Key Points
CDH17 is expressed in human thymic epithelial cells.
CDH17 mutations may be a rare cause of leaky severe combined immune deficiency that can be corrected by HSCT.
Introduction
Cadherin 17 (encoded by the CDH17 gene) is a member of the cadherin superfamily encoding calcium-dependent membrane-associated glycoproteins. CDH17 is developmentally regulated and its transcripts are detected in the fetal but not adult liver and intestines. Somatic CDH17 mutations have been reported in gastrointestinal malignancies and implicated in tumor progression.1-4 CDH17 is also expressed in murine precursor and mature B cells,5 where it contributes to their development and memory B-cell survival.6,7 CDH17 expression has also been demonstrated in mouse thymic medullary epithelial cells (mTECs), independent of their expression of the autoimmune regulator (AIRE), a transcriptional facilitator of promiscuous gene expression required for correct T-cell repertoire selection. In contrast, CDH17 is barely detectable in cortical TECs.8
The consequences of CDH17 mutations for lymphoid development and immune function remain, however, unknown. Here we report on a CDH17-mutated patient with leaky severe combined immunodeficiency, who was successfully treated by hematopoietic stem cell transplantation (HSCT), and discuss the possible role of CDH17 mutations in the pathogenesis of the disease.
Case description
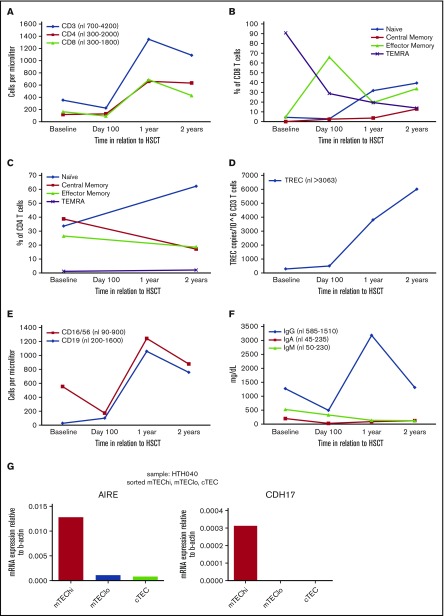
A 5-year-old Saudi Arabian girl presented to us with a long medical history of recurrent sino-pulmonary infections and was noted to have severe T- and B-cell lymphopenia with normal natural killer (NK) cellularity. Thymic output (as quantified by T-cell receptor excision circle [TREC] analysis) and aggregate T-cell function were very low (Table 1). The majority of the peripheral T cells had a memory phenotype, and proliferative response to PHA was low. Tetanus toxoid antibody levels were low. An extensive workup (see Table 1) failed to determine the molecular cause of the observed lymphopenia. During further workup, a stage 4 EBV-positive DLBCL was diagnosed, consistent with reduced immune surveillance. After achieving complete remission following 6 cycles of chemotherapy and surgical resection of tumor tissue in lung and ovaries, the patient underwent myeloablative matched sibling donor HSCT. A radiation containing regimen was chosen given her DLBCL appeared to be only partially responsive to chemotherapy as her ovarian disease was discovered shortly after completing her chemotherapy and her lung lesions still contained active disease after 6 cycles of chemotherapy. Posttransplant, the girl developed grade 2 skin GVHD, CMV viremia/pneumonia, and pulmonary actinomycosis. Remaining in remission from her DLBCL, she had full and stable immune reconstitution by within 2 years following HSCT, and both CD4 and CD8 naïve T-cells, TRECs, T-cell function, and immunoglobulin levels returned to normal values (Table 1; Figure 1). Pre-HSCT peripheral blood WES revealed compound heterozygous mutations in CDH17. Each of her parents had one of the noted and distinct mutations in CDH17 and was healthy with no concern for immune deficiency.
Table 1.
Clinical course and immune function
| Baseline | DLBCL diagnosis | Workup and HSCT | Day 100 post-HSCT | 1 Year post-HSCT | 2 Years post-HSCT | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical course | Multiple lung, sinus, and tonsil infections starting at age 1.5 y requiring frequent hospitalizations and antibiotics SCD with HPFH G6PD |
Diagnosis: Chest CT with bilateral, extensive lung infiltrates Lung biopsy revealed a DLBCL, EBV+ 2A level lymph nodes, palatine tonsil, ovary involved CSF and bone marrow were negative |
Workup (all negative): HIV PCR FISH for DiGeorge Chromosome breakage studies |
Grade 2 skin GVHD treated with topical steroids only CMV viremia and possible pneumonia treated with IV immunoglobulin and ganciclovir |
Pulmonary actinomyces treated with PCN Bronchiectasis |
Clinically well Treatment of actinomyces continues |
||||||||
| Genetic workup (all negative): Rag 1/2 AT mutation analysis Nijmegen Artemis XLP XIAP | ||||||||||||||
| Immune function | ||||||||||||||
| T cell | Numbers (abs): CD3 = 353, CD4 = 118, CD8 = 163 |
Numbers (abs): CD3 = 223, CD4 = 128, CD8 = 92 |
Numbers (abs): CD3 = 1350, CD4 = 662, CD8 = 688 |
Numbers (abs): CD3 = 1089, CD4 = 633, CD8 = 428 |
||||||||||
| TREC (nl >3063): 289 copies/106 CD3 T cells |
TREC (nl >3063): 500 copies/106 CD3 T cells |
TREC (nl >3063): 3800 copies/106 CD3 T cells |
TREC (nl >3063): 6010 copies/106 CD3 T cells |
|||||||||||
| Proliferation: Low response (<5% of control) to PHA, absent responses to candida and TT |
Treatment: Ritux/pred × 1 cycle Ritux/pred/MTX/Doxo × 2 cycles Ritux/ARAC/MTX × 2 cycles Ritux/Cy/Vinc/pred/MTX/Doxo × 2 cycles Surgical resection of hilar lymph nodes and lung masses Surgical resection of ovary and peritoneal mass |
WES Compound heterozygosity for the c.1796+2 T>C mutation and N176S variant in the CDH17 gene Heterozygous variant in the LIG1 gene Homozygous E7V mutation in the HBB gene Heterozygous S188F mutation in the G6PD gene |
Proliferation: Normal response to PHA. PWM, candida, and TT not tested. |
Proliferation: Normal response to PHA and PWM. Absent responses to candida and TT. |
Proliferation: Normal response to PHA and PWM. Candida and TT not tested. |
|||||||||
| TCR diversity: Normal | ||||||||||||||
| Subsets (%): | Subsets (%): | Subsets (%): | Subsets (%): | |||||||||||
|
TNAIVE TCM TEM TEMRA |
CD4 33.6 38.7 26.5 1.19 |
CD8 4.4 0.05 4.85 90.7 |
TNAIVE TCM TEM TEMRA |
CD4 — — — — |
CD8 2.85 2.5 66 28.7 |
TNAIVE TCM TEM TEMRA |
CD4 — — — — |
CD8 31.7 3.6 19.5 45.2 |
TNAIVE TCM TEM TEMRA |
CD4 62.2 17.1 18.5 2.13 |
CD8 39.4 12.9 33.7 13.9 |
|||
| B cell | Numbers (abs): CD19 = 27 |
Numbers (abs): CD19 = 100 |
Numbers (abs): CD19 = 1058 |
Numbers (abs): CD19 = 756 |
||||||||||
| Immunoglobulins*: IgG 1270, IgA 199, IgM 524, IgE <2 |
Immunoglobulins†: IgG 494, IgA 27, IgM 328, IgE <2 |
Immunoglobulins: IgG 3180, IgA 89, IgM 136, IgE 4 |
Immunoglobulins: IgG 1310, IgA 123, IgM 124, IgE 3 |
|||||||||||
| Vaccine titers: Tetanus Ab 0.06 (minimally protective), diphtheria Ab 0.06 (minimally protective), minimal response to all S.pneumo serotypes |
Response: Mixed response to chemotherapy regimens NED at the time of HSCT |
HSCT details: MSD HSCT (sibling had normal lymphocyte counts, no SCD or G6PD) Cy/TBI 1320cGy preparative regimen CSA/MTX for GVHD prophylaxis |
Vaccine titers: Tetanus Ab 0.21 (protective), diphtheria Ab 0.21 (protective), protective response to 9 of 14 S.pneumo serotypes. |
|||||||||||
| NK cell | Numbers (abs): CD16/56 = 526 |
Numbers (abs): CD16/56 = 72 |
Numbers (abs): CD16/56 = 185 |
Numbers (abs): CD16/56 = 120 |
||||||||||
| Activity: Normal |
Activity: Normal |
Activity: Decreased |
Activity: Normal |
|||||||||||
| Chimerism | 100% Donor chimerism in peripheral blood and bone marrow | 100% Donor chimerism in peripheral blood and bone marrow | 100% Donor chimerism in peripheral blood and bone marrow | |||||||||||
Ab, antibody; abs, absolute; ARAC, cytarabine; AT, ataxia telangiectasia; CMV, cytomegalovirus; CSA, cyclosporine; CSF, cerebrospinal fluid; CT, computed tomography; Cy, cytoxan; DLBCL, diffuse large B-cell lymphoma; Doxo, doxorubicin; EBV, Epstein-Barr virus; FISH, florescence in situ hybridization; G6PD, glucose 6-phosphate deficiency; GVHD, graft-versus-host disease; HBB, β hemoglobin; HPFH, hereditary persistence of fetal hemoglobin; MSD, matched sibling donor; MTX, methotrexate; NED, no evidence of disease; PCN, penicillin; PCR, polymerase chain reaction; PHA, phytohemagglutinin; pred, prednisone; PWM, pokeweed mitogen; Ritux, rituximab; SCD, sickle-cell disease; TBI, total body irradiation; TCM, central memory T cells; TEM, effector memory T cells; TEMRA, effector memory T cells that express CD45RA; TNAIVE, naïve T cells; TT, tetanus toxoid; Vinc, vincristine; WES, whole exome sequencing; XIAP, X-linked inhibitor of apoptosis; XLP, X-linked lymphoproliferative disorder.
Previous IV immunoglobulin replacement therapy is uncertain.
Last IV immunoglobulin infusion 1 month prior.
Figure 1.
Immune recovery over time and CDH17 expression in thymic epithelial cells. (A) T-cell numbers. (B) CD8 T-cell subsets. (C) CD4 T-cell subsets. (D) TRECs. (E) B and NK cell numbers. (F) Immunoglobulin levels. (G) CDH17 expression in the human thymus is restricted to the population of mature medullary epithelial cells (mTEChi) as shown by quantitative reverse transcription PCR analysis. mTEChi is a heterogeneous population of thymic epithelial cells that primarily consists of functionally mature, AIRE-expressing cells.
Methods
Patient
The subject was treated on an institutional review board–approved HSCT protocol at the University of Minnesota.
Human thymus samples
Human thymus samples were obtained from children undergoing corrective cardiac surgery at Great Ormond Street Hospital for Children National Health Service trust after parents’ informed consent and approval by the Great Ormond Street Hospital for Children National Health Service trust with a Material Transfer Agreement to the chancellor, masters, and scholars of the University of Oxford.
Isolation of thymic epithelial cells
TECs were isolated from the thymic tissue after mechanical and enzymatic tissue disruption followed by magnetic-activated cell sorting and fluorescence-activated cell sorting according to the protocol published by Stoeckle et al.9
RNA extraction and quantitative PCR analysis
Total RNA was extracted from sorted TECs using the RNeasy Mini Kit (Qiagen) according to the manufacturers’ protocol, including in-column DNase treatment. Complementary DNA was prepared using the SensiFAST kit (Bioline). Complementary DNA was used for quantitative PCR with the relevant forward TTTACATTTTCCCTCGGCAG and reverse CCTCAAACTCTGTGTGCCTG primers (Sigma), using Sensifast SYBR Hi-Rox Kit (Bioline) and StepOnePlus (Applied Biosystems).
Flow cytometry
Standard flow cytometric methods were used for staining of cell-surface proteins. Anti-human monoclonal antibodies to the following molecules, with the appropriate isotype-matched controls were used for staining: CD3 (OKT-3), CD4 (RPA-T4), CD8 (HIT8a), CD19 (HIB19), CD56 (HCD56), CD16 (3G8), CD45RA (HI100), CD45RO (UCHL1), CD31 (390), and CCR7 (4B12), all from BioLegend (San Diego, CA). Data were collected with an LSRFortessa (BD Biosciences, San Jose, CA), cell analyzer and analyzed with FlowJo software (Tree Star Inc., Ashland, OR).
T-cell proliferation
Peripheral blood mononuclear cells were cultured unstimulated or stimulated with 5 μg/mL PHA (Sigma Aldrich; L8754). Proliferation was assayed 4 days later by measuring 3H-thymidine incorporation into genomic DNA added for the last 16 hours of culture.
Results and discussion
Human TECs interrogated by quantitative reverse transcription PCR showed CDH17 expression only in mature medullary TECs (mTEChi), a heterogeneous population of TECs that primarily but not exclusively consists of functionally mature cells, as molecularly identified by their expression of AIRE (Figure 1G). Pre-HSCT peripheral blood WES revealed compound heterozygous CDH17 mutations (Table 1). The c.1796+2 T>C mutation (IVS13+2 T>C) in CDH17 abolishes the splice donor site of intron 13 and is predicted to cause either abnormal transcripts subjected to nonsense-mediated messenger RNA decay or a protein with abnormal sequence. The c.527A>G mutation (N176S) results in a conservative substitution of one neutral, polar amino acid for another. Although the molecular and functional consequence of this mutation is not yet fully established, it is reported at low frequency in the ExAC database (minor allele frequency: 0.0001319), with 16 heterozygous, but no homozygous subjects carrying this variant. Furthermore, given the thymic expression pattern of CDH17 and the robust donor-derived T-cell recovery post-HSCT, the low thymic output noted at initial presentation appeared to the independent of an intrinsic TEC deficiency.
Overall, predictions would suggest that the CDH17 mutations identified in this child lead in aggregate to altered protein expression and/or function though as stated previously, the molecular and functional consequences of these mutations are unknown. At baseline, the patient had a markedly reduced number of circulating B cells. Because of the role played by CDH17 on B-cell development and survival, the CDH17 mutations may be responsible for B-cell lymphopenia observed in the patient at baseline. Furthermore, because CDH17 mutations and loss of expression are associated with malignant tumor progression, the propensity for EBV lymphoproliferative disease progression to DLBCL may have been supported by CDH17 mutations in the malignant B cells. Many primary immune deficiencies are associated with increased risk for malignancies secondary to defective immune surveillance and infection with oncogenic viruses. Non-Hodgkin lymphomas, including DLBCL, are among the most common.10
In summary, our patient was a compound heterozygote for 2 CDH17 mutations without other known immunodeficiency-related genetic mutations, had on presentation severe lymphopenia, and displayed significantly abnormal T-cell function. Our investigations revealed that CDH17 is also expressed on human mTEChi cells, but because T-cell lymphopoiesis was restored post-HSCT in our patient, the TEC defect does not appear to be limiting. Based on experimental data that suggests in the mouse that CDH17 mutations cause B-cell defects,5,6 we postulate that the patient’s compound heterozygosity for CDH17 mutations affected bipotent lymphoid progenitors and impaired their differentiation. We therefore conclude that the compound heterozygous CDH17 mutations identified in this patient constitute a rare cause of leaky severe combined immunodeficiency that can be cured with HSCT. Future studies delineating the consequences of the patient’s mutations are needed to confirm this conclusion.
Acknowledgments
This work was supported by a grant from the National Cancer Institute, National Institutes of Health (P01 CA065493), and by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Authorship
Contribution: A.R.S. wrote the manuscript; L.D.N., G.A.H., and B.R.B. designed the experiments; I.A.R., S.M., and M.J.M. performed the experiments and edited the manuscript; and A.R.S., T.C.L., L.D.N., G.A.H., and B.R.B. analyzed and/or interpreted the results and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angela R. Smith, 420 Delaware St SE; MMC 484, Minneapolis, MN 55455; e-mail: smith719@umn.edu.
References
- 1.Lee NP, Poon RT, Shek FH, Ng IO, Luk JM. Role of cadherin-17 in oncogenesis and potential therapeutic implications in hepatocellular carcinoma. Biochim Biophys Acta. 2010;1806(2):138-145. [DOI] [PubMed] [Google Scholar]
- 2.Grötzinger C, Kneifel J, Patschan D, et al. LI-cadherin: a marker of gastric metaplasia and neoplasia. Gut. 2001;49(1):73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu LX, Lee NP, Chan VW, et al. Targeting cadherin-17 inactivates Wnt signaling and inhibits tumor growth in liver carcinoma. Hepatology. 2009;50(5):1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takamura M, Ichida T, Matsuda Y, et al. Reduced expression of liver-intestine cadherin is associated with progression and lymph node metastasis of human colorectal carcinoma. Cancer Lett. 2004;212(2):253-259. [DOI] [PubMed] [Google Scholar]
- 5.Ohnishi K, Shimizu T, Karasuyama H, Melchers F. The identification of a nonclassical cadherin expressed during B cell development and its interaction with surrogate light chain. J Biol Chem. 2000;275(40):31134-31144. [DOI] [PubMed] [Google Scholar]
- 6.Ohnishi K, Melchers F, Shimizu T. Lymphocyte-expressed BILL-cadherin/cadherin-17 contributes to the development of B cells at two stages. Eur J Immunol. 2005;35(3):957-963. [DOI] [PubMed] [Google Scholar]
- 7.Funakoshi S, Shimizu T, Numata O, Ato M, Melchers F, Ohnishi K. BILL-cadherin/cadherin-17 contributes to the survival of memory B cells. PLoS One. 2015;10(1):e0117566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sansom SN, Shikama-Dorn N, Zhanybekova S, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014;24(12):1918-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoeckle C, Rota IA, Tolosa E, Haller C, Melms A, Adamopoulou E. Isolation of myeloid dendritic cells and epithelial cells from human thymus. J Vis Exp. 2013;79:e50951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Werff ten Bosch J, van den Akker M. Genetic predisposition and hematopoietic malignancies in children: Primary immunodeficiency. Eur J Med Genet. 2016;59(12):647-653. [DOI] [PubMed] [Google Scholar]