Abstract
The PadR family is a large group of transcriptional regulators that function as environmental sensors. PadR negatively controls the expression of phenolic acid decarboxylase, which detoxifies harmful phenolic acids. To identify the mechanism by which PadR regulates phenolic acid-mediated gene expression, we performed structural and mutational studies of effector and operator recognition by Bacillus subtilis PadR. PadR contains an N-terminal winged helix-turn-helix (wHTH) domain (NTD) and a C-terminal homodimerization domain (CTD) and dimerizes into a dolmen shape. The PadR dimer interacts with the palindromic sequence of the operator DNA using the NTD. Two tyrosine residues and a positively charged residue in the NTD provide major DNA-binding energy and are highly conserved in the PadR family, suggesting that these three residues represent the canonical DNA-binding motif of the PadR family. PadR directly binds a phenolic acid effector molecule using a unique interdomain pocket created between the NTD and the CTD. Although the effector-binding site of PadR is positionally segregated from the DNA-binding site, effector binding to the interdomain pocket causes PadR to be rearranged into a DNA binding-incompatible conformer through an allosteric interdomain-reorganization mechanism.
INTRODUCTION
Microorganisms use danger-sensing mechanisms to defend themselves from toxic environments (1,2). Phenolic acids, including p-coumaric acid and ferulic acid, are prevalently found in soil, food and the gut, but they have toxic effects on some bacteria (Supplementary Figure S1) (3,4). For example, high phenolic acid levels in the diet of ruminants were shown to inhibit the growth of enterohemorrhagic Escherichia coli strain O157:H7 in the colon (5). To avoid the toxicity of phenolic acids, some bacterial species, such as Bacillus subtilis, Bacillus pumilus, Pediococcus pentosaceus and Lactobacillus plantarum, express phenolic acid decarboxylase (padC gene product in B. subtilis), which converts antimicrobial phenolic acids into less toxic vinyl derivatives, as a defense mechanism known as the phenolic acid stress response (6–9). The transcription of the padC gene is regulated by a negative transcription factor, phenolic acid decarboxylase regulator (PadR), in a substrate-inducible manner (8,10–13). In the absence of phenolic acids, PadR binds palindromic sequences in the padC promoter and represses the transcription of the padC gene. However, when p-coumaric acid or ferulic acid are present, PadR senses the phenolic acids and dissociates from the padC operator DNA, resulting in the derepression of the padC transcription.
Since PadR was first reported in P. pentosaceus, the PadR family has continued to expand to include at least 9000 PadR-like protein sequences in the Pfam database (10,14). PadR family members function as transcriptional regulators that are involved in various cellular survival processes, such as toxin production, detoxification, multidrug resistance, antibiotic biosynthesis and carbon catabolism (13,15–20). The PadR family interacts with the operator DNA using a winged helix-turn-helix (wHTH) motif, which contains a three-helix motif (α1, α2 and α3) and a β-stranded wing (β1 and β2) (16,19–24) and exhibits a high structural similarity to the multiple antibiotic resistance regulator (MarR) family in the wHTH superfamily (20,25). Moreover, PadR and its homologues commonly form a homodimer as the DNA-binding unit (19,20,23,26). Although the PadR family members share the conserved DNA-binding features, the PadR family is subdivided into subfamily-1 and subfamily-2, which are different in their molecular sizes and dimerization patterns (16). PadR belongs to the subfamily-1, along with Vibrio cholerae AphA, Listeria monocytogenes LadR, Streptomyces argillaceus MtrY and Corynebacterium glutamicum VanR, and the molecular weights of these proteins are in the range of 19–22 kDa (16–18,20). The PadR subfamily-1 contains two domains and dimerizes using the C-terminal domain that is physically separated from the N-terminal wHTH motif (20). In contrast, the subfamily-2 that includes Lactococcus lactis LmrR and Synechococcus elongatus Pex adopts a single-domain structure with a molecular weight of 11–13 kDa and forms a dimer primarily through the C-terminal helix that is appended to the wHTH motif (19,23,24,26,27).
Despite the discovery of a growing number of PadR family members and the increasing interest in their critical functions, the structural basis of effector-mediated gene regulation is poorly understood in the PadR family because of the highly limited structural information concerning the interactions of PadR family members with effectors and operator DNAs. The structure of LmrR, a subfamily-2 member, in complex with its effectors was solely characterized in bacteria (19,22). LmrR contains a pocket between the two subunits of the LmrR dimer where it binds an effector molecule. However, it is questionable if the effector-binding mode of LmrR is similarly employed by PadR subfamily-1 members. Moreover, because of the unavailability of the structures of complexes between PadR family proteins and operator DNAs, we do not understand how PadR family members recognize their operator DNAs and how effectors induce the dissociation of PadR family members from the DNAs. To reveal the structural mechanism by which PadR regulates effector-responsive transcription, we performed comparative structural and mutational analyses of B. subtilis PadR interactions both with phenolic acid effectors and with the operator DNA. PadR binds phenolic acid in the interdomain pocket, and effector binding allosterically induces PadR to dissociate from the DNA through an interdomain-reorganization mechanism.
MATERIALS AND METHODS
Construction of PadR expression vectors
The PadR gene (UniProt E0TW95; residues 1–182) was amplified by PCR from the genomic DNA of B. subtilis using a forward primer (5′-TAAGGATCCGATGAGAGTATTAAAATACGCCATATTAGG-3′, the BamHI restriction-enzyme site in bold) and a reverse primer (5′-TAAGGCGTCGACCTAATCCTCATCTATCATAGCTA AAATCG-3′, the SalI restriction-enzyme site in bold). The PCR product was ligated into a modified pET49b vector that contains an N-terminal His6 tag and a thrombin cleavage site to construct the PadR expression vector (28,29). The ligation product was transformed into E. coli strain DH5α and a correct clone was confirmed by restriction enzyme digestion and DNA sequencing. For PadR overexpression, the PadR expression vector was transformed into E. coli strains BL21 (DE3) and B834 (DE3). The PadR gene was mutated using the QuikChange site-directed mutagenesis protocol (Agilent) with DNA primers containing the mutated sequences. The sequences of the PadR mutants were verified by DNA sequencing.
Expression and purification of PadR protein
Escherichia coli BL21 (DE3) cells transformed with the PadR expression vector were grown in LB medium at 37°C, and PadR overexpression was induced by adding IPTG to a final concentration of 1 mM when the optical absorbance at 600 nm reached 0.6. The cells were further grown for ∼17 h at 18°C and harvested by centrifugation. The cell pellet was resuspended and sonicated in a solution containing 50 mM Tris, pH 8.0, 200 mM NaCl and 1 mM phenylmethylsulfonyl fluoride. The cell lysate was cleared by centrifugation. The resulting supernatant containing the soluble PadR protein was incubated with Ni-NTA resin (Qiagen) in the presence of 10 mM imidazole for 2 h at 4°C. The resin was washed with a solution of 10 mM imidazole, 50 mM Tris, pH 8.0 and 200 mM NaCl. The PadR protein was eluted with a solution of 250 mM imidazole, 50 mM Tris, pH 8.0 and 200 mM NaCl and dialyzed against a solution of 20 mM Tris, pH 8.0 and 150 mM NaCl. The PadR protein was digested with thrombin to remove the His6 affinity tag. The tag-free PadR protein was further purified by anion-exchange chromatography using a Mono Q 10/100 column (GE Healthcare) with a NaCl gradient of 0–500 mM.
Selenomethionine-labeled PadR (SeMet-PadR) protein was expressed in E. coli B834 (DE3) cells using L-SeMet-supplemented M9 minimal medium (Molecular Dimensions) and purified to homogeneity with Ni-NTA affinity chromatography and anion-exchange chromatography, similar to the native protein.
Crystallization and structure determination of PadR
PadR was crystallized at 18°C using the sitting-drop vapor-diffusion method. The native PadR protein was crystallized under the condition of 20% polyethylene glycol (PEG) 3350 and 0.1 M sodium acetate, pH 4.0. SeMet-PadR crystals were produced in a similar condition to the native crystals. To grow diffraction-quality-sized SeMet-PadR crystals, the streak-seeding method was applied by placing crystal seeds in a drop containing 0.5 μl of 10 mg/ml SeMet-PadR protein and 0.5 μl of 20% PEG 3350 and 0.1 M sodium acetate, pH 4.0. PadR–effector complex crystals were obtained via soaking and co-crystallization methods. The PadR crystals were soaked in 10 mM p-coumaric acid (or 10 mM ferulic acid), 22% PEG 3350 and 0.1 M sodium acetate, pH 4.0, for 18 h. For co-crystallization, PadR at 10 mg/ml was mixed with a 3-fold molar excess of p-coumaric acid and crystallized in 12% PEG 3350 and 0.1 M phosphate-citrate, pH 5.5. PadR was also co-crystallized with a 28-bp dsDNA (5′-C1GGAACATGTAAATAGTTACATGATTAC28-3′; subscripts represent base numbers), containing the padC operator sequence (underlined), in a drop comprising 0.5 μl of 10 mg/ml PadR–dsDNA complex and 0.5 μl of 1.4 M ammonium sulfate, 0.1 M MES, pH 6.5 and 0.2 M lithium sulfate. The crystals were flash-cooled at 100 K in the presence of 25% glycerol or 25% ethylene glycol and diffracted at beamlines 5C and 7A at the Pohang Accelerator Laboratory. The X-ray diffraction data were indexed and scaled using the HKL2000 program (30).
The SeMet-PadR structure was determined by single-wavelength anomalous diffraction (SAD) phasing on the X-ray diffraction data collected at the peak wavelength (0.97930 Å) of selenium. The selenium positions were located and the phases were calculated using the AutoSol program of the Phenix suite (31,32). The SeMet-PadR model was constructed and refined using the Coot and Refmac5 programs, respectively (33,34). The crystal structures of native PadR and its complexes with p-coumaric acid, ferulic acid or dsDNA were determined by molecular replacement with the Phaser program using the SeMet-PadR and native PadR structures, respectively, as search models, and the final models were generated through iterative cycles of model building and refinement (33–35).
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed using a MicroCal iTC200 instrument (Malvern). A total of 1 mM p-coumaric acid in a syringe was titrated into the sample cell containing 80 μM PadR protein at 25°C with stirring at 750 rpm. A total of 0.4 μl and 2 μl of p-coumaric acid were injected for the initial injection and the other injections, respectively. Before the ITC experiments, the PadR protein was dialyzed against reaction buffer (20 mM Tris, pH 8.0 and 150 mM NaCl) for 12 h at 4°C and p-coumaric acid was dissolved in the reaction buffer used for dialysis. Data fitting and evaluation were performed with the Origin 7 software (MicroCal) using the one-site binding model (36).
Fluorescence polarization
A fluorescence polarization (FP) assay was performed to determine the dsDNA-binding affinity of wild-type (WT) and mutant PadR (37). The PadR proteins were prepared by dialyzing against FP assay buffer containing 20 mM Tris, pH 8.0 and 100 mM NaCl. The dsDNA was generated by incubating a fluorescein-labeled oligonucleotide (5′-C1GGAACATGTAAATAGTTACATGATTAC28-3′) and its unlabeled complementary counterpart (5′-G1TAATCATGTAACTATTTACATGTTCCG28-3′) at 95°C for 5 min and then slowly cooling the reaction to the room temperature. The dsDNA (1 nM) was incubated with serially diluted PadR dimer (PadRdimer) protein (0.6 nM–4 μM) in the FP assay buffer in 96-well plates (Corning). The FP signals were measured using an Infinite F200 PRO instrument (Tecan; excitation wavelength, 485 nm; emission wavelength, 535 nm) and analyzed using the Prism 5 program (GraphPad).
Electrophoretic mobility shift assay
An electrophoretic mobility shift assay (EMSA) was performed using PadR protein and operator dsDNA in the presence or absence of p-coumaric acid to monitor the PadR–dsDNA interaction and the inhibitory effect of the phenolic acid on this interaction. For the interaction assay, the purified PadRdimer protein (0.5–2 μM) was incubated with the dsDNA (1 μM; 5′-A4ACATGTAAATAGTTACATGAT25-3′) in 20 mM Tris, pH 8.0 and 150 mM NaCl. The samples were electrophoretically separated on 12% polyacrylamide gels in Tris-borate-EDTA (TBE) buffer for 80 min at 120 V. For the phenolic acid-induced inhibition assay, a mixture of 0.6 μM PadRdimer protein and 0.2 μM dsDNA (5′-C1GGAACATGTAAATAGTTACATGATTAC28-3′) was incubated with 0.2–5.0 mM p-coumaric acid. The samples were electrophoretically separated on 5% polyacrylamide gels in TBE buffer for 40 min at 120 V. After electrophoresis, the gels were stained with ethidium bromide or silver to visualize the DNA and protein, respectively.
Gel-filtration chromatography
Gel-filtration chromatography was performed using a Superdex 200 10/300 column in 20 mM Tris, pH 8.0 and 150 mM NaCl for the analysis of PadR dimerization and its interaction with the operator DNA. To determine the oligomeric state of PadR, 300 μl of PadR WT and dimerization-disrupting mutants, Phe104Arg and Leu156Glu, were injected into the column at a monomer concentration of 16 μM. To examine the PadR–dsDNA interaction, 300 μl of PadRdimer (1.3 μM), dsDNA (1.3 μM; 5′-A4ACATGTAAATAGTTACATGAT25-3′) and PadR–dsDNA complex were applied to the column. The elution profiles of the samples were monitored by UV absorption at 280 nm.
Tryptophan fluorescence-based protein denaturation assay
To analyze the protein stabilities of PadR WT and mutants, a tryptophan fluorescence-based protein denaturation assay was performed in the presence of guanidine hydrochloride (GuHCl) as a protein denaturant. The PadR protein (WT or mutants; 20 μg) was incubated in 1.1–4.0 M GuHCl, 20 mM Tris, pH 8.0 and 100 mM NaCl for 10 min at room temperature. Tryptophan fluorescence was measured using a Synergy H1 instrument (BioTek; excitation wavelength, 295 nm; emission wavelength, 335 nm).
RESULTS
Overall structure of PadR
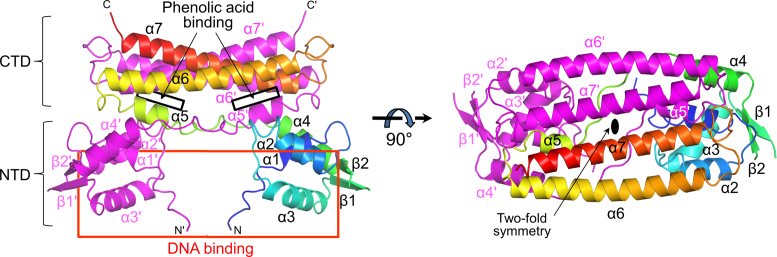
The crystal structure of the apo form of PadR from B. subtilis was determined using SAD phasing of selenomethionine-incorporated PadR protein, and its native structure (PadR1 structure) was refined to 1.7 Å resolution (Supplementary Tables S1 and 2). One PadR polypeptide chain is located in the asymmetric unit of the crystal and forms a homodimer with its crystallographic partner (Figure 1). In the monomer, the PadR structure is separated into two domains, N-terminal and C-terminal domains (NTD and CTD, respectively) (Figure 1; Supplementary Figures S2 and 3). The NTD displays the canonical wHTH domain structure with four α-helices (α1, α2, α3 and α4) and two antiparallel β-strands (β1 and β2) in the order of α1-α2-α3-β1-β2-α4. The NTD is connected to the CTD via an extended 11-residue loop. The CTD consists of one short α-helix (α5) and two longitudinal α-helices (α6 and α7) and presents the signature structure of the PadR subfamily-1 (see below).
Figure 1.
Structure of the PadR dimer. Two PadR chains related by the crystallographic 2-fold symmetry are depicted as ribbons. One chain is rainbow-colored [N-terminus (N), blue; C-terminus (C), red] and the other is colored in magenta. The dsDNA-binding and phenolic acid-binding sites of PadR are schematically shown as red and black boxes, respectively. Secondary structural elements are labeled and a prime symbol indicates the dimerization partner.
PadR is homodimeric as for other wHTH superfamily members. In gel-filtration chromatography, PadR was eluted as a single peak at the size of its dimer (Supplementary Figure S4A). Moreover, in the crystal structure, PadR dimerizes in a similar manner to AphA, displaying the PadR superfamily-1-specific dolmen shape with two angled NTD legs under a plate of two CTDs (Figure 1 and Supplementary Figure S5) (20). Upon dimerization, PadR buries a surface area of ∼3460 Å2 on each side through both polar and apolar interactions (Supplementary Figure S4B). Depending on the domains involved in dimerization, the dimerization interface is segregated into two surfaces: (i) the NTD-CTD′ interface and its 2-fold symmetry-related interface, NTD′-CTD and (ii) the CTD-CTD′ interface (the prime indicates the dimerization partner) (Supplementary Figure S4B). At the NTD-CTD′ dimerization interface, the upper part of the NTD interacts with the N-terminal region of the CTD′. The CTD-CTD′ interface is generated by the symmetrical interaction between the two CTDs of the PadRdimer. Although both the NTD and the CTD participate in dimerization, the CTD is responsible for ∼70% of the dimerization interface and is considered as the primary dimerization domain.
The PadR dimerization interface observed in the PadR structure was verified by a mutational study in solution. PadR Phe104 and Leu156 residues at the NTD-CTD′ and CTD-CTD′ interfaces were replaced with arginine and glutamate residues, respectively. In gel-filtration chromatography, both mutant proteins were eluted as monomers, indicating that PadR dimerization occurs through the dimerization interface that includes the Phe104 and Leu156 residues (Supplementary Figure S4A). The dimerization defect also destabilized the PadR protein. In a tryptophan fluorescence-based protein denaturation analysis, Phe104Arg and Leu156Glu mutations substantially lowered protein stability [denaturant concentrations at half denaturation (C1/2) of 1.81 M and 1.76 M, respectively] than the WT (C1/2, 2.96 M), suggesting that PadR protein is stabilized through dimerization (Supplementary Figure S6).
Interaction of PadR with the operator DNA
As a transcription factor, B. subtilis PadR recognizes palindromic sequences (bsPadR-1 and bsPadR-2 operator sequences shown in Supplementary Figure S7) within the padC promoter region (13). We further probed the interaction of the PadR protein with the bsPadR-1 operator dsDNA using an array of biophysical methods. In an EMSA, the addition of PadR protein altered the mobility of the operator dsDNA in a saturable manner (Supplementary Figure S8A). In gel-filtration chromatography, the mixture of PadR and dsDNA was eluted as a peak that corresponded to a 1:1 PadRdimer:dsDNA complex (Supplementary Figure S8B). An FP assay using a fluorescein-labeled operator DNA revealed the relatively high dsDNA-binding affinity of PadRdimer with an equilibrium constant (Kd) of 8.3 ± 1.8 nM (Supplementary Figure S8C and Table 1). Based on our biophysical studies of the PadR–DNA interaction, one PadR dimer directly interacts with one copy of the bsPadR-1 operator dsDNA.
Table 1. Operator dsDNA-binding capacity of WT and mutant PadR.
| PadR | K d (nM) | K d ratio (mutant/WT) | B max (mP) | B max ratio (mutant/WT) |
|---|---|---|---|---|
| WT | 8.3 ± 1.8 | 269 ± 4 | ||
| Major groove interaction site | ||||
| Arg2Ala | 8.3 ± 0.7 | 1.0 | 229 ± 12 | 0.85 |
| Tyr20Ala | 141.2 ± 15.9 | 17.1 | 269 ± 7 | 1.00 |
| Trp34Ala | 11.9 ± 1.7 | 1.4 | 256 ± 3 | 0.95 |
| Lys37Ala | 13.2 ± 0.8 | 1.6 | 240 ± 5 | 0.89 |
| His38Ala | 27.4 ± 4.9 | 3.3 | 260 ± 6 | 0.97 |
| Ser39Ala | 13.9 ± 0.5 | 1.7 | 253 ± 4 | 0.94 |
| Gln40Ala | 8.7 ± 0.1 | 1.1 | 259 ± 2 | 0.96 |
| Tyr42Ala | >1000 | >100 | ||
| Lys95Ala | 37.0 ± 12.7 | 4.5 | 217 ± 4 | 0.81 |
| Tyr20Ala/His38Ala | 540.6 ± 99.7 | 65.3 | 267 ± 9 | 0.99 |
| His38Ala/Ser39Ala | 83.5 ± 16.3 | 10.1 | 247 ± 8 | 0.92 |
| His38Ala/Tyr42Ala | >1000 | >100 | ||
| Minor groove interaction site | ||||
| Gln61Ala | 10.0 ± 0.8 | 1.2 | 266 ± 2 | 0.99 |
| Lys64Ala | 23.9 ± 2.3 | 2.9 | 242 ± 9 | 0.90 |
| Leu65Ala | 75.0 ± 5.8 | 9.1 | 216 ± 2 | 0.81 |
| Δwing | >1000 | >100 | ||
| Boundary interaction site | ||||
| Lys67Ala | 534.2 ± 96.2 | 64.5 | 264 ± 2 | 0.98 |
| Lys68Ala | 48.8 ± 5.5 | 5.9 | 252 ± 2 | 0.94 |
| Phenolic acid-binding site | ||||
| His154Ala | 17.2 ± 1.8 | 2.1 | 258 ± 1 | 0.96 |
| Arg164Ala | 17.0 ± 1.8 | 2.1 | 262 ± 2 | 0.98 |
| His154Ala/Arg164Ala | 12.9 ± 1.7 | 1.6 | 258 ± 5 | 0.96 |
| Allosteric center | ||||
| Gln32Ala | 7.8 ± 0.2 | 0.9 | 265 ± 6 | 0.99 |
| Gln32Glu | 11.1 ± 0.4 | 1.3 | 264 ± 3 | 0.98 |
The PadR–dsDNA binding affinity was determined using the FP assay and is represented by the dissociation equilibrium constant, Kd. The maximal binding level, Bmax, is also shown. The data represent the means ± S.D. from at least three independent experiments.
PadR–DNA binding interface
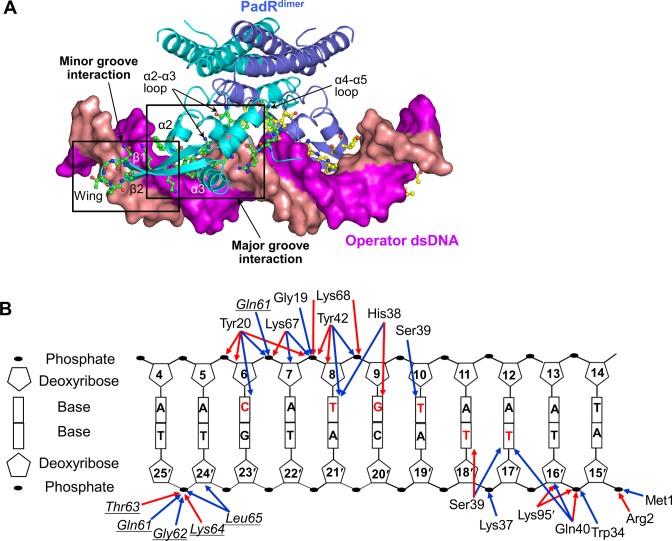
The molecular interaction of PadR with the operator DNA was visualized in the crystal structure of PadR in complex with a 28-bp dsDNA containing the bsPadR-1 operator sequence (PadRdsDNA) (Figure 2 and Supplementary Table S2). In the structure of the complex, the dsDNA diagonally traverses the PadRdimer between the two NTDs under the CTDs. The PadR dimer symmetrically recognizes the palindromic sequence of the dsDNA using the two NTDs, with a 1:1 stoichiometry of PadRdimer to dsDNA. The PadR monomer simultaneously recognizes the major and minor grooves of the dsDNA, forming major and minor groove interaction sites, respectively (Figure 2A).
Figure 2.
PadR–dsDNA interaction observed in the PadRdsDNA structure. (A) Overall PadRdsDNA structure. PadRdimer is depicted as ribbons (cyan and light blue), and the dsDNA is shown as a surface representation (magenta and salmon). PadR residues located in the binding interface are shown as ball-and-stick models (green and yellow). (B) A schematic diagram of the PadR–dsDNA interaction. Red and blue arrows indicate hydrogen bonds and van der Waals interactions, respectively. PadR residues in the minor groove interaction site are shown in italics and are underlined to differentiate them from the residues of the major groove interaction site. DNA bases that PadR makes contacts with are labeled in red.
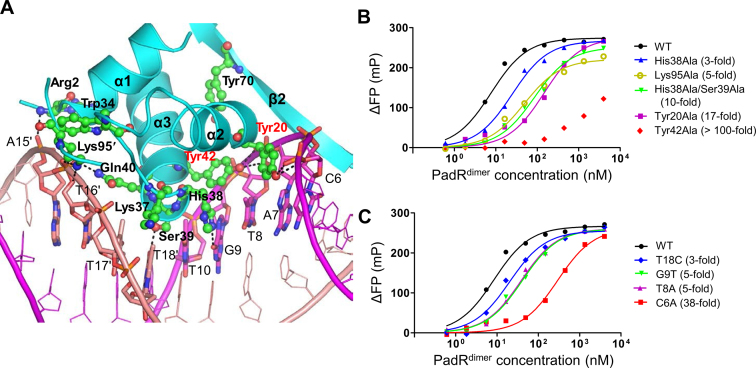
The major groove interaction site is primarily generated by inserting the N-terminal regions of the α2 and α3 helices into the major groove of dsDNA, covering ∼68% of the entire buried surface area (∼1000 Å2 on one PadR monomer) (Figures 2 and 3A). In the major groove interaction site, five PadR residues (Tyr20, His38, Ser39, Gln40 and Tyr42) from the α2 and α3 helices make direct contacts with the bases of C6, T8, G9, T10, T17′ and T18′. The side chains of Tyr20 and Tyr42, which are located at the beginning of the α2 and α3 helices, respectively, form van der Waals interactions with DNA bases at C6 and T8. The side chains of His38 and Ser39 form hydrogen bonds with oxygen atoms at G9 O6 and T18′ O4, respectively, in addition to van der Waals interactions with T8, G9, T10, T17′ and T18′. These direct contacts between the PadR residues and DNA bases are buttressed by DNA backbone interactions located on both sidewalls of the major groove (Figures 2B and 3A). On one side, the Arg2, Trp34, Lys37, Gln40 and Lys95′ residues of PadR make contacts with the DNA backbone at A15′, T16′ and T18′. On the opposite wall of the major groove, the Tyr20 and Tyr42 residues of PadR extensively interact with the DNA backbone at C6-G9 through a network of hydrogen bonds and van der Waals interactions.
Figure 3.
Major groove interaction site and its mutational analysis. (A) Major groove interaction in the PadRdsDNA structure. PadR is shown as cyan ribbons, and its DNA-binding residues are depicted as green ball-and-stick models. Tyr20 and Tyr42, which play a key role in DNA binding, are labeled in red. The dsDNA is colored in magenta and salmon for leading and non-leading strands, respectively, and its PadR-interacting residues are shown as sticks. Hydrogen bonds are represented by dotted lines. (B) FP analysis of the interaction between PadR WT or mutants (Tyr20Ala, His38Ala, His38Ala/Ser39Ala, Tyr42Ala and Lys95Ala) and the operator dsDNA. The affinity differences between the mutants and the WT are shown in parentheses. The data are representative of at least three independent experiments that yielded similar results. (C) FP analysis of the interaction between PadR and the WT or mutated operator DNAs (C6A, T8A, G9T and T18C). The affinity differences between the mutants and the WT are shown in parentheses. The data are representative of at least three independent experiments that yielded similar results.
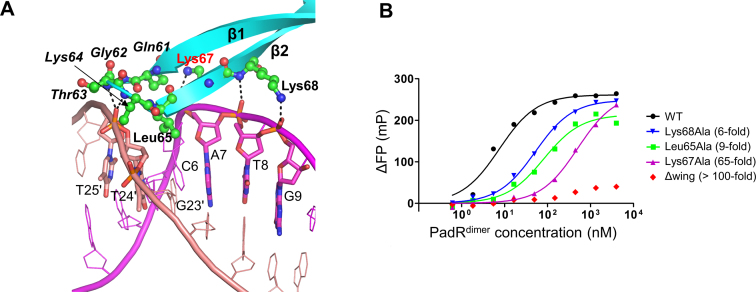
Whereas the major groove interaction is mediated by the helix-turn-helix motif of the PadR NTD, the minor groove interaction occurs by positioning the β-stranded wing of the PadR NTD in the minor groove of the operator dsDNA at C6, G23′, T24′ and T25′ residues (Figures 2 and 4A). The main chain atoms of PadR residues 61–65 from the β1 and β2 strands and their connecting loop primarily interact with the phosphate-ribose backbone of DNA T24′ and T25′. PadR Leu65 is the only residue of which the side chain is inserted into the minor groove toward the DNA bases at C6 and G23′.
Figure 4.
Minor groove and boundary interaction sites and their mutational analysis. (A) Minor groove and boundary interactions in the PadRdsDNA structure. PadR is shown as cyan ribbons, and its DNA-binding residues are depicted as green ball-and-stick models. PadR residues in the minor groove interaction site are shown in italics. Lys67, which plays a key role in DNA binding, is labeled in red. The dsDNA is colored in magenta and salmon for the leading and non-leading strands, respectively, and its PadR-interacting residues are shown as sticks. Hydrogen bonds are represented by dotted lines. (B) FP analysis of the interaction between PadR WT or mutants (Leu65Ala, Lys67Ala, Lys68Ala and Δwing) and the operator dsDNA. The affinity differences between the mutants and the WT are shown in parentheses. The data are representative of at least three independent experiments that yielded similar results.
The β2 strand from the β-stranded wing is also involved in DNA binding using Lys67 and Lys68. The side chains of PadR Lys67 and Lys68 run in opposite directions and stably reside on a ridge between the major and minor grooves of dsDNA (Figures 2B and 4A). The Lys67 and Lys68 side chain atoms form hydrogen bonds with the phosphate oxygen atoms at DNA A7 and G9 and also make van der Waals contacts with the backbone atoms of DNA at A7-G9.
Critical residues of PadR in the PadR–DNA interaction
To evaluate the energetic significance of PadR interface residues in DNA binding, fourteen PadR residues were selected for alanine scanning mutagenesis. Among nine alanine mutations in the major groove interaction site, Tyr20Ala and Tyr42Ala remarkably reduced the DNA-binding affinity of PadR by 17-fold and at least 100-fold, respectively, suggesting that Tyr20 and Tyr42 form a DNA-binding hot spot for the major groove interaction (Figure 3B and Table 1). To exclude the possibility that the mutational defects of PadR in DNA binding were caused by protein misfolding, the protein stability of PadR mutants was assessed using a tryptophan fluorescence-based protein denaturation assay. PadR Tyr20Ala and Tyr42Ala mutants exhibited similar tryptophan fluorescence profiles to the PadR WT in the presence of a protein denaturant, indicating that the Tyr20Ala and Tyr42Ala mutations did not affect the protein stability of PadR (Supplementary Figure S6). Thus, we suggest that the observed DNA-binding defects of the mutants are directly caused by the loss of the interaction of the Tyr20 and Tyr42 side chains with DNA, but not by protein misfolding. Interestingly, Tyr20 and Tyr42 are highly conserved throughout the PadR family, including subfamily-1 and subfamily-2, suggesting that the two tyrosine residues constitute the consensus DNA-recognition motif of the PadR family (Supplementary Figure S2). Notably, in the PadRdsDNA structure, the Tyr20 and Tyr42 residues interact with C6, A7, T8 and G9, of which the conformers deviate from a regular B-form DNA (twist values of 18–25° for DNA C6, A7, T8 and G9 residues; a twist value of 36° for a regular B-form DNA; an average twist value of ∼32° for the 28-bp operator DNA in the PadRdsDNA structure), causing the structure of the operator dsDNA to be slightly bent (38). In addition to mutations at the two tyrosine residues, alanine mutations at His38 and Lys95 reduced the DNA-binding affinity of PadR by 3- to 4-fold. In contrast, a single alanine mutation for PadR Ser39 had a minimal effect on the DNA-binding affinity of PadR (1.7-fold) in solution although Ser39 forms multiple contacts with DNA bases at T10, T17′ and T18′ in the crystal. However, the mutational effect of Ser39Ala would not be negligible due to the cooperativity of Ser39 with His38 in DNA binding. The His38Ala/Ser39Ala double mutation lowered the DNA-binding affinity of PadR by ∼10-fold, whereas the His38Ala and Ser39Ala single mutations lowered the affinity by 3.3-fold and 1.7-fold, respectively. Therefore, Ser39 displays weak positive cooperativity with His38 in DNA binding. Interestingly, in the PadRdsDNA structure, Tyr20, His38, Ser39 and Tyr42 are colocalized at one side of the protruding α2 and α3 helices and are inserted into the concave side of the major groove, contributing to shape complementarity between PadR and dsDNA.
In the minor groove interaction site, alanine substitutions at Leu65 and Lys64 reduced DNA-binding affinity by 9- and 3-fold, respectively (Figure 4B and Table 1). The side chain of Leu65 protrudes into the minor groove and appears to optimize shape complementarity to improve DNA binding. The critical role of the minor groove interaction in PadR–DNA binding was confirmed by a deletion mutant (Δwing) in which PadR residues 60–66 at the β-stranded wing were replaced with a Gly-Ala-Ala tripeptide. The PadR Δwing was severely deficient in DNA binding; the mutant exhibited only ∼15% of the maximal binding level of the PadR WT, even at a PadRdimer concentration of 4 μM (Figure 4B and Table 1). Based on these data, we conclude that the minor groove interaction is required to maximize the PadR–DNA-binding affinity.
In addition to PadR residues at the major and minor groove interaction sites, two lysine residues, Lys67 and Lys68, found at the boundary between the major and minor groove interaction sites also play a critical role in DNA binding (Figure 4A). Lys67Ala and Lys68Ala mutants displayed 65-fold and 6-fold lower DNA-binding affinity, respectively, compared to the WT (Figure 4B and Table 1). In particular, Lys67 is highly conserved as a positively charged residue in both PadR subfamily-1 and subfamily-2, suggesting that in addition to Tyr20 and Tyr42, residue 67 is a signature residue of the PadR family required for DNA recognition (Supplementary Figure S2).
DNA base specificity of PadR
In the PadRdsDNA structure, each PadR monomer makes direct contacts with six nucleotide bases at C6, T8, G9, T10, T17′ and T18′ (Figure 2B). The base specificity of PadR observed in the PadRdsDNA structure was verified by determining the binding affinity of PadR for a 28-bp operator DNA in which the PadR-contacting DNA bases (C6, T8, G9, T10, T17′ and T18′) were individually changed (Figure 3C and Table 2). Base substitution at DNA residues C6 and T8 reduced the PadR-binding affinity of dsDNA by at least 4-fold. The most significant effect of base substitution was observed at C6, of which the changes to adenine, guanine and thymine reduced the PadR-binding affinity of dsDNA by 38-, 24- and 19-fold, respectively, indicating that the cytosine base at DNA residue 6 is indispensable for DNA recognition by PadR (Table 2). The cytosine preference of PadR can be explained by the direct interaction between the DNA C6 base and the PadR Tyr20 side chain in the major groove interaction site and by shape complementarity between the protruding Leu65 side chain and the C6-G23′ base pair in the minor groove interaction site (Figures 3A and 4A). T8A substitution reduced the PadR-binding affinity by 5-fold by disrupting interactions with Tyr42 and His38. G9T and G9A changes decreased the affinity by 5- and 2-fold, respectively, suggesting that PadR displays a weak purine preference potentially due to a steric clash of His38 with pyrimidine bases. T18C substitution lowered the affinity by 3-fold presumably through the loss of interaction with Ser39. Thus, our mutational analysis, combined with the structural study, demonstrated that the PadR dimer symmetrically recognizes 18-bp dsDNA (C6NT8G9NA11NNNNNNT18NC20A21NG23, N representing any nucleotide in the leading DNA strand) containing the two palindromic repeats of 6-bp dsDNA that are separated by a 6-bp spacer.
Table 2. PadR-binding capacity of the operator dsDNA and its sequence variants.
| dsDNA | K d (nM) | K d ratio (mutant/WT) | B max (mP) | B max ratio (mutant/WT) |
|---|---|---|---|---|
| Native operator | ||||
| C6A/C6′A | 311.5 ± 45.5 | 37.6 | 267 ± 3 | 0.99 |
| C6G/C6′G | 200.4 ± 21.7 | 24.2 | 269 ± 3 | 1.00 |
| C6T/C6′T | 160.5 ± 6.9 | 19.4 | 271 ± 3 | 1.01 |
| T8A/T8′A | 41.0 ± 4.0 | 4.9 | 264 ± 3 | 0.98 |
| G9T/G9′T | 37.8 ± 6.7 | 4.6 | 261 ± 1 | 0.97 |
| G9A/G9′A | 15.7 ± 2.9 | 1.9 | 262 ± 5 | 0.97 |
| T10A/T10′A | 11.6 ± 1.3 | 1.4 | 265 ± 6 | 0.99 |
| T17A/T17′A | 9.4 ± 1.4 | 1.1 | 268 ± 10 | 1.00 |
| T18C/T18′C | 20.8 ± 1.9 | 2.5 | 261 ± 9 | 0.97 |
| T18G/T18′G | 9.9 ± 1.7 | 1.2 | 271 ± 12 | 1.01 |
| Bridge operator | 8.9 ± 2.1 | 1.1 | 246 ± 8 | 0.92 |
The PadR–dsDNA binding affinity was determined using the FP assay and is represented by the dissociation equilibrium constant, Kd. The maximal binding level, Bmax, is also shown. The data represent the means ± S.D. from at least three independent experiments.
In the PadRdsDNA crystal, the asymmetric unit contains four PadR dimers (dimers 1–4) and two DNA duplexes, to be inconsistent with the 1:1 molecular stoichiometry of the PadR dimer and 28-bp dsDNA (Supplementary Figure S9). In the crystal, the blunt-ended DNA duplexes are longitudinally aligned through crystal packing to form a pseudo-continuous duplex. Fortuitously, two neighboring DNA duplexes are brought to build another operator site (named as bridge operator DNA) by combining the latter half of one duplex and the former half of the other duplex. Although the bridge operator is partially different in DNA sequence from the native operator (native operator sequence, 5′-C1GGAACATGTAAAT14:A15GTTACATGATTAC28-3′; bridge operator sequence, 5′-A15GTTACATGATTAC28:C1GGAACATGTAAAT14-3′; identical sequences in bold and base specificity sites underlined), the bridge operator satisfies the sequence (C6NT8G9) and 6-bp spacer length requirements of the native operator DNA, and thus the two halves of the bridge operator are clasped by the PadR dimer 2 or dimer 4 in the crystal. In support of this observation, the bridge operator DNA exhibited a similar PadR-binding mode in the PadRdsDNA structure and displayed essentially identical DNA-binding affinity in the FP assay compared to the native operator DNA (Table 2).
Interaction of PadR with phenolic acids
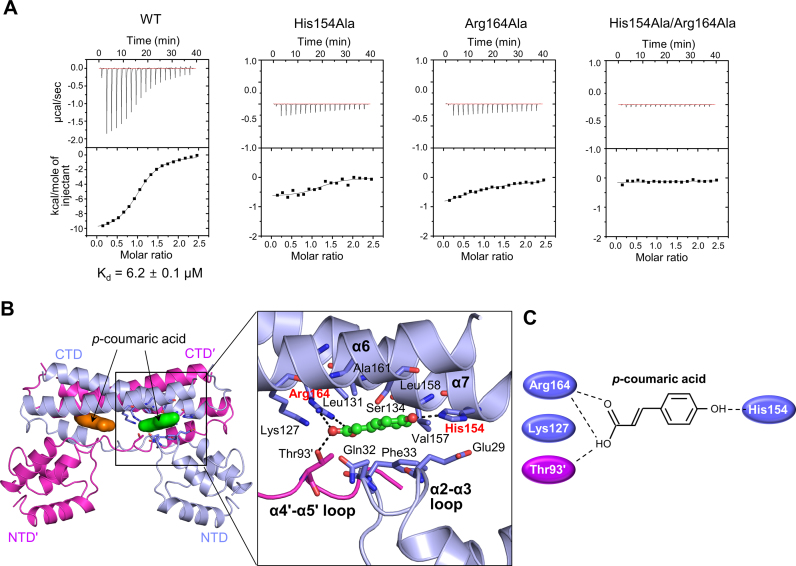
Bacterial transcription factors regulate gene expression by interacting with effector molecules. The direct binding of PadR to the p-coumaric acid effector was analyzed using ITC to understand the first step in the phenolic acid-responsive transcriptional derepression of the padC gene by PadR. The PadR-p-coumaric acid interaction is exothermic and exhibits an intermediate binding affinity (Kd, 6.2 μM) (Figure 5A). The interaction is predominantly enthalpy-driven (ΔH, −10.4 kcal/mol; −TΔS, 3.3 kcal/mol), indicating that the effector-binding energy of PadR is mainly provided by hydrophilic interactions rather than hydrophobic interactions.
Figure 5.
p-coumaric acid recognition by PadR. (A) ITC profiles for the interactions of WT PadR or its mutants (His154Ala, Arg164Ala and His154Ala/Arg164Ala) with p-coumaric acid. The data are representative of two independent experiments that yielded similar results. (B) The PadR-p-coumaric acid interaction observed in the PadR2CA-A structure. PadRdimer is shown as light blue and magenta ribbons. The two p-coumaric acid molecules bound to PadRdimer are depicted as green and orange surfaces (left) or ball-and-stick models (right). The p-coumaric acid-binding residues of PadR are shown as light blue or magenta sticks. His154 and Arg164, which play a key role in effector recognition, are labeled in red. (C) Schematic diagram of the interactions of PadR with the two polar terminal ends of p-coumaric acid.
The effector-recognition mode of PadR was resolved by determining the crystal structures of PadR in complex with p-coumaric acid using two crystal forms (PadR2CA and PadR3CA) (Supplementary Figure S10 and Table S2). The PadR3CA crystal was prepared via a soaking method, and its structure is essentially identical to the apo-PadR (PadR1) structure (root-mean-square deviation for 176 Cα atoms, 0.20 Å), except for the presence of p-coumaric acid. The PadR2CA crystal was generated through PadR-p-coumaric acid cocrystallization, and its asymmetric unit contains three PadR chains (A, B and C). Chain A (PadR2CA-A) is complexed with p-coumaric acid and forms a dimer through a two-fold crystallographic symmetry operation. Chain B (PadR2CA-B) also contains p-coumaric acid and forms a non-crystallographic dimer with chain C (PadR2-C), which lacks p-coumaric acid. Thus, the PadR2CA structure appears to represent both the apo form and the effector-bound form of PadR. Because the three p-coumaric acid-bound PadR structures (PadR2CA-A, PadR2CA-B and PadR3CA) exhibit an identical effector-binding mode, the PadR2CA-A structure will be used for the structural description of the PadR–effector complex unless otherwise specified.
In the PadR-p-coumaric acid complex structure, a PadR monomer binds one p-coumaric acid molecule in a narrow interdomain pocket between the NTD and the CTD from the same subunit (Figure 5B and C). Therefore, one PadR dimer simultaneously binds two p-coumaric acid molecules. Furthermore, p-coumaric acid mainly interacts with the α2-α3 loop of the NTD and the α6 and α7 helices of the CTD and also makes contacts with the α4′-α5′ loop. The apolar portion in the middle of p-coumaric acid is sandwiched by two layers of PadR residues (Gln32 and Phe33 from the α2-α3 loop; Leu131, Ser134, Val157, Leu158, and Ala161 from the α6 and α7 helices). The two polar termini of p-coumaric acid are flanked by four hydrophilic residues (Lys127, His154, Arg164 and Thr93′), which allow PadR to specifically recognize phenolic acids by defining the chemical properties and lengths of the effectors that PadR binds. The oxygen atoms at the carboxy-terminus are electrochemically stabilized by salt bridges with the positively charged Arg164 residue and are also clasped by Thr93′ and Lys127 through a hydrogen bond and a weak ionic interaction, respectively. The hydroxyl end of p-coumaric acid is hydrogen bonded to His154.
To verify the specific interactions observed in the p-coumaric acid-bound PadR structures, mutations were introduced into the p-coumaric acid-binding pocket of PadR. Single alanine mutations at either His154 or Arg164 severely disrupted the PadR-p-coumaric acid interaction, and the His154Ala/Arg164Ala double mutant did not exhibit any detectable binding to p-coumaric acid in the ITC experiment, indicating that His154 and Arg164 play a key role in effector recognition by PadR (Figure 5A). As a result, the mutations disrupted the p-coumaric acid-mediated dissociation of PadR from the operator DNA (Supplementary Figure S11). However, the mutations at His154 and Arg164 did not substantially modulate PadR–DNA binding or PadR protein stability (Supplementary Figure S6 and Table 1). The His154 and Arg164 residues are absolutely conserved across the amino acid sequences of PadR orthologues, suggesting that the proposed effector-recognition mechanism is commonly used by PadR orthologues, regardless of bacterial species (Supplementary Figure S2).
In addition to p-coumaric acid, PadR responds to ferulic acid as an effector molecule (13). To generalize the effector-recognition mode of PadR, we also determined a PadR structure in complex with ferulic acid (PadR3FA) using ferulic acid-soaked PadR crystals (Supplementary Table S2). The PadR3FA structure exhibits a comparable effector-binding mode to the PadR3CA structure due to the similar chemical properties of ferulic acid to p-coumaric acid although small structural differences are observed, such as the additional hydrogen bond of the methoxy group of ferulic acid to Ser134 and a slight shift of ferulic acid compared to p-coumaric acid (Supplementary Figures S1 and 12).
Structural mechanism by which phenolic acids inhibit the PadR–DNA interaction
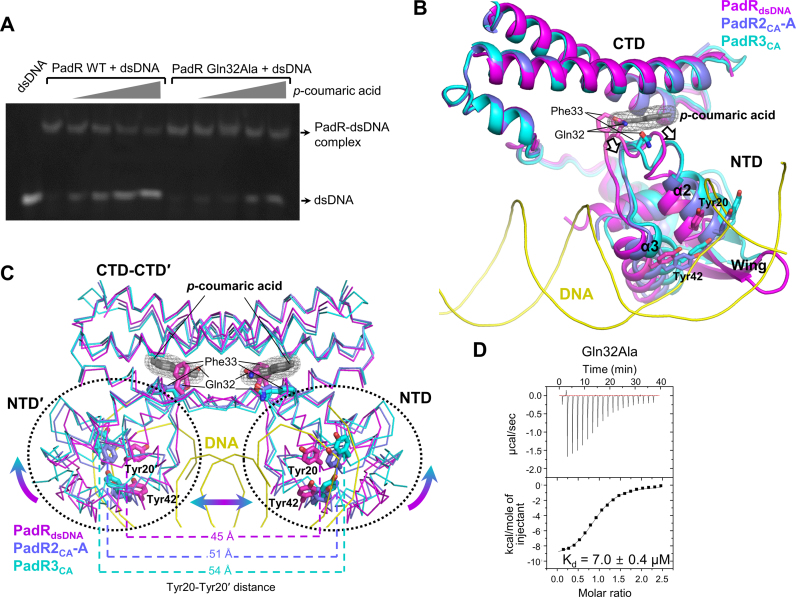
To examine the inhibitory effect of phenolic acids on the PadR–DNA interaction, we performed an EMSA in the presence of p-coumaric acid. Consistent with previous reports, p-coumaric acid dissociated PadR from its operator dsDNA in a dose-dependent manner (Figure 6A) (8,13). Furthermore, we propose a structural mechanism for the p-coumaric acid-induced PadR–DNA dissociation based on the comparative analysis of the DNA-bound and effector-bound PadR structures. When the PadRdsDNA structure is overlaid on the PadR2CA-A and PadR3CA structures using the CTDs, p-coumaric acid is located at least 14 Å away from the dsDNA, indicating that p-coumaric acid does not directly compete with the DNA for access to the PadR protein (Figure 1). Instead, p-coumaric acid binding to the interdomain pocket of PadR shifts residues 29–33 in the α2-α3 loop (Figure 6B). In addition, the NTD of PadR is displaced away from the CTD by translational and rotational movements (Figure 6B and C). Consequently, the DNA-binding residues of PadR in the α2 and α3 helices and the β-stranded wing of the NTD are dislodged and cannot optimally interact with the DNA. Moreover, the NTD-NTD′ distance of the PadR dimer is enlarged to distort the PadR dimer into a more open conformation in which the NTDs do not fit into the major or minor grooves of the operator dsDNA (Figure 6C). Therefore, our comparative structural analysis suggests that p-coumaric acid disrupts the PadR–DNA interaction through an allosteric mechanism in which p-coumaric acid rearranges the interdomain organization of PadR into a conformation incompatible with DNA binding.
Figure 6.
A proposed allosteric mechanism for the phenolic acid-induced PadR–dsDNA dissociation. (A) EMSA for the PadR WT (left) and the PadR Gln32Ala mutant (right). The PadR Gln32Ala mutant shows a defect in the p-coumaric acid-induced PadR–dsDNA dissociation. The data are representative of three independent experiments that yielded similar results. (B) Structural rearrangements of the PadR NTD through the allosteric center of Gln32 and Phe33 upon p-coumaric acid binding. The structures of the PadRdsDNA (magenta ribbons), PadR2CA-A (light blue ribbons) and PadR3CA (cyan ribbons) monomers are superimposed using their CTDs. Because the PadR2CA-B structure is highly similar to the PadR2CA-A structure (root-mean-square deviation for 162 Cα atoms, 0.37 Å), the PadR2CA-B structure is not shown in the figure for clarity. The allosteric center residues (Gln32 and Phe33) and DNA-binding residues (Tyr20 and Tyr42) are depicted as sticks. The dsDNA backbones and p-coumaric acid are represented by yellow coils and by gray molecular meshes and sticks, respectively. The binding of p-coumaric acid induces the movements (arrows) of Gln32, Phe33, the α2 helix, the α3 helix and the wing. (C) The p-coumaric acid-induced structural rearrangement of PadRdimer. The structures of the PadRdsDNA (magenta), PadR2CA-A (light blue) and PadR3CA (cyan) dimers are superimposed. The binding of p-coumaric acid (molecular mesh in gray wires) induces rotational and translational interdomain movements (arrows) in PadR. As a result, the NTD–NTD′ distance of PadR increases and PadR adopts a more open dimeric conformation. The distance between the Cα atoms of Tyr20 and Tyr20′ is shown. (D) Similar p-coumaric acid-binding affinity of the PadR Gln32Ala mutant to the WT PadR. According to ITC experiments, the Gln32Ala mutant binds p-coumaric acid with a Kd value of 7.0 ± 0.4 μM, similar to the WT PadR (Kd, 6.2 ± 0.1 μM; Figure 5A). The data of the PadR Gln32Ala mutant are representative of three independent experiments that yielded similar results.
Among the α2-α3 loop residues of PadR that undergo p-coumaric acid-induced structural rearrangements, the side chain of Gln32 in the PadRdsDNA structure displays steric clashes with the carboxylic group of p-coumaric acid in the PadR2CA-A and PadR3CA structures when the DNA-bound and effector-bound structures are overlaid (Figure 6B). In addition, the conformer of Phe33 in the PadRdsDNA structure would be chemically incompatible with p-coumaric acid because the hydrophobic side chain of Phe33 in the PadRdsDNA structure is in close proximity to the negatively charged carboxylic group of p-coumaric acid. Thus, upon p-coumaric acid binding, Gln32 and Phe33 should be dislodged to make room for p-coumaric acid, providing an explanation for the movement of the α2-α3 loop and interdomain reorganization. Therefore, Gln32 and Phe33 are considered as an allosteric center where the effector-induced structural changes are initiated. The Gln32-mediated allosteric mechanism was verified by the Gln32Ala mutation of PadR. The Gln32Ala mutant exhibited essentially identical affinity for p-coumaric acid and operator DNA as the WT protein (Figure 6D and Table 1). However, the Gln32Ala mutation impaired the ability of PadR to dissociate from DNA in response to phenolic acids (Figure 6A). Higher concentrations of p-coumaric acid were required to dissociate the Gln32Ala mutant from the DNA than for the WT PadR. We anticipate that the Gln32Ala mutation would alleviate the steric clashes between residue 32 and p-coumaric acid because the alanine at residue 32 has a smaller side chain than glutamine and the Gln32Ala mutant would be less efficient in effector-mediated interdomain reorganization. In contrast to Gln32, we could not assess the functional role of Phe33 as an allosteric center by mutation because the Phe33Ala mutant was deficient in p-coumaric acid binding and thus could not be used for the effector-induced PadR–DNA dissociation assay. Instead, the functional importance of Phe33 in the allosteric response of PadR to the effector would be supported by its absolute sequence conservation in PadR orthologs and its close proximity to Gln32 (Supplementary Figure S2). Residue 32 is also conserved, with glutamine residues in B. subtilis PadR and Paenibacillus polymyxa PadR and with glutamate residues in P. pentosaceus PadR and L. plantarum PadR. To verify the conserved role of glutamine and glutamate residues as an allosteric center in PadR orthologs, we mutated Gln32 to a glutamate residue in B. subtilis PadR. Unlike the Gln32Ala mutant, the Gln32Glu mutant exhibited identical p-coumaric acid-induced DNA dissociation to the PadR WT, suggesting that glutamine and glutamate at residue 32 are interchangeable as an allosteric center (Supplementary Figures S6B and 13). Collectively, our structural and mutational studies demonstrate that PadR regulates transcription via the effector-induced allosteric mechanism in which PadR Gln32 and Phe33 function as the allosteric center.
DISCUSSION
PadR represses padC transcription by binding the padC operator DNA. The basic DNA-binding function of the transcription factor is mediated by the wHTH motif. Based on the structural and mutational studies, we demonstrated that Tyr20, Tyr42, Leu65 and Lys67 in the PadR wHTH motif play a key role in DNA binding as a DNA-binding hot spot. In particular, the Tyr20, Tyr42 and Lys67 residues are highly conserved throughout the PadR family and allow PadR family members to converge into a single group. However, the PadR family diverges into two distinct subfamilies (subfamily-1 and subfamily-2) because of their different dimerization modes. PadR superfamily-1 members, including PadR and AphA, present a structure consisting of two separate domains, NTD and CTD. The CTD of the PadR superfamily-1 contains two long α-helices, α6 and α7, that are not observed in the PadR superfamily-2 (Supplementary Figure S14A). The CTD allows PadR to be shaped into a two-legged plate through a unique dimerization mode and to form an interdomain pocket that accommodates an effector molecule. In contrast to the PadR subfamily-1, PadR subfamily-2 members fold into single-domain structures and assemble into a dimer using the N-terminal or C-terminal dimerization helices that continuously extend from the wHTH motif and do not constitute an independent domain.
In addition to the dimerization mode, PadR differs from the PadR subfamily-2 in both the effector-binding region and stoichiometry. Each PadR monomer contains two domains and uses its interdomain pocket to accommodate one phenolic acid molecule. Thus, a PadR dimer binds two effector molecules with a 1:2 stoichiometry (Supplementary Figure S15A). In contrast, LmrR, a member of the PadR subfamily-2, contains only one domain consisting of α1-α4 helices and a β-stranded wing and thus lacks the interdomain pocket observed in PadR (Supplementary Figures S14A and 15B) (19,22). Instead, LmrR has exceptionally long terminal helices, α1 and α4, which provide the dimerization interface. As a result of α1/α4-mediated LmrR dimerization, a unique intersubunit space that can accommodate an effector molecule is created by the α1, α4, α1′ and α4′ helices (Supplementary Figure S15B). In the structure of LmrR in complex with its effector, daunomycin, one LmrR dimer binds only one daunomycin molecule using the intersubunit pocket with a 1:1 stoichiometry. Therefore, we conclude that the subfamily-1 and the subfamily-2 evolved distinct modes of domain organization, dimerization, and effector binding although the three key DNA-binding residues (Tyr20, Tyr42 and Lys67) are conserved in the two groups.
Recently, the structures of an archaeal segregation protein A (AspA) were revealed in complex with DNA (39). AspA was identified as an unusual PadR subfamily-2 member because AspA is not a transcription factor but a centromere-binding protein required for DNA segregation. In addition to the different cellular function, the β-stranded wing of AspA is shorter than that of PadR and does not provide the minor groove interaction that was observed in the PadRdsDNA structure. Despite their distinct features, the bacterial PadR and the archaeal AspA commonly interact with the major groove of dsDNA through the α3 helix. In structural overlays, AspA Tyr41 from the α3 helix mimics PadR Tyr42 by simultaneously interacting with the DNA backbone and base (Supplementary Figure S16). Thus, the DNA-recognition mode of the PadR transcription factor appears to be partially conserved in even the archaeal segregation protein, suggesting that the PadR family evolved from a common ancestor of DNA-binding proteins to transcription factors in bacteria and to segregation factors in archaea.
PadR is the most homologous to the MarR family in the DNA-binding domain among wHTH superfamily members. However, PadR is distinct from MarR family members in dimerization, DNA recognition and effector binding. Unlike PadR, proteins in the MarR family form a triangular shape by dimerizing mainly through two helices (α0 and α5) that are appended to the N-terminal and C-terminal ends of the wHTH domain (Supplementary Figures S14B and 17A). Although PadR Tyr20 and Tyr42 play a key role in DNA binding, the two tyrosine residues are replaced with other types of residues (Lys58 and Thr72 in B. subtilis OhrR, a MarR family member) in the MarR family for DNA recognition (40). Moreover, in contrast to PadR, the effector-binding sites of the MarR family are diversely located within the wHTH domain or in the intersubunit pocket (Supplementary Figure S17) (25,41–43).
Phenolic acid binding to PadR repositions the NTD with respect to the CTD and rearranges the PadR dimer into an open conformer that is incompatible with DNA binding (Figure 6C). This effector-induced allostery has been widely proposed as a transcriptional regulation mechanism in two-domain transcriptional regulators, including NagR, TetR, FadR, OxyR and Fur (44–48). Although the two-domain transcriptional regulators accommodate effectors into positionally diverse sites, effector binding generally induces the allosteric movement of the NTD through interdomain rearrangement or the structural changes of the CTD to regulate DNA-binding ability. Based on the comparative analysis combined with our structure-based mutational and biophysical studies, we conclude that PadR represents a well-coordinated two-domain transcriptional regulator that allosterically induces transcriptional derepression through effector recognition in the interdomain pocket.
Because we have revealed the various structures of PadR in apo forms and in complex with effector and operator DNA molecules, the structural dynamics of PadR would be readily analyzed through structural comparison. DNA-bound and effector-bound PadR proteins adopt mutually exclusive structural states in the conformation of allosteric center residues and the relative arrangement of the NTD and the CTD (Supplementary Figure S18). In the DNA-bound PadR structure, the Gln32 and Phe33 residues protrude toward the α6 and α7 helices of the CTD in upright positions. However, in the effector-bound structures, including PadR2CA-A, PadR2CA-B and PadR3CA, Gln32 and Phe33 consistently prefer squat positions to avoid steric clashes with the effector. Moreover, effector-bound PadR exhibits a more open interdomain conformation compared to DNA-bound PadR. Interestingly, apo-PadR was observed in two distinct forms represented by the PadR1 and PadR2-C structures (Supplementary Figure S18). The PadR1 structure adopts squat positions at the Gln32 and Phe33 residues and exhibits an interdomain orientation similar to the PadR3CA structure. In contrast, the apo form of the PadR2-C structure adopts upright positions of the Gln32 and Phe33 residues in a similar manner to the DNA-bound form with an interdomain arrangement intermediate between the DNA-bound and effector-bound forms. Thus, our comparative analysis suggests that apo-PadR is conformationally diverse in local structures and interdomain orientations.
AVAILABILITY
The atomic coordinates and structure factors for PadR (PDB ID: 5X12) and its complex with dsDNA (PDB ID: 5X11), p-coumaric acid (PDB IDs: 5Y8T and 5X13) and ferulic acid (PDB ID 5X14) have been deposited in the Protein Data Bank (www.pdb.org).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Byung-Chul Oh (Gachon University) for ITC instrument use and the beamline scientists at the Pohang Accelerator Laboratory beamlines 5C and 7A for their assistance with X-ray diffraction.
Author contributions: S.I.Y. conceived and coordinated the study. S.C.P., M.H. and S.I.Y. designed the experiments. S.C.P., Y.M.K., W.S.S. and S.I.Y. performed the experiments and analyzed the data. S.C.P., M.H. and S.I.Y. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea funded by the Ministry of Education [2014R1A1A2053497, 2016R1D1A1B03930540 to S.I.Y.; 2015R1D1A1A01057574 to M.H.]; National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning [2016R1A4A1010115 to S.I.Y.]. Funding for open access charge: Kangwon National University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Grkovic S., Brown M.H., Skurray R.A.. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 2002; 66:671–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Imlay J.A. Transcription factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 2015; 69:93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campos F.M., Couto J.A., Hogg T.A.. Influence of phenolic acids on growth and inactivation of Oenococcus oeni and Lactobacillus hilgardii. J. Appl. Microbiol. 2003; 94:167–174. [DOI] [PubMed] [Google Scholar]
- 4. Licandro-Seraut H., Roussel C., Perpetuini G., Gervais P., Cavin J.F.. Sensitivity to vinyl phenol derivatives produced by phenolic acid decarboxylase activity in Escherichia coli and several food-borne Gram-negative species. Appl. Microbiol. Biotechnol. 2013; 97:7853–7864. [DOI] [PubMed] [Google Scholar]
- 5. Wells J.E., Berry E.D., Varel V.H.. Effects of common forage phenolic acids on Escherichia coli O157:H7 viability in bovine feces. Appl. Environ. Microbiol. 2005; 71:7974–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barthelmebs L., Divies C., Cavin J.F.. Knockout of the p-coumarate decarboxylase gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl. Environ. Microbiol. 2000; 66:3368–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Degrassi G., Polverino De Laureto P., Bruschi C.V.. Purification and characterization of ferulate and p-coumarate decarboxylase from Bacillus pumilus. Appl. Environ. Microbiol. 1995; 61:326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tran N.P., Gury J., Dartois V., Nguyen T.K., Seraut H., Barthelmebs L., Gervais P., Cavin J.F.. Phenolic acid-mediated regulation of the padC gene, encoding the phenolic acid decarboxylase of Bacillus subtilis. J. Bacteriol. 2008; 190:3213–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silva I., Campos F.M., Hogg T., Couto J.A.. Wine phenolic compounds influence the production of volatile phenols by wine-related lactic acid bacteria. J. Appl. Microbiol. 2011; 111:360–370. [DOI] [PubMed] [Google Scholar]
- 10. Barthelmebs L., Lecomte B., Divies C., Cavin J.F.. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 2000; 182:6724–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gury J., Barthelmebs L., Tran N.P., Divies C., Cavin J.F.. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl. Environ. Microbiol. 2004; 70:2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gury J., Seraut H., Tran N.P., Barthelmebs L., Weidmann S., Gervais P., Cavin J.F.. Inactivation of PadR, the repressor of the phenolic acid stress response, by molecular interaction with Usp1, a universal stress protein from Lactobacillus plantarum, in Escherichia coli. Appl. Environ. Microbiol. 2009; 75:5273–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nguyen T.K., Tran N.P., Cavin J.F.. Genetic and biochemical analysis of PadR-padC promoter interactions during the phenolic acid stress response in Bacillus subtilis 168. J. Bacteriol. 2011; 193:4180–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lubelski J., de Jong A., van Merkerk R., Agustiandari H., Kuipers O.P., Kok J., Driessen A.J.. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol. Microbiol. 2006; 61:771–781. [DOI] [PubMed] [Google Scholar]
- 16. Huillet E., Velge P., Vallaeys T., Pardon P.. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol. Lett. 2006; 254:87–94. [DOI] [PubMed] [Google Scholar]
- 17. Florez A.B., Alvarez S., Zabala D., Brana A.F., Salas J.A., Mendez C.. Transcriptional regulation of mithramycin biosynthesis in Streptomyces argillaceus: dual role as activator and repressor of the PadR-like regulator MtrY. Microbiology. 2015; 161:272–284. [DOI] [PubMed] [Google Scholar]
- 18. Morabbi Heravi K., Lange J., Watzlawick H., Kalinowski J., Altenbuchner J.. Transcriptional regulation of the vanillate utilization genes (vanABK Operon) of Corynebacterium glutamicum by VanR, a PadR-like repressor. J. Bacteriol. 2015; 197:959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Madoori P.K., Agustiandari H., Driessen A.J., Thunnissen A.M.. Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition. EMBO J. 2009; 28:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Silva R.S., Kovacikova G., Lin W., Taylor R.K., Skorupski K., Kull F.J.. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J. Biol. Chem. 2005; 280:13779–13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M.. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005; 29:231–262. [DOI] [PubMed] [Google Scholar]
- 22. van der Berg J.P., Madoori P.K., Komarudin A.G., Thunnissen A.M., Driessen A.J.. Binding of the lactococcal drug dependent transcriptional regulator LmrR to its ligands and responsive promoter regions. PLoS One. 2015; 10:e0135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fibriansah G., Kovacs A.T., Pool T.J., Boonstra M., Kuipers O.P., Thunnissen A.M.. Crystal structures of two transcriptional regulators from Bacillus cereus define the conserved structural features of a PadR subfamily. PLoS One. 2012; 7:e48015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arita K., Hashimoto H., Igari K., Akaboshi M., Kutsuna S., Sato M., Shimizu T.. Structural and biochemical characterization of a cyanobacterium circadian clock-modifier protein. J. Biol. Chem. 2007; 282:1128–1135. [DOI] [PubMed] [Google Scholar]
- 25. Alekshun M.N., Levy S.B., Mealy T.R., Seaton B.A., Head J.F.. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 2001; 8:710–714. [DOI] [PubMed] [Google Scholar]
- 26. Isom C.E., Menon S.K., Thomas L.M., West A.H., Richter-Addo G.B., Karr E.A.. Crystal structure and DNA binding activity of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291. BMC Microbiol. 2016; 16:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee C., Kim M.I., Hong M.. Structural and functional analysis of BF2549, a PadR-like transcription factor from Bacteroides fragilis. Biochem. Biophys. Res. Commun. 2017; 483:264–270. [DOI] [PubMed] [Google Scholar]
- 28. Jeon Y.J., Park S.C., Song W.S., Kim O.H., Oh B.C., Yoon S.I.. Structural and biochemical characterization of bacterial YpgQ protein reveals a metal-dependent nucleotide pyrophosphohydrolase. J. Struct. Biol. 2016; 195:113–122. [DOI] [PubMed] [Google Scholar]
- 29. Song W.S., Nam M.S., Namgung B., Yoon S.I.. Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation. Biochem. Biophys. Res. Commun. 2015; 458:843–848. [DOI] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor W.. Processing x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 31. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., Afonine P.V., Zwart P.H., Hung L.W.. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009; 65:582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 34. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 35. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiseman T., Williston S., Brandts J.F., Lin L.N.. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989; 179:131–137. [DOI] [PubMed] [Google Scholar]
- 37. Lundblad J.R., Laurance M., Goodman R.H.. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 1996; 10:607–612. [DOI] [PubMed] [Google Scholar]
- 38. Zheng G., Lu X.J., Olson W.K.. Web 3DNA–a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009; 37:W240–W246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schumacher M.A., Tonthat N.K., Lee J., Rodriguez-Castaneda F.A., Chinnam N.B., Kalliomaa-Sanford A.K., Ng I.W., Barge M.T., Shaw P.L., Barilla D.. Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages. Science. 2015; 349:1120–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hong M., Fuangthong M., Helmann J.D., Brennan R.G.. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005; 20:131–141. [DOI] [PubMed] [Google Scholar]
- 41. Kumarevel T., Tanaka T., Umehara T., Yokoyama S.. ST1710-DNA complex crystal structure reveals the DNA binding mechanism of the MarR family of regulators. Nucleic Acids Res. 2009; 37:4723–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davis J.R., Brown B.L., Page R., Sello J.K.. Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors. Nucleic Acids Res. 2013; 41:3888–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chang Y.M., Chen C.K., Ko T.P., Chang-Chien M.W., Wang A.H.. Structural analysis of the antibiotic-recognition mechanism of MarR proteins. Acta Crystallogr. D Biol. Crystallogr. 2013; 69:1138–1149. [DOI] [PubMed] [Google Scholar]
- 44. Fillenberg S.B., Grau F.C., Seidel G., Muller Y.A.. Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR. Nucleic Acids Res. 2015; 43:1283–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hinrichs W., Kisker C., Duvel M., Muller A., Tovar K., Hillen W., Saenger W.. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science. 1994; 264:418–420. [DOI] [PubMed] [Google Scholar]
- 46. van Aalten D.M., DiRusso C.C., Knudsen J.. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001; 20:2041–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jo I., Chung I.Y., Bae H.W., Kim J.S., Song S., Cho Y.H., Ha N.C.. Structural details of the OxyR peroxide-sensing mechanism. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:6443–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deng Z., Wang Q., Liu Z., Zhang M., Machado A.C., Chiu T.P., Feng C., Zhang Q., Yu L., Qi L. et al. . Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 2015; 6:7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.