Abstract
A bacterial translation factor EF-P alleviates ribosomal stalling caused by polyproline sequence by accelerating Pro-Pro formation. EF-P recognizes a specific D-arm motif found in tRNAPro isoacceptors, 9-nt D-loop closed by a stable D-stem sequence, for Pro-selective peptidyl-transfer acceleration. It is also known that the T-stem sequence on aminoacyl-tRNAs modulates strength of the interaction with EF-Tu, giving enhanced incorporation of non-proteinogenic amino acids such as some N-methyl amino acids. Based on the above knowledge, we logically engineered tRNA’s D-arm and T-stem sequences to investigate a series of tRNAs for the improvement of consecutive incorporation of d-amino acids and an α, α-disubstituted amino acid. We have devised a chimera of tRNAPro1 and tRNAGluE2, referred to as tRNAPro1E2, in which T-stem of tRNAGluE2 was engineered into tRNAPro1. The combination of EF-P with tRNAPro1E2NNN pre-charged with d-Phe, d-Ser, d-Ala, and/or d-Cys has drastically enhanced expression level of not only linear peptides but also a thioether-macrocyclic peptide consisting of the four consecutive d-amino acids over the previous method using orthogonal tRNAs.
INTRODUCTION
In nature, ribosomal translation system exclusively utilizes the 19 proteinogenic l-amino acids and glycine, i.e. their d-form counterparts are excluded from peptide/protein synthesis. However, various artificial methods have been developed for introducing non-proteinogenic amino acids, including d-amino acids, into peptides. For instance, genetic code reprogramming combined with a reconstituted in vitro translation system, such as the flexible in-vitro translation (FIT) system, is one of the most reliable methods for utilizing diverse non-proteinogenic substrates, including d-amino acids, N-methyl amino acids, β-amino acids, and α-hydroxy acids (1–9). Despite the fact that there had been several examples of successful sole incorporation of certain kinds of d-amino acids into peptide chain by such or similar methods (4,5,10–12), consecutive incorporation of d-amino acids had yet faced a major challenge, in contrast to N-methyl amino acids and α-hydroxy acids which were well introduced in a row (6,7,9,13,14).
Recently, we have reported for the first time the successful consecutive incorporation of d-amino acids in a customized FIT system where the respective concentrations of EF-Tu, IF2 and EF-G were optimized, and the tRNAGluE2 pre-charged with the d-amino acids prepared by flexizymes was used (5). In this system, it has been hypothesized that higher concentration of EF-Tu could contribute to enhancing the accommodation rate of d-aminoacyl-tRNA on to ribosome. Elevating the IF2 concentration would also accelerate the initiation event, and lowering the EF-G concentration would reduce undesired peptidyl-tRNA drop-off from the ribosome. tRNAGluE2 has higher binding affinity to EF-Tu than the previously used tRNAAsnE2, and thus accommodation rate of d-aminoacyl-tRNAGluE2 could be improved. By the use of this customized FIT system, translation yield of a peptide containing two d-Ala in a row was improved 5-fold compared to the conventional in-vitro translation (or our FIT) system.
On the other hand, all of the aforementioned refinements did not influence the acceleration of the peptidyl transfer (PT) reaction step, which could be the most critical determinant for the overall rate of peptide synthesis. Indeed, even though the observed enhancement was significant, the overall expression level of the peptide with two consecutive d-Ala was generally less than 0.3 μM, which is several fold less than those of all-l-form peptides (∼1.4 μM). To further improve the translation efficiency of peptides containing consecutive d-amino acids, the enhancement of the peptidyl transfer rate between two d-amino acids on the peptidyl-tRNA and the aminoacyl-tRNA would be crucial.
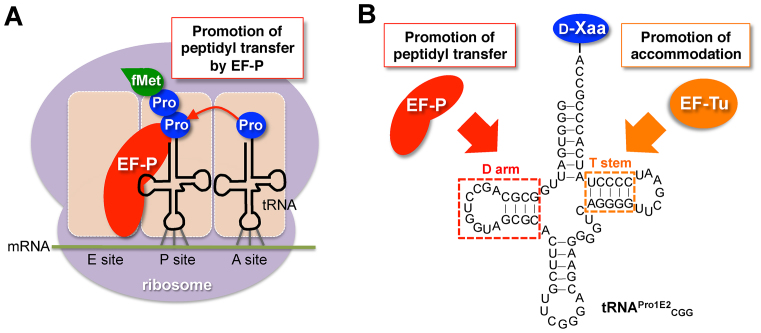
EF-P is a prokaryotic translation factor that accelerates peptide bond formation between two prolines (Pro, Figure 1A) (15–16). The rate of peptide bond formation between Pro and Pro is by far slowest without EF-P among those of the proteinogenic amino acids (16). Hence, EF-P plays a critical role in accelerating the elongation rate of Pro–Pro bond formation, preventing from the ribosomal stalling that tends to induce peptidyl-tRNA drop-off, i.e. premature termination, at the Pro-Pro sequences. We have recently reported (17) that EF-P recognizes the D-arm structure of P-site peptidyl-prolyl-tRNAPro, and thus the rate acceleration of Pro-Pro bond formation depends on tRNAPro. More specifically, the 9-nt (nucleotide) D-loop closed by 4-bp (base pair) stable D-stem is the critical determinant for the recognition of tRNAPro by EF-P. Since EF-P accelerates the inherently slow peptidyl-transfer rate of Pro-Pro, we have wondered if EF-P is able to accelerate other intrinsic slow PT rate, such as d-amino acids (Figure 1B).
Figure 1.
Schematic depiction of promotion of proline and d-amino acid incorporation mediated by EF-P. (A) Role of EF-P in acceleration of consecutive proline incorporation. EF-P binds to ribosome in between E site and P site, and interact with the P-site peptidyl-prolyl-tRNA to accelerate peptidyl transfer between the P-site peptidyl-prolyl-tRNA and the A-site prolyl-tRNA. (B) Construct of d-aminoacyl-tRNA with optimized D-arm and T-stem structures. A specific D-arm structure consisting of 9-nt D-loop closed by a stable D-stem sequence recruits EF-P on to the tRNA and promotes peptidyl transfer between d-amino acids. A specific T-stem structure improves EF-Tu-binding affinity and accommodation rate of the d-aminoacyl-tRNA on to the ribosomal A site.
Here we report that EF-P significantly enhances consecutive incorporation of d-amino acids charged onto tRNAPro1 derivatives, and the observed effect depends on the D-arm structure of the tRNAPro1 constructs. Then, we constructed a new engineered tRNA, referred to as tRNAPro1E2 (Figure 1B, tRNAPro1E2), that has a chimeric structure of the D-arm of tRNAPro1 and the T-stem of tRNAGluE2. Using the series of tRNAPro1E2 derivatives with different anticodons, we have shown ribosomal synthesis of two macrocyclic peptides consisting of four and five consecutive d-amino acids with as high as 9.5-fold improvement compared with the previously reported values using tRNAGluE2. Evidently, this demonstration provides a unique approach that the logical engineering of both D-arm and T-stem in elongator tRNAs have enabled us to enhance the overall PT rate assisted by EF-P and EF-Tu, leading to significant improvement of consecutive incorporation of d-amino acids.
MATERIALS AND METHODS
Preparation of flexizyme and tRNA
Flexizymes (dFx and eFx) and tRNAs used for pre-charging d-amino acids, 2-aminoisobutyric acid and l-proline were transcribed by T7 RNA polymerase in vitro. Template DNAs that have a T7 promoter prior to dFx or tRNA sequence were prepared by extension of forward and reverse extension primer pairs, followed by PCR using forward and reverse PCR primers (Supplementary Table S1). The PCR products were purified by phenol/chloroform extraction and ethanol precipitation, and then used for transcription at 37°C for 16 h in a 250 μl reaction mixture containing 40 mM Tris–HCl (pH 8.0), 22.5 mM MgCl2, 1 mM DTT, 1 mM spermidine, 0.01% Triton X-100, 120 nM T7 RNA polymerase, 0.04 U/μl RNasin RNase inhibitor (Promega) and 3.75 mM NTP mix. In the case of tRNA preparation, 5 mM GMP or CMP was added to the above solution depending on the nucleotide at the 5′-end (G or C). The resulting RNA transcripts were treated with RQ1 DNase (Promega) for 30 min at 37°C, and then purified by 12% (dFx and eFx) or 8% (tRNA) polyacrylamide gel containing 6 M urea.
Aminoacylation of tRNA
Activated amino acids (d-alanine 3,5-dinitrobenzyl ester (d-Ala-DBE), d-serine 3,5-dinitrobenzyl ester (d-Ser-DBE), d-cysteine 3,5-dinitrobenzyl ester (d-Cys-DBE), chloroacetyl d-phenylalanine cyanomethyl ester (d-ClAcF-CME), l-proline 3,5-dinitrobenzyl ester (l-Pro-DBE) and 2-aminoisobutyric acid 3,5-dinitrobenzyl ester (Aib-DBE)) were synthesized by previously reported methods (1,18). Aminoacylation was carried out at 0°C in reaction mixtures containing 50 mM HEPES–KOH (pH 7.5), 600 mM MgCl2, 20% DMSO, 25 μM dFx or eFx, 25 μM tRNA, and 5 mM activated amino acid. eFx was used for d-ClAcF-CME, and dFx for the other amino acids. Reaction time was 2 h for Ala, ClAcF, Pro and Aib or 6 h for Ser and Cys. The aminoacyl-tRNA was recovered by ethanol precipitation, and then the pellet was washed twice with 70% ethanol containing 0.1 M sodium acetate (pH 5.2), once with 70% ethanol and dissolved in 1 mM sodium acetate (pH 5.2).
Preparation of EF-P
Escherichia coli EF-P gene was cloned into a modified pET28a vector that has PreScission protease recognition site instead of thrombin site. E. coli epmA and epmB genes were cloned into pETDuet-1 vector. These vectors were cointroduced into E. coli Rosetta2 (DE3). The cells were cultured in LB medium with 0.5 mM IPTG for 2 h at 37°C, and lysed by sonication. The cell lysate was applied to a Ni-NTA column to purify the histidine-tagged EF-P. The column was washed with buffer A (20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 2 mM imidazole and 1 mM 2-mercaptoethanol), and then the histidine-tagged EF-P was eluted by buffer A containing 300 mM imidazole. Turbo3C protease was added to the eluate for cleaving the histidine-tag, and dialyzed against the buffer A at 4°C overnight. The sample was applied on the Ni-NTA column, and the flow-through and wash fractions were recovered as EF-P without histidine-tag. Then, the protein was concentrated by Amicon ultra 10k centrifugal filter (Merck Millipore). Modification of EF-P was confirmed by mass spectrometric analysis as previously described (17).
Translation of model peptides
Translation of the model peptides was carried out in the modified FIT (Flexible In-vitro Translation) system in which the concentrations of IF2, EF-G and EF-Tu/Ts were optimized for d-amino acid incorporation (3, 0.1 and 20 μM, respectively) (2,17). The composition of the modified FIT system is as follows; 50 mM HEPES-KOH (pH 7.6), 100 mM potassium acetate, 12.3 mM magnesium acetate, 2 mM ATP, 2 mM GTP, 1 mM CTP, 1 mM UTP, 20 mM creatine phosphate, 0.1 mM 10-formyl-5,6,7,8-tetrahydrofolic acid, 2 mM spermidine, 1 mM DTT, 1.5 mg/ml E. coli total tRNA, 1.2 μM E. coli ribosome, 0.6 μM methionyl-tRNA formyltransferase, 2.7 μM IF1, 3 μM IF2, 1.5 μM IF3, 0.1 μM EF-G, 20 μM EF-Tu/Ts, 0.25 μM RF2, 0.17 μM RF3, 0.5 μM RRF, 4 μg/ml creatine kinase, 3 μg/ml myokinase, 0.1 μM inorganic pyrophosphatase, 0.1 μM nucleotide diphosphate kinase, 0.1 μM T7 RNA polymerase, 0.13 μM AspRS, 0.11 μM LysRS, 0.03 μM MetRS, 0.02 μM TyrRS, 0.05 mM [14C]-aspartic acid (Asp), 0.5 mM lysine, 0.5 mM methionine, 0.5 mM tyrosine, 25 μM each pre-charged aminoacyl-tRNA and 0.5 μM DNA template. For translation of rP1 and rP2, 0.09 μM GlyRS and 0.5 mM glycine were also added to the above solution. Translation reactions were carried out in a 2.5-μl solution at 37°C, and stopped by adding an equal volume of stop solution (0.9 M Tris-HCl (pH 8.45), 8% SDS, 30% glycerol and 0.001% xylene cyanol) and incubating at 95°C for 2 min. Then, the samples were analyzed by 15% tricine SDS-PAGE and autoradiography using Typhoon FLA 7000 (GE Healthcare). Peptide yield was normalized by intensity of [14C]-Asp band.
For MALDI-TOF mass spectrometric analysis of peptides, 0.5 mM cold Asp was added to the above solution instead of [14C]-Asp. Translation was carried out at 37°C for 20 min, and then diluted with an equal volume of 2xHBS buffer (100 mM HEPES-KOH (pH 7.6), 300 mM NaCl). Then, the peptide was purified by ANTI-FLAG M2 affinity gel (Sigma). The reaction mixture was added to 5-μl gel beads and incubated for 30 min at 25°C. The gel beads were washed with 25 μl of 1xHBS buffer (50 mM HEPES-KOH (pH 7.6), 150 mM NaCl), and the peptide was eluted from the beads with 15 μl of 0.2% trifluoroacetic acid. Then, the peptide was desalted with SPE C-tip (Nikkyo Technos) and eluted with 1.2 μl of 80% acetonitrile, 0.5% acetic acid solution containing 50%-saturated (R)-cyano-4-hydroxycinnamic acid. MALDI-TOF-MS analysis was performed by using UltrafleXtreme (Bruker Daltonics) in reflector/positive mode. Peptide calibration standard II (Bruker Daltonics) was used for external mass calibration. For drift time ion mobility mass spectrometry, the above translation solution was directly analyzed without FLAG purification by SYNAPT G2-Si HDMS (Waters) using ACQUITY UPLC Peptide BEH300 C18 column (Waters).
RESULTS
Essential D-arm structure of tRNAPro1 recognized by EF-P for promotion of consecutive d-amino acid incorporation
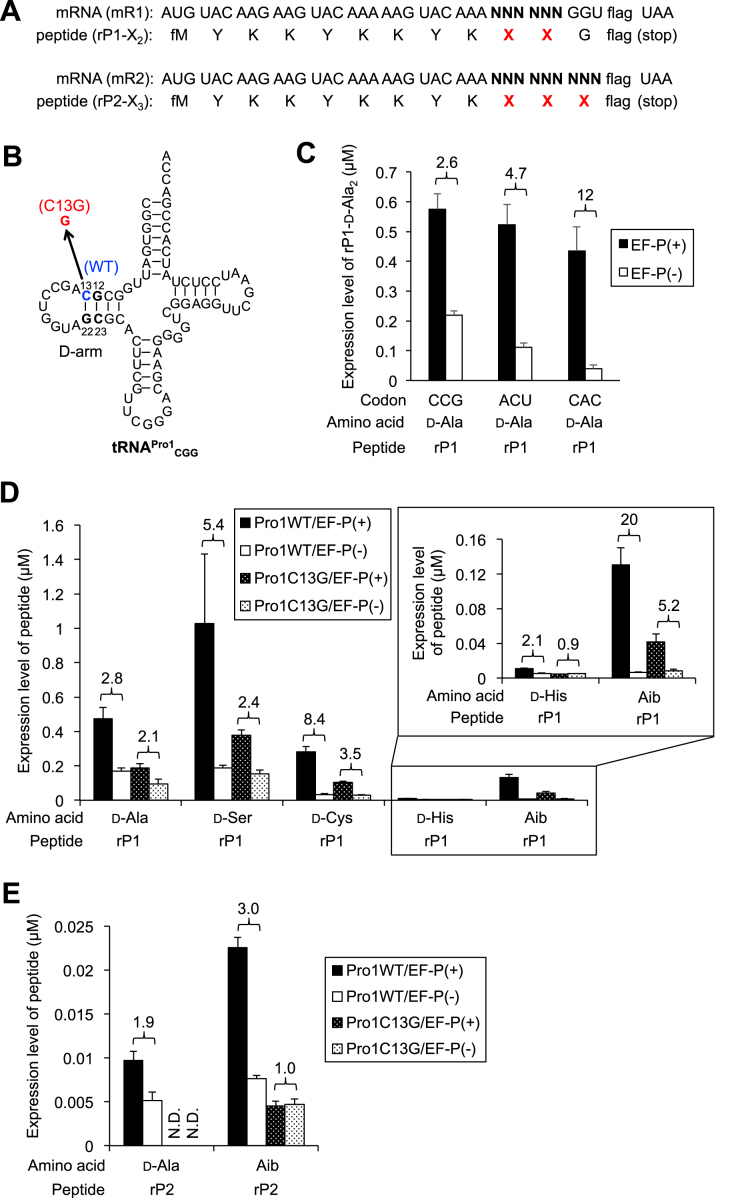
Based on our knowledge of EF-P’s capability of enhancing the slow PT rate in Pro-Pro elongation, we hypothesized that EF-P could accelerate slow consecutive incorporation of some d-amino acids into nascent peptide chain by using tRNAPro. To test this, we first prepared two mRNA constructs (mR1 and mR2) coding for two or three d-amino acids in a row, respectively (Figure 2A). d-amino acids were pre-charged on tRNAPro1 derivatives with corresponding anticodons by means of the flexizyme technology, and introduced into the flexible in-vitro translation (FIT) system containing 5 μM EF-P to synthesize rP1 or rP2 peptide. In this particular FIT system, [14C]-Asp, cold Met, Tyr, Lys, Gly and the corresponding ARSs are added, and the other amino acid/ARS pairs not coded by the mRNAs (mR1 and mR2) were omitted; instead, the d-aminoacyl-tRNA was added to assign the d-amino acid at NNN codon. Concentrations of IF2, EF-Tu and EF-G were set at 3, 20 and 0.1 μM, respectively, which are previously optimized values for consecutive incorporation of d-amino acids (5). Wild-type (WT) tRNAPro1CGG has a C/G base pair at positions 13 and 22 located at the edge of D-stem, whereas mutant tRNAPro1CGG has C-to-G point mutation at the position 13 to disrupt the base-pair formation (Figure 2B). Since EF-P strictly recognizes the 9-nt D-loop closed with 4-bp stable D-stem structure of the WT tRNA, the C13G mutation would diminish the function to EF-P. Thus, tRNAPro1 and the C13G mutant as a negative control were used for decoding the NNN codons that designated a d-amino acid in the mR1 and mR2. Accordingly, the anticodon of tRNAPro1 could be altered to others in order to decode the target codon (Supplementary Table S1).
Figure 2.
Promotion of d-amino acid incorporation by EF-P. (A) mRNA sequences (mR1 and mR2) and the corresponding peptide sequences (rP1 and rP2) used in this experiment. Arbitrarily chosen codons are introduced at the NNN, and used for incorporation of d-amino acids (X) using pre-charged d-aminoacyl-tRNAs. The amino acid sequence of flag is DYKDDDDK. (B) Structure of tRNA used for incorporation of d-amino acids at CCG codon. Wild-type tRNAPro1CGG (WT) has a C/G base pair at positions 13 and 22, whereas mutant tRNAPro1CGG (C13G) has a C-to-G mutation at the position 13 to break the base-pair formation. Sequence of anticodon loop can be arbitrarily changed for decoding different codons. See Supplementary Table S1 for the details. (C) Expression level of peptide rP1 in which two-consecutive d-Ala were introduced. Codons used for the d-Ala incorporation were indicated. Black bars indicate EF-P(+) translation, and white bars EF-P(–) translation. Numbers above the bars indicate relative translation yield calculated as the ratio of EF-P(+) to EF-P(-). Concentration of EF-P in the EF-P(+) translation is 5 μM. Translation time is 15 min. Error bars, S.D. (n = 3). (D, E) Expression levels of rP1 (D) and rP2 (E) containing two or three consecutive d-amino acids or Aib. Aib: 2-aminoisobutyric acid. Wild-type tRNAPro1CGG (plain bars) or the C13G mutant tRNAPro1CGG (dotted bars) were used for incorporation of the d-amino acids or Aib at CCG codons. Black bars indicate EF-P(+) translation, and white bars EF-P(–) translation. Numbers above the bars indicate the ratio of EF-P(+) to EF-P(–). Concentration of EF-P in the EF-P(+) translation is 5 μM. Translation time is 15 min. Error bars, S.D. (n = 3). Enlarged bar graph for incorporation of d-His and Aib is shown in upper right corner of Figure 2D.
We tested three arbitrary codons on mR1 (NNN = CCG, ACU and CAC) for introducing two consecutive d-Ala residues into rP1 (rP1-d-Ala2) using WT d-Ala-tRNAPro1 (Figure 2C, Supplementary Figure S1A). The resulting peptide was separated by tricine SDS-PAGE, and quantified by autoradiography. The addition of EF-P significantly improved the expression level of rP1-d-Ala2 regardless of codons in a similar range (0.4–0.6 μM), although the apparent level of enhancement differed depending on codons due to the difference in the background level of d-Ala incorporation without EF-P (2.6-, 4.7- and 12-fold for CCG, ACU and CAC, respectively). As a positive control, we also performed EF-P enhancement of consecutive l-Pro incorporation at the position of X residues in rP1-l-Pro2 and rP2-l-Pro3 peptides using l-Pro-tRNAPro1 (Supplementary Figure S1B, D, 1.4- and 22-fold improvements, respectively). The molecular weights of these peptides were also confirmed by MALDI-TOF mass spectrometric analysis, confirming the identity of respective peptides (Supplementary Figure S1F). As the molecular weight of rP1-d-Ala2 is identical to that of rP1-l-Ala2, mobility of the same m/z ions derived from respective peptides were analyzed by drift time ion mobility mass spectrometry to confirm that d-Ala could be correctly introduced (Supplementary Figure S1E). rP1-d-Ala2 and rP1-l-Ala2 exhibited identical m/z values (834.4075 and 834.4071, respectively) but different drift times (4.64 and 4.57 ms, respectively), showing their structural difference.
We then decided to study if EF-P was able to enhance d-Ser, d-Cys and d-His charged onto tRNAPro1CGG suppressing CCG codon on mR1 (Figure 2D, Supplementary Figure S1C). The enhancement effect turned out to be 5.4-, 8.4-, and 2.1-fold, indicating that EF-P was able to enhance two consecutive incorporation of these d-amino acids in a 2–10-fold range. On the other hand, the use of the C13G mutant of tRNAPro1CGG reduces the EF-P enhancement effect, giving 2.1-, 2.4-, 3.5- and 0.9-fold for d-Ala, d-Ser, d-Cys and d-His in rP1-d-X2. It should be noted that the absolute expression level was enhanced by more than 2-fold by the use of WT/EF-P over C13G/EF-P, and thus the EF-P effect was unmistakably observed in each case. Accurate expression of these peptides was confirmed by MALDI-TOF mass spectrometric analysis (Supplementary Figure S1F).
We also tested an achiral α, α-dimethyl substituted amino acid (Aib; 2-aminoisobutyric acid) (Figure 2D, Supplementary Figure S1C, F). Since the α,α-disubstitution sets a similar situation in the ribosome PT center like d-amino acid, we have observed very low efficiency of consecutive incorporation of Aib. Thus, we could expect a similar EF-P enhancement. In fact, the EF-P enhancement for the Aib incorporation into rP1-Aib2 was 20-fold by the use of WT tRNAPro1 compared with 5-fold by the C13G mutant. Again, the absolute expression level was enhanced more than 3-fold by the WT/EF-P combination.
We next examined three consecutive incorporations of d-Ala and Aib into the nascent chain of rP2 (Figure 2E, Supplementary Figure S1C, F). The EF-P enhancement of d-Ala into rP2-d-Ala3 was 1.9-fold using WT tRNAPro1CGG whereas the expression using the C13G mutant was undetectable level in both WT and mutant tRNAs. The EF-P enhancement was 3-fold when rP2-Aib3 was expressed using WT tRNAPro1CGG, whereas no effect was observed using the C13G mutant. The absolute expression level was more than 3-fold, indicating that EF-P is able to stimulate the overall expression level of successive incorporations of these amino acids.
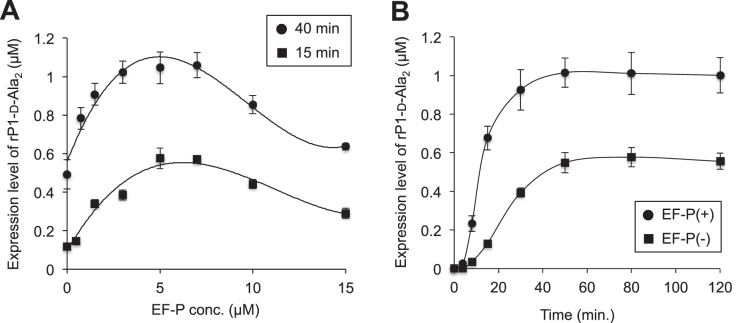
We also titrated the EF-P concentration as a function of the expression level of rP1-d-Ala2 in the presence of WT d-Ala-tRNAPro1CGG (Figure 3A). In the titration experiments at 40 and 15 min time points, 5–7 μM EF-P gave the maximal expression level of rP1-d-Ala2, whereas higher concentrations of EF-P (10 and 15 μM) declined the EF-P effect. This declination could be attribute to that an excess amount of EF-P remains to occupy the E site of ribosome even after the peptidyl transfer reaction completed, resulting in inhibition of the translocation of deacyl-tRNA from P site to E site. A similar trend was also observed in the introduction of consecutive l-Pro in our previous report (17). Considering that the concentration of ribosome in our translation system is 1.2 μM, the optimal concentration of EF-P is about 5-fold of the ribosome concentration. Time course analysis of translation of rP1-d-Ala2 was also carried out in the presence (5 μM) and absence of EF-P. In both conditions, the yield of full-length peptide plateaued at 50 min (Figure 3B). At 50 min, the expression level of rP1-d-Ala2 reached to approximately 1 μM in the presence of 5 μM EF-P, which is comparable to the expression level of a full-l-peptide containing l-Ala2 shown in our previous report (17).
Figure 3.
Titration of EF-P and time course analysis in translation of d-Ala-containing peptide. (A) Titration of EF-P concentration in translation of rP1 peptide containing two consecutive d-Ala at CCG codons. Translation time is 40 min (black circle) or 15 min (black square). Error bars, S.D. (n = 3). (B) Time course analysis of translation of rP1 peptide with two consecutive d-Ala. Black circle and black square indicate EF-P(+) and EF-P(–) translation, respectively. The concentration of EF-P in the EF-P(+) experiment is 5 μM. Error bars, S.D. (n = 3).
Engineering of the T-stem structure of tRNAPro1 further enhances consecutive d-alanine incorporation
It has been reported that the affinity of EF-Tu to l-aminoacyl-tRNA differs depending on the species of l-aminoacyl group and tRNA (19,20). For instance, Dale et al. have reported that ΔG° values of l-Val-tRNAAsn, l-Val-tRNAPro and l-Val-tRNAGlu are –8.8, –9.3 and –11.7 kcal/mol, respectively (20). It has been also reported that the affinity of misacylated Asp-tRNAAsn or Glu-tRNAGln to EF-Tu is lower than that of cognate Asn-tRNAAsn or Gln-tRNAGln, and thus cannot form a stable complex with EF-Tu (21,22). Since the affinities of naturally occurring l-aminoacyl-tRNAs to EF-Tu are tuned to a nearly uniform, they are equally maximized to the accommodation rate to the A site of ribosome (19,23). However, d-aminoacyl-tRNAs very likely possess poorer affinities to EF-Tu than the natural counterparts, and therefore the previously used tRNA species were not necessarily optimized for the accommodation rate. Based on this consideration, we recently devised a new tRNA elongator based on the sequence of tRNAGlu, referred to as tRNAGluE2, whose affinity to EF-Tu increased (5). This tRNA engineering indeed improved the incorporation of d-amino acid and made possible to incorporate d-amino acids successively, which had never been achieved using d-aminoacyl-tRNAAsnE2 in the FIT system previously used (4,5).
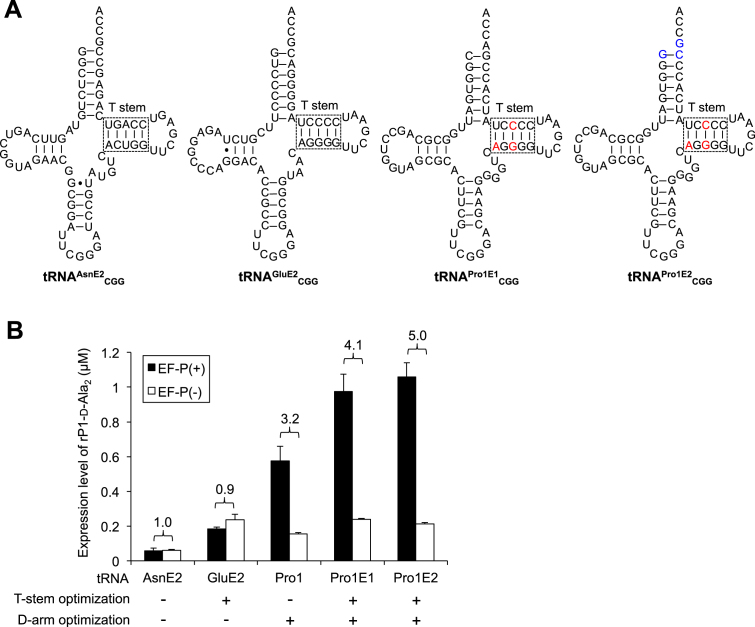
Although tRNAGluE2 had been engineered to be a better affinity to EF-Tu, its D-arm sequence would not be suitable for the EF-P recognition. Because the critical recognition element of EF-Tu to tRNAGluE2 mainly exists in the T-stem structure, we implanted the tRNAGluE2 T-stem into tRNAPro1, creating a chimeric tRNA, referred to as tRNAPro1E1 (Figure 4A, tRNAPro1E1CGG). We also designed another chimeric tRNA, referred to as tRNAPro1E2CGG, containing additional mutations at the discriminator base that disrupted the recognition by ProRS (24). By introducing this mutation, tRNAPro1E2CGG could not be charged with Pro by ProRS, i.e., it is an orthogonal tRNA to ProRS. The WT tRNAPro1CGG, tRNAPro1E1CGG, tRNAPro1E2CGG, tRNAGluE2CGG and tRNAAsnE2CGG (Figure 4A) were used to compare the incorporation efficiency of two consecutive d-Ala into the nascent chain of rP1-d-Ala2 (Figure 4B, Supplementary Figure S2A). Consequently, more than 4-fold enhancement of the peptide expression level was observed by the combination use of EF-P with tRNAPro1E1CGG (4.1-fold) and tRNAPro1E2CGG (5.0-fold). On the other hand, tRNAGluE2CGG and tRNAAsnE2CGG showed no enhancement by addition of EF-P, indicating the importance of the D-arm structure for EF-P binding. To further verify this, more extensive comparison of tRNAPro1E2 and tRNAGluE2 was conducted by introducing d-Ala and d-Phe into peptides rP1-d-Ala2 and rP1-d-Phe2 at UAG, ACU or CCG codon (Supplementary Figure S2B, D, E). Combination of EF-P and tRNAPro1E2 showed 3.3-, 4.4- and 4.3-fold improvement in d-Ala incorporation at UAG, ACU and CCG codons, respectively, and 3.9-fold improvement in d-Phe incorporation at UAG, whereas no significant enhancement was observed in any cases by tRNAGluE2 (0.8–1.1-fold improvement). These results clearly indicate that the chimeric tRNAPro1E2 originating from T-stem of tRNAGluE2 and D-arm of tRNAPro1 is able to enhance the overall PT rate assisted by EF-P and EF-Tu. This has led us perform the next series of experiments to test multiple kinds of d-amino acids for the consecutive incorporation.
Figure 4.
Effect of tRNA structure on the EF-P-dependent acceleration of d-amino acid incorporation. (A) Structure of tRNAs used for d-amino acid incorporation. Red and blue letters in tRNAPro1E1CGG and tRNAPro1E2CGG indicate alteration from the wild-type tRNAPro1CGG sequence. (B) Expression level of peptide rP1 containing two consecutive d-Ala at CCG codons. tRNA used for d-Ala incorporation were indicated. Numbers above the bars indicate the ratio of EF-P(+) to EF-P(–). Translation time is 15 min. Error bars, S.D. (n = 3).
Incorporation of multiple kinds of d-amino acids enhanced by EF-P
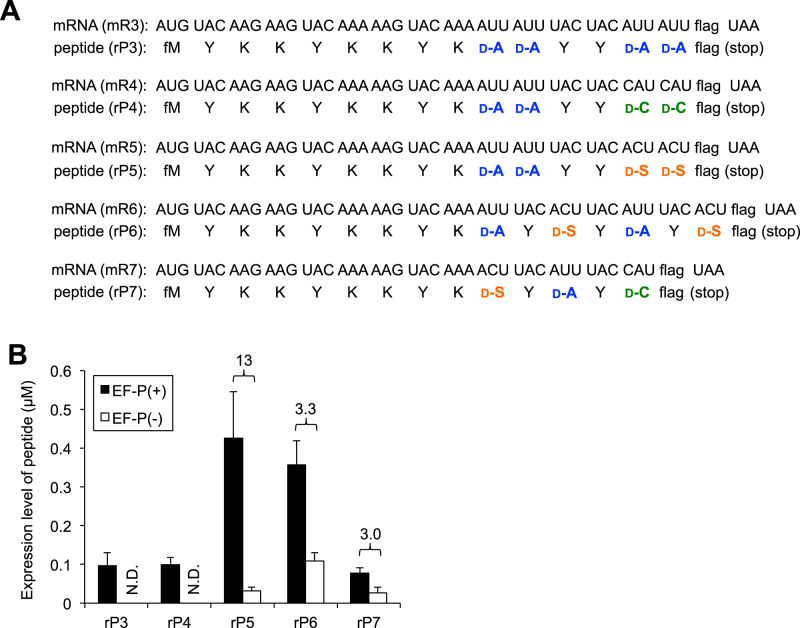
Since the combination of d-Ala-tRNAPro1E2 and EF-P was able to elevate the peptide expression level of rP1-d-Ala2, we extended this concept to the incorporation of consecutive multiple kinds of d-amino acids. We prepared five different types of template mRNAs (Figure 5A, mR3–7) to synthesize corresponding peptides that contain one, two or three different kinds of d-amino acids in their sequences (Figure 5A, rP3–7). In these mRNAs, d-Ala-tRNAPro1E2GAU, d-Cys-tRNAPro1E2GUG and d-Ser-tRNAPro1E2GGU were assigned to AUU, CAU and ACU codons, respectively. These codons are arbitrarily chosen as typical examples. In the presence of 5 μM EF-P an intense band in the respective rP3 and rP4 expression was observed (Supplementary Figure S2C, rP3 and rP4), whereas in its absence no band is detected. Since the rP3 and rP4 contained double sections of two-consecutive d-amino acids (d-Ala2-YY-d-Ala2 or d-Ala2-YY-d-Cys2; Y = l-Tyr), EF-P largely elevated their expression level from the undetectable background level. In the case of rP5 containing double sections of two consecutive d-amino acids (d-Ala2-YY-d-Ser2), the absence of EF-P was able to produce a detectable level of rP5, but the addition of EF-P gave a remarkable 13-fold enhancement (Figure 5B, rP5, Supplementary Figure S2C, rP5). The expression of rP6 and rP7 consisting alternate sequences of l- and d-amino acids was low but yet detectable level in the absence of EF-P; however, the addition of EF-P was able to enhance the expression level by 3.3- and 3.0-fold, respectively (Figure 5B, rP6 and rP7, Supplementary Figure S2C, rP6 and rP7). The MALDI-TOF MS analysis of the respective peptides showed expected molecular masses in the presence of EF-P (Supplementary Figure S2F, rP6 and rP7; note that rP7 was mainly deteced as a β-mercaptoethanol adduct to the d-Cys residue). These results indicate that the combination of EF-P and d-aminoacyl-tRNAPro1E2 gives remarkable impact on increasing the expression level of peptides containing multiple kinds of d-amino acids.
Figure 5.
d-amino acid incorporation into various peptide sequences promoted by EF-P. (A) mRNA sequence (mR3, mR4, mR5, mR6 and mR7) and the corresponding peptide sequence (rP3, rP4, rP5, rP6 and rP7). (B) Expression levels of peptides rP3, rP4, rP5, rP6 and rP7. tRNAPro1E2 variants were used for incorporation of d-amino acids. Numbers above the bars indicate the ratio of EF-P(+) to EF-P(–). Translation time is 15 min. Error bars, S.D. (n = 3).
Ribosomal synthesis of macrocyclic d-peptides stimulated by EF-P
We have recently demonstrated expression of macrocyclic d-peptides using the tRNAGluE2 derivatives using the FIT system, which was not previously possible at all. However, the expression level of the macrocyclic d-peptides was yet 4-fold lower than the macrocyclic l-peptides. We envisioned that the newly developed elongators based on tRNAPro1E2 in this work could improve the expression level for macrocyclic d-peptides. Therefore, we set experiments to see how much EF-P is further able to elevate the expression level of two model d-peptides, rP8 and rP9 (Figure 6A), accessorized with l-amino acid residues facilitating the MALDI-TOF mass spectrum detection.
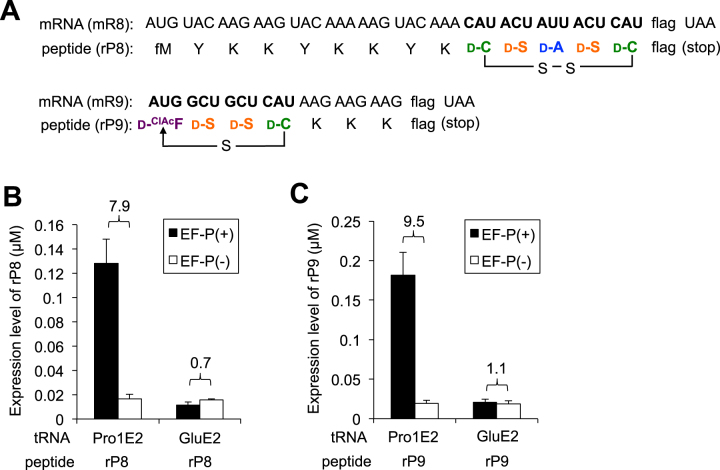
Figure 6.
Translation of macrocyclic d-peptides closed by disulfide or thioether bond. (A) mRNA sequence (mR8 and mR9) and the corresponding peptide sequence (rP8 and rP9). Two d-Cys residues included in the rP8 form a disulfide bond to give a macrocyclic structure. The sulfhydryl group of the d-Cys in rP9 attacks the α-carbon of the N-terminal ClAc group to form a thioether bond and give a macrocyclic structure. (B, C) Expression levels of peptides rP8 (B) and rP9 (C). tRNAPro1E2 or tRNAGluE2 was used for incorporation of d-amino acids. Numbers above the bars indicate the ratio of EF-P(+) to EF-P(–). Translation time is 15 min. Error bars, S.D. (n = 3).
The rP8 consists of d-Cys-d-Ser-d-Ala-d-Ser-d-Cys, which was previously introduced using the respective d-aminoacyl-tRNAGluE2 (Figure 6A, rP8) (5). MALDI-TOF analysis of the expressed rP8 peptide using tRNAPro1E2 with EF-P shows that these d-amino acids were correctly introduced and the two d-Cys residues formed a disulfide bond to give the macrocyclic structure (Supplementary Figure S3C, rP8). The tricine-SDS-PAGE analysis of rP8 revealed that EF-P paired with d-aminoacyl-tRNAPro1E2 was able to enhance nearly 8-fold of the expression level compared with that expressed using d-aminoacyl-tRNAGluE2, which showed no EF-P enhancement due to the lack of the optimal D-arm motif (Figure 6B, Supplementary Figure S3A). In a similar manner, we have designed another model d-peptide, rP9 consisting of a d-ClAcF-d-Ser-d-Ser-d-Cys continuous stretch, of which the N-terminal chloroacetyl group of d-ClAcF spontaneously reacts with the sulfhydryl group of d-Cys to yield thioether macrocyclic scaffold (Figure 6A, rP9). Previously, we analyzed the mobility of rP9 as well as mixed d/l-rP9 peptides containing l-Phe, l-Ser or l-Cys by tricine SDS-PAGE (5), in which the full-d-rP9 peptide showed a unique mobility compared with the mixed d/l-rP9 peptides, indicating that our translation system can synthesize pure full-d-rP9 without contamination of l-Phe, l-Ser nor l-Cys. Again, expression of rP9 was enhanced by nearly 10-fold by EF-P paired with the d-aminoacyl-tRNAPro1E2 over the EF-P-inactive tRNAGluE2 counterparts (Figure 6C), indicating that the thioether-macrocyclic d-peptide could be efficiently produced (Supplementary Figure S3C, rP9).
DISCUSSION
Among the twenty proteinogenic amino acids, l-Pro is the only cyclic secondary amino acid that exhibits the lowest reactivity in the peptide bond formation both as an acceptor and as a donor of nascent peptide chain (25–29). In a recent report that analyzed the structure of ribosome complex with an A-site l-Pro-tRNA and a P-site l-Pro-Pro-tRNA, unfavorable orientation of the α-amino group of the A-site l-Pro-tRNA as well as a bent l-Pro-Pro peptide in the P site were observed, both of which cause inefficient peptidyl transfer reaction (29). Thus, incorporation of two or more consecutive l-Pro residues is unignorably slow, and causes ribosomal stalling. In nature, EF-P alleviates ribosomal stalling caused by such poly-l-Pro stretch by accelerating the PT reaction. We previously reported that EF-P recognizes the conserved D-arm structure of tRNAPro isoacceptors for l-Pro-specific acceleration (17). We and Rodnina's group also reported that incorporation of some Pro-like cyclic secondary amino acids, which were charged onto tRNAPro, were accelerated by EF-P (17,26). Considering that consecutive incorporation of d-amino acids is also extremely inefficient like consecutive l-Pro incorporation, we hypothesized that EF-P might be able to stimulate consecutive incorporation of d-amino acids if appropriately designed tRNA was used. Here we have shown that EF-P is able to promote the incorporation of d-amino acids and an α, α-disubstituted amino acid charged on tRNAPro depending on the conserved D-arm motif (Figure 2D, E, Supplementary Figure S1C), which indicates that EF-P recognizes the P-site peptidyl-d-aminoacyl-tRNA in a similar manner to the recognition of peptidyl-l-Pro-tRNA.
In the case of l-Pro incorporation, EF-P presumably provides a better orientation of the P-site peptidyl-l-Pro, i.e. the catalysis is in an entropic way rather than enthalpic (26). Since the high resolution structure regarding the conformations of d-aminoacyl- and peptidyl-tRNA in the ribosome PTase center was yet unavailable, the mechanism how the peptidyl transfer between d-amino acids is accelerated by EF-P remains to be elucidated. Structural analysis using a ribosome with peptidyl-d-aminoacyl-tRNA on the P site and d-aminoacyl-tRNA on the A site would be helpful for further understanding of the mechanism. Rapid kinetic analysis of peptide bond formation between two d-amino acids should also be required to understand the function of EF-P along with the chimeric tRNA. It would be also worth evaluating the effect of EF-P on incorporation of other kinds of difficult non-proteinogenic amino acids such as β-amino acids and N-methyl amino acids. New insights obtained from such analyses would help to further improve the EF-P-mediated PT activation in incorporation of d- and other kinds of amino acids.
We have also previously reported that tRNAGluE2 variants that have an optimal T-stem motif with higher EF-Tu binding affinity improved accommodation of d-aminoacyl-tRNA and overall expression levels of peptides containing d-amino-acids (5). Considering this result, we logically constructed the chimeric tRNAs implanted with both of the optimal D-arm and T-stem motifs, referred to as tRNAPro1E1 and tRNAPro1E2. These tRNAs successfully acquired EF-P-dependent PT promotion as well as improvement of EF-Tu-dependent accommodation of d-aminoacyl-tRNA (Figure 4, Supplementary Figure S2A, B, D, E). On the other hand, tRNAGluE2 exhibited no EF-P enhancement, showing the importance of the conserved D-arm motif derived from the tRNAPro isoacceptors for EF-P binding. Recently, Huang et al. posted a preliminary result that EF-P enhanced consecutive d-amino acid incorporation by using tRNAGluE2 (doi: https://doi.org/10.1101/125930, as a non-peer-reviewed preprint), in which amber suppressor tRNAGluE2 was used for incorporation of d-Phe at UAG codon. However, as demonstrated in our previous work, the D-arm engineering is essential for the observation of EF-P enhancement (17), and therefore the result reported by Huang et al. was inconsistent with our knowledge. We thus added an additional experiment using d-Phe, which is used in their report, to see if EF-P was able to enhance consecutive elongation of d-Phe charged onto tRNAPro1E2 or tRNAGluE2 suppressing UAG codon (Supplementary Figure S2B, D, E). The result was clear that the EF-P enhancement with 3.9-fold was observed only for d-Phe-tRNAPro1E2, whereas no EF-P enhancement was observed for d-Phe-tRNAGluE2. This result was indeed consistent with the EF-P enhancement observed for d-Ala-tRNAPro1E2 suppressing UAG, ACU and CCG codons. Thus, at least in our hands, we were unable to reproduce the fundamental outcome observed by Huang et al.
Macrocyclization of peptidic scaffold often grants not only peptidase resistance but also higher affinity to binding partner proteins, thus giving more potential as drug leads compared with linear peptides (30). The RaPID (Random nonstandard Peptides Integrated Discovery) system where expression of mass libraries of nonstandard peptides in the FIT system was integrated with mRNA display, enables us to rapidly screen potent macrocyclic peptides containing multiple nonproteinogenic amino acids (31,32). Even though we were able to express macrocyclic peptides containing a single d-amino acid as an initiator and some N-methyl amino acids as elongators, it was still difficult for us to express a peptide library containing multiple d-amino acids as elongators due to their poor incorporation efficiency. However, such macrocyclic d-peptides have a higher potential as drug leads than the l-peptides due to their peptidase resistance. We here have achieved nearly 10-fold improvement in translation of macrocyclic peptides consisting of more than four consecutive d-amino acids closed by a thioether bond, indicating that our translation system coupled with tRNAPro1E2 and EF-P is an ideal platform for constructing a macrocyclic peptide library containing multiple d-amino acids (Figure 6). Integration of such d-peptide library with RaPID system will enable efficient screening of peptide ligands against various therapeutic targets.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nihon Waters K.K. for analysis of peptides by drift time ion mobility mass spectrometry.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Science and Technology Agency (JST) PRESTO of Molecular Technology and Creation of New Functions [JPMJPR14K3]; JST CREST Rising Star Award of Molecular Technology (to T.K.); Grants-in-Aid for JSPS Fellows [26-9576 to Y.I.]; JST CREST of Molecular Technologies [JPMJCR12L2 to H.S.]. Funding for open access charge: JST PRESTO of Molecular Technology and Creation of New Functions [JPMJPR14K3].
Conflict of interest statement. None declared.
REFERENCES
- 1. Murakami H., Ohta A., Ashigai H., Suga H.. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods. 2006; 3:357–359. [DOI] [PubMed] [Google Scholar]
- 2. Goto Y., Katoh T., Suga H.. Flexizymes for genetic code reprogramming. Nat. Protoc. 2011; 6:779–790. [DOI] [PubMed] [Google Scholar]
- 3. Goto Y., Murakami H., Suga H.. Initiating translation with D-amino acids. RNA. 2008; 14:1390–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fujino T., Goto Y., Suga H., Murakami H.. Reevaluation of the D-amino acid compatibility with the elongation event in translation. J. Am. Chem. Soc. 2013; 135:1830–1837. [DOI] [PubMed] [Google Scholar]
- 5. Katoh T., Tajima K., Suga H.. Consecutive elongation of D-amino acids in translation. Cell Chem. Biol. 2017; 24:1–9. [DOI] [PubMed] [Google Scholar]
- 6. Kawakami T., Murakami H., Suga H.. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem. Biol. 2008; 15:32–42. [DOI] [PubMed] [Google Scholar]
- 7. Kawakami T., Ishizawa T., Murakami H.. Extensive reprogramming of the genetic code for genetically encoded synthesis of highly N-alkylated polycyclic peptidomimetics. J. Am. Chem. Soc. 2013; 135:12297–12304. [DOI] [PubMed] [Google Scholar]
- 8. Fujino T., Goto Y., Suga H., Murakami H.. Ribosomal synthesis of peptides with multiple β-amino acids. J. Am. Chem. Soc. 2016; 138:1962–1969. [DOI] [PubMed] [Google Scholar]
- 9. Ohta A., Murakami H., Higashimura E., Suga H.. Synthesis of polyester by means of genetic code reprogramming. Chem. Biol. 2007; 14:1315–1322. [DOI] [PubMed] [Google Scholar]
- 10. Dedkova L.M., Fahmi N.E., Golovine S.Y., Hecht S.M.. Enhanced D-amino acid incorporation into protein by modified ribosomes. J. Am. Chem. Soc. 2003; 125:6616–6617. [DOI] [PubMed] [Google Scholar]
- 11. Dedkova L.M., Fahmi N.E., Golovine S.Y., Hecht S.M.. Construction of modified ribosomes for incorporation of D-amino acids into proteins. Biochemistry. 2006; 45:15541–15551. [DOI] [PubMed] [Google Scholar]
- 12. Achenbach J., Jahnz M., Bethge L., Paal K., Jung M., Schuster M., Albrecht R., Jarosch F., Nierhaus K.H., Klussmann S.. Outwitting EF-Tu and the ribosome: translation with D-amino acids. Nucleic Acids Res. 2015; 43:5687–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Subtelny A.O., Hartman M.C., Szostak J.W.. Ribosomal synthesis of N-methyl peptides. J. Am. Chem. Soc. 2008; 130:6131–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamagishi Y., Shoji I., Miyagawa S., Kawakami T., Katoh T., Goto Y., Suga H.. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem. Biol. 2011; 18:1562–1570. [DOI] [PubMed] [Google Scholar]
- 15. Ude S., Lassak J., Starosta A., Kraxenberger T., Wilson D., Jung K.. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013; 339:82–85. [DOI] [PubMed] [Google Scholar]
- 16. Doerfel L.K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M.V.. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013; 339:85–88. [DOI] [PubMed] [Google Scholar]
- 17. Katoh T., Wohlgemuth I., Nagano M., Rodnina M.V., Suga H.. Essential structural elements in tRNAPro for EF-P-mediated alleviation of translation stalling. Nat. Commun. 2016; 7:11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saito H., Kourouklis D., Suga H.. An in vitro evolved precursor tRNA with aminoacylation activity. EMBO J. 2001; 20:1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. LaRiviere F.J., Wolfson A.D., Uhlenbeck O.C.. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001; 294:165–168. [DOI] [PubMed] [Google Scholar]
- 20. Dale T., Sanderson L.E., Uhlenbeck O.C.. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry. 2004; 43:6159–6166. [DOI] [PubMed] [Google Scholar]
- 21. Becker H.D., Kern D.. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12832–12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stanzel M., Schon A., Sprinzl M.. Discrimination against misacylated tRNA by chloroplast elongation factor Tu. Eur. J. Biochem. 1994; 219:435–439. [DOI] [PubMed] [Google Scholar]
- 23. Schrader J.M., Chapman S.J., Uhlenbeck O.C.. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hasegawa T., Yokogawa T.. Escherichia coli proline tRNA: structure and recognition sites for prolyl-tRNA synthetase. Nucleic Acids Symp. Ser. 2000; 44:7–8. [DOI] [PubMed] [Google Scholar]
- 25. Pavlov M.Y., Watts R.E., Tan Z., Cornish V.W., Ehrenberg M., Forster A.C.. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doerfel L.K., Wohlgemuth I., Kubyshkin V., Starosta A.L., Wilson D.N., Budisa N., Rodnina M.V.. Entropic Contribution of Elongation Factor P to Proline Positioning at the Catalytic Center of the Ribosome. J. Am. Chem. Soc. 2015; 137:12997–13006. [DOI] [PubMed] [Google Scholar]
- 27. Wohlgemuth I., Brenner S., Beringer M., Rodnina M.V.. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 2008; 283:32229–32235. [DOI] [PubMed] [Google Scholar]
- 28. Johansson M., Ieong K.W., Trobro S., Strazewski P., Aqvist J., Pavlov M.Y., Ehrenberg M.. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Melnikov S., Mailliot J., Rigger L., Neuner S., Shin B.S., Yusupova G., Dever T.E., Micura R., Yusupov M.. Molecular insights into protein synthesis with proline residues. EMBO Rep. 2016; 17:1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Driggers E.M., Hale S.P., Lee J., Terrett N.K.. The exploration of macrocycles for drug discovery–an underexploited structural class. Nat. Rev. Drug Discov. 2008; 7:608–624. [DOI] [PubMed] [Google Scholar]
- 31. Hipolito C.J., Suga H.. Ribosomal production and in vitro selection of natural product-like peptidomimetics: the FIT and RaPID systems. Curr. Opin. Chem. Biol. 2012; 16:196–203. [DOI] [PubMed] [Google Scholar]
- 32. Passioura T., Katoh T., Goto Y., Suga H.. Selection-based discovery of druglike macrocyclic peptides. Annu. Rev. Biochem. 2014; 83:727–752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.