Abstract
Background
Previous studies suggest circulating, blood-based microRNAs (miRNAs) may serve as minimally invasive prostate cancer biomarkers, however there is limited data from prospective clinical trials. Here, we explore the role of candidate plasma miRNAs as potential biomarkers in the SWOG 0925 randomized phase II study of androgen deprivation combined with cixutumumab versus androgen deprivation alone in patients with new metastatic hormone-sensitive prostate cancer.
Methods
Correlative biospecimens, including circulating tumor cells (CTCs) and plasma for miRNA analysis, were collected at baseline and after 12 weeks on treatment from 50 patients enrolled on SWOG 0925. Circulating microRNAs were quantified using real time RT-PCR microRNA array that allowed specific analysis of previously identified candidate miRNAs (miR-141, miR-200a, miR-200b, miR-210 and miR-375) as well as discovery analysis to identify new candidate miRNAs. MiRNA levels were correlated to previously reported CTC counts using CellSearch® (Veridex) and with the primary study outcome of 28-week PSA response (≤0.2, 0.2 to ≤4.0, or >4.0 ng/mL), previously shown to correlate with overall survival.
Results
We observed a correlation between baseline circulating miR-141, miR-200a, and miR-375 levels with baseline CTCs. Baseline miR-375 levels were associated with 28-week PSA response (≤0.2, 0.2 to ≤4.0, or >4.0 ng/mL, P=0.007). Using ROC curve analysis, there was no significant difference between baseline miR-375 and baseline CTC in predicting 28-week PSA response (≤0.2 vs. >0.2 ng/mL). To discover novel candidate miRNAs, we analyzed 365 miRNAs for association with the 28-week PSA response endpoint and identified new candidate miRNAs along with the existing candidates miR-375 and miR-200b (P=0.0012, P=0.0046, respectively.
Conclusions
Baseline plasma miR-141, miR-200a, and miR-375 levels are associated with baseline CTC count. Baseline miR-375 was also associated with the trial endpoint of 28-week PSA response. Our results provide evidence that circulating miRNA biomarkers may have value as prognostic biomarkers and warrant further study in larger prospective clinical trials.
Keywords: circulating, microRNA, prostate cancer, miR-375, miR-200, miR-141
INTRODUCTION
Therapeutic options for advanced prostate cancer have changed dramatically in the past decade[1, 2]. Better prognostic and predictive biomarkers are needed to address the clinical challenges of appropriate selection and sequencing of the range of new treatment options. Earlier work demonstrated the prognostic value of circulating tumor cells and led to FDA approval of CTC enumeration to aid in prognosis of men with metastatic castration resistant prostate cancer[3, 4]. Additional studies have indicated the potential value of assessing expression of the androgen receptor splice variant 7 (AR-V7) in circulating tumor cells in the castration resistant setting[5]. However, the lower sensitivity of CTC detection has been an issue, for example, less than half of men in SWOG 0925 had CTCs at baseline[6]. Identifying better prognostic and predictive biomarkers therefore remains a largely unmet need.
Circulating, blood-based microRNAs (miRNAs) are similar to circulating tumor cells (CTCs) in that they serve as promising, minimally invasive prostate cancer biomarker candidates[7–11]. MiRNAs are short (~22 nucleotide) non-protein encoding RNAs that play an important role in regulation of gene expression via modulation of specific messenger RNA (mRNA) targets[12]. A single miRNA may regulate hundreds to thousands of mRNA transcripts and therefore may summarily reflect biological gene expression networks and provide complementary biological information to CTCs. MiRNAs may also be more sensitive and simple to assay than CTCs.
To explore the possibility that miRNAs may be more sensitive and easier to assay than CTCs and to see if miRNAs are predictive of PSA outcomes at 28 weeks, we prospectively collected correlative biospecimens on the randomized Phase II SWOG 0925 clinical trial[8]. We previously reported findings on CTCs and IGF-1R biomarkers, along with association to the primary endpoint of PSA after 28 weeks of therapy. To briefly summarize, of 50 patients that provided CTC samples, 39 had evaluable results and 16/39 (41%) had no detectable CTCs despite being collected from a population of patients with newly diagnosed metastatic prostate cancer.
Here, we report the results of our biomarker analysis of circulating miRNA in association with CTCs as well as with the primary endpoint of the clinical trial: PSA at 28 weeks, an outcome previously shown to correlate with overall survival[13]. We focused on 5 candidate circulating miRNAs which were previously identified via miRNA profiling and validated to be elevated in prostate cancer cases compared to controls: miR-141, miR-200a, miR-200b, miR-210 and miR-375[7]. In addition, we used a wider profiling approach in which we examined plasma levels of 365 miRNAs to identify candidates whose baseline levels associated with the study endpoint of 28-week PSA as novel prognostic biomarker candidates.
PATIENTS AND METHODS
Patients
The randomized, Phase II SWOG S0925 study investigated androgen deprivation (AD) combined with cixutumumab versus AD alone in patients with new metastatic hormone-sensitive prostate cancer (ClinicalTrials.gov NCT01120236) has previously been described[6]. The results of the study did not show a significant difference in outcome between the two treatment arms, so we considered the patients in both arms in aggregate for the purpose of biomarker analyses. Eligible patients for the miRNA analysis were patients enrolled in SWOG 0925, had pathologic confirmation of prostate cancer, PSA ≥5 ng/mL, and at least one radiographic detectable metastasis and had not yet started any form of AD. Fifty patients consented to the biomarker specimen collection and participated in the biomarker sub-study. All study procedures were performed in accordance with the declaration of Helsinki guidelines and with ethics approval from the Institutional Review Boards at participating SWOG sites.
Blood processing and isolation of plasma and CTCs from clinical samples
Plasma and CTCs were collected prior to initiation of AD and again at 12 weeks from 50 patients enrolled in the SWOG 0925 translational science substudy. Plasma samples designated for circulating miRNA analysis were collected into Becton Dickinson lavender top vacutainer tubes containing K2EDTA. Whole blood (7.5ml) was collected for CellSearch® circulating tumor cells assessment (Veridex, Raritan, NJ). All clinical samples obtained were collected and processed locally as previously described[7] and then shipped to the Fred Hutchinson Cancer Research Center within 24 hours of blood draw. Briefly, upon arrival, plasma samples were centrifuged at 3000xg for 5 minutes at room temperature to separate the plasma, aliquoted into cryovials and stored at −80C. CTC measurements were performed using the clinical CellSearch platform. Both timepoints from a single patient were extracted and run in the same batch to minimize batch effect.
Quantification of circulating miRNAs
Total RNA was isolated from 200ul plasma using the miRNeasy RNA isolation kit (Qiagen, Germany) in the following manner: Frozen aliquots were thawed on ice, centrifuged at 3000xg for 3 minutes at room temperature to remove additional cellular debris and insoluble components[28], and 200 μl of plasma supernatant was measured and denatured using 10X volume (1 ml) Qiazol, vortexed and incubated at room temperature for 10 minutes. C. elegans spiked-in oligonucleotides were introduced (as a mixture of 25 fmol of each oligonucleotide in 5 μl total volume per liquid sample) after denaturation, which were used for normalization of variability in RNA isolation across samples as previously described[7]. RNA was extracted using 0.2X volume chloroform (220 μl), and total RNA was isolated following the manufacturer’s protocol. For a given sample, RNA isolated from each 200 μl aliquot was eluted using 50μl RNAse-free water at 1000xg, 3 min, 4°C. RNA was stored at −70°C.
Reverse transcription was performed in triplicate then pooled (to minimize batch effect) using 13 μl of RNA as input for the MirCURY Exiqon Universal RT kit following manufacturer’s protocol. Reaction was incubated at 42°C for 60 minutes followed by 5 minutes at 95C, transfer to non-stick tubes and stored at −20C. Complementary DNA was then combined with SYBR Green Master mix, ROX and 10 μl was aliquoted into each well of a miRNA Ready-to-Use PCR, Human panel I, V1.M qRT-PCR array (Exiqon). Each sample was run in duplicate.
miRNA Detection and Normalization
Missing (i.e., Undetermined) raw cycle threshold (Ct) values were set to 40 (the highest possible raw Ct value in the data). Any miRNA targets with a Ct value of 40 in 60% or more of the samples were filtered out before the normalization and excluded from further analysis. A total of 280 (out of 375) miRNA targets passed the filtering. The raw Ct values for all the 280 miRNA targets were then quantile normalized (using R function “normalize.quantiles”) across all the samples.
The average coefficient of variation was 4.0% (range: 1.1–9.8%) for all miRNAs and 5.6% (range: 3.2–8.0%) for the 5 previously validated miRNAs.
Study population and evaluable samples
Fifty patients participated in the translational science biomarker sub-study (patient demographics shown in Table 1). Among these patients, 1 patient was ineligible for the trial, 6 patients initiated LHRH therapy prior to registration, and 3 patients lacked available samples for miRNA analysis. MiRNA data from the 40 remaining eligible and analyzable patients were used in these analyses. Baseline CTCs were collected for all 40 patients mentioned above, but 4 CTC samples were not assessable, resulting in 36 eligible patients for the baseline CTC analysis.
Table 1.
Patient Characteristics of all Eligible Patients
| N | % | |
|---|---|---|
| AGE | 40 | |
| Median, (range) | 68 (50, 85) | |
| RACE | ||
| White | 36 | 90% |
| Black | 2 | 5% |
| Other | 2 | 5% |
| Zubrod PS | ||
| 0–1 | 40 | 100% |
| 2 | 0 | 0% |
| GLEASON SCORE | ||
| <7 | 3 | 8% |
| 7 | 7 | 18% |
| >7 | 30 | 74% |
| PSA AT ENTRY (ng/ml) | ||
| < 20 ng/mL | 11 | 28% |
| ≥ 20 ng/mL | 29 | 72% |
| Treatment Arm | ||
| Arm 1 - cixutumumab + ADT | 24 | 60% |
| Arm 2 – ADT alone | 16 | 40% |
Statistical Considerations
The Friedman test was used to assess correlations between the five previously validated, normalized miRNA data as ranks (due to lack of normally distributed data) and categories of baseline CTC counts, and normalized miRNA data and PSA response categories at 28 weeks after registration. This nonparametric method accounts for the ordinal nature of baseline CTC categories (0, 1–4, ≥ 5 CTC count per 7.5mL whole blood) and PSA response categories (PSA≤0.2, 0.2<PSA≤4.0, PSA>4.0 ng/mL). A significance level for these validated analyses was specified as ≤ 0.05.
In addition to the five previously identified candidate miRNA markers, we performed a wider miRNA profiling analysis to potentially identify additional miRNAs that may be correlated with baseline CTC categories and/or 28-week PSA response category. Following quantile normalization, an additional 275 miRNAs were analyzed using the same Friedman test. A Bonferroni correction (0.05/275 = .00018) could be used as a guideline for evaluating statistical significance, but if miRNA are correlated, this approach would be overly conservative.
RESULTS
Correlation of five previously validated baseline miRNAs (CTs) and baseline CTC counts
We first examined whether baseline assessments of any of the five candidate circulating miRNAs are associated with baseline CTC counts (Table 2). There was an association between baseline CTC counts and baseline circulating miR-141 (P=0.0006), but not with baseline levels of the other four, miR-200a, miR-200c, miR-210 or miR-375.
Table 2.
Correlation of Baseline miRNAs (CTs) with Baseline Circulating Tumor Cell Categories for All Prespecified miRNA measures and Exploratory miRNA with p< 0.10
| miRNA | Median miRNA for Each CTC group | p-value* | ||
|---|---|---|---|---|
| CTC=0 | CTC = 1–4 | CTC= 5+ | ||
| miR-141 | 32.9 | 32.7 | 31.9 | 0.0006 |
| miR-200a | 34.4 | 34.7 | 33.0 | 0.23 |
| miR-200b | 33.4 | 33.2 | 33.6 | 0.52 |
| miR-210 | 32.5 | 32.7 | 32.1 | 0.39 |
| miR-375 | 32.8 | 32.7 | 30.2 | 0.13 |
| Additional exploratory miRNA candidates | ||||
| miR-188-5p | 34.5 | 37.1 | 34.4 | 0.02 |
| miR-885-5p | 33.2 | 36.0 | 34.1 | 0.04 |
| miR-429 | 36.7 | 34.3 | 33.5 | 0.04 |
| miR-135a | 33.5 | 33.6 | 34.2 | 0.04 |
| miR-483-3p | 37.2 | 37.8 | 39.5 | 0.05 |
| miR-190 | 35.2 | 34.5 | 34.5 | 0.05 |
| miR-370 | 34.6 | 33.9 | 33.0 | 0.05 |
| miR-299-5p | 35.0 | 35.7 | 33.7 | 0.06 |
| miR-199b-5p | 32.1 | 31.7 | 32.5 | 0.06 |
| miR-570 | 35.1 | 35.0 | 36.5 | 0.07 |
Based on 2 degree of freedom chisquare, Friedman test
Correlation of five previously identified baseline miRNA and 28-week PSA response categories
We examined whether baseline assessments of any of the five candidate circulating miRNAs are associated with 28-week PSA response (Table 3), the primary outcome of the clinical study, and thus serve as a prognostic biomarker at time of starting treatment. We observed a significant association between 28-week PSA response categories (PSA≤0.2 ng/mL, 0.2<PSA≤4.0 ng/mL, PSA >4.0 ng/mL) and circulating miR-375 (p=0.001) and miR-200b (p=0.005). In contrast, we did not observe a significant association between 28-week PSA response categories and miR-141 (P=0.06), miR-200a (P=0.32), or miR-210 (P=0.59).
Table 3.
Correlation of Baseline miRNAs (CTs) with 28-week PSA Response Categories for All Prespecified miRNA measures and Exploratory miRNA with p< 0.10
| miRNA | Median miRNA value for Each PSA response group | p-value* | ||
|---|---|---|---|---|
| PSA ≤ 0.2 | 0.2< PSA≤4.0 | PSA > 4.0 ng/ml | ||
| miR-141 | 32.6 | 32.0 | 31.5 | 0.06 |
| miR-200a | 34.5 | 33.8 | 33.8 | 0.32 |
| miR-200b | 33.6 | 33.6 | 32.6 | 0.005 |
| miR-210 | 32.5 | 32.1 | 32.3 | 0.59 |
| miR-375 | 33.0 | 32.6 | 29.5 | 0.001 |
| Additional exploratory miRNA candidates | ||||
| hsa-miR-505 | 32.1 | 32.6 | 31.9 | 0.004 |
| hsa-miR-136 | 32.3 | 30.5 | 32.9 | 0.009 |
| hsa-miR-204 | 34.6 | 36.2 | 35.0 | 0.02 |
| hsa-miR-99a | 32.7 | 34.6 | 32.4 | 0.02 |
| hsa-miR-130a | 32.3 | 31.4 | 31.7 | 0.02 |
| hsa-miR-10a | 35.3 | 37.4 | 35.6 | 0.03 |
| hsa-miR-450a | 35.9 | 39.6 | 36.8 | 0.03 |
| hsa-miR-376b | 30.8 | 29.5 | 31.5 | 0.03 |
| hsa-miR-376c | 30.7 | 29.1 | 31.4 | 0.034 |
| hsa-miR-431 | 33.4 | 31.8 | 33.8 | 0.04 |
| hsa-miR-381 | 35.5 | 34.3 | 36.3 | 0.04 |
| hsa-miR-671-5p | 34.8 | 34.3 | 33.5 | 0.05 |
| hsa-miR-376a | 32.4 | 30.7 | 32.4 | 0.05 |
| hsa-miR-411 | 33.8 | 33.3 | 34.9 | 0.05 |
| hsa-miR-125b | 31.4 | 31.6 | 31.1 | 0.06 |
| hsa-miR-370 | 34.3 | 32.9 | 34.3 | 0.06 |
| hsa-miR-127-3p | 31.6 | 30.3 | 32.1 | 0.06 |
Based on 2 degree of freedom chisquare, Friedman test
Analysis to identify novel miRNA candidates
Following quantile normalization, we evaluated whether the miRNAs may be associated with baseline CTCs. The candidate miRNAs that are most strongly correlated with CTC categories are listed in Table 2 and those most correlated with 28-week PSA response categories are listed in Table 3. All miRNAs are associated with baseline CTC and PSA response in Appendix Tables A and B.
DISCUSSION
In this companion translational biomarker sub-study to the prospective randomized, Phase II SWOG 0925 study, we report our findings examining circulating miRNA biomarkers for treatment response prediction in a multi-center cooperative group clinical trial. This is an important step in demonstrating feasibility of miRNA assessment in the broader context of clinical practice.
Analysis from the SWOG 9346 clinical trial demonstrated that PSA at 7-months (28-week) is prognostic for overall survival in men with newly diagnosed hormone sensitive metastatic prostate cancer[13] and therefore served as the primary endpoint for SWOG 0925. Here we also evaluated the correlation of baseline circulating miRNA with the primary endpoint to determine if similar prognostic information could be gleaned at the start of initiation with AD and before the 7 months (28-week) timepoint. We found that baseline circulating miR-375 was associated with 28-week PSA, in both the candidate analysis and the discovery analysis. That we found the association through both analyses lends greater confidence in the results.
Multiple studies have identified miR-375 as elevated in advanced prostate cancer tumors, in circulating blood[11, 14, 15] and in urine[16]. Recent work suggests that miR-375 is involved in the epithelial-mesenchymal-transition (EMT) signature [17], and that disruption of this regulatory network may result in altered expression of miR-375. Another recent report suggests miR-375 mediated repression of the tumor suppressor CBX7, a member of the Polycomb complex involved in epigenetic regulation, which may be associated with prostate tumorigenesis[18].
We sought to explore candidate miRNAs that we previously identified through differential examination of blood samples from metastatic prostate cancer patients and healthy controls, miR-141, miR-200a, miR-200c, miR-210, miR-375. It is possible that this approach could miss some candidate miRNAs and indeed following the discovery analysis, we identified a number of additional potential candidate miRNAs to be considered for further investigation (Tables 2 and 3). Given that the numbers of patients were relatively small (approximately 34–40) and the number of miRNAs evaluated in parallel was substantially greater (365), we are cautious about overinterpreting the significance of these candidate miRNAs without further studies. Since the number of miRNAs evaluated was large, it is possible that a fraction were calculated to be significant by chance (i.e. false positive). However, through simple mathematical calculation, we would expect about three from the discovery list to be significant and represent true positives. Further study will be needed for validation.
In reviewing the list of candidates identified through discovery analysis, some have been reported in the literature previously, either in association with biological processes such as in vitro studies, or in studies specifically to identify new prostate cancer biomarkers. It is interesting that the miR-200 family (especially miR-141) correlates with CTCs. The miR-200 family has been identified in other studies, and evidence suggests a role in the EMT[7]. A recent study suggests miR-141 may have multiple mechanisms for affecting tumor growth and metastases[19]. Another Phase 2 study of circulating miRNAs in patients with metastatic castration resistant prostate cancer treated with docetaxel identified miR-200 family members.[20]
We and others have reported that miR-375 is elevated in plasma of CRPC patients[11, 21–23]. It is noteworthy that miR-375 is also made by non-prostate tissues including endocrine tissues[24], and it is therefore possible that expression of miR-375 may reflect a tissue state affecting treatment response independent of CTCs. We are encouraged to find that miR-375 has also been reported in both plasma and urine from patient studies[16, 25]. Our findings add to the evidence that circulating miR-375 warrants further study in prospective clinical trials.
We previously reported on CTC enumeration using the CellSearch platform from this study as a prognostic biomarker and here we have compared miRNA assessments to CTCs in order to explore if miRNAs may potentially be similar to, more sensitive than and/or provide complementary data to CTCs. Our sample size was limited by smaller numbers of patients eligible due to the requirement for not have initiated AD at start of biomarker substudy. Moreover, only 59% of patients had a measureable CTC count at baseline, thus limiting our ability to do statistically rigorous comparisons between CTCs and miRNAs. Nevertheless, we found that of the 5 candidate miRNAs, baseline miR-141, miR-200a, and miR-375 appear to be correlated with baseline CTC counts, suggesting that circulating miRNA and CTCs may be released following the same or linked biological mechanisms and/or that circulating miRNAs may be released from CTCs. While more investigations will be needed to further explore whether circulating miRNAs may be more sensitive that CTCs detected by CellSearch, we recognize that there is also ongoing work around improving the sensitivity of CTC enumeration as well as specific molecular assessments of CTCs[26, 27].
In conclusion, we evaluated circulating miRNAs from the SWOG 0925 clinical trial in which 41% of patients had baseline CTC = 0. We found association between candidate miRNAs miR-141, miR-200a, miR-200c and miR-375 levels were significantly correlated with the stratified baseline CTCs as enumerated by CellSearch, but the sample sizes were insufficient to determine whether circulating miRNAs may be more sensitive. Baseline miR-375 level and baseline CTC count were associated with the primary endpoint of 28-week PSA, providing the potential for prognostication at time of initiating AD. Finally, our discovery analysis identified novel potential candidate miRNAs, but more investigation is needed for confirmation.
Eventual survival outcomes from SWOG 0925 will be enlightening, and if our observation holds true in additional studies, baseline circulating miR-375 in combination with other clinical factors and biomarkers could serve to further stratify patients for future clinical trials and/or more intensified therapies. This work substantially adds to evidence for the role of circulating miRNAs as prostate cancer prognostic biomarkers.
Supplementary Material
1A. Scatterplot Display of miRNAs (CTs) with Significant Associations with 28-week PSA Response Category. 1B. Scatterplot of miRNA (CTs) with Significant Association with Baseline CTC Count Category
Supplementary Tables 1A and 1B: All miRNAs w/ p-values
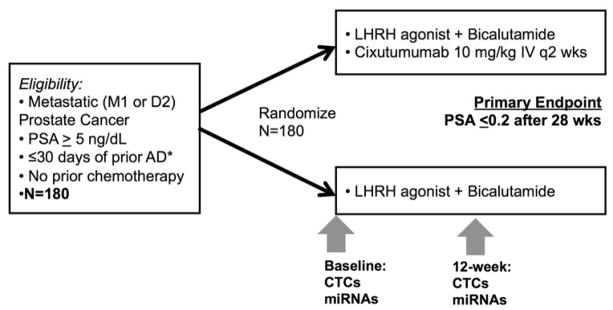
Figure 1. SWOG 0925 Trial Schema with Translational Biomarkers.
*Patients were not eligible for the translational miRNA and CTC analysis unless they had received no prior androgen deprivation (AD). LHRH – Luteinizing hormone releasing hormone.
CTCs – circulating tumor cells. miRNAs – microRNAs.
Acknowledgments
We gratefully acknowledge all of the patients who participated in the SWOG 0925 study. We thank James Yan and Emily Gallichotte for technical assistance and Hui Jiang for assistance with quantile normalization analysis.
FUNDING: Research reported in this publication was supported by the National Institutes of Health under Award Numbers CA180888, CA180819, CA180818, CA180828, CA46368, CA180801, CA180835, CA35421, CA180834, CA142559, CA35281, CA35090, CA37981, CA45807, CA46282, CA180846, CA180830, CA35431, CA58416, CA63848, CA63844, CA12644, CA11083, CA35178, CA67575, CA45808, Pacific Northwest SPORE CA097186, TR01 5R01DK085714 (MT); ImClone Systems (subsidiary of Eli Lilly and Company) and Veridex LLC (Janssen Diagnostics/Johnson & Johnson), and Prostate Cancer Foundation Creativity Award (MT). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Cancer ClinicalTrials.gov Registry Number: NCT01120236
Footnotes
DISCLOSURE: The authors have no disclosures to report.
References
- 1.Crawford ED, et al. Treating Patients with Metastatic Castration Resistant Prostate Cancer: A Comprehensive Review of Available Therapies. J Urol. 2015;194(6):1537–47. doi: 10.1016/j.juro.2015.06.106. [DOI] [PubMed] [Google Scholar]
- 2.Ritch CR, Cookson MS. Advances in the management of castration resistant prostate cancer. BMJ. 2016;355:i4405. doi: 10.1136/bmj.i4405. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 4.Goldkorn A, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32(11):1136–42. doi: 10.1200/JCO.2013.51.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu EY, et al. SWOG S0925: A Randomized Phase II Study of Androgen Deprivation Combined With Cixutumumab Versus Androgen Deprivation Alone in Patients With New Metastatic Hormone-Sensitive Prostate Cancer. J Clin Oncol. 2015;33(14):1601–8. doi: 10.1200/JCO.2014.59.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabris L, et al. The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur Urol. 2016;70(2):312–22. doi: 10.1016/j.eururo.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fendler A, et al. The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol. 2016;13(12):734–752. doi: 10.1038/nrurol.2016.193. [DOI] [PubMed] [Google Scholar]
- 10.Thieu W, et al. The role of microRNA in castration-resistant prostate cancer. Urol Oncol. 2014;32(5):517–23. doi: 10.1016/j.urolonc.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng HH, et al. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS One. 2013;8(7):e69239. doi: 10.1371/journal.pone.0069239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain M, et al. Absolute prostate-specific antigen value after androgen deprivation is a strong independent predictor of survival in new metastatic prostate cancer: data from Southwest Oncology Group Trial 9346 (INT-0162) J Clin Oncol. 2006;24(24):3984–90. doi: 10.1200/JCO.2006.06.4246. [DOI] [PubMed] [Google Scholar]
- 14.Huang X, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen HC, et al. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate. 2013;73(4):346–54. doi: 10.1002/pros.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuopelyte K, et al. The utility of urine-circulating miRNAs for detection of prostate cancer. Br J Cancer. 2016;115(6):707–15. doi: 10.1038/bjc.2016.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selth LA, et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene. 2017;36(1):24–34. doi: 10.1038/onc.2016.185. [DOI] [PubMed] [Google Scholar]
- 18.Pickl JM, et al. Ago-RIP-Seq identifies Polycomb repressive complex I member CBX7 as a major target of miR-375 in prostate cancer progression. Oncotarget. 2016;7(37):59589–59603. doi: 10.18632/oncotarget.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat Commun. 2017;8:14270. doi: 10.1038/ncomms14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin HM, et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br J Cancer. 2017;116(8):1002–1011. doi: 10.1038/bjc.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, et al. Analysis of circulating miRNAs 21 and 375 as potential biomarkers for early diagnosis of prostate cancer. Neoplasma. 2016;63(4):623–8. doi: 10.4149/neo_2016_417. [DOI] [PubMed] [Google Scholar]
- 22.Kachakova D, et al. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015;34(3):189–200. doi: 10.1089/dna.2014.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watahiki A, et al. Plasma miRNAs as biomarkers to identify patients with castration-resistant metastatic prostate cancer. Int J Mol Sci. 2013;14(4):7757–70. doi: 10.3390/ijms14047757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eliasson L. The small RNA miR-375 - a pancreatic islet abundant miRNA with multiple roles in endocrine beta cell function. Mol Cell Endocrinol. 2017 doi: 10.1016/j.mce.2017.02.043. [DOI] [PubMed] [Google Scholar]
- 25.Foj L, et al. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate. 2017;77(6):573–583. doi: 10.1002/pros.23295. [DOI] [PubMed] [Google Scholar]
- 26.Antonarakis ES, et al. Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J Clin Oncol. 2017:JCO2016701961. doi: 10.1200/JCO.2016.70.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josefsson A, et al. Circulating Tumor Cells as a Marker for Progression-free Survival in Metastatic Castration-naive Prostate Cancer. Prostate. 2017;77(8):849–858. doi: 10.1002/pros.23325. [DOI] [PubMed] [Google Scholar]
- 28.Cheng HH, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8(6):e64795. doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1A. Scatterplot Display of miRNAs (CTs) with Significant Associations with 28-week PSA Response Category. 1B. Scatterplot of miRNA (CTs) with Significant Association with Baseline CTC Count Category
Supplementary Tables 1A and 1B: All miRNAs w/ p-values