Abstract
Fungal tRNA ligase (Trl1) is an essential enzyme that repairs RNA breaks with 2′,3′-cyclic-PO4 and 5′-OH ends inflicted during tRNA splicing and non-canonical mRNA splicing in the fungal unfolded protein response. Trl1 is composed of C-terminal cyclic phosphodiesterase and central polynucleotide kinase domains that heal the broken ends to generate the 3′-OH,2′-PO4 and 5′-PO4 termini required for sealing by an N-terminal ligase domain. Trl1 enzymes are found in all human fungal pathogens and are promising targets for antifungal drug discovery because their domain compositions and biochemical mechanisms are unique compared to the mammalian RtcB-type tRNA splicing enzyme. A distinctive feature of Trl1 is its preferential use of GTP as phosphate donor for the RNA kinase reaction. Here we report the 2.2 Å crystal structure of the kinase domain of Trl1 from the fungal pathogen Candida albicans with GDP and Mg2+ in the active site. The P-loop phosphotransferase fold of the kinase is embellished by a unique ‘G-loop’ element that accounts for guanine nucleotide specificity. Mutations of amino acids that contact the guanine nucleobase efface kinase activity in vitro and Trl1 function in vivo. Our findings fortify the case for the Trl1 kinase as an antifungal target.
INTRODUCTION
Fungal tRNA ligase Trl1 is an essential agent in the repair of programmed tRNA and mRNA breaks with 2′,3′-cyclic phosphate and 5′-OH ends that are generated during tRNA splicing and the unfolded protein response (1,2). Trl1 performs three distinct RNA repair reactions catalyzed by three distinct protein domains: (i) the 2′,3′-cyclic phosphate (>p) end is hydrolyzed to a 3′-OH,2′-PO4 by a C-terminal cyclic phosphodiesterase (CPD) module that belongs to the 2H phosphoesterase superfamily; (ii) the 5′-OH end is phosphorylated by a central NTP-dependent polynucleotide kinase module of the P-loop phosphotransferase superfamily and (iii) the 3′-OH,2′-PO4 and 5′-PO4 ends are sealed by an N-terminal ATP-dependent RNA ligase module, of the covalent nucleotidyltransferase superfamily, to form an unconventional 2′-PO4, 3′-5′ phosphodiester at the splice junction (3–9).
Fungal Trl1 enzymes are potential therapeutic targets because their domain structures and biochemical mechanisms are unique compared to the RtcB-type tRNA repair systems elaborated by metazoa, archaea, and many bacteria (10–18). RtcB is a GTP-dependent RNA ligase that splices 3′-PO4 and 5′-OH ends via a different chemical mechanism vis à vis Trl1. RtcB is absent from the proteomes of most fungi and mammalian proteomes have no homologs of the ligase domain of fungal Trl1. Moreover, there is no 5′ kinase step in the RtcB pathway of RNA repair. Whereas polynucleotide kinases are widely distributed in nature, the kinase domains of fungal Trl1 enzymes are unique in that they have a strong preference for GTP as the phosphate donor (5,8,9,19). This raises the prospect that the fungal kinase donor site could be targeted by a small molecule inhibitor that interacts with the guanine specificity determinant(s) of Trl1, but does not bind to the donor sites of the many other cellular P-loop phosphotransferases, which either have no NTP donor preference or prefer ATP as substrates.
To fortify the case for fungal tRNA ligase as a drug target, we need to understand the properties of Trl1 enzymes produced by fungi that cause human disease. To that end, we previously characterized Trl1 from the human fungal pathogens Aspergillus fumigatus and Coccidioides immitis (9). Aspergillus fumigatus causes invasive pulmonary disease in immunocompromised individuals, especially those with hematological malignancies or who have undergone bone marrow or solid organ transplantation. Coccidioides immitis is the agent of San Joaquin Valley Fever, a disease prevalent in the US desert Southwest region. The Aspergillus and Coccidioides Trl1 enzymes both strongly prefer GTP as the NTP phosphate donor for their 5′ kinase reactions (9).
Despite the centrality of tRNA ligase to fungal protein synthesis and the unfolded protein response, and the mechanistic insights gained from biochemical studies of Trl1 catalysis and substrate specificity, further progress is hindered by the absence of an atomic structure of a fungal tRNA ligase or any of its component domains. Here, we fill in part of this knowledge gap, by characterizing and solving the 2.2 Å crystal structure of an autonomous kinase domain of Candida albicans Trl1 in complex with GDP and magnesium. Candida albicans is a significant human pathogen, causing a spectrum of illnesses ranging from local infections of oral and genital mucosa to invasive systemic infections with significant morbidity and mortality in immunocompromised hosts. The Candida kinase structure reveals a distinctive ‘G-loop’ motif, conserved in other fungal tRNA ligases, that accounts for guanine nucleotide specificity. Effects of G-loop mutations and nucleotide analogs on kinase activity highlight guanine O6 recognition as a key determinant of NTP specificity.
MATERIALS AND METHODS
CalTrl1 domain purifications
Recombinant Candida Trl1(1–400), Trl1(1–410), Trl1(401–832) and Trl1(401–636) proteins were purified from soluble extracts of 4-liter cultures of IPTG-induced Escherichia coli BL21-CodonPlus(DE3) pET28b-His10Smt3-Trl1 strains by sequential nickel-affinity, tag-cleavage, tag removal by second nickel-affinity and gel filtration steps as described previously for Aspergillus and Coccidioides Trl1 proteins (9). Protein concentrations were determined by using the Biorad dye reagent with bovine serum albumin as the standard.
Preparation of 3′ 32P-labeled RNA>p and RNAp substrates
HORNA3’p oligonucleotides labeled with 32P at the penultimate phosphate were prepared by T4 Rnl1-mediated addition of [5′-32P]pCp to 19-mer or 9-mer synthetic oligoribonucleotides. The 20-mer HORNA3’p oligonucleotide was treated with E. coli RNA 3′-terminal phosphate cyclase (RtcA) and ATP to generate the 2′,3′-cyclic phosphate derivative, HORNA>p. The labeled RNAs were gel-purified, eluted from an excised gel slice, and recovered by ethanol precipitation.
Crystallization and structure determination
CalTrl1 KIN crystals were grown at 22°C by sitting drop vapor diffusion. A 1 μl solution of 0.1 mM KIN (2.5 mg/ml), 10 mM MgCl2 and 2 mM GTP was mixed with an equal volume of precipitant solution containing 0.2 M sodium dihydrogen phosphate, 20% PEG3350. Crystals appeared after 1 day and were frozen directly in liquid nitrogen. Alternatively, crystals were soaked for 2 h in precipitant solution containing 10 mM HgCl2 prior to freezing. X-ray diffraction data were collected from single crystals at the Advanced Photon Source beamline 24ID-C. Indexing and merging of the diffraction data were performed in HKL2000 (20). The phases were obtained using single-wavelength anomalous dispersion (SAD) data from a single crystal of Hg derivative with SHELX (21). About 90% of the polypeptide chain was traced using SAD data in Phenix (22). The partial model was used to solve the native structure by molecular replacement. Interactive model building was performed using O (23). Refinement was accomplished with Phenix. Data collection and refinement statistics are summarized in Supplementary Table S1.
Mutagenesis
Mutations were introduced into the Trl1(401–636) ORF of the pET28b-His10Smt3-Trl1(401–636) plasmid by two-step overlap extension PCR. The plasmid inserts were sequenced to verify that no unwanted coding changes were introduced during PCR amplification and cloning. The mutant proteins were purified along with the wild-type protein following the same procedure used for CalTrl1 domain purifications, except that the gel filtration step was omitted. Instead, the tag-free KIN proteins were concentrated by centrifugal ultrafiltration to 3 mg/ml, then dialyzed against buffer C (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 10% glycerol) and stored at –80°C.
S. cerevisiae trl1Δ complementation
We tested trl1Δ complementation by plasmid shuffle in a S. cerevisiae trl1Δ p(CEN URA3 TRL1) strain (5), which is unable to grow on medium containing 0.75 mg/ml FOA (5-fluoroorotic acid), a drug that selects against the URA3 plasmid. trl1Δ cells were transfected with CEN LEU2 plasmids harboring wild-type or mutated CalTRL1 under the control of the yeast TPI1 promoter (9). Individual Leu+ transformants were selected and streaked on agar medium containing FOA. CalTRL1 alleles that failed to give rise to FOA-resistant colonies after incubation for 7 days at 30°C were deemed to be lethal. Individual FOA-resistant CalTRL1 colonies were grown to mid-log phase in YPD broth and adjusted to A600 of 0.1. Aliquots (3 μl) of serial 10-fold dilutions were spotted to YPD agar plates, which were incubated at 20, 25, 30 and 37°C.
RESULTS
Candida albicans Trl1
Candida albicans (Cal) Trl1 is an 832-amino acid polypeptide composed of N-terminal ligase (LIG), central kinase (KIN), and C-terminal CPD domains (Supplementary Figure S1A). CalTrl1 shares 42% amino acid identity with S. cerevisiae Trl1 and expression of CalTRL1 complements the lethality of a trl1Δ knockout in S. cerevisiae (9). We initially split CalTrl1 into two component fragments: LIG (two versions, from amino acids 1–400 and 1–410) and KIN-CPD (amino acids 401–832). We produced recombinant LIG1–400, LIG1–410, and KIN-CPD in E. coli as His10Smt3 fusions and purified them from soluble extracts by sequential Ni-affinity chromatography/imidazole elution, removal of the His10Smt3 tag by treatment with Ulp1 protease, recovery of the tag-free Trl1 proteins in the flow-through of a second Ni-affinity column, and a final Superose-200 gel-filtration step. To assay the composite end-healing and end-sealing pathway, we used a 20-mer RNA with 5′-OH and 2′,3′-cyclic phosphate (>p) ends and a single radiolabel between the 3′-terminal and penultimate nucleosides (Supplementary Figure S1B). The HORNA>p substrate (20 nM) was reacted with the LIG and KIN-CPD domains, singly or together, in the presence of 10 mM Mg2+, 0.1 mM ATP and 0.1 mM GTP and the products were analyzed by urea-PAGE. The mixture of LIG and KIN-CPD domains converted the HORNA>p substrate into ligated products, these being a circular RNA formed by intramolecular ligation (which migrated ahead of the substrate strand) and RNA multimers formed via intermolecular end joining (which migrated well behind the substrate strand) (Supplementary Figure S1B). Reaction of the isolated KIN-CPD domain with the 20-mer HORNA>p substrate resulted in its conversion to a 5′-PO4 product that migrated slightly ahead of the input 20-mer, Reaction of HORNA>p with the isolated LIG domains elicited no change in its electrophoretic mobility.
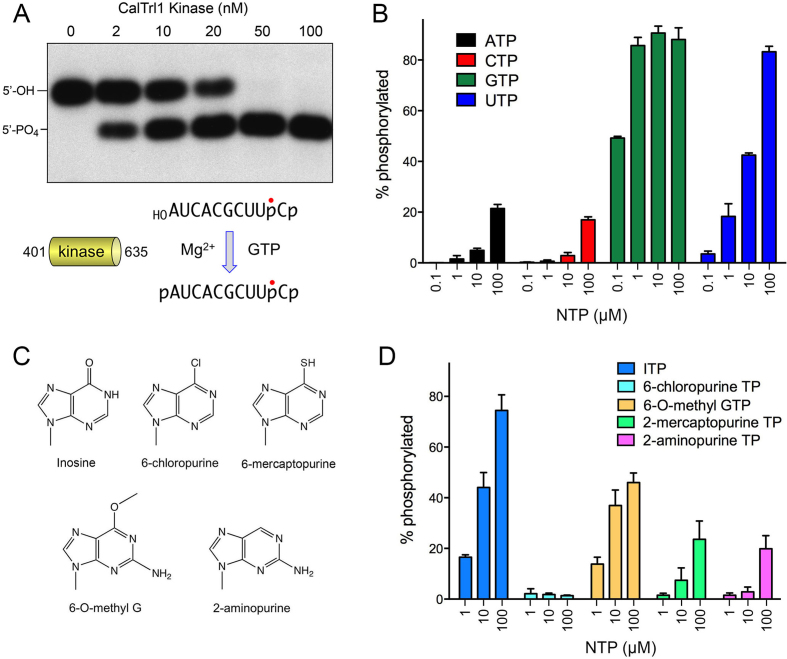
An autonomous kinase domain of CalTrl1 prefers GTP as the phosphate donor
We produced and purified a recombinant segment of CalTrl1 from aa 401 to 635 as a candidate stand-alone kinase module. Reaction of the 50–100 nM KIN domain with a 10-mer HORNAp substrate (radiolabeled between the 3′-terminal and penultimate nucleosides) in the presence of magnesium and GTP resulted in conversion of all of the input 5′-OH RNA into a more rapidly migrating 5′-phosphorylated product (Figure 1A). The extent of 5′ phosphorylation increased with KIN concentration in the range of 2–20 mM enzyme (Figure 1A). No 5′ phosphorylation was detected in the absence of exogenous GTP. To test NTP substrate preference, 100 nM KIN domain was reacted with 20 nM 3′ 32P-labeled 10-mer HORNA>p substrate and magnesium for 10 min in the presence of 0.1, 1, 10 or 100 μM ATP, GTP, CTP or UTP (Figure 1B). Varying the GTP concentration from 100 to 1 μM had no effect on the yields of phosphorylated RNA. The fact that the kinase reaction was half-saturated at 0.1 μM GTP (i.e. 5-fold molar excess over the input RNA substrate and equimolar to input KIN domain) attests to the efficiency with which GTP is used as the phosphate donor. By contrast, the extents of RNA phosphorylation with 100 μM ATP or CTP were less than that seen with 0.1 μM GTP, signifying that ATP and CTP were at least three orders of magnitude less effective as phosphate donors. Lower concentrations of ATP and CTP were ineffective in supporting kinase activity (Figure 1B). Whereas UTP was an effective kinase substrate at the highest concentration tested (100 μM), back titration progressively diminished the extent of phosphorylation. The finding that 10 μM UTP supported a similar level of product formation as 0.1 μM GTP highlights a two order of magnitude difference in preference for GTP versus UTP. These and other recent experiments (9) show that GTP donor preference is a feature shared by Trl1 enzymes from three human fungal pathogens.
Figure 1.
An autonomous kinase domain of CalTrl1 prefers GTP as the phosphate donor. (A) Reaction mixtures (10 μl) containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 100 μM GTP, 20 nM 32P-labeled 10-mer HORNA3′p (shown at bottom, with the 32P-label indicated by •), and 0, 2, 10, 20, 50 or 100 nM CalTrl1 KIN domain were incubated at 22°C for 20 min. The reactions were quenched with an equal volume of 90% formamide, 30 mM EDTA. The labeled RNAs were resolved by urea-PAGE. An autoradiogram of the gel is shown. The positions of the 5′-OH RNA substrate and 5′-PO4 RNA product are indicated on the left. (B–D) Reaction mixtures (10 μl) containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 20 nM 10-mer HORNA3′p, 100 nM KIN, and either 0.1, 1, 10 or 100 μM ATP, CTP, GTP or UTP (panel B) or 1, 10 or 100 μM purine NTP analogs (panel D; analog structures shown in panel C) were incubated at 22°C for 2 min. The extents of RNA phosphorylation as a function of NTP concentration are plotted in bar graph format. Each datum is the average of three separate experiments ± SEM.
Test of GTP analogs as phosphate donors
We gained insights to the nucleobase specificity of the Candida kinase by testing purine ribonucleoside triphosphate analogs (Figure 1C) as substrates for HORNAp phosphorylation. The NTPs were included at 1, 10 and 100 μM concentration and the extents of phosphorylation in a 10 min reaction with 100 nM KIN were quantified (Figure 1D). Inosine triphosphate was the most effective substrate among analogs tested, implying that the exocyclic 2-NH2 moiety of GTP is not an essential determinant of nucleobase recognition per se. By contrast, 2-aminopurine triphosphate was a much less effective substrate for the kinase reaction, i.e. 100 μM 2-aminopurine triphosphate was as active as 1 μM ITP (Figure 1D). This result highlights the importance of the 6-oxo atom of guanine for phosphate donor function. This conclusion was fortified by the inactivity of 100 μM 6-chloropurine triphosphate and the feeble activity of 2-mecaptopurine triphosphate (Figure 1D). The finding that 6-O-methyl guanosine triphosphate was almost as effective as ITP suggests that the 6-oxo atom acts as a hydrogen-bond acceptor, presumably from an amino acid side chain hydrogen donor or a main chain amide donor on the kinase enzyme. This scenario would account for the ability of UTP to serve as an alternative phosphate donor (Figure 1B), by virtue of its pyrimidine 4-oxo group accepting the putative hydrogen bond in lieu of the guanine 6-oxo.
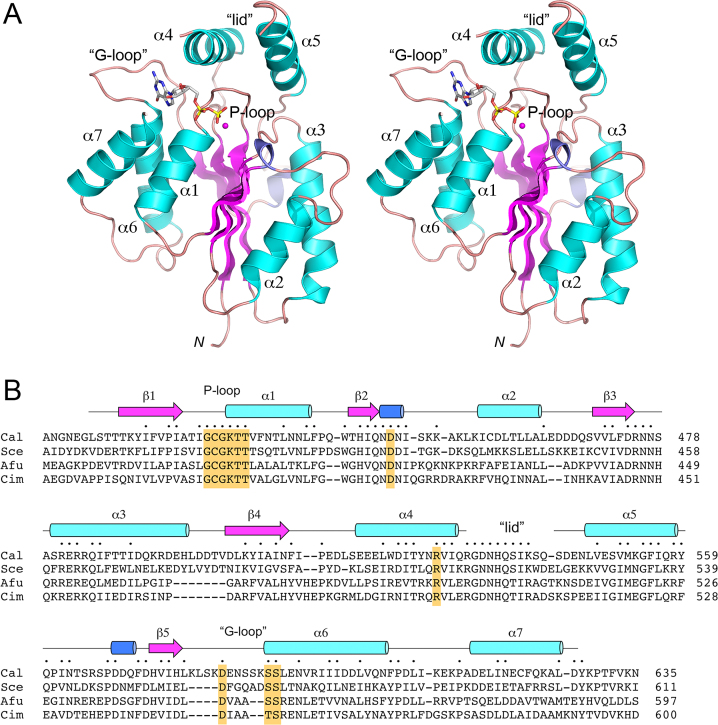
Structure of the CalTrl1 kinase domain
Crystals of the Candida KIN domain were grown by hanging drop vapor diffusion after mixing a sample of the protein solution containing 0.6 mM kinase, 2 mM GTP and 10 mM MgCl2 with an equal volume of precipitant solution containing 0.2 M sodium dihydrogen phosphate, 20% PEG3350. The crystals diffracted to 2.2 Å resolution and belonged to space group P212121. Hg-SAD phasing was accomplished using diffraction data from a crystal that had been soaked in 10 mM HgCl2 (Supplementary Table S1). The refined model of the native KIN at 2.2 Å resolution (Rwork/Rfree 0.187/0.252; Supplementary Table S1) comprised a kinase monomer that had GDP and Mg2+ bound in the active site (Figure 2A). The tertiary structure consists of a central 5-stranded parallel β-sheet with topology β2↑β3↑β1↑β4↑β5↑, proceeding from front to back in the view in Figure 2A. The central sheet is flanked by seven α helices and two short 310 helices (Figure 2A and B). The P-loop motif (422GCGKTT427) is located between the β1 strand and the α1 helix. GDP is bound within a crescent-shaped groove formed by the P-loop and an overlying ‘lid’ domain composed of helices α4 and α5. The 532RGNNHQSIKSQ542 peptide loop connecting the lid helices was disordered (Figure 2A and B). A main-chain trace of the kinase tertiary structure, colored according to the B-factors of the Cα atoms highlights that the central β-sheet and the α1 helix are the most well-ordered structural elements (Supplementary Figure S2). The highest B-factors applied to the amino acids at the junction between the disordered lid-loop and helix α5. A DALI search (24) with the KIN structure identified numerous members of the P-loop phosphotransferase superfamily as homologs. Top DALI hits were: E. coli gluconate kinase (pdb 1KO1; Z score 13.7; 2.3 Å rmsd at 142 Cα positions with 13% amino acid identity) and Penicillium chrysogenum adenosine 5′-phosphosulfate kinase (pdb 1M7G; Z score 13.3, 2.8 Å rmsd at 154 Cα positions with 15% amino acid identity). DALI also recovered hits to other polynucleotide kinases, including Clostridium thermocellum Pnk (pdb 4QM6; Z score 12.5; 2.7 Å rmsd at 142 Cα positions with 15% amino acid identity), mammalian DNA 5′-OH kinase (pdb 1YJ5; Z score 11.7; 2.8 Å rmsd at 133 Cα positions with 14% amino acid identity), bacteriophage T4 Pnk (pbd 2IA5; Z score 10.5; 2.7 Å rmsd at 123 Cα positions with 17% amino acid identity), and Capnocytophaga gingivalis Pnk (pdb 4XRU; Z score 8.5; 2.8 Å rmsd at 115 Cα positions with 14% amino acid identity).
Figure 2.
Structure of the CalTrl1 kinase domain. (A) Stereo view of the kinase tertiary structure, depicted as a ribbon model with magenta β strands, cyan α helices (numbered sequentially), and blue 310 helices. The N-terminus (Leu407) is indicated by N. The GDP in the phosphate donor site is rendered as a stick model. Mg2+ is depicted as a magenta sphere. The phosphate-binding P-loop and guanine-binding ‘G-loop’ are indicated. (B) Secondary structure elements (colored as in panel A) are displayed above the CalTrl1 KIN primary structure, which is aligned to the primary structures of the KIN domains of S. cerevisiae (Sce), A. fumigatus (Afu) and C. immitis (Cim) Trl1. Positions of amino acid side chain identity or similarity in all four proteins are indicated by dots above the Cal sequence. Gaps in the alignment are indicated by dashes. Conserved P-loop, aspartate general acid, lid, and G-loop elements are highlighted in gold shading.
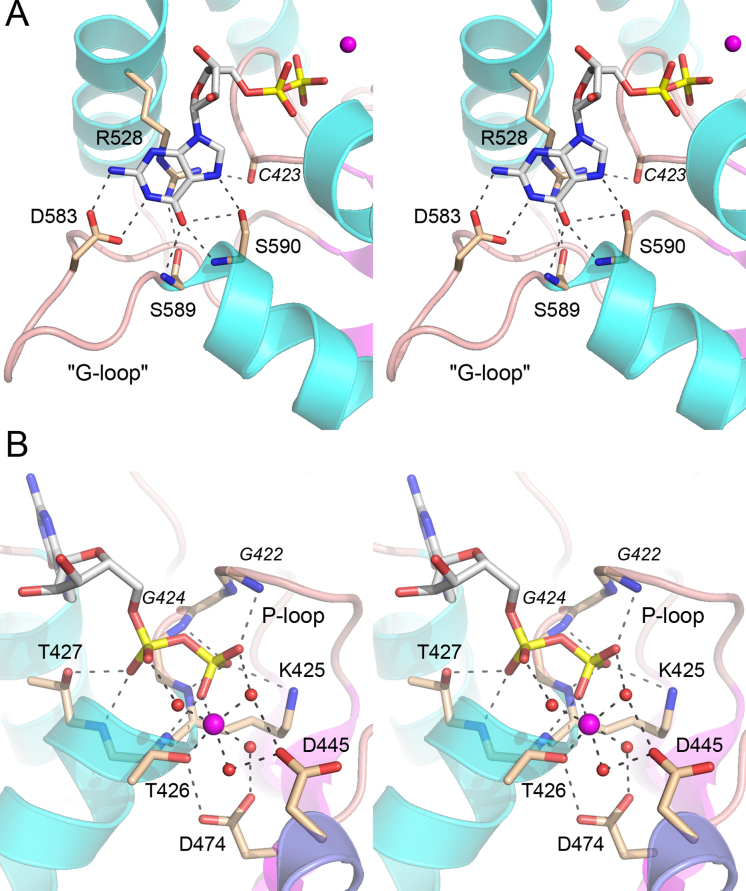
Interactions with GDP phosphates and magnesium
A simulated annealing omit map revealed electron density for GDP in the active site (Supplementary Figure S3), suggesting that the γ phosphate of the added GTP was hydrolyzed during crystal growth. Alternatively, the donor site was pre-filled with GDP during enzyme production in E. coli and that GDP remained tightly bound during the KIN purification. A detailed view of the atomic contacts to GDP, and an associated Mg2+ ion, is shown in Figure 3B. Main-chain amide nitrogens of P-loop residues Gly422, Gly424, Lys425 and Thr426 donate hydrogen bonds to the β phosphate of GTP, which is also coordinated by the P-loop Lys425-Nζ. The Thr427 hydroxyl and main-chain amide donate hydrogen bonds to the GDP α phosphate. One of the sites in the octahedral coordination complex of the Mg2+ ion is occupied by a GDP β phosphate oxygen. The other sites are filled by the Thr426 Oγ atom and four waters. Thus, the KIN protein makes only a single direct atomic contact to the metal cofactor. An outer shell of atomic contacts to the waters in the metal complex includes the Asp445 and Asp474 side chains and the GDP α and β phosphates (Figure 3B). KIN Asp445 is a conserved component of the kinase active site (Figure 2B) that, in C. thermocellum Pnk (CthPnk), serves as a general base catalyst in de-protonating the 5′-OH of the polynucleotide phosphate acceptor for its nucleophilic attack on the NTP γ phosphate (25). The disordered lid loop of the KIN domain includes two amino acids, Arg532 and His535, that are conserved among fungal Trl1 enzymes (Figure 2B); the equivalent Arg511 and His515 residues in S. cerevisiae Trl1 are essential for kinase activity in vitro and Trl1 function in vivo (7). The corresponding basic side chains in the lid loop of CthPnk coordinate the NTP γ phosphate (25); thus it is possible that the disorder of the lid loop reflects the absence of a γ phosphate group in the GDP-bound Trl1 KIN domain.
Figure 3.
Interactions with the guanine nucleobase, GDP phosphates, and magnesium. (A) Stereo view of the G-loop highlighting KIN interactions with the guanine nucleobase. (B) Stereo view of the P-loop highlighting contacts to the GDP phosphates and magnesium. Amino acids and GDP are shown as stick models with beige and gray carbons, respectively. Mg2+ is depicted a magenta sphere in the center of an octahedral coordination complex. Waters in the metal coordination complex are denoted by red spheres. Atomic contacts are indicated by dashed lines.
Structural basis for guanine specificity
The distinctive feature of the Trl1 KIN domain structure, vis à vis other P-loop phosphotransferases and other polynucleotide kinases, is an extended loop between strand β5 and helix α6 that abuts the guanine nucleobase of GDP (Figure 2A). A close-up view of this ‘G-loop’ and the protein contacts to the guanine nucleobase is shown in Figure 3A. (Note, there are no contacts to the ribose sugar of the guanosine nucleoside.) The conserved Arg528 side chain of the lid α4 helix makes a π-cation stack on the base, an interaction that is not specific for guanine. The lid arginine π-cation interaction with the NTP base (be it A, G. C or U) is a feature shared with CthPnk (26). Whereas the arginine contributes to donor NTP affinity in CthPnk (26), it does not explain the GTP specificity of the fungal Trl1 kinase. Our structure reveals that the guanine nucleobase is engaged by a network of hydrogen bonds from G-loop amino acids Asp583 (to guanine N1 and N2 from the Asp carboxylate), Ser589 (to guanine O6 from the main chain amide), and Ser590 (to guanine O6 and N7, from the main chain amide and Oγ, respectively) (Figure 3A). The N1, N2 and O6 contacts of the G-loop are all guanine-specific and explain why CalTrl1 does not accept ATP as the phosphate donor.
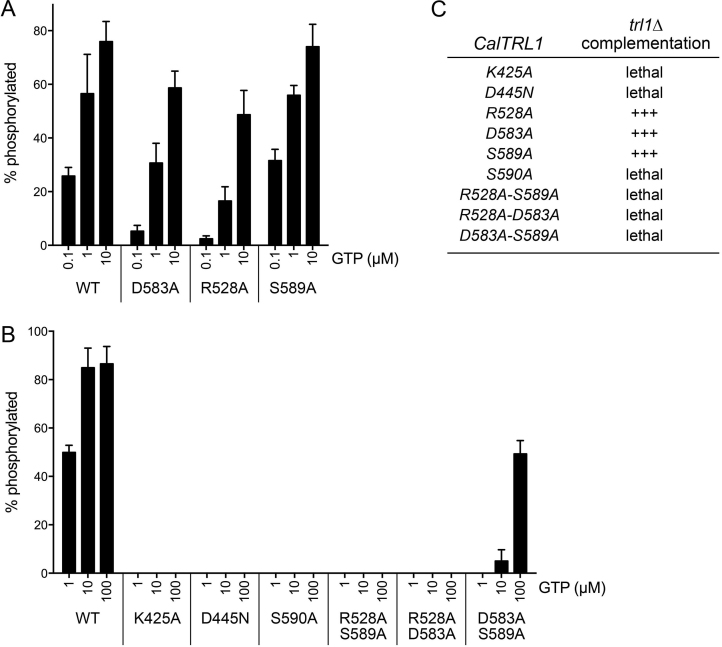
Structure-guided mutational analysis
To interrogate the contributions of the guanine-interacting amino acids to CalTrl1 function, we mutated Arg528, Asp583, Ser589 and Ser590 to alanine. As putative positive controls for loss of function, we also mutated two constituents of the active site: the P-loop Lys425 was changed to alanine and the general base Asp445 was changed to asparagine. The mutations were introduced into the bacterial expression construct for the KIN domain, and the recombinant mutant KIN proteins were purified (Supplementary Figure S4) and assayed for kinase activity as a function of GTP concentration (Figure 4). The P-loop K425A, general base D445N, and G-loop S590A mutations effaced RNA kinase activity in vitro, even at 100 μM GTP (Figure 4B). By contrast, mutants R528A, D583A, and S589A retained kinase activity (Figure 4A). Thinking that there might be functional redundancy among the three inessential G-loop amino acids, we purified and tested a set of double-alanine mutants: R528A-S589A, R528A-D583A and D583A-S589A. The R528A-S589A and R528A-D583A mutations did away with kinase activity in vitro (Figure 4B). The D583A-S589A mutant displayed activity at 100 μM GTP roughly equal to the activity of wild-type KIN at 1 μM GTP. The activity of D583A-S589A declined sharply at 10 μM GTP and was undetectable at 1 μM GTP (Figure 4B), suggesting that the loss of the Asp583 and Ser589 side chains affected kinase affinity for the GTP phosphate donor.
Figure 4.
Mutational analysis. (A, B) Reaction mixtures (10 μl) containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 20 nM 32P-labeled 10-mer HORNA3′p, 100 nM KIN (wild-type or mutant as indicated), and GTP as specified were incubated at 22°C for 10 min. The extents of RNA phosphorylation as a function of NTP concentration are plotted in bar graph format. Each datum is the average of three separate experiments ± SEM. (C) The indicated CalTRL1 alleles on CEN plasmids were tested for S. cerevisiae trl1Δ complementation as described under Methods. +++ signifies growth as well as the wild-type CalTRL1 control at 20, 25, 30 and 37°C.
The same KIN mutations were introduced into a yeast expression plasmid for full-length CalTRL1 and the mutant alleles were tested by plasmid shuffle for complementation of S. cerevisiae trl1Δ. There was an excellent correlation between mutational effects on Trl1 function in vitro and in vivo, to wit: (i) kinase-active mutants R528A, D583A, and S589A complemented trl1Δ and (ii) kinase-dead alleles K425A, D445N, S590A, R528A-S589A and R528A-D583A were lethal in vivo, as was the kinase-defective allele D583A-S589A (Figure 4C).
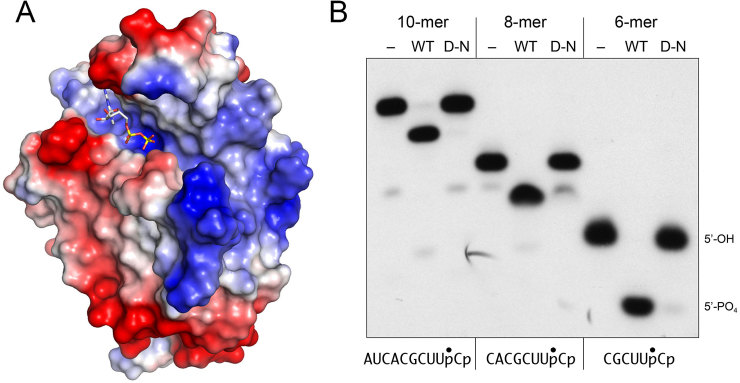
Surface electrostatics and minimized 5′-OH RNA substrates for the KIN domain
A surface electrostatic model of the KIN domain prepared in Pymol (Figure 5A) highlights a broad swath of positive potential to the right of the guanine nucleotide donor site that provides, in principle, a favorable interface with the negatively charge phosphodiester backbone of the cleaved tRNA substrate for 5′ end-healing. Indeed, there are two deep grooves on this surface that could potentially accommodate the duplex segment and single-stranded ends of the anticodon stem-loop. Because the KIN domain does not require a stem-loop configuration in the phospho-acceptor, we wanted to define a minimal length of RNA suitable for phosphorylation, e.g. by serially removing nucleotides from the 5′ end of the 10-mer substrate used here for kinase assays. We found that the KIN domain quantitatively phosphorylated 8-mer and 6-mer substrates (Figure 5B), as well as a 4-mer CUUpCp (not shown). Activity on the short RNAs was abolished by the D474N mutation (Figure 5B).
Figure 5.
Surface electrostatics and minimized 5′-OH RNA substrates for the KIN domain. (A) A surface electrostatic model of the KIN domain was generated in Pymol. GDP in the phosphate donor site is depicted as a stick model. (B) Reaction mixtures (10 μl) containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 100 μM GTP, 20 nM 32P-labeled HORNA3’p of varying length as specified (depicted at bottom, with the 32P-label indicated by •) and either 100 nM wild-type KIN domain (WT), D445N mutant (D–N), or no enzyme (–) were incubated at 22°C for 20 min. The labeled RNAs were resolved by urea–PAGE. An autoradiogram of the gel is shown. The positions of the 6-mer 5′-OH RNA substrate and 5′-PO4 RNA product are indicated on the right.
DISCUSSION
The present study extends our knowledge of tRNA splicing enzymology to the fungal pathogen C. albicans and provides an atomic structure of the Candida Trl1 kinase catalytic domain (the first structure reported of any domain of a fungal tRNA ligase) that accounts for its strong preference for GTP as the phosphate donor. GTP specificity is a feature shared by multiple fungal Trl1 kinases that have been interrogated, including those of the yeasts S. cerevisiae and K. lactis (5,8,19) and the pathogenic fungi A. fumigatus and C. immitis (9). This GTP preference distinguishes the fungal Trl1 enzymes from the plant tRNA ligase AtRNL, which is mechanistically and structurally homologous to yeast Trl1, but is active with either ATP, GTP, CTP or UTP as the phosphate donor in the plant kinase reaction (27).
A primary structure alignment of the Candida, Saccharomyces, Aspergillus and Coccidioides Trl1 KIN domains highlights 90 positions of amino acid side chain identity/similarity in the four fungal enzymes (Figure 2B). Protein segments of greatest local conservation—including the P-loop, the peptide motif embracing the aspartate general base, and the lid—are those that form the active site of phosphoryl transfer. For example, the P-loop lysine and the general base aspartate are both essential for Candida kinase activity in vitro and in vivo. Although disordered in the Cal KIN structure, the conserved lid-loop arginine and histidine residues are essential for Saccharomyces kinase activity in vitro and in vivo (7).
The distinctive structural feature of the Candida kinase is the G-loop that engages the guanine nucleobase. Located between the β5 strand and the α6 helix, the G-loop is variable in length among fungal Trl1 kinases ((7); Figure 2B) and had not been recognized as a functional motif because of limited amino acid conservation (7). However, with the benefit of the Cal KIN structure, we can appreciate that: (i) the vicinal hydroxyamino acids Ser589–Ser590 (which engage in main chain hydrogen bonds with guanine O6 and a side chain hydrogen bond to guanine N7) are conserved in the four kinases aligned in Figure 2B; and (ii) the G-loops of the Saccharomyces, Aspergillus and Coccidioides Trl1 kinases include an aspartate residue that might contact guanine N1 and N2 à la Asp583 in the Cal KIN structure (Figure 3A). Our mutagenesis underscores that the G-loop and the conserved α4 arginine collaborate to bind and establish preference for GTP as the kinase donor, via a combination of π-cation contacts to the purine ring from Arg528 and base edge contacts from the G-loop. Whereas single alanine substitutions for Arg528, Asp583 or Ser589 do not seriously compromise the Candida kinase, any combination of two alanine mutations cripples the enzyme. The peculiar G-loop and guanine-binding pocket of Candida KIN presents a plausible target for a small molecule that could selectively inhibit the fungal Trl1 kinase without affecting the many other P-loop superfamily phosphotransferases that do not share the guanine specificity of the fungal Trl1 kinase domains.
AVAILABILITY
Structural coordinates have been deposited in Protein Data Bank under accession code 5U32.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institutes of Health [GM42498 to S.S.]; Geoffrey Beene Cancer Research Center (to S.S.); National Cancer Institute [P30-CA008748; to support the MSKCC structural biology core laboratory]; National Institutes of Health [P41GM103403, HEI-S10RR029205; supporting the APS synchrotron]; Department of Energy [DE-AC02-06CH11357; supporting the APS synchrotron]. Funding for open access charge: NIGMS [GM42498].
Conflict of interest statement. None declared.
REFERENCES
- 1. Abelson J., Trotta C.R., Li H.. tRNA splicing. J. Biol. Chem. 1998; 273:12685–12688. [DOI] [PubMed] [Google Scholar]
- 2. Sidrauski C., Cox J.S., Walter P.. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996; 87:405–413. [DOI] [PubMed] [Google Scholar]
- 3. Greer C.L., Peebles C.L., Gegenheimer P., Abelson J.. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983; 32:537–546. [DOI] [PubMed] [Google Scholar]
- 4. Apostol B.L., Westaway S.K., Abelson J., Greer C.L.. Deletion analysis of a multifunctional yeast tRNA ligase polypeptide: identification of essential and dispensable functional domains. J. Biol. Chem. 1991; 266:7445–7455. [PubMed] [Google Scholar]
- 5. Sawaya R., Schwer B., Shuman S.. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 2003; 278:43298–43398. [DOI] [PubMed] [Google Scholar]
- 6. Wang L.K., Shuman S.. Structure-function analysis of yeast tRNA ligase. RNA. 2005; 11:966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang L.K, Schwer B., Englert M., Beier H., Shuman S.. Structure-function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res. 2006; 34:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Remus B.S., Shuman S.. Distinctive kinetics and substrate specificities of plant and fungal tRNA ligases. RNA. 2014; 20:462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Remus B.S., Schwer B., Shuman S.. Characterization of the tRNA ligases of pathogenic fungi Aspergillus fumigatus and Coccidioides immitis. RNA. 2016; 22:1500–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka N., Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011; 286:7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka N., Chakravarty A.K., Maughan B., Shuman S.. A novel mechanism of RNA repair by RtcB via sequential 2’,3’-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011; 286:43134–43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka N., Meineke B., Shuman S.. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 2011; 286:30253–30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chakravarty A.K., Subbotin R., Chait B.T., Shuman S.. RNA ligase RtcB splices 3’-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. USA. 2012; 109:6072–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakravarty A.K., Shuman S.. The sequential 2′,3′ cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res. 2012; 40:8558–8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Englert M., Xia S., Okada C., Nakamura A., Tanavde V., Yao M., Eom S.H., Koningsberg W.H., Söll D., Wang J.. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desai K.K., Bingman C.A., Phillips G.N., Raines R.T.. Structure of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry. 2013; 52:2518–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C.L., Lürhmann R., Söll D. et al. . HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011; 331:760–764. [DOI] [PubMed] [Google Scholar]
- 18. Kosmaczewski S.G., Edwards T.J., Han S.M., Eckwahl M.J., Meyer B.I., Peach S., Hesselberth J.R., Wolin S.L., Hammarlund M.. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 2014; 15:1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Westaway S.K., Belford H.G, Apostol B.L., Abelson J., Greer C.L.. Novel activity of a yeast ligase deletion polypeptide: evidence for GTP-dependent tRNA splicing. J. Biol. Chem. 1993; 268:2435–2443. [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 21. Sheldrick G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Cryst. 2010; D66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M.. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991; A47:110–119. [DOI] [PubMed] [Google Scholar]
- 24. Holm L., Kaariainen S., Rosenstrom P., Schenkel A.. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008; 24:1780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Das U., Wang L.K., Smith P., Jacewicz A., Shuman S.. Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP•Mg2+ and 5’-OH oligonucleotide and a product complex with GDP•Mg2+ and 5’-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition. Nucleic Acids Res. 2014; 42:1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das U., Wang L.K., Smith P., Shuman S.. Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial Pnkp•Hen1 RNA repair system. Biochemistry. 2013; 52:4734–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Remus B.S., Shuman S.. A kinetic framework for tRNA ligase and enforcement of a 2′-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA. 2013; 19:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.