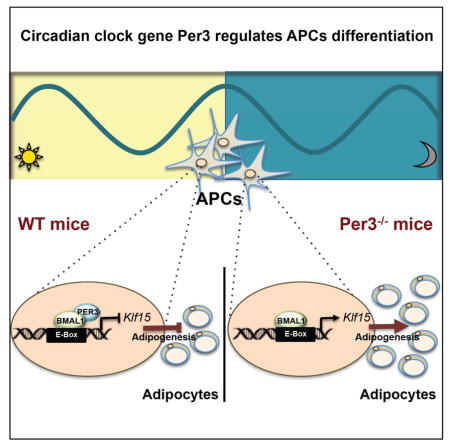
Summary
The generation of new adipocytes from precursor cells (adipogenesis) has implications for systemic metabolism and is a commonly used model for studying the process of cell differentiation in vitro. Previous studies from us, and others, suggested that the peripheral circadian clock can influence adipogenesis in vitro, but the mechanisms driving this activity and the relevance for adipogenesis in vivo are unknown. Here we reveal that mouse adipocyte precursor cells (APCs) contain a circadian clock that oscillates in vivo. We expose context-specific features of the clock in APCs: expression of the canonical core clock component Per1 does not significantly oscillate, whereas the lesser-understood paralog Per3 has a prominent rhythm. We discovered that deletion of Per3 promotes adipogenesis in vivo by a clock output pathway in which PER3 and BMAL1 directly regulate Klf15 expression. These findings demonstrate that Per3 has a major role in the APC clock and regulates adipogenesis in vivo.
Keywords: adipocyte precursor cells, circadian clock, adipogenesis, Per3, Klf15
Graphical Abstract
Introduction
The circadian clock is an ancient intricate molecular network of feedback loops that cycle with a daily rhythm and entrains organisms to the ~24 hour light/dark cycles of the day (Bass and Lazar, 2016; Reppert and Weaver, 2002). The first circadian clock component identified in animals was the Period gene (Per) in drosophila melanogaster and flies with mutations in this gene have altered periodicity of their circadian rhythm (Hardin et al., 1990; Konopka and Benzer, 1971). Mammals have three Per paralog family members (Per1-3) that function as major elements of the circadian clock. Per1 and Per2 are established core components of the negative arm of the circadian clock (Bae et al., 2001). The role of Per3 in the circadian clock is less well understood but it is not thought to be a core clock component, in part, because deletion of Per3 has a modest effect on circadian behavior in mice (Bae et al., 2001; Shearman et al., 2000).
In mammals, the circadian clock was first identified in the central nervous system (CNS) where it receives entrainment signals that originate in retinal photoreceptors that sense the day/night light cycles (Reppert and Weaver, 2002). Peripheral cells can also contain a circadian clock and receive entrainment cues from the central oscillator (Schibler et al., 2015). However, there are context-specific characteristics, as well as tissue-specific inputs, which distinguish peripheral clocks from the central clock in the CNS (Balsalobre et al., 2000; Bass and Lazar, 2016). Peripheral circadian clocks have a significant influence over systemic metabolism (reviewed in (Asher and Schibler, 2011; Bass and Takahashi, 2010; Feng and Lazar, 2012)). For example, circadian oscillations in the liver are highly responsive to dietary changes and disruption of these oscillations in hepatocytes is implicated in the pathogenesis of metabolic disease (Hatori et al., 2012). Adipose tissue is also an influential regulator of metabolism and mature adipocytes contain a circadian clock (Henriksson and Lamia, 2015; Otway et al., 2009; Yang et al., 2006). Furthermore, expression of circadian clock components in adipocytes is connected to adipose tissue modulation of metabolism (Kettner et al., 2015).
In addition to roles in adipocyte physiology, evidence suggests that the circadian clock also affects the highly regulated process of adipogenesis from precursor cells (Costa et al., 2011; Grimaldi et al., 2010; Kawai et al., 2010; Shimba et al., 2005a). However, prior studies examining a putative role for the circadian clock in adipogenesis were forced to rely on in vitro assays, with surrogate cell types modeling adipocyte precursor cells. Attempts to validate putative connections between the circadian clock and adipogenesis identified using these approaches could not be observed in vivo using the available tools (Grimaldi et al., 2010); leaving the relevance of the circadian clock and its components to physiological induction of adipogenesis unclear. Recent advances in stem cell biology have identified endogenous adipose-depot resident bona fide adipocyte precursor cells (APCs) enabling in vivo studies on adipogenesis (Rodeheffer et al., 2008; Tang et al., 2008). Therefore, we applied these new approaches to investigate whether the circadian clock is present in primary APCs, relevant to adipogenesis in vivo and, if so, elucidate the mechanisms by which physiologically induced adipogenesis is regulated by the circadian clock.
In this study, we demonstrate that primary APCs contain a functioning circadian clock. Strikingly, we discovered that the poorly understood Per3 paralog gene has robust circadian oscillations in expression in APCs and deletion of the Per3 gene significantly increases adipogenesis in vivo. We elucidated that the mechanism of this activity occurs by a previously unrecognized complex containing PER3 and BMAL1 that regulates the expression of Kruppel-Like factor 15 (Klf15). Our study establishes that a PER3 mediated output pathway from the circadian clock crosstalks with Klf15 to regulate adipogenesis in APCs.
Results
Endogenous and primary APCs contain a functional circadian clock
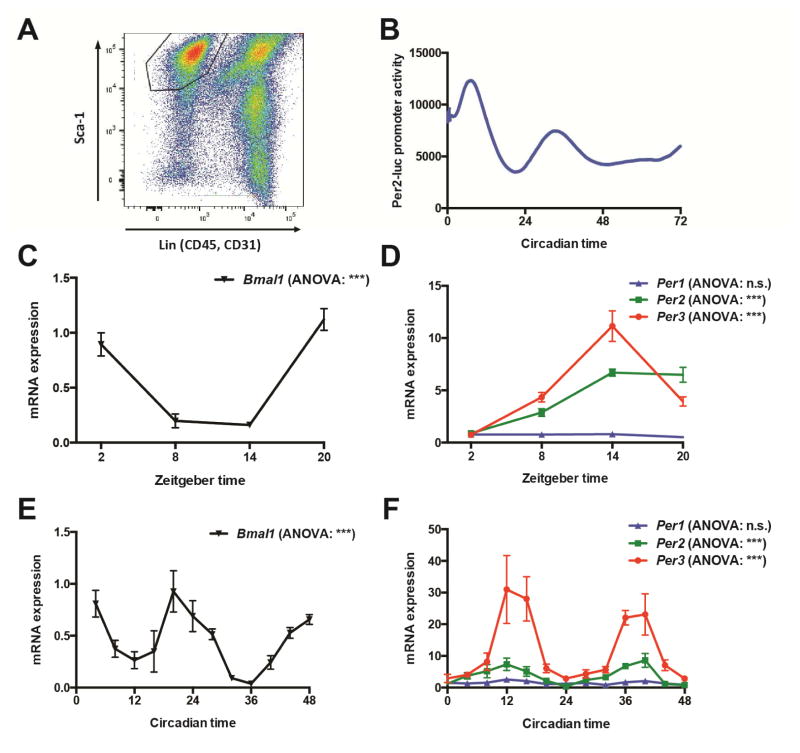
In order to investigate if bona fide APCs contain a circadian clock, we used mice that have a luciferase reporter transgene gene knocked-in to the Per2 locus resulting in the production of luciferase protein fused to the C-terminus of PER2 (mPer2Luc)(Yoo et al., 2004). As the Per2 gene codes for the expression of a core circadian clock component, expression of luciferase under Per2 regulation is a validated reporter of core circadian clock activity in cells (Liu et al., 2007). We performed fluorescence activated cell sorting (FACS) to purify a defined population of APCs from the subcutaneous adipose depot that is positive for stem cell antigen (Sca1+) and negative for cell lineage markers, CD45 and CD31 (Figure 1A) from mPer2Luc mice as previously described (Krueger et al., 2014; Rodeheffer et al., 2008). Using continuous real-time monitoring of luciferase activity in the purified APCs, we established that primary APCs contain a functional circadian clock (Figure 1B).
Figure 1. APCs contain a circadian clock that oscillates in vivo.
(A) FACS plot with gating on population of primary APCs from adipose tissue.
(B) Real-time continuous monitoring of luciferase activity in primary APCs from mPer2Luc mice.
(C, D) Quantification by RT-qPCR of the expression levels of circadian clock genes in APCs immediately lysed following isolation by FACS from WT mice across an in vivo time-course (N = 3–8 mice per time-point). Data analyzed by one-way analysis of variance (ANOVA) and post hoc Bonferroni.
(E, F) Quantification by RT-qPCR of the expression levels of circadian clock genes in SVF immediately lysed from WT mice housed in constant darkness across an in vivo time-course (N = 3 mice per time-point). Data analyzed by one-way ANOVA and post hoc Bonferroni.
***P <0.001. n.s. non-significant. Error bars represent ± SEM.
See also Figure S1
In order to test if the circadian clock is oscillating in APCs in vivo, we performed a time-course experiment isolating APCs by FACS from wild-type mice (WT) at different zeitgeber (ZT) times followed by immediate cell lysis and RNA isolation. Using reverse transcription and quantitative polymerase chain reaction (RT-qPCR) to measure the expression levels of circadian clock components throughout the time-course, we found compelling evidence that the clock is oscillating in vivo in APCs (Figures 1C and 1D). Bmal1 expression levels were significantly down-regulated during Zeitgeber times 8–14 [ZT8-14], and increased during ZT20-ZT2 (Figure 1C). As expected, Per2 and Per3 expression levels oscillated anti-phasic with Bmal1 (Figure 1D). However, surprisingly, Per1 expression did not significantly oscillate by ANOVA analysis in APCs in vivo (Figure 1D), but did oscillate in the liver (Figure S1), indicating context-specificity of the clock components cycling in APCs. In order to perform a high-resolution analysis of the circadian clock as well as examine in vivo circadian oscillations in the adipose progenitor cells under constant darkness (to analyze the APC clock independent from entrainment cues from the central clock), we took advantage of the fact that the stromal vascular fraction (SVF) of adipose tissue in very young mice is significantly enriched for APCs. Using SVF enabled us to isolate cells every four hours from the subcutaneous depots with significantly less manipulation of the cells. These studies confirmed the presence of a circadian clock as well as the robustness of oscillations in Per3 expression during constant darkness (Figures 1E and 1F).
Per3 regulates adipogenesis in APCs in vivo
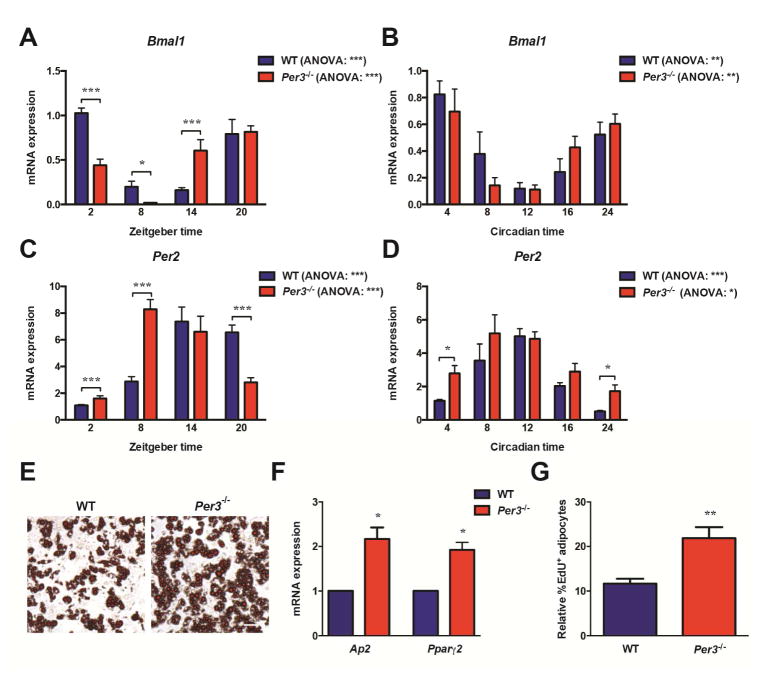
The unexpected absence of significant oscillations in Per1 expression coupled with the particularly robust oscillations in Per3 we discovered in APCs inspired us to hypothesize that the expression profile of the Per paralog genes has specific functional relevance for APC activity in vivo. Per2 is an established core circadian clock component but significantly less is known about the role of Per3 in circadian biology. We used a mouse line with the Per3 gene deleted (Per3−/−) (Shearman et al., 2000) to elucidate the impact of Per3 on core circadian clock oscillation in APCs in vivo. We found that the deletion of Per3 causes alterations in the expression of core circadian clock components Bmal1 and Per2 in APCs and in the SVF of mice housed in constant darkness (Figures 2A–2D), but oscillations were still significant by ANOVA. The effect of Per3 deletion on Per1 expression was not significant (Figure S2A).
Figure 2. Per3 regulates adipogenesis in APCs in vivo.
(A, C) Quantification by RT-qPCR of the expression levels of circadian clock genes Bmal1 and Per2 in APCs immediately lysed following isolation by FACS from WT and Per3−/− mice across an in vivo time-course (N = 5 mice per time-point for each genotype). Data within a genotype was analyzed by one-way ANOVA; the effect of genotype and time was tested by two-way ANOVA followed by post hoc Bonferroni.
(B, D) Quantification by RT-qPCR of the expression levels of circadian clock genes Bmal1 and Per2 in SVF immediately lysed from WT and Per3−/− mice housed in constant darkness across an in vivo time-course (N = 3 mice per time-point for each genotype). Data within a genotype was analyzed by one-way ANOVA; the effect of genotype and time was tested by two-way ANOVA followed by post hoc Bonferroni.
(E) Levels of spontaneous adipogenesis in APCs isolated from Per3−/− mice compared to WT mice stained with Oil Red O. Scale bar represents 100 μm.
(F) Levels of spontaneous adipogenesis in APCs isolated from Per3−/− mice compared to WT mice quantified by measuring the expression levels of Ap2 and Pparγ2 using RT-qPCR. Data analyzed by Student’s t-test.
(G) Flow cytometry quantifying the relative percentage of EdU+ adipocytes after an in vivo pulse-chase to measure the level of adipogenesis in Per3−/− mice compared to WT littermates (N = 5 mice per genotype). Data analyzed by Student’s t-test.
*P <0.05, **P <0.01, ***P <0.001. Error bars represent ± SEM.
See also Figure S2
In order to test for a circadian clock output pathway mediated by Per3 that is functionally germane to APC activity, we investigated if deletion of Per3 alters the levels of adipogenesis in APCs. First, we monitored the levels of spontaneous adipogenesis in primary APCs sorted from Per3−/− mice compared to APCs from WT littermate controls. We found that APCs from Per3−/− mice had significantly higher levels of spontaneous adipogenesis as visualized by Oil Red O staining (Figure 2E) and quantified by measuring the expression levels of markers of adipogenesis (Figure 2F). Conversely, enforced overexpression of Per3, by infecting APCs with adenovirus that expresses Per3, blocked adipogenesis compared to APCs infected with control adenovirus expressing GFP (Figure S2B). Performing time-course experiments monitoring the expression of Pparγ (a transcription factor ‘master regulator’ of adipogenesis), confirmed that the Per3 enforced block in differentiation occurs early in the differentiation process (Figure S2C).
Next, we monitored for differences in adipogenesis rates in vivo in WT and Per3−/− mice. For these studies, we used a pulse-chase protocol with an analog of thymidine, 5-ethynyl-2’-deoxyuridine (EdU) pulse-chase protocol that we (Wong et al., 2016), and others (Jeffery et al., 2015), have previously developed and validated to measure the rate of adipogenesis in vivo (see Supplemental Experimental Procedures). We pulsed Per3−/− and WT littermate control mice with EdU for 3 days to label APCs followed by a two-week chase period. At the end of the chase period, the adipose depots were harvested and the mature adipocytes were isolated. The number of EdU labeled (EdU+) adipocytes was quantified by flow cytometry (Figure S2D). As mature adipocytes do not proliferate, EdU+ adipocytes arise from APCs that were labeled with EdU during the pulse and differentiated during the chase period. Using this approach, we monitored the rate of adipogenesis in vivo and found that Per3−/− mice had significantly higher rates of adipogenesis than the WT littermate controls (Figure 2G).
PER3 regulates the expression of Klf15 in APCs
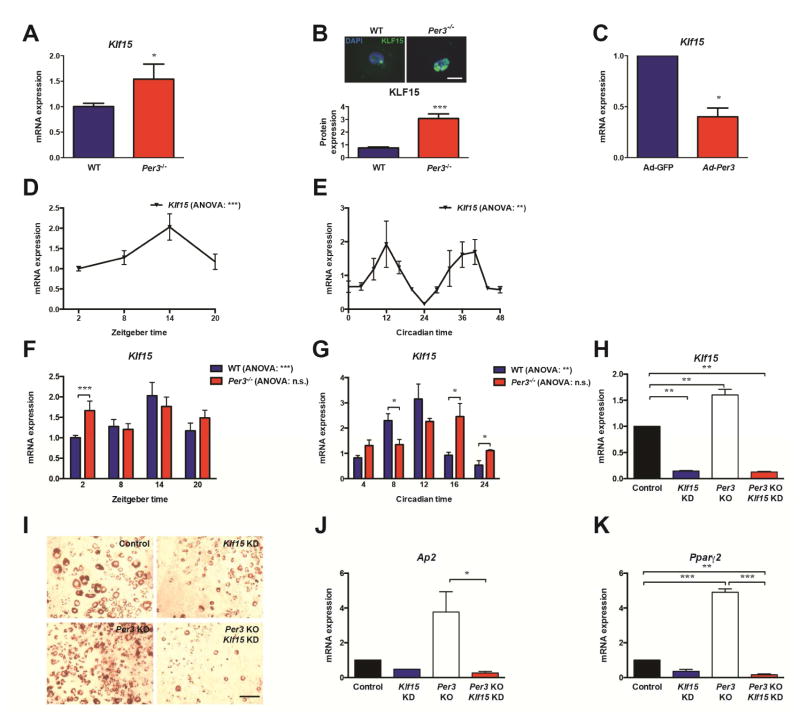
In order to identify the mechanism by which Per3 regulates differentiation of APCs in vivo, we performed genome-wide expression profiling comparing freshly isolated APCs from Per3−/− to WT littermate mice using massive parallel sequencing of the transcriptomes (RNA-Seq) at ZT2 (Figure S3). Interestingly, these studies revealed that the expression level of Klf15, a gene that displays circadian oscillations in expression in cardiac and liver cells (Han et al., 2015; Jeyaraj et al., 2012), and promotes adipogenesis in 3T3-L1 cells (Mori et al., 2005), is significantly increased in endogenous APCs from Per3−/− compared to WT APCs. In independent experiments, we confirmed the differential expression of Klf15 in APCs from Per3−/− compared to WT littermate mice by RT-qPCR (Figure 3A). Using immunohistochemistry on primary APCs following their isolation from the adipose tissues by FACS, we found that Per3−/− APCs also express higher levels of KLF15 protein compared to WT littermates (Figure 3B). Reciprocally, we found that overexpression of Per3 in APCs inhibits Klf15 expression (Figure 3C). Together, these studies reveal that Per3 regulates the expression of Klf15 in APCs.
Figure 3. PER3 regulates the expression of Klf15.
(A) Quantification by RT-qPCR of the expression of Klf15 in APCs from Per3−/− mice compared to WT littermate controls (N = 7 mice for each genotype). Data analyzed by Student’s t-test.
(B) Images (upper) and quantification (lower) of immunocytochemistry for KLF15 (green) and DAPI (blue) in APCs after isolation by FACS from Per3−/− mice compared to WT littermate controls (N = 3 mice per genotype). Scale bar represents 20 μm. Data analyzed by Student’s t-test.
(C) Quantification by RT-qPCR of the expression of Klf15 in APCs isolated from WT mice and infected with adenovirus expressing either GFP or Per3. Data analyzed by Student’s t-test.
(D) Quantification by RT-qPCR of the expression levels of Klf15 in APCs immediately lysed following isolation by FACS from WT mice across an in vivo time-course (N = 7 mice per time-point). Data analyzed by one-way ANOVA and post hoc Bonferroni.
(E) Quantification by RT-qPCR of the expression levels of Klf15 in SVF immediately lysed from WT mice housed in constant darkness across an in vivo time-course (N = 3 mice per time-point for each genotype). Data analyzed by one-way ANOVA and post hoc Bonferroni.
(F) Quantification by RT-qPCR of the expression levels of Klf15 in APCs immediately lysed following isolation by FACS from WT and Per3−/− mice across an in vivo time-course (N = 7 mice per time-point for each genotype). Data within a genotype was analyzed by one-way ANOVA; the effect of genotype and time was tested by two-way ANOVA followed by post hoc Bonferroni.
(G) Quantification by RT-qPCR of the expression levels of Klf15 in SVF immediately lysed from WT and Per3−/− mice housed in constant darkness across an in vivo time-course (N = 3 mice per time-point for each genotype). Data within a genotype was analyzed by one-way ANOVA; the effect of genotype and time was tested by two-way ANOVA followed by post hoc Bonferroni.
(H – K) RT-qPCR quantification of the expression levels of Klf15 (H), Ap2 (J) and Pparγ2 (K) and Oil red O staining (I) in APCs isolated from WT or Per3−/− mice infected with adenoviruses expressing shRNA-scramble control (Control) or shRNA-Klf15 vectors. Data analyzed by Student’s t-test.
*P <0.05, **P <0.01, ***P <0.001. n.s. non-significant Error bars represent ± SEM.
See also Figures S3 and S4
Our discovery that Per3 regulates Klf15 expression in the above static assays inspired us to test if the expression of Klf15 is circadian in APCs in vivo and, if so, would deletion of Per3 alter the rhythmicity of this expression pattern. Therefore, we monitored Klf15 expression over a time-course in APCs isolated from adipose tissue by FACS and immediately lysed for RNA extraction. These studies revealed that the expression of Klf15 oscillates in endogenous APCs in WT mice (Figure 3D). This cycling pattern is also present in the SVF of mice housed in constant darkness (Figure 3E). Intriguingly, deletion of Per3 altered the expression pattern of Klf15, resulting in the loss of significant oscillations in Klf15 expression (Figures 3F and 3G).
We next tested if Klf15 is functionally relevant to Per3 regulation of adipogenesis. Specifically, we tested if Klf15 is necessary for the increased level of adipogenesis observed in Per3−/− APCs. Importantly, we found that shRNA knockdown of Klf15 expression (Figure 3H) restored adipogenesis in Per3−/− APCs to approximately wild-type levels (Figures 3I–3K). Reciprocally, we found that blocking the repression of Klf15 expression by Per3 is sufficient to rescue APCs from Per3 mediated inhibition of adipogenesis (Figure S4A–D).
Klf15 is directly regulated by PER3 and BMAL1
The disruption of Klf15 oscillations in Per3−/− APCs suggests that a circadian clock output pathway, coordinated by Per3, regulates Klf15 expression in APCs. To begin to test this, we searched the DNA sequence of the Klf15 gene and proximal regions for evidence of canonical circadian clock regulatory elements. Our in silico analysis identified several putative E-box elements in the Klf15 gene. E-boxes are key cis-regulatory elements that can be occupied by circadian clock components to modulate the expression of target genes (Ko and Takahashi, 2006). We found that the DNA sequences of two of the putative E-boxes within the Klf15 gene were conserved in humans, suggesting functional importance (So et al., 2008). One of the sites is located in intron 1, ~1.2kb downstream of the transcriptional start site (TSS) and the other is located in exon 1, ~4.5kb downstream of the TSS (designated as E-box 1 and E-box 2, respectively). Therefore, we tested if these sites are relevant for PER3 regulation of Klf15 expression.
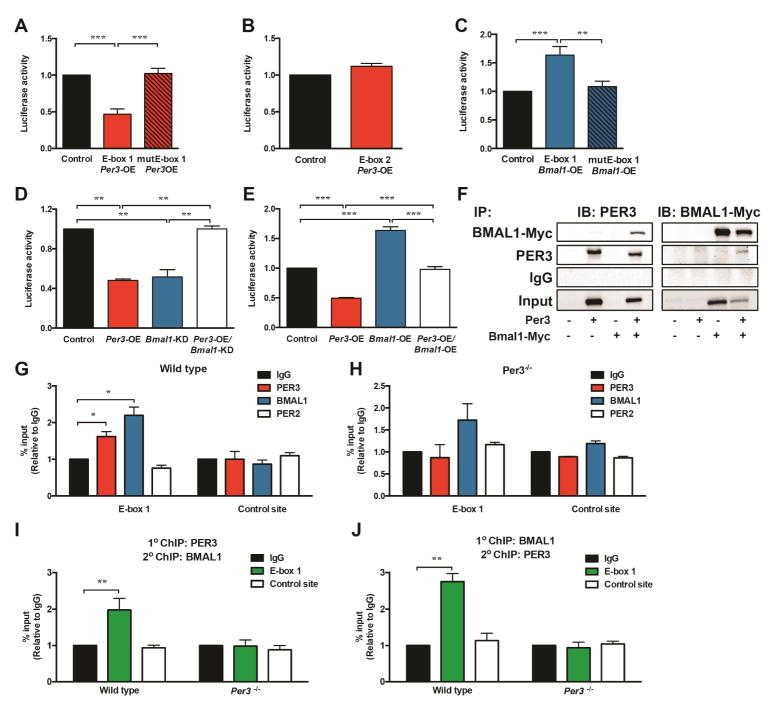
First, we generated luciferase reporter constructs containing DNA fragments that span these elements and performed luciferase assays on cells co-transfected with the reporter constructs and a Per3 expression plasmid or empty control plasmid. Interestingly, we found that Per3 inhibited the expression of luciferase activity only in cells transfected with the reporter construct containing E-box 1 (Figure 4A) and not the reporter construct containing E-box 2 (Figure 4B). To confirm that E-box 1 was the response element recognized by PER3 to regulate expression, we disrupted the canonical E-box 1 sequence in the reporter construct by performing site-directed mutagenesis. Mutating E-box 1 abolished Per3 regulation of the reporter activity (Figure 4A), indicating that this E-box is a critical site for PER3 regulation of Klf15 expression. PER proteins tether to response elements by forming a complex with other clock components (Papazyan et al., 2016). BMAL1 is a core clock component that directly occupies E-boxes (Darlington et al., 1998) and, therefore, we tested if BMAL1 could also affect the expression of the reporter construct that contains E-box 1. Interestingly, we found that Bmal1 induced expression of the reporter construct (Figure 4C). This induction was abolished when the E-box 1 sequence was mutated in the reporter construct (Figure 4C). Together, these data indicate that both Per3 and Bmal1 could use E-box 1 as a DNA response element but have opposite effects on transcription. Therefore, we tested for functional interactions between Per3 and Bmal1 in regulating expression of the Klf15 reporter construct. Intriguingly, we found that Bmal1 is required for Per3 mediated repression of expression of the reporter (Figure 4D) and, reciprocally, Per3 blocks Bmal1 induction of expression (Figure 4E). These results suggest that PER3 might form a complex with BMAL1 at E-box 1 that regulates expression. To test if PER3 can form a complex with BMAL1, we performed immunoprecipitation studies. We discovered that immunoprecipitating PER3 results in co-precipitation of BMAL1 and, reciprocally, immunoprecipitating BMAL1 results in co-precipitation of PER3 (Figure 4F).
Figure 4. Klf15 is directly regulated by PER3 and BMAL1.
(A) Luciferase assays on cells transfected with reporter plasmids containing the Klf15 wild-type E-box 1 (E-box 1) or a reporter plasmid with the E-box 1 sequence mutated by site directed mutagenesis (mutE-box 1) co-transfected with a Per3 expression plasmid. Luciferase activity in cells co-transfected with the reporter and Per3 expression plasmids (Per3-OE) were compared to activity in cells co-transfected with the E-box 1 containing reporter plasmid and empty control vector (Control). Data analyzed by Student’s t-test.
(B) Luciferase assays on cells transfected with a reporter plasmid containing the Klf15 E-box 2 co-transfected with a Per3 expression plasmid compared to the luciferase activity in cells co-transfected with the E-box 2 containing reporter plasmid and empty control vector (Control). Data analyzed by Student’s t-test.
(C) Luciferase assays on cells transfected with reporter plasmids containing the Klf15 wild-type E-box 1 (E-box 1) or a reporter plasmid with the E-box 1 sequence mutated by site directed mutagenesis (mutE-box 1) co-transfected with a Bmal1 expression plasmid (Bmal1-OE). Luciferase activity in cells co-transfected with the reporter and Bmal1 expression plasmids were compared to activity in cells co-transfected with the E-box 1 containing reporter plasmid and empty control vector (Control). Data analyzed by Student’s t-test.
(D) Luciferase assays on cells transfected with reporter plasmids containing the Klf15 wild-type E-box 1 co-transfected with Per3, shRNA-Bmal1 (Bmal1-KD) plasmids or both. Luciferase activities were compared to activity in cells co-transfected with the Klf15 reporter plasmid and scrambled control and empty plasmids (Control). Data analyzed by ANOVA followed by post hoc Bonferroni.
(E) Luciferase assays on cells transfected with reporter plasmids containing the Klf15 wild-type E-box 1 co-transfected with Per3, Bmal1 expression plasmids or both, Per3 and Bmal1 expression plasmids. Luciferase activities were compared to activity in cells co-transfected with the Klf15 reporter plasmid and empty control plasmids (Control). Data analyzed by ANOVA followed by post hoc Bonferroni.
(F) Immunoblots (IB) from Immunoprecipitation (IP) of BMAL1-MYC and PER3 from cells transfected with Per3 and/or Bmal1-Myc expression plasmids or control (untransfected) cells.
(G, H) Chromatin immunoprecipitation (ChIP) using antibodies against PER2, PER3, BMAL1 or IgG control antibodies at E-box 1 and a non-E-box control site in the Klf15 gene in SVF isolated form WT (G) and Per3−/− (H) mice. Data analyzed by Student’s t-test.
(I, J) ChIP of PER3 followed by re-ChIP of BMAL1 or IgG control (I) and BMAL1 ChIP followed by re-ChIP of PER3 or IgG control (J) at the E-box 1 site and a non-E-box control site in the Klf15 gene in SVF isolated from WT and Per3−/− mice. Data analyzed by Student’s t-test.
*P <0.05, **P <0.01, ***P <0.001. Error bars represent ± SEM.
Given the above results, we next decided to interrogate the endogenous Klf15 gene for the enrichment of PER3 and BMAL1 at the E-box 1 site. Using primary SVF isolated from WT mice, we performed chromatin immunoprecipitation (ChIP) of PER3 at E-box 1. We found that PER3 is enriched at E-box 1 relative to immunoprecipitation of the same region with control IgG antibodies, as well as compared to another DNA region proximal to the E-box 1 in the Klf15 gene (Figure 4G). In addition, we performed ChIP using chromatin isolated from Per3−/− mice as control, to confirm that the PER3 antibodies we used specifically immunoprecipitated PER3 (Figure 4H). Importantly, we also found that occupancy of this site is specific for PER3, as ChIP with PER2 antibodies revealed that PER2 is not enriched on the E-box (Figures 4G and 4H).
Using antibodies to BMAL1 that are validated for ChIP (DiTacchio et al., 2011), we found that, like PER3, BMAL1 is significantly enriched at E-box 1 in the Klf15 gene (Figure 4G). Finally, we tested if PER3 and BMAL1 co-occupy the endogenous E-box 1 site in the Klf15 gene. By performing ChIP-re-ChIP in both directions (both PER3 ChIP followed by re-ChIP of the DNA precipitated in the PER3 ChIP with BMAL1 as well as ChIP of BMAL1 followed by re-ChIP of the DNA precipitated in the BMAL1 ChIP with PER3), we revealed that PER3 and BMAL1 co-occupy the endogenous E-box 1 in the Klf15 gene (Figures 4I and 4J). We are not aware of prior reports identifying a regulatory complex containing PER3 and BMAL1. Therefore, our discovery of this interaction on an endogenous response element represents a previously unknown mechanism for circadian clock components to regulate the expression of target genes.
Discussion
Peripheral circadian clocks can influence body composition (Henriksson and Lamia, 2015). For example, deletion of core circadian clock components in mature adipocytes results in reduced energy expenditure, promoting an increase in adipose tissue mass (Paschos et al., 2012). This process is similar to the pathophysiological response observed when the circadian timing of caloric intake is perturbed (Arble et al., 2009; Chaix and Zarrinpar, 2015). While the effects mediated by mature adipocytes presumably primarily affect adipocyte size, adipose tissue can also expand by increasing cell number. A substantial amount of research has identified a number of candidate genes in preadipocyte model cell lines that regulate adipogenesis (reviewed in (Rosen and MacDougald, 2006) and (Cristancho and Lazar, 2011)), yet it remains unclear which of these are physiologically relevant. Intriguing, primarily using immortalized cell lines, prior studies implicated the circadian clock in the regulation of adipogenesis (Guo et al., 2012; Shimba et al., 2005b), suggesting a role for this intriguing system in the regulation of adipocyte number, in addition to the role it has in metabolism. Indeed, Pparγ, considered a ‘master regulator’ of adipogenesis, and the PPARγ co-factor PGC-1α were both found to have circadian oscillations in expression levels (Yang et al., 2006). However, the mechanisms by which the circadian clock mediates this process are poorly understood.
We applied advances in stem cell biology to study the circadian clock-adipogenesis connection and discovered mechanisms by which Per3 regulates adipogenesis in bona fide APCs. It is important to note that we chose to focus our studies on subcutaneous adipose tissue because the tools for identifying and monitoring bona fide APCs in this tissue are robustly validated (Rodeheffer et al., 2008); it is possible that there are context-specific regulation of APC activities in different adipose depots and types of adipocytes (Sanchez-Gurmaches et al., 2016). Future studies are needed to probe this question in detail when the tools for examining other adipose tissues are refined. It will also be of great interest to elucidate if the mechanism of the Per3 mediated circadian clock output pathway we identified is observed more broadly or represents a context-specific deployment of clock components in APCs.
In summary, our study demonstrates that the circadian clock has a critical role in the regulation of adipogenesis. Furthermore, we discovered that the poorly understood PER3 circadian clock component has an unanticipated central role in mediating an output pathway from the clock which crosstalks with the Krüppel-like family member Klf15 to regulate adipogenesis in endogenous APCs. Intriguingly, the regulatory complex containing PER3 and BMAL1 we discovered on the Klf15 gene had not previously been observed in circadian biology. It is tempting to speculate about the physiological implications of embedding circadian clock control into a pathway that regulates adipogenesis. One potential implication is that the rhythm of Per3 serves to gate the initiation of adipogenesis, restricting differentiation to occur in a subpopulation of APCs where Per3 oscillations are synchronized with other cell-intrinsic, proximal and systemic signals.
Methods
Mice
C57BL/6, Per3−/− (B6.129S4-Per3tm1Drw/J; 010493), and mPer2Luc (B6.129S6-Per2 tm1Jt/J; 006852) mice were obtained from Jackson Laboratory and maintained under a normal diurnal cycle with access to food and water ad libitum.
For experiment’s conducted in 12:12 light:dark conditions, ZT0 is defined as the time when the lights are turned on. For circadian experiments in constant darkness, mice were maintained in constant darkness for 60 hours prior to the start of the experiment. All experiments were conducted in age-matched mice using wild-type littermates as controls where appropriate. Stanford University’s Administrative Panel on Laboratory Animal Care Committee approved all studies involving animals.
Adipocyte Precursor Cells isolation and FACS
For a detailed description, see Supplemental Experimental Procedures
Plasmid Constructs and Vectors
For a detailed description, see Supplemental Experimental Procedures
Cell Culture
For a detailed description, see Supplemental Experimental Procedures
Real-time Circadian Monitoring
For a detailed description, see Supplemental Experimental Procedures
Immunohistochemistry
For a detailed description, see Supplemental Experimental Procedures
RNA Extraction and RT-qPCR
For a detailed description, see Supplemental Experimental Procedures
In vivo EdU Pulse Chase
For a detailed description, see Supplemental Experimental Procedures
Transient Transfections and Reporter Assays
For a detailed description, see Supplemental Experimental Procedures
Adipogenesis Assays
For a detailed description, see Supplemental Experimental Procedures
Chromatin Immunoprecipitation (ChIP) and ChIP-re-ChIP
ChIP assays were performed as previously described (Ji et al., 2016). Briefly, SVF cells were fixed in 1% formaldehyde for 10 minutes followed by sonication to shear DNA using a Bioruptor (Diagenode). Immunoprecipitation (IP) was performed using antibodies against PER2, PER3, BMAL1 (Santa Cruz Biotechnology) or IgG. ChIP grade protein A/G magnetic beads (Thermo Fisher Scientific) were used in the immunoprecipitation and the precipitated DNA fragments were isolated using the ChIP DNA clean and concentrator kit (Zymo Research). The amount of precipitated DNA was quantified by qPCR using the following primers: E-box 1 (Fwd: CCGGGAGGATAAACAGCA G, Rev: ACCCTGGGGACACTGAAGTA), and Control site (Fwd: GGTCCTTTTCTGGGTCCTGG, Rev: GTGCAACCTCAGGCAAATGG). For the ChIP-re-ChIP assays, complexes from the primary ChIP were re-suspended with 10 mM dithiothreitol (DTT) for 30 minutes prior to the re-ChIP.
Co-Immunoprecipitation (Co-IP)
For a detailed description, see Supplemental Experimental Procedures
RNA-Seq
Total RNA was isolated from APCs isolated by FACS from the inguinal subcutaneous adipose depots of 4-week old Per3−/− and littermate WT control mice at ZT 2. Libraries of cDNA were generated using oligo d(T) priming. Sequencing was performed on an Illumina HiSeq2000, with around 40 million paired-end reads from each sample. Using the DNANexus pipeline, raw RNA-Seq reads were aligned to the mm9 reference genome using STAR v2.4.0 (Dobin et al., 2013) with default parameters. After alignment, an estimation of transcript abundance was calculated by fragments per kilobase of exon per million aligned fragments (FPKM) values using Cufflinks (Trapnell et al., 2013) in the Tuxedo protocol.
Accession Numbers
RNA-Seq data described in this study have been deposited into the GEO database under accession number GSE92762.
Statistical Analysis
Statistical analyses were performed using Prism GraphPad (GraphPad Software Inc.). For the analysis of circadian experiments with a single cohort, one-way analysis of variance (ANOVA) and post hoc Bonferroni was performed to test for oscillations across time points. Two-way ANOVA and post hoc Bonferroni for multiple corrections were used to compare cohorts across the circadian timecourses. Statistical differences between individual groups was performed using Student’s t-tests. P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The research was funded by an NIH Director’s New Innovator Award (DP2 OD006740) and R01 DK114217 and Stanford SPARK Translational Research Program at Stanford to B.J.F. A.A. and M.J.C. were supported in part by the Lucile Packard Foundation for Children’s Health, Stanford NIH-NCATS-CTSA UL1 TR001085, and the Child Health Research Institute of Stanford University. B.R.G. was supported by Alfonso Martin Escudero Foundation in Spain. B.J.F. is the Bechtel Endowed Faculty Scholar in Pediatric Translational Medicine.
Footnotes
Author Contributions
A.A. performed the real-time circadian experiments, Klf15 and Per3 adipogenesis assays, luciferase reporter assays and the ChIP and ChIP-re-ChIP experiments. M.J.C. conducted the in vivo adipogenesis experiments, the circadian-time course experiments in APCs and assisted with the RNA-seq and validation. B.R.G. performed FACS and analysis, Co-IP experiments, and real-time circadian experiments. L.J. assisted with the ChIP assays and S.L.M. assisted with the generation of constructs used in the luciferase assays. All authors participated in the analysis of the data. B.J.F. designed experiments, analyzed results and wrote the manuscript with input from all the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354:994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian Integration of Metabolism and Energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix A, Zarrinpar A. The effects of time-restricted feeding on lipid metabolism and adiposity. Adipocyte. 2015;4:319–324. doi: 10.1080/21623945.2015.1025184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MJ, So AYL, Kaasik K, Krueger KC, Pillsbury ML, Fu YH, Ptacek LJ, Yamamoto KR, Feldman BJ. Circadian Rhythm Gene Period 3 Is an Inhibitor of the Adipocyte Cell Fate. Journal of Biological Chemistry. 2011;286:9063–9070. doi: 10.1074/jbc.M110.164558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Lazar MA. Clocks, Metabolism, and the Epigenome. Molecular Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B, Bellet M, SK, Astarita G, Hirayama J, Amin R, Granneman J, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Chatterjee S, Li L, Kim JM, Lee J, Yechoor VK, Minze LJ, Hsueh W, Ma K. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012;26:3453–3463. doi: 10.1096/fj.12-205781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Zhang R, Jain R, Shi H, Zhang L, Zhou G, Sangwung P, Tugal D, Atkins GB, Prosdocimo DA, et al. Circadian control of bile acid synthesis by a KLF15-Fgf15 axis. Nat Commun. 2015;6:7231. doi: 10.1038/ncomms8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E, Lamia KA. Adipose Clocks: Burning the Midnight Oil. J Biol Rhythms. 2015;30:364–373. doi: 10.1177/0748730415581234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Church CD, Holtrup B, Colman L, Rodeheffer MS. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat Cell Biol. 2015;17:376–385. doi: 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483:96–99. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Gupta M, Feldman BJ. Vitamin D Regulates Fatty Acid Composition in Subcutaneous Adipose Tissue Through Elovl3. Endocrinology. 2016;157:91–97. doi: 10.1210/en.2015-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci U S A. 2010;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner NM, Mayo SA, Hua J, Lee C, Moore DD, Fu L. Circadian Dysfunction Induces Leptin Resistance in Mice. Cell Metab. 2015;22:448–459. doi: 10.1016/j.cmet.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger KC, Costa MJ, Du H, Feldman BJ. Characterization of Cre recombinase activity for in vivo targeting of adipocyte precursor cells. Stem Cell Reports. 2014;3:1147–1158. doi: 10.1016/j.stemcr.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, et al. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280:12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- Otway DT, Frost G, Johnston JD. Circadian rhythmicity in murine pre-adipocyte and adipocyte cells. Chronobiol Int. 2009;26:1340–1354. doi: 10.3109/07420520903412368. [DOI] [PubMed] [Google Scholar]
- Papazyan R, Zhang Y, Lazar MA. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol. 2016;23:1045–1052. doi: 10.1038/nsmb.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy Kvβ, Friedman JM. Identification of White Adipocyte Progenitor Cells In Vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Hung CM, Guertin DA. Emerging Complexities in Adipocyte Origins and Identity. Trends Cell Biol. 2016;26:313–326. doi: 10.1016/j.tcb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Gotic I, Saini C, Gos P, Curie T, Emmenegger Y, Sinturel F, Gosselin P, Gerber A, Fleury-Olela F, et al. Clock-Talk: Interactions between Central and Peripheral Circadian Oscillators in Mammals. Cold Spring Harb Symp Quant Biol. 2015;80:223–232. doi: 10.1101/sqb.2015.80.027490. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci U S A. 2008;105:5745–5749. doi: 10.1073/pnas.0801551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JC, Krueger KC, Costa MJ, Aggarwal A, Du H, McLaughlin TL, Feldman BJ. A glucocorticoid- and diet-responsive pathway toggles adipocyte precursor cell activity in vivo. Sci Signal. 2016;9:ra103. doi: 10.1126/scisignal.aag0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.