Abstract
Alternative end-joining (alt-EJ) repair of DNA double-strand breaks is associated with deletions, chromosome translocations, and genome instability. Alt-EJ frequently uses annealing of microhomologous sequences to tether broken ends. When accessible pre-existing microhomologies do not exist, we have postulated that new microhomologies can be created via limited DNA synthesis at secondary-structure forming sequences. This model, called synthesis-dependent microhomology-mediated end joining (SD-MMEJ), predicts that differences between DNA sequences near double-strand breaks should alter repair outcomes in predictable ways. To test this hypothesis, we injected plasmids with sequence variations flanking an I-SceI endonuclease recognition site into I-SceI expressing Drosophila embryos and used Illumina amplicon sequencing to compare repair junctions. As predicted by the model, we found that small changes in sequences near the I-SceI site had major impacts on the spectrum of repair junctions. Bioinformatic analyses suggest that these repair differences arise from transiently forming loops and hairpins within 30 nucleotides of the break. We also obtained evidence for ‘trans SD-MMEJ,’ involving at least two consecutive rounds of microhomology annealing and synthesis across the break site. These results highlight the importance of sequence context for alt-EJ repair and have important implications for genome editing and genome evolution.
INTRODUCTION
Non-homologous end-joining repair of DNA double-strand breaks (DSBs), while normally accurate, can also be an error-prone process, creating small insertions, deletions, and chromosome translocations (1–4). Classical non-homologous end joining (C-NHEJ) involves binding of the broken ends by the Ku70/Ku80 heterodimer, recruitment of processing factors such as nucleases and polymerases, and ligation by a complex containing DNA ligase 4 (Lig4) (5–12). While C-NHEJ can repair blunt-ended breaks, it also utilizes small microhomologies during alignment of broken ends (13). In the absence of Ku or Lig4, an alternative end-joining mechanism predominates (14–17). Frequently referred to as A-NHEJ or alt-EJ, alternative end joining is highly mutagenic and is often distinguished from C-NHEJ by the formation of large deletions (18). Many of these deletions are thought to be created by DNA resection and subsequent annealing of microhomologous sequences found in the resulting single-stranded DNA (19,20). For this reason, alt-EJ is sometimes referred to as microhomology-mediated end joining (MMEJ) (19,21). However, alt-EJ does not always appear to involve annealing at microhomologies, suggesting that alt-EJ might actually comprise several repair mechanisms.
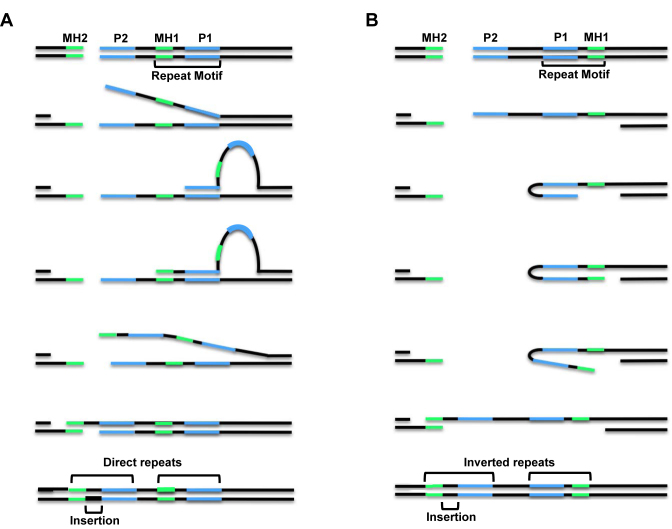
In Drosophila melanogaster, alt-EJ is commonly used to repair DSBs, even in the presence of fully functional C-NHEJ (22–24). Drosophila alt-EJ frequently results in a type of repair class termed indels, in which deletions are accompanied by insertions of one to tens of base pairs (bp). Previously, we showed that many types of DSB repair junctions that occur in wild-type Drosophila and flies lacking Lig4, including indels, simple deletions, and apparent blunt-ended joins, can all be explained by a model called synthesis-dependent microhomology-mediated end joining (SD-MMEJ) (25). In the SD-MMEJ model, single-stranded DNA that is formed by resection or duplex unwinding at DSB sites forms transient secondary structures, such as hairpins or loops, via annealing at microhomologous sequences called ‘primer repeats’ (Figure 1). These secondary structures can involve annealing between misaligned direct repeats in two single-stranded DNA molecules (referred to as ‘loop-out’ mechanisms, Figure 1A) or annealing between two inverted repeats within a single-strand of DNA (referred to as ‘snap-back’ mechanisms, Figure 1B). The 3′ DNA end in these structures then uses the ssDNA as a template for non-processive synthesis. Unwinding of the extended secondary structure and subsequent annealing of the nascent DNA with microhomologous sequences found on the other side of the break (‘microhomology repeats’) allows for completion of repair.
Figure 1.
The SD-MMEJ model for alternative end joining repair. (A) Loop-out mechanism with DNA unwinding prior to loop formation. (B) Snap-back mechanism with DNA resection prior to hairpin formation. Both mechanisms utilize annealing of break-proximal primer repeats (P2) to break-distal primer repeats (P1), which primes nascent synthesis that can lead to insertions (black) and the creation of new microhomologous sequences (MH1, green). For loop-out SD-MMEJ, P1 and P2 are direct repeats, while for snap-back SD-MMEJ they are inverted repeats. Repair concludes with unwinding of secondary structures, annealing of nascent microhomologies with MH2 sequences on the other side of the break, fill-in synthesis, and ligation. For the repair events shown here, the inserted sequence becomes part of new, longer direct or inverted repeats. Not shown are the trimming of non-homologous flap intermediates when P2 and MH2 are not directly adjacent to the break site, or deletion junctions with no net insertion that are formed when P1 and MH1 are directly adjacent to each other.
Repair products consistent with the SD-MMEJ model contain repeat motifs that are composed of the P1 primer repeat, the MH1 microhomology repeat, and any DNA between these repeats. In other words, the repeat motif is composed of the outermost primer repeat utilized during secondary structure formation and DNA that is subsequently synthesized to create the microhomology used to span the break. Depending on the amount of DNA found between the primer repeats and microhomology repeats, the resulting junction can be an indel, simple deletion/microhomology join, or apparent blunt join (25–27).
Previous studies have demonstrated that DNA sequences flanking a DSB can influence which type of repair is preferentially used. In both Saccharomyces cerevisiae and mammalian cells, longer direct repeats are used more frequently than shorter repeats during MMEJ, but not all long repeats are utilized equally and short repeats are sometimes favored. This indicates that factors besides repeat length influence MMEJ repair (28–32). Studies using S. pombe showed that alt-EJ can be affected by DNA sequences directly adjacent to an endonuclease-induced break (33). Similarly, repair of Cas9-induced breaks in human cells is highly dependent upon the primary DNA sequence flanking the break site (34). However, the exact mechanisms by which sequences flanking double-strand breaks influence repair and determine preferred repair outcomes are poorly characterized.
Our initial characterization of SD-MMEJ suggested that it might depend on the presence of secondary-structure forming sequences located in DNA flanking the break site (25). To test this hypothesis, we have now carried out a fine-level study of targeted sequence changes to characterize how flanking sequences impact the frequency and types of SD-MMEJ repair junctions. Our results demonstrate that secondary-structure forming sequences on both sides of an endonuclease-induced DSB can promote SD-MMEJ and have a strong influence on the mechanisms by which alt-EJ proceeds in Drosophila. Surprisingly, we find that increasing the length of the stem beyond four base pairs in hairpin-forming sequences does not result in their increased usage as primer repeats, suggesting that SD-MMEJ proceeds most efficiently with repeats that form transient and easily-disrupted secondary structures. Furthermore, our data provide evidence that secondary structure-forming sequences located up to 30 bp distal to the break site can be used in SD-MMEJ. Finally, we show that SD-MMEJ is a remarkably plastic process, with small changes in DSB-flanking sequences having large effects on the types of repair junctions that are recovered.
MATERIALS AND METHODS
Preparation of plasmids
The plasmid containing part of the Iw7 construct (35) was created by cloning a 1 kb amplified fragment containing the I-SceI recognition site into pBlueScript. Single base pair changes in Iw7 were created by site-directed mutagenesis with Phusion polymerase (NEB) to create plasmids M1–M5. Purified plasmids were obtained using the Macherey-Nagel NucleoBond Xtra Midi kit and diluted to 250 ng/μl in injection buffer (1 mM sodium phosphate and 50mM potassium chloride).
Fly Stocks
The genotype of the fly stock used for injections was w1118; P{Ubiq::I-SceI¸w+}, Sp / CyO, P{Ubiq::I-SceI¸w+}, in either a wild-type or lig4169a mutant background (22,23). Oregon-R flies were used as a no I-SceI control. All flies were maintained in bottles containing a cornmeal-agar medium in a 25°C incubator on a 12 h light–dark cycle. Freshly eclosed flies were placed in cages and fed yeast paste on a grape agar substrate to promote embryo laying. Embryos <2 h old were recovered and dechorionated for two minutes in a 50% bleach solution. Embryos (n = 60 per experiment) were aligned uniformly on double-stick tape attached to a glass cover slip for injections of plasmids.
Microinjection and DNA Recovery
Embryos were desiccated for one minute and coated in halocarbon oil to prevent rupturing of the membrane when injected. The microinjections were done using a Zeiss compound microscope fitted with injection apparatus and a Parker-Hanfin Picospritzer II. Embryos were incubated at 25°C for 4 h to allow for I-SceI cutting and repair of the ensuing double-strand breaks. Halocarbon oil was removed using a 1% sodium dodecyl sulfate solution in 0.7% sodium chloride buffer.
Plasmid DNA was extracted by grinding embryos with a disposable pestle in 200 μl Buffer A (100 mM Tris–HCl, pH 7.5, 100 mM EDTA, 100 mM NaCl, 0.5% SDS). After incubating at 65°C for 30 min, 800 μl of LiCL/KAc solution (1 part 5 M KAc:2.5 parts 6 M LiCL) was added and tubes were incubated on ice for 10 min. The solution was centrifuged for 10 minutes and plasmids were precipitated from the supernatant with isopropanol.
DNA purification and sequencing
For Sanger sequencing, plasmid DNA recovered from embryos was transformed into XL1-Blue competent Escherichia coli and purified by alkaline lysis. Sanger sequencing was performed by Eton Bioscience Inc., using a primer approximately 200 bp from the I-SceI recognition site.
For high-throughput amplicon sequencing, approximately 300 bp of sequence flanking the I-SceI site was amplified from recovered plasmids by PCR with Q5 polymerase (NEB) for 19 cycles using an Eppendorf Vapo Protect thermocycler with a pooled set of primers containing one, two or three random bases at the 5′ end. AMPure bead purification was performed on the PCR products and the purified DNA was subjected to a second PCR to attach indices for amplicon sequencing. A final AMPure purification step was performed to remove all products less than 100 bp. The samples were pooled with 5% PhiX DNA and sequenced on an Illumina MiSeq platform using a version 3 chip with 2 × 300 paired-end reads.
Bioinformatic analysis
Raw reads from paired-end sequencing were trimmed to the amplicon primer sequence and overlapping read pairs were merged into single consensus reads as FASTQ files (CLC Bio Genomics Workbench). Junctions lacking 10 bp of reference sequence at both the 5′ and 3′ ends of each amplicon were removed as PCR artifacts as they failed to span both sides of the break. Reads were trimmed to common starting and end positions, mapped to reference sequences corresponding to the original Iw7 or M1-M5 constructs with Geneious alignment software (BioMatters) and exported as SAM files for analysis. The structure of repair junctions was determined by using the CIGAR string to identify sequences matching the 5′ and 3′ ends of the amplicon, deleted sequences relative to the I-SceI cut site, and inserted sequences (Supplementary Table S2).
Junctions with <30 reads and appearing in both uncut control and the experimental samples were removed from the data set. Following removal of these junctions, the normalized percentage of inaccurate reads per junction was calculated by dividing the number of reads per junction by the total number of inaccurate reads. Deletion junctions (containing only linear sequence present in the original construct) were characterized as apparent blunt joins (ABJ) or microhomology junctions (MHJ). These were analyzed to determine SD-MMEJ consistency using a novel pure Python suffix tree library, which searched for a break-spanning region that contained both microhomology and primer junctions. Insertion sequences (containing novel sequences not present in the original construct) were analyzed using an in house R-script to determine SD-MMEJ consistency, as defined in the text.
RESULTS
Development of a high-throughput SD-MMEJ assay
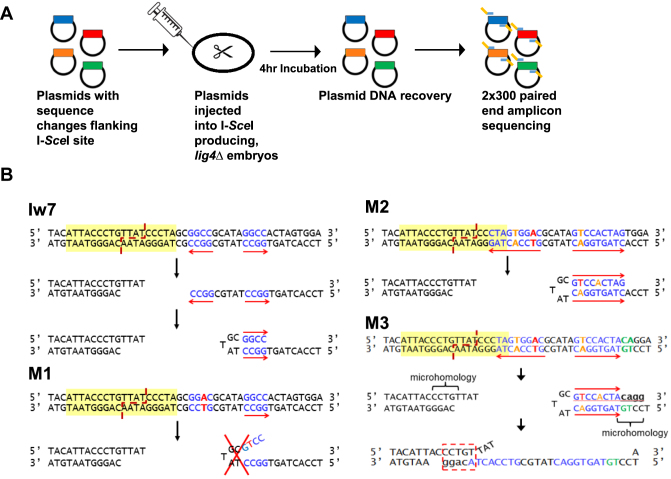
Our previous SD-MMEJ study (25) utilized a single I-SceI endonuclease recognition site positioned on chromosome 2 (Iw7; (35)). Because this system required that independent repair events be recovered in the progeny of individual males, we developed a plasmid injection system to allow for rapid, high throughput characterization of I-SceI repair products (Figure 2A). Purified plasmid DNA was injected into 0–2 h-old, C-NHEJ deficient lig4 mutant embryos expressing the I-SceI endonuclease. The embryos were incubated for four hours at 25°C to allow for cutting and repair to take place, after which the plasmids were recovered and transformed into bacteria. DNA from individual colonies, each representing an independent repair event, was purified and Sanger sequenced. Comparison of the repair events recovered from the plasmid system to our previously published results with Iw7 located on chromosome 2 (25) suggests that similar types of inaccurate repair products are recovered regardless of whether the construct is located on a plasmid or in the genome, with roughly equivalent percentages of apparent blunt joins, microhomology junctions, and indels (Supplementary Table S1).
Figure 2.
A high-throughput SD-MMEJ assay. (A) In vivo extract system. Purified plasmid constructs are injected into dechorionated embryos expressing I-SceI. Following incubation to allow for cutting and repair, plasmids are recovered and sequenced directly by Sanger sequencing (not shown) or prepared for high throughput amplicon sequencing. (B) Diagrams showing the predicted effects of right-side flanking sequence changes on SD-MMEJ repair. Yellow highlighting indicates the I-SceI recognition site. In Iw7, blue text corresponds to a GGCC direct and inverted repeat that can participate in snap-back SD-MMEJ (shown) or loop-out SD-MMEJ (not shown). M1 has a change in the first GGCC repeat (red) that is postulated to prevent formation of the hairpin (shown) or loop (not shown) used during the creation of a prominent SD-MMEJ product. M2 has two additional alterations (orange) that are postulated to promote the formation of a 9-nt inverted repeat that can be used to prime snap-back synthesis during SD-MMEJ. M3 has two additional changes (green) that are predicted to act as the start of a microhomology primer. Following synthesis of 4 nt, the nascent DNA could anneal to a CCTGT microhomology on the left side of the break.
To further increase the number of repair junctions that we could analyze, we employed high-throughput amplicon sequencing. Cut and repaired plasmids recovered from injected embryos were subjected to PCR, using primers that amplified the repair junction plus approximately 100 bp of flanking sequence (Figure 2A). The products were then sequenced in multiplexed, paired-end MiSeq reactions. A ‘no-cut’ control in which plasmids were injected into embryos not expressing I-SceI was also included. The sequencing reads were trimmed and aligned to the Iw7 reference sequence. Inaccurate repair events that were observed in the ‘no-cut’ control were assumed to be sequencing artifacts and were removed from all data sets. The remaining reads were binned according to similarity and repair junctions with more than 30 total reads were selected for further analysis (Supplementary Table S2).
Comparison of the inaccurate repair events obtained from the Iw7 construct showed that of the 64 unique deletions we obtained from Sanger sequencing, 59 (92%) were also recovered in the amplicon data set, plus an additional 71 novel deletions, demonstrating the utility of the high-throughput approach. While only 25 out of 105 unique insertion repair events from Sanger sequenced plasmids appeared in the amplicon collection, this is likely due to the greater potential diversity of insertion repair events. Importantly, plasmid repair events with large insertions were recovered at similar frequencies with both Sanger and high-throughput amplicon sequencing protocols, indicating that amplicon sequencing does not bias against the recovery of these types of events.
In order to analyze the effects of flanking sequence upon SD-MMEJ repair outcomes, we designed additional constructs with single or multiple base pair changes relative to the original Iw7 sequence (Figure 2B). In all of the constructs, the original 18 bp I-SceI recognition sequence remains intact, ensuring efficient cutting. Our logic in the design of these constructs was to first impair the formation of a prominent SD-MMEJ product, and then to create new primer and microhomology repeats that could act as drivers of other SD-MMEJ products. Because the most frequently utilized primer repeats from our initial study were located to the right of the I-SceI recognition site, we focused first on the right side. The M1 construct contains a single base-pair change in a prominently utilized GGCC direct and inverted repeat that we have previously identified as being used in 20–40% of all SD-MMEJ consistent repair products (Figure 2B) (25). We anticipated that this change would abolish any repair products that utilize this repeat during loop-out and snap-back SD-MMEJ. The M2 construct contains two additional base-pair changes that we predicted would enhance two specific SD-MMEJ products: one involving loop-out SD-MMEJ with a 4-nucleotide (nt) TAGT direct repeat and the other utilizing snap-back SD-MMEJ with a 9-nt inverted GTCCACTAG repeat. The M3 construct changes two base pairs downstream of the 9-nt inverted repeat, 30 bp away from the break site. We hypothesized that synthesis resulting from the formation of this snap-back intermediate should create a 5-nt microhomology that could anneal with single-stranded sequence on the opposite side of the break.
Single base pair changes in sequences prone to form secondary structures affect repair outcomes
We measured the sizes of the deletions and insertions for all inaccurate repair junctions recovered from the amplicon sequencing. The vast majority of junctions had deletions of fewer than 20 bp; the mean deletion size for all four constructs was between 9 and 11 bp (Supplementary Figure S1A). Overall, the distribution of insertion sizes was similar between the four different constructs, with most insertions being less than 15 bp (Supplementary Figure S1B). Thus, it appears that the M1-M3 mutations do not have a large effect on the relative sizes of the repair products.
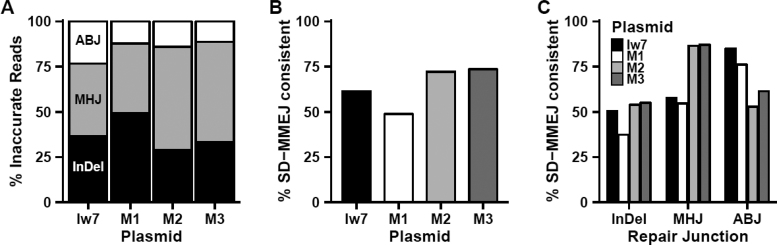
In order to facilitate further analysis, we designed custom programs in Python and R that can identify repeat motifs and primer and microhomology repeats potentially utilized during SD-MMEJ (see materials and methods). We classified repair events recovered from each of the four sets of injected embryos as deletions with apparent blunt joins, deletions with microhomologies, or indels (insertions with or without a deletion). While we did observe variation in the types of repair products recovered for each of the four constructs, the only statistically significant difference was an increase in the percentage of microhomology junctions recovered for the M2 construct, compared to both M1 and Iw7 (Figure 3A, P < 0.05, ANOVA, Tukey's HSD test). The single base-pair change made in M1 resulted in a twelve percent increase in the percentage of repair junctions with insertions, but this increase in indels was not significant and was not observed in the M2 and M3 repair products.
Figure 3.
Effects of single nucleotide changes in I-SceI flanking sequences on SD-MMEJ outcomes. (A) Types of inaccurate repair junctions. ABJ = apparent blunt join; MHJ = microhomology junction; InDel = insertion/deletion junction. (B) Overall SD-MMEJ consistency. Each bar represents the percentage of reads for each junction determined to be SD-MMEJ consistent. (C) SD-MMEJ consistency by repair junction type.
In addition, we determined whether each repair event was consistent with the SD-MMEJ model. For the latter analysis, each junction had to meet three criteria to qualify as SD-MMEJ consistent: (1) the repair event had to contain a repeat motif (P1 + MH1 + intervening sequence) of at least 4 bp; (2) the break-distal end of the repeat motif had to occur within 30 bp of the site of break rejoining; and (3) the repeat motif had to contain a primer repeat of at least 1 bp and a microhomology repeat of at least 1 bp. Repair events that failed to meet all three criteria were classified as SD-MMEJ inconsistent. Interestingly, the overall percentage of SD-MMEJ consistency for all three types of repair junctions decreased with the M1 mutation and increased with the M2 and M3 mutations, particularly for the microhomology and indel repair events (Figure 3B and C). Although these changes were not statistically significant, they suggest that sequences adjacent to a double-strand break can influence the efficiency of SD-MMEJ repair.
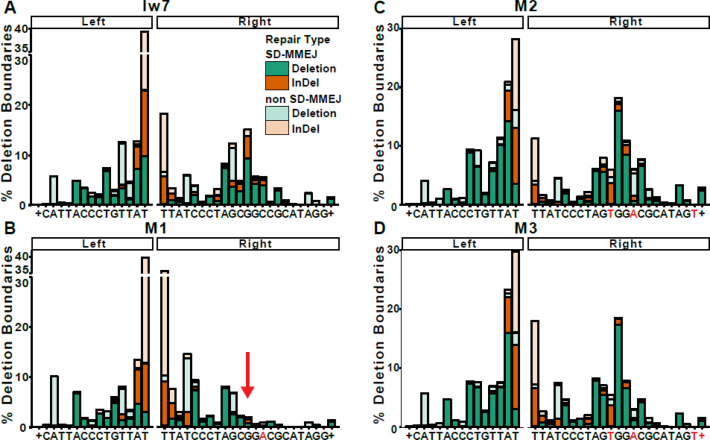
Furthermore, the distribution of right-side deletion boundaries (endpoints), as illustrated in Figure 4, was shifted towards the break site in the M1 repair products and accompanied by an increase in the percentage of non-SD-MMEJ consistent junctions. The deletion boundary at the GGCC direct repeat in SD-MMEJ consistent junctions, while readily apparent in the Iw7 sequence, was conspicuously absent in the M1 mutant junctions (Figure 4B, see arrow). In contrast, the deletion boundaries for the M2 and M3 repair junctions were similar to those for Iw7 repair products. Interestingly, flanking sequence changes made in M1-M3 had little effect on the left-side deletion boundaries. Based on these observations, we conclude that the single mutation in M1 decreases SD-MMEJ consistent repair, while the M2 and M3 mutations promote SD-MMEJ, likely by providing new secondary structure-forming sequences.
Figure 4.
Changes in I-SceI flanking sequence alter the deletion boundaries. Deletion boundaries for inaccurate repair events for (A) Iw7, (B) M1, (C) M2 and (D) M3. Deletion boundaries are defined as the first non-ambiguous base accurately aligned to the left and right of the I-SceI break. SD-MMEJ consistent junctions are represented by darker colors. + indicates all bases to the right or left of the indicated sequence. The arrow in (B) shows the effect of the C→A mutation on the deletion boundary at a prominently used primer repeat.
Comparison of repeat motifs in deletion junctions highlights preferred primers used during SD-MMEJ
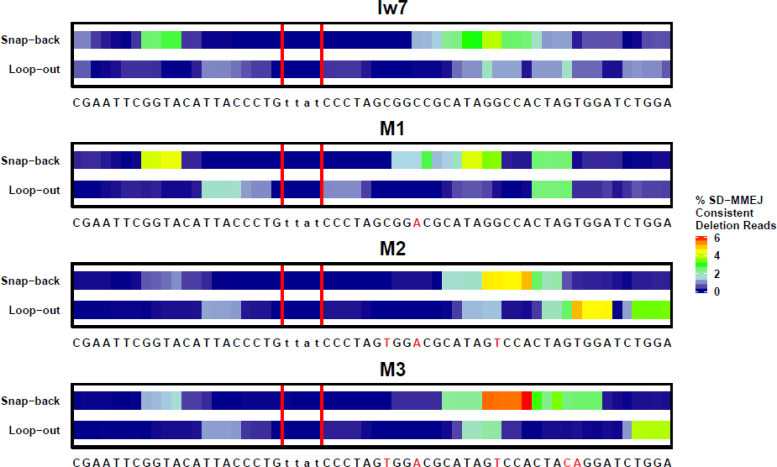
To further characterize the sequences that might mediate SD-MMEJ in the different contexts, we first focused on the SD-MMEJ consistent deletion junctions. We conducted computational analysis to identify repeat motifs for SD-MMEJ consistent deletions. A single repeat motif comprises the outermost primer repeat utilized during secondary structure formation and DNA that is subsequently synthesized to create the microhomology used to span the break (Figure 1).The overall distribution of these repeat motifs are shown for each of the four sequences in Figure 5, with bases that are found in more commonly utilized repeat motifs indicated by warmer colors.
Figure 5.
The prevalence of repeat motifs utilized in SD-MMEJ consistent deletion junctions depends on the I-SceI flanking sequence. Repeat motifs contain the P1 primer repeat, the adjacent MH1 microhomology repeat, and any intervening sequence. Colors correspond to the frequency that each base is found in a repeat motif, with warmer colors indicating greater frequencies. Red vertical lines indicate the TTAT/AATA overhangs produced by I-SceI cutting.
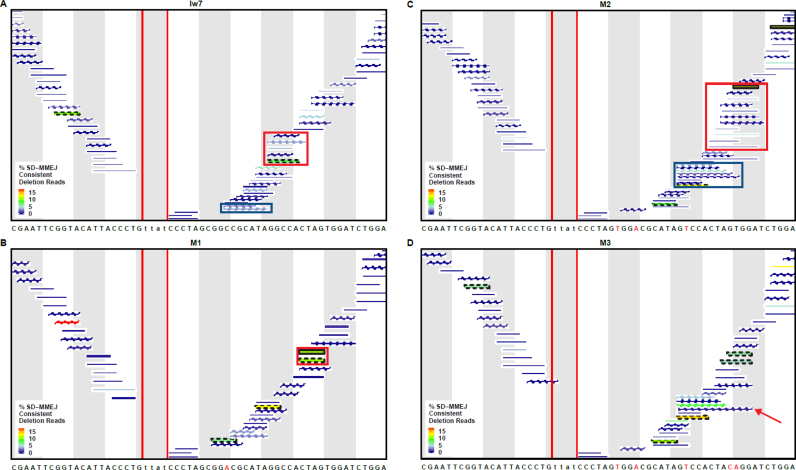
Interestingly, we observed that a sizeable number of repair junctions from all four plasmids were consistent with the frequent utilization of a GGTA/TACC inverted repeat for snap-back synthesis on the left side of the break (Figures 5 and 6A–D). This was one of the four most common primer repeats identified in our previous, chromosome-based assay (25). In contrast, repeat motifs identified on the right side of the break differed between the different constructs, especially at the locations where mutations were made. This indicates that sequence changes made on the right side of the break did not affect the primer repeats utilized on the left side and suggests that the two sides act independently of each other during the secondary structure formation phase of SD-MMEJ repair.
Figure 6.
Changes in I-SceI flanking sequence influence the secondary structures utilized during SD-MMEJ repair. Individual repeat motifs in SD-MMEJ consistent deletion junctions, with their relative prevalence indicated by color. Percent SD-MMEJ consistent deletion reads was calculated by dividing the total number of reads for each repeat motif by the total number of SD-MMEJ consistent deletion reads. Repeat motifs corresponding to loop-out mechanisms are indicated by straight lines, while snap-back mechanisms are indicated by hatched lines. Red vertical lines indicate the TTAT/AATA overhangs produced by I-SceI. (A) The red box highlights snap-back synthesis products utilizing a GGCC direct repeat, while the blue box highlights loop-out synthesis using the GCGG/CCGC inverted repeat. (B) The red box highlights a CTAG repeat motif created by both snap-back and loop-out repair mechanisms. (C) The red box highlights repeat motifs resulting from snap-back synthesis utilizing part/all of a 6 nt TCCACT/AGTGGA inverted repeat or an 8 nt CTAGTGGA direct repeat. The blue box indicates repeat motifs resulting from snap-back synthesis using part/all of a 9 nt CTAGTGGAC/GTCCACTAG inverted repeat. (D) The arrow indicates a repeat motif with a CAGG microhomology that anneals with CCTG on the left side of the break.
In the Iw7 repair products, we saw strong evidence for snap-back synthesis utilizing a GGCC direct repeat and GCGG/CCGC inverted repeat (Figure 6A, red box and blue box, respectively). The loss of these repeats in M1 resulted in the increased prevalence of a CTAG repeat motif that could be created through either snap-back or loop-out repair mechanisms (Figure 6B, red box). Thus, a single mutation in the flanking sequence ten base pairs from a double-strand break can have a large impact on secondary structures that are utilized during SD-MMEJ repair.
Analysis of the most prevalent repeat motifs in the M2 repair products suggests that the two new mutations created the potential for new secondary structures to form and drive SD-MMEJ. For example, a repeat motif containing all or part of the CTAGTGGA sequence located 30 bp from the break (Figure 6C, red box) could result from either snap-back synthesis utilizing either a 6 nt AGTGGA/TCCACT inverted repeat or an 8 nt CTAGTGGA direct repeat. Both of these repeats depend on the two new mutations found in the M2 flanking sequence. Furthermore, as predicted, we saw evidence for the utilization of the 9 nt CTAGTGGAC/GTCCACTAG inverted repeat in repair products predicted to be generated by snap-back synthesis (Figure 6C, blue box). Interestingly, many of the M2 repair junctions did not appear to utilize the entire 9 nt inverted repeat, suggesting that repeats of this length may be disfavored during SD-MMEJ.
Mutation of the GT to the CA in the M3 sequence abolished usage of the 8 nt CTAGTGGA direct repeat observed in M2, as would be expected (Figure 6D). An 8 nt TAGTGGAC/GTCCACTA inverted repeat was still frequently used and there was evidence for annealing of a newly synthesized CAGG with a CCTG microhomology on the left side of the break, as had been postulated during the design of M3 (Figure 6D, red arrow). Thus, using the SD-MMEJ model we were able to predict some of the new deletion junctions that were observed in the M2 and M3 sequence contexts.
Analysis of repeat motifs and primer plots for indel events provides insight into preferred SD-MMEJ characteristics
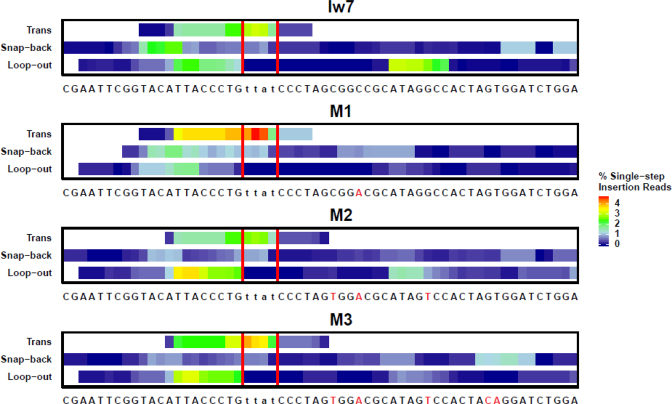
SD-MMEJ provides a unifying mechanism to describe the formation of both simple deletion and indel repair events. However, the deletion junctions that we recovered could also be explained by a simple MMEJ mechanism, in which pre-existing microhomologies simply anneal and repair is completed by fill-in synthesis. Thus, we next analyzed the indels, whose formation can only be explained by synthesis of new DNA prior to annealing. We computationally identified prominent repeat motifs in SD-MMEJ consistent repair products that could be explained by single-step insertions, which can occur via a single round of annealing at a primer repeat, synthesis, and dissociation followed by annealing at newly synthesized microhomologies. These repeat motifs are represented in heat map plots (Figure 7 and Supplementary Figure S2). Most of the repeat motifs identified in the indel repair products were located close to the break site, with a preference for the left side. This contrasts with the repeat motifs in the simple deletion products, which were located primarily on the right side (compare Figures 5–7).
Figure 7.
The prevalence of repeat motifs utilized in SD-MMEJ consistent single-step insertions depends on the I-SceI flanking sequence. Repeat motifs contain the P1 primer repeat, the adjacent MH1 microhomology repeat, and any DNA between these repeats (the insertion). Colors correspond to the frequency that each base in found in a repeat motif, with warmer colors indicating greater frequencies. Red vertical lines indicate the TTAT/AATA overhangs produced by I-SceI cutting. Trans SD-MMEJ occurs with microhomology annealing across the break site, followed by synthesis, dissociation of the nascent DNA, and reannealing at new microhomologies prior to completion of repair.
As with the deletion junctions, we observed striking differences for the indels in the most highly represented repeat motifs between the different plasmids. For example, the GGCC direct repeat was frequently identified in the Iw7 loop-out repeat motifs but was absent in M1 loop-out repair products (Supplementary Figure S2A and S2B, for example see Figure 9A). Interestingly, the snap-back SD-MMEJ products with the GCGG/CCGC inverted repeat were less commonly observed in the indels than in the simple deletions isolated from the Iw7 background (compare Figure 6A to Supplementary Figure S2A, for an example see Supplementary Figure S3A).
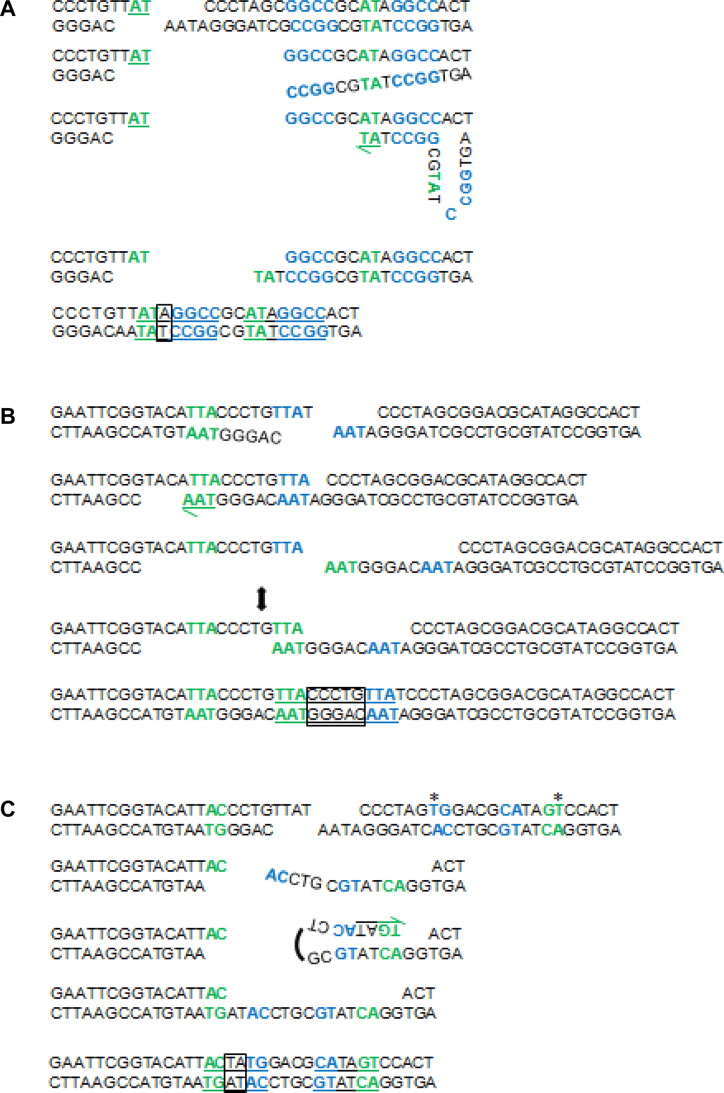
Figure 9.
Models for SD-MMEJ consistent indel junctions. Shown are primer repeats (blue), microhomology repeats (green), insertions (boxes), and final repeat motif (underlined). (A) Iw7 loop-out event. Unwinding/resection of DNA on the right side of the break allows primer repeats to anneal, followed by limited synthesis, dissociation, and annealing of microhomology repeats. Endonuclease/flap cleavage events are not shown for simplicity. (B) M1 trans SD-MMEJ event. The two-ended arrow indicates transition of the TTA primer repeat to a microhomology repeat. This junction can also be explained by a snap-back mechanism on the left side of the break. (C) M2 snap back event. Asterisks indicate engineered sequence changes utilized during repair. Note that 3′→5′ resection or endonuclease activity following the annealing of primer repeats is required prior to initial synthesis.
In addition to snap-back and loop-out repair products, we were able to identify a group of indels which could only be created through annealing of single-stranded DNA across the junction, synthesis, dissociation, and reannealing at a new microhomology junction. We refer to these types of junctions as ‘trans’ SD-MMEJ junctions (for an example, see Figure 9B). The beginning of the repeat motifs for most of the trans junctions occurred very close to the break site and extended for a short distance to the left, suggesting that the initial annealing occurred within the TTAT overhang and synthesis proceeded using single-stranded DNA on the left side of the break as a template.
To gain further insight into SD-MMEJ sequence preferences, we determined the primer repeats that could have been used for the indel repair events from each of the four plasmids. Plots of these potential primers for the Iw7 sequence and M1-M3 constructs are shown in Figure 8 and Supplementary Figure S4. In these plots, the DNA sequences between the primers form the loops in the proposed secondary structures; inspection of these sequences shows the potential for additional base pairing within the loops. Notably, primer two (p2, the primer most proximal to the break) is usually located within 15 bp of the I-SceI cut site, with the exception of a TGGATC direct repeat located 30 bp from the break (Figure 8). Individual junctions with the greatest number of reads utilize primer repeat pairs that are 3–4 nt in length and separated by fewer than 10 bp (Supplementary Figure S5). However, the total number of junctions using primer repeats of 1–2 nt is greater than the total number of junctions using longer primer repeats, presumably because more of the shorter primers are present in the flanking sequence. Remarkably, some of the indel junctions can be explained by SD-MMEJ mechanisms in which single nucleotides pair across a distance of more than 25 bp. For these junctions, the most frequently used short primer repeats are not randomly distributed, suggesting that these repair events are also influenced by the sequences flanking the primer repeats.
Figure 8.
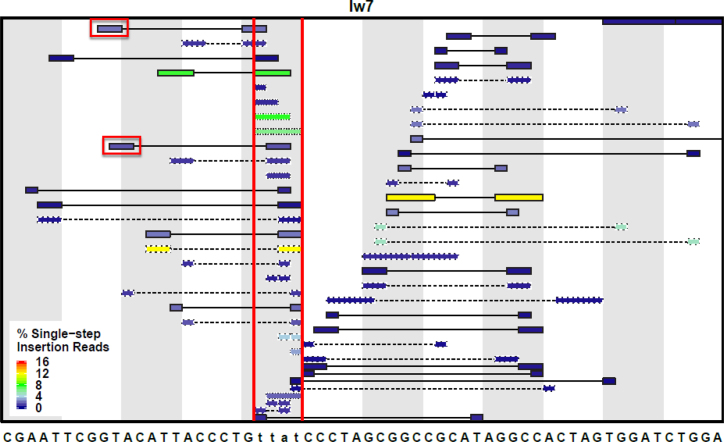
Specific primer repeats are preferentially used during SD-MMEJ for single-step insertions. All possible primer pairs (P1 and P2) are shown for the Iw7 construct, along with the type of SD-MMEJ event for each primer set. Primer pairs do not consider overall synthesis length and microhomology formation, therefore, different junctions may have the same primer pair. Colors correspond to the frequency that each primer pair is utilized, with warmer colors indicating greater frequencies. Loop-out = solid outline and connecting line; snap-back = dashed outline and connecting line; trans = dotted outline. Red vertical lines indicate the TTAT/AATA overhangs produced by I-SceI cutting. Red boxes indicate the loop-out priming sites ablated in the M4 plasmid construct.
Inspection of the primer plots reveals secondary structures that might form in each of the four sequence contexts to promote SD-MMEJ. Some of these are common to all contexts, particularly primer repeats highly utilized on the left side of the break. For example, the most frequent SD-MMEJ consistent indels involve snap-back with annealing between a TA/AT inverted repeat or loop-out with annealing between a TTA direct repeat. Both of these primer repeats are located directly to the left of the break site (Figure 8 and S4, for examples see Supplementary Figure S4B and S4C).
However, in many instances the pattern of primer usage directly correlates with the sequence changes made in each of the plasmids. For example, in M1, a unique TCC/GGA inverted primer repeat is frequently utilized during snap-back SD-MMEJ (Supplementary Figure S4A, red box). Similarly, in M2 the eight nucleotide CTAGTGGA direct primer repeat and nine nucleotide CTAGTGGAC/GTCCACTAG inverted primer repeat are utilized during loop-out and snap-back SD-MMEJ, similar to what was observed with the simple deletion junctions (Supplementary Figure S4B). Finally, a unique class of SD-MMEJ consistent indels frequently observed in the M2 and M3 contexts is best explained by snap-back synthesis using a TG/CA inverted primer repeat (Supplementary Figure S4B and C, red boxes, see Figure 9C for an example).
In addition to the unambiguous trans priming events shown in Figures 7, 8 and Supplementary Figure S2, all loop-out events represented by the repeat motifs in Figure 7 could also be explained by a trans mechanism. One example of this is shown in Supplementary Figure S3D, where a TACCCT insertion could be created either via annealing of a GT/CA microhomology in a trans priming event or via annealing of an AT direct repeat in a loop-out mechanism. Because our models classify these ambiguous junctions as loop-out events, our analysis may actually be underestimating the percentage of trans SD-MMEJ consistent events that occur with priming across the break site. Because the repeat motifs for the trans SD-MMEJ events tend to cluster on the left side of the break (Figure 7), this could also explain the leftward bias of repeat motifs that we observe for the SD-MMEJ consistent loop-out indels compared to the simple deletions.
Sequence changes to the left of the I-SceI cut site also affect SD-MMEJ in predictable ways
The primer repeat utilization plots shown in Figures 8 and S4 indicate that changes made to the I-SceI flanking sequences in the M1-M3 constructs had the greatest effect on the usage of primer repeats on the right side. To test whether we could also affect left-flanking repeat motifs in SD-MMEJ consistent repair events, we generated two constructs with single base pair changes to the left of the I-SceI recognition site, using logic similar to that employed with the M1-M3 constructs. In the M4 construct, a T–A base pair just outside of the I-SceI recognition site was changed to an A-T base pair, ablating two 2-nt loop-out priming sites that were utilized in 1–2% of the single-step insertion reads in the original Iw7 sequence (Supplementary Figure S6 and Figure 8). In the M5 construct, an A–T base pair was swapped with a T-A base pair, creating a potential 4-bp GTTA priming site for loop-out synthesis (Supplementary Figure S6).These plasmids were injected into I-SceI expressing embryos, recovered, and subjected to amplicon sequencing as before. For these experiments, we utilized both wild-type and lig4 mutant embryos, to determine whether primer utilization during SD-MMEJ changes when C-NHEJ repair is available.
Comparison of inaccurate repair junctions recovered from the M4 and M5 constructs showed slight variation in the types of inaccurate repair (Supplementary Figure S7A). The percentage of SD-MMEJ consistent repair for the M4 construct was similar to the original Iw7 construct and increased for the M5 construct. Interestingly, for all types of repair junctions the SD-MMEJ consistent repair percentage stayed the same or increased in LIG4+ embryos that were capable of carrying out C-NHEJ (Supplementary Figure S7B and C). Both the deletion and insertion length distributions and the deletion boundaries for the M4 and M5 constructs were similar to those measured for the Iw7 construct and were not affected by Lig4 status (Supplementary Figures S1, S8 and S9).
Inspection of the repeat motif plots for M4 SD-MMEJ consistent deletion junctions showed very little variance on the right side of the break compared to the Iw7 construct (Supplementary Figures S10 and S11). The largest discernible difference was the utilization of new primer repeats in snap-back synthesis reactions on the left side of the break (Supplementary Figure S11, red box); these repeats utilized the engineered T→A base change in both wild-type and lig4 genetic backgrounds. Similarly, the sequence change in M4 promoted frequent recovery of new SD-MMEJ consistent snap-back insertion products, especially in the absence of Lig4 (Supplementary Figures S12, S13, red box).
As predicted, the single base pair change in M5 promoted use of the GTTA primer for loop-out synthesis in both SD-MMEJ consistent deletion and insertion junctions (Supplementary Figures S10–S14, blue boxes in Supplementary Figures S11, S13 and S14). In contrast, the base change did not have major effects on the utilization of repeat motifs to the right of the I-SceI site. Finally, the SD-MMEJ consistent repair product spectrum in the M5 background was similar in both wild-type and lig4 embryos. We conclude that changing the flanking sequences within thirty base pairs of either side of a double-strand break can have major effects on SD-MMEJ repair, regardless of whether C-NHEJ is available.
DISCUSSION
Alternative end joining is frequently associated with the use of microhomologies to tether double-strand break ends during the repair process. However, much remains to be learned regarding the parameters that govern microhomology usage and the source of templated insertions during alt-EJ. One of the more remarkable aspects of alt-EJ in Drosophila, and likely in other organisms, is that the use of pre-existing microhomologies for simple MMEJ is often eschewed in favor of a more complex process that involves multiple rounds of microhomology annealing and synthesis. One implication of this observation is that alt-EJ is a highly dynamic and versatile process, adaptable to differences in local sequence context and repair factor availability.
Previous studies from our lab and others found that secondary structure-forming sequences can exert strong influences during alt-EJ and led us to propose the SD-MMEJ model. According to the model, SD-MMEJ can produce a variety of repair junction types, including apparent blunt joins that are normally associated with C-NHEJ repair and microhomology junctions that are commonly attributed to MMEJ. Importantly, the SD-MMEJ model can also provide mechanistic insight into the generation of complex indels that represent extreme examples of alt-EJ repair.
What sequence contexts promote SD-MMEJ?
In this study, we used a plasmid injection/recovery assay in fly embryos and next-generation amplicon sequencing to obtain an expanded collection of alt-EJ repair junctions. Analysis of over 1000 different common repair products recovered from six different constructs demonstrates that making single nucleotide changes as far as 30 bp from an I-SceI break can radically alter the spectrum of alt-EJ repair outcomes. In some cases, we were able to predict the repair outcomes based on an analysis of single-strand sequences likely to form hairpins and loops. For example, changing the identity of bases most commonly utilized as primer repeats in the Iw7 sequence eliminated the expected repair junctions. Similarly, creation of new primer repeats in the M2, M3 and M5 constructs resulted in recovery of repair products that were consistent with SD-MMEJ mechanisms utilizing these repeats.
Overall, the most highly represented repair junctions demonstrated a preference for secondary structures involving primer repeats of 3–4 nt, separated by an inter-repeat distance of fewer than 10 base pairs. However, we also observed many examples of SD-MMEJ consistent junctions that utilized primer repeats of only a single nucleotide, along with primer repeats separated by up to 25 base pairs. Because annealing at these distant primer repeats would be predicted to be rare and highly transient, it seems likely that the annealing and subsequent extension may be protein mediated (see below).
In our analysis of the M2 and M3 junctions, we found few SD-MMEJ consistent products that appeared to utilize the newly-created 8- and 9-bp primer repeats. Instead, the most frequently recovered repair junctions contained smaller portions of the repeats, ranging from 3–6 bp. Our interpretation of this result is that the primer repeats most likely to result in effective SD-MMEJ repair are those that form quasi-stable and easily disrupted secondary structures, in line with a view of SD-MMEJ as a highly flexible and dynamic repair process. A corollary to this finding is that one or more exonucleases or endonucleases must act on the single-stranded substrate either before or after formation of the secondary structures, in order to generate a base-paired 3′ DNA end that can serve as a template for synthesis. The identity of these nuclease(s) remains to be determined.
Results obtained using the M3 construct showed that sequence changes up to 30 bp distal from the I-SceI cut site resulted in an altered spectrum of novel, SD-MMEJ consistent junctions. While we have yet to establish a distance for which single nucleotide mutations no longer influence SD-MMEJ repair, we expect that the maximum distance may be limited by the amount of naked single-stranded DNA at the break site. In eukaryotes, the footprint for the RPA trimer, which binds single-stranded DNA and prevents secondary structure formation, has been shown to be approximately 28–30 nt (36–38). Thus, we hypothesize that the mutations in the M3 plasmid represent the outer limit of where sequences flanking a DSB may influence SD-MMEJ outcomes. Interestingly, RPA has been shown to be inhibitory to MMEJ in yeast by preventing annealing of single-stranded microhomologous sequences (39,40).
Several aspects of our analysis suggest that each side of the DSB acts independently during the initial stages of SD-MMEJ, when hairpins and loops form on either side of the break and prime synthesis in cis. Specifically, alterations in flanking sequences to the right of the I-SceI break for M1-M3 affect the usage of primer repeats on the right side but not the left, while changes in left-side flanking sequences for M4 and M5 have the opposite effect. However, at some point during repair, annealing of microhomologous sequences across the break must occur. If this happen early in the repair process, a trans SD-MMEJ process may occur. In this scenario, insertions result from annealing of ssDNA across the break, limited synthesis, dissociation, and reannealing at a newly created microhomology. These trans repair events have been previously reported in another study in mammals, suggesting that this type of repair is conserved (41,42).
While some of our junctions are unarguably created through a trans mechanism (Figure 7), all of the postulated loop-out junctions could also be explained by initial microhomology annealing in trans, although many of these would require annealing to internal microhomologies and non-homologous tail clipping. Furthermore, because multiple plasmids are injected into each embryo in our system, some of the postulated snap-back junctions could arise through an intermolecular trans-SD-MMEJ reaction. Thus, it will be informative to test the relative proportions of different SD-MMEJ mechanisms in a chromosomal context.
Does DNA polymerase theta drive SD-MMEJ?
In mammals, translesion DNA polymerase theta (Pol θ) promotes MMEJ and insertions during alt-EJ (43–46). Purified Pol θ polymerase domain, which crystallizes as a dimer, can also align microhomologous sequences and promote synthesis in MMEJ-like reactions in vitro (47,48). Pol θ possesses an N-terminal helicase-like domain with DNA-dependent ATPase activity and a C-terminal polymerase domain (49); the cooperation of these two domains could be extremely beneficial during SD-MMEJ repair.
Previous work from our lab showed that depletion of Drosophila Pol θ decreases both overall alt-EJ repair (50) and the percentage of SD-MMEJ consistent junctions following creation of a chromosomal DSB by I-SceI (25). Furthermore, the few indels recovered from flies expressing very low levels of Pol θ had little synthesis and were less complex than wild-type indels. It will be interesting to repeat these experiments in Pol θ domain-specific mutants to determine the extent to which SD-MMEJ depends on the different domains of Pol θ.
In the C. elegans germline, alt-EJ of Cas9-induced breaks is entirely dependent on Pol θ (51). Residual alt-EJ exists in Pol θ-deficient Drosophila and mammals, suggesting that the overall contribution of Pol θ to SD-MMEJ and alt-EJ may vary between different organisms (44,50). Interestingly, two recent investigations into the genetic requirements for random integration of exogenous DNA demonstrated that Pol θ was responsible for all C-NHEJ independent integration events in mammals (52,53). Furthermore, loss of human Pol θ abolished Cas9-induced DSB repair events involving microhomologies or templated insertions, strongly arguing that, in certain contexts, Pol θ is solely responsible for alt-EJ (52).
Implications of SD-MMEJ
Alt-EJ has been implicated in the formation of oncogenic chromosome translocations in a variety of human cancers, with many translocation junctions bearing sequence motifs characteristic of SD-MMEJ repair (54–58). Thus, SD-MMEJ, with its inherent mutagenic potential, could drive the formation of genome rearrangements that represent the first step towards cellular transformation. However, SD-MMEJ could also serve as a beneficial evolutionary mechanism allowing for the creation of novel SNPs and promoting the expansion and diversification of short sequence repeats. Such a mechanism was recently shown to operate at tandem repeats following creation of a DSB using TALENs in zebrafish (59). Similarly, SD-MMEJ repair could drive the expansion of small repeats in gene promoters, thereby creating a rapid mechanism by which to alter gene expression (60–62).
Finally, with the growing use of targetable nucleases, especially Cas9 and Cpf1, for gene editing purposes, a better understanding of how sequence context affects repair pathway choice is needed (63). Recognition that certain secondary-structure forming sequences may promote inaccurate SD-MMEJ repair is particularly important for researchers using site-specific nucleases to create gene-inactivating frameshifts and indels (64,65). Relevant to human genome editing, efficient SD-MMEJ/alt-EJ may inhibit processes involving gene modification by homologous recombination. Thus, our findings highlight the importance of considering flanking sequence context when choosing DSB sites for genome editing.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Connor Clairmont for making improvements to the SD-MMEJ consistency program, Erik Kastman and Benjamin Wolfe for assistance with statistics, Alexander Ferazzoli for sequencing help, Sergei Mirkin for scientific insight, and members of the McVey lab past and present for helpful discussions.
Footnotes
Present address: Amy Marie Yu, Department of Biology, University of Wisconsin-La Crosse, 1725 State Street, La Crosse, WI 54601, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [MCB-1716039 to M.M.]; National Institutes of Health [R01-GM092866 to M.M., R00-ES022633 to S.A.R.]; Department of Defense [Breast Cancer Breakthrough Award BC141727 to S.A.R.]. Funding for open access charge: National Science Foundation [MCB-1716039].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lieber M.R., Ma Y., Pannicke U., Schwarz K.. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003; 4:712–720. [DOI] [PubMed] [Google Scholar]
- 2. Betermier M., Bertrand P., Lopez B.S.. Is non-homologous end-joining really an inherently error-prone process. PLoS Genet. 2014; 10:e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lieber M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008; 283:1–5. [DOI] [PubMed] [Google Scholar]
- 4. Chiruvella K.K., Liang Z., Wilson T.E.. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 2013; 5:a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lieber M.R., Lu H., Gu J., Schwarz K.. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008; 18:125–133. [DOI] [PubMed] [Google Scholar]
- 6. Ma Y., Lu H., Schwarz K., Lieber M.R.. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005; 4:1193–1200. [DOI] [PubMed] [Google Scholar]
- 7. Rulten S.L., Grundy G.J.. Non-homologous end joining: common interaction sites and exchange of multiple factors in the DNA repair process. Bioessays. 2017; 39:doi:10.1002/bies.201600209. [DOI] [PubMed] [Google Scholar]
- 8. Menon V., Povirk L.F.. End-processing nucleases and phosphodiesterases: An elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair. DNA Repair (Amst). 2016; 43:57–68. [DOI] [PubMed] [Google Scholar]
- 9. Emerson C.H., Bertuch A.A.. Consider the workhorse: nonhomologous end-joining in budding yeast. Biochem. Cell Biol. 2016; 94:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams G.J., Hammel M., Radhakrishnan S.K., Ramsden D., Lees-Miller S.P., Tainer J.A.. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst.). 2014; 17:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ochi T., Wu Q., Blundell T.L.. The spatial organization of non-homologous end joining: from bridging to end joining. DNA Repair (Amst.). 2014; 17:98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grundy G.J., Moulding H.A., Caldecott K.W., Rulten S.L.. One ring to bring them all–the role of Ku in mammalian non-homologous end joining. DNA Repair (Amst.). 2014; 17:30–38. [DOI] [PubMed] [Google Scholar]
- 13. Pannunzio N.R., Li S., Watanabe G., Lieber M.R.. Non-homologous end joining often uses microhomology: implications for alternative end joining. DNA Repair (Amst.). 2014; 17:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iliakis G., Murmann T., Soni A.. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015; 793:166–175. [DOI] [PubMed] [Google Scholar]
- 15. Ceccaldi R., Rondinelli B., D’Andrea A.D.. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodgers K., McVey M.. Error-prone repair of DNA double-strand breaks. J. Cell Physiol. 2016; 231:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frit P., Barboule N., Yuan Y., Gomez D., Calsou P.. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst.). 2014; 17:81–97. [DOI] [PubMed] [Google Scholar]
- 18. Boulton S.J., Jackson S.P.. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996; 24:4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sfeir A., Symington L.S.. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 2015; 40:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Decottignies A. Alternative end-joining mechanisms: a historical perspective. Front. Genet. 2013; 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McVey M., Lee S.E.. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008; 24:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McVey M., Radut D., Sekelsky J.J.. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics. 2004; 168:2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Preston C.R., Flores C.C., Engels W.R.. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics. 2006; 172:1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei D.S., Rong Y.S.. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics. 2007; 177:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu A.M., McVey M.. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010; 38:5706–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sinha S., Villarreal D., Shim E.Y., Lee S.E.. Risky business: Microhomology-mediated end joining. Mutat Res. 2016; 788:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kostyrko K., Mermod N.. Assays for DNA double-strand break repair by microhomology-based end-joining repair mechanisms. Nucleic Acids Res. 2016; 44:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma J.L., Kim E.M., Haber J.E., Lee S.E.. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell Biol. 2003; 23:8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee K., Lee S.E.. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007; 176:2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Villarreal D.D., Lee K., Deem A., Shim E.Y., Malkova A., Lee S.E.. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012; 8:e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang L., Deng L., Chen Y., Li G.C., Shao C., Tischfield J.A.. Modulation of DNA end joining by nuclear proteins. J. Biol. Chem. 2005; 280:31442–31449. [DOI] [PubMed] [Google Scholar]
- 32. Sharma S., Javadekar S.M., Pandey M., Srivastava M., Kumari R., Raghavan S.C.. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015; 6:e1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li P., Li J., Li M., Dou K., Zhang M.J., Suo F., Du L.L.. Multiple end joining mechanisms repair a chromosomal DNA break in fission yeast. DNA Repair (Amst.). 2012; 11:120–130. [DOI] [PubMed] [Google Scholar]
- 34. van Overbeek M., Capurso D., Carter M.M., Thompson M.S., Frias E., Russ C., Reece-Hoyes J.S., Nye C., Gradia S., Vidal B. et al. . DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell. 2016; 63:633–646. [DOI] [PubMed] [Google Scholar]
- 35. Rong Y.S., Golic K.G.. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics. 2003; 165:1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fanning E., Klimovich V., Nager A.R.. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006; 34:4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wyka I.M., Dhar K., Binz S.K., Wold M.S.. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry. 2003; 42:12909–12918. [DOI] [PubMed] [Google Scholar]
- 38. Cai L., Roginskaya M., Qu Y., Yang Z., Xu Y., Zou Y.. Structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry. 2007; 46:8226–8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deng S.K., Chen H., Symington L.S.. Replication protein A prevents promiscuous annealing between short sequence homologies: Implications for genome integrity. Bioessays. 2015; 37:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng S.K., Gibb B., de Almeida M.J., Greene E.C., Symington L.S.. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014; 21:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Black S.J., Kashkina E., Kent T., Pomerantz R.T.. DNA polymerase theta: a unique multifunctional end-joining machine. Genes (Basel). 2016; 7:E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kent T., Mateos-Gomez P.A., Sfeir A., Pomerantz R.T.. Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife. 2016; 5:e13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wood R.D., Doublie S.. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair (Amst.). 2016; 44:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wyatt D.W., Feng W., Conlin M.P., Yousefzadeh M.J., Roberts S.A., Mieczkowski P., Wood R.D., Gupta G.P., Ramsden D.A.. Essential roles for polymerase theta-mediated end joining in the repair of chromosome breaks. Mol. Cell. 2016; 63:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yousefzadeh M.J., Wyatt D.W., Takata K., Mu Y., Hensley S.C., Tomida J., Bylund G.O., Doublie S., Johansson E., Ramsden D.A. et al. . Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014; 10:e1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Schendel R., van Heteren J., Welten R., Tijsterman M.. Genomic scars generated by polymerase theta reveal the versatile mechanism of alternative end-joining. PLoS Genet. 2016; 12:e1006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kent T., Chandramouly G., McDevitt S.M., Ozdemir A.Y., Pomerantz R.T.. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 2015; 22:230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zahn K.E., Averill A.M., Aller P., Wood R.D., Doublie S.. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015; 22:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seki M., Marini F., Wood R.D.. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003; 31:6117–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chan S.H., Yu A.M., McVey M.. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010; 6:e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Schendel R., Roerink S.F., Portegijs V., van den Heuvel S., Tijsterman M.. Polymerase Theta is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun. 2015; 6:7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saito S., Maeda R., Adachi N.. Dual loss of human POLQ and LIG4 abolishes random integration. Nat. Commun. 2017; 8:16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zelensky A.N., Schimmel J., Kool H., Kanaar R., Tijsterman M.. Inactivation of Pol theta and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 2017; 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Byrne M., Wray J., Reinert B., Wu Y., Nickoloff J., Lee S.H., Hromas R., Williamson E.. Mechanisms of oncogenic chromosomal translocations. Ann. N. Y. Acad. Sci. 2014; 1310:89–97. [DOI] [PubMed] [Google Scholar]
- 55. Soni A., Siemann M., Pantelias G.E., Iliakis G.. Marked contribution of alternative end-joining to chromosome-translocation-formation by stochastically induced DNA double-strand-breaks in G2-phase human cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015; 793:2–8. [DOI] [PubMed] [Google Scholar]
- 56. Hromas R., Williamson E., Lee S.H., Nickoloff J.. Preventing the Chromosomal Translocations That Cause Cancer. Trans. Am. Clin. Climatol. Assoc. 2016; 127:176–195. [PMC free article] [PubMed] [Google Scholar]
- 57. Nussenzweig A., Nussenzweig M.C.. A backup DNA repair pathway moves to the forefront. Cell. 2007; 131:223–225. [DOI] [PubMed] [Google Scholar]
- 58. Chiarle R., Zhang Y., Frock R.L., Lewis S.M., Molinie B., Ho Y.J., Myers D.R., Choi V.W., Compagno M., Malkin D.J. et al. . Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011; 147:107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang W., Zheng J., He Y., Luo C.. Tandem repeat modification during double-strand break repair induced by an engineered TAL effector nuclease in zebrafish genome. PLoS One. 2013; 8:e84176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abe H., Gemmell N.J.. Evolutionary footprints of short tandem repeats in avian promoters. Sci. Rep. 2016; 6:19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lang M., Juan E.. Binding site number variation and high-affinity binding consensus of Myb-SANT-like transcription factor Adf-1 in Drosophilidae. Nucleic Acids Res. 2010; 38:6404–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ohadi M., Mohammadparast S., Darvish H.. Evolutionary trend of exceptionally long human core promoter short tandem repeats. Gene. 2012; 507:61–67. [DOI] [PubMed] [Google Scholar]
- 63. Carroll D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 2014; 83:409–439. [DOI] [PubMed] [Google Scholar]
- 64. Singh V., Braddick D., Dhar P.K.. Exploring the potential of genome editing CRISPR-Cas9 technology. Gene. 2017; 599:1–18. [DOI] [PubMed] [Google Scholar]
- 65. Schiml S., Puchta H.. Revolutionizing plant biology: multiple ways of genome engineering by CRISPR/Cas. Plant Methods. 2016; 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.