Abstract
The protein–RNA interactions within the flavivirus replication complex (RC) are not fully understood. Our structure of dengue virus NS3 adenosine triphosphatase (ATPase)/helicase bound to the conserved 5′ genomic RNA 5′-AGUUGUUAGUCU-3′ reveals that D290 and R538 make specific interactions with G2 and G5 bases respectively. We show that single-stranded 12-mer RNA stimulates ATPase activity of NS3, however the presence of G2 and G5 leads to significantly higher activation. D290 is adjacent to the DEXH motif found in SF2 helicases like NS3 and interacts with R387, forming a molecular switch that activates the ATPase site upon RNA binding. Our structure guided mutagenesis revealed that disruption of D290–R387 interaction increases basal ATPase activity presumably as a result of higher conformational flexibility of the ATPase active site. Mutational studies also showed R538 plays a critical role in RNA interactions affecting translocation of viral RNA through dynamic interactions with bases at positions 4 and 5 of the ssRNA. Restriction of backbone flexibility around R538 through mutation of G540 to proline abolishes virus replication, indicating conformational flexibility around residue R538 is necessary for RNA translocation. The functionally critical sequence-specific contacts in NS3 RNA binding groove in subdomain III reveals potentially novel allosteric anti-viral drug targets.
INTRODUCTION
Dengue, a predominantly Aedes aegypti borne infection, is increasing at an alarming rate globally. World Health Organization estimates that half of the world’s population is at risk of infection, with children being most vulnerable to severe dengue diseases and suffering the highest rate of mortality (1,2). Recent outbreaks of Zika virus (ZIKV) (3), associated with congenital defects in fetuses and Guillain–Barré syndrome (4), and Yellow Fever virus (YFV) (5), resulting in liver dysfunction and hemorrhagic fever (6), have enhanced global concern about this family of viruses. A tetravalent dengue vaccine based on the yellow fever 17-D backbone was recently approved in several countries, even though its efficacy against dengue virus (DENV) serotypes 1 and 2 is lower in young children (7). Moreover, following vaccination, higher rates of hospital admissions have been observed in previously uninfected individuals younger than 9 years (8). There is no specific antiviral treatment for dengue fever at present (9) and clinical studies with re-purposed drugs have not identified any efficacious compounds so far. Therefore, the search for targets amenable to anti-viral drug development, based on detailed structural and mechanistic studies remains an urgent priority, to treat infections provoked by DENV or ZIKV.
DENV belongs to the family Flaviviridae and contains a positive sense capped single strand RNA genome of ∼11 kb in size encoding ten proteins as a single open reading frame that is flanked by 5′ and 3′ untranslated region (UTR) regions that regulate translation and replication (10). Following direct translation, the polyprotein is co- and post-translationally processed by host and viral proteases to generate three structural and seven non-structural (NS) proteins NH3+ C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 COO−. NS3 and NS5 contain all the enzymatic activities required for viral life cycle and have been extensively studied (11–14). Here we focus on NS3, a multi-functional protein endowed with protease, helicase, adenosine triphosphatase (ATPase) and RNA 5′ triphosphatase (RTPase) activities (11,15–17). NS3 shares 77% amino-acid sequence identity across the four DENV serotypes and 67 and 50% identity with NS3 from ZIKV and YFV, respectively. The N-terminal region of NS3 of DENV consists of 167 amino acids, which, together with the central 40 amino-acid region of the membrane-associated NS2B co-factor, constitute the serine protease that cleaves the viral polyprotein (18–23). The C-terminal domain (residues 172–618) of NS3 (NS3H), comprises three subdomains and possesses RNA helicase/NTPase/RTPase activities (24–26). NS3H belongs to the superfamily 2 of RNA helicases/NTPases having Walker A, GK(S/T) and Walker B, DEx(D/H) motifs, along with other superfamily-characteristic conserved sequence motifs present in subdomains I and II (which possess the fold of the RecA protein). Subdomain III, which has a fold unique to the viral NS3 protein, forms the ‘roof’ of a ssRNA-binding groove that runs roughly across the middle of the flat triangular-shaped NS3H protein (Figure 1A). Helicase activity of NS3 is responsible for unwinding duplex RNA utilizing the chemical energy derived from ATP hydrolysis (27). Functional activity of helicase and NTPase of NS3 has been very well characterized for several members of the family Flaviviridae (24,28–30) including the Hepatitis C virus (HCV) NS3 protein (31–36).
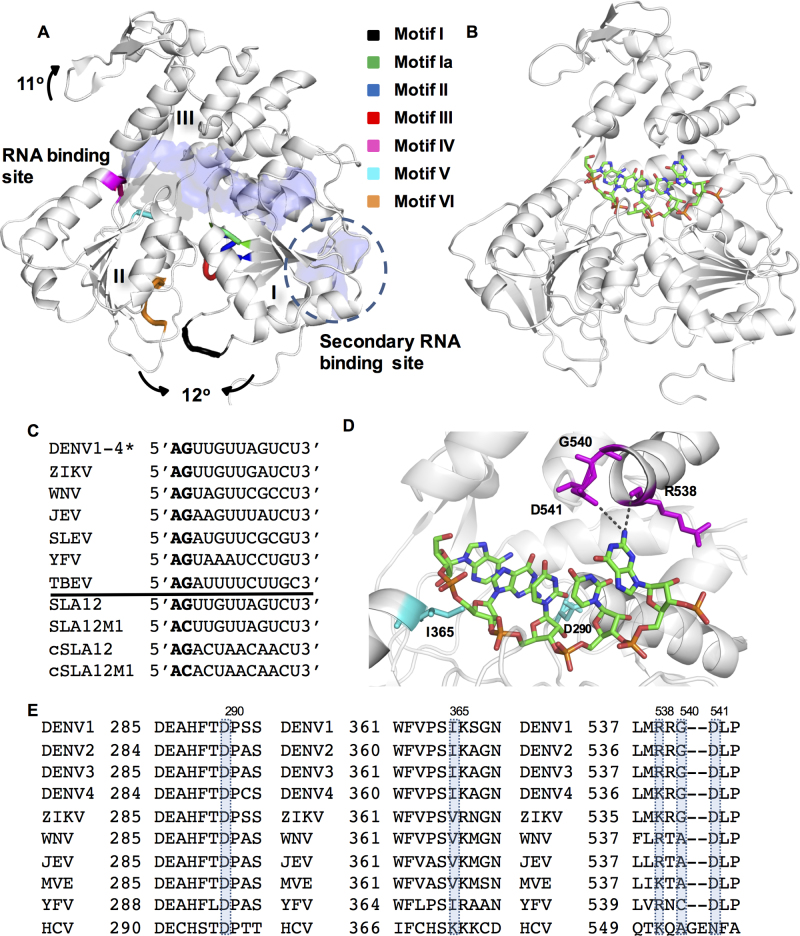
Figure 1.
Crystal structures of DENV NS3H and interactions with ssRNA. (A) Three-dimensional (3D) structural model of the helicase domain of NS3. The motifs I–VI typically found in superfamily 2 helicases are shown in NS3H (PDB ID: 2JLQ (11)), with the RNA-binding regions identified by (11) (‘RNA-binding site’) and (19) (‘secondary RNA-binding site’) and structural changes on RNA binding shown with arrows. (B) Crystal structure of DENV4 NS3H bound SLA12. NS3H is shown in white, while bound SLA12 shown as sticks and colored according to atoms. (C) Sequence alignment of 12-mer RNA from 5′ UTR of flaviviruses showing the 5′ conserved AG sequence and sequences of 12-mer RNAs used in this study (*DENV serotypes 1–4 have identical sequence), Genbank accession numbers are DENV1:EU081230, DENV2:EU081177, DENV3:EU081190, DENV4:GQ398256, ZIKV:KX80026, WNV:M12294, JEV:M55506, SLEV: DQ525916, YFV:X15062 and TBEV: KC835595. (D) Magnified SLA12 RNA binding shows interactions of D290 (subdomain I) and I365 (subdomain II) with G2 in cyan (also observed with cSLA12), and interactions with the R538 main chain and D541 (subdomain III) with G5 (only observed with SLA12). G540 which confers flexibility to the region is also shown in magenta. (E) These residues are conserved throughout evolution as shown in sequence alignments, with Genbank accession numbers as (C) in addition to MVE:AF161266 and HCV:KC191671.
Biochemical and structural studies of NS3H identified specific residues and conformations associated with nucleotide hydrolysis and RNA binding (11,15). The ATPase active site sits at the interface of subdomains I and II of NS3H and consists of the P-loop (motif I), DEAH motif (motif II) and motif VI (Figure 1A) that are present in all SF2 helicases (37). Upon ssRNA binding the ATPase site rearranges from an ‘open’ less active state to a ‘closed’ activated state with the P-loop shifted towards the protein core and reordering of an extra turn at the N-terminal of α1 helix yielding optimum activity (11). The RNA-binding groove of DENV NS3H separates subdomains I and II from subdomain III, and was crystallized with 12-mer RNA, previously referred to as ‘RNA12’ (11) which represents the reverse complement of stem loop A region and is identified as ‘cSLA12’ in this work. Another viral RNA-binding site has been mapped to an arginine patch at the surface of subdomain II of NS3H although it has so far not been characterized structurally (Figure 1A) (19). RNA binding is further allosterically regulated by the presence of the N-terminal protease domain of NS3 which significantly enhances both ATPase and helicase activities for dengue (16,17) and West Nile virus NS3, which also require the protease domain for RNA recognition (38). Despite revealing atomic interactions and conformational changes during the ATP hydrolysis cycle, the exact mechanism of translocation along the RNA is still unresolved for flaviviruses. Moreover, interesting comparisons can be drawn based on studies with HCV (39–41).
With the aim to elucidate the translocation mechanism of DENV NS3, we determined the crystal structure of DENV4 NS3 helicase in complex with the highly conserved 5′ gRNA region (the first 12 nt including the strictly conserved AG dinucleotide present in all flavivirus genomic RNA) which we refer to as ‘SLA12’. Importantly, the present study is distinct from our previously reported studies that used the reverse complement ‘RNA12’ sequence. The rationale for the present work is based on a recent structural study which revealed that the AG dinucleotide at the 5′ end is absolutely essential for RNA genome capping by the NS5 methyltransferase domain (42). Using structure-based mutations, we have confirmed the importance of evolutionary conserved amino acids D290 and R538 of NS3H that directly interact with bound RNA. While the importance of conformational flexibility of the peptide backbone around R538 in subdomain III was demonstrated for the enzymatic activities in recombinant NS3 as well as in mutant viruses constructed in this study.
MATERIALS AND METHODS
Cell lines
BHK-21 (baby hamster kidney fibroblast cells, ATCC) was maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin at 37°C in a humidified CO2 incubator. C6/36, an Aedes albopictus cell line (ATCC), was cultured in RPMI 1640 medium with 10% FBS, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and 1% P/S at 28°C in the absence of CO2.
Site-directed mutagenesis of DENV4 NS2B18NS3FL (NS3FL)
The construct encoding the wild-type (WT) DENV4 NS2B18NS3FL protein described previously (16) was used as a template for generating mutants. Site-directed mutagenesis was carried-out using the Quick Change Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. For D290A mutation, forward primer (FP) 5′-GAAGCACATTTCACCGCTCCTTCTAGTGTC-3′ and reverse primer (RP) 5′-GACACTAGAAGGAGCGGTGAAATGTGCTTC-3′, for R538A mutation, FP 5′-TTTGTGGAATTAATGGCGAGAGGAGACCTT-3′ and RP 5′-AAGGTCTCCTCTCGCCATTAATTCCACAAA-3′ and for G540P mutation, FP 5′-GAATTAATGAGGAGACCAGACCTTCCGGTG-3′ and 5′-CACCGGAAGGTCTGGTCTCCTCATTAATTC-3′ were used. All desired mutations were confirmed by nucleotide sequencing.
Protein expression and purification
For crystallization an NS3 helicase construct (NS3H), comprising residues 172–618, was used as described previously (11) whereas for functional studies full-length NS3 (NS3FL), NS2B18-G4SG4-NS3, was utilized which we have previously shown (16) to be necessary for soluble protein expression. Expression of NS3FL and mutant proteins has been described previously (11). There was no difference in growth rate of cells harboring mutant constructs as compared with WT. All the mutant proteins expressed to the same level as WT.
Purification has been described previously (43), briefly cells resuspended in His Buffer A (20 mM phosphate buffer, pH 7.4, 1 M NaCl, 40 mM imidazole) were lysed by repeated freeze/thaw cycles, sonicated and clarified by centrifugation. The supernatant was then loaded onto a His Buffer A pre-equilibrated HisTrapFF column (GE Healthcare Lifesciences) and eluted by a linear gradient of imidazole from 40 to 500 mM. Fractions containing Trx-His-tagged protein were then buffer-exchanged over a PD-10 desalting column (GE Healthcare) into desalting buffer (200 mM NaCl, 10 mM phosphate buffer, 2.7 mM KCl, pH 7.4) and the tags cleaved by thrombin at 4°C for 18 h. The cleavage mixture was passed over a HisTrapFF column equilibrated with desalting buffer prior to anion exchange chromatography; tag-free protein was diluted with Anion Buffer (20 mM Tris, 50 mM NaCl pH 7.6 for NS3FL and pH 8.7 for NS3H) and loaded onto a HiTrapQ column and eluted with an increasing salt concentration up to 1 M. Final polishing size exclusion chromatography was performed using a superdex 200 column in S200 buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 1 mM 1,4-dithiothreitol (DTT), 5% glycerol). Purified proteins were confirmed to be >95% pure by Sodium dodecyl sulphate-polyacrylamide gel electrophoresis and concentrated to 20 mg/ml.
Crystallization and data collection
Crystallization and data collection performed as described previously (11). NS3H in complex with SLA12 was crystallized and cryo-protected as per the cSLA12 conditions and D290A NS3H mutant crystallized and cryo-protected as per the free NS3H (PDB ID: 2JLQ; (11)) conditions. Following cryoprotection, data collection was performed as per (11) rapidly cooling crystals in a nitrogen gas stream (Oxford Cryosystems) and collecting on an R-AXIS IV + + imaging plate detector at Nanyang Technological University (Singapore). Intensities were integrated, scaled and merged using MOSFLM and Scala programs (44).
Structure solution, refinement and analysis
Both structures are isomorphous to published structures (11). Refinement was carried out using iterative cycles of phenix.refine (45) and model building in Coot (46). A summary of the refinement statistics and stereochemistry analysis is given in Table 1. Figures were prepared using the program Pymol.
Table 1. Data collection and refinement statistics for NS3H-SLA12 (PDB ID: 5XC6) and D290A (PDB ID: 5XC7).
| NS3H + SLA12 | NS3HD290A | |
|---|---|---|
| Wavelength (Å) | 1.5418 | 1.5418 |
| Resolution range (Å) | 19.86–2.90 | 29.43–2.10 |
| Space group | C 1 2 1 | P 21 21 21 |
| Unit cell (Å, °) | 132.9, 105.4, 72.7, 90, 116.4, 90 | 53.05, 92.17, 102.52 |
| Unique reflections* | 19 928 (1996) | 30 117 (2430) |
| Multiplicity | 3.6 (3.6) | 5.5 (5.4) |
| Completeness (%) | 99.7 (100) | 100 (100) |
| Mean I/sigma (I) | 8.00 (2.3) | 15.88 (2.5) |
| Wilson B-factor (Å2) | 44.79 | 33.65 |
| R-merge | 0.159 (0.573) | 0.069 (0.666) |
| R-work | 0.20 | 0.19 |
| R-free | 0.25 | 0.23 |
| Number of non-hydrogen atoms | 7444 | 3852 |
| Macromolecules | 7434 | 3631 |
| Ligands | 10 | 13 |
| Protein residues | 902 | 451 |
| RMS (bonds) (Å) | 0.009 | 0.011 |
| RMS (angles) (°) | 1.23 | 1.11 |
| Ramachandran | ||
| Favoured (%) | 95.10 | 97.80 |
| Allowed (%) | 4.90 | 2.20 |
| Outliers (%) | 0.00 | 0.00 |
| Average B-factor (Å2) | 41.71 | 40.33 |
| Macromolecules | 41.69 | 40.30 |
| Ligands | 58.53 | 41.43 |
| Solvent | 40.82 |
*High resolution shell statistics shown in parentheses.
ATPase assay
Enzyme activity was measured by quantifying the release of free phosphate following ATP hydrolysis by malachite green assay (47). Sequences of all RNA oligonucleotides used in this study are shown in Figure 1C. The assay was performed in a final volume of 50 μl in a costar 96 well half-area micro-titer plate (Corning, NY, USA). Purified protein at a concentration of 2.5 nM was pre-incubated with 5 μM single stranded RNAs in a reaction buffer (50 mM Tris–HCl, pH7.5; 2 mM MgCl2; 1.5 mM DTT; 0.05% Tween 20; 0.25 ng/μl bovine serum albumin) of 40 μl for 15 min at 30°C. The ATPase reaction was started by adding 10 μl of ATP and incubated further for 30 min at 30°C. A total of 10 μl malachite green reagent (BioAssay systems) was added and incubated at room temperature for 30 min to form a complex with molybdate and free orthophosphate that is quantified by measuring absorbance. The plate was read in the Tecan Infinite M200 microplate reader (Tecan, Durham, NC, USA) at 635 nm. Background absorbance correction of reaction mixtures to account for non-enzymatic hydrolysis of ATP was obtained in the absence of the enzyme. For determination of the kinetic constants, the same assay was used at various concentrations of one substrate and fixed concentration of the others. In all cases, initial velocity was measured and expressed as picomoles of inorganic phosphate released during the reaction per minute. Data were fitted to the Michaelis–Menten equation (v = Vmax[S]/Km+[S]) to calculate apparent Km and Vmax by non-linear regression using GraphPad Prism (version 5.00, GraphPad Software, San Diego, CA, USA).
Molecular beacon helicase assay
To monitor ATP-dependent duplex RNA unwinding helicase activity of purified NS3 protein, an improved fluorescence based molecular beacon assay developed previously was used (43). The steady-state kinetic parameters for ATP and duplex RNA substrate in the helicase assay were measured to understand the impact of the mutations on the RNA unwinding activity and determine the interdependence between the ATPase and helicase activities. The Km for ATP was obtained by varying its concentration up to 2 mM at a fixed duplex RNA concentration of 10 nM. Similarly, the Km for duplex RNA was obtained by varying its concentration up to 10 nM at a fixed ATP concentration of 2 mM. In all cases, initial velocity was measured and expressed as pmoles of product formed per minute. The kinetic parameters determined for various substrates and proteins are summarized in Table 2. Data were fitted to the Michaelis–Menten equation to calculate apparent Km and Vmax by non-linear regression using GraphPad Prism (version 5.00, GraphPad Software, San Diego, CA, USA).
Table 2. Summary table of kinetic constants determined for different substrates on ATPase activity and helicase activity with WT and mutant proteins.
| Kinetic parameters | Biochemical assay | RNA substrates | Proteins | |||
|---|---|---|---|---|---|---|
| WT | D290A | R538A | G540P | |||
| Apparent Km for ATP (μM) | ATPase assay | SLA12 | 248 ± 4.0 | 129 ± 0.7 | 112 ± 1.4 | 163 ± 2.0 |
| cSLA12 | 82 ± 5.0 | 170 ± 6.0 | 140 ± 12 | 253 ± 7.0 | ||
| molecular beacon helicase assay (MBHA) | Duplex RNA | 530 ± 33 | 1597 ± 132 | 2629 ± 81 | 277 ± 4.0 | |
| Apparent Km for RNA (μM) | ATPase assay | SLA12 | 0.5 ± 0.03 | 0.1 ± 0.02 | 0.3 ± 0.01 | 0.9 ± 0.1 |
| cSLA12 | 0.86 ± 0.05 | 0.4 ± 0.06 | 0.3 ± 0.04 | 1.1 ± 0.06 | ||
| MBHA | Duplex RNA (×10−2) | 3.5 ± 0.3 | 0.9 ± 0.07 | 2.3 ± 0.1 | 0.9 ± 0.07 | |
| Apparent Vmax (pmoles/min) | ATPase assay | SLA12 | 103 ± 3.0 | 90 ± 2.0 | 44 ± 1.4 | 46 ± 2.0 |
| cSLA12 | 62 ± 1.4 | 88 ± 1.4 | 34 ± 1.4 | 32 ± 1.4 | ||
| MBHA | Duplex RNA (×10−2) | 3 ± 0.4 | 0.7 ± 0.07 | 0.7 ± 0.001 | 0.4 ± 0.07 | |
| Apparent kcat for RNA (s−1) | ATPase assay | SLA12 | 14 ± 0.4 | 12 ± 0.3 | 6 ± 0.2 | 6 ± 0.3 |
| cSLA12 | 8 ± 0.2 | 12 ± 0.2 | 5 ± 0.2 | 4 ± 0.2 | ||
| MBHA | Duplex RNA (×10−4) | 1 ± 0.1 | 0.2 ± 0.002 | 0.2 ± 0.001 | 0.1 ± 0.0001 | |
| Apparent kcat/Km for RNA (s−1μM−1) | ATPase assay | SLA12 | 28 ± 1.9 | 120 ± 24 | 20 ± 0.94 | 6.7 ± 0.81 |
| cSLA12 | 9.3 ± 0.59 | 30 ± 4.5 | 17 ± 2.3 | 3.6 ± 0.27 | ||
| MBHA | Duplex RNA (×10−3) | 3 ± 0.4 | 2 ± 0.2 | 0.9 ± 0.04 | 1 ± 0.09 | |
Fluorescence-based thermal shift assay
The assay was performed in 96 well plates using 10 μl of RNA substrate added initially to a final concentration of 25 μM for SLA12 and 25 nM for duplex RNA. Next, 5 μM protein and 20× Sypro Orange (Invitrogen) were added in a volume of 10 μl. The plate was then sealed with an optical seal and centrifuged. The thermal scan ranged from 20–95°C with a temperature ramp rate of 1.0°C/min. The fluorescence intensity upon binding of Sypro Orange was measured with excitation\emission wavelength of 533–580 nm in Light Cycler480 II (Roche Applied Science). The data analysis and report generation were performed by the instrument software. The Tm was calculated manually from the negative derivative plot at the point of inflection of the curve (the midpoint for protein unfolding).
Site-directed mutagenesis of NS3 DENV2 full length cDNA clone, in vitro transcription and transfection
The NS3 mutations, D290A, R538A, G540P, D541A and D541N were introduced into the genome-length DENV2–3295 (GenBank accession: EU081177.1) cDNA clone using QuikChange II XL site-directed mutagenesis kit (Stratagene) according to manufacturer’s instructions. Mutations were confirmed by automated DNA sequencing. The cDNA clones of WT and NS3 mutants were subjected to in vitro transcription using T7 mMESSAGE mMACHINE kit (Ambion). The in vitro transcribed RNA was transfected into BHK-21 cells as previously described (48) and monitored for virus replication profile analysis since this cell line is most commonly used for efficient viral RNA transfection. For comparison, the replication-defective NS3:N570A mutant that was previously characterized (48) was included as a control. Samples were harvested every 24 h post-transfection until 120 h. Supernatants were collected for plaque quantification and extracellular viral RNA quantification by real-time reverse transcriptase-polymerase chain reaction (RT-PCR). Cells were washed once with phosphate buffered saline prior to lysing with Trizol reagent (Invitrogen) for cellular viral RNA quantification.
Real-time RT-PCR and immunofluorescence assay (IFA)
Viral RNA was extracted from the supernatants using QIAamp Viral RNA Mini Kit (Qiagen) according to manufacturer’s instructions. Viral load in the supernatants was measured by RT-qPCR in Bio-Rad real time thermal cycler CFX96 using iTaq Universal SYBR green one step kit (Bio-Rad) with primers (FP 5′-CAGGCTATGGCACTGTCACGAT-3′ and RP 5′-CCATTTGCAGCAACACCATCTC-3′) targeting the DENV2 E gene. Plasmid fragment containing the E gene fragment of DENV2–3295 (GenBank accession EU081177.1) was used to generate standard curve for quantification of viral genome copy number and results were reported as absolute viral RNA genome copy per mililiter of supernatant. The detection limit of RT-PCR assay is indicated as gray line in graph.
The intracellular detection of both positive and negative strand was previously described (48). The viral genome copy numbers were normalized to actin expression followed by normalization to expression levels at 6 h post-transfection. The rate of replication was reported as the sum of absolute number of both positive and negative viral RNA genome copy per microgram of RNA used for RT-qPCR.
Immunofluorescence assay against dsRNA using anti-dsRNA mouse antibody J2 (Scicons) and NS3 protein by anti-NS3 human antibody 3F8 were performed as previously described (49). Digitized images were captured using Zeiss LSM 710 upright confocal microscope (Carl Zeiss, Germany) at 64× magnification. Image processing performed with ImageJ software (50).
Virus recovery in C6/36 cells
Day 3 post-transfection BHK-21 supernatants of the various NS3 mutants were used to infect a confluent T25 flask of C6/36 cells. The infection amount is normalized to the genome equivalents (GE) that were calculated from real-time RT-PCR as described above. An inoculum containing ∼1 × 107 GE was used to infect C6/36 for 1 h. The inoculums were removed and replaced with 2% FBS maintenance RPMI 1640 medium. Small quantity of the supernatant (∼100 μl) was collected from the flask at 24, 48 and 72 h post-infection for plaque quantification using standard BHK-21 plaque assay.
Statistical analysis
The error bars in the figures represent the standard error of mean of more than one measurement using GraphPad Prism software. The statistical significance of differences between groups using different conditions was evaluated by the two-tailed unpaired t-test in GraphPad Prism software. P-values ≤ 0.05 were considered statistically significant.
Data deposition
The structural data from this publication has been deposited to the RCSB protein data bank (PDB) and assigned the identifiers 5XC6 (NS3H with SLA12) and 5XC7 (NS3H D290A).
RESULTS
The NS3H:SLA12 structure reveals sequence-specific helicase–RNA interactions
In light of recent reports disclosing sequence specific recognition of genomic viral RNA by both the NS3 and NS5 proteins (42,51,52), we determined the crystal structure of NS3H in complex with SLA12, the first 12 residues of the 5′ UTR of DENV, to assess whether any base-specific contact are formed. Crystals of NS3H bound to SLA12 diffracted to 2.9 Å, and data collection and refinement statistics are presented in Table 1. Clear continuous electron density resolved residues 168–618 of the two independent NS3H molecules. The phosphodiester backbone linking nucleotides 1–6 of SLA12 (5′AGUUGU3′) as well as bases 1–5 were well ordered. The sixth uracil base was poorly resolved in the density as shown in Supplementary Figure S1. This is similar to the cSLA12 (5′AGACUAACAACU3′)-NS3H structures we reported earlier (PDB IDs: 2JLU, 2JLV, 2JLX, 2JLY, 2JLZ (11)) where the adenine bases of the sixth and seventh nucleotides are mobile (Supplementary Figure S1). The single stranded RNA SLA12 (Figure 1B) is bound in an extended conformation in the groove separating the ATP-binding subdomains I and II from its subdomain III similar to the binding of cSLA12 (see Figure 1C for sequence alignment) (11). A comparison of the atomic contacts established between SLA12 or cSLA12 with NS3H revealed a number of conserved interactions with both the sugar–phosphate backbone and the RNA bases (Figure 1C and D). Of note, NS3H was also previously crystallized bound to RNA13 5′UAGACUAACAACU3′ (PDB ID: 2JLW (11)) and there is good agreement in all three complexes in terms of overall RNA positioning in the NS3H-binding tunnel. It is worth noting that the adenine at position 2 of RNA13 does not make a hydrogen bond with D290. For both SLA12 and cSLA12 sequences, protein–RNA interactions are made through residues from subdomain I and II contacting the sugar–phosphate backbone either directly or through water molecules. The hydrogen bond between D290 and G2 base, seen for NS3H bound to cSLA12, is also observed for the SLA12 complex, with a distance of 2.5 Å. In addition to the D290–G2 interaction, the G2 phosphodiester moiety interacts with the main chain amide of I365. Previous studies have highlighted the importance of this residue whose hydrophobic interaction appears to be required for helicase activity but not for ATPase activity (53). The hydrophobic character of this residue is conserved in NS3 across all flaviviruses, but not in the case the HCV NS3 where it is a lysine (Figure 1E), and may assist positioning the RNA entering the helicase. The structure of NS3H in complex with SLA12 also revealed additional NS3–RNA interactions as the sequence of the 12-mer RNA is different from cSLA12 (Figure 1C and D). This is seen for the G5 base of SLA12 (U5 in cSLA12) which interacts with the D541 carboxylic acid side chain and with main chain atoms of R538 (Figures 1D and 2A).
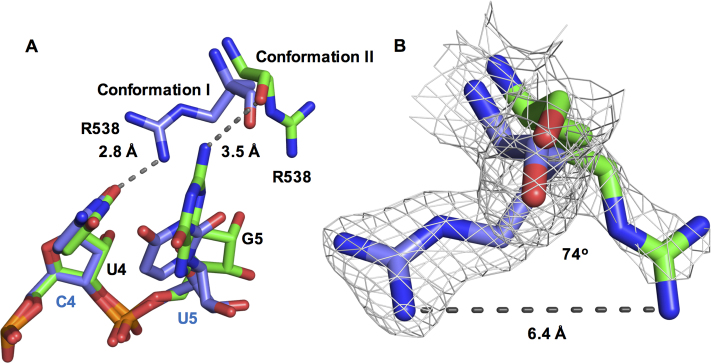
Figure 2.
The RNA interactions of Arg538 across NS3H structures. (A) Arg538 is shown in two conformations. In conformation I (cSLA12, PDB ID: 2JLU (11), chain B) the side chain (blue) interacts with the C4 base (cSLA12) and the position occupied by U4 base in SLA12 is indicated in green. In conformation II (SLA12 structure chain A) Arg538 main chain interaction with G5 in SLA12 is shown in green. (B) electron density for Arg538 in conformation I (blue) and conformation II (green) with the guanidinium groups 6.4 Å apart.
R538 interacts with G5 and might play a role in ssRNA translocation
We also note that the R538 side-chain displays different conformations in each independent monomer of the SLA12-bound structure, which was also observed in the complex with cSLA12 (Figure 2A and B) (11). A view of the electron density for each conformation is shown in Supplementary Figure S1. In both NS3H–SLA12 and NS3H–cSLA12 complexes, electron density is present only for the C-α of one molecule, whilst R538 is fully resolved in the other molecule. Comparison of the two conformations of R538 revealed a shift between main and side chain interactions with the ssRNA (Figure 2A and B). The guanidinium group of R538 is hydrogen bonding with the O2 atom of U4 (cSLA12) whereas in the SLA12-bound structure, the R538 main chain is hydrogen bonding to the N2 atom of G5 (Figure 2A). This differential binding induces a significant displacement of 6.4 Å and a rotation by 74° of the side-chain of R538 (Figure 2B) suggesting R538 undergoes dynamic interactions with RNA.
These two protein–guanine base interactions involving D290–G2 and R538–G5 are of particular interest, especially given the strictly conserved nature of the 5′AG dinucleotide found in all flavivirus genomes (Figure 1C). We note that the DENV, JEV, WNV, SLEV and ZIKV genomic 5′ ends all have G5, but not YFV and TBEV (Figure 1C), potentially providing a mechanism for the specific recognition of the 5′ UTR in a large subgroup of flaviviruses. Conversely, at the protein level, a sequence-based alignment of these flaviviruses revealed that residues D290 and R538 have been conserved throughout evolution (Figure 1E).
Comparison with ZIKV NS3H
Recently the structure of ZIKV NS3H was determined in complex with a 7-mer ssRNA, ‘AGAUCAA’ (PDB ID: 5GJB; (54)) and 9-mer ssRNA, ‘AGACUCCAU’ (PDB ID: 5MFX; unpublished), with the first five bases resolved in each structure. Interestingly, the ssRNA oligonucleotides used contains the conserved 5′AG dinucleotide. However, similar to cSLA12, these ssRNA do not contain the G5 base. The AMPPNP-DENV (PDB ID: 2JLR; (11)) and the ATP–ZIKV (PDB ID: 5GJC; (54)) NS3H structures adopt the same conformation with an root-mean-square deviation (RMSD) of 0.41 Å. Differences were observed only in the flexible loop regions. In the RNA-bound state these two enzymes adopt different conformations with an RMSD of 2.17 Å after superposition. The key interacting residues and their differences between ZIKV NS3H ssRNA and DENV NS3H ssRNA were highlighted by Tian et al. (54). Strong conservation of NS3H between the viruses (72% sequence identity) results in the conservation of RNA-binding residues (Figure 3A and B). The conserved D290 residue (D291 in ZIKV) is hydrogen bonding with the G2 base in each structure (highlighted in Figure 3A and B, with conserved residues shown in Figure 3C). However, K537 from ZIKV NS3H (equivalent to DENV R538), is seen to interact with the 2′-OH group of the ribose sugar. Interactions between ZIKV NS3H and the phosphate backbone are primarily conserved, however T409, D410 and R388 are each displaced toward the 5′ end of the ssRNA (Figure 3A and B). This movement is accompanied by a shift of subdomain II (blue) and the RNA position by 4.2 Å approximately equal to 1 nt distance (Figure 3C) agreeing with the 3′-5′ directionality of SF2 helicases (37). Taken together, the conserved nature of these NS3H protein residues and their interactions with the RNA bases point to an important role for D290 and R538 for RNA translocation and for the unwinding activity of NS3. Guided by these conserved NS3H–RNA interactions across these major flavivirus pathogens, we next designed mutant NS3 proteins having D290A, R538A and G540P mutations, and also 12-mer RNAs with various changes introduced as shown in Figure 1C, and used these various proteins and RNA for the functional studies of NS3 described below.
Figure 3.

Conserved DENV and ZIKV NS3H ssRNA interactions. Schematic representation of nucleotide binding by (A) DENV (SLA12) and (B) ZIKV NS3H (PDB ID: 5GJB (54)). RNA presented 5′-3′ with ribose sugars numbered accordingly, subdomains are represented by colored labels (subdomain I: orange; subdomain II: blue; subdomain III: black) and residues identified within this study highlighted with blue boxes. (C) These structures were superimposed, using NS3H subdomain I, to generate a cartoon representation showing the shift of RNA, distance shown, between DENV (gray) and ZIKV (colored) with interacting residues, calculated using LigPlot, labeled and shown as sticks.
The ATPase activity of NS3 can be stimulated by shorter RNA in a sequence dependent manner
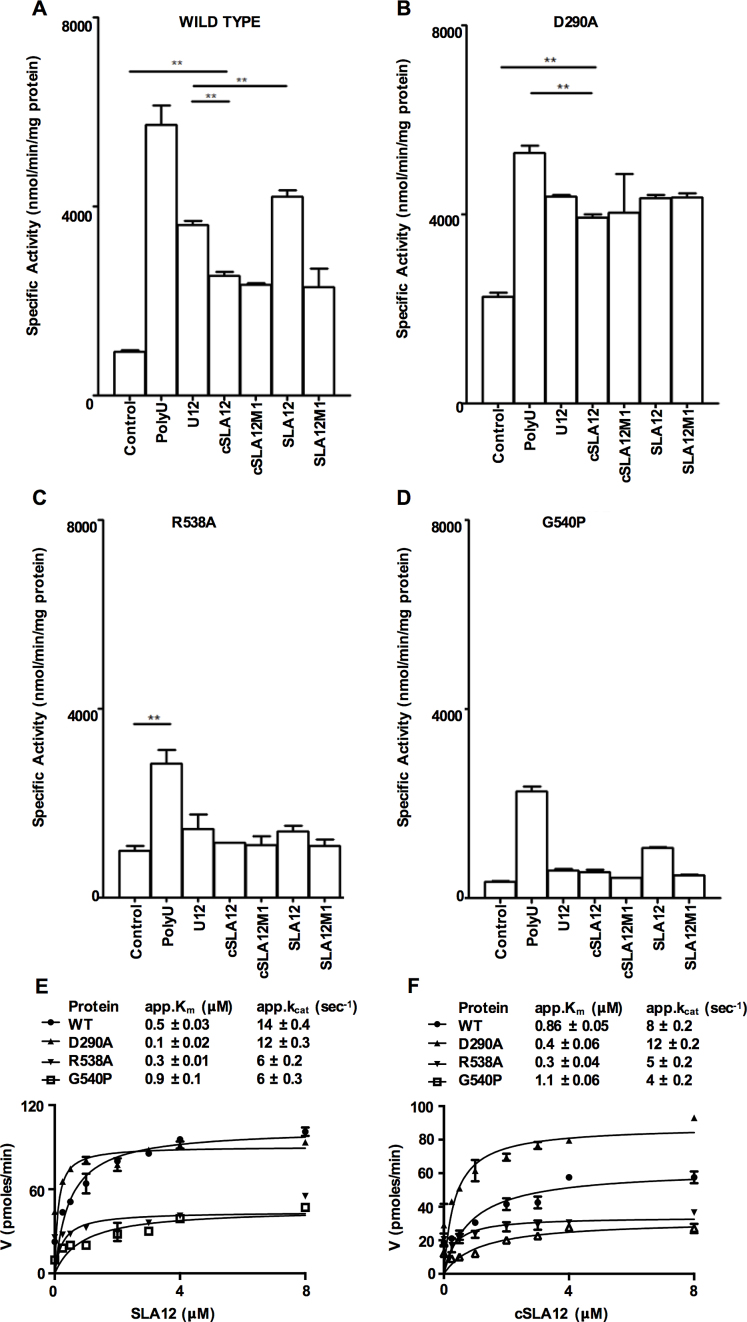
Given the specific interactions between RNA bases and NS3H residues described above, we set out to biochemically characterize a possible sequence-dependent RNA recognition by NS3. Therefore, we studied mutant proteins where RNA interacting residues had been individually mutated to Ala. We utilized the full length NS3 (NS3FL) protein construct for these studies as previously described (17), in order to evaluate the impact of the changes on the enzyme activities of NS3 and its mutants (see ‘Materials and Methods’ section). The addition of polyU (300–3000 bases long; Amersham) resulted in ∼6-fold stimulation of the ATPase activity compared to the basal activity for the WT NS3FL as observed previously (15,19,53,55) (Figure 4A). Next, to investigate if the short 12-mer RNAs (Figure 1C) show allosteric activation of ATPase activity, the polyU RNA was replaced in the ATPase assay with U12, cSLA12, cSLA12M1, SLA12 or SLA12M1 respectively (Figure 1C). These assays showed that all RNAs tested were able to stimulate the ATPase activity (Figure 4A). The level of stimulation, however, varied among the RNAs tested, and in all cases they were lower than the stimulation observed with polyU. Among the 12-mer RNAs SLA12 showed the highest level of stimulation followed by U12 and the remaining three cSLA12, cSLA12M1 and SLA12M1 stimulated NS3 to about the same level (Figure 4A). The second nucleotide ‘G’ was replaced with ‘C’ in SLA12 or cSLA12 to give rise to SLA12M1 or cSLA12M1 (Figure 1C). Previous reports ascribed the higher level of stimulation by polyU to NS3 binding at both the RNA-binding tunnel and also at the positive patch at the surface of subdomain I (19) (Figure 1A). Interestingly the level of ATPase stimulation by U12 is higher compared to cSLA12 even though the later had ‘AG’ in its sequence. By varying the concentration of either ATP or the 12-mer RNAs, we determined their enzyme kinetic constants for SLA12 and cSLA12. The overall summary of the kinetic parameters determined for various substrates and proteins is presented in Table 2. NS3FL showed a preference for SLA12 with a Km of 0.5 μM and kcat/Km of 28 s−1μM−1 compared to cSLA12 that has a Km of 0.86 μM and kcat/Km of 9.3 s−1μM−1 (Table 2). Together these data suggest that occupancy of the RNA-binding cleft of NS3 by a short RNA has an allosteric effect on the ATPase activity, that SLA12 RNA is the preferred substrate and that there is a sequence-dependent recognition of RNA by NS3 of DENV (Figure 4A).
Figure 4.
Specific activity of WT or mutant NS3 proteins in ATPase assay. (A) WT NS3, (B) NS3 D290A, (C) NS3 R538A and (D) NS3 G540P ATPase activity in the absence or presence of various RNA were measured as indicated in ‘Materials and Methods’ section and significant differences for activity are shown by the bars within the plots with asterisks (**P ≤ 0.05) with n ≥ 4. The kinetic plot and parameter for (E) SLA12 and (F) cSLA12 with WT and mutant NS3 proteins. Error bars represent the standard error of mean of duplicate measurements.
ATPase activity stimulation of NS3 D290A mutant by 12-mer RNA is not sequence dependent
Having shown that short 12-mer RNA substrate can stimulate the ATPase activity of NS3 in a sequence-specific manner, we next addressed the significance of the side chains that directly interact with the bases in the RNA binding tunnel. NS3FL with mutations at D290, R538 and G540 were tested for their effect on ATPase activity with short 12-mer RNA substrates.
Mutation of D290 to Ala disrupts the formation of the H bond with G2 of both SLA12 and cSLA12 and showed a higher basal ATPase activity than the WT protein. We speculate that this activation may be the result of disruption of intramolecular interactions between D290 (subdomain I) and R387 (subdomain II) which forms part of the switch for RNA stimulation of ATPase activity. We emphasize that only part of the RNA stimulation switch is abolished because the increased basal (RNA independent) ATPase activity of the D290A mutant is still sensitive to RNA binding (see thermal shift data below). In fact, all the 12-mer RNAs tested (Figure 4B) stimulated NS3FL D290A to a similar level. The kcat/Km toward SLA12 and cSLA12 is increased 4- and 3-fold respectively (Table 2) when compared with the WT NS3FL suggesting higher specificity of D290A for both 12-mer RNAs. To explore this further the NS3H carrying the D290A mutation was purified and crystallized. The crystals diffracted to 2.1 Å resolution (Table 1). This structure adopted a similar conformation to the free NS3H structure with an RMSD of 0.23 Å over 399 residues and the mutant structure also reveals the disruption of the ionic interaction between D290 and R387, which is observed in all NS3H (without bound RNA) as well as NS3FL structures solved (Supplementary Figure S2). Possible reasons for the increased basal activity of D290A will be discussed later.
The R538 and G540 residues located within subdomain III of the NS3H domain showed a more drastic effect on the ATPase activity when mutated to Ala and proline respectively. In the case of R538A, the ATPase activity was similar to the basal level observed for WT NS3 and the presence of SLA12 or the other 12-mer RNAs did not stimulate the activity (Figure 4C and D). The G540P mutant shows lower basal activity with reduced catalytic efficiency with both SLA12 and cSLA12 as is evident from their kcat/Km values of 6.7 and 3.6 s−1μM−1 respectively (Figure 4E and F) compared to WT (kcat/Km of 28 and 9.3 s−1μM−1 respectively). SLA12 stimulates the ATPase activity but nevertheless the stimulated activity is similar to the basal activity in WT NS3 (Figure 4D). One key conclusion from our studies so far is that despite their remote location away from the ATPase active site located in the interface between subdomains I and II, residues R538 and G540 in subdomain III have a profound effect on the ATP hydrolysis activity, possibly through allosteric conformational/dynamical changes transmitted to the active site through the protein backbone (56).
SLA12 is able to compete with duplex RNA binding
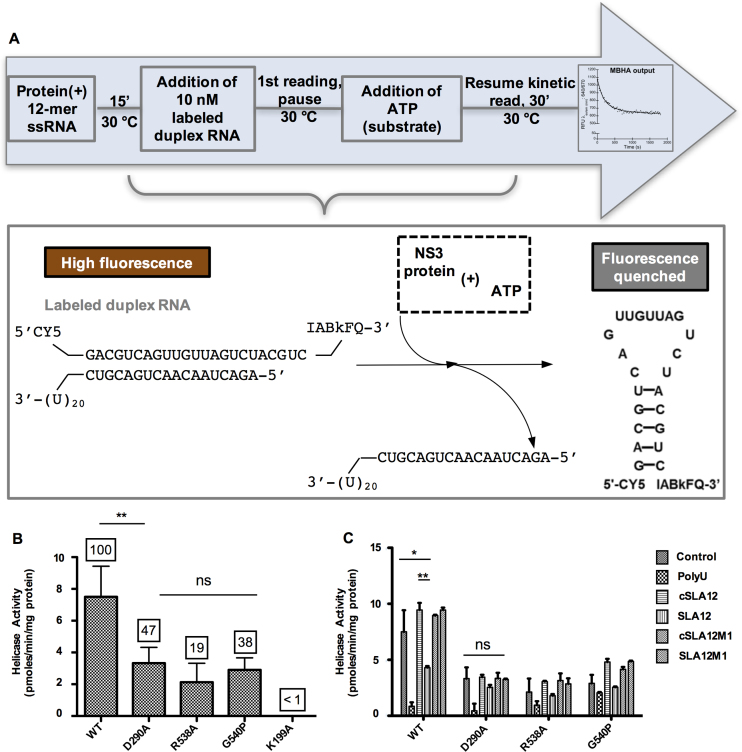
In order to assess the effect of the mutations within the RNA binding tunnel of NS3FL on the helicase activity of NS3FL, we used a previously developed non-radioactive molecular beacon helicase assay (MBHA) to continuously monitor helicase catalyzed unwinding (Figure 5A) (43). The helicase activity of NS3FL was compared to the D290A, R538A or G540P mutants. In addition, K199A (located within the Walker A motif) mutant was included as a negative control, as it was previously shown to be completely inactive in ATP hydrolysis and 5′ RNA-triphosphatase activities (24) and was inactive as a helicase in the MBHA. Compared to the helicase activity observed for WT NS3FL, the mutants showed 47, 19 or 38% activity for D290A, R538A or G540P respectively (Figure 5B).
Figure 5.
Helicase activity of WT or mutant NS3 proteins determined by the MBHA. (A) Schematic depiction of MBHA. Duplex RNA used in the assay contains one strand labeled with a cyanine 5 (Cy5) fluorophore at the 5′ end and an Iowa Black FQ (IABkFQ) quencher at the 3′ end. A complementary strand has a U20 overhang for the loading/recruitment of the helicase. ATP-dependent unwinding results in the release of the fluorophore/quencher labeled RNA that forms a hairpin to quench the fluorescence emitted by the fluorophore. (B) Specific activity of NS3 mutant proteins as compared to WT protein. The numbers on top of the bars represent percentage of helicase activity based on WT NS3 which is denoted as 100%. (C) Competition of 12-mer RNA with duplex RNA in the helicase assay as measured for WT, D290A, R538A and G540P proteins. In order to understand the interplay between duplex RNA and ATP binding, we investigated the impact of adding the various 12-mer RNAs or polyU to the MBHA mixture for measuring unwinding activity of the WT and mutant NS3 proteins. A control reaction without any 12-mer RNA was included to compare the activity level. In order to understand the interplay between duplex RNA and ATP binding, we investigated the impact of adding the various 12-mer RNAs or polyU to the MBHA mixture for measuring unwinding activity of the WT and mutant proteins. Error bars represent the standard error of mean of duplicate measurements and significant differences are shown by the bars within the plots with asterisks (*P ≤ 0.10, **P ≤ 0.05 and ns = no significant difference).
As for the steady-state kinetic parameters for ATP and duplex RNA substrate in the helicase assay, WT NS3 displayed an apparent Km of 0.035 and 530 μM for duplex RNA and ATP respectively, with an apparent Vmax of 0.03 pmoles/min and kcat/Km of 3 × 10−3 s−1μM−1. The D290A mutant protein shows ∼4-fold reduction in helicase activity and displayed a 4-fold lower Km compared to WT NS3 but its Km for ATP was 3-fold higher. Despite sacrificing over 50% of helicase activity, the D290A mutant retains almost 70% unwinding efficiency (kcat/Km) for duplex RNA relative to WT. The R538A mutant retained only 19% of WT helicase activity with a 3-fold reduced unwinding efficiency for duplex RNA (kcat/Km of 0.9 × 10−3 s−1μM−1), due to a lower apparent Km for RNA (0.023 μM) and a 5-fold higher Km for ATP (2629 μM). The G540P mutation showed reduced apparent Km value for RNA and ATP by 4- and 2-fold respectively, resulting in a reduced kcat/Km of 1 × 10−3 s−1μM−1 for duplex RNA unwinding when compared with WT.
To measure the helicase inhibition, the NS3 protein was incubated with 12-mer ssRNA prior to the addition of duplex RNA to start the helicase activity. A reduction in helicase activity was interpreted as the 12-mer RNA being able bind to the RNA groove and compete/prevent duplex RNA binding. Approximately 50% reduction in helicase activity was observed in the presence of SLA12 but not cSLA12, cSLA12M1 or SLA12M1. The Asp at position 290 plays an important role since the competition by SLA12 is no longer significant when G2 is mutated to C in SLA12M1 (Figure 5C). That the 12-mer RNAs were unable to significantly compete with the duplex RNA for mutant protein binding to decrease the helicase activity, except for polyU which binds an allosteric site (Figure 5C), suggests that sequence specific interactions with the RNA observed in the crystal structure are important for ssRNA binding in solution.
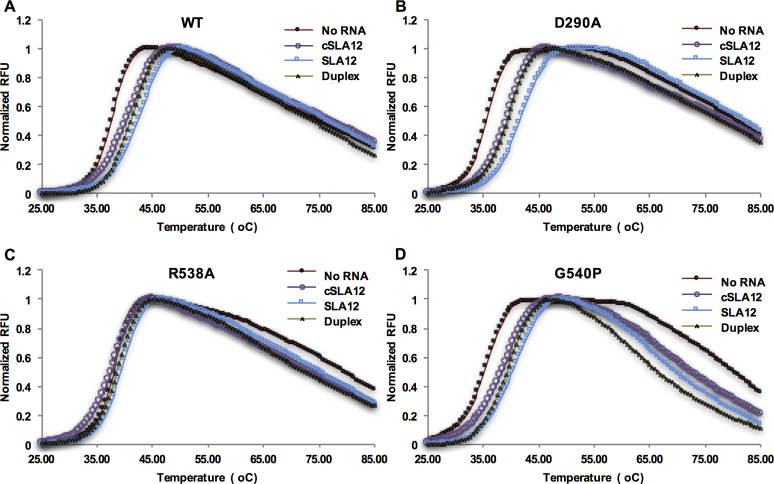
Thermal shift assays indicate stabilization of NS3FL by SLA12
Taking another approach thermal shift assays were conducted for WT NS3 and mutant proteins to evaluate the impact of RNA binding in the presence of SLA12, cSLA12 or duplex RNA, as binding has previously been correlated with stabilization in the thermal shift assay (57). Although not quantitative thermal shift data has been shown to correlate with results from other biophysical methods (58). The melting temperature (Tm) values determined for the different RNAs and proteins are summarized in Table 3. The WT protein showed a Tm of 37.5°C in the absence of any RNA. In the presence of SLA12, the Tm increased by 4.5°C while addition of cSLA12 or duplex RNA only increased the Tm by 2.8 and 2.5°C respectively (Figure 6A). This data suggest that SLA12 makes the maximum contacts with NS3 to stabilize the molecule presumably through G2 and G5 base positions as observed in the structure. The D290A mutant protein which displayed a higher level of basal ATPase activity than WT protein showed an Tm of 35.5°C without any RNA. This suggests that the mutation did not make NS3 more stable although the basal ATPase activity is higher than WT. Furthermore the reduction in stability may be speculated to result from the abolishment of the intramolecular interactions between D290 (subdomain I) and R387 of subdomain II, liberating a molecular ‘lock’ between the two subdomains (Supplementary Figure S2). This charge interaction is also disrupted upon RNA binding (shown in Figure 3A) allowing for the rearrangement of the ATPase site into the more active conformation. Addition of SLA12 resulted in an Tm increase of 5.6°C while both cSLA12 and duplex RNA showed an Tm increase of ∼3.8°C (Figure 6B). Interestingly despite the differences in Tm, the RNA stimulated ATPase activity for D290A were somewhat similar for SLA12 and cSLA12.
Table 3. Summary table of melting temperatures as derived from thermal shift assays.
| no RNA | cSLA12 | SLA12 | Duplex RNA | |
|---|---|---|---|---|
| WT | 37.5 ± 0.021 | 40.5 ± 0.071 | 42.0 ± 0.16 | 41.1 ± 0.19 |
| D290A | 35.5 ± 0.11 | 39.2 ± 0.050 | 41.1 ± 0.49 | 39.4 ± 0.11 |
| R538A | 38.0 ± 0.071 | 37.7 ± 0.049 | 38.9 ± 0.15 | 38.7 ± 0.18 |
| G540P | 35.2 ± 0.18 | 38.4 ± 0.12 | 39.9 ± 0.071 | 39.3 ± 0.28 |
Figure 6.
Thermal shift assays representing the melting temperature of WT and mutant NS3 proteins. Melting curves show increase in fluorescence upon binding of Sypro-Orange dye to (A) WT, (B) D290A, (C) R538A or (D) G540P protein when subjected to thermal ramping from 20 to 95°C in the presence of ssRNA (SLA12, cSLA12) or duplex RNA (dsSLA12).
The R538A mutant displayed an Tm closer to WT (38.0°C) however the addition of the RNAs did not significantly change the Tm value suggesting that this mutant probably does not bind RNA in the RNA-binding groove, separating subdomains I and II from subdomain III (Figure 6C). This is in agreement with the ATPase activity data (Figure 4C) that is not stimulated above the basal activity by any of the 12-mer RNAs tested. The melting profile for the G540P mutant showed an Tm of 35.2°C without any RNA. Interestingly an increase in Tm in the presence of RNA in a similar fashion to the WT protein is shown (Figure 6D) even though the basal ATPase activity of the mutant is much lower than that of the WT protein (Figure 4D). Overall the data shows that the base specific interactions seen in the crystal structure with SLA12 leads to the highest stability in NS3 and an increase in its ATPase specific activity. The data appears to suggest that D290 may transduce to the ATPase active site the RNA-binding signal sensed by R538 through conformational/dynamical changes transmitted to the active site through the protein backbone (56).
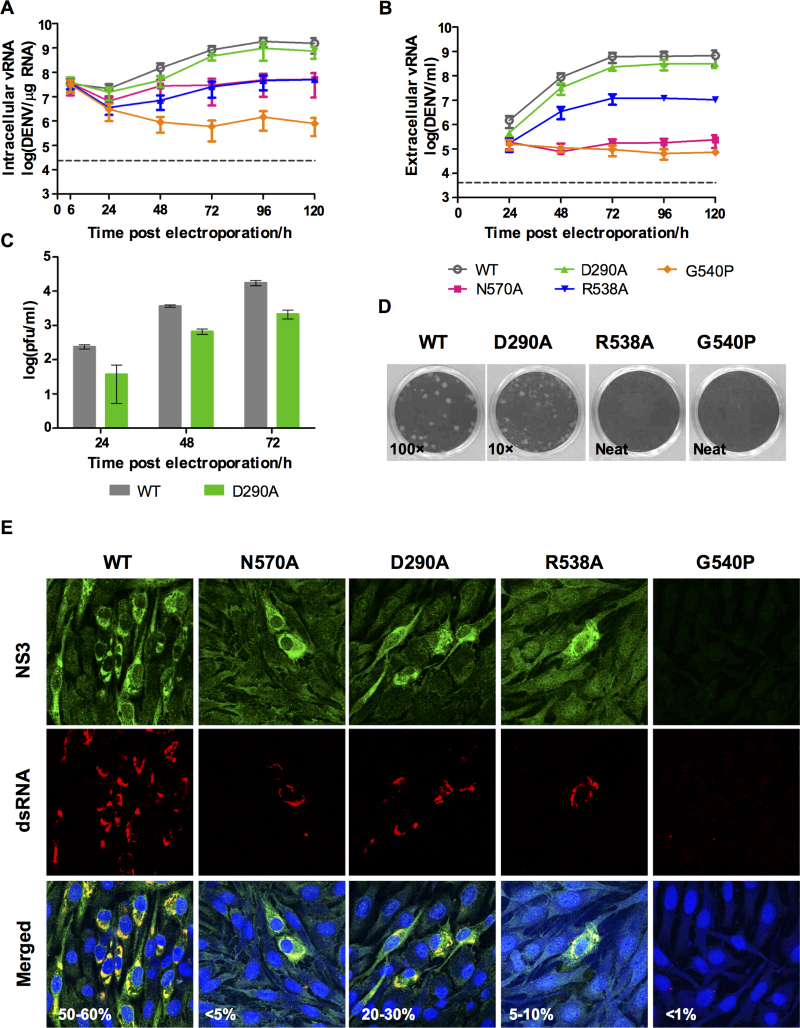
Sequence-specific interactions modulate viral genome replication
In order to determine the impact of these NS3 RNA-binding groove mutations on viral replication, infectious cDNA clones harboring D290A, R538A or G540P mutations were generated by site-directed mutagenesis. Assessment of viral RNA synthesis by real-time RT-PCR showed that the D290A mutation resulted in a ∼2-fold lower viral RNA replication rate compared to WT (Figure 7A). This is also in agreement with the level of extracellular viral RNA level which is ∼2-fold lower than WT (Figure 7B). The slower replication rate of NS3:D290A mutant is also reflected in >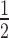 log reduction in infectious virus production (Figure 7C) and smaller plaques detected by standard BHK-21 plaque assay (Figure 7D). A lower percentage of infection (20–30%) compared to WT is also detected by immunofluorescence staining of the transfected cells with anti-NS3 antibody (3F8; (49)) and dsRNA (Figure 7E). These results suggest that the D290A mutation reduces both virus RNA replication and infectious virion production probably due to the reduced helicase activity. The R538A mutant on the other hand exhibited severe attenuation in viral genome replication, with >1 log reduction in viral RNA synthesis as compared to WT (Figure 7A). Extracellular RNA can be detected in supernatant from R538A transfected cells suggesting that plaques in the BHK-21 plaque assay are too tiny to be visualized (Figure 7B–D). The G540P mutation was also severely impaired with an even lower level of intracellular RNA than that observed for the replication defective negative control N570A (48). Extracellular RNA detection was similar to that of N570A but no plaques could be observed although some staining of dsRNA can be observed in the immunofluorescence staining even though no NS3 can be detected.
log reduction in infectious virus production (Figure 7C) and smaller plaques detected by standard BHK-21 plaque assay (Figure 7D). A lower percentage of infection (20–30%) compared to WT is also detected by immunofluorescence staining of the transfected cells with anti-NS3 antibody (3F8; (49)) and dsRNA (Figure 7E). These results suggest that the D290A mutation reduces both virus RNA replication and infectious virion production probably due to the reduced helicase activity. The R538A mutant on the other hand exhibited severe attenuation in viral genome replication, with >1 log reduction in viral RNA synthesis as compared to WT (Figure 7A). Extracellular RNA can be detected in supernatant from R538A transfected cells suggesting that plaques in the BHK-21 plaque assay are too tiny to be visualized (Figure 7B–D). The G540P mutation was also severely impaired with an even lower level of intracellular RNA than that observed for the replication defective negative control N570A (48). Extracellular RNA detection was similar to that of N570A but no plaques could be observed although some staining of dsRNA can be observed in the immunofluorescence staining even though no NS3 can be detected.
Figure 7.
Characterization of DENV2 WT and NS3 mutants D290A, R538A and G540P. BHK-21 cells were transfected with 10 μg of in vitro transcribed RNA and the replication kinetics of WT and NS3 mutants was followed over 5 days. The DENV2 N570A (NS3–NS5 interaction mutant, (48)) was included as a control. (A) Total vRNA synthesis measured by real time RT-PCR. Gray dotted line represents the detection level of uninfected control. (B) Extracellular vRNA as detected by real time RT-PCR. Gray dotted line represents the detection level of uninfected control. (C) Virus titer in the supernatants as measured by plaque assay. (D) Plaque morphologies of the WT and NS3 mutants at the indicated dilution factor (inset). (E) Confocal images showing localization of NS3 and dsRNA for WT and the mutant viruses. The images were taken at 72 h post-transfection using 64× oil immersion lens and the percentage of infection was indicated.
We then quantified the intracellular positive- and negative-strand viral RNA with strand-specific real-time PCR as described previously (48). The result shows that compared to WT and D290A mutant which show a positive slope for negative-strand RNA synthesis until 24 h post-transfection, R538A and G540P show a steep negative slope similar to what was observed for N570A (Supplementary Figure S3). We believe that the negative slope was indicative of insufficient negative-strand RNA synthesis for supporting viral RNA replication (48). The G540P mutant unlike the other mutants continued to show a reduced level of negative strand RNA synthesis until 48 h post-transfection. This probably explains why the total intracellular RNA for G540P mutant is lower than for N570A mutant (Figure 7A). The detection of 5–10% infectivity for R538A is consistent with the pattern of negative strand RNA synthesis in comparison with N570A which shows around 5% infectivity at day 3 post-transfection. Infection of BHK-21 cells with neat supernatant did not recover detectable plaques (Figure 7D) for R538A and G540P. To examine this further we used the day 3 post-transfection BHK-21 supernatants to infect C6/36 for virus amplification and recovery (48). Infectious virus could be recovered from R538A mutant transfected BHK-21 supernatant after infection in C6/36 (Supplementary Figure S4). The infectious titer for R538A mutant is ∼1 log lower than WT (Supplementary Figure S4B), that is consistent with the slower genomic RNA synthesis observed in BHK-21 cells after RNA transfection (Figure 7A). The R538A mutant displayed very small plaque morphology (Supplementary Figure S4B).
The G540P mutant was found to be lethal, hence no resulting virus could not be expanded in C6/36 cells. Overall the mutational study confirm that the residues tested are important for RNA replication, demonstrating a correlation between the in vitro and in vivo results, revealed in the rate of replication and production of infectious virus particles. Similarly, the introduction of mutations into motifs I, II and VI abolishes both helicase activity and replication of BVDV (59) and DENV2 (60,61), indicating that impaired helicase activity in turn results in a reduced rate of virus replication.
DISCUSSION
The NS3 protein of DENV is a multi-functional protein where the C-terminal helicase may act to resolve the secondary structures within the gRNA or unwind the duplex replicative form (RF) using the energy derived from ATP hydrolysis (62,63). There is limited information on the RNA–protein interactions during flavivirus replication. In this work, we have used X-ray crystallography, biochemical assays and reverse genetics to elucidate the specific recognition of the gRNA 5′ end by NS3H and its functional implications in enzymatic activities as well as virus replication. Previous studies with RNA12, the reverse complement sequence of the 5′ UTR of DENV genome which is identified as cSLA12 in this work, showed specific interactions with the AG dinucleotide at the 5′ of the sequence. Recently, we showed that the DENV 5′ end AG dinucleotide is absolutely essential for integral steps in RNA capping as revealed in the crystal structure of 5′-m7G0pppA1G2U3U4G5U6U7–3′ with DENV3 NS5, where the AG dinucleotide was captured in a conformation where it was poised for the 2′O methylation reaction (42). While the potential of SF2 helicases to display sequence specificity has been speculated (37), other examples of helicase specificity can be found in members of the DEAD-box protein A family (64,65), it was shown that the HCV NS3 recognizes the 3′ UTR (66). The strong 5′ UTR sequence conservation among flaviviruses and the requirement for both NS3 and NS5 to interact with the 5′ end of the gRNA to construct the type 1 RNA cap formed the basis for our investigation of the RNA sequence specificity of NS3. The crystal structure of SLA12 that is identical to the 12 conserved nucleotides of the 5′ UTR of DENV1–4 forms specific interactions through the bases at positions G2 and G5 with NS3 residues D290 and R538, respectively. The hydrogen bond between the side chain carboxylic group of D290 and N2 atom of G2 is disrupted in the D290A mutant. Hence, this mutation reduces the sequence specificity for the RNA and is consistent with our observation that the mutant shows less differential stimulation of its ATPase activity by the various 12-mer RNAs. The structure of D290A mutant in NS3H does not reveal major differences compared to WT although D290 and R387 are located in subdomains I and II respectively, and the mutation disrupted the ionic interaction with R387. In theory, it is plausible that the disruption of the D290–R387 charge interaction due to the elimination of the carboxylic side chain in the D290A mutant may increase the ATPase site free energy and lower the activation energy required for ATPase activity. This may also lead to some level of futile ATPase cycling if the energy from the hydrolyzed ATP is not used for RNA translocation which accounts for the higher basal activity in the absence of RNA. Indeed, other studies with unrelated enzymes have shown that mutations in allosteric sites can impact on the conformational dynamics of residues in the active site freeing the enzyme from the need for activation by allosteric modulators (56,67). It has also been suggested that mutations of the DExH (motif II) in HCV NS3 result in electronic changes in the local environment that impact the ATPase activity (68). Therefore the slight attenuation of the D290A mutant virus in this study may be due to effects of lowered ATP concentration within membrane vesicles containing the replication complex (RC) as a result of increased basal ATP consumption. Furthermore, the important regulatory role of D290 in NS3 function within the RC is suggested by the isolation of a D290N compensatory mutation that conferred fitness to a replication-impaired DENV2 recombinant virus containing chimeric DENV4 MTase and DENV2 RdRp NS5 (69).
The interaction of G5 with R538 main chain carbonyl group of the amide bond was uniquely observed with SLA12. However, the importance of the different conformations of this residue and the adjacent residues in RNA translocation can also be gleaned from the cSLA12 interaction between C4 base with the side chain of R538 (11). The R538A mutant virus replication is more attenuated than D290A virus and has a small-plaque phenotype. No compensatory second site mutations could be detected for R538A (data not shown) which suggests that the observed virus attenuation may be due to the impaired NS3 helicase activity. Overall the data suggest that R538 plays an important role in both translocation and communicating RNA presence in the RNA-binding tunnel to the ATPase site (∼23 Å between R538 and ATPase site) through a series of dynamic interactions across the C-α backbone. The G5 base of SLA12 interacts with a conserved D541 residue in addition to the main chain amide of R538. However, the D541A or D541N mutant viruses that were made as part of this work (Supplementary Figure S5) showed similar replication as WT at the early stage (first 24 h) and resulted only in an ∼3-fold reduction at 48 h post-transfection. This is not as pronounced as the effect observed for R538A mutation. Nevertheless, the RNA interaction of D541 and R538 suggests that flexibility of the protein backbone in this region may be important. In fact, the rigidification of the flexible loop by mutation of glycine at position 540 to proline was lethal to the virus. Given this region is located on the ‘roof’ of the RNA binding groove, unique to flaviviral helicases, of NS3 provides potential new targets for the design of antiviral drugs unique to the viral NS3.
In terms of the mechanics of viral RNA replication in the RC, the role of the sequence recognition by NS3 of the conserved 5′ UTR requires further studies. However, based on previous studies (70) and our current data for sequence specific binding we can propose the following working model viral genomic RNA replication (Figure 8). The replication of the initial gRNA requires the recognition of the 5′ sequence element by NS5 so that replication of the negative strand RNA can be initiated and carried. NS3 would be recruited to resolve RNA secondary structures. The affinity of NS3 for the 5′ sequence element is speculated to result in its binding at the free 5′ region. This affinity is also reflected in the subsequent replication of the double-stranded RF form so that the displaced strand is marked by a NS3 bound to its 5′ sequence element to be guided out of the RC for packaging into newly formed virion. This is supported by observation that NS3 is involved in viral RNA packaging of YFV (71) but it is not packaged in the virion within the nucleocapsid. Interestingly, the HCV helicase appears to recognize the 3′ UTR rather than the 5′ UTR (66), in line with the 3′–5′ directionality of SF2 helicases and possibly hinting at different mechanisms for replication. The resolution of the events in the RC require more biophysical and biochemical methods in conjunction with mutants to unravel the critical protein–protein and protein–RNA interactions.
Figure 8.
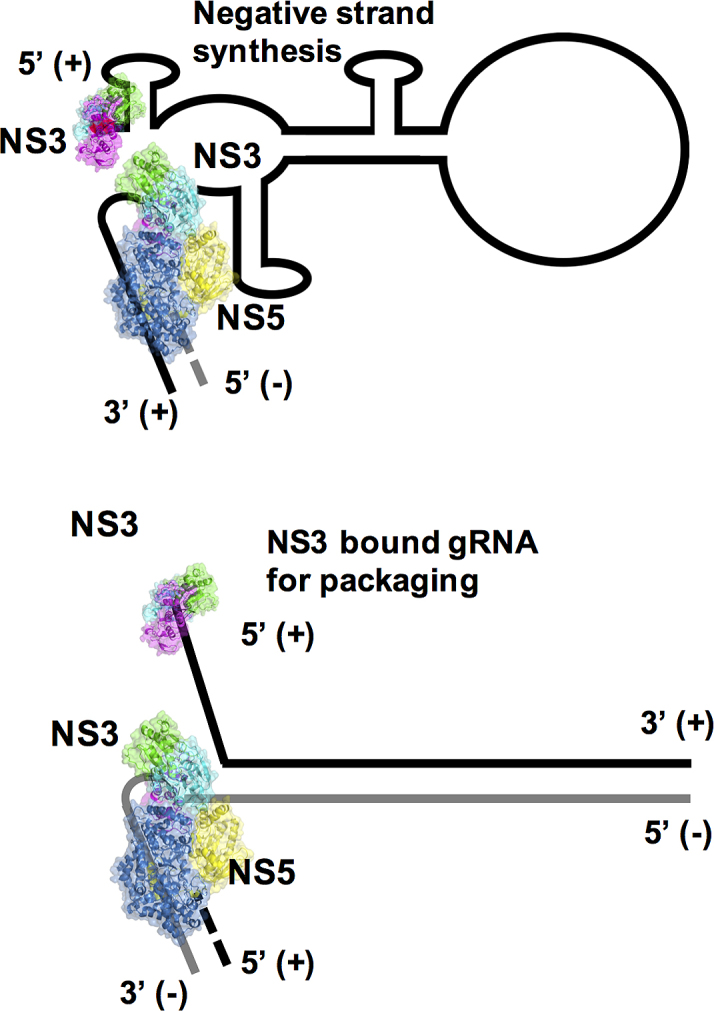
A working model for RNA replication involving the NS3 and NS5 proteins. Based upon recent findings (48,51) we propose that the specificity of NS3 for SLA12 may allow NS3 to separate duplex RNA. Allowing NS3 to bind and unwind the duplex in a 3′→5′ direction feeding the 3′ end of the negative strand to NS5 which generates further positive stranded daughter RNA in a 5′→3′ direction. Negative RNA strand is shown in gray.
CONCLUDING REMARKS
Recent outbreaks in members of the genus flavivirus have increased global concern over their potential impact creating a more urgent need for the development of anti-virals and the identification of anti-viral targets. Proteins NS3 and NS5 represent prime targets for inhibition of the replication of flaviviruses. The structure-guided characterization of NS3 mutants D290A, R538A and G540P has elucidated the important roles of these residues in RNA recognition, translocation and ATPase coordination. The mutation of these residues impairs viral replication and infection as shown through reduced RNA production, plaque formation in plaque assays and infectivity of the virus. Our studies show that the RNA interaction with subdomain III could form a target for the design of new antiviral allosteric drugs or alternatively, for the structure-based design of attenuated viruses as vaccines. Many questions still remain on the mechanism of flavivirus RNA replication. Here, we have proposed a working model for that process that can be further tested to validate and discover the precise molecular events in more details.
AVAILABILITY
The structural data from this publication has been deposited to the RCSB protein data bank (PDB) and assigned the identifiers 5XC6 (NS3H with SLA12) and 5XC7 (NS3H D290A).
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues from the Vasudevan lab, in particular Drs Moon Tay and Danny Doan, for their technical contributions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Health, Singapore TDuke-NUS Signature Research Program; National Medical Research Council, Singapore (http://www.nmrc.gov.sg) [NMRC/ MOHIAFCat1/0018/2014, NMRC/CBRG/0073/2014, NMRC/CBRG/0103/2016]. Funding for open access charge: National Medical Research Council, Singapore [NMRC/CBRG/0103/2016].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O. et al. The global distribution and burden of dengue. Nature. 2013; 496:504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guzman M.G., Harris E.. Dengue. Lancet. 2015; 385:453–465. [DOI] [PubMed] [Google Scholar]
- 3. Manrique-Hernandez E.F., Fernandez-Nino J.A., Idrovo A.J.. Global performance of epidemiologic surveillance of Zika virus: rapid assessment of an ongoing epidemic. Public Health. 2017; 143:14–16. [DOI] [PubMed] [Google Scholar]
- 4. Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L., Guimaraes K.P., Benazzato C., Almeida N., Pignatari G.C., Romero S. et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016; 534:267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vasconcelos P.F., Monath T.P.. Yellow fever remains a potential threat to public health. Vector Borne Zoonotic Dis. 2016; 16:566–567. [DOI] [PubMed] [Google Scholar]
- 6. Chen Z., Liu L., Lv Y., Zhang W., Li J., Zhang Y., Di T., Zhang S., Liu J., Li J. et al. A fatal yellow fever virus infection in China: description and lessons. Emerg. Microbes Infect. 2016; 5:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scott L.J. Tetravalent dengue vaccine: a review in the prevention of dengue disease. Drugs. 2016; 76:1301–1312. [DOI] [PubMed] [Google Scholar]
- 8. Aguiar M., Stollenwerk N., Halstead S.B.. The risks behind Dengvaxia recommendation. Lancet Infect. Dis. 2016; 16:882–883. [DOI] [PubMed] [Google Scholar]
- 9. Low J.G., Ooi E.E., Vasudevan S.G.. Current status of dengue therapeutics research and development. J. Infect. Dis. 2017; 215:S96–S102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindenbach B.D., Rice C.M.. Molecular biology of flaviviruses. Adv. Virus Res. 2003; 59:23–61. [DOI] [PubMed] [Google Scholar]
- 11. Luo D., Xu T., Watson R.P., Scherer-Becker D., Sampath A., Jahnke W., Yeong S.S., Wang C.H., Lim S.P., Strongin A. et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008; 27:3209–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lim S.P., Wang Q.Y., Noble C.G., Chen Y.L., Dong H., Zou B., Yokokawa F., Nilar S., Smith P., Beer D. et al. Ten years of dengue drug discovery: progress and prospects. Antiviral Res. 2013; 100:500–519. [DOI] [PubMed] [Google Scholar]
- 13. Selisko B., Wang C., Harris E., Canard B.. Regulation of Flavivirus RNA synthesis and replication. Curr. Opin. Virol. 2014; 9:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luo D., Vasudevan S.G., Lescar J.. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral Res. 2015; 118:148–158. [DOI] [PubMed] [Google Scholar]
- 15. Xu T., Sampath A., Chao A., Wen D., Nanao M., Chene P., Vasudevan S.G., Lescar J.. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J. Virol. 2005; 79:10278–10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luo D., Xu T., Hunke C., Gruber G., Vasudevan S.G., Lescar J.. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 2008; 82:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo D., Wei N., Doan D.N., Paradkar P.N., Chong Y., Davidson A.D., Kotaka M., Lescar J., Vasudevan S.G.. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 2010; 285:18817–18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yusof R., Clum S., Wetzel M., Murthy H.M., Padmanabhan R.. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J. Biol. Chem. 2000; 275:9963–9969. [DOI] [PubMed] [Google Scholar]
- 19. Li H., Clum S., You S., Ebner K.E., Padmanabhan R.. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 1999; 73:3108–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chambers T.J., Nestorowicz A., Amberg S.M., Rice C.M.. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J. Virol. 1993; 67:6797–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Falgout B., Pethel M., Zhang Y.M., Lai C.J.. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 1991; 65:2467–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wengler G., Czaya G., Farber P.M., Hegemann J.H.. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J. Gen. Virol. 1991; 72:851–858. [DOI] [PubMed] [Google Scholar]
- 23. Assenberg R., Mastrangelo E., Walter T.S., Verma A., Milani M., Owens R.J., Stuart D.I., Grimes J.M., Mancini E.J.. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol. 2009; 83:12895–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benarroch D., Selisko B., Locatelli G.A., Maga G., Romette J.L., Canard B.. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004; 328:208–218. [DOI] [PubMed] [Google Scholar]
- 25. Yon C., Teramoto T., Mueller N., Phelan J., Ganesh V.K., Murthy K.H., Padmanabhan R.. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 2005; 280:27412–27419. [DOI] [PubMed] [Google Scholar]
- 26. Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M.. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989; 17:4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singleton M.R., Wigley D.B.. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002; 184:1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borowski P., Niebuhr A., Mueller O., Bretner M., Felczak K., Kulikowski T., Schmitz H.. Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol. 2001; 75:3220–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Warrener P., Tamura J.K., Collett M.S.. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 1993; 67:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Utama A., Shimizu H., Morikawa S., Hasebe F., Morita K., Igarashi A., Hatsu M., Takamizawa K., Miyamura T.. Identification and characterization of the RNA helicase activity of Japanese encephalitis virus NS3 protein. FEBS Lett. 2000; 465:74–78. [DOI] [PubMed] [Google Scholar]
- 31. Gwack Y., Kim D.W., Han J.H., Choe J.. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 1996; 225:654–659. [DOI] [PubMed] [Google Scholar]
- 32. Jin L., Peterson D.L.. Expression, isolation, and characterization of the hepatitis C virus ATPase/RNA helicase. Arch. Biochem. Biophys. 1995; 323:47–53. [DOI] [PubMed] [Google Scholar]
- 33. Kanai A., Tanabe K., Kohara M.. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 1995; 376:221–224. [DOI] [PubMed] [Google Scholar]
- 34. Kim D.W., Gwack Y., Han J.H., Choe J.. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 1995; 215:160–166. [DOI] [PubMed] [Google Scholar]
- 35. Preugschat F., Averett D.R., Clarke B.E., Porter D.J.. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 1996; 271:24449–24457. [DOI] [PubMed] [Google Scholar]
- 36. Tai C.L., Chi W.K., Chen D.S., Hwang L.H.. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 1996; 70:8477–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Linder P., Jankowsky E.. From unwinding to clamping–the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. [DOI] [PubMed] [Google Scholar]
- 38. Chernov A.V., Shiryaev S.A., Aleshin A.E., Ratnikov B.I., Smith J.W., Liddington R.C., Strongin A.Y.. The two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J. Biol. Chem. 2008; 283:17270–17278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Appleby T.C., Anderson R., Fedorova O., Pyle A.M., Wang R., Liu X., Brendza K.M., Somoza J.R.. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J. Mol. Biol. 2011; 405:1139–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dumont S., Cheng W., Serebrov V., Beran R.K., Tinoco I. Jr, Pyle A.M., Bustamante C.. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006; 439:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gu M., Rice C.M.. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao Y., Soh T.S., Lim S.P., Chung K.Y., Swaminathan K., Vasudevan S.G., Shi P.-Y., Lescar J., Luo D.. Molecular basis for specific viral RNA recognition and 2′-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5). Proc. Natl. Acad. Sci. U.S.A. 2015; 112:14834–14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Basavannacharya C., Vasudevan S.G.. Suramin inhibits helicase activity of NS3 protein of dengue virus in a fluorescence-based high throughput assay format. Biochem. Biophys. Res. Commun. 2014; 453:539–544. [DOI] [PubMed] [Google Scholar]
- 44. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lanzetta P.A., Alvarez L.J., Reinach P.S., Candia O.A.. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979; 100:95–97. [DOI] [PubMed] [Google Scholar]
- 48. Tay M.Y., Saw W.G., Zhao Y., Chan K.W., Singh D., Chong Y., Forwood J.K., Ooi E.E., Gruber G., Lescar J. et al. The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J. Biol. Chem. 2015; 290:2379–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moreland N.J., Tay M.Y., Lim E., Rathore A.P., Lim A.P., Hanson B.J., Vasudevan S.G.. Monoclonal antibodies against dengue NS2B and NS3 proteins for the study of protein interactions in the flaviviral replication complex. J. Virol. Methods. 2012; 179:97–103. [DOI] [PubMed] [Google Scholar]
- 50. Collins T.J. ImageJ for microscopy. Biotechniques. 2007; 43:25–30. [DOI] [PubMed] [Google Scholar]
- 51. Hodge K., Tunghirun C., Kamkaew M., Limjindaporn T., Yenchitsomanus P.T., Chimnaronk S.. Identification of a conserved RNA-dependent RNA polymerase (RdRp)-RNA interface required for Flaviviral replication. J. Biol. Chem. 2016; 291:17437–17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen C.J., Kuo M.D., Chien L.J., Hsu S.L., Wang Y.M., Lin J.H.. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 1997; 71:3466–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sampath A., Xu T., Chao A., Luo D., Lescar J., Vasudevan S.G.. Structure-based mutational analysis of the NS3 helicase from dengue virus. J. Virol. 2006; 80:6686–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tian H., Ji X., Yang X., Zhang Z., Lu Z., Yang K., Chen C., Zhao Q., Chi H., Mu Z. et al. Structural basis of Zika virus helicase in recognizing its substrates. Protein Cell. 2016; 7:562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suzich J.A., Tamura J.K., Palmer-Hill F., Warrener P., Grakoui A., Rice C.M., Feinstone S.M., Collett M.S.. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 1993; 67:6152–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jimenez-Oses G., Osuna S., Gao X., Sawaya M.R., Gilson L., Collier S.J., Huisman G.W., Yeates T.O., Tang Y., Houk K.N.. The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat. Chem. Biol. 2014; 10:431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hew K., Dahlroth S.L., Venkatachalam R., Nasertorabi F., Lim B.T., Cornvik T., Nordlund P.. The crystal structure of the DNA-binding domain of vIRF-1 from the oncogenic KSHV reveals a conserved fold for DNA binding and reinforces its role as a transcription factor. Nucleic Acids Res. 2013; 41:4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ericsson U.B., Hallberg B.M., Detitta G.T., Dekker N., Nordlund P.. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 2006; 357:289–298. [DOI] [PubMed] [Google Scholar]
- 59. Gu B., Liu C., Lin-Goerke J., Maley D.R., Gutshall L.L., Feltenberger C.A., Del Vecchio A.M.. The RNA helicase and nucleotide triphosphatase activities of the bovine viral diarrhea virus NS3 protein are essential for viral replication. J. Virol. 2000; 74:1794–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Matusan A.E., Pryor M.J., Davidson A.D., Wright P.J.. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 2001; 75:9633–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chiang P.Y., Wu H.N.. The role of surface basic amino acids of dengue virus NS3 helicase in viral RNA replication and enzyme activities. FEBS Lett. 2016; 590:2307–2320. [DOI] [PubMed] [Google Scholar]
- 62. Miller S., Sparacio S., Bartenschlager R.. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 2006; 281:8854–8863. [DOI] [PubMed] [Google Scholar]
- 63. Bartenschlager R., Miller S.. Molecular aspects of Dengue virus replication. Future Microbiol. 2008; 3:155–165. [DOI] [PubMed] [Google Scholar]
- 64. Hardin J.W., Hu Y.X., McKay D.B.. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J. Mol. Biol. 2010; 402:412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang S., Hu Y., Overgaard M.T., Karginov F.V., Uhlenbeck O.C., McKay D.B.. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006; 12:959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Banerjee R., Dasgupta A.. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 2001; 75:1708–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adamczyk A.J., Cao J., Kamerlin S.C., Warshel A.. Catalysis by dihydrofolate reductase and other enzymes arises from electrostatic preorganization, not conformational motions. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14115–14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Frick D.N., Rypma R.S., Lam A.M., Frenz C.M.. Electrostatic analysis of the hepatitis C virus NS3 helicase reveals both active and allosteric site locations. Nucleic Acids Res. 2004; 32:5519–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Teramoto T., Boonyasuppayakorn S., Handley M., Choi K.H., Padmanabhan R.. Substitution of NS5 N-terminal domain of dengue virus type 2 RNA with type 4 domain caused impaired replication and emergence of adaptive mutants with enhanced fitness. J. Biol. Chem. 2014; 289:22385–22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V.. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006; 20:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Patkar C.G., Kuhn R.J.. Yellow Fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 2008; 82:3342–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.