Abstract
DNA damage response (DDR) is essential for genome stability and human health. Recently, several RNA binding proteins (RBPs), including fused-in-sarcoma (FUS), have been found unexpectedly to modulate this process. The role of FUS in DDR is closely linked to the pathogenesis of amyotrophic lateral sclerosis (ALS), a progressive neurodegenerative disease that affects nerve cells in the brain and the spinal cord. Given that RBM45 is also an ALS-associated RBP, we wondered whether RBM45 plays any function during this process. Here, we report that RBM45 can be recruited to laser microirradiation-induced DNA damage sites in a PAR- and FUS-dependent manner, but in a RNA-independent fashion. Depletion of RBM45 leads to abnormal DDR signaling and decreased efficiency in DNA double-stranded break repair. Interestingly, RBM45 is found to compete with histone deacetylase 1 (HDAC1) for binding to FUS, thereby regulating the recruitment of HDAC1 to DNA damage sites. A common familial ALS-associated FUS mutation (FUS-R521C) is revealed to prefer to cooperate with RBM45 than HDAC1. Our findings suggest that RBM45 is a key regulator in FUS-related DDR signaling whose dysfunction may contribute to the pathogenesis of ALS.
INTRODUCTION
Human genome is constantly exposed to multiple endogenous and exogenous genotoxic assaults that usually cause DNA damage. To combat this threat, cells have evolved a sophisticated system, termed DNA-damage response, to detect DNA lesions, signal their presence and promote their repair (1). Dysfunction of DDR has been shown to affect diverse cellular processes, implicating its biological significance in preventing human diseases (2,3).
In recent years, several RNA-binding proteins (RBPs), known as splicing and alternative splicing factors, such as NONO, RBMX, FUS (4–7), have been found to play important roles in DDR. FUS is a member of the FET (TAF15, EWS and TLS) family of RNA-binding proteins (RBPs), whose pathological aggregation within cytoplasmic inclusion bodies is a hallmark of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). FUS can be recruited to double-stranded breaks (DSBs) in a PAR-dependent manner, and promote DSB repair through both homologous recombination (HR) and non-homologous end-joining (NHEJ) (6). Consistently, FUS knockout mice manifest a severe deficiency in spermatogenesis and enhanced radio-sensitivity (8), which are closely connected with DSB repair defects. The recruitment of FUS to DSBs is an early event in DDR. It is believed that the function of FUS in DDR is mediated, at least partially, by promoting the recruitment of HDAC1 through their associations (9). Notably, human familial ALS (fALS) patients with FUS-R521C or FUS-P525L mutation exhibit evidence of DNA damage in cortical neurons and spinal motor neurons (9). Interestingly, several FUS proteins that harbor fALS mutations were found to exhibit an aberrant interaction with HDAC1 in vivo and to be defective in DDR and repair. However, the underlying mechanism(s) responsible for the abnormal protein interaction remain enigma.
RBM45, also named drb1, is recently found to be a FUS-interacting RBP (10). Although RBM45 localizes predominately in the nucleus via a C-terminal nuclear-localization sequence (NLS) (11), it has also been reported to distribute within TAR DNA-binding protein 43 (TDP43)-positive cytoplasmic inclusions in ALS, FTLD-TDP and Alzheimer's disease (AD) patients (12). Additionally, a recent immunoprecipitation and mass spectrometry study indicates that RBM45 associates with several ALS-linked RBPs and likely contributes to neurodegeneration in ALS (12). Although the pathological aggregation of RBM45 with TDP43 in the cytoplasm may confer cellular toxicity (12), we speculate that aggregation-induced loss of normal RBM45 function might also play an important role in ALS pathogenesis. Unfortunately, the role of RBM45 in vivo remains largely unknown.
In this study, we have identified a novel role of RBM45 in DDR. We found that RBM45 is recruited to chromatin and to laser-induced sites of DNA damage through the Linker and RRM3 domains in a PAR-dependent manner. Meanwhile, this recruitment is promoted by FUS. Depletion of RBM45 results in an excessive recruitment of HDAC1 to the chromatin after X-ray irradiation, causing an impaired DSB repair and increased cellular sensitivity to X-ray. Our results suggest that RBM45 serves as a negative regulator to prevent FUS-mediated excessive recruitment of HDAC1 to the sites of DNA damage.
MATERIALS AND METHODS
Cell culture and reagents
Human HeLa, U2OS and 293T cells were obtained from the American Type Culture Collection (Rockville, MD, USA). All cell lines were grown in Dulbecco Modified Eagle medium (DMEM) at 37°C, 5% CO2 with 10% fetal bovine serum. Under indicated situations, 10 μM ATM inhibitor (KU55933), 20 μM DNA-PK inhibitor (NU7026), or 50 μM PARP inhibitor (ABT-888) were applied to cells 1 h prior to laser microirradiation. Full-length RBM45, HDAC1 and FUS cDNAs were cloned into pEGFP-C3 (Clontech), MC-Flag-pCS2, MC-HA-pCS2, or pNTAP expression vectors as indicated to generate EGFP, Flag, HA or SBP fusion proteins, respectively. Anti-Flag M2 agarose affinity gel was purchased from Sigma (A2220). Streptavidin Sepharose High Performance was from GE healthcare (17-5113-01). Cell transfections with plasmids or siRNAs were performed by using PEI (Sigma) or Lipofectamine RNAiMax (Invitrogen), respectively, following the manufacturer's instructions. Cells were analyzed 48–72 h later after transfection. siRNAs were obtained from GenePharma (Shanghai, China). The gene-specific target sequences were as follows:
siRBM45-1: CCUUCAUUGAUGAUGGAAGU
siRBM45-2: UGGGCUACGUACGAUACUUAA
siRNA-FUS-1: ATGAATGCAACCAGTGTAAGG
siRNA-FUS-2: CAATTCCTGATCACCCAAGGG
siRNA-PARP1-1: GCATGATTGACCGCTGGTA
siRNA-PARP1-2: GATAGAGCGTGAAGGCGAA
The negative control siRNA (siNC) sequence was: UUCUCCGAACGUGUCACGU.
Antibodies used in this study included rabbit polyclonal antibodies against RBM45 (Abcam, Ab123912), HA-tag (Biolegend, 902302), Flag (Sigma, 7425), H3.1 (Abmart, p30266); mouse monoclonal antibodies against HDAC1 (Abcam, Ab31263), phosphor-H2AX (Millipore, 05-636-AF555), FUS (Santa Cruz, sc-4771), PARP1 (Santa Cruz, sc8007), β-actin (Protein tech, 00001-1), Biotin (Terminal)-PAR (Trevigen, 4336-100-02).
Collection of chromatin fractions
HeLa cells irradiated with X-ray (10 Gy) were harvested and fractionated as previously described (13–15). The collected chromatin fractions were analyzed through western blotting.
Clonogenic assay
Cell survival assay after genotoxic treatments was performed as described previously (14). Cells were treated with the indicated dosage of X-ray and further incubated in complete medium for 14 days. Colonies were fixed and counted. Survival fraction was calculated as the number of colonies in the test condition divided by the number of colonies in the control and plotted.
Laser microirradiation and imaging
The microirradiation was performed with a pulsed nitrogen laser (Spectra-Physics; 365 nm, 10 Hz pulse) as previously described (16). For quantitating the percentage of cells with RBM45 accumulation at sites of laser irradiation, GFP-RBM45 expressing cells were selected for laser microirradiation followed by treatment with 0.05% triton X-100 to distinguish the accumulation of RBM45 along the line of irradiation (Figure 1B, C, F, Figure 2A, Figure 3A, B, Supplementary Figure S4A). Pretreating cells with Hoechst 33342 (1 μg/ml) also promotes the RBM45 foci visible (Figure 1D, Figure 2B). To measure the damage response to laser microirradiation, in each experiment, damage response in over thirty cells was examined. Standard deviations (SDs) were derived from at least two independent experiments. For the kinetic analysis of RBM45 recruitment at laser-irradiated sites, the mean intensity of the focus was obtained after subtraction of the background intensity in the irradiated cell. The data are presented as means and standard errors from 10 individual cells.
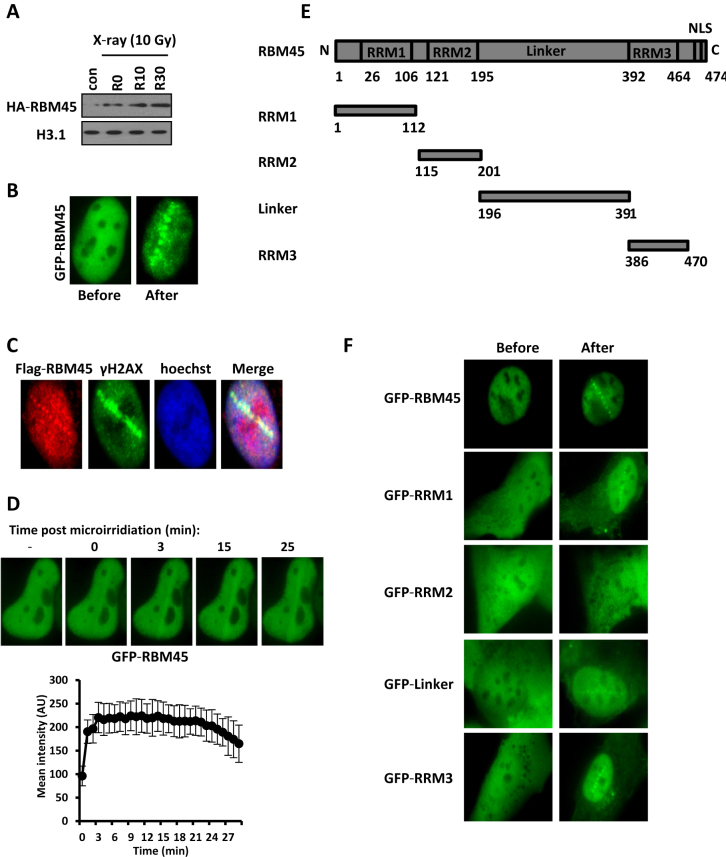
Figure 1.
Accumulation of RBM45 at DNA damage sites. (A) HeLa cells transfected with HA-RBM45 were irradiated with X-ray (10 Gy). The Triton-insoluble fractions were harvested at different recovery time points: R0 (0 min), R10 (10 min) and R30 (30 min). The levels of HA-RBM45 were detected by immunoblotting with anti-HA antibody. H3.1: loading control. (B) Detection of GFP-RBM45 at laser-induced DNA damage sites. U2OS cells expressing GFP-RBM45 were laser micro-irradiated, followed by an immediate treatment with 0.05% Triton X-100. Cell images before microirradiation and after treatments were captured. (C) U2OS cells transfected with Flag-RBM45 were fixed immediately after microirradiation and stained with anti-Flag and anti-γH2AX antibodies. The nuclei were labeled with Hoechst 33342. (D) U2OS cells transfected with GFP-RBM45 were pretreated with Hoechst 33342 followed by laser micro-irradiation. Cell images were captured and the average intensity of the accumulated RBM45 at laser-irradiated sites was quantified. Error bars represent standard errors based on 10 independent measurements. (E) Schematic representation of RBM45 domains. RRM: RNA recognition motif. (F) U2OS cells transfected with a series of truncated GFP-RMB45 were micro-irradiated followed by treatment with 0.05% triton X-100. Cell images were recorded as in (B).
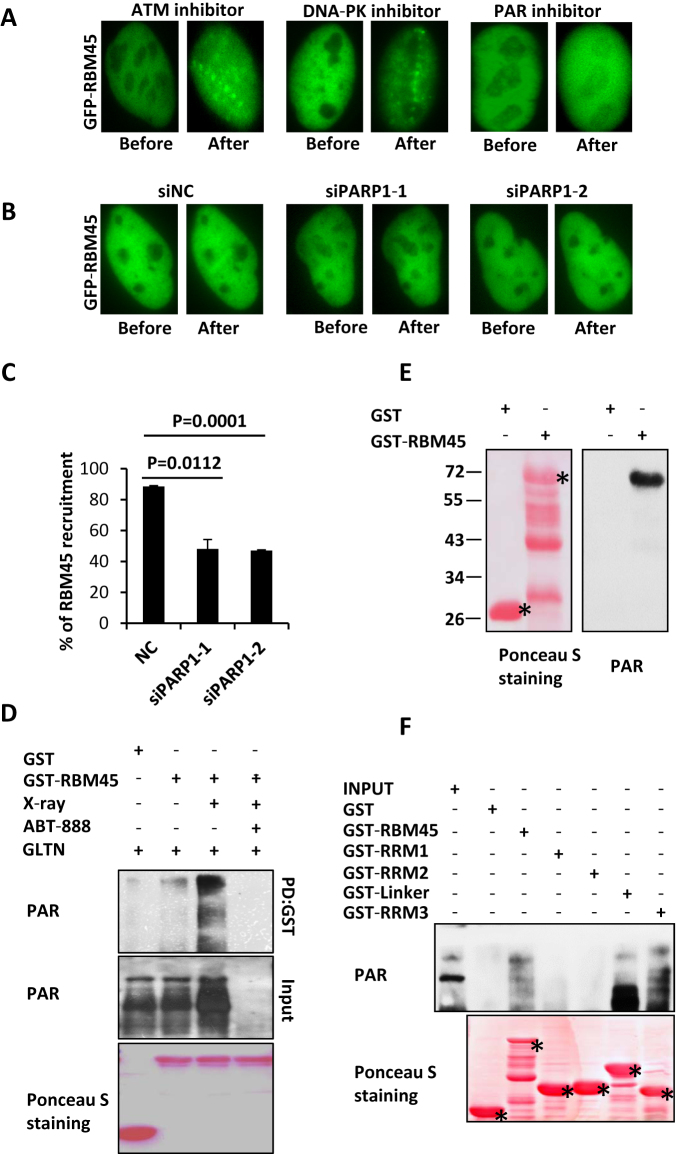
Figure 2.
RBM45 accumulates at DNA damage sites in a PAR-dependent manner and binds to PAR directly. (A) PARP inhibitor abrogates the accumulation of RBM45 at DNA damage sites. U2OS cells transfected with GFP-RBM45 were pretreated with ATM inhibitor (KU55933), DNA-PK inhibitor (NU7026), or PARP inhibitor (ABT-888) for 1 h prior to laser microirradiation. Micro-irradiated cells were then treated by 0.05% triton X-100. (B) Depletion of PARP1 inhibits the recruitment of RBM45. 48 h after transfection with siNC or siPARP1, U2OS cells expressing GFP-RBM45 were pretreated with Hoechst 33342 followed by laser micro-irradiation. NC: negative control. (C) Percent of cells with GFP-RBM45 accumulation after PARP1 knockdown. Data are presented as mean ± SD. n = 2; 30–40 irradiated cells per experiment. (D) Recombinant RBM45 bound to PAR from cell lysate. 293T cells with or without X-ray treatment were used in GST pull down assay to detect binding ability of RBM45 to PAR. All of the groups were pretreated with gallotannin (GLTN) to make X-ray induced PAR visible. (E) Recombinant RBM45 binds to biotin-labeled PAR chain directly. GST-RBM45 transferred onto PVDF membrane was incubated with biotin-labeled PAR. Biotin signal was examined. Asterisks mean specific bands. (F) Mapping domain of RBM45 binding to PAR. GST-tagged RBM45 truncations were incubated with 293T cell lysate. Antibody against PAR was used to detect the signal. Asterisks mean specific bands.
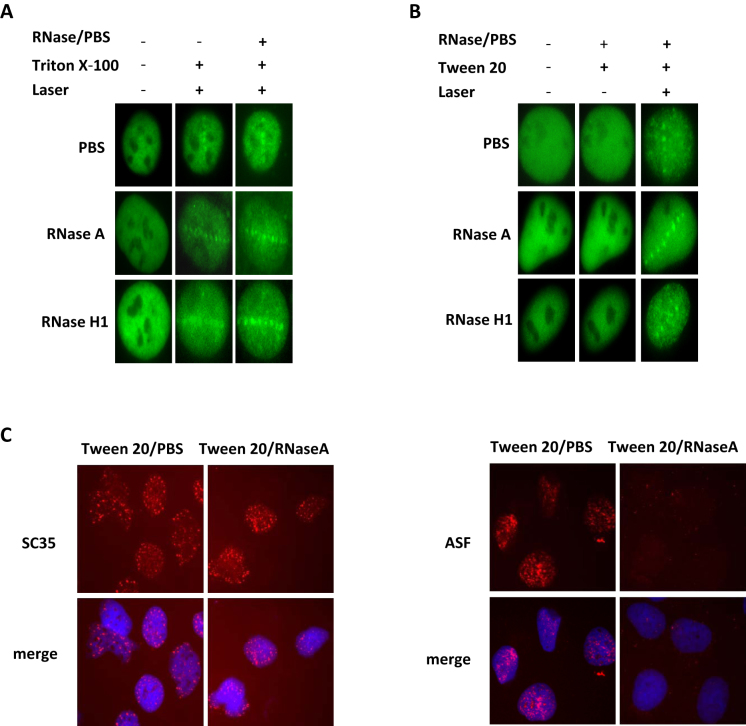
Figure 3.
RNA is not essential for the recruitment of RBM45. (A) U2OS cells expressing GFP-RBM45 were micro-irradiated and treated with Triton X-100 (0.05%), then incubated with RNase A (1 mg/ml) or RNase H (10 U/ml) for 15 min at room temperature. Images were captured. (B) U2OS cells expressing GFP-RBM45 were treated with 2% Tween 20 for 10 min followed by incubation with either PBS, RNase A (1 mg/ml), or RNase H (10 U/ml) for 15 min. Cells were then laser-irradiated. And images were captured immediately. (C) Confirmation the effect of RNase A by staining of SC35 and ASF. After incubation with 2% Tween 20, cells were treated with PBS or RNase A followed by immunostaining with SC35 or ASF antibody.
RNase treatment
To check whether the assembly of RBM45 at laser-damaged sites is RNA-dependent, U2OS cells were permeabilized with Tween 20 (2%) for 10 min, followed by treatment with RNase H (10 U/ml) or RNase A (1 mg/ml) for 15 min at room temperature. After that cells were laser-irradiated, and imaged immediately.
To examine whether the residence of RBM45 at laser-damaged sites is RNA-dependent, U2OS cells were irradiated and permeabilized with 0.05% Triton X-100 to distinguish the foci of RBM45, then further treated with RNase A (1 mg/ml), RNase H (10 U/ml), or PBS for 15 min.
Co-immunoprecipitation and western blotting
293T cells transfected with HA-RBM45 or SBP-RBM45 and Flag-FUS were harvested and lysed with NETN buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40), and the whole cell lysates were immunoprecipitated with anti-Flag M2 beads. To detect the interaction between HDAC1 and FUS or RBM45, cells transfected with HA-HDAC1 and Flag-FUS or Flag-RBM45 were harvested, and the whole cell lysates were immunoprecipitated with an antibody against HA. For RNA- or DNA-independent protein interactions at the indicated situations, RNase A (0.1 mg/ml) or DNA intercalator ethidium bromide (EtBr) (200 μg/ml) was used in immunoprecipitation assay. Samples were separated by SDS-PAGE and detected by immunoblotting with the indicated antibodies.
GST fusion protein purification and GST pull down assay
GST fusion proteins were prepared using the E. coli BL21 strain. Transformed BL21 cells were cultured (200 ml) to the log phase (OD600: 0.6) and protein expression was induced overnight with 0.2 mM IPTG. The cell pellets were sonicated in PBS buffer (20 ml) with 1 mM PMSF, 1 mM DTT, 10 mM EDTA, then spun at 13 000 g for 30 min at 4°C. The supernatant was incubated with GST beads (GE healthare, 17-0756-01) for 1 h at 4°C. After washed twice with PBS and high salt buffer (20 mM Imidazole pH 6.8, 1 M NaCl, 1 mM EDTA, 1 mM DTT), the beads were incubated with the indicated cell lysates for at least 4 h followed by washing with NETN buffer. The binding proteins were separated by SDS-PAGE followed by western blotting as described (17).
HR and NHEJ assay
To examine the effect of RBM45 depletion on HR efficiency, 293T cells transfected with siRNAs targeting NC or BRCA1 or RBM45 were further transfected with the DR-GFP reporter and I-SceI-IRES-DsRedNLS expression vector by using Lipofectamine 2000 as described (18). Two days later, the frequency of HR-mediated repair events was calculated by analyzing GFP positive cells out of the DsRed-positive cells in flow cytometry analysis (BD FACS Aria). The efficiency of HR in cells transfected with siBRCA1 or siRBM45 was shown relative to the one in cells with siNC. To examine the effect of RBM45 overexpression on HR efficiency, 293T cells were transfected with HA-vector/ HA-RBM45, DR-GFP reporter and I-SceI-IRES-DsRedNLS expression vector. The extent of repair in cells transfected with HA-RBM45 was shown relative to that in cells transfected with HA-vector. Data from three independent experiments were used to generate the histogram.
The NHEJ reporter plasmid was digested with HindIII (Thermo scientific) and purified with an Omega gel extraction kit. To examine the effect of RBM45 depletion on NHEJ efficiency, 293T cells transfected with siRNAs targeting NC or Ku70 or RBM45 were transfected with the linearized NHEJ reporter along with pCherry to serve as a transfection control by using Lipofectamine 2000 as described (19). 24 h after transfection, cells were harvested and analyzed by flow cytometry (BD FACS Aria). The efficiency of NHEJ in cells transfected with siKu70 or siRBM45 was shown relative to the one in siNC-transfected cells. To examine the effect of RBM45 overexpression on NHEJ efficiency, 293T cells were transfected with HA-vector/ HA-RBM45, NHEJ reporter, and pCherry. The extent of repair in cells transfected with HA-RBM45 was shown relative to that in cells transfected with HA-vector. Data from three independent experiments were used to generate the histogram.
Micrococcal nuclease digestion
After digested with 0.25% trypsin, HeLa cells (60 mm dish) transfected with siNC or siRBM45 were permeabilized with 0.5% Triton X-100 for 10 min, followed by spun at 2000 rpm for 3 min at 4°C to collect the nuclei. Then the nuclei were washed, resuspended in 150 μl digestion buffer (50 mM Tris–HCl pH7.9, 5 mM CaCl2, 100 μg/ml bovine serum albumin, protease inhibitors), and aliquoted into three tubes. Prior to digestion, one tube was added with 0.5 M (1.5 μl) EDTA and set aside as the undigested control. Another two tubes were added with 0.25 units of micrococcal nuclease (Thermo Scientific, EN0181) and incubated at 37°C for 4 or 8 min, respectively. The digestion was stopped by adding 0.5 M (1.5 μl) EDTA. DNA was isolated by a TIANamp genomic DNA kit, adjusted to the same concentration and subjected to 2% agarose gel electrophoresis. The intensity of each lane was consecutively quantified by using Quantity One software (Bio-Rad) as previous described (20).
PAR-binding assay
PAR-binding properties of purified proteins were analyzed as described (21). Briefly, 500 ng of the indicated protein were transferred onto a PVDF membrane following separation on a 10% SDS-PAGE gel. Subsequently, the membrane was incubated with 100 nM biotin-PAR (Trevigen, 4336-100-02), or 100 nM biotin for negative control. And Horseradish Peroxidase Streptavidin was used to detect the signal. Biotin-PAR can also be examined directly by antibody against PAR.
Subcellular fractionation
Cells (60 mm dish) transfected Flag-FUS/FUS-R521C and HA-RBM45 were washed with ice-cold PBS and resuspended in 500 μl hypotonic buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 1 mM NaF and 1 mM Na3VO4). The cells were incubated for 15 min on ice followed by the addition of 0.5% NP-40. The cells were spun at 1000 × g for 5 min at 4°C and the supernatants were collected as cytoplasmic fractions. The nuclei were resuspended in 150 μl extraction buffer (20 mM HEPES pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 1 mM NaF and 1 mM Na3VO4) for 15 min followed by centrifuged at 12 000 × g for 10 min at 4°C. The supernatants were collected as nuclear extracts.
Comet assay
The alkaline comet assay was carried out as previously (14). Briefly, after exposed to X-ray irradiation on ice, cells were harvested immediately. Then cells (1 × 104) were mixed with 0.8% low melting agarose and layered onto agarose-coated slides. Cells on the slides were then lysed with a lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris pH 10.0, 1% N-lauroylsarcosine and 1% Triton X-100) for 1 h at 4°C. After lysis, slides were incubated for 20 min with electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH > 13) followed by electrophoresis (20 min, 25 V, 300 mA). Then slides were placed into 100% ethanol and air-dried. After stained with 5 μg/ml ethidium bromide (Sigma), images were taken using a fluorescent microscope (Leica). Average Tail Moment was analyzed (100 cells/slide) by using Comet Assay Software Project Casp-1.2.2 (University of Wroclaw, Poland). The reported Tail Moments were the mean values±standard deviation of two independent experiments.
RESULTS
RBM45 can be recruited to sites of DNA damage
Recently, the ALS-associated protein FUS has been reported to play an important role in DDR. Given that RBM45 interacts with FUS, we were wondering whether RBM45 has some function(s) in this process. We first harvested the chromatin fractions in cells treated with or without X-ray and examined whether the chromatin loading of RBM45 is affected. We found that X-ray irradiation quickly promoted RBM45 chromatin accumulation through western blotting (Figure 1A). In line with it, when U2OS cells expressing GFP-RBM45 were micro-irradiated followed by permeabilization with Triton X-100, a robust accumulation of GFP-RBM45 at DNA damage tracks was rapidly detected (Figure 1B). To exclude the possibility that the recruitment was related to GFP tag, U2OS cells were transfected with Flag-tagged RBM45 followed by microirradiation. Immunofluorescence results showed that Flag-RBM45 could still be recruited to the damage sites marked by γH2AX (Figure 1C). We further found that the recruitment of GFP-RBM45 at laser damage sites was also clearly detectable in Hoechst-sensitized cells (Figure 1D). We then determined the recruitment kinetics of RBM45 in living cells for up to 30 min. GFP-RBM45 could accumulate at DNA damage sites immediately after microirradiation (Figure 1D). The average fluorescence intensity of the accumulated RBM45 could reach a peak around 5 min after irradiation and then persisted for >20 min (Figure 1D).
To check which domain of RBM45 is responsible for its recruitment to DNA damage sites, we constructed several truncated GFP-RBM45 expression vectors (Figure 1E) and transfected them into U2OS cells. We found that, similar to the full-length RBM45, all mutants except GFP-RRM2 could be recruited to laser-induced damaged sites (Figure 1F), indicating that RBM45 is targeted to DNA damage sites through multiple motifs.
RBM45 is recruited to DNA damage sites in a PAR-dependent manner
Previous studies show that several RNA binding proteins, such as FUS, RBMX and NONO, are recruited to DNA damage sites in a PAR-dependent manner (4–6,22). We were wondering whether PAR is also required for RBM45 recruitment. RBM45-expressing U2OS cells were incubated with a PARP inhibitor, ABT-888, for 1 h prior to laser microirradiation as described (23). Intriguingly, no GFP-RBM45 accumulation could be detected in the presence of ABT-888 (Figure 2A, Supplementary Figure S1A). We then treated the cells with either KU55933 or NU7441 prior to microirradiation, and found that neither the ATM inhibitor nor the DNA-PK inhibitor could block the recruitment of GFP-RBM45 (Figure 2A, Supplementary Figure S1B). Given that 75–90% of the PAR-chains were the products of PARP1 (24), the effect of PARP1 on the recruitment of RBM45 was then examined. Analogous to PAR inhibitor, depletion of PARP1 dramatically decreased the enrichment of RBM45 after laser irradiation (Figure 2B, C, Supplementary Figure S1C). These results indicate that PARP activity is important for RBM45 to localize to the damage sites.
Since PARP activity can catalyze the formation of PAR chains, which are platforms mediating PAR-binding proteins accumulation, we next examined whether RBM45 can interact with PAR chains. The cells were pretreated with gallotannin (GLTN), a cell-permeable PARG inhibitor that suppresses PAR degradation in vivo. Then the cell lysates were harvested immediately after X-ray irradiation and incubated with recombinant GST-RBM45. GST pull-down result showed that RBM45 weakly associated with PAR under unperturbed situation (Figure 2D). While X-ray treatment significantly enhanced their interaction as more PAR chains were synthesized. Meanwhile, pre-incubation with ABT-888 which diminishes PAR chain formation, blocked the association between PAR and GST-RBM45 (Figure 2D), confirming a specific binding between them. In line with that, treatment of cells with H2O2 also enhanced the interaction between PAR chain and RBM45 (Supplementary Figure S2A).
We then determined whether RBM45 associates with PAR directly. GST-RBM45 was transferred onto a PVDF membrane after separation on SDS-PAGE gel. Then the membrane was incubated with biotin-labeled PAR-chain followed by incubation with the antibody against biotin. As shown in Figure 2E, a specific band of biotin-PAR chain was detected on the position of GST-RBM45, but not GST or other degradation bands of RBM45. In addition, specific band on the position of RBM45 also can be detected by antibody against PAR (Supplementary Figure S2B). However, there was no biotin signal could be observed if the membrane incubated with biotin, indicating that RBM45 could not bind to biotin (Supplementary Figure S2C). All of these data support a direct association between RBM45 and PAR.
We also purified a series of truncated GST-RBM45 peptides and incubated them with 293T cell lysates. We found that RRM1 displayed a faint interaction with PAR, while the Linker and RRM3 motifs manifested a much stronger association with PAR. Notably, RRM2 had no detectable association with PAR (Figure 2F), which might explain why RRM2 domain failed to be recruited to the sites of damage.
RNA is not essential for the recruitment of RBM45 to DNA damage sites
Since RBM45 is a RNA binding protein, we wanted to know whether RNA binding is essential for the recruitment or residence of RBM45 at laser-damaged sites. We utilized two different modes of RNase treatments to answer the question. First, cells expressing GFP-RBM45 were micro-irradiated followed by Triton X-100 permealization and RNase A or H treatments as described previously (25). The recruitment of RBM45 was not obviously changed in the presence of RNase A or H (Figure 3A), suggesting that the residence of RBM45 on the DNA damage site is RNA-independent. Next, cells expressing GFP-RBM45 were pretreated with Tween 20 and RNase, prior to microirradiation as described previously (26,27) to determine whether RNA is essential for the initial assemble of RBM45 on the DNA damage site. The recruitment of RBM45 was still detectable (Figure 3B). While under the same condition, the localization of ASF (alternative splicing factor 1 or splicing factor 2) but not SC35 (serine/arginine-rich splicing factor 2) in nuclear speckles was significantly abrogated (Figure 3C). Given the fact that the localization of ASF but not SC35 in nuclear speckles is sensitive to RNase treatment, these results confirmed the efficacy of RNase treatment. Therefore, RNA binding is unnecessary for the assembly and residence of RBM45 at the DNA damage sites.
RBM45 is important for DDR and efficient DNA repair
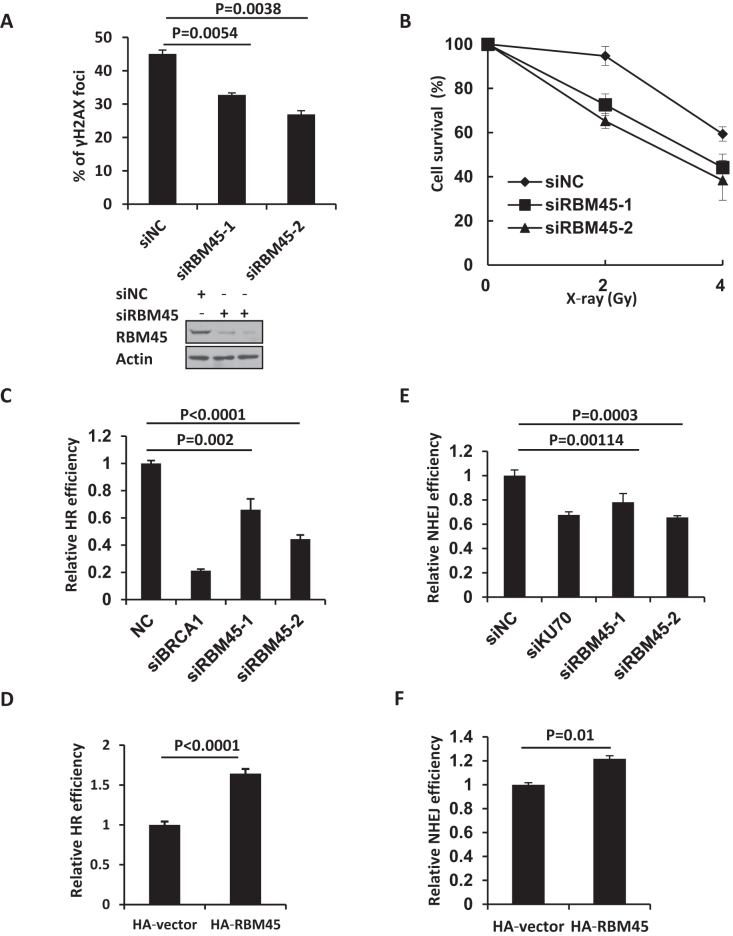
Given that RBM45 can be recruited to laser-induced DNA damage sites, we next examined whether it plays any role in DDR. Phosphorylation of the histone variant H2AX (γH2AX) is an early event in the cellular response to DSBs. We monitored the levels of X-ray-induced γH2AX in HeLa cells transfected with siRBM45. We found that depletion of RBM45 significantly reduced the number of cells with more than ten γH2AX foci at 30 min after X-ray irradiation (Figure 4A, Supplementary Figure S1D). We further harvested the X-ray-irradiated HeLa cells and performed alkaline comet assay. The result showed that X-ray irradiation induced a similar extent of DNA strand breaks in the control and RBM45-depleted cells (Supplementary Figure S2D, S2E), excluding the possibility that the reduced γH2AX level at the early stage resulted from less DNA damage. These data suggest a role of RBM45 in γH2AX foci formation after DNA damage. We also examined the effect of RBM45 depletion on cellular survival after IR treatment. The clonogenic assay revealed that knockdown of RBM45 sensitized the cells to IR killing (Figure 4B).
Figure 4.
RBM45 participates in DDR and is important for efficient DNA repair. (A) HeLa cells expressing siRNA targeting NC or RBM45 were stained with γH2AX antibody 30 min later after X-ray (1 Gy) irradiation. Cells with more than ten γH2AX foci were counted. P values from unpaired t test were included. Western blot analysis verified the efficiency of siRNAs targeting RBM45 in HeLa cells. NC: negative control. (B) RBM45 depletion confers modest radio-sensitivity in HeLa cells. Cells expressing siNC or siRBM45 were irradiated and colony formation assay was performed. Values are presented as means ± SD, n = 3. siNC: negative control. (C) Detection of DSB repair efficiency mediated by homologous recombination (HR). 293T cells expressing siRNAs for NC, RBM45, or BRCA1 were cotransfected with I-SceI and DR-GFP. 48 h later cells were harvested and analyzed by FACS. siBRCA1 was used as a positive control. siNC: negative control. (D) RBM45 overexpression enhances the HR efficiency. 48 h after HA-RBM45 or HA-vector cotransfection with I-SceI and DR-GFP, cells were harvested and analyzed by FACS. (E) Detection of DSB repair efficiency mediated by NHEJ. 293T cells expressing siRNAs for NC, RBM45, or Ku70, were co-transfected with pCherry and pEGFP-Pem1-Ad2 plasmids. 24 h later cells were harvested and analyzed by FACS. siKu70 was used as a control. siNC: negative control. (F) RBM45 overexpression enhances the NHEJ efficiency. 24 h after HA-RBM45 or HA-vector cotransfection with pCherry and pEGFP-Pem1-Ad2 plasmids, cells were harvested and analyzed by FACS. P values from unpaired t test were included.
To further address the functional significance of the recruitment of RBM45 to DSB sites, we next examined whether RBM45 regulated NHEJ or HR, two major DSB repair pathways. The role of RBM45 on HR was evaluated in 293T cells transfected with DR-GFP reporter and I-SceI-IRES- DsRedNLS plasmid. Cleavage of the I-SceI sites leads to the restoration of the GFP gene through HR (Supplementary Figure S3A). We observed that similar to BRCA1 depletion, knockdown of RBM45 led to a significant reduction of GFP positive cells (Figure 4C, Supplementary Figure S3B). Meanwhile, no obvious changes in the cell cycle profile could be detected when RBM45 was depleted (Supplementary Figure S3C), indicating that the observed HR defect was not indirectly caused by a change in the cell cycle distribution. In line with that, overexpression of RBM45 could markedly enhance HR efficiency (Figure 4D). For the NHEJ assay, pEGFP-Pem1-Ad2 plasmid was linearized with HindIII (NEB). A pCherry plasmid was co-transfected with linearized pEGFP-Pem1-Ad2 as a control for transfection efficiency (Supplementary Figure S3D). Similar to Ku70 depletion, knockdown of RBM45 led to a reduction in NHEJ efficiency (Figure 4E, Supplementary Figure S3E). Consistently, overexpression of RBM45 enhanced NHEJ efficiency (Figure 4F). These results suggest that RBM45 is important for HR and NHEJ.
FUS promotes RBM45 recruitment
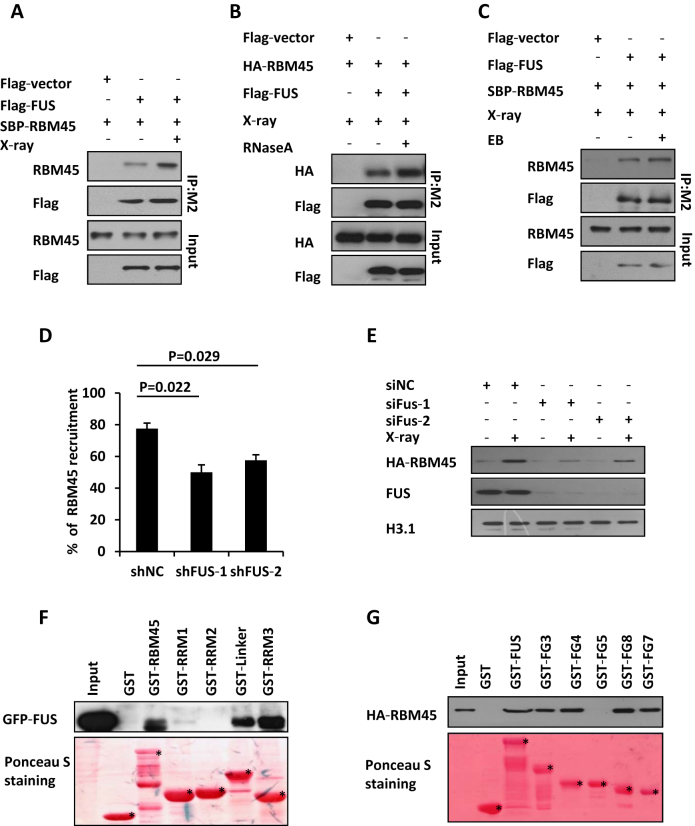
Given that RBM45 interacts with FUS under unperturbed condition (10,28), we wondered whether they still associate with each other after DNA damage treatment. Notably, Co-IP experiment showed that the interaction between RBM45 and FUS was markedly enhanced after X-ray treatment (Figure 5A). Considering FUS and RBM45 are both RNA binding proteins, which can also bind to RNA or DNA, we checked whether their interaction was mediated by RNA or DNA. We found that RBM45 still associated with FUS in the presence of RNase A or EB (Figure 5B, Figure 5C), indicating that this interaction is not mediated by RNA or DNA. Next, we examined whether FUS affected RBM45 accumulation after DNA damage. FUS-depleted U2OS cells were transfected with GFP-RBM45 and exposed to laser microirradiation. We noted that depletion of FUS significantly decreased the proportion of cells with GFP-RBM45 enrichment compared to shNC-treated control cells (Figure 5D, Supplementary Figure S4A). Moreover, depletion of FUS also impaired the chromatin loading of RBM45 upon exposure to X-ray irradiation (Figure 5E). Conversely, knockdown of RBM45 did not affect FUS recruitment to DNA damage sites (Supplementary Figure S4B, C). These results suggest that RBM45 functions downstream of FUS after DNA damage.
Figure 5.
RBM45 interacts with FUS which promotes its recruitment. (A) Detection the interaction of RBM45 and FUS by Immunoprecipitation. 293T cells co-transfected with Flag-FUS and SBP-RBM45 were lysed immediately upon 10 Gy of X-ray exposure. Anti-M2 Flag beads were used for immunoprecipitation. The Immunoprecipitates (IP) and inputs were immunoblotted with antibodies against RBM45 or flag, respectively. (B) RNA is not essential for the interaction between RBM45 and FUS. 293T cells co-transfected with Flag-FUS and HA-RBM45 were lysed immediately upon 10 Gy of X-ray exposure. 100 μl RNase A (1 mg/ml) or PBS was added to 900 μl 293T lysate before immunoprecipitation with M2 beads. Antibodies against HA or Flag were used for immunoblotting. (C) DNA is not essential for the interaction between RBM45 and FUS. 293T cells were treated as in (A). The lysates were supplemented with or without EB (200 μg/ml) prior to immunoprecipitation with anti-Flag M2 beads. (D) FUS promotes the recruitment of RBM45 to DNA damage sites. 48 h after expressing the indicated mCherry-shRNA, U2OS cells were transfected with GFP-RBM45. Then cells were micro-irradiated. NC: negative control. Data are presented as mean ± SD. n = 2; 30–40 irradiated cells per experiment. P values from unpaired t test were included. (E) Immunoblotting the chromatin fractions from cells expressing siNC or siFUS to examine the enrichment of HA-RBM45 on chromatin after DNA damage. (F) Mapping the functional domains of RBM45 that interact with FUS by GST pull down assay. Purified GST-RBM45 fragments were incubated with the lysates of 293T cells expressing GFP-FUS followed by immunoblotting with anti-GFP antibody. GST-fusion proteins were shown by Ponceau S staining of the PVDF membrane. Asterisks mean specific bands. (G) GST pull down to map the FUS domains essential for RBM45-FUS interaction as in (F). Asterisks mean specific bands.
To characterize the domain(s) of RBM45 required for its association with FUS, we purified a series of truncated GST-RBM45 fragments and performed GST-pull down assay with GFP-FUS expressing cell lysates. We found that the FUS-binding domains overlapped with its PAR-binding motifs. Namely, both the Linker and RRM3 motifs interacted with FUS strongly, while RRM1 exhibited a weak interaction and RRM2 displayed no interaction with FUS (Figure 5F).
We also mapped the domains in FUS mediated its interaction with RBM45. A series of truncated GST-FUS peptides (Supplementary Figure S4D) were purified and incubated with HA-RBM45 expressing lysates. All of the peptides except FG5 which includes the RRM motif were found to interact with RBM45 (Figure 5G), indicating that FUS associates with RBM45 through multiple domains.
RBM45 competes with HDAC1 to interact with FUS, and prevents HDAC1 from excessive recruitment
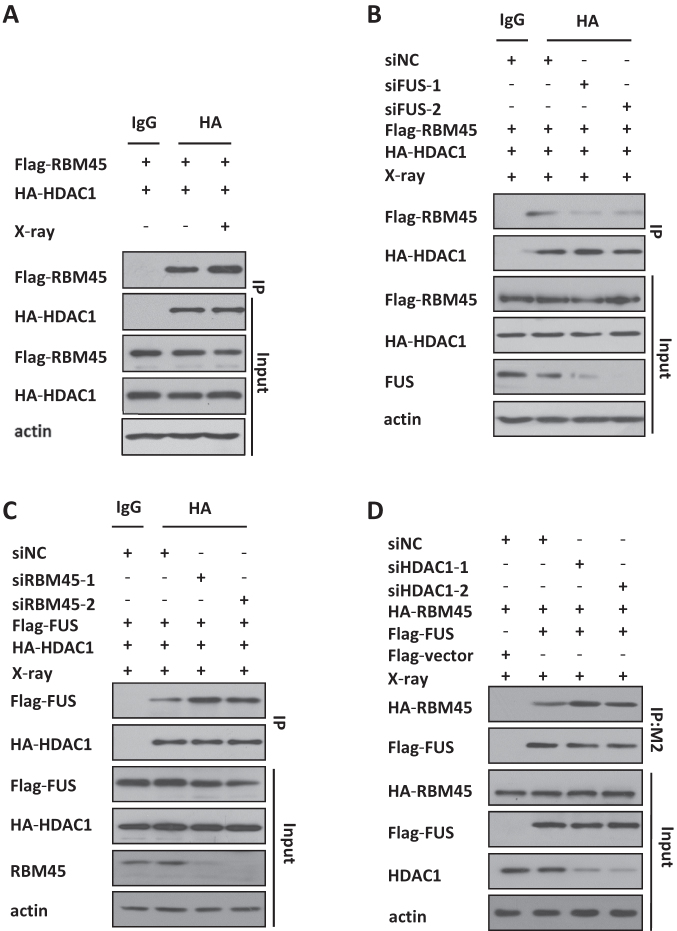
FUS is known to associate with HDAC1 and promote the recruitment of HDAC1 to DNA damage sites (9). Our finding that FUS is also required for RBM45 recruitment prompt us to check the relationship among these proteins. First, through Co-IP we found that RBM45 also associated with HDAC1 under physiological condition, and X-ray exposure enhanced their interaction (Figure 6A). Interestingly, the RBM45/HDAC1 interaction was dramatically decreased when FUS was knock down with siRNA (Figure 6B), indicating that FUS might mediate their interaction. Then we examined the effect of RBM45 on the association of FUS and HDAC1. Intriguingly, the binding of FUS and HDAC1 was obviously increased after depletion of RBM45 (Figure 6C). Similarly, knockdown of HDAC1 also enhanced the interaction between FUS and RBM45 (Figure 6D). These results revealed that RBM45 competed with HDAC1 to interact with FUS.
Figure 6.
RBM45 competes with HDAC1 for binding with FUS. (A) RBM45 interacts with HDAC1. After transfected with the indicated plasmids, cells were lysed and immunoprecipitated by antibodies against HA or IgG, and the bound proteins were analyzed by immunoblotting with anti-Flag monoclonal antibody. (B) Depletion of FUS decreases the interaction between RBM45 and HDAC1. 48 h after expressing siNC or siFUS, 293T cells were cotransfected with Flag-RBM45 and HA-HDAC1, then lysed and immunoprecipitated by anti-HA antibody. The bound proteins were analyzed by immunoblotting with anti-Flag. (C) RBM45 knockdown enhances the cooperation between FUS and HDAC1. 293T cells were transfected with siNC or siRBM45 before co-transfected with flag-FUS and HA-HDAC1. Anti-HA and anti-Flag antibodies were used for precipitation and blotting, respectively. (D) HDAC1 knockdown increases the interaction between RBM45 and FUS. 293T cells expressing siRNA were co-transfected with flag-FUS and HA-RBM45. Anti-Flag M2 beads were used for immunoprecipitation, and anti-HA antibody was used for immunoblotting.
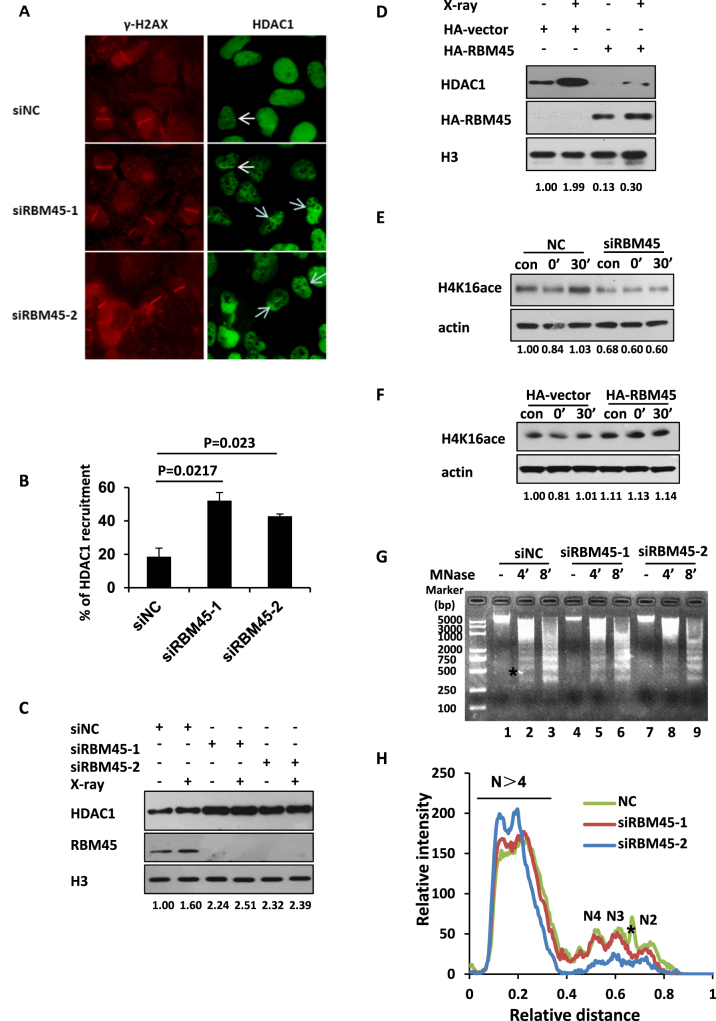
Next, we set out to determine whether RBM45 influences the recruitment of HDAC1 to the sites of DNA damage. RBM45-depleted U2OS cells were exposed to microirradiation and the recruitment of endogenous HDAC1 was checked. We noted that depletion of RBM45 led to an overt increase in the recruitment of HDAC1 to the damage sites (Figure 7A, B, Supplementary Figure S5A). Additionally, the chromatin loading of HDAC1 was also remarkably increased upon RBM45 depletion under physical condition or X-ray irradiation (Figure 7C). In contrast, western blotting analysis showed that RBM45 overexpression caused a reduced chromatin loading of HDAC1 (Figure 7D). Therefore, RBM45 negatively regulates the recruitment of HDAC1.
Figure 7.
RBM45 regulates the recruitment of HDAC1. (A and B) Depletion of RBM45 increases the percentage of cells with HDAC1 accumulation. U2OS cells were transfected with siNC or siRBM45. 48 h later, cells were laser micro-irradiated, fixed and stained with HDAC1 and γH2AX antibodies. Cells with HDAC1 recruited to the laser-induced sites of damage were counted. Data are presented as mean ± SD. n = 2; 30–40 irradiated cells per experiment. P values from an unpaired t test were included. (C) RBM45 knockdown enhances the enrichment of HDAC1 on chromatin. Chromatin fractions from siNC- or siRBM45-treated HeLa cells were extracted at 30 min after X-ray irradiation followed by immunoblotting with anti-HDAC1 antibody. (D) Overexpression of RBM45 inhibits the binding of HDAC1 to chromatin. Chromatin fractions from HeLa cells expressing HA-vector or HA-RBM45 were extracted followed by western blotting with anti-HDAC1 antibody. (E) Depletion of RBM45 affects H4K16ac. 72 h after transfected with siNC or siRBM45, cells were irradiated with X-ray (10 Gy). 0 or 30 min later, cells were harvested for immunoblotting. (F) Overexpression of RBM45 affects H4K16ac. 24 h after transfected with either HA or HA-RBM45, cells were treated as in (E). (G, H) RBM45 affects the chromatin structure. HeLa cells expressing siNC or siRBM45 were digested with micrococcal nuclease (MNase, MN) for 4 or 8 min. The genomic DNA was analyzed using agarose gel electrophoresis (G). The position of size markers were shown at the left. The intensities of lane 2, 5, 8 were consecutively quantified by using Quantity One software (Bio-Rad). N2–N4 reflect the number of nucleosomes in each oligonucleosome (H). The asterisk is a nonspecific signal.
HDAC1 is known to regulate the acetylation of histone H4 at lysine 16 (H4K16ac) (29), we test if depletion of RBM45 affects the level of H4K16ac. Western blotting analysis revealed that the level of H4K16ac displayed a dynamic change within 30 min after IR exposure, which was decreased immediately upon exposure to X-ray irradiation, and recovered 30 min later (Figure 7E). Intriguingly, knockdown of RBM45 resulted in an obvious reduction at the level of H4K16ac under unperturbed condition (Figure 7E). Additionally, the IR-triggered dynamic change of H4K16ac level observed in siNC-treated cells was hardly detected in RBM45-depleted cells (Figure 7E). Meanwhile, overexpression of RBM45 increased the level of H4K16ac in the absence or presence of X-ray irradiation (Figure 7F). Considering H4K16ac can regulate chromatin structure (30–32), which is important for efficient DNA damage response, we examined the effect of RBM45 depletion on chromatin structure. The nuclei of RBM45-depleted cells were isolated and digested with micrococcal nuclease (MNase) to evaluate its effect on nucleosome compaction. We found that the nuclei from the siNC control was more sensitive to MNase treatment relative to those from RBM45-depleted group (Figure 7G and H), indicating that the global chromatin structure is more ‘close’ in the absence of RBM45.
FUS mutant R521C prefers to interact with RBM45 than HDAC1
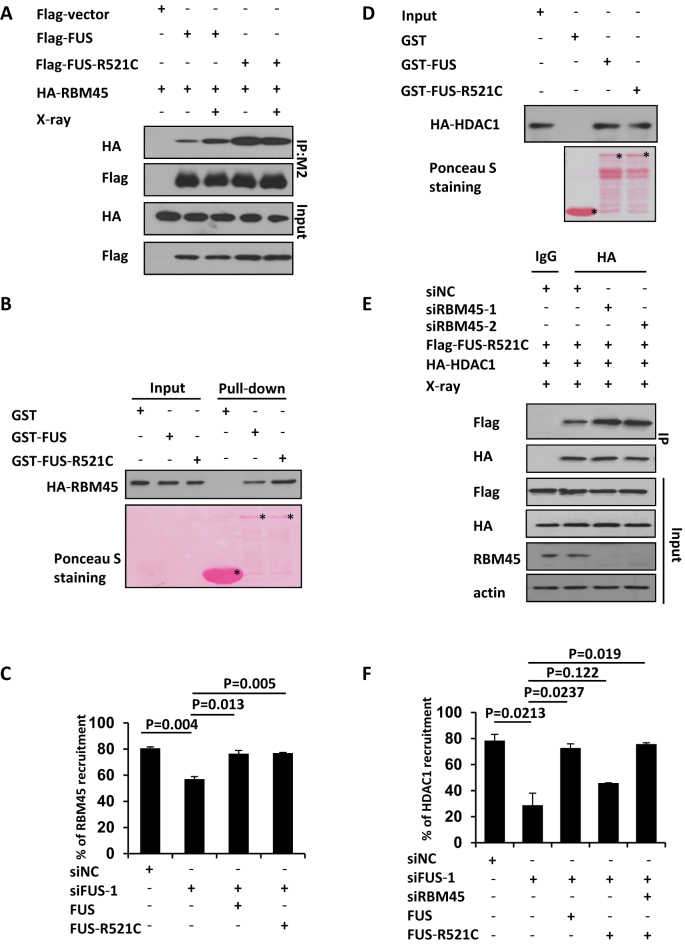
FUS-R521C mutation is a common fALS-associated mutation, and FUS-R521C mice exhibits evidence of DNA damage (33,34). FUS mutant R521C is reported to have a reduced interaction with HDAC1 in vivo (9). Thus we assessed whether FUS-R521C mutant still associates with RBM45. Intriguingly, our Co-IP result revealed that the interaction between FUS-R521C and RBM45 was dramatically increased compared to that between WT FUS and RBM45 under unperturbed condition and X-ray irradiation (Figure 8A). FUS R521C mutation is known to mildly impair nuclear import (9), we therefore isolated the cytoplasmic and nuclear fractions to check where the enhanced interaction occurs. The results showed that FUS-R521C and FUS mainly localized in nuclei, where FUS-R521C exhibited enhanced interaction with RBM45 (Supplementary Figure S5D). Consistently, GST-FUS-R521C also exhibited an enhanced association with RBM45 relative to GST-FUS in vitro (Figure 8B), indicating that R521C mutation confers FUS an increased affinity to RBM45. This finding prompted us to check whether FUS-R521C could promote RBM45 recruitment to the sites of damage. We complemented FUS-depleted cells with siRNA-resistant WT or R521C FUS expression constructs. Interestingly, both WT and R521C mutant FUS could rescue the defective RBM45 recruitment in siFUS-treated cells after microirradiation (Figure 8C, Supplementary Figure S5B). Considering FUS mutant R521C is defective in association with HDAC1 in vivo, we wondered whether mutation of R521C directly interferes with the physical binding between FUS and HDAC1. We performed a GST pull down assay and found that the ability of FUS-R521C to interact with HDAC1 was comparable with that of WT FUS (Figure 8D). This result prompted us to wonder the decreased interaction between FUS-R521C and HDAC1 in vivo was caused by a competitively enhanced association between FUS-R521C and RBM45. To confirm that, we harvested the RBM45-depleted cell lysates for co-IP assay. In line with our expectation, knockdown of RBM45 promoted an enhanced interaction between FUS-R521C and HDAC1 (Figure 8E). Furthermore, FUS-R521C could also completely rescue the impaired accumulation of HDAC1 to the damage site in siFUS-treated cells when RBM45 was depleted (Figure 8F, Supplementary Figure S5C).
Figure 8.
FUS mutant R521C prefers to interact with RBM45 than HDAC1. (A, B) FUS- R521C mutation in fALS enhances its interaction with RBM45. (A) Cells co-transfected with Flag-FUS or Flag-FUS-R521C with HA-RBM45 were harvested for immunoprecipitation using M2 beads. Antibodies against HA or Flag were used for blotting. (B) Purified GST-FUS or GST-FUS-R521C was incubated with cell lysate expressing HA-RBM45. Antibody against HA was used for blotting. Asterisks mean specific bands. (C) FUS-R521C mutation promotes RBM45 recruitment to the damage sites. 48 h after expressing siNC or siFUS, cells were transfected with Flag-FUS or Flag-FUS-R521C as well as GFP-RBM45, followed by microirradiation. Data are presented as mean ± SD. n = 2; 30–40 irradiated cells per experiment. P values from unpaired t test were included. (D) FUS-R521C mutation does not affect its interaction with HDAC1 in vitro. Purified GST-FUS or GST-FUS-R521C was incubated with cell lysates expressing HA-HDAC1. Antibody against HA was used for blotting. Asterisks mean specific bands. (E) Depletion of RBM45 stimulates the association between FUS-R521C and HDAC1 in vivo. 293T cells transfected with siNC or siRBM45 were co-transfected with Flag-FUS-R521C and HA-HDAC1. Anti-HA was used for immunoprecipitation. (F) RBM45 knockdown makes FUS-R521C functional in promoting HDAC1 recruitment. 48 h after transfection of siRNAs targeting FUS and RBM45, cells were transfected with FUS or FUS-R521C followed by microirradiation. Cells were then stained with antibody against HDAC1. Data are presented as mean ± SD. n = 2; 30–40 irradiated cells per experiment.
DISCUSSION
RBM45, a member of neural RNA-binding proteins, has been shown to play an important role in DDR in our study. We have found that RBM45 is recruited to the damage sites in a PAR- and FUS-dependent manner, and it promotes DSB repair by preventing HDAC1 from excessive recruitment (Figure 9A). FUS-R521C mutation in ALS patients enhances its interaction with RBM45, which likely accounts for the diminished interactions between FUS-R521C and HDAC1 in vivo, and thereby decreases the recruitment of HDAC1 to sites of damage (Figure 9B). It is known that an impairment of HDAC1 recruitment can decrease the efficiency of NHEJ (35), a primary mechanism for DNA DSB repair in postmitotic neurons, thereby causing increased DNA damage in ALS patients. Therefore, our work provides a further explanation for the pathogenesis of ALS disease.
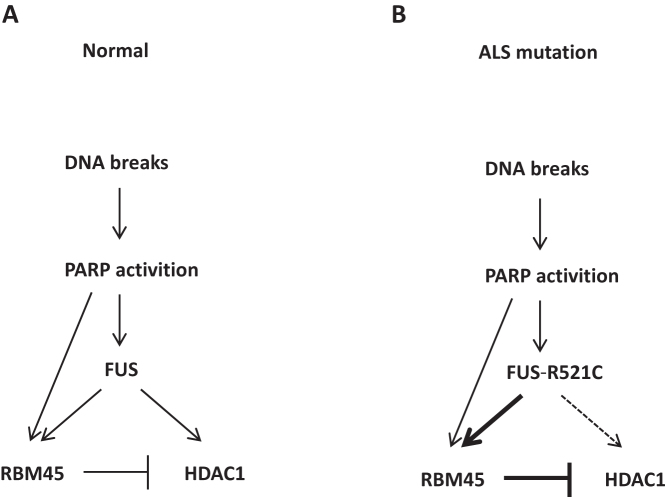
Figure 9.
Models of RBM45 in DNA damage response under normal and ALS mutation conditions. (A) RBM45 relocalizes to the sites of DNA damage in a way dependent on PAR and FUS to prevent excessive recruitment of HDAC1. (B) FUS-R521C, a mutant in fALS patients, enhances its interaction with RBM45, which decreases its association with HDAC1 and diminishes the recruitment of HDAC1.
Protein poly ADP-ribosylation (PARylation) is a widespread post-translational modification at or near DNA lesions, which modulates a number of biological processes including chromatin reorganization and DDR (36,37). PARylation appears rapidly at DNA damage sites and serves as an initial sensor to mediate the early recruitment of DNA damage repair machineries. Here, we showed that, analogous to FUS, RBM45 interacts with PAR chain directly and is recruited to sites of damage in a PAR-dependent manner. Meanwhile, FUS is also required for efficient RBM45 recruitment after DNA damage. It is reported that RNA and PAR chain share similar structures (38). RBM45 consists of three RRM domains and a Linker domain. We found that all of them, except RRM2 domain, could be recruited to the DNA damage sites. Consistently, GST-pull down assay showed that RRM2 domain failed to interact with PAR or FUS. To our surprise, Linker domain manifested an obvious recruitment after laser microirradiation and a strong interaction with PAR chain. Structure analysis reveals that there is a potential RRM motif in the Linker domain, which itself can interact with RNA directly like RRM domains (39). These information provide an explanation for the PAR-dependent recruitment of the RBM45 Linker domain.
Growing evidence suggests that RNA binding or splicing proteins play major roles in DDR (40), while some of them function in DDR through association with RNA (41). Recently, small RNAs have been described to have an unanticipated direct role in the control of DDR activation at the sites of DNA damage (42–45). Additionally, lncRNAs have also been reported to play a role in DNA damage response (46–48). It is necessary to check whether the recruitment of RBM45 is RNA-dependent. Interestingly, RNA component seems not essential for both the assembly and stable retention of RBM45 at the sites of DNA damage in our experiments. However, given that RBM45-interacting proteins are mainly enriched in the nuclear RNA processing process in the Gene Ontology analysis (28), we could not exclude the possibility that the RNA binding of RBM45 might be involved in other aspects in DDR, which deserve further exploration in the future.
Changes of histone acetylation are likely to occur in a biphasic manner following DSB induction, with a rapid deacetylation on sites such as H4K16 occurring to promote NHEJ, possibly by generating a less dynamic ‘repressed’ chromatin state, then followed by histone acetylations that enhance HR by making chromatin more ‘open’. HDAC1 which belongs to Class I HDACs, has been reported to be recruited to the sites of DNA damage to regulate the acetylation of histone H3 lysine 56 (H3K56) and H4K16 (35). Depletion of HDAC1 has been shown to lead to a sustained DNA damage signaling and defects especially in NHEJ (35). Given that a coordinated chromatin relaxation and compaction is essential for an efficient DDR in physiological setting (49), the recruitment of HDAC1 to the sites of DNA damage must be precisely controlled. It has been reported that FUS interacts with HDAC1, and promotes its efficient recruitment after DNA damage (9). However, how to prevent an excessive HDAC1 recruitment to the sites of damage remains unclear. In this study, we revealed that RBM45 competed with HDAC1 for binding to FUS. Consistently, depletion of RBM45 led to an increased accumulation of HDAC1 to the sites of DNA damage and a more compact chromatin structure. Conversely, overexpression of RBM45 increased the level of H4K16ac. We also mapped the domains of FUS essential for FUS/RBM45 interaction (FUS-FG3, FG4, FG7, FG8 fragments) and found that they covered the motifs mediating FUS to associate with HDAC1 (FUS-FG4, FG7 fragments). Therefore, RBM45 functions as a brake to prevent the FUS-dependent excessive recruitment of HDAC1 and thereby fine-tuning the chromatin compaction of damaged region upon DNA damage.
An impairment of HDAC1 recruitment is known to decrease the NHEJ-mediated DSB repair (35). Given that NHEJ is considered to be the primary mechanism for DSB repair in postmitotic neurons, it will be detrimental for the neurons if the recruitment of HDAC1 is affected. Recently, it has been reported that fALS FUS-R521C mutant shows a reduced association with HDAC1 and a decreased HDAC1 recruitment, causing a marked reduction in NHEJ (9). However, the underlying mechanism responsible for the altered interaction between fALS FUS-R521C and HDAC1 remains unclear. In this study, we found that FUS WT and FUS-R521C were comparable in their abilities to interact with HDAC1, while the interaction between FUS-R521C and RBM45 was dramatically enhanced compared to that between WT FUS and RBM45. Intriguingly, depletion of RBM45 stimulated the interaction between FUS-R521C and HDAC1, and rescued the defective recruitment of HDAC1 to the damage sites in the presence of FUS-R521. Additionally, although fALS FUS-R521C has a negative effect on HDAC1 recruitment, we found that it does not interfere with the efficient RBM45 enrichment to the sites of damage. Therefore, the enhanced interaction between FUS-R521C and RBM45 leads to a decreased cooperation between FUS-R521C and HDAC1 and thereby diminishes the HDAC1 recruitment in vivo.
Collectively, we reveal that RBM45, which can form inclusion body in ALS patients, plays an important role in DDR. Given that RBM45 closely associates with FUS and TDP-43, two other ALS-linked RBPs whose cellular distribution are modulated by proteins with established roles in DNA damage signaling, it is possible that a dysfunctional DDR of RBM45 also contributes to the pathogenesis of ALS, which deserves further researches in the future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Huadong Pei (Beijing Proteome Research Center) for providing HR and NHEJ reporter plasmids, Dr. Yungui Yang (Beijing Institute of Genomics) for providing instructions on RNase A treatment, Tong Zhao (Institute of Microbiology) for help on FACS experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
NSFC81630078, MOST2017YFC1001001, NSFC31670822, NSFC31471331, NSFC91519324, NSFC31570816, NSFC81371415; CAS Strategic Priority Research Program [XDB14030300]; State Key Laboratory of Membrane Biology. Funding for open access charge: CAS/SAFEA International Partnership Program for Creative Research Teams, National Natural Science Foundation of China [91519324, 81630078].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yuan F., Qian L., Zhao X., Liu J.Y., Song L., D’Urso G., Jain C., Zhang Y.. Fanconi anemia complementation group A (FANCA) protein has intrinsic affinity for nucleic acids with preference for single-stranded forms. J. Biol. Chem. 2012; 287:4800–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo K., Li L., Li Y., Wu C., Yin Y., Chen Y., Deng M., Nowsheen S., Yuan J., Lou Z.. A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev. 2016; 30:2581–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y.. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012; 40:10287–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J.. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012; 14:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mastrocola A.S., Kim S.H., Trinh A.T., Rodenkirch L.A., Tibbetts R.S.. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 2013; 288:24731–24741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salton M., Lerenthal Y., Wang S.Y., Chen D.J., Shiloh Y.. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010; 9:1568–1576. [DOI] [PubMed] [Google Scholar]
- 8. Kuroda M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T. et al. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J. 2000; 19:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang W.Y., Pan L., Su S.C., Quinn E.J., Sasaki M., Jimenez J.C., Mackenzie I.R., Huang E.J., Tsai L.H.. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 2013; 16:1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y., Collins M., Geiser R., Bakkar N., Riascos D., Bowser R.. RBM45 homo-oligomerization mediates association with ALS-linked proteins and stress granules. Scientific Rep. 2015; 5:14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mashiko T., Sakashita E., Kasashima K., Tominaga K., Kuroiwa K., Nozaki Y., Matsuura T., Hamamoto T., Endo H.. Developmentally regulated RNA-binding protein 1 (Drb1)/RNA-binding motif protein 45 (RBM45), a nuclear-cytoplasmic trafficking protein, forms TAR DNA-binding protein 43 (TDP-43)-mediated cytoplasmic aggregates. J. Biol. Chem. 2016; 291:14996–15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins M., Riascos D., Kovalik T., An J., Krupa K., Hood B.L., Conrads T.P., Renton A.E., Traynor B.J., Bowser R.. The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients. Acta Neuropathol. (Berl.). 2012; 124:717–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo C., Tang T.S., Bienko M., Parker J.L., Bielen A.B., Sonoda E., Takeda S., Ulrich H.D., Dikic I., Friedberg E.C.. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 2006; 26:8892–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang X., Lv L., Chen Q., Yuan F., Zhang T., Yang Y., Zhang H., Wang Y., Jia Y., Qian L. et al. Mouse DNA polymerase kappa has a functional role in the repair of DNA strand breaks. DNA Repair (Amst.). 2013; 12:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Z., Huang M., Ma X., Li H., Tang T., Guo C.. REV1 promotes PCNA monoubiquitylation through interacting with ubiquitylated RAD18. J. Cell Sci. 2016; 129:1223–1233. [DOI] [PubMed] [Google Scholar]
- 16. Yang Y., Liu Z., Wang F., Temviriyanukul P., Ma X., Tu Y., Lv L., Lin Y.F., Huang M., Zhang T. et al. FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress. Nucleic Acids Res. 2015; 43:8325–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu H., Li X., Ning G., Zhu S., Ma X., Liu X., Liu C., Huang M., Schmitt I., Wullner U. et al. The Machado-Joseph disease deubiquitinase Ataxin-3 regulates the stability and apoptotic function of p53. PLoS Biol. 2016; 14:e2000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang H., Shao Z., Shi L.Z., Hwang P.Y., Truong L.N., Berns M.W., Chen D.J., Wu X.. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 2012; 287:21471–21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seluanov A., Mittelman D., Pereira-Smith O.M., Wilson J.H., Gorbunova V.. DNA end joining becomes less efficient and more error-prone during cellular senescence. PNAS. 2004; 101:7624–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang D., Zhou J., Liu X., Lu D., Shen C., Du Y., Wei F.Z., Song B., Lu X., Yu Y. et al. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. PNAS. 2013; 110:5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gagne J.P., Haince J.F., Pic E., Poirier G.G.. Affinity-based assays for the identification and quantitative evaluation of noncovalent poly(ADP-ribose)-binding proteins. Methods Mol. Biol. 2011; 780:93–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rulten S.L., Rotheray A., Green R.L., Grundy G.J., Moore D.A., Gomez-Herreros F., Hafezparast M., Caldecott K.W.. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014; 42:307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun Y., Yang Y., Shen H., Huang M., Wang Z., Liu Y., Zhang H., Tang T.S., Guo C.. iTRAQ-based chromatin proteomic screen reveals CHD4-dependent recruitment of MBD2 to sites of DNA damage. Biochem. Biophys. Res. Commun. 2016; 471:142–148. [DOI] [PubMed] [Google Scholar]
- 24. Wang Z., Wang F., Tang T., Guo C.. The role of PARP1 in the DNA damage response and its application in tumor therapy. Front. Med. 2012; 6:156–164. [DOI] [PubMed] [Google Scholar]
- 25. Pryde F., Khalili S., Robertson K., Selfridge J., Ritchie A.M., Melton D.W., Jullien D., Adachi Y.. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 2005; 118:2043–2055. [DOI] [PubMed] [Google Scholar]
- 26. Britton S., Coates J., Jackson S.P.. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 2013; 202:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abu-Zhayia E.R., Khoury-Haddad H., Guttmann-Raviv N., Serruya R., Jarrous N., Ayoub N.. A role of human RNase P subunits, Rpp29 and Rpp21, in homology directed-repair of double-strand breaks. Scientific Rep. 2017; 7:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y., Collins M., An J., Geiser R., Tegeler T., Tsantilas K., Garcia K., Pirrotte P., Bowser R.. Immunoprecipitation and mass spectrometry defines an extensive RBM45 protein-protein interaction network. Brain Res. 2016; 1647:79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson D.P., Spitz G.S., Tharkar S., Quayle S.N., Shearstone J.R., Jones S., McDowell M.E., Wellman H., Tyler J.K., Cairns B.R. et al. HDAC1,2 inhibition impairs EZH2- and BBAP-mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma. Oncotarget. 2015; 6:4863–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L.. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006; 311:844–847. [DOI] [PubMed] [Google Scholar]
- 31. Tse C., Sera T., Wolffe A.P., Hansen J.C.. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998; 18:4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shogren-Knaak M., Peterson C.L.. Switching on chromatin: mechanistic role of histone H4-K16 acetylation. Cell Cycle. 2006; 5:1361–1365. [DOI] [PubMed] [Google Scholar]
- 33. Qiu H., Lee S., Shang Y., Wang W.Y., Au K.F., Kamiya S., Barmada S.J., Finkbeiner S., Lui H., Carlton C.E. et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J. Clin. Invest. 2014; 124:981–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009; 323:1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P.. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010; 17:1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei H., Yu X.. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics. 2016; 14:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Min W., Bruhn C., Grigaravicius P., Zhou Z.W., Li F., Kruger A., Siddeek B., Greulich K.O., Popp O., Meisezahl C. et al. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat. Commun. 2013; 4:2993. [DOI] [PubMed] [Google Scholar]
- 38. Gibson B.A., Kraus W.L.. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012; 13:411–424. [DOI] [PubMed] [Google Scholar]
- 39. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wickramasinghe V.O., Venkitaraman A.R.. RNA processing and genome stability: cause and consequence. Mol. Cell. 2016; 61:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. d’Adda di Fagagna F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014; 24:171–178. [DOI] [PubMed] [Google Scholar]
- 42. Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y.. A role for small RNAs in DNA double-strand break repair. Cell. 2012; 149:101–112. [DOI] [PubMed] [Google Scholar]
- 43. Yang Y.G., Qi Y.. RNA-directed repair of DNA double-strand breaks. DNA Repair (Amst.). 2015; 32:82–85. [DOI] [PubMed] [Google Scholar]
- 44. Gao M., Wei W., Li M.M., Wu Y.S., Ba Z., Jin K.X., Liao Y.Q., Adhikari S., Chong Z., Zhang T. et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014; 24:532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francia S., Michelini F., Saxena A., Tang D., de Hoon M., Anelli V., Mione M., Carninci P., d’Adda di Fagagna F.. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012; 488:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma V., Khurana S., Kubben N., Abdelmohsen K., Oberdoerffer P., Gorospe M., Misteli T.. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep. 2015; 16:1520–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeng Q., Wang Q., Chen X., Xia K., Tang J., Zhou X., Cheng Y., Chen Y., Huang L., Xiang H. et al. Analysis of lncRNAs expression in UVB-induced stress responses of melanocytes. J. Dermatol. Sci. 2016; 81:53–60. [DOI] [PubMed] [Google Scholar]
- 48. Lees-Miller S.P., Beattie T.L., Tainer J.A.. Noncoding RNA joins Ku and DNA-PKcs for DNA-break resistance in breast cancer. Nat. Struct. Mol. Biol. 2016; 23:509–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burgess R.C., Burman B., Kruhlak M.J., Misteli T.. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014; 9:1703–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.