+ cell conditions and the corresponding random data set. *P < .001. TSS, transcriptional start site.
In both H/F-LV transduced unstimulated and stimulated CD34+ cells, integration was favored within genes and transcriptionally active units as called by the RefSeq and UniGene databases, in accordance with previous studies2,4,51 (Figure 4A; supplemental Figure 6). Much lower frequencies of ISs to gene 5′ ends (<10 kb distance to transcriptional start sites) and intragenic regions (<10 kb RefSeq genes) were detected (Figure 4B). Integration was enriched near sites of DNAse I cleavage and CpG islands (Figure 4B). This reflects the fact that over long intervals, DNAse I sites and CpG islands can be considered surrogate markers for gene-rich regions, favored for retroviral integration, whereas over shorter intervals, these markers are enriched near gene promoters, disfavored for VSV-G-LV integration. Of note, for unstimulated CD34+ cells, integration frequency within gene-rich regions was increased compared with cytokine-activated CD34+ cells (supplemental Figure 6). Interestingly, we found a significantly higher number of ISs for unstimulated CD34+ cells in chromosomes 17 and 19 (supplemental Figure 7). Overall, we can conclude that H/F-LV integration in the unstimulated and cytokine-activated CD34+ cells resembles the classical LV integration pattern.3,4,51-53
H/F-LVs promote high-level transduction of prestimulated and resting HSCs
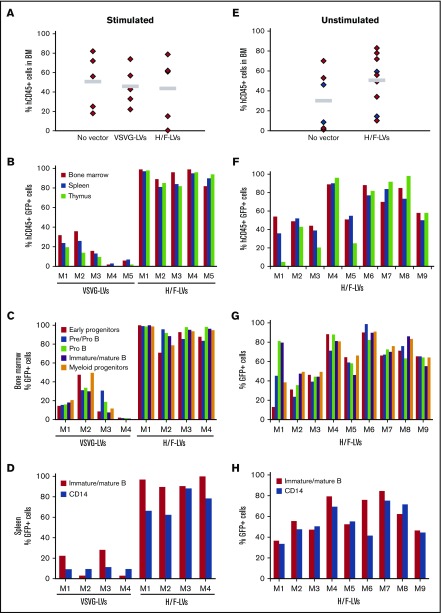
To evaluate the long-term reconstitution capacity of H/F-LV–transduced hCD34+ cells in vivo, we transplanted prestimulated hCD34+ cells transduced with either H/F-LVs or VSV-G-LVs into NSG mice. A comparable level of hCD45+ cell engraftment was detected for LV transduced and untransduced hCD34+ cells (Figure 5A), indicating that their migration and engraftment were not impaired.
Figure 5.
High-level hCD34+-cell transduction by H/F-LV is maintained in myeloid and lymphoid lineages in primary NSG recipient mice. hCD34+ cells were prestimulated with a cytokine cocktail (TPO + SCF + Flt3-L) for 16 hours (A-D) or not (E-H) and transduced with H/F-LVs (MOI, 10) or VSV-G-LVs (MOI, 50) in the presence of retronectin for 36 hours. Subsequently, the cells (2 × 105) were injected into the liver of irradiated newborn NOD/SCID γc−/− mice (NSG mice). Upon reconstitution for 12 weeks, the BM, spleen, and thymus of these primary engrafted mice were analyzed. (A,E) Total human cell engraftment analyzed by anti-hCD45 staining using FACS; no significant differences in reconstitutions with H/F-LVs vs VSV-G-LVs and vs untransduced were detected (paired Student t test). Gray line indicates mean % hCD45 reconstitution. (B,F) The percentage of transduced human cells (GFP+hCD45+ cells) was analyzed by FACS in the different hematopoietic tissues (BM, spleen, and thymus). (C,G) Transduction levels of the different hematopoietic cell lineages in the BM of NSG reconstituted mice. The percentage of GFP+ immature early progenitor cells (CD34+CD19–CD10–), pre/pro B cells (CD34+CD19+CD10–), pro B cells (CD34+CD19+CD10+), immature and mature B cells (CD34–CD19+CD10+CD20+), and myeloid progenitors (CD13+) are shown. (D,H) Transduction levels of different cell lineages in the spleen of NSG reconstituted mice. The percentages of GFP+ mature B cells (CD34–CD19+CD20+), monocytes, and granulocytes (CD14+) are shown. M1 to M9 represent individual reconstituted NSG mice for independent transductions with the indicated vectors.
Overall, VSV-G-LVs allowed a much lower transduction of SCID repopulating cells compared with H/F-LVs. H/F-LV–transduced hCD34+ cells resulted in the highest transduction levels of engrafted human cells in the BM (up to 100% hCD45+GFP+ cells). These high transduction levels were sustained in the primary recipient mice in all hematopoietic tissues (Figure 5B). Analysis of the BM revealed equivalent high-level H/F-LV transduction for different B-cell subpopulations, myeloid progenitors, monocytes, and early progenitors, but this was not the case for VSV-G-LVs (Figure 5C). Consistent with these results, splenocytes and thymocytes revealed a similar H/F-LV–transduction profile (Figure 5D; supplemental Figure 8).
To evaluate whether H/F-LVs transduced long-term reconstituting HSCs in the unstimulated hCD34+ cell population, we applied a short transduction protocol (18-24 hours) at low vector doses (MOI, 10) without cytokine stimulation before injection into NSG mice. High-level reconstitution with hCD45+ cells was detected for H/F-LV–transduced G0 hCD34+ cells (Figure 5E). Interestingly, we identified 1 low-level transduction group (5 of 9 mice) and 1 with high (>80%) transduction efficiencies (4 of 9 mice; Figure 5F). In contrast, for VSV-G-LV–transduced G0 hCD34+ cells, low transduction levels were obtained (supplemental Table 2). For both groups, BM revealed equivalent high-level H/F-LV transduction for the different B-cell subpopulations, myeloid progenitors, monocytes, and importantly, early progenitors (Figure 5G). Splenocytes showed a similar transduction profile (Figure 5H). In agreement, the vector copy number per cell was low for VSV-G LVs compared with H/F-LVs (supplemental Table 2).
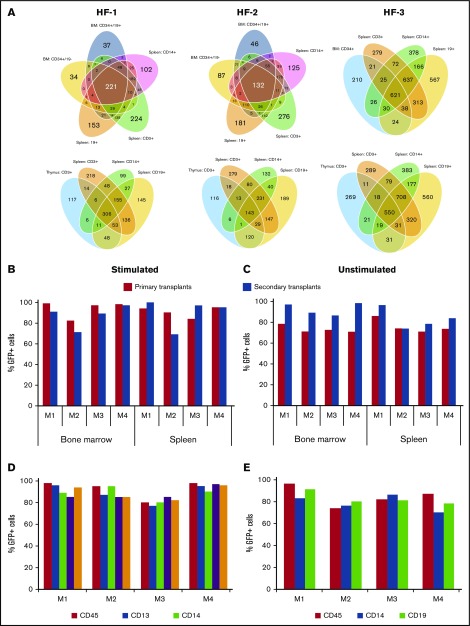
To confirm that true HSCs were gene modified by H/F-LVs in the resting CD34+ cells, we transduced these cells (18-24 hours) with H/F-LVs encoding for a 32-bp-long barcode library54 at low vector doses (MOI, 5) without cytokine stimulation before injection into NSG mice. After the mice had been reconstituted for 4 months, we isolated different hematopoietic lineages in the BM, spleen, and thymocytes and identified the barcodes present in the different lineages (Figure 6A). For 3 independent H/F-LV preparations in reconstituted mice, we identified 221, 132, and 621 common barcodes among 5 and 4 different lineages, respectively, in the BM (Figure 6A, upper diagrams) and 306, 143, and 550 common barcodes among 4 different lineages in the spleen (Figure 6A, lower diagrams). This indicates that HSCs were transduced. To further reinforce these results, secondary reconstitutions of NSG mice were performed with hCD34+ cells isolated from BM from primary recipients (Figure 6B-E). These BM hCD34+ cells were cultured in SCF and IL-6 before injection to maintain their homing capacity.55 At 12 weeks after transplantation, the percentage of CD45+GFP+ cells in the spleen and BM was maintained or even increased in the secondary recipients (Figure 6B-C). Indeed, for resting CD34+ cells, the percentage of GFP+hCD45+ cells in the BM was increased in secondary recipient mice (4 of 4) compared with primary recipient mice (Figure 6C). Importantly, high and equivalent levels of CD34+GFP+, lymphoid, and myeloid cells were detected in the BM of the secondary recipient mice (Figure 6D-E). We concluded that H/F-LVs allowed high-level genetic modification of resting hHSCs without cell cycle entry resulting in long-term engraftment and differentiation into all the different cell lineages in vivo.
Figure 6.
H/F-LVs transduce long-term reconstituting HSCs at levels approaching 100%. (A) Unstimulated hCD34+ cells were transduced with different H/F-LV preparations (H/F-1, H/F-2, H/F-3) encoding for a library of barcodes (32 bp long) and were injected into the livers of irradiated newborn NSG mice. Upon reconstitution, the bone marrow and spleen of these primary engrafted mice were analyzed for GFP+CD45+ cells in the BM and the spleen. An MOI of 5 was chosen. Upon 12 weeks of reconstitution, mice were euthanized and different populations from the BM (CD34+CD19– early progenitors, CD34+CD19+ B-cell progenitors, CD13+ myeloid progenitors), spleen (CD19+, CD3+, CD14+ cells), and CD3+ thymocytes were isolated. The integrated bar codes in each of these subpopulations were determined and the shared bar codes between the different lineages are indicated in a Venn diagram. (B-E) Transduced hCD34+ cells were injected into the liver of irradiated newborn NSG mice. Upon reconstitution, the bone marrow and spleen of these primary engrafted mice (first transplantation) were analyzed for GFP+CD45+ cells in the BM and the spleen. Subsequently, the hCD34+ cells, isolated from the BM of these mice, were injected (following the same procedure as for the primary NSG engraftments) into secondary recipient mice. At 12 weeks from engraftment, these secondary NSG mice were analyzed for the percentage of transduced human cells (GFP+hCD45+) in the BM and spleen. Analysis of engraftment of transduced human cells (GFP+hCD45+) in the BM and spleen of primary (black bars) and secondary (white bars) transplanted mice reconstituted with hCD34+ cells (B) prestimulated with a cytokine cocktail (TPO + SCF + Flt3-L) or (C) unstimulated and transduced with H/F-LVs. Detailed phenotypic analysis of transduced human cells in the BM of secondary transplanted mice reconstituted with (D) prestimulated or (E) unstimulated H/F-LV transduced hCD34+ cells. Analysis of transduced human cells (GFP+) in the myeloid progenitors (CD13+), monocytes (CD14+), progenitor cells (CD34+), and B cells (CD19+) in secondary recipient mice.
H/F-LVs outperform VSV-G-LVs for correction of hCD34+ cells in unfractionated BM and from FA patients
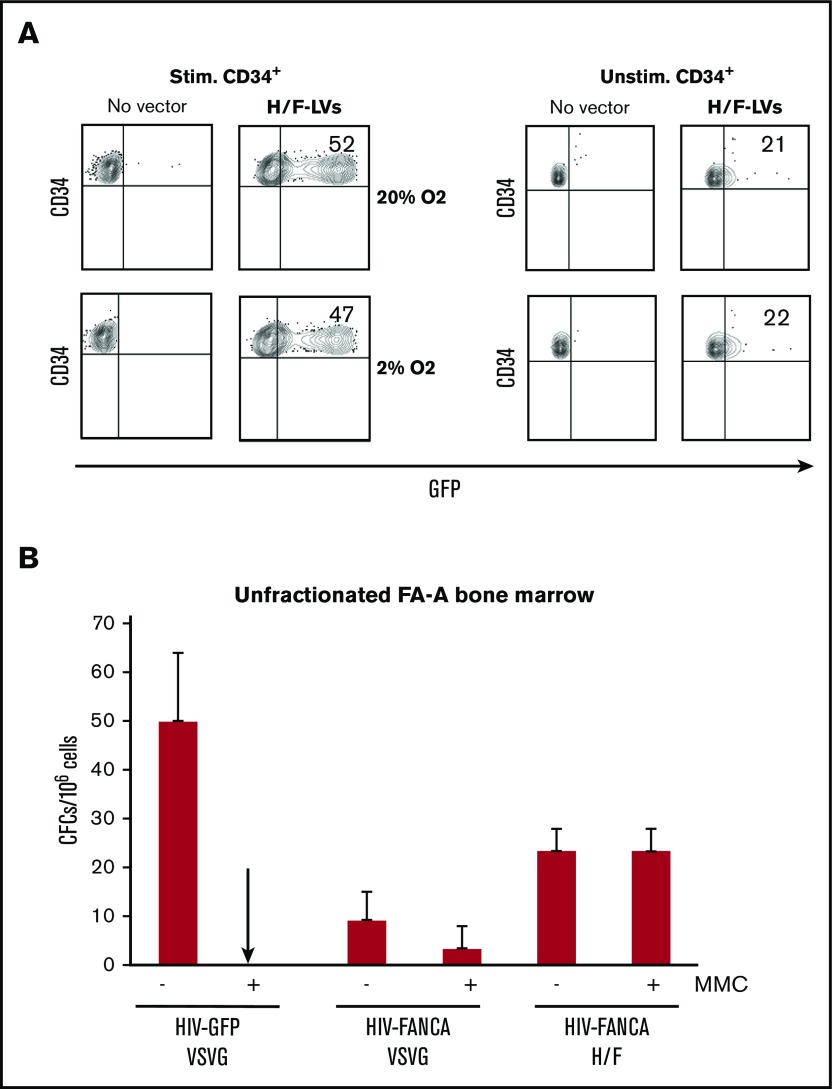
FA is a rare genetic syndrome characterized by BM failure. Gene therapy by infusion of FA-corrected autologous HSCs may offer a potential cure (reviewed in Tolar et al56). However, collecting hCD34+ cells in FA patients is especially challenging because of the reduced numbers of hCD34+ cells present in their BM or mPB.39,57,58 In addition, the FA genetic defect makes the HSCs more fragile.15,59 Therefore, it would be beneficial to transduce hCD34+ cells directly in unfractionated FA BM aspirates. To preserve FA HSCs, they are currently transduced under low oxygen conditions before being reinfused into patients.39 In view of a potential clinical application for FA, we first confirmed that H/F-LV transduction for both cytokine-stimulated and unstimulated healthy hCD34+ progenitors was equivalent at 20% and 2% oxygen (Figure 7A).
Figure 7.
Efficient correction of hematopoietic progenitors in total BM from FA-A patients with H/F-LVs encoding for FANCA. (A) Cytokine prestimulated hCD34+ cells were transduced with GFP encoding H/F-LVs at an MOI of 5 in 20% or 2% oxygen culture conditions. Three days after transduction, the percentage of GFP+ cells was determined by FACS. Data are representative of 3 independent experiments. (B) Cells from an unfractionated FA-A bone marrow sample (n = 3) were prestimulated for 18 hours and transduced with the GFP- or FANCA-encoding LVs as indicated in the presence of retronectin at an MOI of 5 and in low oxygen conditions (5%). CFC assays were performed 24 hours later in the absence or presence of mytomycin C (MMC).
Next, we evaluated whether an H/F-LV encoding the FANCA gene would allow correction of FA CD34+ cells under low oxygen conditions in unfractionated BM from FA-A patients with two LVs: 1 encoding EGFP (HIV-GFP) and 1 coding for the correcting FANCA complementary DNA (HIV-FANCA), both under the control of a PGK promoter. HIV-FANCA vector design is identical to that of the vector used in an ongoing FA clinical trial.7 Importantly, FA cells that regain FANCA expression become resistant to mitomycin C, a classical DNA interstrand cross-linker that is highly toxic for FA cells. Remarkably, in the case of correction of total BM with H/F-HIV-FANCA, 100% of the colonies were corrected, whereas only 25% of the colonies for the VSV-G-FANCA-LV group survived the mitomycin C treatment (Figure 7B). Thus, a low dose of H/F-LVs conferred therapeutic FANCA gene transfer and corrected unfractionated BM progenitor cells from FA patients under low oxygen conditions bypassing the inherent problems of FA hCD34+ cell isolation and long-term culture. This procedure would facilitate the infusion of high numbers of corrected functional HSCs in FA patients and is a very significant step forward in the treatment of this disease.
Discussion
Highly efficient gene delivery without compromising HSC function is the quintessential goal in HSC-based gene therapy. Here we show that H/F-LVs transduce HSCs not exposed to any exogenous stimulation to an extent not reported before (40% to 70% transduction). These results were strengthened by the conservation of high transduction levels in all hematopoietic lineages of primary NSG recipients. Remarkable enhancement of transduction was shown in the whole hematopoietic lineage in 100% of the secondary NSG recipients, indicating that true HSCs were genetically altered. This means that the H/F-LVs overcome an important barrier for clinical gene therapy using VSV-G-LVs, which demonstrate limited efficiency in particular in resting HSCs.12,35,60,61 It is essential to transduce HSCs without changing their capacity for self-renewal, homing, and differentiation, which is supported by minimal or no stimulation of these targets. Moreover, cytokine stimulation induces HSCs to exit their quiescent state accompanied by induction of DNA damage in these cells.15 In addition, culture of HSCs in cytokine cocktails required for VSV-G-LV transduction leads to loss of engraftment potential. Thus, H/F-LV mediates high transduction efficiency without compromising HSC engraftment, homing, and differentiation capacity.
Wang and colleagues61 showed that rapamycin strongly facilitated efficient VSV-G-LV transduction in prestimulated hCD34+ cells that reached 100% and enhanced marking frequency of long-term engrafting SCID repopulating cells. However, the addition of rapamycin during VSV-G-LV transduction of resting hCD34+ cells did not significantly increase transduction.61,62 This is in accordance with the absence of the VSV receptor low-density lipoprotein on resting hCD34+ cells.12 The authors suggest that rapamycin leads to increased levels of LV cytoplasmic uptake, reverse transcription, and genomic integration. Thus, it is tempting to speculate that H/F-LV transduction of resting hCD34+ cells might still be increased by the addition of rapamycin. However, caution is warranted because of the known cell cycle blockage this drug induces.61
We identified CD46 as the MV receptor that allows this spectacular H/F-LV transduction efficiency for as well as quiescent as cytokine-stimulated HSCs. This is reinforced by the fact that retronectin increased H/F-LV transduction of resting CD34+ cells by tightening Hgp-CD46 binding. The mechanism that explains this high-level H/F-LV entry and productive transduction needs to be investigated, but for T and B cells, we might speculate that CD46 binding induces an alternative cell entry (eg, macropinocytosis).21
During engraftment, transplanted HSCs home to the BM niche, where they initiate both differentiation and self-renewal. There, they reside in a hypoxic niche, where they acquire dormancy and maintain stemness.63,64 Therefore, it might be crucial to not overexpose the hCD34+ HSCs to cytokines, which are currently required for high-level transduction with VSV-G-LVs.13,14 Indeed, recent clinical trials1-4 still use strong cytokine cocktails, although this can affect the integrity of the HSC pool.65-67 Importantly, compared with hCD34+ cells in G0, cells in G1 or SG2M phases of the cell cycle demonstrated diminished engraftment.44,66,68,69 Very recently we demonstrated that a new LV pseudotype carrying the Baboon envelope gp’s at their surface (BaEV-LVs) outperformed VSV-G-LVs for hHSC transduction.43 BaEV-LVs and H/F-LVs manage to transduce resting hHSCs up to 30% and 70%, respectively. H/F-LVs achieved transduction levels never before reached in these resting targets. This will be of significance in the clinic. Thus, H/F-LVs improve resting HSC transduction and conserve HSC potential, a combination needed for treating diseases, in which gene-corrected repopulating cells have no selective advantage (eg, hemoglobinopathies). FA patients would in particular benefit from resting hCD34+ cell transduction. Indeed, the FA genetic defect causes fragility of the hCD34+ cells, which are strongly reduced in numbers in patients and difficult to isolate.7,39,70 Cytokine stimulation and culture can induce increased FA hCD34+ cell apoptosis.15 Therefore, minimizing hCD34+ cell cytokine stimulation and manipulation, combined with efficient gene transfer would be invaluable for FA gene therapy.7,39,71 We have demonstrated here that H/F-LVs target gene transfer to hCD34+ cells in unfractionated BM of FA patients, which would clearly benefit FA gene therapy trials. It is expected that consistent with data obtained from mosaic FA patients,72-74 genetically corrected HSCs will develop a marked proliferation advantage over uncorrected cells.72 Clinical use of H/F-LVs for total BM transduction seems close because Ozog et al75 reported H/F-LV titer improvement using 293T knockout for the CD46 receptor.
In both unstimulated and stimulated hCD34+ cells, integrations were favored in transcriptionally active and gene-dense genomic regions. The classical LV integration pattern, which is generally accepted to be safer than that of murine leukemia virus–based vectors in hCD34+ cells, is conserved in H/F-LV transduction in the absence of cytokine stimulation (G0 CD34+ cells).3,4,51-53,76 A recent study determined the VSV-G-LV IS pattern in thymidine-treated CD34+ cells inducing a blockage into the G0/G1a phase of the cell cycle.77 Of interest, they revealed ISs primarily in RefSeq genes, in agreement with our results.
Taken together, the efficient transduction of resting HSCs by H/F-LVs has major translational implications for more complex diseases such as FA and for many other diseases. These H/F-LVs have the potential to significantly improve clinical efficacy of gene delivery to HSCs, which allows the use of low vector doses, short exposure, and omission of cytokine stimulation while maintaining high transduction efficiency. Therefore, H/F-LVs could pave the way to future research in hematopoiesis and clinical applications still out of reach today.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the staff of the animal care facility (Plateau de Biologie Expérimentale de la Souris) at the Ecole Normale Supérieure de Lyon and the flow cytometry and vectorology platform (UMS3444/US8, Lyon, France).
This work was supported by grants from Agence Français dystrophie musculaire, Agence nationale de recherche sur les sida, Agence de recherche sur le cancer, and the European Community (FP7-HEALTH-2007-B/222878 “PERSIST”), ERA-Net for Research Programmes on Rare Diseases E-RARE grant “GETHERTHALPLUS,” and Lentigen Inc Miltenyi.
Authorship
Contribution: E.V. coordinated the project, designed and performed experiments, analyzed and discussed the data, and wrote the manuscript; C.L., F.A., and A. Girard-Gagnepain designed and performed experiments, analyzed the data, and discussed results; C.F., C.C., O.B., A. Gutierrez-Guerrero, and D.N. performed experiments and analyzed data; F.J.R.-R., J.A.B., and P.R. performed the transduction of Fanconi anemia bone marrow cells and analyzed data; K.C. and L.T. provided the bar code technology and performed the bar code identification and bioinformatic analysis in the different lineages; N.T. and I.C. performed experiments and provided critical discussion and technical advice; L.S.V. and R.G. performed integration site analysis and provided critical discussion and technical advice; and F.-L.C. provided critical discussion and reading of the manuscript.
Conflict-of-interest disclosure: P.R. and J.A.B. received funding from Rocket Pharma. J.A.B. is a consultant for Rocket Pharma. The remaining authors declare no competing financial interests.
Correspondence: Els Verhoeyen, International Center for Infectiology Research, Enveloped Viruses, Vectors and Innate Responses, Ecole Normale Supérieure de Lyon, 46 Allée d'Italie, 69364 Lyon Cedex 07, France; e-mail: els.verhoeyen@ens-lyon.fr or els.verhoeyen@unice.fr.
References
- 1.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. . Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326(5954):818-823. [DOI] [PubMed] [Google Scholar]
- 2.Cavazzana-Calvo M, Payen E, Negre O, et al. . Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467(7313):318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffi A, Montini E, Lorioli L, et al. . Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341(6148):1233158. [DOI] [PubMed] [Google Scholar]
- 4.Aiuti A, Biasco L, Scaramuzza S, et al. . Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341(6148):1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton BC, Booth C, Thrasher AJ. Lentivirus technologies for modulation of the immune system. Curr Opin Pharmacol. 2015;24:119-127. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee S, Thrasher AJ. Gene therapy for PIDs: progress, pitfalls and prospects. Gene. 2013;525(2):174-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tolar J, Becker PS, Clapp DW, et al. . Gene therapy for Fanconi anemia: one step closer to the clinic. Hum Gene Ther. 2012;23(2):141-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton RE, Reitsma MJ, Uchida N, Brown PO. Transduction of human progenitor hematopoietic stem cells by human immunodeficiency virus type 1-based vectors is cell cycle dependent. J Virol. 1999;73(5):3649-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Case SS, Price MA, Jordan CT, et al. . Stable transduction of quiescent CD34(+)CD38(-) human hematopoietic cells by HIV-1-based lentiviral vectors. Proc Natl Acad Sci USA. 1999;96(6):2988-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchida N, Sutton RE, Friera AM, et al. . HIV, but not murine leukemia virus, vectors mediate high efficiency gene transfer into freshly isolated G0/G1 human hematopoietic stem cells. Proc Natl Acad Sci USA. 1998;95(20):11939-11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci USA. 2013;110(18):7306-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amirache F, Lévy C, Costa C, et al. . Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123(9):1422-1424. [DOI] [PubMed] [Google Scholar]
- 13.Ahmed F, Ings SJ, Pizzey AR, et al. . Impaired bone marrow homing of cytokine-activated CD34+ cells in the NOD/SCID model. Blood. 2004;103(6):2079-2087. [DOI] [PubMed] [Google Scholar]
- 14.Peled A, Petit I, Kollet O, et al. . Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283(5403):845-848. [DOI] [PubMed] [Google Scholar]
- 15.Walter D, Lier A, Geiselhart A, et al. . Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549-552. [DOI] [PubMed] [Google Scholar]
- 16.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. . Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. . A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348(3):255-256. [DOI] [PubMed] [Google Scholar]
- 18.Frecha C, Costa C, Lévy C, et al. . Efficient and stable transduction of resting B lymphocytes and primary chronic lymphocyte leukemia cells using measles virus gp displaying lentiviral vectors. Blood. 2009;114(15):3173-3180. [DOI] [PubMed] [Google Scholar]
- 19.Frecha C, Costa C, Nègre D, et al. . Stable transduction of quiescent T cells without induction of cycle progression by a novel lentiviral vector pseudotyped with measles virus glycoproteins. Blood. 2008;112(13):4843-4852. [DOI] [PubMed] [Google Scholar]
- 20.Lévy C, Frecha C, Costa C, et al. . Lentiviral vectors and transduction of human cancer B cells. Blood. 2010;116(3):498-500. [DOI] [PubMed] [Google Scholar]
- 21.Frecha C, Lévy C, Costa C, et al. . Measles virus glycoprotein-pseudotyped lentiviral vector-mediated gene transfer into quiescent lymphocytes requires binding to both SLAM and CD46 entry receptors. J Virol. 2011;85(12):5975-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431-455. [DOI] [PubMed] [Google Scholar]
- 23.Aversa G, Carballido J, Punnonen J, et al. . SLAM and its role in T cell activation and Th cell responses. Immunol Cell Biol. 1997;75(2):202-205. [DOI] [PubMed] [Google Scholar]
- 24.Cocks BG, Chang CC, Carballido JM, Yssel H, de Vries JE, Aversa G. A novel receptor involved in T-cell activation. Nature. 1995;376(6537):260-263. [DOI] [PubMed] [Google Scholar]
- 25.Mühlebach MD, Mateo M, Sinn PL, et al. . Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480(7378):530-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Swart RL, Ludlow M, de Witte L, et al. . Predominant infection of CD150+ lymphocytes and dendritic cells during measles virus infection of macaques. PLoS Pathog. 2007;3(11):e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Vries RD, Lemon K, Ludlow M, et al. . In vivo tropism of attenuated and pathogenic measles virus expressing green fluorescent protein in macaques. J Virol. 2010;84(9):4714-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109-1121. [DOI] [PubMed] [Google Scholar]
- 29.Yilmaz OH, Kiel MJ, Morrison SJ. SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood. 2006;107(3):924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larochelle A, Savona M, Wiggins M, et al. . Human and rhesus macaque hematopoietic stem cells cannot be purified based only on SLAM family markers. Blood. 2011;117(5):1550-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manchester M, Smith KA, Eto DS, Perkin HB, Torbett BE. Targeting and hematopoietic suppression of human CD34+ cells by measles virus. J Virol. 2002;76(13):6636-6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sintes J, Romero X, Marin P, Terhorst C, Engel P. Differential expression of CD150 (SLAM) family receptors by human hematopoietic stem and progenitor cells. Exp Hematol. 2008;36(9):1199-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boussaad I, Varagnolo L, Hornich V, et al. . Wild-type measles virus interferes with short-term engraftment of human CD34+ hematopoietic progenitor cells. J Virol. 2011;85(15):7710-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lévy C, Amirache F, Costa C, et al. . Lentiviral vectors displaying modified measles virus gp overcome pre-existing immunity in in vivo-like transduction of human T and B cells. Mol Ther. 2012;20(9):1699-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frecha C, Fusil F, Cosset FL, Verhoeyen E. In vivo gene delivery into hCD34+ cells in a humanized mouse model. Methods Mol Biol. 2011;737:367-390. [DOI] [PubMed] [Google Scholar]
- 36.González-Murillo A, Lozano ML, Alvarez L, et al. . Development of lentiviral vectors with optimized transcriptional activity for the gene therapy of patients with Fanconi anemia. Hum Gene Ther. 2010;21(5):623-630. [DOI] [PubMed] [Google Scholar]
- 37.Verhoeyen E, Wiznerowicz M, Olivier D, et al. . Novel lentiviral vectors displaying “early-acting cytokines” selectively promote survival and transduction of NOD/SCID repopulating human hematopoietic stem cells. Blood. 2005;106(10):3386-3395. [DOI] [PubMed] [Google Scholar]
- 38.Verhoeyen E, Relouzat F, Cambot M, et al. . Stem cell factor-displaying simian immunodeficiency viral vectors together with a low conditioning regimen allow for long-term engraftment of gene-marked autologous hematopoietic stem cells in macaques. Hum Gene Ther. 2012;23(7):754-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacome A, Navarro S, Río P, et al. . Lentiviral-mediated genetic correction of hematopoietic and mesenchymal progenitor cells from Fanconi anemia patients. Mol Ther. 2009;17(6):1083-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornils K, Thielecke L, Hüser S, et al. . Multiplexing clonality: combining RGB marking and genetic barcoding. Nucleic Acids Res. 2014;42(7):e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn PA, Morris JC, Bukovsky AA, et al. . Lentivirus-mediated gene transfer into hematopoietic repopulating cells in baboons. Gene Ther. 2002;9(21):1464-1471. [DOI] [PubMed] [Google Scholar]
- 42.Frecha C, Costa C, Nègre D, et al. . A novel lentiviral vector targets gene transfer into human hematopoietic stem cells in marrow from patients with bone marrow failure syndrome and in vivo in humanized mice. Blood. 2012;119(5):1139-1150. [DOI] [PubMed] [Google Scholar]
- 43.Girard-Gagnepain A, Amirache F, Costa C, et al. . Baboon envelope pseudotyped LVs outperform VSV-G LVs for gene transfer into early-cytokine-stimulated and resting HSCs. Blood. 2014;124(8):1221-1231. [DOI] [PubMed] [Google Scholar]
- 44.Gothot A, van der Loo JC, Clapp DW, Srour EF. Cell cycle-related changes in repopulating capacity of human mobilized peripheral blood CD34(+) cells in non-obese diabetic/severe combined immune-deficient mice. Blood. 1998;92(8):2641-2649. [PubMed] [Google Scholar]
- 45.Passegué E, Wagers AJ. Regulating quiescence: new insights into hematopoietic stem cell biology. Dev Cell. 2006;10(4):415-417. [DOI] [PubMed] [Google Scholar]
- 46.Kremer JR, Brown KE, Jin L, et al. . High genetic diversity of measles virus, World Health Organization European Region, 2005-2006. Emerg Infect Dis. 2008;14(1):107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogalska J, Santibanez S, Mankertz A, Makowka A, Szenborn L, Stefanoff P. Spotlight on measles 2010: An epidemiological overview of measles outbreaks in Poland in relation to the measles elimination goal. Euro Surveill. 2010;15(17). [DOI] [PubMed]
- 48.Vongpunsawad S, Oezgun N, Braun W, Cattaneo R. Selectively receptor-blind measles viruses: Identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J Virol. 2004;78(1):302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seki F, Takeda M, Minagawa H, Yanagi Y. Recombinant wild-type measles virus containing a single N481Y substitution in its haemagglutinin cannot use receptor CD46 as efficiently as that having the haemagglutinin of the Edmonston laboratory strain. J Gen Virol. 2006;87(Pt 6):1643-1648. [DOI] [PubMed] [Google Scholar]
- 50.Tahara M, Takeda M, Seki F, Hashiguchi T, Yanagi Y. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J Virol. 2007;81(6):2564-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suerth JD, Maetzig T, Brugman MH, et al. . Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol Ther. 2012;20(5):1022-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell RS, Beitzel BF, Schroder AR, et al. . Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2(8):E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521-529. [DOI] [PubMed] [Google Scholar]
- 54.Thielecke L, Aranyossy T, Dahl A, et al. . Limitations and challenges of genetic barcode quantification. Sci Rep. 2017;7:43249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai S, Wang H, Bailey B, et al. Differential secondary reconstitution of in vivo-selected human SCID-repopulating cells in NOD/SCID versus NOD/SCID/ γ chain mice. Bone Marrow Res 2011;2011:252953. [DOI] [PMC free article] [PubMed]
- 56.Tolar J, Adair JE, Antoniou M, et al. . Stem cell gene therapy for fanconi anemia: report from the 1st international Fanconi anemia gene therapy working group meeting. Mol Ther. 2011;19(7):1193-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croop JM, Cooper R, Fernandez C, et al. . Mobilization and collection of peripheral blood CD34+ cells from patients with Fanconi anemia. Blood. 2001;98(10):2917-2921. [DOI] [PubMed] [Google Scholar]
- 58.Larghero J, Marolleau JP, Soulier J, et al. . Hematopoietic progenitor cel harvest and functionality in Fanconi anemia patients. Blood. 2002;100(8):3051. [DOI] [PubMed] [Google Scholar]
- 59.Cumming RC, Lightfoot J, Beard K, Youssoufian H, O’Brien PJ, Buchwald M. Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat Med. 2001;7(7):814-820. [DOI] [PubMed] [Google Scholar]
- 60.Girard-Gagnepain A, Amirache F, Costa C, et al. . Baboon envelope pseudotyped LVs outperform VSV-G-LVs for gene transfer into early-cytokine-stimulated and resting HSCs. Blood. 2014;124(8):1221-1231. [DOI] [PubMed] [Google Scholar]
- 61.Wang CX, Sather BD, Wang X, et al. . Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124(6):913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrillo C, Cesana D, Piras F, et al. . Cyclosporin a and rapamycin relieve distinct lentiviral restriction blocks in hematopoietic stem and progenitor cells. Mol Ther. 2015;23(2):352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shima H, Takubo K, Iwasaki H, et al. . Reconstitution activity of hypoxic cultured human cord blood CD34-positive cells in NOG mice. Biochem Biophys Res Commun. 2009;378(3):467-472. [DOI] [PubMed] [Google Scholar]
- 64.Shima H, Takubo K, Tago N, et al. . Acquisition of G0 state by CD34-positive cord blood cells after bone marrow transplantation. Exp Hematol. 2010;38(12):1231-1240. [DOI] [PubMed] [Google Scholar]
- 65.Tisdale JF, Hanazono Y, Sellers SE, et al. . Ex vivo expansion of genetically marked rhesus peripheral blood progenitor cells results in diminished long-term repopulating ability. Blood. 1998;92(4):1131-1141. [PubMed] [Google Scholar]
- 66.Jetmore A, Plett PA, Tong X, et al. . Homing efficiency, cell cycle kinetics, and survival of quiescent and cycling human CD34(+) cells transplanted into conditioned NOD/SCID recipients. Blood. 2002;99(5):1585-1593. [DOI] [PubMed] [Google Scholar]
- 67.Glimm H, Oh IH, Eaves CJ. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0). Blood. 2000;96(13):4185-4193. [PubMed] [Google Scholar]
- 68.Gothot A, Pyatt R, McMahel J, Rice S, Srour EF. Functional heterogeneity of human CD34(+) cells isolated in subcompartments of the G0 /G1 phase of the cell cycle. Blood. 1997;90(11):4384-4393. [PubMed] [Google Scholar]
- 69.Habibian HK, Peters SO, Hsieh CC, et al. . The fluctuating phenotype of the lymphohematopoietic stem cell with cell cycle transit. J Exp Med. 1998;188(2):393-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ceccaldi R, Parmar K, Mouly E, et al. . Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Müller LU, Milsom MD, Kim MO, Schambach A, Schuesler T, Williams DA. Rapid lentiviral transduction preserves the engraftment potential of Fanca(-/-) hematopoietic stem cells. Mol Ther. 2008;16(6):1154-1160. [DOI] [PubMed] [Google Scholar]
- 72.Gregory JJ Jr, Wagner JE, Verlander PC, et al. . Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci USA. 2001;98(5):2532-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gross M, Hanenberg H, Lobitz S, et al. . Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98(2-3):126-135. [DOI] [PubMed] [Google Scholar]
- 74.Mankad A, Taniguchi T, Cox B, et al. . Natural gene therapy in monozygotic twins with Fanconi anemia. Blood. 2006;107(8):3084-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ozog S, Minder P, Timberlake N, Garijo O, Torbett B. CD46 receptor knockout in packaging cell lines improves production of measles glycoprotein pseudotyped lentiviral vectors and transduction efficiency of human CD34+ cells. In: Proceedings from the American Society of Gene & Cell Therapy 20th Annual Meeting; 10-13 May 2017; Washington, DC. Abstract 769. [Google Scholar]
- 76.Cattoglio C, Pellin D, Rizzi E, et al. . High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 2010;116(25):5507-5517. [DOI] [PubMed] [Google Scholar]
- 77.Papanikolaou E, Paruzynski A, Kasampalidis I, et al. . Cell cycle status of CD34(+) hemopoietic stem cells determines lentiviral integration in actively transcribed and development-related genes. Mol Ther. 2015;23(4):683-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.