Abstract
Progress in aging research is constrained by the time-requirement of measuring lifespans. Even the most rapid model for eukaryotic aging, the replicative lifespan of Saccharomyces cerevisiae, is technically limited to only several lifespan measurements each day. Here, we report a 384-well plate based technique to measure replicative lifespan, termed High-Life. Using the High-Life technique, a single researcher can compare lifespan for more than 1000 conditions per day. We validated the technique with long-lived mutant strains and the lifespan-extending compound ibuprofen. We also applied this technique to screen a small compound library for lifespan extension. Two hits, terreic acid and mycophenolic acid, were validated on our single-cell Replicator device, and found to extend mean replicative lifespan by 15% and 20%, respectively. Together, we report a technique for high-throughput lifespan measurement, and identify two lifespan-extending compounds. Our technique could be used to efficiently drive early-stage discovery of pro-longevity therapeutics.
Graphical Abstract
INTRODUCTION
Aging is the greatest risk factor for morbidity and mortality throughout the developed world (Harman 1991). It is therefore desirable to modulate the aging process to extend healthy lifespan. However, only several interventions that extend lifespan in a conserved manner have been described, including mTOR inhibition (Medvedik et al. 2007; Miller et al. 2011; Bjedov et al. 2010; Robida-Stubbs et al. 2012) and dietary restriction (McCay, Crowell, and Maynard 1935; Klass 1977; Lakowski and Hekimi 1998; Lin, Defossez, and Guarente 2000; Kaeberlein et al. 2006). While translation of existing interventions to human therapeutics has great potential to improve health in old age, further discoveries are necessary to fully minimize the health-effects of age-related disease.
One of the greatest impediments to the progress of aging research is the fundamental time-requirement of longitudinal aging studies. The lifespan of model organisms can range from years in mammals to several days in the yeast Saccharomyces cerevisiae. Throughput limitations have been partially addressed with massive parallel studies in the moderately long-lived organism Caenorhabditis elegans (Petrascheck, Ye, and Buck 2009) or technology that enables rapid, but not scalable, experiments in short-lived models (Liu, Young, and Acar 2015; Jo et al. 2015). These approaches are constrained in that they permit either large-scale or quick turn-around, but not both.
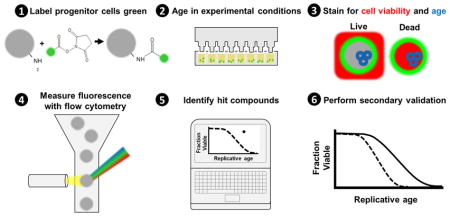
Here we introduce a technique (High-Life: High throughput replicative Lifespan measurement) that boasts the benefit of both approaches: a massively multiplexed method to measure replicative lifespan in the short-lived model organism S. cerevisiae. Our protocol uses green-fluorescent labeling to identify progenitor cells, red-fluorescent labeling to differentiate non-viable cells, and blue-fluorescent labeling of bud scars to determine replicative age (Fig. 1). Each parameter is measured using a flow cytometer. Using a plate-based autosampler with our optimized sampling rate of 1 well/minute, throughput exceeds 1000 wells per day. Each well can contain a different strain or media condition.
Fig. 1. Workflow of High-Life experiments.
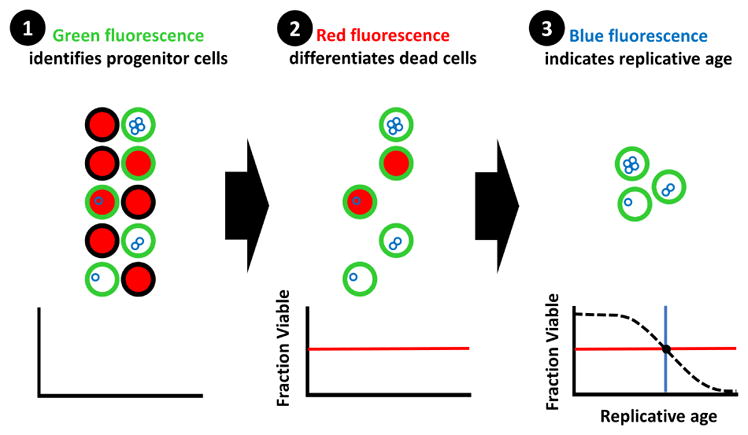
A visual depiction of the work-flow for High-Life experiments. The progenitor cell population of interest is persistently labeled with a green NHS-Ester fluorescein conjugate, which asymmetrically segregates to the mother cell during division. The fraction of cells viable within the progenitor population is then determined using propidium iodide; live cells exclude the red dye. Finally, the replicative age of viable progenitor cells is measured using wheat germ agglutinin conjugated to a blue fluorophore, which labels bud scars left behind with each division. A complete lifespan curve can be constructed using serial measurements taken over the course of 2–3 days.
RESULTS
Validation of the High-Life Technique
To achieve high throughput, we decided that our measurement system should be automated, and that our assay be performed in 384-well plates. We opted to perform our assay using an autosampler-equipped flow cytometer, in a volume of 100 μL. Unfortunately, growth of even a single cell and its progeny in such a small volume will result in nutrient starvation before the natural replicative lifespan is exhausted. To circumvent this issue, High-Life experiments are performed in the background of the Mother Enrichment Program (MEP) (Lindstrom and Gottschling 2009). MEP strains express a CRE recombinase fused to an estrogen-binding domain for only a short time after birth. In the presence of β-estradiol, the recombinase translocates to the nucleus where it can excise two essential genes that have been modified to contain the exogenous LoxP sequence. Addition of β-estradiol to the media thus renders newborn daughters inviable without affecting existing mother cells, preventing exponential growth of the cell population and nutrient depletion (Fig. 2a).
Fig. 2. Validation of the High-Life technique components.
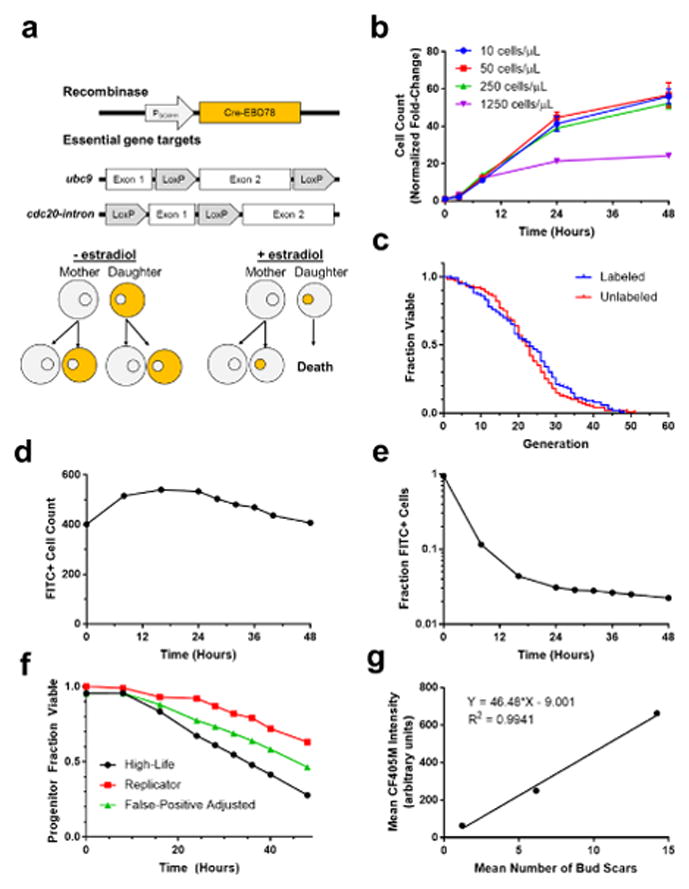
a. Schematic representation of the MEP. β-estradiol inducible Cre-recombinase is expressed under the control of a daughter cell specific promoter, PSCW11. LoxP sites are integrated surrounding components of the essential genes ubc9 and cdc20. b. The fold-change in MEP cells inoculated at different initial densities is plotted over time, with three-hour culture prior to the addition of β-estradiol. Error bars are S.E.M. for 6 independent replicates. c. Viability plotted against replicative age for fluorescein-labeled versus unlabeled cells. N = 100 cells for each group; mean lifespans were 22.9 and 22.4, respectively. d. Total number of cells that fall into the fluorescein-positive fraction at various times during a representative High-Life experiment. e. The fraction of all cells that fall into the fluorescein-positive fraction at various times during a representative High-Life experiment. f. The fraction of progenitor cells viable is plotted against time, beginning after either birth of the cell (Replicator) or initiation of the culture (High-Life), for representative experiments. A third line (False-Positive Adjusted) represents that fraction of viable cells in the High-Life environment after correcting for the false-positive rate observed with propidium iodide staining in the Replicator device. Correction was performed by extrapolating a linear trend of false-positives between cells in the Replicator device stained with propidium iodide after 16 or 40 hours of culture, and multiplying the observed High-Life viability by 1 minus the calculated false-positive fraction. 100 cells were considered for the Replicator experiment. The High-Life experiment shows the mean of 48 replicate wells. g. The mean number of bud scars observed on fluorescein-labeled cells after 0, 8, and 24 hours of culture, plotted against the CF405M fluorescence intensity observed at the same timepoint. For bud scar counting, 20 cells were analyzed at each timepoint. CF405M intensity values are the mean of 48 replicate wells.
Throughput of a flow cytometry-based assay is dependent on the cell density, as cell density is positively correlated with the sample processing speed in each well. Since it is desirable to complete processing of the entire 384-well plate as fast as possible, we sought to determine the maximum cell density we could use without causing nutrient depletion. We induced the MEP with β-estradiol and cultured the cells at different densities, then measured total cell number at various times up to 48 hours later. No growth rate defect was observed for inoculation densities of up to 250 cells/μL (Fig. 2b). To reduce the risk of partial nutrient depletion, we performed subsequent High-Life experiments with ≤20 cells/μL.
Replicative lifespan has two fundamental parameters: replicative age in the population of interest and the fraction of cells viable at that age. We aimed to measure these lifespan parameters in an unmonitored liquid culture by breaking down our effort into three steps: (1) differentiate the progenitor cells of interest from their progeny, (2) identify the viable fraction of progenitor cells, and (3) determine the replicative age of the viable progenitor cells.
Asymmetric segregation of the cell wall between mother and daughters has previously enabled magnetic sorting of a progenitor cell population (Smeal et al. 1996). We adapted this technique to label the progenitor population cell wall with a fluorescein conjugated N-HydroxySuccinimide-ester (NHS-ester) (Fig. 1, step 1). It is important that the label not alter the cells’ natural lifespan. We confirmed that labeling does not affect replicative lifespan using our previously published Replicator device (Liu, Young, and Acar 2015) (Fig. 2c). We also confirmed that the fluorescent label was retained by mother cells, and not passed to their daughters. When cultured, the total number of labeled cells (mothers) increased after initiation, and subsequently declined gradually (Fig. 2d). The initial increase in labeled cells is consistent with separation of cells that were partially budded during labeling; the decline may be explained by fragmentation of dead cells such that they no longer triggered the flow cytometer. We also observed a decrease in the fraction of labeled cells over time (Fig. 2e), representing the generation of unlabeled daughters. Overall, these results indicate that we are able to track the population of progenitor cells in an unmonitored liquid culture without impacting cell health.
Once the progenitor cell fraction is identified, it is necessary to determine which fraction of these cells is viable (Fig. 1, step 2). For this purpose, we used the well-characterized viability dye propidium iodide (Deere et al. 1998) by labeling, culturing, and staining cells. Flow cytometry revealed a time-dependent decline in progenitor cell viability, consistent with expectations for an aging population (Fig. 2f). To assess if the rate of decline was the same as is observed using other lifespan measurement methods, we used our medium-throughput, single-cell Replicator device (Liu, Young, and Acar 2015). This technology enabled us to collect images of trapped mother cells throughout their entire lifespan. Analyzing the image series, we measured replicative lifespan and the length of each budding interval. We found that the rate of viability decline measured with propidium iodide exceeded that seen in our Replicator device (Fig. 2f). To further understand the source of this error we used the Replicator device to measure fluorescence intensity of cells introduced to propidium iodide after 16 or 40 hours in culture. After 16 hours, 2% of live cells fell above an intensity threshold constructed to approximate the flow cytometry experiment’s gate. By 40-hours, 20% of live cells fell into the dead region, largely due to dead daughter cells that failed to separate, indicating that a small fraction of aged but live cells will stain as non-viable using propidium iodide. Based on an assumption of linearity between these points, we projected a false-positive rate over the entire time-course of a High-Life experiment, and plotted a corrected viability curve (Fig. 2f). While this curve does not precisely match that observed using the Replicator device, we suggest that limitations in the intrinsic ability to relate microscopic fluorescence intensity data to flow cytometric data may underlie the remaining difference.
As the third and final step, we sought to measure the replicative age of the viable progenitor cells we identified (Fig. 1, step 3). A bud scar is left on the mother cell wall with each division, and therefore the number of bud scars is directly proportional to replicative age (Barton 1950). The protein lectin wheat germ agglutinin (WGA) has been demonstrated to bind specifically to bud scars (Chen et al. 2003). We tested whether WGA conjugated to the blue fluorophore CF405M could be used to measure replicative age with a flow cytometer by labeling cells, culturing, and staining simultaneously with CF405M-WGA and propidium iodide. We compared CF405M intensity measured with a flow cytometer to the number of bud scars observed microscopically on cells cultured for the same period of time, and observed a directly proportional relationship (R2 = 0.9941) (Fig. 2g). This finding confirmed our ability to accurately measure replicative age using CF405M-WGA and a flow cytometer.
High-Life Detects Lifespan Extension from Pharmacologic and Genetic Interventions
Taken together, our validation experiments showed that we could measure all parameters necessary to construct a replicative lifespan curve. We next sought to answer two questions: can our method be used to detect lifespan extension, and does our method deliver reproducible results day to day? Ibuprofen extends replicative lifespan via inhibition of tryptophan import (He et al. 2014). We performed two independent High-Life experiments in the presence or absence of ibuprofen. For this, we labeled cells, cultured them in the presence of β-estradiol and +/− ibuprofen, stained them with propidium iodide and CF405M-WGA at multiple later time points, and acquired readings with a flow cytometer. We observed an increase in replicative lifespan in the presence of ibuprofen, confirming the ability of our method to detect lifespan extension (Fig. 3a–b). To assess the sensitivity and specificity of High-Life, we fit a trend-line and 95% confidence interval to the untreated condition (Fig. 3b). Using this confidence interval as a cutoff, we measured the fraction of ibuprofen-treated samples that fell within the confidence interval (false-negatives) and fraction of untreated samples that fell outside the confidence interval (false-positives), for each timepoint (Fig. 3c). We found that the rate of false-negatives and false-positives was lowest for measurements taken after 24-hours of culture, and recommend this length of culture for comparative measurements of replicative lifespan.
Fig. 3. Detection of lifespan extension using High-Life.
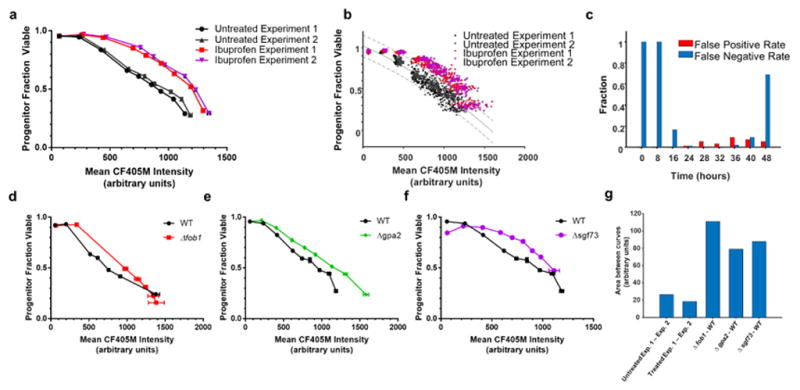
a. Fraction of progenitor cells viable plotted against the corresponding blue fluorescence intensity, with timepoints taken at 0, 8, 16, 24, 28, 32, 36, 40, and 48 hours after labeling. Two separate experiments are shown, each comparing ibuprofen treated samples to an untreated control. Error bars are S.E.M. of 48 replicate wells. b. A plot of the individual replicate points that compose the mean values shown in Figure 3a. The solid line is the result of second order polynomial fitting on pooled data from both untreated experiments; the dashed lines denote a 95% confidence interval. c. The fraction of wells in the ibuprofen condition that fall below the upper boundary of the 95% confidence interval (false negatives), and fraction of untreated wells that fall above the upper boundary of the 95% confidence interval (false positives) at each timepoint sampled. d–f. Fraction of progenitor cells viable plotted against the corresponding blue fluorescence intensity for wild-type, Δfob1 (d), Δgpa2 (e), and Δsgf73 (f) strains sampled at various times after labeling. Error bars are S.E.M. of 12 independent replicates. g. Areas between the curve for High-Life experiments performed under the same conditions (left), or with strains expected to exhibit lifespan differences (right). For consistent comparison, areas between the respective curves were computed with Mean CF405M Intensity ranging from 70 to 1100.
After testing our technique in the ibuprofen environment, we asked whether this technique could be used to identify increases in lifespan from genetic interventions. For this goal, we used the technique to create replicative lifespan curves for three strains harboring gene deletions previously demonstrated to extend replicative lifespan: fob1, gpa2, and sgf73. All three strains showed an increase in replicative lifespan compared to the wild-type control (Fig. 3d–f). While the two-axis uncertainty inherent to High-Life limits the application of traditional statistical techniques, we sought to quantitively assess the difference between wild-type and long-lived strain measurements. For this purpose, we measured the area between curves in the wild-type, ibuprofen-treated, and long-lived strain conditions (Fig. 3g). This area was 3–5x greater when comparing conditions with an expectation for a lifespan difference, versus for two experiments performed for the same condition. This data indicates that we can reliably and reproducibly detect extension of replicative lifespan using our technique.
A High-Life Screen Identifies Lifespan Extending Compounds
High-Life enables rapid assessment of lifespan in a high-throughput manner. We therefore sought to test whether it was suitable to screening for compounds that extend replicative lifespan. For this purpose, we selected several libraries containing 2640 compounds, including kinase inhibitors, FDA-approved compounds, and compounds which had failed clinical development. We assayed the effect of these compounds on High-Life readings after 24 hours in culture at 10 μM concentration. As a positive control, we used ibuprofen. Replicates of ibuprofen treatment were reproducibly distinguishable from negative control points (Fig. 4a). To test the analytical specificity of our screen, we selected for follow-up 99 compounds which qualitatively deviated from the control (Fig. 4a). We remained blinded to the identity of these compounds until our original results were repeated: a second 24-hour High-Life measurement was conducted with 3–4 replicate wells for each compound to differentiate random variation from genuine lifespan extension. The average readings for 12 compounds fell at least slightly above controls (Fig. 4b), and their identities were unblinded. One compound was rapamycin, which has previously been demonstrated to extend replicative lifespan (Medvedik et al. 2007). For the remaining 11 compounds, we found no literature to indicate a role in lifespan extension.
Fig. 4. Plate-based screening using High-Life.
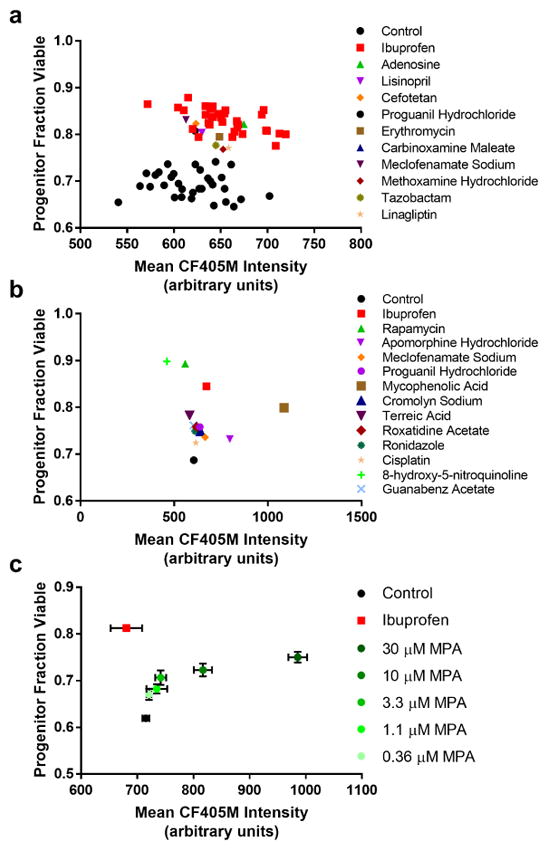
a. A plot of progenitor fraction viable versus CF405M intensity for individual wells of a 384 well plate based screen. Shown for a single plate are all untreated (DMSO) negative control samples, all ibuprofen-treated positive control samples, and all compounds from this plate that were selected for follow-up. b. A plot of progenitor fraction viable versus CF405M intensity for the averages of all wells for control, ibuprofen, and confirmed hit compounds. Error bars are not shown, as S.E.M. bars were generally smaller than the points themselves. Control points are the average of 28 replicate wells; compound points are the average of 3–4 replicate wells. c. Dose-response for cells treated with mycophenolic acid (MPA) during 24 hours of culture, chosen as a representative of the three compounds with clear concentration-dependent effects selected for follow-up validation. Error bars are S.E.M. for four or more replicate wells. See Supplementary Figure 1 for dose-response of additional compounds.
Fresh samples for 7 of these 11 compounds were obtained from a secondary source, and subjected to a dose-response experiment. Three compounds exhibited a concentration-dependent increase in cell survival (Fig. 4c), while the remainder continued to show only mild deviation from the control. To differentiate artifactitious High-Life readings from lifespan extension, we performed a secondary validation experiment for the three compounds: we measured replicative lifespan on a single-cell level in the presence of 10 μM compound using our previously published Replicator device (Supplementary Movie 1) (Liu, Young, and Acar 2015). This platform closely recapitulates results from micromanipulator studies, and 98% of cells that were tracked in our control experiments were able to be followed from birth to death. One compound, 8-hydroxy-5-nitroquinoline, was toxic and caused most cells to arrest immediately (data not shown). However, terreic acid and mycophenolic acid exhibited 15% and 20% extension of mean replicative lifespan, respectively, compared to untreated cells (Fig. 5).
Figure 5. Secondary Validation of Screening Hits.
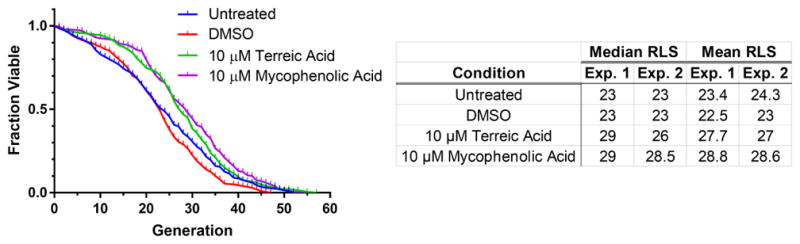
Survival curves and lifespan characteristics for wild-type, haploid cells grown in the absence (untreated) or presence of DMSO vehicle-control, 10 μM terreic acid, or 10 μM mycophenolic acid. In each experiment, lifespan measurements were made on a single-cell level for 100 cells in each condition using our microfluidic Replicator device. Each curve contains pooled data from two independent experiments.
DISCUSSION
Together, our results demonstrate a technique for measuring the replicative lifespan of S. cerevisiae. It improves upon previous methods through its massively increased throughput and by avoiding a requirement for microfluidic or microdissection equipment to measure lifespan. We show that this technique is reliable and reproducible, and can be used to detect lifespan extension from both genetic and pharmacologic interventions. Furthermore, we demonstrate its potential by identifying two compounds which extend replicative lifespan. One of the compounds we identified, terreic acid, is a quinone epoxide inhibitor of Bruton’s tyrosine kinase (Kawakami et al. 1999). Mycophenolic acid, on the other hand, is an immunosuppressant used to prevent organ rejection after transplantation, and an inhibitor of inosine monophosphate dehydrogenase (Escobar-Henriques and Daignan-Fornier 2001). The exact mechanisms through which these two compounds extend replicative lifespan are not known.
Our technique elicits the possibility of large-scale screening for compounds that extend replicative lifespan. Such compounds frequently exhibit conserved effects in higher organisms (Medvedik et al. 2007; Miller et al. 2011; Bjedov et al. 2010; Robida-Stubbs et al. 2012). Therefore, this technique could serve as an important discovery platform for pre-clinical leads that might slow age-related decline. In addition to compound screening, there exists a need for rapid testing of genetic manipulations’ effects on lifespan. The method as described here is not suited for large-scale genetic screening, since it must be performed in the genetic background of the MEP. However, our labeling procedure is based on a technique used for magnetic sorting of wild-type mother cells. We suggest nutrient depletion could be avoided by magnetic sorting rather than genetic modification, and that this modification to the technique could permit high-throughput genetic screening.
For decades, the time-requirement of longitudinal aging studies has limited progress in this field of research. Here we describe a high-throughput, rapid technique to measure replicative lifespan. Such methods have the potential to transform aging research by increasing the speed of progress while reducing associated costs.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Culture Conditions
All experiments were conducted in a BY4741 strain background (TransOMIC TKY0002). Strains containing the genetic modifications of the Mother Enrichment Program (Lindstrom and Gottschling 2009) were constructed by lithium acetate transformation (Gietz and Schiestl 2007) with PCR products derived from MEP strain UCC8773. Deletion strains were prepared similarly, with transformation DNA from PCR on the genomic DNA of corresponding strains from the yeast deletion library (Giaever et al. 2002) (GE Dharmacon).
Synthetic media (CSM 2% glucose) was used for all experiments. Cells were maintained in aerobic conditions at 30 °C, in either 50 mL con ical tubes (Becton Dickinson F2070) or 384-well plates (Greiner Bio-One 781201). Cultures in tubes were performed in an Innova-42 shaker (New Brunswick Scientific) at 225 rpm. Plate-based cultures were performed in a humidified incubator kept at 95% relative humidity, and the plates were covered with a breathable membrane (Thermo Scientific 241205) to prevent evaporation. Agitation was provided by a microplate shaker (Union Scientific 9779-TC) at an amplitude of 0.04 inches.
Compounds
As our positive control for lifespan extension, we used ibuprofen (Sigma I-1892). The following compound libraries, obtained from the Yale Center for Molecular Discovery, were screened for this manuscript: (1) 320/355 compounds in the Selleckchem Kinase Inhibitor Library, (2) the Enzo-640 FDA-approved drugs catalog, (3) the Enzo Kinase Inhibitor Library, and (4) the Microsource Pharmakon 1600 library.
Determining Maximum Cell Density to Avoid Nutrient Depletion
10 mL of cells were grown overnight for 16 hours to mid-log phase, then diluted to the indicated densities in ice-cold media. 80 μL of cell suspension was aliquoted to 12 wells of four 384-well plates for each cell density. The plates were covered with a breathable membrane and placed on a shaking platform in a humidified incubator for 3 hours. Each well was then treated with 20 μL pre-warmed, 30 °C 5 μM β-estradiol (Sigma E8875) in media, and the plates were returned to the incubator. For the 0-hour timepoint, this addition was instead performed immediately after initially aliquoting the plate. After 0, 8, 24, and 48 hours from the time cells were aliquoted to the plate, the total cell count was measured using a flow cytometer.
NHS-Fluorescein Labeling
10 mL of cell culture was grown overnight for 18 hours to mid-log phase. The cells were then spun down at 1000xg for 3 minutes at room temperature. The supernatant was poured off, and the cells were resuspended in 1 mL 3.5 mg/mL NHS-Fluorescein (Life Technologies 46410) in 10x PBS (Life Technologies 14200075). The cells were then placed on a rocking platform in the dark for 15 minutes at room temperature. They were then diluted to 50 mL in ice-cold 1x PBS (Life Technologies 141901144), mixed, and spun down at 1000xg for 2 minutes at 4 °C. The supernatant was discarded, and this wash step was repeated. Afterward, the supernatant was discarded, and the cells were resuspended in 1 mL ice-cold media.
High-Life Experiments
Cells were labeled with NHS-fluorescein as described above, then diluted to 20 cells/μL or less in media. 80 μL/well was aliquoted to 384-well plates, which were covered with a breathable membrane and placed on a shaker platform in a humidified incubator. After 3 hours, the plates were removed from the incubator and each well was treated with 20 μL pre-warmed, 30 °C 5 μM β-estradiol in media. In the case of ibuprofen or compound-treated wells, the compound was diluted in this volume at a 5x concentration to achieve a final concentration of 10 μM, or 100 μM for ibuprofen. The plates were then returned to the incubator. At indicated times, one plate was removed from the incubator and placed on an autosampler cooled to 8 °C attached to a Stratedigm flow cytometer. The cytometer was set to automatically add and mix 20 μL of aqueous solution containing 60 μg/mL CF405M-WGA (Biotium 29028) and propidium iodide (Sigma P4864), then incubate for 30 minutes prior to acquiring 80 μL of sample for each well.
Replicator Experiments
To obtain the single-cell level data for the age and generation-durations of replicatively aging mother cells, we re-analyzed data from our previously published Replicator paper (Liu, Young, and Acar 2015). We used data collected in the same media condition (CSM 2% glucose) as in the current work. The experimental protocols used for secondary validation with our Replicator device are described in detail in our previous publication (Liu, Young, and Acar 2015). Cells were grown for ~24 hours in CSM 2% glucose prior to loading to the microfluidic device. Once cells were loaded, media was swapped to provide CSM 2% glucose control media (untreated), media containing DMSO as a vehicle control (American Bio AB00435), media containing 10 μM terreic acid (Sigma SML0480), or media containing 10 μM mycophenolic acid (Sigma M5255). An automated microscope was used to track individual mother cells, and replicative lifespan was later determined by counting the number of daughters produced before death. In the case of compound validation, only newborn cells were included in the lifespan experiment. To compare the replicative lifespan of labeled and unlabeled cells, only cells that were present within the traps at the start of the experiment were included. For these experiments, cells were loaded to the microfluidic device at an increased rate of 100 μL/min to increase the number of trapped mother cells. Green fluorescent images were also taken at the start of the experiment to confirm that the cells were visibly labeled.
Bud Scar Staining
Cells were prepared as described in the “High-Life Experiments” section above, except diluted to 100 cells/μL prior to loading on a plate. After 0, 8, and 24 hours, all cells from a single plate were transferred to a 50 mL conical tube, and pelleted at 1000xg for 3 minutes. The supernatant was aspirated, and the cells were resuspended in 900 μL of sterile water and transferred to a 1.5 mL tube. 100 μL of 1 mg/mL Fluorescent Brightener 28 (Sigma F3543) in water was added, and the solution was incubated at room temperature in the dark for 5 minutes. Next, the solution was pelleted at 13000xg for 30 seconds, the supernatant was aspirated, and the cells were resuspended in 1 mL sterile water. This wash step was then repeated once. Finally, the cells were resuspended in 10 μL sterile water, and stored on ice in the dark until imaged. Z-stack brightfield and fluorescent images with 0.2 μm spacing were acquired for each sample on a confocal microscope. For green-fluorescent mother cells, the number of bud scars in the blue fluorescent channel were counted manually.
Confidence Interval Determination and Computing Areas Between Curves
In Fig. 3b, second-order polynomial fitting was performed on the pooled data set obtained from the two untreated experiments. Both the fitting and the 95% confidence interval computation were performed using MATLAB’s curve fitting toolbox.
Areas under respective curves were computed using trapezoidal numerical integration by calling MATLAB’s trapz function. The numerical integrations were performed over the same Mean CF405M Intensity range from 70 to 1100. Corresponding fractions of progenitor cells viable at the starting (70) and the ending point (1100) were computed through linear interpolation for each curve. Area differences between respective pairs of curves were then computed to generate Fig. 3g.
Supplementary Material
Related to Figure 2 and Figure 5. A representative imaging location from a wild-type control experiment performed on the Replicator device. Each lifespan curve is constructed of data pooled from multiple imaging locations within one or more device lanes.
Acknowledgments
The authors thank Acar laboratory members for useful discussions, D. Gottschling for providing the yeast strain UCC8773, P. Gareiss for help with the preparation of the compound library tested, and S. Katrancha for critical reading of the manuscript. EAS acknowledges support through an NSF Graduate Research Fellowship and Gruber Science Fellowship. MA acknowledges funding through a New Scholar in Aging Award from t he Ellison Medical Foundation (AG-NS-1015-13) and NIH Director’s New Innovator Award (1DP2AG050461-01).
Footnotes
AUTHOR CONTRIBUTIONS
EAS and MA contributed to project planning, interpretation of the data and results, and design and preparation of the manuscript. EAS and PL contributed to strain construction, data collection, data analysis, and preparation of the figures. EAS, PL, and MA read and approved the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: EAS and MA have filed a provisional patent application on the screening technology and compounds introduced and described in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barton AA. Some Aspects of Cell Division in Saccharomyces Cerevisiae. Journal of General Microbiology. 1950;4(1):84–86. doi: 10.1099/00221287-4-1-84. [DOI] [PubMed] [Google Scholar]
- Bjedov Ivana, Toivonen Janne M, Kerr Fiona, Slack Cathy, Jacobson Jake, Foley Andrea, Partridge Linda. Mechanisms of Life Span Extension by Rapamycin in the Fruit Fly Drosophila Melanogaster. Cell Metabolism. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Cuiying, Dewaele Sylviane, Braeckman Bart, Desmyter Liesbeth, Verstraelen Jan, Borgonie Gaetan, Vanfleteren Jacques, Contreras Roland. A High-Throughput Screening System for Genes Extending Life-Span. Experimental Gerontology. 2003;38(10):1051–63. doi: 10.1016/S0531-5565(03)00186-4. [DOI] [PubMed] [Google Scholar]
- Deere Daniel, Shen Jian, Vesey Graham, Bell Philip, Bissinger Peter, Veal Duncan. Flow Cytometry and Cell Sorting for Yeast Viability Assessment and Cell Selection. Yeast. 1998;14(2):147–60. doi: 10.1002/(SICI)1097-0061(19980130)14:2<147::AID-YEA207>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques M, Daignan-Fornier B. The Journal of Biological Chemistry. 2. Vol. 276. American Society for Biochemistry and Molecular Biology; 2001. Transcriptional Regulation of the Yeast Gmp Synthesis Pathway by Its End Products; pp. 1523–30. [DOI] [PubMed] [Google Scholar]
- Giaever Guri, Chu Angela M, Ni Li, Connelly Carla, Riles Linda, Véronneau Steeve, Dow Sally, et al. Nature. 6896. Vol. 418. Nature Publishing Group; 2002. Functional Profiling of the Saccharomyces Cerevisiae Genome; pp. 387–91. [DOI] [PubMed] [Google Scholar]
- Gietz R Daniel, Schiestl Robert H. Nature Protocols. 1. Vol. 2. Nature Publishing Group; 2007. High-Efficiency Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method; pp. 31–34. [DOI] [PubMed] [Google Scholar]
- Harman D. Proceedings of the National Academy of Sciences of the United States of America. 12. Vol. 88. National Academy of Sciences; 1991. The Aging Process: Major Risk Factor for Disease and Death; pp. 5360–63. http://www.ncbi.nlm.nih.gov/pubmed/2052612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Chong, Tsuchiyama Scott K, Nguyen Quynh T, Plyusnina Ekaterina N, Terrill Samuel R, Sahibzada Sarah, Patel Bhumil, et al. PLoS Genetics. 12. Vol. 10. Public Library of Science; 2014. Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import; p. e1004860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Myeong Chan, Liu Wei, Gu Liang, Dang Weiwei, Qin Lidong. Proceedings of the National Academy of Sciences of the United States of America. 30. Vol. 112. National Academy of Sciences; 2015. High-Throughput Analysis of Yeast Replicative Aging Using a Microfluidic System; pp. 9364–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein Tammi L, Smith Erica D, Tsuchiya Mitsuhiro, Linnea Welton K, Thomas James H, Fields Stanley, Kennedy Brian K, Kaeberlein Matt. Aging Cell. 6. Vol. 5. Blackwell Publishing Ltd; 2006. Lifespan Extension in Caenorhabditis Elegans by Complete Removal of Food; pp. 487–94. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Hartman SE, Kinoshita E, Suzuki H, Kitaura J, Yao L, Inagaki N, et al. Terreic Acid, a Quinone Epoxide Inhibitor of Bruton’s Tyrosine Kinase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(5):2227–32. doi: 10.1073/pnas.96.5.2227. http://www.ncbi.nlm.nih.gov/pubmed/10051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass Michael R. Aging in the Nematode Caenorhabditis Elegans: Major Biological and Environmental Factors Influencing Life Span. Mechanisms of Ageing and Development. 1977;6:413–29. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Proceedings of the National Academy of Sciences of the United States of America. 22. Vol. 95. National Academy of Sciences; 1998. The Genetics of Caloric Restriction in Caenorhabditis Elegans; pp. 13091–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez Pa, Guarente L. Requirement of NAD and SIR2 for Life-Span Extension by Calorie Restriction in Saccharomyces Cerevisiae. Science (New York, NY) 2000;289(5487):2126–28. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lindstrom Derek L, Gottschling Daniel E. The Mother Enrichment Program: A Genetic System for Facile Replicative Life Span Analysis in Saccharomyces Cerevisiae. Genetics. 2009;183(2):413–22. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Ping, Young Thomas Z, Acar Murat. Yeast Replicator: A High-Throughput Multiplexed Microfluidics Platform for Automated Measurements of Single-Cell Aging. Cell Reports. 2015 doi: 10.1016/j.celrep.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell Mary F, Maynard LA. The Effect of Retarded Growth Upon the Length of Life Span and Upon the Ultimate Body Size. The Journal of Nutrition. 1935;10(1):63–79. [PubMed] [Google Scholar]
- Medvedik Oliver, Lamming Dudley W, Kim Keyman D, Sinclair David A. MSN2 and MSN4 Link Calorie Restriction and TOR to Sirtuin-Mediated Lifespan Extension in Saccharomyces Cerevisiae. PLoS Biology. 2007;5(10):2330–41. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller Richard A, Harrison David E, Astle CM, Baur Joseph A, Boyd Angela Rodriguez, deCabo Rafael, Fernandez Elizabeth, et al. The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 2. Vol. 66. Oxford University Press; 2011. Rapamycin, but Not Resveratrol or Simvastatin, Extends Life Span of Genetically Heterogeneous Mice; pp. 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrascheck Michael, Ye Xiaolan, Buck Linda B. Annals of the New York Academy of Sciences. 1. Vol. 1170. Blackwell Publishing Inc; 2009. A High-Throughput Screen for Chemicals That Increase the Lifespan of Caenorhabditis Elegans; pp. 698–701. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs Stacey, Glover-Cutter Kira, Lamming Dudley W, Mizunuma Masaki, Narasimhan Sri Devi, Neumann-Haefelin Elke, Sabatini David M, Keith Blackwell T. TOR Signaling and Rapamycin Influence Longevity by Regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metabolism. 2012;15(5):713–24. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeal Tod, Claus James, Kennedy Brian, Cole Francesca, Guarente Leonard. Loss of Transcriptional Silencing Causes Sterility in Old Mother Cells of S. Cerevisiae. Cell. 1996;84(4):633–42. doi: 10.1016/S0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 2 and Figure 5. A representative imaging location from a wild-type control experiment performed on the Replicator device. Each lifespan curve is constructed of data pooled from multiple imaging locations within one or more device lanes.


