Summary
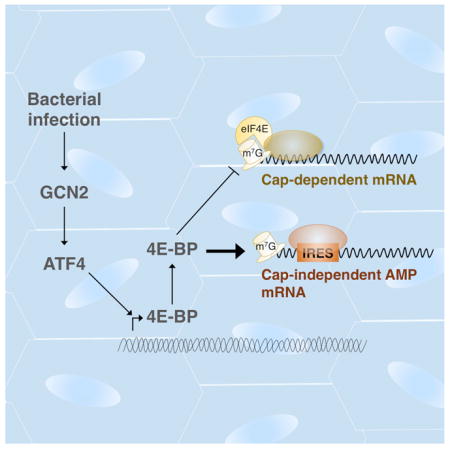
Bacterial infection often leads to suppression of mRNA translation, but hosts are nonetheless able to express immune response genes through as yet unknown mechanisms. Here, we use a Drosophila model to demonstrate that antimicrobial peptide (AMP) production during infection is paradoxically stimulated by the inhibitor of cap-dependent translation, 4E-BP (encoded by the Thor gene). We found that 4E-BP is induced upon infection with pathogenic bacteria by the stress-response transcription factor, ATF4, and its upstream kinase, GCN2. Loss of gcn2, atf4 or 4e-bp compromised immunity. While AMP transcription is unaffected in 4e-bp mutants, AMP protein levels are substantially reduced. The 5’ untranslated regions (UTRs) of AMPs score positive in cap-independent translation assays, and this cap-independent activity is enhanced by 4E-BP. These results are corroborated in vivo using transgenic 5’UTR reporters. These observations indicate that ATF4-induced 4e-bp contributes to innate immunity by biasing mRNA translation toward cap-independent mechanisms, thus enhancing AMP synthesis.
eTOC blurb
The cap-dependent translation inhibitor, 4E-BP, is induced during bacterial infection in Drosophila and animals lacking 4E-BP are immunocompromised. How an effective innate immune response is mounted in a translation suppressive environment, and why 4E-BP is required in this process was unknown. Vasudevan et al demonstrate that antimicrobial peptides, which are key components of the Drosophila innate immune response, are translated cap-independently via IRES-like elements in their 5’UTRs and that their translation is enhanced by 4E-BP-mediated suppression of cap-dependent translation.
Introduction
Various types of stresses dampen protein synthesis by reducing the availability of critical components of translation initiation. One such example is mediated by stress-activated kinases that phospho-inhibit eIF2α, which is part of the ternary complex (eIF2-GTP-Met-tRNAiMet) that delivers initiator methionine to the translation preinitiation complex. Remarkably, even when translation initiation is inhibited by phosphorylation of eIF2α, a subset of transcripts containing overlapping upstream ORFs (uORFs) in their 5’UTR, such as ATF4 (encoded by cryptocephal or crc in Drosophila), is favorably synthesized. ATF4 responds to cellular stress by transcriptionally inducing various stress responsive transcripts. As metazoans have multiple eIF2α kinases that active this pathway, these signaling pathways are often collectively referred to as the ‘integrated stress response’ (ISR)(Harding et al., 2003; Ron and Walter, 2007).
Translational inhibition mechanisms associated with viral infection were first observed over fifty years ago in ascites-tumor cells infected with encephalomyocarditis virus and since then this phenomenon has been extended to most viral infections (Mohr and Sonenberg, 2012). Such translational inhibition is mediated in part by PKR, one of the four eIF2α kinases in mammals that are activated by dsRNAs. While not as well recognized, literature reports that pathogenic bacterial infection also causes translation inhibition in various infection models from C. elegans to mammals (reviewed comprehensively in (Lemaitre and Girardin, 2013)). In Drosophila, GCN2, an eIF2α kinase that responds to amino acid deprivation, triggers an mRNA translational block in the host when infected with Pseudomonas entomophila (P.e). Other studies have reported the induction of the cap-dependent translational inhibitor 4e-bp (Thor in Drosophila) in response to bacterial pathogens (Bernal and Kimbrell, 2000; Chakrabarti et al., 2012; Rodriguez et al., 1996). Inhibition of translation initiation by 4E-BP targets at the 5’-cap of eukaryotic mRNAs, where the 7-methylguanosine moiety is recognized by eIF4E (as part of the eIF4F complex) to recruit other translation initiation factors, and eventually the 40S ribosomal subunit. 4E-BP, or eIF4E-binding protein, imposes translation inhibition by binding to eukaryotic initiation factor 4E (eIF4E), thereby specifically inhibiting cap-dependent translation. As 4e-bp mutant flies are immune compromised (Bernal and Kimbrell, 2000; Rodriguez et al., 1996), it is thought that induction of 4e-bp in response to bacterial infection is a host adaptation mechanism. The hypothesis that translation inhibition contributes to a host defense mechanism raises two important questions: First, how does translational inhibition aid the host in mounting an immune response that relies on antimicrobial peptide (AMP) expression? Second, what signaling pathways mediate 4e-bp induction in response to bacterial infection? Here, we identify the GCN2-ATF4 signaling as the mediator of 4e-bp induction in Drosophila during infection. We also show that 4E-BP paradoxically stimulates AMP synthesis as part of the innate immune response to bacterial infection in Drosophila. In a broader sense, we demonstrate a mechanism that explains one of the long-standing issues of how Drosophila AMPs can be expressed under conditions of translational inhibition.
Results
4e-bp is transcriptionally induced by ATF4 during infection in the fat body and gut
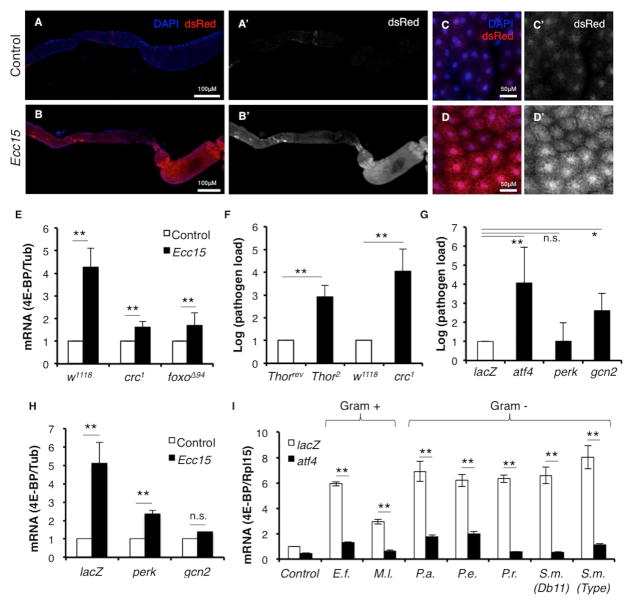
Independent studies have established that bacterial and fungal infection induce 4e-bp transcription in Drosophila (Bernal and Kimbrell, 2000; Levitin et al., 2007). However, the signaling pathway(s) responsible for this induction remains unknown. There are two known transcriptional regulators of 4e-bp, the best characterized of which is the stress-responsive transcription factor, FOXO (Jünger et al., 2003; Puig et al., 2003). Recently, we reported that the eIF2α-kinase-responsive transcription factor, ATF4, regulates 4e-bp transcription in Drosophila through an intronic element in the 4e-bp locus (Kang et al., 2017; Yamaguchi et al., 2008). Based on this regulation, we developed the 4E-BPintron-dsRed reporter that faithfully reports ATF4 activity in Drosophila tissues (Kang et al., 2017) and this led us to examine the possible role of eIf2α kinases in 4e-bp induction during infection. We specifically tested if ATF4 is active in the context of infection, by orally infecting 3rd instar larvae with the non-lethal gram-negative bacterial pathogen, Ecc15. In response to infection, dsRed expression was elevated in the larval gut (Fig. 1A’, B’) and also in the fat body (Fig. 1C’, D’), which is known to be an auxiliary immune-response tissue. We verified by qPCR that 4E-BPintron-dsRed induction reflected induction of 4E-BP itself, and found that 4e-bp mRNA was upregulated in response to Ecc15 infection (Fig. 1E), as reported previously (Bernal and Kimbrell, 2000). This induction was suppressed in the background of the homozygous atf4 hypomorphic mutant, crc1 (Fig. 1E) indicating that ATF4 mediates 4e-bp induction during infection. 4e-bp induction was also suppressed in foxo mutants (Fig. 1E), indicating that multiple transcription factors can regulate 4E-BP induction during infection.
Fig. 1. GCN2-ATF4 signaling induces 4E-BP during infection.
1A–D. 4E-BPintron-dsRed expression in uninfected larva (A, A’, C, C’) and in larva infected with Ecc15 (B, B’, D, D’). Shown are the gut (A, A’, B, B’) and the fat body (C, C’, D, D’). A’, B’, C’, D’ are dsRed channel only images.
1E. qPCR analysis of 4E-BP expression from control larvae (w1118), atf4 mutants (crc1 homozygotes) and foxo mutants (foxoΔ94 homozygotes) infected with Ecc15.
1F. Systemic pathogen load assay measuring Ecc15 levels in 4e-bp mutants (Thor2 homozygotes) with 4e-bp revertants (Thorrev) as control, and in homozygotic crc1 with w1118 as control.
1G. Systemic pathogen load assay on larvae expressing UAS-lacZRNAi (negative control), -atf4RNAi, -perkRNAi, -gcn2RNAi driven by a fat body specific driver (Dcg-Gal4).
1H. qPCR analysis of 4E-BP induction in larvae expressing UAS-lacZRNAi or –gcn2RNAi driven by Dcg-Gal4.
1G. qPCR analysis of 4E-BP induction in larvae expressing UAS-lacZRNAi or atf4RNAi driven by Dcg-Gal4 infected with various gram-positive and -negative pathogens. (E.f. = Enterococcus faecalis, M.l. = Micrococcus luteus, S.m. = Serratia marcescens, P.a. = Pseudomonas aeruginosa, P.e. = Pseudomonas entomophila, P.r. = Providencia rettgrei).
Error bars indicate standard error from at least 3 independent experiments. *=p<0.05, **=p<.01, n.s.=not significant.
4e-bp mutants were previously shown to be immune-compromised. Since we identified ATF4 as a regulator of 4e-bp expression during infection, we predicted that atf4 mutants would be similarly immune-compromised. We tested this using systemic pathogen load assays to measure Ecc15 levels in the larvae after infection. When compared to isogenic controls, crc1 homozygotic mutants had higher systemic pathogen load (Fig. 1F). Consistent with previously published data, homozygous null 4e-bp mutants, Thor2, were similarly immune compromised when compared to its respective isogenic control, Thorrev (see materials and methods for details).
GCN2 signaling primarily activates ATF4-mediated 4e-bp induction in the fat body
Upon further examination of the transcriptional induction of 4e-bp post-infection, we see increased 4e-bp transcripts both in the fat body and in the gut (Fig. S1). Since the fat body is the primary site of AMP synthesis in response to infection (Imler, 2014), we sought to examine the signaling pathways upstream of 4e-bp in this tissue. ATF4 can be activated by either of two known eIF2α-kinases in Drosophila: the ER stress-responsive kinase PERK, or the amino acid deprivation activated kinase GCN2. To determine which of these two kinases lies upstream of ATF4 during infection, we knocked down gcn2 and perk with the fat body-specific Dcg-Gal4 driver. Depletion of gcn2, but not perk, in the fat body resulted in an increased systemic pathogen load upon Ecc15 infection (Fig. 1G). While depletion of gcn2 in the fat body led to near complete suppression of 4e-bp induction during Ecc15 infection, depletion of perk also resulted in a reduction of 4e-bp, albeit to a lesser extent (Fig. 1H). To further examine the role of the kinases, we tested the effect of gcn2 and perk depletion in the fat body 4E-BPintron-dsRed induction during infection. We observed that knockdown of gcn2 in the fat body had a stronger effect on 4E-BPintron-dsRed than that caused by the knockdown of perk (Fig S1B). Together these data suggest that GCN2 is the major modulator of ATF4 in the context of 4e-bp induction by pathogenic bacterial infection, with a minor role for PERK. Notably, AMP mRNA levels were also induced when cultured Drosophila S2 cells were deprived of amino acids (Fig. S1C). However, this is likely to be mediated through an ATF4-independent mechanism as Tunicamycin treatment, which also stimulates ATF4 induction, did not induce AMP mRNAs (Fig. S1D).
We next examined if ATF4-mediated 4e-bp induction occurs in response to infection by other bacterial strains. Several Drosophila pathogens were tested, including gram-positive bacteria (Enterococcus faecalis, Micrococcus luteus) and other gram-negative bacteria (Serratia marcescens, Pseudomonas aeruginosa, Pseudomonas entomophila, Providencia rettgrei). We observed 4e-bp induction with all pathogens we tested, albeit to different extents (Fig. 1I). Knockdown of atf4 with a fat body specific driver, Dcg-Gal4, resulted in suppression of 4e-bp induction with all pathogens tested (Fig. 1I), suggesting that ATF4/4E-BP axis activation occurs as part of a general immune response to pathogenic bacteria.
Levels of AMP proteins, but not transcripts, are reduced in 4e-bp mutants
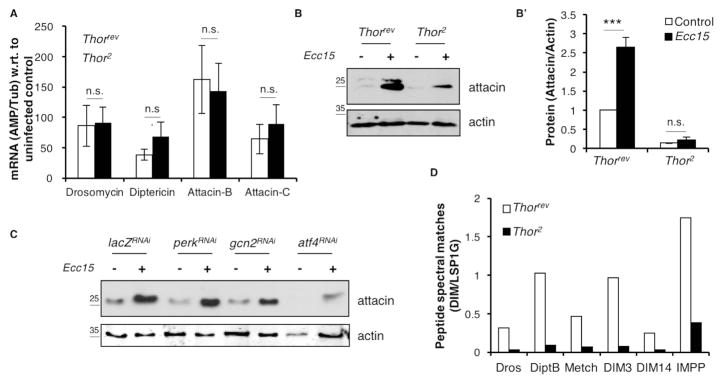
To gain insight into why 4e-bp mutants were immune-compromised, we asked whether 4E-BP regulated the canonical innate immune response pathways. In response to infection, two branches of the NF-κB signaling involving the Toll and immune deficiency (IMD) receptors are activated and initiate a signaling cascade which culminates in the induction of transcripts encoding AMPs (Buchon et al., 2014). AMPs are required for combating pathogen load with different classes of AMPs targeting different aspects of pathogen physiology. We examined whether 4E-BP affected the Toll or IMD signaling by using AMP transcriptional induction upon infection as readouts for pathway activity. We observed that the transcriptional induction of AMPs such as Drosomycin (downstream of Toll), Diptericin A and Attacins (downstream of IMD) is unaffected in Thor2 homozygotic mutants (Fig. 2A). Therefore, we concluded that 4E-BP is not required for signaling events leading to AMP transcriptional induction upon infection.
Fig. 2. Levels of AMP protein, but not transcripts, are negatively affected in 4E-BP mutants.
2A. qPCR analysis of AMP induction in homozygotic Thorrev(white) and Thor2 (blue) infected with Ecc15. Data were normalized to the uninfected control of the respective genotype.
2B. Western blot analysis of hemolymph collected from homozygotic Thorrev and Thor2 infected with Ecc15 with Attacin A antibody (top panel) and a loading control, actin (bottom panel).
2B’ shows quantification of the blot with all values normalized to Thorrev control. Fig. S2A shows individual blots used to generate the graph in 2B’.
2C. Western blot analysis of hemolymph collected from control and infected larvae expressing RNAi targeting perk, gcn2 and atf4 driven by Dcg-Gal4.
2D. Mass spectrometry analysis of hemolymph collected from Thorrevand Thor2 infected with Ecc15. Data represent the peptide spectral matches (PSM) for each AMP or DIM normalized to larval serum protein 1G (LSP1G). (Dros = Drosocin, DiptB = Diptericin B, Metch = Metchnikowin, DIM = Drosophila immune molecule, IMPP = Immune induced peptide precursor). Also see Fig. S2D and Table S4.
For 2A, B, error bars indicate standard error from at least 3 independent experiments.. ***=p<0.001. n.s.=not significant.
However, western blotting of hemolymph collected from infected and uninfected Thor2 homozygotic larvae showed a significant reduction in the protein levels of Attacins in comparison to Thorrev control larvae. (Fig. 2B, B’, S2A). Consistent with data in Figure 1, knockdown of gcn2 and atf4 in the fat body also resulted in reduced attacin protein levels in the hemolymph (Fig. 2C). This was corroborated by mass spectrometric analysis of hemolymph from Thor2 larvae, which indicated a reduction in the levels of other AMPs such as Drosocin, Diptericin B and Metchnikowin, and other Drosophila immune molecules (DIMs) (Fig. 2D, S2D, D’) when normalized to larval serum protein. Thus, 4e-bp mutant animals display normal induction of AMP mRNA but lower levels of AMP proteins. As these data could not rule out the alternative possibility that reduced Attacin levels were due to proteasomal degradation in Thor2 mutants, we fed larvae with 10 μM of a proteasomal inhibitor, bortezomib (Fig. S2B), a condition that readily activates the proteasome inhibition response in flies (Tsakiri et al., 2013). These conditions did not restore the reduced Attacin levels in the hemolymph (Fig. S2C). Together, these data suggested that 4e-bp mutants were immune-compromised because of reduced AMP synthesis.
The 5’UTRs of Drosomycin, Attacin A and 4e-bp support cap-independent translation
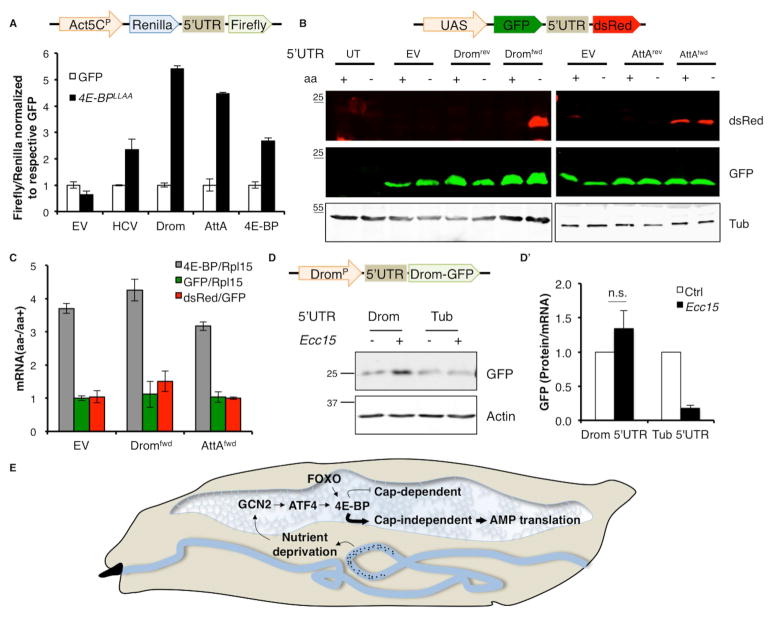
Since 4E-BP is an inhibitor of cap-dependent translation, we expected loss of 4e-bp to enhance cap-dependent translation but our data shows that AMP synthesis was reduced under these conditions. Moreover, the transcriptional induction of 4e-bp during infection suggests that AMP mRNAs are capable of being translated in the presence of 4e-bp. These observations prompted us to examine the possibility that AMPs are translated through an eIF4E-independent mechanism. There are several known mechanisms of translation that can bypass eIF4E requirement. One is through the presence of an internal ribosomal entry site (IRES) element in the 5’UTR of mRNAs (Komar and Hatzoglou, 2011), which allow those transcripts to recruit the translation machinery independent of eIF4E and thus can bypass translational regulation by 4E-BP (Marr et al., 2007). To test the 5’UTRs of the AMPs for possible IRES elements we performed bicistronic assays in Drosophila S2 cells using Renilla and Firefly luciferase reporters (schematic, Fig. 3A). These two reporters are transcribed as a single mRNA, where translation of Renilla luciferase is dependent on cap-recognition, but Firefly luciferase translation is dependent on the intervening sequence being able to recruit ribosomes independent of cap-recognition (schematic, Fig. 3A). We used the Hepatitis C virus (HCV) IRES as a positive control (Fig. 3A). We did not see a significant change in the Renilla luciferase activities of the reporters tested, thus the ratio of Firefly to Renilla luciferase activity represents the ability of the 5’UTRs tested to support cap-independent translation. S2 cells expressing bicistronic reporters with the 5’UTRs of Drosomycin and Attacin A inserted in the forward orientation allowed the translation of the Firefly luciferase (Fig. 3A). Interestingly, we also found that the 5’UTR of 4e-bp itself scored positively in the bicistronic assay (Fig. 3A). Surprisingly, the cap-independent activities of these 5’UTRs were higher than that of the well-characterized HCV IRES.
Fig. 3. 5’UTRs of Drosomycin, Attacin A and 4E-BP can be translated cap-independently.
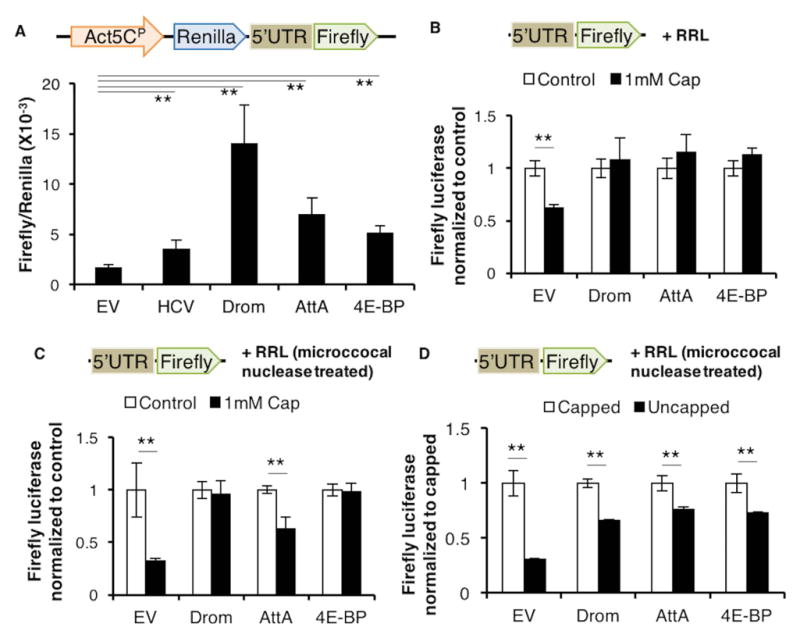
3A. Schematic showing the bicistronic construct (top). (Bottom) Luciferase activity from S2 cells expressing bicistronic constructs containing the 5’UTRs from Drosomycin (Drom), Attacin A (AttA) and 4e-bp, with the empty vector (EV) without any 5’UTR inserted (EV) as a negative control and, Hepatitis C virus (HCV) IRES inserted as a positive control. The mini-Actin 5C promoter (Act5CP) drives reporter expression. Firefly luciferase activity of each sample is normalized to its respective Renilla luciferase activity.
3B. 5’-capped Firefly monocistronic reporter mRNA (schematic on top) translated in control RRL (white) or RRL treated with excess cap-complex (black). Data for each 5’UTR are normalized to respective control samples.
3C. Monocistronic assay as performed in 3B but in RRL treated with microccocal nuclease to eliminate competing capped transcripts.
3D. Comparison of translation efficiency of capped versus uncapped monocistronic mRNA in microccocal nuclease-treated RRL (as performed in 3B).
Error bars indicate standard error from at least 3 independent experiments. **=p<.01. Fold changes are not significant unless indicated otherwise.
To further validate the 5’ cap-requirement of the AMP transcripts we performed in vitro cap competition assays. We measured translation of 5’-capped, monocistronic Firefly luciferase reporter mRNA (schematic, Fig. 3B) in vitro using rabbit reticulocyte lysates (RRL) in the presence of excess cap analog (m7GpppG). Excess cap compound competes with cap-dependent transcripts for eIF4E complexes but should not affect cap-independent transcripts. While translation of the control luciferase reporter mRNA was negatively affected by the addition of excess m7GpppG, reporter transcripts containing the 5’UTR of Drosomycin, Attacin A and 4e-bp were indifferent to the presence of the excess m7G (Fig. 3B). We also performed these assays in RRL treated with micrococcal nuclease, to eliminate effects of competing mRNAs (Fig. 3C). Drosomycin and 4e-bp remain resistant to excess cap while the Attacin A 5’UTR reporter showed a small but significant decrease in translation. Nevertheless, the Attacin A 5’UTR reporter’s resistance to excess cap remained higher than the control mRNA (Fig. 3C). To further investigate the requirement for an m7G cap we performed monocistronic assays with uncapped mRNA. We observed that the 5’UTRs of Drosomycin, Attacin A and 4e-bp supported translation with similar efficiencies, retaining roughly 75% activity without a cap. By contrast the control transcripts retained only 30% activity when uncapped (Fig. 3D). Together these data support the idea that certain AMP transcripts can be translated independent of eIF4E.
Cap-independent translation bias imposed by 4E-BP drives AMP synthesis
Data from Fig. 2 showing that AMP protein levels are reduced in 4e-bp mutants, and Fig. 3 showing that AMPs can be translated cap-independently together suggest a role for 4E-BP in biasing cellular translation to cap-independent mechanisms. To test the idea that 4E-BP is required for favoring cap-independent translation of AMPs during infection, we examined how its expression affected translation of the bicistronic reporter in S2 cells (Fig. 3A). We used a constitutively active mutant of 4e-bp, 4E-BPLLAA, which cannot be inactivated by the 4E-BP kinase TOR, and binds tightly to eIF4E therefore inhibiting eIF4E consistently when overexpressed (Miron et al., 2003). The cap-independent translation of Firefly luciferase in bicistronic reporters (schematic, Fig 4A) containing the 5’UTRs of Drosomycin, Attacin A and 4e-bp was enhanced over 5, 4 and 2 fold respectively in the presence of 4E-BPLLAA in comparison to cells expressing GFP as a control (Fig. 4A). Strikingly, the enhancement of AMP translation in the presence of 4E-BPLLAA exceeded that seen with the HCV IRES (over 2 fold). Since in the context of infection, we saw that 4e-bp is activated by GCN2/ATF4, we asked whether GCN2 activation by amino-acid deprivation was sufficient to enhance cap-independent activity of AMP 5’UTRs. We tested this using a second bicistronic reporter with GFP reporting cap-dependent translation and dsRed reporting cap-independent translation (schematic, Fig 4B). S2 cells expressing the Drosomycin 5’UTR bicistronic reporter in the forward orientation showed an enhanced dsRed expression when subjected to amino acid deprivation to activate GCN2 (Fig. 4A). While the Attacin A 5’UTR supported cap-independent translation of dsRed, this was not enhanced significantly upon GCN2 activation. Drosomycin and Attacin A 5’UTRs in the reverse orientation did not yield cap-independent translation. GFP levels remained unchanged with both reporters, most likely due to the persistence of GFP that was synthesized prior to the relatively short amino acid deprivation treatment (4 h). To eliminate the possibility that the reporter may be transcriptionally regulated due to any cryptic promoter activity in the 5’UTR, we measured the ratio of dsRed mRNA to GFP mRNA in amino acid deprived S2 cells expressing the fluorescent bicistronic reporters (Fig. 4C). We saw no change in relative levels of GFP and dsRed qPCR signals suggesting that there is no cryptic promoter element that may respond to amino acid deprivation. We also did not detect any change in the transcript levels of the bicistronic reporter itself upon starvation (Fig. 4C). Consistent with previous observations, we saw an induction of 4e-bp in response to amino acid deprivation (Fig. 4C).
Fig. 4. 4E-BP enhances the translation of AMPs.
4A. Bicistronic reporter (schematic on top) assays with 5’UTRs indicated on X-axis in S2 cells overexpressing either GFP as a control (white) or 4E-BPLLAA (black). Firefly luciferase activity of each sample is normalized to Renilla luciferase activity and all samples are normalized to their respective control (GFP). Error bars indicate standard deviation.
4B. Western blot analysis of the fluorescent bicistronic reporter (schematic on top) with Drosomycin (Drom) and Attacin A (AttA) 5’UTR in forward (fwd) or reverse (rev) orientation in S2 cells grown in either complete media (control) or media lacking amino acids (aa). (UT= untransfected control, EV= empty vector).
4C. qPCR analysis of S2 cells expressing the Dromfwd and AttAfwd 5’UTR fluorescent bicistronic reporter (as in 4B) subjected to amino acid (aa) deprivation. Data represents fold changes in aa deprived cells w.r.t. cells grown in complete media, with mRNA normalized as indicated in legend.
4D. Western blot of hemolymph from transgenic larvae expressing monocistronic reporters (schematic on top) driven by the Drosomycin promoter (DromP) with indicated 5’UTRs (Drom= Drosomycin, Tub= Tubulin) infected with Ecc15. The Drosomycin-GFP (Drom-GFP) fusion protein is detected using an anti-GFP antibody. 4D’ quantifies translation of Drom-GFP.relative to Drom-GFP mRNA levels.
4E. A model for 4E-BP activation and function during enteric infection in larvae. In response to enteric infections by pathogens (indicated by black dots), GCN2 is activated in the fat body likely due to nutrient deprivation. GCN2 activation in the fat body leads to ATF4 synthesis and subsequent transcriptional induction of 4e-bp. FOXO also contributes to 4e-bp induction. Once induced, 4E-BP itself is translated cap-independently and blocks cap-dependent translation of most cellular transcripts. In doing so, it promotes (indicated by bold arrow) cap-independent translation of AMP transcripts. Such bias in cellular translation by 4E-BP is required to drive AMP synthesis during the infection, thus explaining the role of 4E-BP in the innate immune response.
Error bars indicate standard error from 3 independent experiments.
To test the cap-independent features of the AMP 5’UTRs in vivo, we generated transgenic flies bearing monocistronic reporters (schematic, Fig. 4D). These reporters utilized the Drosomycin promoter (DromP) to drive the expression of Drosomycin-GFP bearing either the Drosomycin 5’UTR or Tubulin 5’UTR as a negative control. To allow for a fair comparison of their expression, the two reporters were targeted into the same attP landing site in the genome. As expected, both versions of the reporter were transcriptionally induced in response to Ecc15 infection, since the Drosomycin promoter is driving their expression (Fig. S3A). We then tested the expression of these two reporters in control and infected larva by western blotting for GFP to detect the Drosomycin-GFP fusion protein in the hemolymph. Our data showed that while the level of Drosomycin-GFP bearing the Drosomycin 5’UTR increased substantially in response to infection, the reporter bearing the Tubulin 5’UTR did not show a significant increase (Fig. 4D). Normalizing the GFP protein levels for each reporter to the respective GFP mRNA levels indicates that while the Drosomycin 5’UTR reporter sees a substantial increase in translation upon infection, the translation of the Tubulin 5’UTR reporter is relatively suppressed (Fig 4D’). Taken together, these data show that increased 4E-BP levels during infection result in preferential translation of cap-independent transcripts, such as Drosomycin, over cap-dependent transcripts.
Discussion
Much research has focused on the transcriptional response to infection mediated by the innate immune response pathways (Buchon et al., 2014). Although translational inhibitors such as GCN2 and 4E-BP have been implicated in the antibacterial response, the incongruence of translation inhibition and the need for AMP synthesis had not been addressed experimentally. Here, we provide results that resolve this discrepancy and establish that AMPs have evolved mechanisms to bypass translation inhibition imposed by pathogenic bacteria via 4E-BP activation. We show that the ISR pathway promotes innate immunity against pathogenic bacteria by mediating 4e-bp induction, which in turn biases cellular translation towards cap-independent mechanisms that favor AMP synthesis. The newly identified role of the ISR pathway in the innate immune response is likely coordinated with the established roles of other innate immune response pathways, such as those mediated by FOXO, IMD and Dorsal/Dif.
In addition to regulation by GCN2/ATF4 and FOXO, 4E-BP is also famously regulated post-translationally by another amino-acid sensitive kinase, TOR. While under steady state conditions, TOR phospho-inactivates 4E-BP, TOR itself is inactivated in response to amino acid deprivation. Thus 4E-BP newly synthesized in response to infection-mediated amino acid deprivation will likely not be subject to inactivation by TOR.
Our observations are consistent with the positive effects of 4e-bp induction against a non-lethal pathogen such as Ecc15 (Bernal and Kimbrell, 2000). However, infection by a severe pathogen such as P.e. results in a GCN2-mediated translation block in the gut and is detrimental to the host immune response (Chakrabarti et al., 2012). Why GCN2 mediates such different outcomes remains unresolved, but it could be due to factors other than ATF4 that are downstream of GCN2.
It is worth noting that GCN2 engages two different translation inhibition mechanisms: 1) phospho-eIF2α, which induces ATF4 and, 2) 4e-bp, which is transcriptionally induced by ATF4 (Kang et al., 2017; Yamaguchi et al., 2008). We primarily focused on 4E-BP because the effect of phospho-eIF2α on translation may be temporary since ATF4 induces the expression of an eIF2α phosphatase subunit, GADD34, thereby stimulating eIF2α dephosphorylation. We speculate that eIF2α phosphorylation may not serve as a long-term deterrent for AMP translation, while the second translational block by 4E-BP may persist longer. In addition, our data shows that 4e-bp itself is synthesized favorably by cap-independent translation conferred by its 5’UTR (Fig. 3, 4A), suggesting that this inhibition mechanism is self-sustaining.
Intriguingly, the 5’UTRs of Drosomycin and Attacin A, which confer cap-independent translation capabilities to their respective mRNA (Fig. 3, 4A), are relatively small at 63 and 29 bases respectively. To the best of our knowledge, these are the smallest characterized cap-independent translation elements known. While there are no known consensus sequences for IRESes, scoring positively in biochemical assays such as bicistronic assays and cap competition assays has been recognized to be a reliable indicator for them (Dever et al., 1992; Young et al., 2015; Zhou et al., 2008). While both Drosomycin and Attacin A are likely translated cap-independently via IRES-like elements in their 5’UTRs, they may utilize slightly different mechanisms. Though both 5’UTRs confer cap-independence which is enhanced by 4E-BP (Fig. 3, 4A, B), the Attacin A 5’UTR reporter shows basal translation of the reporter even in the absence of 4E-BP while the Drosomycin 5’UTR relies heavily on 4E-BP (Fig 4B). Additionally, eliminating competing transcripts in the presence of excess m7GpppG also had a small but significant effect on Attacin A 5’UTR reporter mRNA (Fig 3B, C), suggesting that there is an additional layer of regulatory mechanisms acting on these 5’ UTRs.
Since suppression of cap-dependent translation by 4E-BP appears to stimulate the synthesis of AMPs (Fig. 4), we speculate that the AMP 5’UTRs do not compete well with other mRNAs for ribosomes and initiation factors. Thus, in addition to the established role of 4E-BP in the inhibition of cap-dependent translation, our data shows a more nuanced role for 4E-BP as a promoter of cap-independent translation required for driving the synthesis of essential immune response proteins. Interestingly, it had been previously suggested that eIF2α-kinases such as GCN2 could promote cap-independent translation (Fernandez et al., 2001, 2002), although the effectors of such regulation were unknown. Based on our work here showing that 4E-BP downstream of GCN2/ATF4 signaling regulates AMP translation, we can now surmise that these previously observed instances of cap-independent translation downstream of phospho-eIF2α are also likely mediated by ATF4-induced 4E-BP. Additionally, given the conservation of translation inhibition during infection in mammals, it would be interesting to examine whether the mammalian innate immune response is aided by any of the three known mammalian 4E-BPs and if first-response cytokines are synthesized cap-independently.
Methods
Fly stocks and S2 cell culture
All Drosophila stocks were reared on standard cornmeal medium at room temperature. Stocks used are listed in Table S1.
S2 cells were grown on standard Schneider’s medium (Life Technologies) supplemented with 10% FBS and 1% penicillin/streptomycin. For luciferase reporter assays, DNA was transfected at a 5:1 ratio of expression plasmid to reporter plasmid using PEI or Effectene (Qiagen).
Larval infection
Bacteria were grown in LB broth in an overnight culture and pelleted. All bacteria tested were grown at 37°C except Ecc15 and S.marcescens, which were grown at 25°C. 4-day old larvae orally infected with food mixed 5:1 by weight with the bacterial pellet for 4 hours.
Systemic pathogen load
After infection, the larvae were briefly washed in 70% ethanol to surface sterilize them. 3–4 larvae per condition were homogenized using a pestle in 100 μl of PBS and serially diluted homogenate was plated on LB agar. Colonies were counted from the same dilution for different conditions and genotypes and normalized to the control.
Immunofluorescence (IF) and Western blotting (WB)
Larval guts and fat body were dissected in PBS and fixed in 4% PFA, PBT (0.1% Tween) for 20 minutes. The tissues were then washed 3x in PBT and stained with respective antibodies. Samples were imaged using a LSM700 Zeiss microscope at 20X magnification unless otherwise specified. Details for antibodies can be found in Supplemental Information.
Hemolymph collection for WB and mass spectrometry (MS)
Hemolymph from larvae was collected by making a small incision in the cuticle and bleeding them into PBS. For WB, hemolymph from 10–20 larvae was mixed with sample buffer and analyzed by SDS-PAGE. For mass spectrometry, hemolymph samples were prepared from 75 larvae (see Supplemental Information for details) and the peptide spectral matches (PSM) for each AMP was normalized to the PSM for controls used in Fig. 2D, S2D and S2D’.
Bicistronic assays
Molecular cloning details for reporters and assay buffer composition can be found in the Supplemental Information. 36 hours after transfection with the luciferase bicistronic reporter, cells were lysed in passive lysis buffer (Promega). Firefly expression was measured in FF assay buffer. Renilla expression was measured by addition of an equal volume of Ren assay buffer.
For experiments in Fig. 4A, cells were co-transfected with an expression construct driven by the mini-Actin5C promoter driving either GFP or 4E-BPLLAA either (pMA6-GFP or pMA6-4E-BPLLAA) cells were harvested for luciferase assays as described above. For experiments in Fig 4B, cells transfected with the fluorescent bicistronic reporter were starved using amino acid-free Schneider’s growth medium (US Biologicals) and harvested in lysis buffer containing 1% NP40, 1x TBS, protease inhibitor cocktail (Roche), and subjected to western blotting.
In vitro Transcription and mRNA reporter preparation
Transcription templates for monocistronic reporters were created by PCR with a template specific forward primer containing either a T7 or T3 promoter sequence and a vector specific reverse primer. Templates were transcribed using either T7 or T3 RNA polymerase and purified using LiCl precipitation. Transcripts were capped using Vaccinia Virus Capping enzyme (New England Biolabs) and purified by phenol/chloroform extraction and isopropanol precipitation. Transcripts were poly(A) tailed using E.coli poly(A)polymerase (New England Biolabs) and purified by phenol/chloroform extraction and isopropanol precipitation.
In vitro translation assays
Translation assays were performed in rabbit reticulocyte Lysate (Green Hectares, McFarland, WI) (treated with Micrococcal nuclease for experiments in 3C, D) and mRNA reporter prepared as described above (see Supplemental Information for buffer composition details). Translation reactions were incubated at 37°C for 30 min and luc iferase activity was measured as described above. For experiments with excess m7G cap, a cap structure analogue (New England Biolabs, #S1407S) was added to a final concentration of 1 mM.
Supplementary Material
Mass spectrometric metrics for the peptides utilized in the analysis of AMP protein levels (Related to Figure 2)
Highlights.
4E-BP is transcriptionally induced by GCN2-ATF4 signaling in response to bacterial infection
4E-BP mutants show unaltered antimicrobial peptide (AMP) transcript levels, but have reduced AMP translation
AMP 5’UTRs support cap-independent translation
Translation bias by 4E-BP drives cap-independent AMP translation
Acknowledgments
We thank Drs. Mimi-Shirasu Hiza and Brian Lazzaro (for providing the pathogens used in this project); Drs. Neil Silverman and Donggi Park (for providing the anti-Attacin antibody); Drs. Marc Amoyel, Erika Bach, Jessica Treisman, Deborah Kimbrell, and Ian Mohr (for discussions and advice); the Bloomington and Vienna Drosophila Stock Centers for fly lines. This work was supported by grants from the National Institutes of Health (R01EY020866) and the March of Dimes (FY#13-204) to H.D.R., the National Institutes of Health (R01GM11703) to M.T.M. and Beatrix Ueberheide (R41 GM103362), and the American Heart Association fellowship (17POST33420032) to D.V.
Footnotes
Author contributions: D.V. and H.D.R. conceived the project, designed the experiments, analyzed the data, and wrote the paper. N.C. and M.T.M. designed and performed the in vitro IRES assays and edited the paper. J.S. provided technical assistance for all fly experiments. V.C. and B.U. generated the mass spectrometry data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Bernal A, Kimbrell DA. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc Natl Acad Sci U S A. 2000;97:6019–6024. doi: 10.1073/pnas.100391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology. Nat Rev Immunol. 2014;14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, Lemaitre B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 2012;12:60–70. doi: 10.1016/j.chom.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Fernandez J, Yaman I, Mishra R, Merrick WC, Snider MD, Lamers WH, Hatzoglou M. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J Biol Chem. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
- Fernandez J, Yaman I, Sarnow P, Snider MD, Hatzoglou M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem. 2002;277:19198–19205. doi: 10.1074/jbc.M201052200. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Imler JL. Overview of Drosophila immunity: a historical perspective. Dev Comp Immunol. 2014;42:3–15. doi: 10.1016/j.dci.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Jünger MA, Rintelen F, Stocker H, Wasserman JD, Végh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Vasudevan D, Kang K, Kim K, Park JE, Zhang N, Zeng X, Neubert TA, Marr MT, Ryoo HD. 4E-BP is a target of the GCN2-ATF4 pathway during Drosophila development and aging. J Cell Biol. 2017;216:115–129. doi: 10.1083/jcb.201511073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Girardin SE. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nat Rev Microbiol. 2013;11:365–369. doi: 10.1038/nrmicro3029. [DOI] [PubMed] [Google Scholar]
- Levitin A, Marcil A, Tettweiler G, Laforest MJ, Oberholzer U, Alarco AM, Thomas DY, Lasko P, Whiteway M. Drosophila melanogaster Thor and response to Candida albicans infection. Eukaryotic Cell. 2007;6:658–663. doi: 10.1128/EC.00346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, D’Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 2007;21:175–183. doi: 10.1101/gad.1506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M, Lasko P, Sonenberg N. Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol Cell Biol. 2003;23:9117–9126. doi: 10.1128/MCB.23.24.9117-9126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell Host Microbe. 2012;12:470–483. doi: 10.1016/j.chom.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Zhou Z, Tang ML, Meller S, Chen J, Bellen H, Kimbrell DA. Identification of immune system and response genes, and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics. 1996;143:929–940. doi: 10.1093/genetics/143.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Tsakiri EN, Sykiotis GP, Papassideri IS, Terpos E, Dimopoulos MA, Gorgoulis VG, Bohmann D, Trougakos IP. Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging Cell. 2013;12:802–813. doi: 10.1111/acel.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Ishihara H, Yamada T, Tamura A, Usui M, Tominaga R, Munakata Y, Satake C, Katagiri H, Tashiro F, et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 2008;7:269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Young SK, Willy JA, Wu C, Sachs MS, Wek RC. Ribosome Reinitiation Directs Gene-specific Translation and Regulates the Integrated Stress Response. J Biol Chem. 2015;290:28257–28271. doi: 10.1074/jbc.M115.693184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectrometric metrics for the peptides utilized in the analysis of AMP protein levels (Related to Figure 2)