Abstract
Recent evidence indicates a link between Parkinson's Disease (PD) and the expression of a-synuclein (SNCA) isoforms with different 3′ untranslated regions (3′UTRs). Yet, the post-transcriptional mechanisms regulating SNCA expression are unknown. Using a large-scale in vitro /in silico screening we identified RNA-binding proteins (RBPs) that interact with SNCA 3′ UTRs. We identified two RBPs, ELAVL1 and TIAR, that bind with high affinity to the most abundant and translationally active 3′ UTR isoform (575 nt). Knockdown and overexpression experiments indicate that both ELAVL1 and TIAR positively regulate endogenous SNCA in vivo. The mechanism of regulation implies mRNA stabilization as well as enhancement of translation in the case of TIAR. We observed significant alteration of both TIAR and ELAVL1 expression in motor cortex of post-mortem brain donors and primary cultured fibroblast from patients affected by PD and Multiple System Atrophy (MSA). Moreover, trans expression quantitative trait loci (trans-eQTLs) analysis revealed that a group of single nucleotide polymorphisms (SNPs) in TIAR genomic locus influences SNCA expression in two different brain areas, nucleus accumbens and hippocampus. Our study sheds light on the 3′ UTR-mediated regulation of SNCA and its link with PD pathogenesis, thus opening up new avenues for investigation of post-transcriptional mechanisms in neurodegeneration.
INTRODUCTION
Parkinson's disease (PD) is the second most common human neurodegenerative disorder, after Alzheimer disease. PD is a multifactorial disorder in which different factors such as aging, genetic susceptibility and environmental insults converge to cause neurodegeneration. Idiopathic PD represents over 90% of PD cases, while genetic PD, caused by mutations in one or more of the PD-associated loci, is only 10% of the cases (1).
PD initiates in the central nervous system and spreads to the peripheral and enteric parts of the nervous system. The most important feature in the brains of PD patients is the selective loss of dopaminergic pigmented neurons within the substantia nigra and, to a lesser extent, neurons residing in the ventral tegmental and retrorubral areas (2). The neuropathological hallmark of Parkinson's disease is the presence of eosinophilic inclusion in the soma of neurons known as Lewy bodies (LBs), as well as in the neurites where the inclusions are called Lewy neurites. LBs are composed of a mixture of lipids, neuromelanin and up to several hundred individual proteins, including ubiquitin, heat-shock proteins, dj-1, sod1 and 2, synphilin-1, tau, tyrosine hydroxylase, and many others, but the key component is α-synuclein (SNCA gene), a small protein enriched at the presynaptic terminals (3).
The post-transcriptional mechanisms controlling SNCA expression are at present unknown, although we previously observed that UTR-mediated regulation could play a key role in controlling α-synuclein protein abundance (4). Specific SNCA transcript isoforms with different 3′UTR lengths have been found enriched in cerebral cortex samples of post-mortem PD patients (5). Recently, in a cohort of 202 cases of de novo motor PD, significantly lower levels of SNCA mRNA with extended 3′UTRs have been quantified by digital expression analysis (6). These findings are particularly relevant to PD etiopathogenesis: a switch of alternative polyadenylation to favour expression of specific SNCA 3′UTR isoforms enables the binding of a number of trans-acting factors that alter protein production, localisation and function.
Post-transcriptional networks induce changes in expression levels that are about one order of magnitude smaller than those caused by transcription factors, but α-synuclein is highly concentrated at the pre-synaptic terminals of neurons (70–140 uM) and even small fluctuations of its concentration can indeed induce aggregation (7). In the present work we address the question of which RNA-binding proteins (RBPs) are able to bind the different 3′UTRs of SNCA mRNA and control its expression. Following the unbiased discovery of protein interactors by means of a large-scale in vitro / in silico screening, we prioritised two RBPs, ELAVL1 and TIAR, that target SNCA 3′UTRs and their influence stability and translation efficiency. Our study suggests that TIAR and ELAVL1 are key regulators of α-synuclein intracellular concentration and function.
MATERIALS AND METHODS
Human protein array
The RNA sequence corresponding to SNCA 3′UTR short (575 bp), medium (1.07 kb) and long (2.5 kb) were cloned in pBluescript-SK(+) empty plasmid. The RNA probes of the SNCA long 3′UTR (3′UTRL) were synthesized by in vitro transcription (IVT) with T7 and T3 RNA polymerase (Agilent) for the sense and antisense strand respectively, and fluorescently labeled with Label IT μArray Cy5 labeling kit (Mirus) applying minor modifications to manufacturer's instructions. RNA concentration and labeling density were measured with Nanodrop 1000 spectrophotometer (Thermo Scientific). RNA integrity was verified by Agilent 2100 Bioanalyzer. The two probes corresponding to the SNCA 3′UTR long sense and antisense sequence were loaded on a 1% agarose denaturing gel (2.2 M formaldehyde). Both the size and the integrity of the probes were checked.
ProtoArray® Human Protein Microarrays v5.2 (Life Technologies), containing 9546 spotted proteins, were probed with the Cy5 labeled RNA of interest as previously reported (8). The dry slides were scanned within 2 h of the completion of the hybridization using a GenePix 4000B Microarray scanner (Molecular Devices). The intensity of the signal at 635 nm wavelength at each spotted protein location was determined with GenePix Pro 6.1 software (Molecular Devices). Data was filtered based on signal to background ratio for each of the duplicate spots to be >2.5-fold and Z-score ≥ 3 from the global mean signal from all the spotted proteins.
Luciferase gene reporter assay
SNCA short 3′UTR (570 bp), medium (1.07 kb) and long (2.5 kb) were sub-cloned in a pGL4-TK-Firefly luciferase (pGL4-TK-FL) plasmid vector at the 3′end of the Firefly luciferase coding sequence. To guarantee the expression of the long or medium 3′UTR isoforms only, the proximal polyadenylation sites (PAS), present within the sequence of the 3′UTRs, were deleted at positions 262–267 nt, 468–473 nt, 529–553 nt and 1054–1059 nt (taken from (9)).
HeLa cells at 80% of confluency were transfected with with Lipofectamine 2000 (Life Technologies) following manufacturer's instructions. 130 fmol of pGL4-TK-FL empty plasmid or pGL4-TK-FL carrying the sequence of the short medium or long 3′UTR, were co-transfected with 50 ng of pGL4-TK-Renilla luciferase plasmid (pGL4-TK-RL), used as internal control to normalize for differences in transfection efficiency.
Forty eight hours after transfection, the dual luciferase activity was measured at a Tecan Infinite M-200 plate reader, using Dual-Luciferase reporter assay system (Promega) following manufacturer's instructions. Firefly luciferase activity was normalized with Renilla luciferase activity and then normalized Firefly luciferase activity of pGL4-TK-FL-SNCA 3′UTRs constructs was compared to the activity of pGL4-TK-FL vector.
In the case of TIAR and ELAVL1 knock-downs, the RNA corresponding to each of the aforementioned constructs were in vitro transcribed using MEGAscript T7 transcription kit (Ambion) in presence of modified G-cap nucleotide. HeLa control and knockdown cells were seeded in 48-well plates and Firefly and Renilla RNA were co-transfected using TransMessenger transfection reagent (QIAgen) following manufacture's instructions. Four hours after transfection cells were lysed and the dual luciferase activity was measured as described before.
RNA affinity purification
The biotinylated RNA probes of SNCA short 3′UTR (3′UTRS) and long (3′UTRL) were synthesized by in vitro transcription from linearized pBSK plasmid vector by using a T7 MegaScript kit (Ambion) with addition of biotin-14-CTP (Life Technologies). RNA concentration and integrity were verified by NanoDrop 1000 spectrophotometer (Thermo Scientific) and by denaturing agarose gel electrophoresis of the RNA. Successful biotinylation was checked using the Chemiluminescent Nucleic Acid Detection Module kit (Thermo Scientific).
RNA affinity purification was performed as described by Hammerle M. with some adaptations (10) (see Supplementary Materials for details).
RNA electrophoretic mobility shift assay
Fragments A (1–192 nt), B (193–390 nt) and C (391–575 nt) of SNCA 3′UTR short were obtained by deletion of pBSK(+)-3′UTR short with Gibson cloning approach. Corresponding radiolabeled RNA probes were synthesized by IVT with T7 Megascript kit (Ambion) and incorporation of α32P-UTP (Perkin Elmer). Size and integrity of the probe were verified on a 8% TBE-Urea gel, while probe radioactivity was measured with a scintillation counter (Perkin Elmer). N-terminal GST-tagged TIAR and ELAVL1 proteins were overexpressed in Escherichia coli from pGEX2T and pDEST15 vectors, respectively, and purified using glutathione sepharose beads. The RNA electrophoretic mobility shift assay and the competition assay were performed as previously described (11) (see Supplementary Materials for details).
RNA stability assay
Hela cells (control, TIAR and ELAVL1 knockdown) were seeded in 12-wells plate at a density of 1.6 × 105 cells per well. The general transcription inhibitor actinomycin D (Sigma Aldrich) was added at a concentration of 5 g/ml for 0, 4, 8 and 12 h. Total RNA was isolated with Maxwell 16 LEV simply RNA cells kit (Promega) and analyzed by RT-qPCR. ACTB was used as a reference gene for normalization (ΔCt), and the ΔCt of each time point was compared to time 0 h (ΔΔCt). For each experiment the average of triplicate wells per condition was calculated and used to determine mean values and standard deviation of three biological replicates. A Student's t-test was performed to assess for significant differences of RNA decay rate between the three conditions.
Measurement of α-synuclein, TIAR and ELAVL1 levels in motor cortex and primary fibroblasts of Parkinson's disease (PD) and Multiple System Atrophy (MSA) patients
The human post-mortem motor cortex brain tissue stored at –80°C was obtained from the Brainbank from Institut d’Investigacions Biomèdiques August Pi I Sunyer (IDIBAPS)-Biobank, with the corresponding informed consent signed by donors or relatives. Approval of the local ethics committee was requested for the use of brain tissue and for access to medical records for research purposes. The tissue samples were dissected and used in a manner compliant with the Declaration of Helsinki. Brain tissue of five Parkinson's disease (PD) patients, five Multiple System Atrophy (MSA) patients and four control individuals (without neurological affectation) was used in this study. The three groups were matched for age of onset and age of death, gender and severity of the disease (Supplementary Table S1).
Primary cultured fibroblasts (∼5 million cells/pellet) were obtained from skin biopsy of idiopathic PD, LRRK2-associated PD, MSA patients and healthy control individuals (Supplementary Table S2).
Total proteins were extracted by lysing with RIPA buffer supplemented with Protease inhibitors (Roche). After sonication with bioruptor sonifier (Branson) samples were centrifuged at 16 000 g for 15 min and supernatants were measured with Bradford reagent (Sigma Aldrich). α-synuclein, TIAR, ELAVL1, tubulin and β-actin levels were measured by western blot as described previously. Statistical significance was calculated by Wilcoxon's test.
RNA 3′end sequencing, polysome profiling, TIAR and ELAVL1 knockdown and overxpression
See Supplementary Materials for detailed description of the protocol.
RESULTS
Large-scale identification of 3′ UTR interactions
Human SNCA mRNA is expressed with five isoforms of different 3′UTR lengths (ranging from 290 nt to 2500 nt) depending on the choice of the polyadenylation site (5).
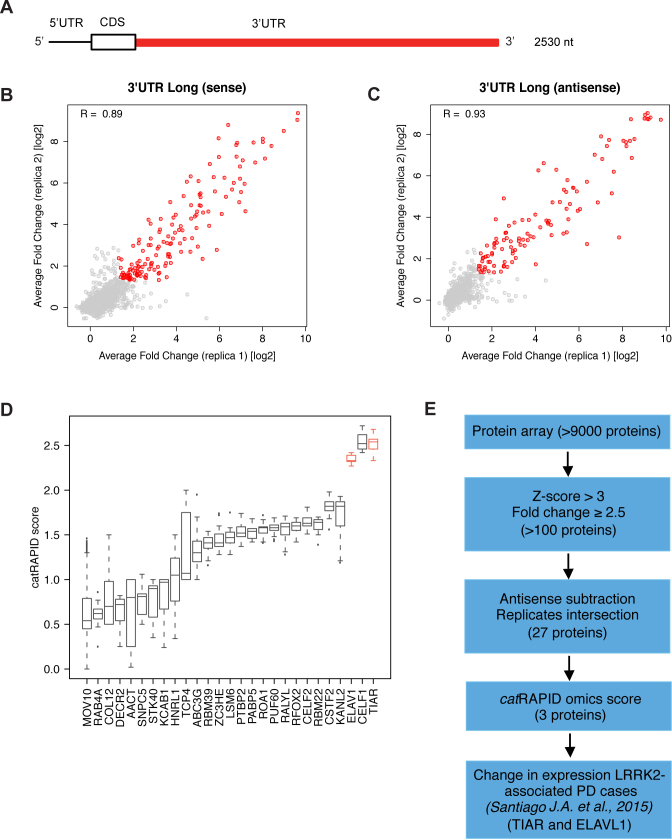
We probed the largest SNCA 3′UTR (2500 nt, containing sequences of the other isoforms, Figure 1A) and its antisense RNA (negative control) on three separate protein arrays (8,12) obtaining high reproducibility between replicates (Pearson's correlations of >0.90; Figure 1B and C; Material and Methods; Supplementary Materials). To identify significant binding events, we selected proteins with fold change (signal to background ratio) >2.5 and associated Z-score >3, obtaining around 100 proteins per experiment (8,12). By considering the intersection of three independent replicates, and removing interactions in common between sense and antisense RNAs, we discarded unspecific binders. Out of 9546 candidates, we identified 27 proteins binding to SNCA 3′UTRs (Figure 1D).
Figure 1.
Large scale screening of SNCA 3′UTR protein interactors. (A) SNCA 3′UTR long (3′UTR L) used in our experiments in vitro; Pearson's correlation between two protein array replicates probed with (A) 3′UTR L sense and (B) antisense (right) RNAs. (sense: R = 0.89; (C) antisense: R = 0.93). Selected binders with fold change (signal to background ratio) >2.5 and associated Z-score > 3 are represented as red dots, while less significant binders are reported in grey. (D) Median value of catRAPID score of SNCA 3′UTR top interactors: 27 proteins; TIAR (also known as TIAL1) and ELAVL1 (ELAV1) are highlighted in red). (E) Sketch of the in vitro / in silico procedure followed to identify TIAR and ELAVL1 interactions.
Following a recently published approach (13), we used the catRAPID algorithm (14) to prioritize candidates for experimental characterization (Supplementary Materials). catRAPID estimates RNA–RBP affinities exploiting physico-chemical properties of nucleotide and amino acid chains including secondary structure, hydrogen bonding, and van der Waals’ propensities to predict protein–RNA associations (15). Importantly, catRAPID calculations of RBP interactions with SNCA 3′UTR show high agreement with protein array results (strong signal RBPs are predicted with an Area under the ROC curve >0.80; Supplementary Materials; Supplementary Figure S1A).
Our approach indicates that TIAR, CELF1 and ELAVL1 have the highest interaction propensities (catRAPID interaction score > 2; Figure 1D). We selected TIAR and ELAVL1 for a more in depth investigation as they are significantly down-regulated in PD patients (differently from CELF1; Figure 1E) (16). Moreover, literature evidence indicates that TIAR and ELAVL1 bind to similar sequences and regulate mRNA stability and translation (17–22), thus suggesting common mechanisms of action on SNCA 3′UTR. Comparison of our RBP candidates with interactions reported in the UTR database AURA (23,24) reveals that TIAR and ELAVL1 have the largest number of binding sites in SNCA 3′UTR (respectively: 6 for TIAR and 12 for ELAVL1), while other proteins are associated with lower signal intensities (AGO1; catRAPID score = 1.39; 1 binding site) or interact with control RNA (TIA1; catRAPID score = 2.46; 4 binding sites; Supplementary Figure S1B).
SNCA with short 3′UTR is predominant in SH-SY5Y and HeLa cells and has increased protein translation
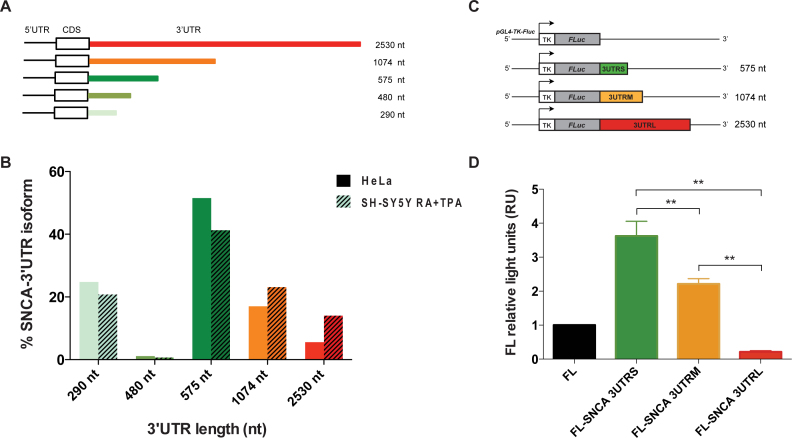
To quantify the abundance of the different 3′UTR isoforms of SNCA mRNA (Figure 2A), we performed a modified sequencing protocol to which we refer as gene specific-3′end RNA sequencing (Material and Methods; Supplementary Materials).
Figure 2.
Measurement of SNCA 3′UTR isoforms expression in HeLa and in vitro differentiated SH-SY5Y cell lines and effect of 3′UTR length on gene reporter activity. (A) Details of the five isoforms of SNCA transcript with different lengths of the 3′UTR, ranging from 290 nt (light green) to 2.5 kb (red). (B) Percentage of each of the five SNCA transcript isoforms in HeLa cells and in vitro differentiated SH-SY5Y cells measured by gene-specific 3′end RNA sequencing. (C) Dual luciferase gene reporter assay. Scheme of the three constructs carrying the sequence of SNCA 3′UTR S (575 nt), M (1074 nt) and L (2530 nt) at the 3′ of luciferase coding sequence. (D) Firefly relative to Renilla luciferase activity of the three constructs (FL-SNCA 3′UTRS, FL-SNCA 3′UTRM and FL-SNCA 3′UTRL) compared to control empty vector FL. * P-value < 0.05 (Wilcoxon's test).
We used two different cell lines, HeLa and human neuroblastoma cells (SH-SY5Y). SH-SY5Y cells were in vitro differentiated towards a dopaminergic neuron-like phenotype by treatment with retinoic acid (RA) and 12-O-tetradecanoyl-phorbol-13-acetate (TPA), in order to recapitulate the biological context of substantia nigra dopaminergic neurons that are mainly affected in Parkinson's disease (25). We found that the isoform carrying the 575 nt 3′UTR is the most abundant accounting for 51.5% and 41.3% of total SNCA mRNA in HeLa and in vitro differentiated SH-SY5Y respectively (Figure 2B). The least abundant isoforms correspond to those carrying 480 nt and 2.5 kb long 3′UTR (Figure 2B). The isoform carrying the 2.5 kb long 3′UTR is instead expressed 2.5 times more in SH-SY5Y than HeLa cells, in agreement with the lengthening of 3′UTRs observed in the brain (Figure 2B) (26).
Moreover, the relative firefly luciferase activity of construct FL-SNCA 3UTRS (575 nt) was more than 3.5 and 17.5 times higher than the firefly luciferase empty vector and FL-SNCA 3UTRL (2.5 kb) respectively (Figure 2C and D; P-value < 0.05; Wilcoxon test). A similar trend is observed at the RNA level, with FL-SNCA 3UTRS being two times more abundant than the control vector carrying only the luciferase coding sequence (Supplementary Figure S2). Our finding suggests that the long 3′UTR contains a higher number of sequence elements targeted by trans-acting factors, such as microRNAs and destabilizing RBPs, which promote RNA degradation or inhibit its translation.
Taken together, our experiments indicate that the isoform carrying the short 3′UTR is mostly contributing to protein production and we proceeded to further characterise TIAR and ELAVL1 binding and their action.
TIAR and ELAVL1 are able to bind specific regions of SNCA 3′UTR in vitro and in vivo
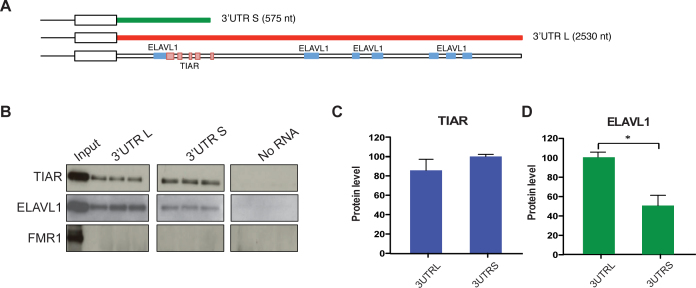
Using an RNA affinity purification assay we confirmed that TIAR and ELAVL1 bind to both SNCA ‘long’ (2.5 kb) and ‘short’ (575 nt) 3′UTR isoforms (Figure 3A; Material and Methods; Supplementary Materials).
Figure 3.
In vitro validation of TIAR and ELAVL1 binding to SNCA 3′UTR. (A) CLIP binding sites along the sequence of SNCA 3′ UTR as reported in the Atlas of UTR regulatory activity (AURA) database (http://aura.science.unitn.it/). TIAR cluster of binding sites (291–318, 353–367, 429–437, 476–511, 578–584 nt) are represented as pink boxes and ELAVL1 binding sites (230–315, 1224–1276, 1546–1567, 1650–1690, 2057–2102, 2113–2153 and 2229–2269 nt) are represented as blue boxes. (B) RNA affinity purification assay performed in technical triplicates. (C) TIAR and (D) ELAVL1 are co-purified with the in vitro synthesized RNA of SNCA 3′UTR long and short (Western Blot). FMR1 is used as negative control.
As shown in Figure 3B–D, ELAVL1 interacts preferably with the long 3′UTR, in agreement with previously published CLIP data that report the presence of multiple binding sites all along the 2.5 kb of the 3′UTR (27–29) (Figure 3A). TIAR interacts with both 3′UTRs (Figure 3B and C) in accordance with CLIP binding sites mapped in the first 600 nt of the UTR region (30) (Figure 3A). FMR1 was chosen as a negative control on the basis of protein microarray results (fold change < 1.7). As shown in Figure 3B, FMR1 does not bind to the two RNA sequences tested. TIAR and ELAVL1 binding was also confirmed in absence of UV cross-linking (Supplementary Figure S3).
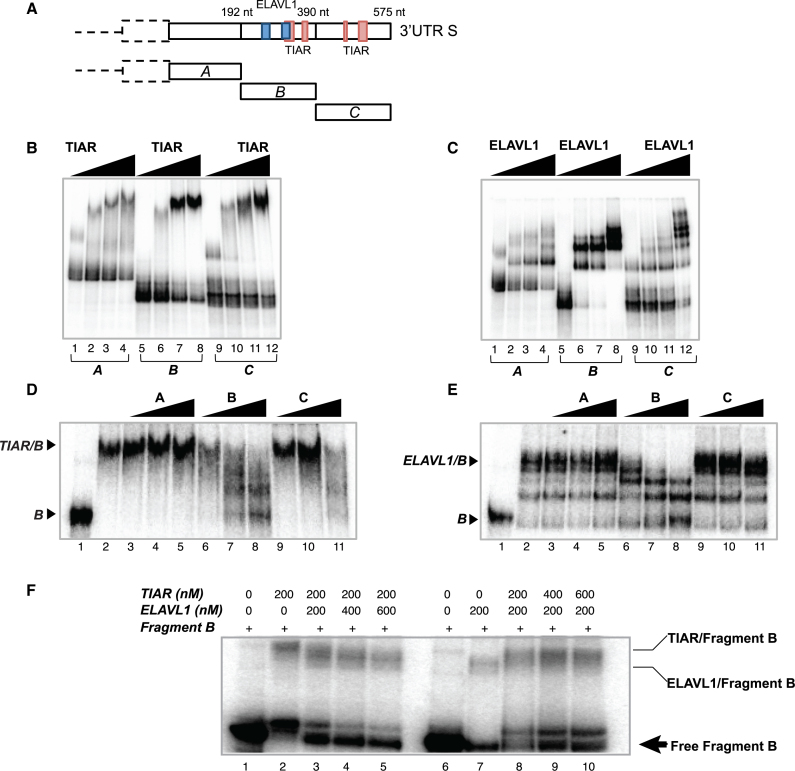
We next defined the region of SNCA short 3′UTR (575 nt) that interacts with TIAR and ELAVL1 by measuring electrophoretic mobility of nucleotides 1–192 (A), 193–390 (B) and 391–575 (C; Figure 4A) upon incubation with increasing concentrations of the GST-tagged TIAR and ELAVL1 proteins (Figure 4B and C). As shown in Figure 4B and C, both proteins have a binding ability to the three fragments but the highest affinity was observed for fragment B (Supplementary Figure S5). Indeed, TIAR and ELAVL1 bind specifically to fragment B, as seen by using fragments A, B and C as unlabeled competitors (Figure 4D and E). We also observed higher molecular complexes in the case of ELAVL1, corresponding to dimers and multimers forming upon incubation with high ELAVL1 concentrations (Figure 4C). To further investigate TIAR and ELAVL1 binding to fragment B, we incubated the RNA with a fixed amount of one protein and increasing amounts of the other. As shown in Figure 4F, the two proteins compete for their binding to fragment B and increasing amounts of TIAR or ELAVL1 show clear displacement of the protein/RNA complex.
Figure 4.
In vitro characterization of TIAR and ELAVL1 binding sites of SNCA 3′UTR short. (A) Schematic representation of the sequence of SNCA 3′UTR S and TIAR/ELAVL1 binding sites previously identified by CLIP. The sequence of the 3′UTR S is divided into three fragments (A, B and C) of ∼190 nt each to test the binding ability of TIAR and ELAVL1. (B) RNA electrophoretic mobility shift assay (REMSA) of fragments A, B and C of SNCA 3′UTR S upon incubation with increasing concentrations of GST-tagged TIAR protein (0, 100, 200, 500 nM). (C) RNA electrophoretic mobility shift assay (REMSA) of fragments A, B and C of SNCA 3′UTR S upon incubation with increasing concentrations of GST-tagged ELAVL1 protein (0, 100, 200, 500 nM). (D) Competition gel shift assay showing binding specificity of TIAR protein for fragment B. Lane 1. Probe B alone, Lane 2. Probe B with 200 nM TIAR, Lane 3–11. Probe B with 200 nM TIAR in presence of 0.1, 1 and 10 nM of cold probe A, B and C. (E) Competition gel shift assay showing binding specificity of ELAVL1 protein for fragment B RNA. Lane 1. Probe B alone, Lane 2. Probe B with 100 nM ELAVL1, Lane 3–11. Probe B with 100 nM ELAVL1 in presence of 0.1, 1 and 10 nM of cold probe A, B and C. (F) Competition assay. Labeled fragment B was incubated with 200 nM of TIAR (lane 2) or 200 nM of ELAVL1 (lane 7). Competition between TIAR and ELAVL1 was performed by incubating fragment B with constant amount of TIAR (200nM) and increasing amount of ELAVL1 (lanes 3, 4 and 5) or constant amount of ELAVL1 (200nM) and increasing amounts of TIAR (lanes 8, 9 and 10).
These experiments indicate that nucleotides 193–390 (fragment B) interact with TIAR and ELAVL1 proteins with the highest binding affinity and sequence specificity and that the two proteins compete for the binding.
Post-transcriptional regulation of SNCA by TIAR and ELAVL1
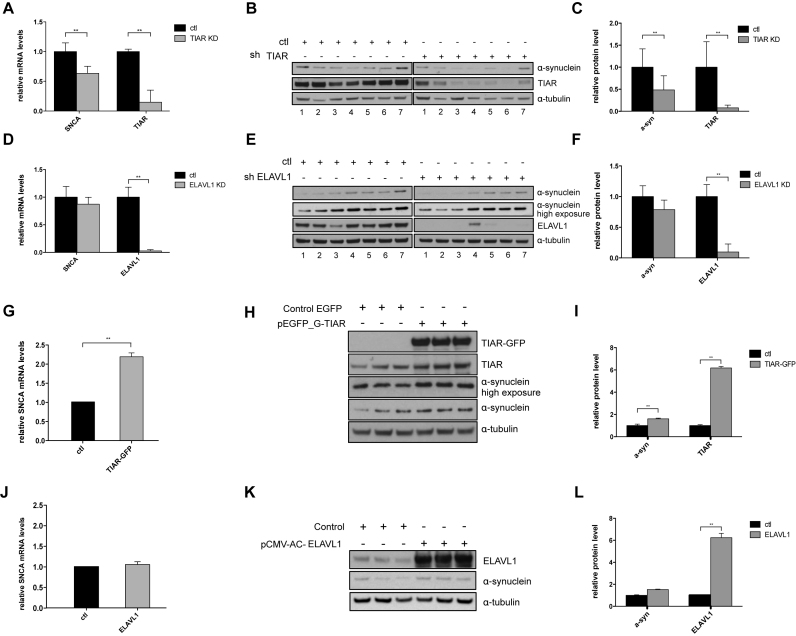
TIAR and ELAVL1 knockdown and overexpression in HeLa cells indicate that the two RBPs play a positive regulatory role on SNCA expression (Material and Methods; Supplementary Materials). This is particularly evident in the case of TIAR whose expression significantly influences the levels of both SNCA RNA and protein (Figure 5A–C, Figure 5G–I) inducing changes in expression that are compatible with those observed in idiopathic Parkinsonism (31,32). More specifically, upon TIAR knockdown we observed a significant decrease of endogenous α-synuclein expression at the RNA and protein level with a 0.63 and 0.5 fold-change respectively (Student's t-test, P-value < 0.01, Figure 5A–C). TIAR overexpression induces a 2.2 and 1.6 fold increase in α-synuclein RNA and protein, respectively. By contrast, ELAVL1 showed a milder effect and mainly on SNCA protein levels (Figure 5D–F, Figure 5J–L). Indeed, upon ELAVL1 knockdown we observed ∼0.8-fold decrease of α-synuclein RNA and protein levels, while ELAVL1 overexpression only increases the protein levels of 1.52-fold with no significant change of the mRNA levels with respect to control.
Figure 5.
Analysis of TIAR and ELAVL1 regulatory role on α-synuclein expression in HeLa cells. (A) Relative SNCA mRNA levels measured by qPCR upon stable knockdown of TIAR in HeLa cells compared to control cells. (B) α-synuclein protein down-regulation measured by Western Blot upon stable knockdown of TIAR protein in HeLa cells. (C) Average and standard deviation of seven biological replicates are reported (**P-value < 0.01, Student's t-test). (D) Fold change of SNCA mRNA upon KD of ELAVL1. (E) α-synuclein protein down-regulation measured by western blot upon stable knockdown of ELAVL1 in HeLa cells. (F) Average and standard deviation of seven biological replicates are reported (**P-value < 0.01, Student's t-test) (G–I) Relative SNCA mRNA and α-synuclein protein up-regulation in HeLa cells overexpressing GFP-tagged TIAR protein compared to control cells (**P-value < 0.01, Student's t-test). (J-L) Relative SNCA mRNA and α-synuclein protein levels in HeLa cells overexpressing ELAVL1 protein compared to control cells (*P-value < 0.05, Student's t-test).
The decrease of SNCA mRNA upon knockdown of TIAR and, to a lesser extent, of ELAVL1, suggest that one of the possible mechanisms of regulation orchestrated by these proteins could be the stabilisation of the mRNA through direct binding at the 3′UTR and interference with RNA degradation factors such as microRNAs and other RBPs. As a matter of fact, the role of ELAVL1 as an RNA stabilizing factor is extensively documented in the literature (33,34). In the case of TIAR, the protein is also known for its effect on translation regulation (35–37) but we cannot exclude that the protein might play other functions in mRNA metabolism.
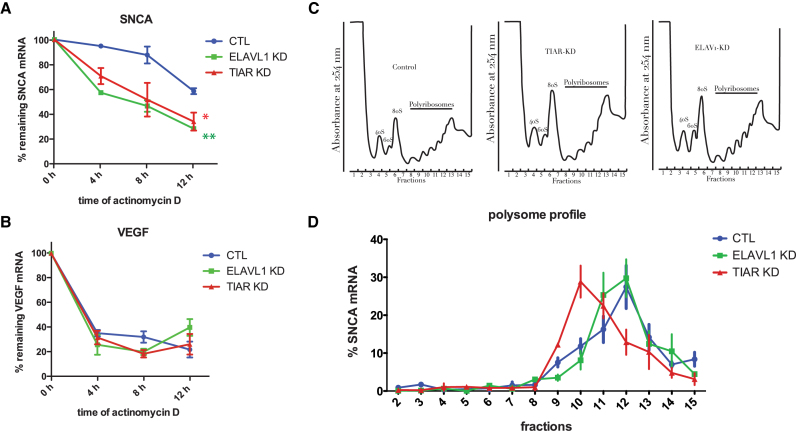
We used the transcription inhibitor actinomycin D to test that the two proteins could stabilize SNCA mRNA. We measured a decay rate of 30% and 43% in TIAR and ELAVL1 knockdown cells respectively while only 5% decay rate was observed in control cells at 4 hours of actinomycin D treatment (Figure 6A and B), showing that both RBPs protect SNCA mRNA from degradation.
Figure 6.
TIAR and ELAVL1 role in SNCA mRNA stability and translation. (A) Percentage of remaining SNCA mRNA measured at 0, 4, 8 and 12 h after transcription inhibition with actinomycin D in control, TIAR knockdown and ELAVL1 knockdown HeLa cells. Mean and standard deviation of three independent experiments are shown (Kolmogorov-Smirnov test with respect to control curve, *P-value < 0.1; **P-value < 0.01). (B) VEGF mRNA is a control for the effectiveness of the actinomycin D treatment. (C) Polysome profile of HeLa control, TIAR knockdown and ELAVL1 knockdown cells. (D) Percentage of SNCA mRNA distribution across polysome gradient in HeLa control, TIAR knockdown and ELAVL1 knockdown conditions. Data shown are mean with standard deviation of three independent experiments.
As TIAR and ELAVL1 are reported to modulate mRNA translation (35–37), we decided to test whether they could participate to the translation of SNCA mRNA. To do so, we traced the distribution of SNCA mRNA across different fractions of the polysome profile of HeLa control, TIAR and ELAVL1 knockdown cells (Figure 6C). In all three conditions SNCA mRNA was distributed (70–75%) between fractions 3 and 7 or more (10–12,38) ribosomes. We observed a substantial shift of SNCA mRNA in cells depleted for TIAR protein, from the more translationally active high molecular weight (HMW) polysomal fraction 12, corresponding to six-seven ribosomes, to the less translationally active low molecular weight (LMW) fraction 10, which corresponds to three ribosomes (Figure 6D). Indeed, only 12.9% of SNCA mRNA co-sedimented with fraction 12 in TIAR knockdown cells compared to 27.5% of SNCA mRNA present in the same fraction in control cells. In contrast, no relevant change was observed upon knockdown of ELAVL1 (Figure 6D).
We conclude that, in addition to stabilizing SNCA mRNA, TIAR also promotes SNCA mRNA translation through a mechanism that needs to be further elucidated. By contrast, ELAVL1 seems to play a main role in SNCA mRNA stabilization, in agreement with its very well documented function in RNA metabolism. Yet, it does not have any relevant effect on its translation efficiency.
TIAR and ELAVL1 regulation is mediated by SNCA 3′UTR
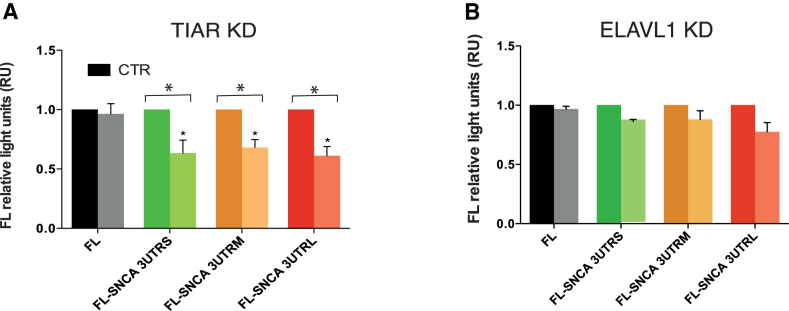
To test whether the regulation observed endogenously (Figure 5) requires the binding to SNCA 3′UTRs, we measured the dual firefly luciferase activities of the constructs FL-SNCA 3UTRS, FL-SNCA 3UTRM and FL-SNCA 3UTRL upon TIAR or ELAVL1 knockdown. We obtained a significant decrease of the relative firefly luciferase activity of 3′UTRS, 3′UTRM and 3′UTRL in HeLa cells depleted of TIAR (Figure 7A). More specifically, we measured a fold decrease of 0.63, 0.68 and 0.61 for FL-SNCA 3′UTRS, FL-SNCA 3′UTRM and FL-SNCA 3′UTRL. No significant change was instead observed for the construct carrying the sequence of the firefly luciferase gene, indicating that the decrease is specific for the constructs containing SNCA 3′UTRs. Our result indicates that the positive regulatory role of TIAR is indeed achieved through the direct binding of the RBP to the 3′UTR of SNCA mRNA, in agreement with our initial hypothesis. As for ELAVL1 KD, only a mild decrease of the signal is observed in all three RNAs containing SNCA 3′UTRs with respect to control HeLa cells with an average fold change of 0.86, 0.88 and 0.77 respectively Figure 7B).
Figure 7.
Dual firefly luciferase activity of SNCA 3′ UTRs upon TIAR or ELAVL1 knockdown. We measured luciferase activities of SNCA 3′UTRs with respect to control empty vector FL. (A) TIAR KD. We observed a fold decrease of 0.63, 0.68 and 0.61 for FL-SNCA 3′UTRS, FL-SNCA 3′UTRM and FL-SNCA 3′UTRL. No significant change was instead observed for the construct carrying the sequence of the firefly luciferase gene (B). ELAVL1 KD. Mild decrease of the signal is observed for FL-SNCA 3′UTRS, FL-SNCA 3′UTRM and FL-SNCA 3′UTRL with an average fold change of 0.86, 0.88 and 0.77 respectively. * P-value < 0.05 (Wilcoxon's test).
MicroRNAs do not compete with TIAR and ELAVL1 binding to SNCA 3′ UTR in HeLa cells
Previous reports indicate that ELAVL1 regulates microRNA expression by intervening in pri- or pre-microRNAs processing of miR-7 and miR-16 (39,40). As miR-7 has been implicated in down-regulation of SNCA expression (41), it is possible that a regulatory loop involving ELAVL1 inhibition of miR-7 causes release of SNCA from miR-7 producing an increase of α-synuclein expression. Such a scenario is compatible with our observation that ELAVL1 depletion induces SNCA down-regulation, which could be mediated by miR-7 up-regulation. However, when we measured miR-7 expression in ELAVL1 in both knockdown and control HeLa cells, we found that the general level of expression of miR-7 is very low (PCR cycle threshold Ct of 33, while U6 control shows Ct of 27) and we therefore ruled out its involvement.
Similarly, the overlap between TIAR binding site (Figure 3B–C) and the target region of miR-153, which was reported to down-regulate SNCA expression (42), suggested a possible competition between the two trans-factors. However, miR-153 expression is almost undetectable in HeLa cells (PCR cycle threshold Ct of 33, while U6 control shows Ct of 22), therefore its participation in α-synuclein down-regulation upon TIAR depletion is very unlikely to occur in our cellular system. Yet, we cannot exclude that miR-153 could play a role in TIAR regulation in the nervous system.
We then used the multiMiR algorithm (43) to systematically investigate other microRNAs interacting with the long SNCA 3′UTR (containing all other 3′UTRs). We collected a total of 142 predicted and 9 experimentally validated microRNAs (MultiMiR score > 0.7) and manually retrieved their binding sites from the microRNA.org server. Using the PhastCons score to measure evolutionary conservation of sequence blocks across multiple vertebrates (44), we separated the microRNAs in two classes: conserved (PhastCons score > 0.57 corresponding to conservation across all mammals) and non-conserved (PhastCons score < 0.57).
We specifically focused on microRNAs having binding sites overlapping with TIAR and ELAVL1 SNCA 3′UTR contacts annotated in AURA database (23,24). We found a total of 9 microRNAs co-localizing with ELAVL1 binding regions, but only one of them, miR-539–5p, was found to be conserved. As for TIAR, two non-conserved microRNAs, miR-126–5p and miR-3134, showed contacts overlapping with the binding sites of the protein. We proceeded to test the expression levels of the microRNAs in our HeLa cells by means of a high throughput approach (Supplementary Material). After signal normalization, we found around 432 miRNAs that passed the expression threshold. However, none of those miRNAs overlap with either TIAR or ELAVL1 binding sites. In accordance with this result, an external dataset taken from a published high-throughput small RNA sequencing analysis (45) reveals nearly undetectable levels of expression of candidate microRNAs (Supplementary Table S3).
Overall, our analysis indicates that microRNAs levels of expression are poor in two independent HeLa cell lines, as also directly observed for miR-7 and miR-153 by TaqMan assay. We therefore ruled out that these microRNAs might have a role in TIAR- and ELAVL1-dependent SNCA regulation in the cell model used for our functional assays.
TIAR and ELAVL1 expressions are altered in frontal motor cortex and in primary fibroblasts from patients affected by PD and MSA synucleinopathies
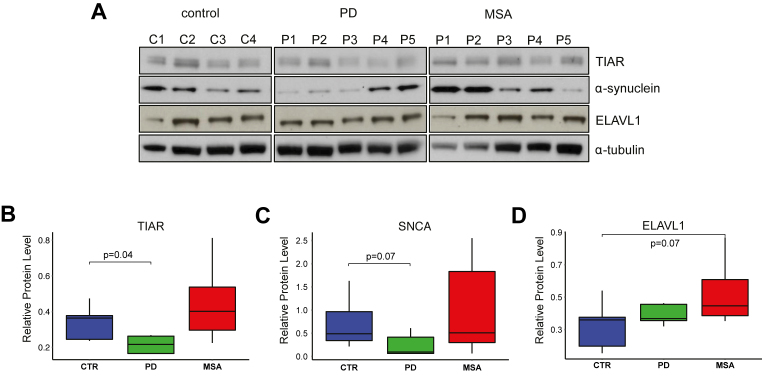
To address implications of TIAR or ELAVL1 in the onset of neurodegeneration, we investigated whether their abundances are altered in conditions associated with changes in α-synuclein expression: PD and another synucleinopathy, Multiple System Atrophy (MSA). PD and MSA belong to a group of synucleinopathies characterized by abnormal accumulation of α-synuclein. Differently from PD, where α-synuclein positive aggregates appear to be largely neuronal, oligodendroglia inclusion prevail in MSA (46,47). The pathogenic mechanisms leading to neurodegeneration in MSA are not well understood, but partially differ from PD since MSA patients show resistance to L-dopa treatment (48,49). In contrast to PD, disease-causative genetic mutations responsible of monogenic forms of MSA have so far not been identified (50–52). Neurodegeneration typically occur with a multisystemic distribution that partially differs from the one observed in PD. For this reason, we decided to examine the frontal motor cortex, which is generally affected with similar degree in both PD and MSA at the late stages (Supplementary Table S1; Material and Methods; Supplementary Materials). We found that TIAR was down-regulated in PD with respect to control individuals with a fold-change of 0.65 (P-value = 0.09, Wilcoxon's test, Figure 8A and B; four controls, five PD and five MSA cases). No alteration of ELAVL1 expression was observed in PD, while a 1.5-fold increase was measured in MSA when compared to controls (P-value = 0.15, Wilcoxon's test, Figure 8A–D). The results correlated with α-synuclein protein expression that was found decreased in PD with respect to controls (fold-change = 0.49, P-value = 0.15, Wilcoxon's test, Figure 8A–C) and slightly increased in MSA. In agreement with these results, Neystat et al. showed reduction of SNCA expression in substantia nigra of PD affected individuals (53) and another group reported 50% reduction at the cellular level in substantia nigra neurons and frontal cortex neurons in PD (54). Also in accordance with our findings, oligodendrocytes isolated from MSA brains expressed elevated levels of SNCA compared to control (55) and a recent strand-specific RNA-sequencing analysis of MSA brain transcriptome reported a moderate increase of SNCA expression in frontal cortex of MSA patients compared to normal individuals (56).
Figure 8.
TIAR, ELAVL1 and α-synuclein protein levels in motor cortex post-mortem samples. (A) α-synuclein TIAR and ELAVL1 protein level in motor cortex tissue of post-mortem control individuals, PD and MSA patients measured by western blot. Normalized average values of (B) TIAR, (C) α-synuclein and (D) ELAVL1 in the three groups (Wilcoxon's test P-values are reported).
Analysis of primary cultured fibroblast obtained from human skin biopsies of sporadic PD (sPD), LRRK2-associated PD (L2PD), MSA and healthy control individuals (Supplementary Table S2) revealed that α-synuclein is also very poorly expressed in the tissue (Supplementary Figure S6A). Nevertheless, we could measure a significant down-regulation of TIAR in PD and MSA patients with respect to control individuals (Supplementary Figure S6A-B). Results were similar when the two PD groups (sPD and L2PD) were analyzed together or separately (Supplementary Figure S6B-D), with the exception of ELAVL1 for which we observed a statistically significant increase specific of the L2PD group (Supplementary Figure S6E).
Overall, our results show an alteration of TIAR and ELAVL1 levels in two different tissues of patients affected by PD or MSA. Due to intra-group variability and small sample size we cannot exclude that the observed changes are due to other causes. Nevertheless, TIAR down-regulation is consistent in both tissues and is accompanied by α-synuclein decrease in motor cortex, in agreement with our functional assays.
Trans-expression quantitative trait loci analysis reveals functional association between TIAR and SNCA
To investigate if naturally occurring mutations in SNCA RNA could affect ELAVL1 and TIAR binding, we downloaded all the single nucleotide polymorphisms (SNPs) annotated in 1000 Genomes (hg19) within the genomic coordinate of the 3′UTR (4:90, 645, 249–90, 647, 778).
The SNPs were ranked using CADD (57) to select functional and deleterious variants based on characteristics such as evolutionary conservation, type of substitution and presence of regulatory elements. The degree of the deleteriousness of SNPs was measured using PHRED (score > 10 indicates significant implication). Out of 67 SNPs, 5 SNPs fall into ELAVL1 binding sites, 3 in TIAR binding sites and 2 are located in ELAVL1/TIAR shared binding regions. 6 of these SNPs overlapping with TIAR/ELAVL1 binding sites have a PHRED score >10, suggesting possible effect on SNCA expression and functionality (Supplementary Table S4).
In addition, we investigated 6 SNPs associated to PD (58) and found that one of them, rs356165, is located in SNCA 3′UTR. The other 5 SNPs are in a genomic region >20KB downstream SNCA 3′UTR. These PD-associated SNPs are not highly ranked by CADD (score < 10), however two of them (rs356182 and rs356219) were detected by other studies as associated to PD (59,60).
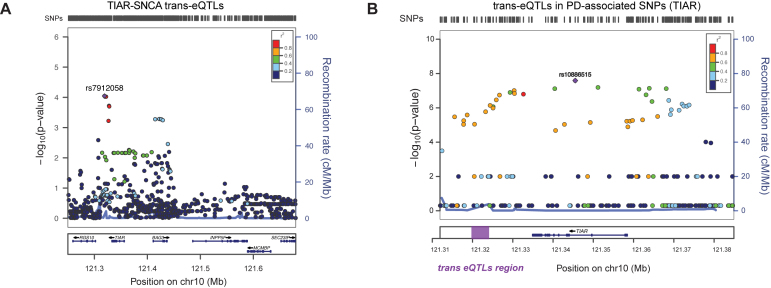
As observed in previous reports (61,62), RBPs and target RNAs are expressed in tissue-specific patterns that are altered in pathological conditions. Thus, it is possible that SNPs in TIAR and ELAVL1 loci affect their RBP activities modifying SNCA expression. To test this hypothesis, we computed trans expression Quantitative Trait Loci (trans-eQTLs) of TIAR and ELAVL1 SNPs in relation to SNCA using 10 brain tissues available from GTEx database (Supplementary Materials) (63). Remarkably, hippocampus and nucleus accumbens basal ganglia showed highly significant SNCA trans-eQTL P-values (<1.9e−4) associated with polymorphisms in TIAR locus. The link is particularly evident in hippocampus, where polymorphisms rs7912058, rs113562141, rs4751739 and rs4751740 (R2 = 1; D’ = 1 between them) located few kilobases dowstream TIAR, reach a SNCA trans-eQTL P-value of 9e−5, showing a singularly strong association between the two loci (Figure 9A). In addition, we found that the minor allele of the leading SNP rs7912058-G is associated with decreased expression of SNCA (Supplementary Figure S7). Our result is further supported by a trans-eQTLs analysis of 22 818 tag-SNPs from 100 random genes showing low interaction score in our protein array experiments. Indeed, out of 22 818 only three tag-SNPs (rs72983079 in GNA15, rs3782119 in BET1L and rs116911351 in C20orf166 gene) achieved significant P-values in the order of magnitude of e−4, which further highlights the strength of the association with TIAR (binomial test P-value = 0.02).
Figure 9.
SNCAtrans expression Quantitative Trait Loci (trans-eQTLs) analysis of TIAR and ELAVL1. (A) SNCAtrans-eQTLs analysis in the region chr10:121250246–121682830 (hg19) spanning from 84 kb upstream TIAR gene to 94 kb downstream neighboring INPP5F gene. The analysis, performed using GTEX data available for human hippocampus tissue (n = 81), shows highly significant SNCA trans-eQTLs P-values (P-values < 10−9) for a group of SNPs downstream TIAR gene (red and purple dots). A locus zoom graph shows that the significant SNCA trans-eQTLs in TIAR region present the typical pattern of linkage disequilibrium (LD) decay of association signals, with a top-associated SNP and other surrounding associated SNPs in progressively decaying LD values (‘Recombination rate’ on right y-axes). (B) PD-associated SNPs from previous GWAS studies (58) in the genomic region of chromosome 10 including TIAR gene. The –log(P-value) is represented on the left y-axes, while the ‘recombination rate’ is represented on the right y-axes. The region downstream TIAR locus where significant SNCA trans-eQTLs map is highlighted with a purple rectangle.
The SNPs in the trans-eQTLs TIAR region (Figure 9A) present the typical pattern of linkage disequilibrium (LD) decay of GWAS association signals, with a dominant SNP surrounded by variants with lower P-values. Interestingly, using PLINK algorithm (64), we calculated that our trans-eQTLs are in moderate LD (R2 ∼ 0.4; D’ = 1) with significantly PD-associated SNPs in TIAR locus found in a previous work (58) (Figure 9B). Overall, this indicates that the association of TIAR with PD might come from its regulatory role in SNCA expression. The same genomic region hosts cis-eQTLs for TIAR locus (rs11199019, rs185650278, rs4636576, rs6585553, rs4752328; tibial nerve tissue) suggesting that this sequence exerts a regulatory role on TIAR gene expression.
As for the ELAVL1 gene, we could not find any significant association with PD, nor significant trans-eQTLs for SNCA gene.
DISCUSSION
In the last twenty years many efforts have been undertaken to study the biochemical features of PD-associated α-synuclein from aggregation and post-translational modification to turnover and interaction with cellular components (64–66). However, recent studies (5,6) revealed the importance of transcriptional and post-transcriptional regulations in the context of pathology (67). For the first time, we presented here a study on the post-transcriptional regulation of α-synuclein by RBPs targeting the 3′ untranslated region of its mRNA.
Using a large-scale in vitro / in silico screening, we discovered RBPs interacting with SNCA 3′ UTR. Two interactors, ELAVL1 and TIAR, bind to different SNCA isoforms generated by alternative polyadenylation mechanisms. Among 5 different known transcript isoforms, the one carrying the short 3′UTR (575 bases) was found to be the most abundant in SH-SY5Y and HeLa cells (∼50% of total SNCA mRNA). Using a gene reporter assay, we observed that the short 3′UTR isoform is more active than longer isoforms (1.07 and 2.5 kb 3′UTR) and highly contributing to protein synthesis in vivo. The short 3′UTR contains high-affinity binding sites for both RBPs in a region comprised between 190–575 nt.
Knockdown and overexpression experiments in HeLa cells show that ELAVL1 and TIAR positively regulate endogenous α-synuclein. The mechanism through which the two RBPs regulate α-synuclein expression implies mRNA stabilization, as shown by the RNA decay assay, and enhancement of translation in the case of TIAR, as demonstrated by polyribosome profiling experiments. This result is compatible with the well-documented role of ELAVL1 as RNA stabilization factor. Indeed, ELAVL1 interferes with ARE-dependent mRNA degradation by protecting the body of the mRNA from decay, rather than slowing down mRNA deadenylation (34). While it is generally assumed that ELAVL1 stabilizes mRNAs primarily by competing with proteins designated to mRNA degradation such as AUF-1, KSRP or TTP (21,68,69), other mechanisms, such a direct competition with microRNAs or regulation of microRNAs expression, might be involved (70,71). It would be of great interest to investigate the role of microRNAs in TIAR- and ELAVL1-dependent regulation of SNCA. However, our analysis indicates that the level of expression of microRNAs targeting TIAR and ELAVL1 binding sites is poor in two independent HeLa cell lines. We therefore excluded that microRNAs might have any relevant role in TIAR and ELAVL1 dependent SNCA regulation in our functional assays.
As for the effect observed on SNCA mRNA translation, our results show that TIAR promotes protein production by shifting SNCA mRNA towards the high molecular weight polysomal fractions. Although TIAR was previously reported as a translational inhibitor in presence of stress conditions by induction of stress granules formation (72), it is possible that it has a dual role in RNA translation. Indeed, as described for other RBPs, many factors contribute to determine the final regulatory effect on target RNAs, from the sequence and structure of binding regions to the availability of interacting partners (18). Further characterization of the molecular complexes formed by TIAR and ELAVL1 will be needed to elucidate the mechanism of regulation of SNCA. Notably, TIAR, together with other RBPs markers of stress granules, was found in pathological lesions of several neurological conditions, such as Alzheimer disease, prion disease, tauopathies and Huntington disease (73–76). This observation suggests that the aberrant assembly of ribonucleoprotein granules could alter the expression of a number of target genes leading to toxicity (77).
Interestingly, TIAR and ELAVL1 protein levels are altered in post-mortem brain tissues from patients affected by PD and MSA. In correlation with α-synuclein levels, we observed a down-regulation of TIAR in PD and an increase of ELAVL1 in MSA. This result suggests that the two RBPs have a key role in PD and MSA. Indeed, ELAVL1 was recently shown to play a neuroprotective role in mice hippocampal neurons following strong glutamatergic excitation through the post-transcriptional orchestration of specialized genes involved in mitochondrial dysfunction, oxidative damage and programmed cell death (78). Similarly, TIAR was found to target a number of genes associated with inflammation and mitochondrial metabolism, such as TNF-alpha, COX-2 and mitochondrial cytochrome C (79–81). In agreement with these observations, a previous study reported significant increase in the level of innate immune components including complement and cytokines (e.g., IL-1, IL-2, IL-6 and TNF) in the substantia nigra and CSF of PD patients (82). Moreover, microglial activation was reported to parallel neuronal degeneration in MSA (83) and increased expression of proinflammatory genes was observed in post-mortem tissue from the rostral pons of MSA patients (84). Supporting these findings, polymorphisms in proinflammatory genes such as IL-1β, IL-1α or TNF-α were reported to be associated with increased MSA risk (85,86). Therefore, we speculate that the two RBPs might coordinately regulate the expression of entire sets of genes involved in pathways characteristics of PD and MSA neurodegeneration.
Notably, TIAR and ELAVL1 were identified in a group of 2000 significantly down-regulated genes in a network-based meta-analysis of four independent microarray studies sporadic and LRRK2-associated PD cases (16). Additional indications of TIAR association to PD via regulation of α-synuclein, come from the GWAS and trans-eQTLs analysis on the genomic locus comprising TIAR region. Importantly, Nalls et al. (58) found a group of frequent variants in TIAR locus with significant association to PD. This finding is in strong agreement with our trans-eQTLs analysis that identifies a group of SNPs in the same genomic locus influencing SNCA expression in two different brain areas, nucleus accumbens and hippocampus.
Fundamental questions about the mechanism through which TIAR- and ELAVL1-mediated regulation of SNCA can lead to disease remain to be addressed. To what extent α-synuclein overexpression caused by the increase of TIAR and ELAVL1 activity can induce protein aggregation and cell toxicity? Is α-synuclein the only PD-associated target of these two RBPs?
From a more clinical perspective, testing TIAR and ELAVL1 levels in human biospecimen (urine, blood, serum or cerebrospinal fluid) would be of great interest for improving the diagnosis and prognosis of different synucleinopathies. Indeed, distinguishing MSA from PD can be difficult owing to PD-like features in MSA, including occasionally a transient L-dopa response in some patients. Moreover, MSA is characterized by a prognosis of a markedly shorten life span with respect to PD (87,88), so a high level of accuracy in the diagnosis is needed but not yet available (89).
In conclusion, further studies would be needed to obtain deeper insights into the role of TIAR and ELAVL1 in the development of PD and other synucleinopathies, which will help to evaluate the impact of post-transcriptional events on disease.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Fátima Gebauer, Juan Valcarcel and Benedetta Bolognesi for interesting discussions and advice. We thank Silvia Rodríguez, Joana Ribeiro and Iona Gelabert for their technical support. We also thank Diego Garrido and Claudia Giambartolomei for helpful discussion on trans-eQTLs analysis. The authors thank the brain donors and their families for the neurological tissues used in our research.
Authors contributions: D.M. and E.B. performed all the experiments. D.C., C.M.L. and J.A.R. performed the computational analysis. R.F.S., M.E. and M.J.M provided clinical samples. D.M., E.B. T.B.O. and G.G.T conceived the hypothesis and designed the experiments. D.M. and G.G.T. wrote the manuscript.
APPENDIX
MEMBERS OF THE CMSAR
Asunción Ávila (MD; PhD)2, Àngels Bayés (MD; PhD)3, Teresa Botta-Orfila (PhD)4, Núria Caballol (MD)5, Matilde Calopa (MD)6, Jaume Campdelacreu (MD; PhD)6, Yaroslau Compta (MD; PhD)1, Mario Ezquerra (PhD)1, Oriol de Fàbregues (MD; PhD)7, Rubén Fernández-Santiago (PhD)1, Darly Girado (PhD)1, Jorge Hernández-Vara (MD; PhD)7, Serge Jaumà (MD)6, Domenica Marchese (PhD)4, Maria J. Martí (MD; PhD)1, Javier Pagonabarraga (MD; PhD)8, Pau Pastor (MD; PhD)9, Lluís Planellas (MD)1, Claustre Pont-Sunyer (MD, PhD)10, Víctor Puente (MD)11, Montserrat Pujol (MD)12, Josep Saura (PhD)13, Gian Gaetano Tartaglia (PhD)4, Eduard Tolosa (MD; PhD)1, Francesc Valldeoriola (MD; PhD)1
Parkinson's disease and Movement Disorders Unit, Neurology Service, ICN, Hospital Clínic, IDIBAPS, CIBERNED, University of Barcelona, Barcelona, Catalonia, Spain
Neurology Service Hospital General de l’Hospitalet, Consorci Sanitari Integral, L’Hospitalet de Llobregat, Barcelona, Catalonia, Spain
Parkinson's disease and Movement Disorders Unit, Clínica Teknon, Barcelona, Catalonia, Spain
Gene function and evolution group; Centre for genomic regulation (CRG), Institucio Catalana de Recerca i Estudis Avançats (ICREA) Universitat Pompeu Fabra (UPF), Barcelona, Catalonia, Spain
Department of Neurology, Hospital Sant Joan Despí Moisès Broggi.Consorci Sanitari Integral, Barcelona, Catalonia, Spain
Parkinson's disease and Movement Disorders Unit, Neurology Service, Hospital de Bellvitge, Hospitalet de Llobregat, Catalonia, Spain
Parkinson's disease and Movement Disorders Unit, Neurology Service, Hospital del Vall d’Hebron, Barcelona, Catalonia, Spain
Parkinson′s disease and movement disorders unit. Neurology Service. Hospital de la Creu i Sant Pau, Barcelona, Catalonia, Spain
Neurology Service, Hospital Mútua de Terrassa, Terrassa, Catalonia, Spain
Neurology Service, Hospital General de Granollers, Catalonia, Spain
Neurology Service, Hospital del Mar, Barcelona, Catalonia, Spain
Neurologia Service, Hospital de Santa Maria, Lleida, Catalonia, Spain Biochemistry and Molecular Biology Unit, School of Medicine, Neuroradiology Section, Magnetic Resonance Unit, Centre de IDIBAPS, University of Barcelona, Barcelona, Catalonia, Spain
Contributor Information
Catalan MSA Registry (CMSAR):
Asunción Ávila, Àngels Bayés, Teresa Botta-Orfila, Núria Caballol, Matilde Calopa, Jaume Campdelacreu, Yaroslau Compta, Mario Ezquerra, Oriol de Fàbregues, Rubén Fernández-Santiago, Darly Girado, Jorge Hernández-Vara, Serge Jaumà, Domenica Marchese, Maria J Martí, Javier Pagonabarraga, Pau Pastor, Lluís Planellas, Claustre Pont-Sunyer, Víctor Puente, Montserrat Pujol, Josep Saura, Gian Gaetano Tartaglia, Eduard Tolosa, and Francesc Valldeoriola
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’; CERCA Programme / Generalitat de Catalunya; European Research Council [RIBOMYLOME_309545]; Spanish Ministry of Economy and Competitiveness [BFU2014–55054-P]; ‘Fundació La Marató de TV3’ [PI043296]. Funding for open access charge: European Research Council (RIBOMYLOME_309545) Fundació la Marató de TV3 [PI043296].
Conflict of interest statement. None declared.
REFERENCES
- 1. de Lau L.M.L., Breteler M.M.B.. Epidemiology of Parkinson's disease. Lancet Neurol. 2006; 5:525–535. [DOI] [PubMed] [Google Scholar]
- 2. Dauer W., Przedborski S.. Parkinson's disease: mechanisms and models. Neuron. 2003; 39:889–909. [DOI] [PubMed] [Google Scholar]
- 3. Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M.. Alpha-synuclein in Lewy bodies. Nature. 1997; 388:839–840. [DOI] [PubMed] [Google Scholar]
- 4. Zanzoni A., Marchese D., Agostini F., Bolognesi B., Cirillo D., Botta-Orfila M., Livi C.M., Rodriguez-Mulero S., Tartaglia G.G.. Principles of self-organization in biological pathways: a hypothesis on the autogenous association of alpha-synuclein. Nucleic Acids Res. 2013; 41:9987–9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rhinn H., Qiang L., Yamashita T., Rhee D., Zolin A., Vanti W., Abeliovich A.. Alternative α-synuclein transcript usage as a convergent mechanism in Parkinson's disease pathology. Nat. Commun. 2012; 3:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Locascio J.J., Eberly S., Liao Z., Liu G., Hoesing A.N., Duong K., Trisini-Lipsanopoulos A., Dhima K., Hung A.Y., Flaherty A.W. et al. . Association between α-synuclein blood transcripts and early, neuroimaging-supported Parkinson's disease. Brain. 2015; 138:2659–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Raaij M.E., van Gestel J., Segers-Nolten I.M.J., de Leeuw S.W., Subramaniam V.. Concentration dependence of alpha-synuclein fibril length assessed by quantitative atomic force microscopy and statistical-mechanical theory. Biophys. J. 2008; 95:4871–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siprashvili Z., Webster D.E., Kretz M., Johnston D., Rinn J.L., Chang H.Y., Khavari P.A.. Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics. 2012; 13:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sotiriou S., Gibney G., Baxevanis A.D., Nussbaum R.L.. A single nucleotide polymorphism in the 3′UTR of the SNCA gene encoding alpha-synuclein is a new potential susceptibility locus for Parkinson disease. Neurosci. Lett. 2009; 461:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hämmerle M., Gutschner T., Uckelmann H., Ozgur S., Fiskin E., Gross M., Skawran B., Geffers R., Longerich T., Breuhahn K. et al. . Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatol. Baltim. MD. 2013; 58:1703–1712. [DOI] [PubMed] [Google Scholar]
- 11. Bechara E.G., Didiot M.C., Melko M., Davidovic L., Bensaid M., Martin P., Castets M., Pognonec P., Khandjian E.W., Moine H. et al. . A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009; 7:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J. et al. . Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013; 493:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cirillo D., Blanco M., Armaos A., Buness A., Avner P., Guttman M., Cerase A., Tartaglia G.G.. Quantitative predictions of protein interactions with long noncoding RNAs. Nat. Methods. 2016; 14:5–6. [DOI] [PubMed] [Google Scholar]
- 14. Agostini F., Zanzoni A., Klus P., Marchese D., Cirillo D., Tartaglia G.G.. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinforma. Oxf. Engl. 2013; 29:2928–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bellucci M., Agostini F., Masin M., Tartaglia G.G.. Predicting protein associations with long noncoding RNAs. Nat. Methods. 2011; 8:444–445. [DOI] [PubMed] [Google Scholar]
- 16. Santiago J.A., Potashkin J.A.. Network-based metaanalysis identifies HNF4A and PTBP1 as longitudinally dynamic biomarkers for Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2257–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cok S.J., Acton S.J., Morrison A.R.. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 2003; 278:36157–36162. [DOI] [PubMed] [Google Scholar]
- 18. Izquierdo J.M. Control of the ATP synthase beta subunit expression by RNA-binding proteins TIA-1, TIAR, and HuR. Biochem. Biophys. Res. Commun. 2006; 348:703–711. [DOI] [PubMed] [Google Scholar]
- 19. Kim H.S., Wilce M.C.J., Yoga Y.M.K., Pendini N.R., Gunzburg M.J., Cowieson N.P., Wilson G.M., Williams B.R.G., Gorospe M., Wilce J.A.. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011; 39:1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subramaniam K., Kandasamy K., Joseph K., Spicer E.K., Tholanikunnel B.G.. The 3′-untranslated region length and AU-rich RNA location modulate RNA-protein interaction and translational control of β2-adrenergic receptor mRNA. Mol. Cell. Biochem. 2011; 352:125–141. [DOI] [PubMed] [Google Scholar]
- 21. Suswam E.A., Nabors L.B., Huang Y., Yang X., King P.H.. IL-1beta induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3′ untranslated region: Involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int. J. Cancer. 2005; 113:911–919. [DOI] [PubMed] [Google Scholar]
- 22. Wigington C.P., Jung J., Rye E.A., Belauret S.L., Philpot A.M., Feng Y., Santangelo P.J., Corbett A.H.. Post-transcriptional regulation of programmed cell death 4 (PDCD4) mRNA by the RNA-binding proteins human antigen R (HuR) and T-cell intracellular antigen 1 (TIA1). J. Biol. Chem. 2015; 290:3468–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dassi E., Malossini A., Re A., Mazza T., Tebaldi T., Caputi L., Quattrone A.. AURA: Atlas of UTR Regulatory Activity. Bioinforma. Oxf. Engl. 2012; 28:142–144. [DOI] [PubMed] [Google Scholar]
- 24. Dassi E., Re A., Leo S., Tebaldi T., Pasini L., Peroni D., Quattrone A.. AURA 2: Empowering discovery of post-transcriptional networks. Transl. Austin Tex. 2014; 2:e27738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Korecka J.A., van Kesteren R.E., Blaas E., Spitzer S.O., Kamstra J.H., Smit A.B., Swaab D.F., Verhaagen J., Bossers K.. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PloS One. 2013; 8:e63862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miura P., Shenker S., Andreu-Agullo C., Westholm J.O., Lai E.C.. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 2013; 23:812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M.. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011; 8:559–564. [DOI] [PubMed] [Google Scholar]
- 28. Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M., Tuschl T., Ohler U., Keene J.D.. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011; 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., Rajewsky N.. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol. Cell. 2011; 43:340–352. [DOI] [PubMed] [Google Scholar]
- 30. Wang Z., Kayikci M., Briese M., Zarnack K., Luscombe N.M., Rot G., Zupan B., Curk T., Ule J.. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010; 8:e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chartier-Harlin M.-C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M. et al. . Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet Lond. Engl. 2004; 364:1167–1169. [DOI] [PubMed] [Google Scholar]
- 32. Chiba-Falek O., Nussbaum R.L.. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum. Mol. Genet. 2001; 10:3101–3109. [DOI] [PubMed] [Google Scholar]
- 33. Brennan C.M., Steitz J.A.. HuR and mRNA stability. Cell. Mol. Life Sci. CMLS. 2001; 58:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng S.S., Chen C.Y., Xu N., Shyu A.B.. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998; 17:3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kedersha N.L., Gupta M., Li W., Miller I., Anderson P.. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999; 147:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mazan-Mamczarz K., Lal A., Martindale J.L., Kawai T., Gorospe M.. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 2006; 26:2716–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Podszywalow-Bartnicka P., Wolczyk M., Kusio-Kobialka M., Wolanin K., Skowronek K., Nieborowska-Skorska M., Dasgupta Y., Skorski T., Piwocka K.. Downregulation of BRCA1 protein in BCR-ABL1 leukemia cells depends on stress-triggered TIAR-mediated suppression of translation. Cell Cycle Georget. Tex. 2014; 13:3727–3741. [DOI] [PubMed] [Google Scholar]
- 38. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu F., Zhang X., Lei Y., Liu X., Liu Z., Tong T., Wang W.. Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J. Cell. Biochem. 2010; 111:727–734. [DOI] [PubMed] [Google Scholar]
- 40. Choudhury N.R., de Lima Alves F., de Andrés-Aguayo L., Graf T., Cáceres J.F., Rappsilber J., Michlewski G.. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013; 27:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Junn E., Lee K.-W., Jeong B.S., Chan T.W., Im J.-Y., Mouradian M.M.. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13052–13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 2010; 285:12726–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ru Y., Kechris K.J., Tabakoff B., Hoffman P., Radcliffe R.A., Bowler R., Mahaffey S., Rossi S., Calin G.A., Bemis L. et al. . The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014; 42:e133–e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. et al. . Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grolmusz V.K., Tóth E.A., Baghy K., Likó I., Darvasi O., Kovalszky I., Matkó J., Rácz K., Patócs A.. Fluorescence activated cell sorting followed by small RNA sequencing reveals stable microRNA expression during cell cycle progression. BMC Genomics. 2016; 17:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cykowski M.D., Coon E.A., Powell S.Z., Jenkins S.M., Benarroch E.E., Low P.A., Schmeichel A.M., Parisi J.E.. Expanding the spectrum of neuronal pathology in multiple system atrophy. Brain J. Neurol. 2015; 138:2293–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Halliday G.M., Holton J.L., Revesz T., Dickson D.W.. Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. (Berl.). 2011; 122:187–204. [DOI] [PubMed] [Google Scholar]
- 48. Churchyard A., Donnan G.A., Hughes A., Howells D.W., Woodhouse D., Wong J.Y., Kalnins R.M., Mendelsohn F.A., Paxinos G.. Dopa resistance in multiple-system atrophy: loss of postsynaptic D2 receptors. Ann. Neurol. 1993; 34:219–226. [DOI] [PubMed] [Google Scholar]
- 49. Wenning G.K., Ben Shlomo Y., Magalhães M., Daniel S.E., Quinn N.P.. Clinical features and natural history of multiple system atrophy. An analysis of 100 cases. Brain J. Neurol. 1994; 117:835–845. [DOI] [PubMed] [Google Scholar]
- 50. Al-Chalabi A., Dürr A., Wood N.W., Parkinson M.H., Camuzat A., Hulot J.-S., Morrison K.E., Renton A., Sussmuth S.D., Landwehrmeyer B.G. et al. . Genetic variants of the alpha-synuclein gene SNCA are associated with multiple system atrophy. PloS One. 2009; 4:e7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Houlden H., Bettencourt C., Chelban V.. Updates on potential therapeutic targets in MSA. ACNR. 2016; 15:8–11. [Google Scholar]
- 52. Scholz S.W., Houlden H., Schulte C., Sharma M., Li A., Berg D., Melchers A., Paudel R., Gibbs J.R., Simon-Sanchez J. et al. . SNCA variants are associated with increased risk for multiple system atrophy. Ann. Neurol. 2009; 65:610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neystat M., Lynch T., Przedborski S., Kholodilov N., Rzhetskaya M., Burke R.E.. Alpha-synuclein expression in substantia nigra and cortex in Parkinson's disease. Mov. Disord. Off. J. Mov. Disord. Soc. 1999; 14:417–422. [DOI] [PubMed] [Google Scholar]
- 54. Kingsbury A.E., Daniel S.E., Sangha H., Eisen S., Lees A.J., Foster O.J.F.. Alteration in alpha-synuclein mRNA expression in Parkinson's disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2004; 19:162–170. [DOI] [PubMed] [Google Scholar]
- 55. Asi Y.T., Simpson J.E., Heath P.R., Wharton S.B., Lees A.J., Revesz T., Houlden H., Holton J.L.. Alpha-synuclein mRNA expression in oligodendrocytes in MSA. Glia. 2014; 62:964–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mills J.D., Ward M., Kim W.S., Halliday G.M., Janitz M.. Strand-specific RNA-sequencing analysis of multiple system atrophy brain transcriptome. Neuroscience. 2016; 322:234–250. [DOI] [PubMed] [Google Scholar]
- 57. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M. et al. . Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 2014; 46:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pickrell J.K., Berisa T., Liu J.Z., Ségurel L., Tung J.Y., Hinds D.A.. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016; 48:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lill C.M., Roehr J.T., McQueen M.B., Kavvoura F.K., Bagade S., Schjeide B.-M.M., Schjeide L.M., Meissner E., Zauft U., Allen N.C. et al. . Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: the PDGene Database. PLOS Genet. 2012; 8:e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cirillo D., Marchese D., Agostini F., Livi C.M., Botta-Orfila T., Tartaglia G.G.. Constitutive patterns of gene expression regulated by RNA-binding proteins. Genome Biol. 2014; 15:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cirillo D., Livi C.M., Agostini F., Tartaglia G.G.. Discovery of protein–RNA networks. Mol. Biosyst. 2014; 10:1632–1642. [DOI] [PubMed] [Google Scholar]
- 63. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J. et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lautenschläger J., Kaminski C.F., Kaminski Schierle G.S.. α-Synuclein - Regulator of Exocytosis, Endocytosis, or Both. Trends Cell Biol. 2017; 27:468–479. [DOI] [PubMed] [Google Scholar]
- 66. Wong Y.C., Krainc D.. α-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nat. Med. 2017; 23:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rocha E.M., De Miranda B., Sanders L.H.. Alpha-synuclein: pathology, mitochondrial dysfunction and neuroinflammation in Parkinson's disease. Neurobiol. Dis. 2017; doi:10.1016/j.nbd.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 68. Cok S.J., Acton S.J., Sexton A.E., Morrison A.R.. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J. Biol. Chem. 2004; 279:8196–8205. [DOI] [PubMed] [Google Scholar]
- 69. Dai W., Zhang G., Makeyev E.V.. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 2012; 40:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lu Y.-C., Chang S.-H., Hafner M., Li X., Tuschl T., Elemento O., Hla T.. ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages. Cell Rep. 2014; 9:2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Srikantan S., Tominaga K., Gorospe M.. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012; 13:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 2012; 7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ash P.E.A., Vanderweyde T.E., Youmans K.L., Apicco D.J., Wolozin B.. Pathological stress granules in Alzheimer's disease. Brain Res. 2014; 1584:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Goggin K., Beaudoin S., Grenier C., Brown A.-A., Roucou X.. Prion protein aggresomes are poly(A)+ ribonucleoprotein complexes that induce a PKR-mediated deficient cell stress response. Biochim. Biophys. Acta. 2008; 1783:479–491. [DOI] [PubMed] [Google Scholar]
- 75. Vanderweyde T., Apicco D.J., Youmans-Kidder K., Ash P.E.A., Cook C., Lummertz da Rocha E., Jansen-West K., Frame A.A., Citro A., Leszyk J.D. et al. . Interaction of tau with the RNA-Binding Protein TIA1 Regulates tau Pathophysiology and Toxicity. Cell Rep. 2016; 15:1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., Wanker E.E.. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell. 2001; 12:1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bolognesi B., Lorenzo Gotor N., Dhar R., Cirillo D., Baldrighi M., Tartaglia G.G., Lehner B.. A Concentration-Dependent Liquid Phase Separation Can Cause Toxicity upon Increased Protein Expression. Cell Rep. 2016; 16:222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Skliris A., Papadaki O., Kafasla P., Karakasiliotis I., Hazapis O., Reczko M., Grammenoudi S., Bauer J., Kontoyiannis D.L.. Neuroprotection requires the functions of the RNA-binding protein HuR. Cell Death Differ. 2015; 22:703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V.. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 1999; 274:2322–2326. [DOI] [PubMed] [Google Scholar]
- 80. Dixon D.A., Balch G.C., Kedersha N., Anderson P., Zimmerman G.A., Beauchamp R.D., Prescott S.M.. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 2003; 198:475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kawai T., Lal A., Yang X., Galban S., Mazan-Mamczarz K., Gorospe M.. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 2006; 26:3295–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu B., Hong J.-S.. Role of microglia in inflammation-mediated neurodegenerative diseases: mechanisms and strategies for therapeutic intervention. J. Pharmacol. Exp. Ther. 2003; 304:1–7. [DOI] [PubMed] [Google Scholar]
- 83. Ishizawa K., Komori T., Sasaki S., Arai N., Mizutani T., Hirose T.. Microglial activation parallels system degeneration in multiple system atrophy. J. Neuropathol. Exp. Neurol. 2004; 63:43–52. [DOI] [PubMed] [Google Scholar]
- 84. Langerveld A.J., Mihalko D., DeLong C., Walburn J., Ide C.F.. Gene expression changes in postmortem tissue from the rostral pons of multiple system atrophy patients. Mov. Disord. Off. J. Mov. Disord. Soc. 2007; 22:766–777. [DOI] [PubMed] [Google Scholar]
- 85. Infante J., Llorca J., Berciano J., Combarros O.. Interleukin-8, intercellular adhesion molecule-1 and tumour necrosis factor-alpha gene polymorphisms and the risk for multiple system atrophy. J. Neurol. Sci. 2005; 228:11–13. [DOI] [PubMed] [Google Scholar]
- 86. Nishimura M., Kawakami H., Komure O., Maruyama H., Morino H., Izumi Y., Nakamura S., Kaji R., Kuno S.. Contribution of the interleukin-1beta gene polymorphism in multiple system atrophy. Mov. Disord. Off. J. Mov. Disord. Soc. 2002; 17:808–811. [DOI] [PubMed] [Google Scholar]
- 87. Jecmenica-Lukic M., Poewe W., Tolosa E., Wenning G.K.. Premotor signs and symptoms of multiple system atrophy. Lancet Neurol. 2012; 11:361–368. [DOI] [PubMed] [Google Scholar]
- 88. Petrovic I.N., Ling H., Asi Y., Ahmed Z., Kukkle P.L., Hazrati L.-N., Lang A.E., Revesz T., Holton J.L., Lees A.J.. Multiple system atrophy-parkinsonism with slow progression and prolonged survival: a diagnostic catch. Mov. Disord. Off. J. Mov. Disord. Soc. 2012; 27:1186–1190. [DOI] [PubMed] [Google Scholar]
- 89. Compta Y., Giraldo DM., Munoz E., Antonelli F., Fernandez M., Bravo P., Soto M., Camara A., Ferran T., Marti MJ. on behalf of Catalan MSA Registry (CMSAR) . Cerebrospinal fluid levels of coenzyme Q10 are reduced in multiple system atrophy. Parkinsonism and Related Disorders. 2017; doi:10.1016/j.parkreldis.2017.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.