Abstract
Until recently, NADPH oxidase (NOX) enzymes were thought to be a property of multicellularity, where the reactive oxygen species (ROS) produced by NOX acts in signaling processes or in attacking invading microbes through oxidative damage. We demonstrate here that the unicellular yeast and opportunistic fungal pathogen Candida albicans is capable of a ROS burst using a member of the NOX enzyme family, which we identify as Fre8. C. albicans can exist in either a unicellular yeast-like budding form or as filamentous multicellular hyphae or pseudohyphae, and the ROS burst of Fre8 begins as cells transition to the hyphal state. Fre8 is induced during hyphal morphogenesis and specifically produces ROS at the growing tip of the polarized cell. The superoxide dismutase Sod5 is co-induced with Fre8 and our findings are consistent with a model in which extracellular Sod5 acts as partner for Fre8, converting Fre8-derived superoxide to the diffusible H2O2 molecule. Mutants of fre8Δ/Δ exhibit a morphogenesis defect in vitro and are specifically impaired in development or maintenance of elongated hyphae, a defect that is rescued by exogenous sources of H2O2. A fre8Δ/Δ deficiency in hyphal development was similarly observed in vivo during C. albicans invasion of the kidney in a mouse model for disseminated candidiasis. Moreover C. albicans fre8Δ/Δ mutants showed defects in a rat catheter model for biofilms. Together these studies demonstrate that like multicellular organisms, C. albicans expresses NOX to produce ROS and this ROS helps drive fungal morphogenesis in the animal host.
Author summary
We demonstrate here that the opportunistic human fungal pathogen Candida albicans uses a NADPH oxidase enzyme (NOX) and reactive oxygen species (ROS) to control morphogenesis in an animal host. C. albicans was not previously known to express NOX enzymes as these were thought to be a property of multicellular organisms, not unicellular yeasts. We describe here the identification of C. albicans Fre8 as the first NOX enzyme that can produce extracellular ROS in a unicellular yeast. C. albicans can exist as either a unicellular yeast or as multicellular elongated hyphae, and Fre8 is specially expressed during transition to the hyphal state where it works to produce ROS at the growing tip of the polarized cell. C. albicans cells lacking Fre8 exhibit a deficiency in elongated hyphae during fungal invasion of the kidney in a mouse model for systemic candidiasis. Moreover, Fre8 is required for fungal survival in a rodent model for catheter biofilms. These findings implicate a role for fungal derived ROS in controlling morphogenesis of this important fungal pathogen for public health.
Introduction
Reactive oxygen species (ROS) including superoxide anion and hydrogen peroxide play diverse roles in biology. ROS can inflict severe oxidative damage to cellular components, but when carefully controlled, ROS can also be used to combat infection and act in cell signaling processes. A well-studied example of controlled ROS production involves NADPH oxidase (NOX) enzymes [1]. These heme and flavin containing enzymes use electrons from NADPH to reduce molecular oxygen to superoxide [1]. In macrophages and neutrophils, NOX enzymes generate bursts of superoxide in the extracellular milieu or phagolysosomal compartments to assault microbial pathogens. In non-immune cells, ROS from NOX enzymes are widely used in cell signaling pathways to promote growth, development and differentiation [1]. As membrane proteins, NOX enzymes can vectorially release superoxide inside the cell or extracellularly and in either case, the superoxide can react with neighboring superoxide dismutase (SOD) enzymes that disproportionate superoxide to oxygen and hydrogen peroxide. In fact, NOX enzymes often partner with SODs in signaling processes, whereby SOD converts the cell impermeable superoxide to the diffusible hydrogen peroxide signaling molecule [1–5]. NOX-SOD interactions are also prevalent during infection where the microbial pathogen uses its arsenal of extracellular SODs to combat the oxidative burst of host NOX enzymes [6].
The opportunistic fungal pathogen Candida albicans has evolved with a family of three extracellular SOD enzymes (Sod4, Sod5, Sod6) believed to protect the fungus from the attack of host NOX-derived superoxide [7, 8]. We recently reported that these extracellular SODs represent a novel class of Cu-only SOD enzymes that are unique to the fungal kingdom and oomycetes [9, 10]. Much of what is known about fungal Cu-only SODs has emerged from studies on C. albicans Sod5. Sod5 can react with superoxide at rates limited only by diffusion [9, 10], and can effectively degrade superoxide radicals derived from macrophage and neutrophil NOX enzymes [11, 12].
Curiously C. albicans Sod5 appears specific to the filamentous form of the fungus [7, 13]. C. albicans is a polymorphic fungus that can transition from unicellular yeast-like form to pseudo hyphal and true hyphal filamentous states [14, 15]; Sod5 is evidently absent in the yeast-form of C. albicans. The rationale for selective expression of Sod5 during morphogenesis was not clear, as both yeast and hyphal forms exist in the animal host, are subject to immune surveillance and are essential for virulence [15–17]. Moreover, SOD5 is induced in filamentous C. albicans in the absence of any insult from the host [7, 13]. This raises the possibility that filamentous C. albicans witness a source of superoxide not seen in the yeast-like form.
Certain multicellular fungi are capable of generating superoxide themselves using fungal NOX enzymes as part of signaling during differentiation [18–20]. However, unicellular yeasts were believed to not express NOX, as NOX was characterized as a property of multicellular differentiation [21, 22]. This dogma of no NOX in unicellular fungi was recently challenged by the identification of Saccharomyces cerevisiae Yno1, a NOX that localizes to the endoplasmic reticulum and generates intracellular (not extracellular) superoxide [23]. Other than Yno1, there has been no evidence for NOX enzymes in evolutionarily related yeasts including C. albicans.
Here we provide the first evidence for a NOX enzyme in the opportunistic fungal pathogen C. albicans. This NOX, known as Fre8, produces a burst of extracellular ROS in filamentous but not yeast-form cells. We demonstrate that Fre8-superoxide serves as substrate for Sod5, providing a rationale for inducing this extracellular SOD during morphogenesis. Strikingly, the ROS from Fre8 is concentrated at the growing tip of C. albicans hyphae and can promote formation and/or maintenance of elongated hyphae in vitro as well as in infected kidneys during a mouse model for disseminated candidiasis. Moreover, Fre8 enhances C. albicans survival in a rat venous catheter model of candidiasis. These studies show that host NOX is not the only source of ROS at the host-pathogen interface; C. albicans makes its own ROS for hyphal morphogenesis through Fre8.
Results
C. albicans hyphae produce a burst of ROS that serves as substrate for extracellular Sod5
Previously, Schroter et al. demonstrated that C. albicans produces ROS during the transition from the yeast form to the hyphal state [24]. To probe this fungal ROS, we used luminol, a chemiluminescence probe typically used to measure ROS bursts in macrophages and neutrophils [11, 12, 25, 26]. Consistent with findings by Schroter et al [24], C. albicans cells induced to form hyphae by serum treatment exhibited a burst in luminol chemiluminesence not seen in yeast-form cells (Fig 1A). We observed that this ROS is not unique to serum stimulation but is also seen when morphogenesis is induced by elevated amino acid concentrations in IMDM medium and alkaline pH conditions (Fig 1A) [14, 27].
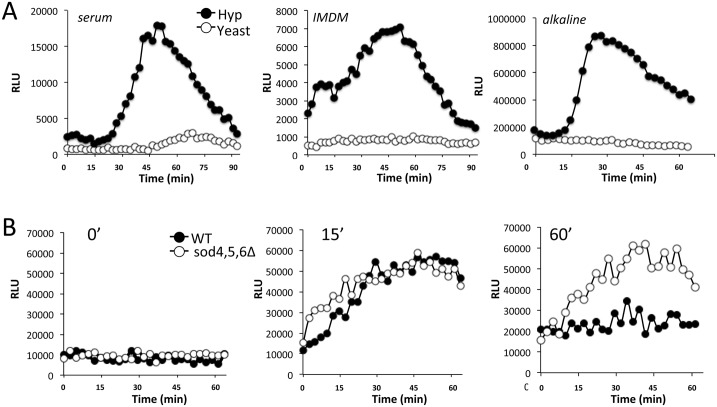
Fig 1. The ROS burst of C. albicans during morphogenesis.
(A) WT strain SC5314 was grown to mid log phase in YPD media at 30°C to obtain the yeast/budding form (“yeast”). Where indicated, cells were induced to form hyphae (“Hyp”) at 37°C for 1 hr with 10% FBS, IMDM or alkaline medium. Both forms of cells were subjected to ROS analysis by luminol chemiluminescence as described in Materials and Methods. (B) WT CAIF100 or the isogenic sod4Δ/Δ sod5Δ/Δ sod6Δ/Δ strain were induced to form hyphae in alkaline medium for the indicated times and subjected to ROS measurements by lucigenin chemiluminescence. Results were recorded as relative luminescence units (RLU) as described in Materials and Methods and plotted in intervals of whole minutes.
The luminol probe used in Fig 1A is not expected to penetrate the fungal cell wall, and should therefore only detect extracellular ROS. This notion of extracellular ROS was corroborated using the chemiluminescence probe lucigenin that cannot cross cell membranes, and is specific for superoxide compared to luminol which can detect both superoxide and hydrogen peroxide [25, 28]. As seen in Fig 1B, cells induced to form hyphae exhibited a defined lucigenin signal within 15 min of morphogenesis. However, in WT cells the lucigenin signal often declined at later times points (60 min, Fig 1B), and we tested whether this reflected induction of extracellular SOD enzymes. Indeed the superoxide signal from lucigenin was enhanced in sod4Δ/Δ sod5Δ/Δ sod6Δ/Δ cells lacking all three extracellular SODs (60 min, Fig 1B) and the same was true with luminol chemiluminescence (Fig 2B). Of the three extracellular SODs, deletion of SOD5 alone was sufficient to enhance ROS during morphogenesis stimulated by serum (Fig 2A) or IMDM (Fig 2B), consistent with the notion that Sod5 is the major extracellular SOD induced during hyphal formation [7]. Compared to effects of sod5Δ/Δ mutations, there was no change in luminol ROS in sod1Δ/Δ mutants lacking the major intracellular Sod1 (Fig 2C). Together these studies demonstrate that cells undergoing morphogenesis produce a burst of extracellular ROS including superoxide that can serve as substrate for extracellular Sod5.
Fig 2. The effect of sod5Δ/Δ mutations and NOX enzyme inhibition on the ROS burst of C. albicans.
The indicated C. albicans strains were induced to form hyphae with either 10% serum (A) or IMDM (B, C, D) and ROS production monitored by luminol as in Fig 1. The indicated strains used are (A) WT SC5314 or the isogeneic sod5Δ/Δ cell; (B, C) WT CAIF100 or the isogenic sod5Δ/Δ, sod1Δ/Δ or sod4Δ/Δ sod5Δ/Δ sod6Δ/Δ strains; (D) WT SC5314. Where indicated, assays were conducted in the presence of the designated concentrations of the NOX inhibitor diphenylene iodonium (DPI) or with 0.5 μM DPI (C).
In multicellular organisms, extracellular ROS is derived from NOX enzymes, although C. albicans was not previously known to express NOX. To address whether the ROS burst of morphogenesis was derived from a NOX enzyme, we used DPI (diphenylene iodonium), a classical inhibitor of NOX enzymes [29–31]. As seen in Fig 2D, there was a dose response inhibition of the ROS burst of C. albicans using DPI, with full inhibition at 0.5 μM. DPI also eliminated the enhanced ROS of sod5Δ mutants (Fig 2C). These studies suggested that C. albicans expresses a NOX enzyme for extracellular ROS during morphogenesis. However, since DPI can also inhibit other flavin containing enzymes [32], we applied molecular genetic approaches to examine the source of the ROS burst.
C. albicans FRE8 encodes a NOX enzyme
In the fungal kingdom, NOX enzymes are part of an expanded family of NADPH oxidoreductases that use electrons from NADPH to either reduce oxygen to superoxide (NOX enzymes) or reduce ferric or cupric metal ions (FRE enzymes) [33]. NOX and FREs are highly similar and it is difficult to predict functionality based on sequence analysis alone [23, 33, 34]. Yeasts are generally thought to only express FRE, not NOX [21, 22] although as mentioned above, this dogma was challenged by identification of S. cerevisiae Yno1 as an endoplasmic reticulum NOX [23]. In C. albicans, there are at least 17 genes annotated as FREs [35, 36], three of which are known cupric or ferric reductases (Fre1, Fre7, Fre10 (S1 Fig, [37–39]); the remainder have uncharacterized functions. By qRT-PCR, we identified a number that are induced during early stages of hyphal morphogenesis coincident with the ROS burst (S1 Fig). The C. albicans orthologue to S. cerevisiaeYNO1 [18] was not among the FREs induced with morphogenesis (S1 Fig). The most highly induced hyphal specific gene was FRE8, also known as CFL11 or C. albicans CR_06670W, orf19.701 (S1 Fig). FRE8 was additionally reported as the most abundantly induced FRE during C. albicans invasion of the kidney [40]. We chose to focus on FRE8 as a potential NOX enzyme.
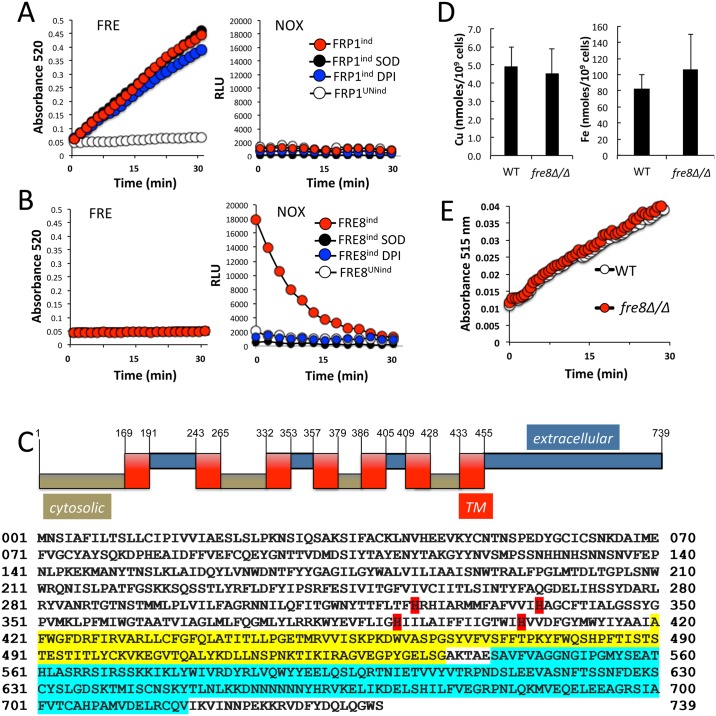
Recombinant versions of codon optimized FRE8 were expressed in Pichia pastoris under control of the methanol inducible AOX2 promoter. An identical procedure has been used to express and analyze activity of mammalian NOX enzyme complexes [41]. In parallel, we expressed a second unknown member of the FRE family, namely FRP1, that is only moderately induced by hyphal stimulation (S1 Fig). P. pastoris cells expressing recombinant FRE8 and FRP1 were assessed for ferric reductase activity and ROS production. As seen in Fig 3A left, Pichia cells expressing FRP1 exhibited clear ferric reductase activity when FRP1 expression was induced with methanol. These cells however, exhibited no ROS that could be detected by luminol (Fig 3A right), indicating that FRP1 encodes a metalloreductase, and not a NOX enzyme. The opposite profile was obtained with Pichia cells expressing FRE8. These cells exhibited no methanol-inducible ferric reductase activity (Fig 3B left), but produced a strong methanol-inducible ROS signal (Fig 3B right). This ROS was eliminated by addition of 0.5 μM of the NOX inhibitor DPI or by addition of exogenous SOD enzyme (bovine SOD1). By contrast the ferric reductase activity of recombinant Frp1 was not altered by exogenous SOD or by DPI (Fig 3A left). These results indicated that FRP1 encodes a metalloreductase while FRE8 encodes a NOX.
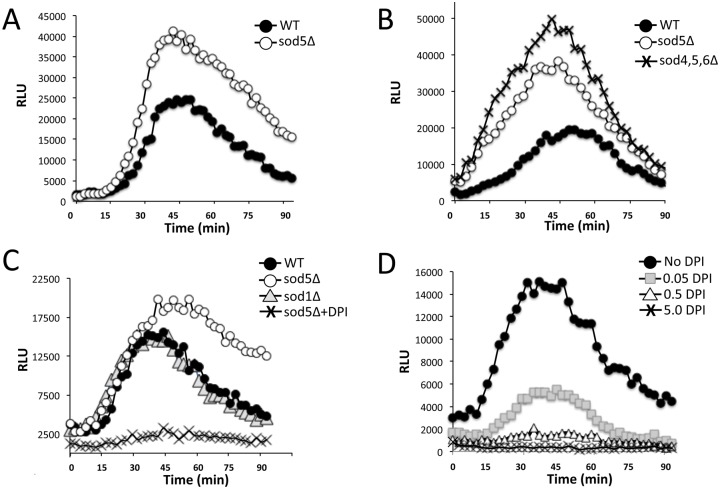
Fig 3. C. albicans FRE8 as a candidate NOX enzyme.
(A, B) P. pastoris strains expressing recombinant C. albicans FRP1 (A) or C. albicans FRE8 (B) under control of the AOX2 promoter were cultured with either methanol to induce gene expression (“ind”) or with glycerol (“UNind”) to prevent expression of recombinant FRP1 or FRE8. Samples were analyzed for ferric reductase (“FRE”) activity (A, B left) or for NOX-like activity (“NOX”) through ROS production by luminol chemiluminescence (A,B right) as described in Materials and Methods. Compared to C. albicans, the luminol substrate appears rapidly depleted in P. pastoris expressing high levels of FRE8. Where indicated, assays were supplemented with 0.1 unit bovine Cu/Zn SOD1 to remove extracellular superoxide or 50 nM DPI to inhibit NOX activity. Although DPI is predicted to inactivate other flavin requiring enzymes [32], this dose of DPI does not inhibit the ferric reductase activity of recombinant Frp1. (C) TOP- predicted transmembrane domain of C. albicans Fre8 based on TMHMM hydropathy plot analysis where brown and blue bars are predicted cytosolic and extracellular/luminal domains respectively and red are transmembrane (TM). BOTTOM–amino acid sequence of C. albicans Fre8 showing predicted histidine ligands for heme (red), FAD (yellow) and NAD (aqua) binding as determined by InterPro and PFAM databases. (D, E) WT SC5314 and isogenic fre8Δ/Δ C. albicans cells induced to form hyphae for 1 hour were analyzed for copper and iron accumulation by ICP-MS (D) or for ferric reductase activity (E) as described in Materials and Methods. ICP-MS results represent the averages of three biological replicates where error bar equals standard deviation. Based on T-test there were no significant differences with either copper (P = 0.73) or iron (P = 0.47) accumulation.
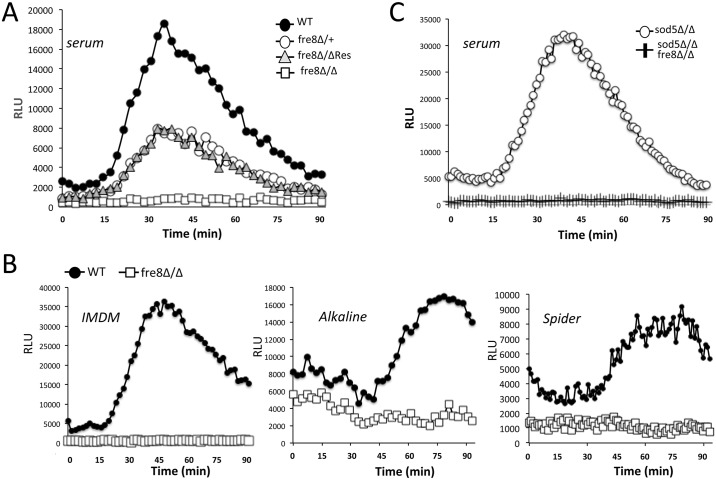
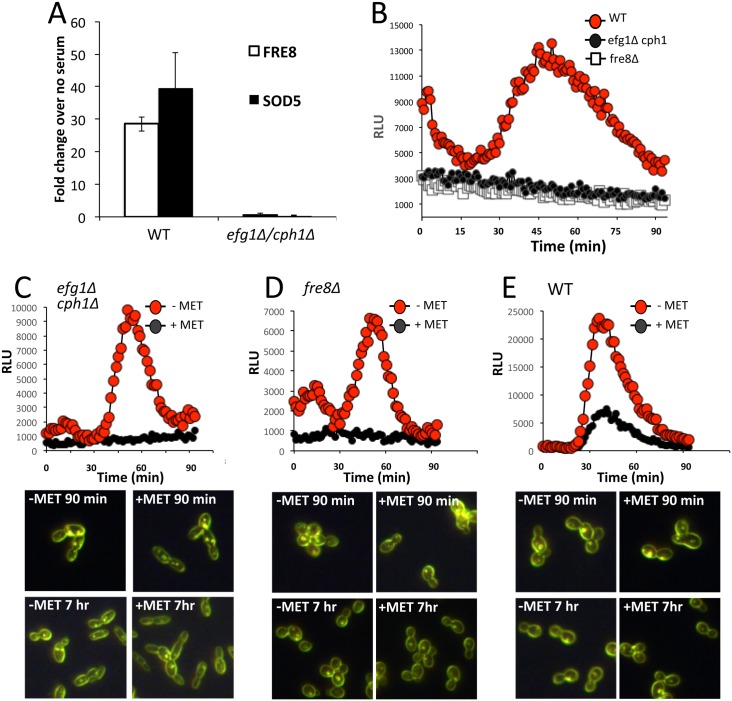
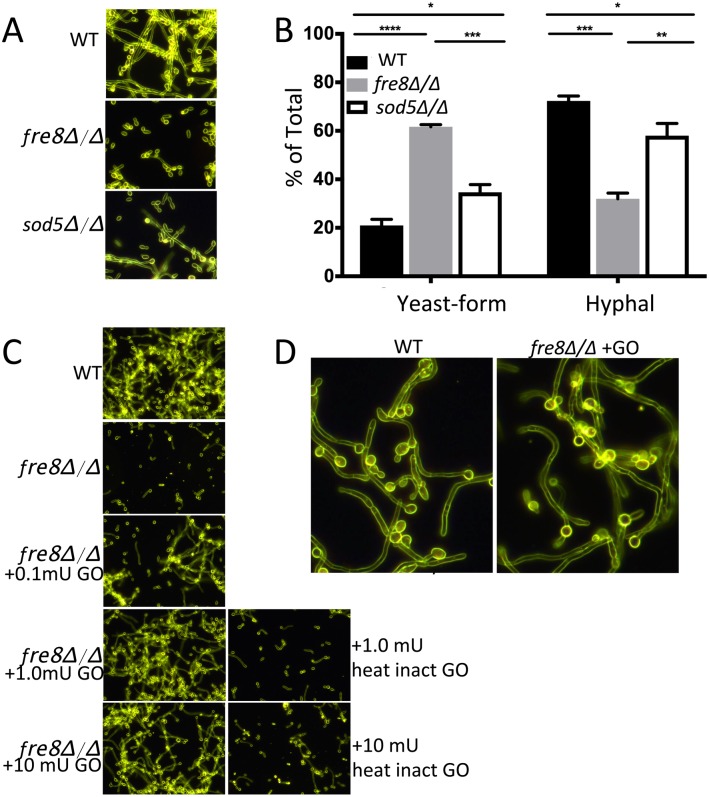
Fre8 contains all the features predicted for a member of the NADPH family of oxidoreductases, including seven transmembrane domains and sequences for binding heme, FAD and NADPH (Fig 3C). To assess the function of Fre8 in vivo, we deleted both copies of genomic FRE8 in C. albicans and tested effects on metals and ROS formation. We observed no impact of fre8Δ/Δ mutations on accumulation of copper or iron (Fig 3D) or on whole cell ferric reductase activity (Fig 3E). However, the fre8Δ/Δ strain was completely defective in generating ROS, and this result held true regardless of the stimuli for morphogenesis including serum (Fig 4A), alkaline medium, IMDM and spider medium (Fig 4B). The heterozygous fre8Δ/+ strain retaining a single genomic copy of FRE8 exhibited roughly a 50% reduction in ROS generation and the same haploinsufficiency was seen with the fre8Δ/Δ strain complemented with a single copy of FRE8 (Fig 4A). As previously mentioned (Fig 2), the ROS burst of hyphal cells is enhanced in a sod5Δ/Δ mutant, and as shown in Fig 4C, this elevated ROS is Fre8-mediated, as no ROS is detected in a double sod5Δ/Δ fre8Δ/Δ mutant. The ROS burst emitted by C. albicans during the morphogenic switch is clearly Fre8-dependent.
Fig 4. The ROS burst of C. albicans morphogenesis is eliminated by fre8Δ/Δ mutations.
The indicated strains were induced to form hyphae by either 10% serum (A,C) or by IMDM, alkaline or Spider medium (B) as described in Materials and Methods and ROS formation by luminol was monitored as in Fig 1. The fre8 heterozygous “fre8Δ/+” or fre8Δ/Δ complemented with a single copy of FRE8 (“fre8Δ/ΔRes”) exhibit haploinsufficiency with regard to ROS formation. All strains were in the background of SC5314.
We observed that FRE8 mRNA is induced within 1 hour of serum treatment (Fig 5A), congruent with the ROS burst (Fig 1A) and the induction of SOD5 (Fig 5A). In C. albicans, morphogenesis involves complex signaling pathways that converge on the Efg1 and Cph1 transcription factors, and efg1Δ/Δ cph1Δ/Δ null cells are incapable of forming hyphae [14, 42]. We observed that efg1Δ/Δ cph1Δ/Δ mutations block induction of FRE8 and SOD5 by serum (Fig 5A) and accordingly, the ROS burst is also eliminated (Fig 5B). SOD5 has previously been shown to fall under control of Efg1 [13]. SOD5 and FRE8 were not identified by ChIP as direct targets of Efg1 or Cph1 [43] [44]; both are subject to chromatin remodeling control by Hir1 that works in concert with Efg1 to control genes for morphogenesis [45].
Fig 5. C. albicans ROS production as a function of FRE8 expression.
(A,B) The indicated strains were induced to form hyphae with 10% FBS for 1 hour and were tested for (A) FRE8 and SOD5 mRNA by qRT-PCR or (B) ROS formation by luminol chemiluminescence as in Fig 1. (A) mRNA levels were compared to that of yeast-form cells prior to serum addition. Results represent the averages of 4 samples over two independent trials, error bars equal standard error. (C-E) The indicated strains engineered to express FRE8 from the MET3 promoter were grown to mid log phase in SC medium either containing methionine (+MET) to repress FRE8, or lacking methionine (-MET) to de-repress FRE8 expression for either 1 hour (Top) or the indicated time points (Bottom). Cells were examined for (Top) ROS production by luminol chemiluminescence and (Bottom) dark field microscopy for cell morphology at 40X magnification. Images were taken of cells following the indicated times of de-repressing FRE8 expression. All strains are in the background of SC5314.
Is hyphal formation required for ROS production? We uncoupled FRE8 expression from morphogenesis through ectopic expression of FRE8 under control of the repressible MET3 promoter in cph1Δ/Δ efg1Δ/Δ cells, fre8Δ/Δ cells and WT C. albicans. Strains were grown under conditions favoring yeast-only growth, and then FRE8 expression was de-repressed by removal of methionine. As seen in Fig 5C–5E there was a burst of ROS from all cells in which FRE8 expression was de-repressed. Importantly, these cells remained in the budding yeast-form, with no evidence of hyphal forms or germ tubes (Fig 5C–5E bottom). Thus the ROS burst is not dependent on morphogenesis, but rather the expression of FRE8, regardless of the morphogenic state. This study also demonstrated that Fre8-ROS is not sufficient to induce hyphal formation.
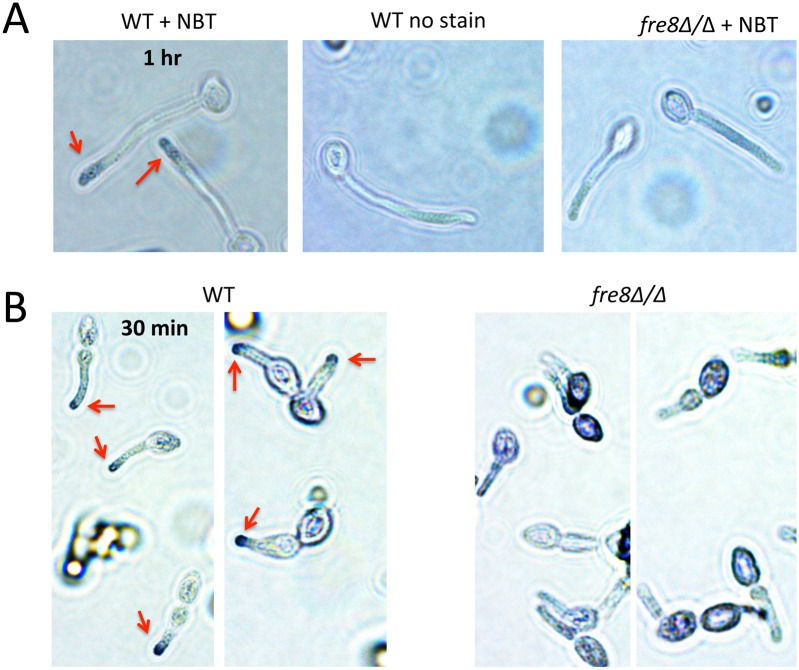
We next examined localization of Fre8-dependent ROS using nitrobluetetrazolium (NBT) which has been used to localize NOX superoxide in multicellular fungi [46, 47]. NBT is reduced by superoxide, forming a purple formazan precipitate. As seen in Fig 6, intense NBT staining was observed at the tip of elongating germ tubes in WT cells but not fre8Δ/Δ mutants (Fig 6A). This staining was discernable within 30 min of stimulating morphogenesis (Fig 6B). Thus, Fre8-dependent ROS is specific to the growing tip of the polarized cell.
Fig 6. Evidence for ROS production by Fre8 at the growing tip of C. albicans hyphae.
The designated cells were induced to form hyphae in 10% serum as in Fig 1A at a density of 4 x 106 cells/ml for either 60 (A) or 30 minutes (B). Cells were stained with nitroblue tetrazolium (NBT) as described in Materials and Methods and subjected to light microscopy at a magnification of 100X.
A role for Fre8 in hyphal development in vitro
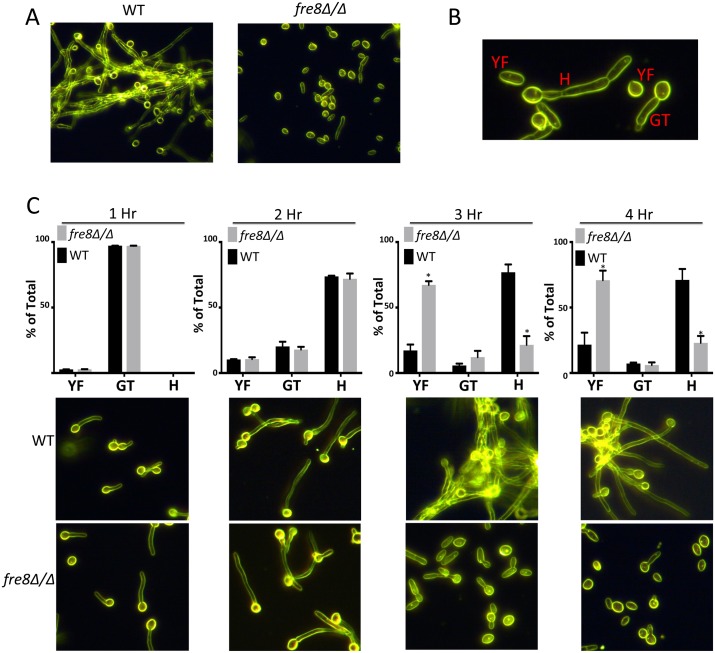
Why would C. albicans produce ROS during hyphal formation? We considered whether this ROS might act as a signal to modulate morphogenesis, as has been shown for ROS modulating development in differentiating fungi [18–20]. Although no detectable change in morphology could be noted in the NBT experiment of Fig 6, these experiments were conducted at early time points when cells were initially forming germ tubes. However, at later time points when cell formed elongated hyphae, a fre8Δ/Δ deficiency could be observed (Fig 7A). This fre8Δ/Δ defect was complemented in the FRE8 re-integrant (Fig 8A top) and appeared completely specific to later stages of hyphal development. After 3–4 hours when WT cells were assembling into elongated hyphae, fre8Δ/Δ cells accumulated abundant yeast-forms (Fig 7C).
Fig 7. Hyphal defect of fre8Δ/Δ cells.
(A) WT SC5314 or the isogeneic fre8Δ/Δ strain were seeded at 6 x 107 cells/ml and induced to form hyphae at 37°C with 10% serum. Following 4 hours (A) or the indicated time points (C), cells were photographed by dark field microscopy at 40X magnification. Shown are accurate representatives of 4–8 images. (B) Examples of the different morphological forms identified and quantified in the graphs of C, top. “Y”, yeast-form including rounded, oval or short oblong morphologies; GF, germ-tubes; H, hyphal. (C top) Roughly 350 cells for each time point were counted over two experimental trials and were classified according to morphological shapes defined in part B. The differences in yeast-form and hyphal cells in fre8Δ/Δ versus WT SC5314 cells is statistically significant (*p<0.05) by two tailed t-test.
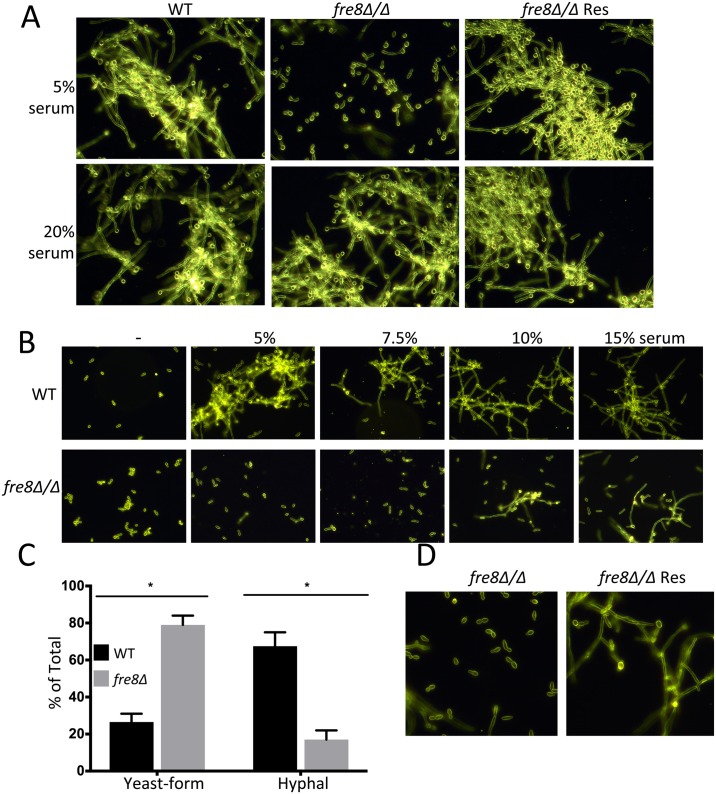
Fig 8. Impact of serum on morphogenesis involving Fre8.
WT, fre8Δ/Δ and the FRE8 re-integrant “fre8Δ/Δ Res” were seeded at either 6 x 107 cells/ml (A) or 4 x 106 cells/ml (B-D) and induced to form hyphae at 37°C or 34°C respectively with the indicated levels of serum (A,B) or with 7.5% serum (C,D). Following 4 hours, cells were photographed (A,B,D) and enumerated (C) as in Fig 7. (C) 300 to 460 WT and fre8Δ/Δ cells over two experimental trials were classified into yeast-form and hyphae as in Fig 7C. The difference between WT and fre8Δ/Δ cells is statistically significant as determined by two tailed t-test, *p = ≤0.03.
The experiments of Fig 7 employed cultures seeded at 6 x 107 cells/ml, a relatively high density where quorum sensing is prominent [48]. However, quorum sensing cannot explain the fre8Δ/Δ defect, as these mutants showed no increased sensitivity to the quorum sensing molecule farnesol (S2 Fig parts A,B), and high density cultures of fre8Δ/Δ cells exhibited WT-like quorum sensing properties [48] (S2 Fig part C and legend). Most importantly, the fre8Δ/Δ defect in hyphal development can also be seen in low density cultures not subject to quorum sensing, e.g., when 4 x 106 cells/ml are stimulated with serum at 34°C (Fig 8B–8D). It is important to note that the requirement for Fre8 in hyphal formation is not absolute, and can be bypassed by potent stimuli for morphogenesis, e.g., high levels of serum (Fig 8A bottom) or when low density cells are shifted to temperatures ≥37°C (S2 Fig Part C). We conclude that Fre8 can act as a modifier of hyphal development, but is not unconditionally essential for the process.
Since Sod5 reacts with Fre8 superoxide to produce H2O2, we tested whether sod5Δ/Δ mutations likewise affect hyphal morphogenesis. As seen in Fig 9A and 9B, sod5Δ/Δ mutants exhibited a defect in hyphal development that was less pronounced than that of fre8Δ/Δ cells. Such an intermediate effect could be expected since Fre8 superoxide may also be converted to H2O2 through spontaneous disproportionation [49] or through extracellular Sod4 and Sod6. Even so, the parallel trends seen with sod5 and fre8 mutations would imply that H2O2 (and not superoxide) underlies the Fre8-defect. To more definitely test this, we addressed whether exogenous H2O2 could bypass the requirement for Fre8 in hyphal development. Previous studies have shown that mM concentrations of H2O2 can induce hyperpolarized buds in C. albicans [50, 51], or pseudohyphae [52, 53], neither of which resemble true C. albicans hyphae. Rather than using a single bolus of H2O2 as was previously done, we used glucose oxidase (GO) to continuously generate exogenous H2O2. As seen in Fig 9C, as little as 0.1 mU of GO (generates 100 pmoles H2O2/min) was able to restore hyphal development to fre8Δ/Δ cells while heat-inactivated GO was without effect. It is important to note that the hyphae formed with GO treated fre8Δ/Δ cells were indistinguishable from that of WT cells (Fig 9D), unlike the elongated buds and pseudohyphae reported for C. albicans treated with mM H2O2 [50–53]. Together, our in vitro studies of Figs 7–9 support a model in which the H2O2 produced by Fre8 can act as a modifier of hyphal morphogenesis.
Fig 9. Evidence for a role for H2O2 in Fre8 control of morphogenesis.
The indicated strains were seeded at 6 x 107 cells/ml and induced to form hyphae for 4 hours with 5% serum. (A,B) Shown are comparisons of WT, fre8Δ/Δ and sod5Δ/Δ cells where photographs (A) are representative of 15 images over three experimental trials and quantification (B) involved classification of 250–400 cells for each of three trials. The comparisons are all statistically significant as determined by one-way ANOVA with Tukey post-test, ****p<0.0001, ***p≤0.0005, **p = 0.004, *p≤0.049. C) The effects of exogenous glucose oxidase (GO) on hyphal formation of fre8Δ/Δ cells. Shown are amounts GO added to a 1 ml culture where 1 mU GO catalyzes the production of 1 nmole H2O2/min. Where indicated, GO was subject to heat inactivation by boiling for 10 min prior to addition to cultures. Results are representative of 6–10 images over two experimental trials. D) Enlargement showing WT-like hyphal development in fre8Δ/Δ cells treated with 1 mU GO.
The role of Fre8 in rodent models of candidiasis
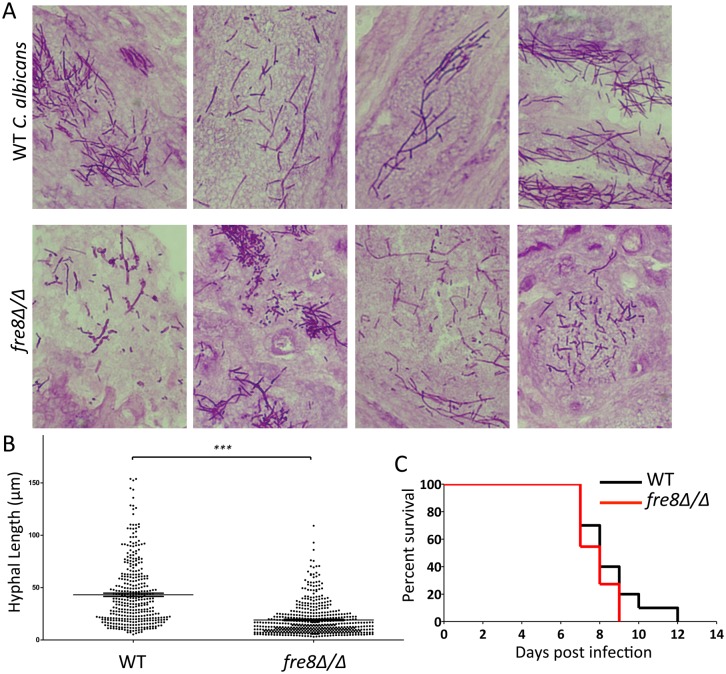
It was important to examine the impact of Fre8 in vivo, as the animal host is the only natural environment for C. albicans. One model examined was the mouse model for disseminated candidiasis where kidney is the target organ. In late stages of infection, kidneys were harvested from mice infected with WT versus fre8Δ/Δ strains and subjected to histological analysis of invading fungi by PAS staining. As seen in Fig 10A top, WT C. albicans predominantly showed elongated hyphal filaments in the infected kidney. By comparison, the fre8Δ/Δ mutant from 4 independent mice produced a mixture of morphological forms with a much larger proportion of shorter filaments or yeast form cells in fre8Δ/Δ mutants compared to WT C. albicans (Fig 10A and 10B). These findings are similar to fre8Δ/Δ effects on morphology in vitro (Figs 7–9). Yet in spite of the changes in morphology observed in vivo, there was no overall impact on pathogenesis with fre8Δ/Δ mutants. fre8Δ/Δ mutants showed no deficiency in virulence (Fig 10C), and markers of host inflammation [54, 55] were similar between SC5314 and fre8Δ/Δ infected kidneys (S3 Fig part A). Colony forming units (CFUs) in the kidney 48 hours post infection were modestly (2–3 fold) lower in fre8Δ/Δ mutations relative to WT (S3 Fig part B). All morphological forms are thought to contribute to infection and invasion [14], and our data with fre8Δ/Δ mutants supports this view.
Fig 10. Effect of fre8Δ/Δ mutations on fungal invasion of the kidney and virulence in a mouse model of disseminated candidiasis.
Mice were infected with either WT SC5314 or the isogenic fre8Δ/Δ mutant by the lateral tail vein injection as described in Materials and Methods. (A) Following 7 days of infection, kidneys were harvested from surviving mice and analyzed for fungal morphology by PAS staining as in Materials and Methods. Shown are the individual kidney sections from 4 independent mice for each group. (B) Quantification of the hyphal length from WT versus fre8Δ/Δ fungi invading the kidney. Results represent 375 and 503 fungal cells from 6 WT-infected and 4 fre8Δ/Δ-infected mice, respectively. The short oblong morphological forms that were prevalent with fre8Δ/Δ cells were included in this analysis, but not the occasional rounded yeast-forms. The difference in hyphal length between WT and fre8Δ/Δ cells is statistically significant as determined by t-test, ***p<0.0001. (C) Survival curves of infected mice including 10 mice from each group. There was no statistical difference between mice infected with WT C. albicans versus the fre8Δ/Δ mutant as determined by the log-rank (Mantel-Cox) test.
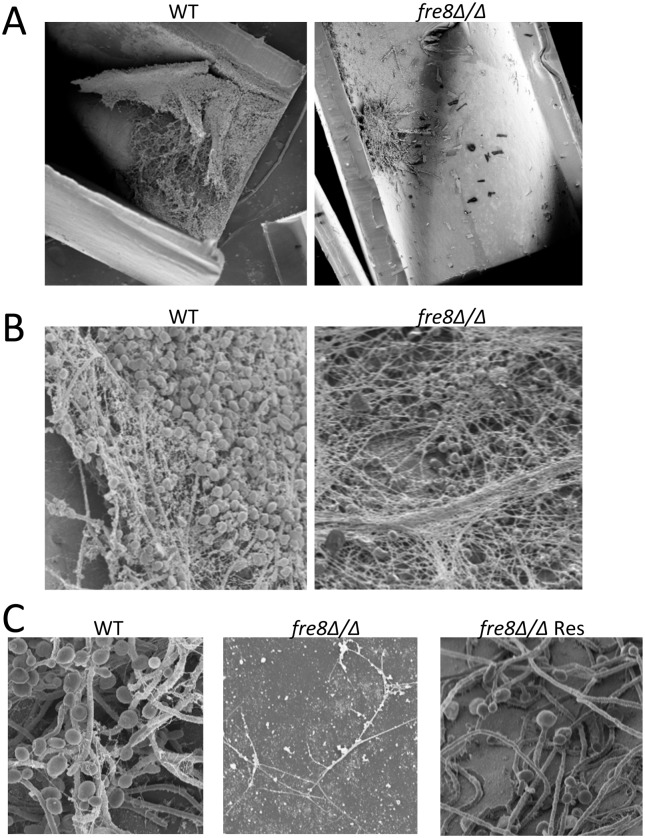
We additionally tested the fre8Δ/Δ mutant in a rodent model of catheter biofilms. C. albicans is capable of forming surface adherent aggregates of biofilms on either biological surfaces (e.g., epithelial cells) or on medical implant devices, and such dense fungal communities are highly tolerant to antifungals [56]. One of the most common clinical biofilm infections involves venous catheter implants and a rodent vascular catheter model faithfully recapitulates the human disease [56]. In this model, WT SC5314 C. albicans forms robust biofilms within 24 hours of injection into the animal catheter (Fig 11A–11C). Over three independent trials, fre8Δ/Δ cells exhibited deficiencies whereby the biofilms were either sparse in number (Fig 11A) or undetected (Fig 11C), and when present, biofilms were often attenuated with few elongated hyphae (Fig 11B). This defect was partially reversed with the Fre8 re-integrant (Fig 11C), consistent with the haploinsufficiency and partial restoration of ROS formation in vitro (Fig 4A).
Fig 11. Effect of fre8Δ/Δ mutations in a rat model of biofilm formation.
SC5314 or the isogenic fre8Δ/Δ strain or the complemented fre8Δ/Δ harboring a single FRE8 allele (“fre8Δ/Δ Res”) were tested for biofilm formation in the rat venous catheter model as described in Materials and Methods. Results in A-C are from three independent experimental trials. SEM images shown were taken at 80X (A), 1000X (B) and 2000X (C) magnification.
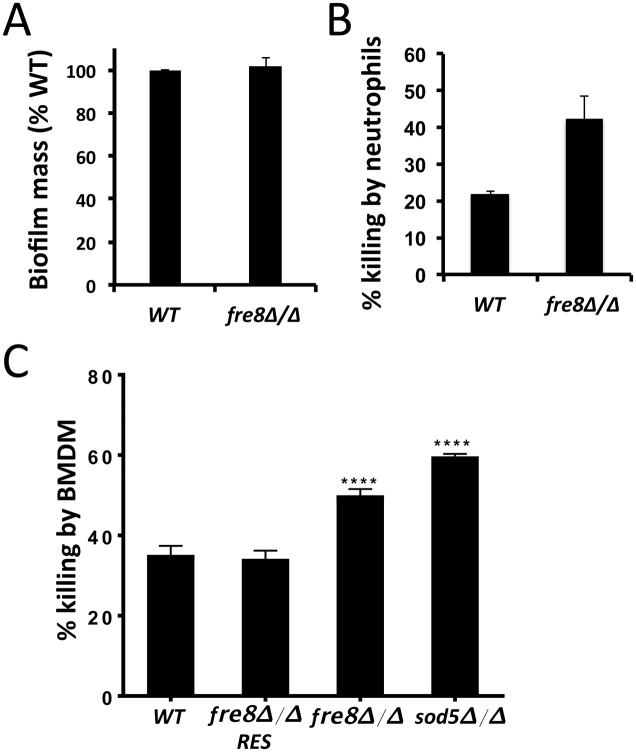
The fre8Δ/Δ defect in biofilms in vivo may very well reflect changes in morphogenesis similar to what we observed in vitro (Figs 7–9). Yet host factors may also contribute. Neutrophils represent the primary leukocytes of Candida biofilms in catheters [57] and we tested whether fre8Δ/Δ cells were more sensitive to neutrophil killing. As seen in Fig 12A, the total mass of C. albicans biofilms in vitro in the absence of any host cells was unchanged in fre8Δ/Δ cells, indicating that there is no primary defect in biofilm formation including adherence. By comparison, fre8Δ/Δ biofilms exhibited a consistent increase in killing by neutrophils (Fig 12B). Thus, the fre8Δ/Δ defect with in vivo biofilms may not reflect a deficit in biofilm formation but rather increased clearance by host immune mechanisms including neutrophils.
Fig 12. Susceptibility of fre8Δ/Δ cells to killing by neutrophils and macrophages.
(A) SC5314 WT or the isogenic fre8Δ/Δ mutant biofilms were grown for 24 hours in 96 well plates and the relative fungal burden was estimated using the XTT assay as described in Materials and Methods. Results were normalized to SC5314 biofilm allowing for averaging of 3 independent experiments. (B) 24 hour SC5314 or fre8Δ/Δ biofilms were incubated with or without human neutrophils (E:T ratio 1:2) for 4 hours and an XTT metabolic assay was used to estimate fungal burden. No biofilm controls were included to estimate neutrophil contribution to XTT assays. Neutrophil contributions were subtracted from biofilm XTT assays to calculate fungal inhibition. Results represent the averages of 3 independent experimental trials. The increase in inhibition of fre8Δ/Δ cells by neutrophils was statistically significant (P ≤ 0.04 by T-test). (C) WT SC5314 or the isogenic fre8Δ/Δ, sod5Δ/Δ or the FRE8 complemented fre8Δ/Δ (fre8Δ/Δ Res) were tested for killing by BMDM as described in Materials and Methods. Results represent the averages of 9 samples over two experimental trials. The increased killing of sod5Δ/Δ and fre8Δ/Δ cells compared to WT or to the fre8Δ/Δ Res was statistically significant ****p<0.0001 by one-way ANOVA with Tukey post-test. There was no statistically significant difference between WT and fre8Δ/Δ Res, while the increased killing in sod5Δ/Δ compared to fre8Δ/Δ was significant (p = 0.004) by one-way ANOVA with Tukey post-test.
We tested whether this increased killing was unique to neutrophils or could be extended to other phagocytes, e.g., macrophages. Our control for macrophage studies was the sod5Δ/Δ C. albicans strain that has been previously shown to be sensitive to macrophage killing due to an inability to degrade host superoxide [11]. We observe that both sod5Δ/Δ and fre8Δ/Δ mutants show statistically significant increases in killing by bone marrow derived macrophages (BMDM) (Fig 12C). The impact of Fre8 derived ROS appears to extend beyond the hyphal morphology effects seen in fungal-only cultures and interactions with host cells are also important.
Discussion
NOX enzymes have evolved to intentionally produce ROS, and until recently, were believed to be a property of multicellular differentiation [21, 22]. The discovery of Yno1 in S. cerevisiae demonstrated that a unicellular fungi can produce ROS through NOX, although the ROS in this case was found to be intracellular [23, 34]. Here we describe C. albicans Fre8 as the second example of NOX in an organism that can grow as a unicellular yeast and the first for this opportunistic fungal pathogen. Moreover, unlike S. cerevisiae Yno1, C. albicans Fre8 is capable of producing extracellular ROS, akin to NOX enzymes in multicellular organisms [1]. In animal cells, NOX enzymes can partner with extracellular SODs that convert the extracellular superoxide free radical to the diffusible H2O2 molecule [1–5]. Likewise C. albicans Fre8 appears to partner with extracellular Sod5, providing a rationale for expressing Sod5 only in hyphal cells [7]. These studies also provide a new twist to Sod5 function at the host-pathogen interface. While Sod5 can clearly react with superoxide from macrophage and neutrophil NOX enzymes [11, 12], our studies here with C. albicans Fre8 indicate that the superoxide for Sod5 is not just coming from the host. It is conceivable that when hyphal cells interact with neutrophils or macrophages, that a “superoxide superstorm” ensues with ROS coming from both the sides of the host-pathogen axis, and with Sod5 operating in the middle.
Why does C. albicans produce ROS during hyphal morphogenesis? Lessons may be taken from multicellular fungi or fruiting body fungi that use NOX derived ROS for morphogenesis and differentiation [18–20]. For example, ROS from these fungal NOX have been implicated in calcium signaling [58], MAP kinase and Rac1 GTPase signaling [46, 59] and cell re-modeling involving cytoskeleton effects [23, 60]. We show that Fre8 derived H2O2 can modulate morphogenesis. Work in other systems has shown that NOX derived H2O2 targets multiple redox sensitive molecules, including protein tyrosine phosphatases, casein kinases with peroxide sensitive degrons; even actin itself can be modulated by oxidation [1, 5, 61, 62]. Since Fre8 ROS is specifically seen at the growing tip of developing hyphae, the H2O2 produced may act locally on one or more redox sensitive targets that promote polarized growth. The precise mechanism of Fre8 control of hyphal biology is the subject of ongoing investigations.
In addition to the morphology defects of fre8Δ/Δ mutants in vitro and in vivo, we observed that these cells are more sensitive to killing by neutrophils and macrophages in vitro. It is possible that the morphological changes in fre8Δ/Δ may somehow render these fungal cells more susceptible to attack by phagocytes. As an alternative possibility, the ROS from Fre8 may help condition cells for the oxidative attack by immune cells. It has been proposed that low dose exposures of C. albicans to H2O2 or to ROS from macrophages can induce adaptive mechanisms to guard against subsequent oxidative insults [63–66]. FRE8 ROS may promote such adaptation against neutrophil and macrophage attack. In future studies, it will be important to determine the impact of fre8Δ/Δ mutations in immunocompromised settings.
Is Fre8 the only NOX of C. albicans? This organism has evolved with a very large family of 17 NOX/FRE enzymes, four of which are metalloreductases (including Frp1 characterized here), leaving 12 with unknown functions [35–39]. The extracellular ROS burst studied here is completely eliminated in fre8Δ/Δ cells suggesting that Fre8 is the only NOX for extracellular ROS in C. albicans at least under these in vitro conditions. During C. albicans invasion of the kidney, Fre8 is the most abundantly expressed member of the FRE/NOX family [40]. Even so, it is possible that other members of this family induced during fungal infection and hyphal morphogenesis such as Fre2 (S1 Fig) may similarly function in a NOX capacity, perhaps secondary to Fre8 [40]. C. albicans may also express NOX enzymes for intracellular ROS analogous to Yno1 of S. cerevisiae. With such a large family of NOX/FRE enzymes, we speculate that additional NOX enzymes will come to light as mediators of ROS signaling in C. albicans. Regardless, it will be of interest to integrate Fre8-ROS signaling into known pathways of hyphal regulation.
Materials and methods
Yeast strains and growth medium
Cultures of Candida albicans cells were typically maintained at 30°C in a yeast extract, peptone based medium (YPD) with 2% (wt/vol) glucose, conditions which support the budding yeast-form of the fungus. The C. albicans fre8Δ/Δ and sod5Δ/Δ strains used in these studies grew identical to WT SC5314 in the yeast-form (S4 Fig). To induce hyphal morphogenesis, yeast-form cells were harvested, starved for 30 min in sterile H2O at 30°C, followed by harvesting and induction of hyphal formation by incubating at 37°C (or 34°C, see below) in various media known to stimulate hyphal formation, including Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco), alkaline YPD (50 mM glycine, pH 9.5), spider medium (1% nutrient broth, 1% mannitol, 11.5 mM potassium phosphate, pH 7.2) or YPD with 5–20% fetal bovine serum (heat inactivated, Corning/Cellgro). Hyphal morphogenesis was stimulated in either “low density” (optical density, OD600 = 0.1–0.2) or “high density” (OD600 = 3.0) conditions. In studies of hyphal morphology, yeast-form cells were cultured to OD600 ≈ 8.0, conditions where all cells grew identically (S4 Fig), and were stimulated to form hyphae with YPD-serum. The level of serum used to investigate the fre8Δ/Δ defect ranged from 5–15% depending on the lot of serum, with low density cultures typically requiring less serum and temperatures of 34°C to demonstrate a dependence on serum for hyphal morphogenesis. Where indicated, cultures were supplemented with 0.1–10 mUnits glucose oxidase (Type II Sigma#G6125) to bypass the fre8Δ/Δ defect in hyphal development. Experiments involving yeast-form cells expressing FRE8 under the MET3 promoter used a synthetic complete (SC) based medium containing 0.67% yeast nitrogen base lacking cysteine and either containing or lacking 85.6 mg/L methionine. Cell were seeded at OD600 = 0.1 and grown for 1–7 hours.
All C. albicans strains used in this study were isogenic to SC5314 or its derivative CA-IF100 (arg4Δ/arg4Δ, leu2Δ/leu2Δ::cmLEU2, his1Δ/his1Δ::cdHIS1, URA3/ura3Δ). The sod1Δ/Δ, sod5Δ/Δ and sod4Δ/Δ sod5Δ/Δ sod6Δ/Δ strains derived from CA-IF100 were kind gifts of Karl Kuchler as previously described [11]. The cph1Δ/Δ efg1Δ/Δ strain derived from SC5314 was a gift from Gerald Fink [42]. Mutations in FRE8 and SOD5 were introduced in SC5314 using the SAT1-flipper cassette method [67]. Deletion in a single FRE8 allele was achieved using plasmid pJGFRE8LKO, in which FRE8 regions -926 to -581 and +2402 to +2802 were inserted into the Kpn1 and XhoI and the NotI and SacI sites respectively of pSFS2 [67]. Following liberation of the cassette by KpnI and SacI digestion and transformation of SC5314 by electroporation, accurate deletion of a single FRE8 allele was verified by PCR, generating the fre8Δ/+ mutant strain CA-JG201. The second FRE8 allele was deleted similarly using a pSFS2 construct, pJGFRE8SKO, containing FRE8 sequences -587 to -3 and +2075 to +2427, creating the fre8Δ/fre8Δ strain CA-JG211. Homozygous sod5Δ/Δ mutations were introduced in either SC5314 (generating strain CA-JG201) or CA-JG211 (generating CA-JG221) using a construct containing SOD5–492 to +53 and +808 to +1253 inserted into the Kpn1 and XhoI and the Not I and SacI sites respectively of pSFS2 [67]. Deletion of both copies of sod5Δ/Δ in strain CA-JG201 and CAJG-221 was verified by PCR. A single copy of FRE8 was introduced into the fre8Δ/Δ strain CA-JG211 as follows: FRE8 sequences -926–+2802 were inserted into the KpnI and XhoI sites of pJGFRE8LKO. Integration into the FRE8 locus at position -926 to +2802 was achieved by transformation of the cassette liberated by digestion with KpnI and SacI generating the fre8Δ/Δ:FRE8 re-integrant strain CA-JG231. To create the construct for expressing FRE8 under control of the MET3 repressible promoter, C. albicans MET3 sequences -1643 to -1 were inserted into Sph1 and Nhe1 sites engineered at FRE8 position -1 in the pJGFRE8LKO re-integrant plasmid described above. Following digestion with Kpn1 and Sac1, the MET3-FRE8 containing cassette was used to transform the cph1Δ/Δ efg1Δ/Δ strain, the fre8Δ/Δ strain CA-JG211 and SC5314 by electroporation. Accurate integration at the FRE8 locus -926 to +2802 was verified by PCR.
Expression of recombinant FRE8 and FRP1 in Pichia pastoris used the PichiaPink Strain 1: ade2 (Thermo Fisher Scientific). P. pastoris cells were maintained in YP-Gal medium (1% yeast extract, 2% peptone, 2% galactose). Protein expression experiments used a buffered YP medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base, 0.00004% biotin) that was supplemented with either 0.5% methanol to induce protein expression under the AOX2 promoter or with 1% glycerol for non-inducing conditions. The plasmid for expressing FRE8 or FRP1 under the P. pastoris AOX2 promoter represented a modified version of pPINK α-HC (Thermo Fisher Scientific) in which a 10X HIS tag was introduced downstream of the α-factor pre-sequence (plasmid pRPp718, kind gift of Ryan Peterson). Following the insertion of a Afe1 site downstream of the HIS tag, FRP1 sequences +1 to +1665 and FRE8 +1 to +2220 were inserted into the Afe1 and Fse1 sites of this expression plasmid, creating in-frame fusions to the N-terminus secretion sequence and HIS tag. The CTG codons from both genes were altered to TCG for optimal expression in P. pastoris; plasmids were linearized by digestion with Spe1 and integrated into the TRP2 locus by transformation.
Biochemical assays
For luminol and lucigenin measurements of ROS, either yeast-form cells (grown in YPD to OD600 of 1.0–2.0) or cells induced to form hyphae as described above were used. Cells were harvested, washed, and suspended in a final OD600 of 0.2 in Hanks buffered saline solution (HBSS) containing 0.2 mM luminol (Cayman chemicals) and 0.5 units/ml horseradish peroxidase. In studies with lucigenin, cells were first washed in 25 mM glycine pH 9.5, 0.5% glucose prior to resuspending in 200 μl of the same alkaline buffer containing 5 μM lucigenin. Samples were analyzed for luminol or lucigenin chemiluminescence in 96 well plates using a BioTek Synergy HT plate reader. Analysis was carried out over 1.5 hours at 37°C with a gain setting at 120–135 and integration time of 1.0 second. Results were plotted according to relative luminescence units (RLU) per 0.04 OD600 units of cells. With experiments involving DPI, 5 μl of DMSO containing the indicated amount of DPI (or no DPI as control) was added to the reaction at time zero.
For qRT-PCR analysis of fungal-only cultures, 50 ml cultures of cells were induced to form hyphae for 1 hr by growth in YPD-10% FBS (as described above); these early hyphal cells or the control yeast-form were harvested, washed and RNA prepared by the hot acid phenol method [68]. cDNA was prepared using the Maxima H Minus First Strand cDNA Synthesis Kit (ThermoFisher Scientific) and qRT PCR carried out using iTaq Universal SYBR Green Supermix (Bio-Rad). Values were normalized to TUB2 and graphed according to the fold change in FRE8 and SOD5 expression in early hyphal versus yeast-form cells. Amplicons of ≈150 residues were prepared using primers as described in S1 Table.
For analysis of inflammatory mRNA markers (TNF-α, IL-17a, IL-6), RNA from whole kidneys was extracted as previously described [69]. cDNA prepared from 5.0 μg of RNA was diluted 1:50 prior to PCR analysis as above. Values were normalized to ActB and graphed according to the fold change over uninfected controls. Primers for host mRNA analyses are listed in S1 Table.
For ferric reductase and NOX activity analyses in P. pastori transformants, cells were grown overnight in 10 ml YP-Gal, washed twice in either glycerol or methanol containing buffered media (described above) and resuspended at an OD600 of 0.1 in 15 mls of the same medium. Following growth at 30°C for 6 hrs, cells were harvested and washed in either HBSS for the luminol assay or 50 mM citrate, pH 6.6, 5% glucose for the ferric reductase assay. Cells were subjected to luminol chemilumiscence precisely as described above for C. albicans. Compared to C. albicans assays, the luminol substrate appears rapidly depleted in P. pastoris expressing high levels of FRE8. For the ferric reductase assay, cells at a OD600 of 0.5 were incubated in 200 μl of a reaction containing 1 mM FeCl3 and 1 mM bathophenanthrolinedisulfonic acid (BPS) in 50 mM citrate, pH 6.6, 5% glucose. Absorbance at 515 or 520 nm was read in 96 well plates on a BioTek Synergy HT plate reader over 1.5 hours at 30°C. Where designated, 0.1 U of bovine Cu/Zn SOD1 or 50 nM DPI were added to the luminol or ferric reductase assay at t = 0. Ferric reductase measurements in C. albicans cells was conducted similarly, using cells induced to form hyphae in IMDM for 1 hr as described above and assayed for ferric reductase using the same conditions described for P. pastoris except C. albicans cells were assayed at OD600 of 0.1.
Total cellular accumulation of copper and iron was measured by inductively coupled plasma mass spectrometry (ICP-MS) using C. albicans cells induced to form hyphae for 1 hr in 10% FBS as described above. Cells were washed twice with 10 mM Tris, 1 mM EDTA, pH 8 and twice with MiliQ deionized water. Cell pellets containing 10.0 OD600 units of cells were resuspended in 500 μl of 20% nitric acid and digested by incubation at 90°C overnight. Samples were diluted 10-fold in MiliQ deionized water and subjected to elemental analysis on a Agilent 7700x ICP-MS instrument.
In vitro biofilm model and fungal killing by neutrophils and macrophages
In vitro biofilms were grown in the wells of 96-well microtiter plates, as previously described [70]. Briefly, C. albicans resuspended in RPMI-MOPS at 1.5 x 106 cells/ml (200μL/well) was added, and incubated for 24 hours at 37°C with 5% CO2. To assess biofilm burden an XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide) assay was performed as an estimate of viable burden, as previously described [70].
For assays involving neutrophils, human neutrophils were collected as follows: Blood was obtained from volunteer donors with written informed consent through a protocol approved by the University of Wisconsin Internal Review Board (IRB). Primary human neutrophils were purified by negative antibody selection using the MACSxpress Neutrophil Isolation and MACSxpress Erythrocyte Depletion kits (Miltenyi Biotec Inc., Auburn, CA), as previously described [71]. Experiments with neutrophils were performed in RPMI 1640 (without phenol red) supplemented with 2% heat-inactivated fetal bovine serum (FBS) and glutamine (0.3 mg/ml). Incubations were at 37°C with 5% CO2. An adaptation of the XTT metabolic assay was used to estimate C. albicans viability following co-culture with neutrophils [71]. Following a 24 h incubation period, biofilms were washed with DPBS and neutrophils were added at 1.5 x 106 cells/ml, which represented an effector:target of 1:2. Following a 4 h incubation, 90 μL of 9:1 XTT working solution (0.75 mg/ml XTT in DPBS with 2% glucose: phenazine methosulfate 0.32 mg/ml in ddH2O) was added to each well. After a 25 minutes incubation, samples were transferred to a Falcon 96 well U bottom plate and centrifuged at 1,200×g for three minutes to pellet cells. Supernatants (110 μl) were then transferred to a 96 well flat bottom plate for absorption reading at 492 nm. A neutrophil only control was used to subtract their contribution to the XTT values. To determine percent killing, values were compared to wells without neutrophils after subtraction of the baseline absorbance.
Macrophage infection assays used bone-marrow derived macrophages (BMDM) isolated from the marrow of hind leg bones of 5- to 8-wk-old C57BL-6 female mice. For differentiation, cells were seeded in 100 mm treated cell culture dishes (Corning, Corning, NY) in Dulbecco’s Modified Eagle medium (DMEM; Corning) with 20% L-929 cell-conditioned medium, 10% FBS (Atlanta Biologicals, Flowery Branch, GA), 2mM Glutamax (Gibco, Gaithersburg MD), 1% nonessential amino acids (Cellgro, Manassas, VA), 1% HEPES buffer, 1% penicillin-streptomycin and 0.1% 2-mercaptoethanol for 6–7 days at 37°C with 9.5% CO2. 105 BMDM were seeded on 96 well plates and activated by incubating overnight using 100 U/ml of IFN-γ (Roche, Indianapolis, IN). C. albicans obtained from overnight cultures in YPD (OD600 = 8.0) and starved in water as for hyphal morphogenesis studies (see above) were washed twice with PBS and incubated for 30 min at 37°C with Guinea pig complement (MP biomedicals, LLC, OH) for opsonization. The fungus was then added to macrophages at a multiplicity of infection (MOI) ratio of 1:10 for 4 hours. After incubation, the media was removed and macrophages lysed in water. Fungal viability was assessed by the XTT assay according to Pierce et al [72]. The same XTT assay was used to determined fungal viability following farnesol treatment.
Rodent infection studies
For the murine model of disseminated candidiasis, ten male BALB/c mice (10 weeks old) per strain were inoculated with 2x105 C. albicans cells of WT SC5314, the fre8Δ/Δ strain or the fre8Δ/Δ strain complemented by FRE8 by lateral tail vein injection. Moribund mice were sacrificed by CO2 asphyxiation and immediately dissected for harvesting kidneys for histology (see below). Fungal burden and host inflammatory markers were analyzed following 48 hours of infection. The spleen and one kidney was processed for CFUs as previously described [69]. The other kidney was placed in 500 μL Trizol and frozen at -80°C for subsequent RNA analyses (see above). Mouse survival was plotted using a log rank test (Mantel Cox) to query any statistical difference.
A jugular vein rat central venous catheter biofilm infection model was used as previously described [73]. Briefly, 24 h following surgical implantation of a jugular venous catheter, C. albicans at 106 cells/ml was instilled in the catheter lumen and flushed after 6 h. After 24 h biofilm growth period, catheters were harvested and fixed overnight (4% formaldehyde, 1% glutaraldehyde, in PBS). They were then washed with PBS, treated with 1% osmium tetroxide, and washed again. Samples were dehydrated through series of ethanol washes followed by critical point drying and mounted on aluminum stubs. Following sputter coating with platinum, samples were imaged in a scanning electron microscope (LEO 1530) at 3kV.
Microscopic visualization of C. albicans in vitro and in infected kidneys
For NBT staining and microscopic analyses of cell morphology, C. albicans cells were induced to form hyphae as described above using YPD containing 10% FBS (in the case of NBT staining). Cells were harvested, washed once with HBSS and resuspended in 1 ml HBSS containing 0.05% nitroblue tetrazolium (NBT). Following an incubation for 30 min in the dark at 37°C, cells were washed 1X with HBSS, 1X with 70% Ethanol and resuspended in 200 μl 50% Glycerol/HBSS. Cells were visualized by light microscopy at 100x magnification on a Zeiss Axio ImagerA2 microscope. For analysis of hyphal morphogenesis, dark field microscopy of live C. albicans cells was accomplished using a Nikon Infinity 1 microscope at 40x magnification. Where indicated, enumeration of cells was carried with culture aliquots first passed through 26 gauge and 31 gauge needles to help break up dense aggregates and enhance visualization of individual cells. Passage through these needles did not affect integrity of the individual cells.
To analyze C. albicans morphology in infected kidneys, freshly harvested kidneys from infected mice were flash frozen in Tissue Tek O.C.T. compound in a dry ice/ethanol bath and were sectioned to 20 μM thickness by cryotome. Tissue slices were adhered to Superfrost Plus Microscope Slides (Fisherbrand Cat. No. 12-550-15) and subjected to Periodic Acid Schiff (PAS) staining by treatment with 0.5% periodic acid (Sigma) for 5 minutes, rinsing briefly with distilled water, then staining 5 minutes with Schiff’s Reagent (Sigma Aldrich). Following a 5 min rinse with water, the mounted tissue was dehydrated using successive 2 min treatments with 50%, 70%, 80%, and twice 95% and 100% ethanol, followed by three 2 min treatment with xylene isomer mixture (Sigma Aldrich) to remove residual ethanol. Cover slips were then mounted with Permount (Fisher) and slides then imaged on a microscope at 40X magnification.
Ethics statement
All experiments involving animals were approved by the Johns Hopkins University (protocols # MO16M168 and MO15H134) and University of Wisconsin (protocol # DA0031, MV1947) Institutional Animal Care and Use Committees according to guidelines established by the Animal Welfare Act, The Institute of Laboratory Animal Resources Guide for the Care and Use of Laboratory Animals, and the Public Health Service Policy. Experiments involving neutrophils were approved by IRB (protocol #2013 1758) and involved cells isolated from healthy human adult donors in which written informed consent was obtained at the time of blood draw, following the guidelines and approval of the University of Wisconsin-Madison Center for Health Sciences Human Subjects Committee.
Supporting information
(XLSX)
Expression of the various members of the FRE family listed by ORF designation were examined by qRT-PCR as described in Materials and Methods. Shown is the fold change in expression after 1 hour stimulation of hyphal morphogenesis by IMDM compared to yeast-form cells (cultured as in Fig 1). Results represent the averages of triplicate cultures. Genes that were previously characterized as cupric or ferric metalloreductases [37–39] or genes induced during fungal invasion of the kidney [40] are indicated by + marks. The C. albicans orthologue to S. cerevisiae Yno1 [18] is indicated.
(TIF)
(A,B) WT SC5314 and isogenic fre8Δ/Δ cells were induced to form hyphae by culturing cells seeded at 4 x 106 cells/ml at 37°C with 5% serum in the presence of the indicated levels of farnesol or methanol vehicle. Following four hours, cells were either (A) photographed or (B) assayed for viability by XTT as described in Materials and Methods where results represent the averages of biological triplicates. The decrease in cell mass/viability with 300 and 400 μM farnesol is statistically significant as determined by ANOVA with Tukey post-test; ****p<0.0001. (C) SC5314 cells seeded at 4 x 106 cells/ml (low density) were cultured for four hours at 37°C in YPD supplemented with or without 5% serum or with conditioned 5% serum media derived from high density WT or fre8Δ/Δ cultures (6 x 107 cells/ml). The conditioned medium was obtained by removing cells from the high density cultures through centrifugation. Results show that low density SC5314 forms hyphae at 37°C even in the absence of serum (-serum), but hyphal formation is blocked by conditioned medium from high density WT and fre8Δ/Δ cultures, indicative of quorum sensing [48]. Photographs are representative of 5–10 images over 2 experimental trials.
(TIF)
Mice were infected with either C. albicans WT SC5314 or the isogenic fre8Δ/Δ or the FRE8 complemented fre8Δ/Δ (fre8Δ/Δ Res) strain by lateral tail vein injection. Following 48 hours of infection, kidney and spleen were harvested and examined for (A) RNA markers of inflammation in the kidney by qRT-PCR as described in Materials and Methods, and (B) CFUs. Results are from 7–8 mice from each group. (A) The mRNA levels of the indicated inflammatory markers is shown as a fold change over uninfected controls. In all three infected strains, the increases in TNF- α, IL-6 and IL-17A are statistically significant compared to uninfected controls (****p<0.0001; ***p<0.0007). There is no statistically significant difference between WT and fre8Δ/Δ for any samples as determined by a one-way ANOVA with a Tukey post-test. There was a small (<2 fold) increase in expression of IL-17 and IL6 in the fre8Δ/Δ RES compared to WT, but the significance of this small variation is uncertain. (B) CFUs are shown as a function of tissue wet weight. The difference between fre8Δ/Δ and the fre8Δ/Δ strain complemented with FRE8 (fre8Δ/Δ RES) is significant (*p = 0.039). There is no statistically significant difference in CFUs obtained from spleen.
(TIF)
The indicated yeast strains were seeded at OD600 = 0.001 and grown at 30°C in YPD where growth by OD600 was either monitored continuously (TOP) or following a 16 hour period (BOTTOM). Results represent the averages of triplicate cultures (TOP) or of two to five experimental trials of hyphal morphogenesis (BOTTOM).
(TIF)
Acknowledgments
We thank Dr. Angelique Besold for informative discussions and for assistance with statistical analyses. We are also grateful to Dr. Ryan Peterson for plasmids.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institutes of Health Grants RO1 GM 50016 (to VCC), RO1 AI 119949 (to VCC), RO1 AI 073289 (to DRA), RO1 HL 059842 (to CC), KO8 AI108727 (to JEN), and F31 DK111114 (to EMC). Funding was also provided by a grant from Burroughs Wellcome Fund 1012299 (to JEN). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283(25):16961–5. Epub 2008/04/19. doi: 10.1074/jbc.R700045200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer G. Targeting Extracellular ROS Signaling of Tumor Cells. Anticancer Res. 2014;34(4):1467–82. [PubMed] [Google Scholar]
- 3.Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med. 2003;35(3):236–56. Epub 2003/07/30 . [DOI] [PubMed] [Google Scholar]
- 4.Sudhahar V, Urao N, Oshikawa J, McKinney RD, Llanos RM, Mercer JF, et al. Copper transporter ATP7A protects against endothelial dysfunction in type 1 diabetic mice by regulating extracellular superoxide dismutase. Diabetes. 2013;62(11):3839–50. Epub 2013/07/26. doi: 10.2337/db12-1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oshikawa J, Urao N, Kim HW, Kaplan N, Razvi M, McKinney R, et al. Extracellular SOD-derived H2O2 promotes VEGF signaling in caveolae/lipid rafts and post-ischemic angiogenesis in mice. PLoS One. 2010;5(4):e10189 Epub 2010/04/28. doi: 10.1371/journal.pone.0010189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broxton CN, Culotta VC. SOD Enzymes and Microbial Pathogens: Surviving the Oxidative Storm of Infection. PLoS Pathog. 2016;12(1):e1005295 Epub 2016/01/08. doi: 10.1371/journal.ppat.1005295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martchenko M, Alarco AM, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol Biol Cell. 2004;15(2):456–67. doi: 10.1091/mbc.E03-03-0179 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, et al. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56(2):397–415. doi: 10.1111/j.1365-2958.2005.04557.x . [DOI] [PubMed] [Google Scholar]
- 9.Gleason JE, Galaleldeen A, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, et al. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc Natl Acad Sci U S A. 2014;111(16):5866–71. Epub 2014/04/09. doi: 10.1073/pnas.1400137111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson RL, Galaleldeen A, Villarreal J, Taylor AB, Cabelli DE, Hart PJ, et al. The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases. Journal of Biological Chemistry. 2016;291(40):20911–23. doi: 10.1074/jbc.M116.748251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol. 2009;71(1):240–52. doi: 10.1111/j.1365-2958.2008.06528.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miramon P, Dunker C, Windecker H, Bohovych IM, Brown AJ, Kurzai O, et al. Cellular Responses of Candida albicans to Phagocytosis and the Extracellular Activities of Neutrophils Are Critical to Counteract Carbohydrate Starvation, Oxidative and Nitrosative Stress. PLoS One. 2012;7(12):e52850 Epub 2013/01/04. doi: 10.1371/journal.pone.0052850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierce JV, Dignard D, Whiteway M, Kumamoto CA. Normal Adaptation of Candida albicans to the Murine Gastrointestinal Tract Requires Efg1p-Dependent Regulation of Metabolic and Host Defense Genes. Eukaryot Cell. 2013;12(1):37–49. Epub 2012/11/06. doi: 10.1128/EC.00236-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9(10):737–48. Epub 2011/08/17. doi: 10.1038/nrmicro2636 . [DOI] [PubMed] [Google Scholar]
- 15.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119–28. doi: 10.4161/viru.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2(5):1053–60. doi: 10.1128/EC.2.5.1053-1060.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman J, Sudbery PE. Candida Albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3(12):918–30. doi: 10.1038/nrg948 . [DOI] [PubMed] [Google Scholar]
- 18.Breitenbach M, Weber M, Rinnerthaler M, Karl T, Breitenbach-Koller L. Oxidative stress in fungi: its function in signal transduction, interaction with plant hosts, and lignocellulose degradation. Biomolecules. 2015;5(2):318–42. Epub 2015/04/10. doi: 10.3390/biom5020318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takemoto D, Kamakura S, Saikia S, Becker Y, Wrenn R, Tanaka A, et al. Polarity proteins Bem1 and Cdc24 are components of the filamentous fungal NADPH oxidase complex. Proc Natl Acad Sci U S A. 2011;108(7):2861–6. Epub 2011/02/02. doi: 10.1073/pnas.1017309108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heller J, Tudzynski P. Reactive Oxygen Species in Phytopathogenic Fungi: Signaling, Development, and Disease. Annu Rev Phytopathol. 2011;49:369–90. doi: 10.1146/annurev-phyto-072910-095355 [DOI] [PubMed] [Google Scholar]
- 21.Aguirre J, Rios-Momberg M, Hewitt D, Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13(3):111–8. Epub 2005/03/02. doi: 10.1016/j.tim.2005.01.007 . [DOI] [PubMed] [Google Scholar]
- 22.Lalucque H, Silar P. NADPH oxidase: an enzyme for multicellularity? Trends Microbiol. 2003;11(1):9–12. Epub 2003/01/16. . [DOI] [PubMed] [Google Scholar]
- 23.Rinnerthaler M, Buttner S, Laun P, Heeren G, Felder TK, Klinger H, et al. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc Natl Acad Sci U S A. 2012;109(22):8658–63. Epub 2012/05/16. doi: 10.1073/pnas.1201629109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroter C, Hipler UC, Wilmer A, Kunkel W, Wollina U. Generation of reactive oxygen species by Candida albicans in relation to morphogenesis. Arch Dermatol Res. 2000;292(5):260–4. Epub 2000/06/27. . [DOI] [PubMed] [Google Scholar]
- 25.Maghzal GJ, Krause KH, Stocker R, Jaquet V. Detection of reactive oxygen species derived from the family of NOX NADPH oxidases. Free Radic Biol Med. 2012;53(10):1903–18. Epub 2012/09/18. doi: 10.1016/j.freeradbiomed.2012.09.002 . [DOI] [PubMed] [Google Scholar]
- 26.Dahlgren C, Karlsson A. Respiratory burst in human neutrophils. J Immunol Methods. 1999;232(1–2):3–14. Epub 2000/01/05. . [DOI] [PubMed] [Google Scholar]
- 27.Maidan MM, Thevelein JM, Van Dijck P. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem Soc Trans. 2005;33(Pt 1):291–3. Epub 2005/01/26. doi: 10.1042/BST0330291 . [DOI] [PubMed] [Google Scholar]
- 28.Muller-Peddinghaus R. In vitro determination of phagocyte activity by luminol- and lucigenin-amplified chemiluminescence. Int J Immunopharmacol. 1984;6(5):455–66. Epub 1984/01/01. . [DOI] [PubMed] [Google Scholar]
- 29.Gianni D, Nicolas N, Zhang H, Der Mardirossian C, Kister J, Martinez L, et al. Optimization and Characterization of an Inhibitor for NADPH Oxidase 1 (NOX-1). 2010. Epub 2012/07/27. NBK98925 [bookaccession]. . [PubMed] [Google Scholar]
- 30.Lee NK, Choi YG, Baik JY, Han SY, Jeong DW, Bae YS, et al. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood. 2005;106(3):852–9. Epub 2005/04/09. doi: 10.1182/blood-2004-09-3662 . [DOI] [PubMed] [Google Scholar]
- 31.Egan MJ, Wang ZY, Jones MA, Smirnoff N, Talbot NJ. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci U S A. 2007;104(28):11772–7. Epub 2007/06/30. doi: 10.1073/pnas.0700574104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulton P, Martin H, Ainger A, Cross A, Hoare C, Doel J, et al. The inhibition of flavoproteins by phenoxaiodonium, a new iodonium analogue. Eur J Pharmacol. 2000;401(2):115–20. Epub 2000/08/05. . [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Krause KH, Xenarios I, Soldati T, Boeckmann B. Evolution of the ferric reductase domain (FRD) superfamily: modularity, functional diversification, and signature motifs. PLoS One. 2013;8(3):e58126 Epub 2013/03/19. doi: 10.1371/journal.pone.0058126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Detry N, Choi J, Kuo HC, Asiegbu FO, Lee YH. In silico sequence analysis reveals new characteristics of fungal NADPH oxidase genes. Mycobiology. 2014;42(3):241–8. Epub 2014/10/28. doi: 10.5941/MYCO.2014.42.3.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baek YU, Li M, Davis DA. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell. 2008;7(7):1168–79. Epub 2008/05/27. doi: 10.1128/EC.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almeida RS, Wilson D, Hube B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009;9(7):1000–12. Epub 2009/10/01. doi: 10.1111/j.1567-1364.2009.00570.x . [DOI] [PubMed] [Google Scholar]
- 37.Hammacott JE, Williams PH, Cashmore AM. Candida albicans CFL1 encodes a functional ferric reductase activity that can rescue a Saccharomyces cerevisiae fre1 mutant. Microbiology. 2000;146 (Pt 4):869–76. Epub 2000/04/28. doi: 10.1099/00221287-146-4-869 . [DOI] [PubMed] [Google Scholar]
- 38.Knight SA, Lesuisse E, Stearman R, Klausner RD, Dancis A. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology. 2002;148(Pt 1):29–40. Epub 2002/01/10. doi: 10.1099/00221287-148-1-29 . [DOI] [PubMed] [Google Scholar]
- 39.Jeeves RE, Mason RP, Woodacre A, Cashmore AM. Ferric reductase genes involved in high-affinity iron uptake are differentially regulated in yeast and hyphae of Candida albicans. Yeast. 2011;28(9):629–44. Epub 2011/08/09. doi: 10.1002/yea.1892 . [DOI] [PubMed] [Google Scholar]
- 40.Amorim-Vaz S, Tran VDT, Pradervand S, Pagni M, Coste AT, Sanglard D. RNA Enrichment Method for Quantitative Transcriptional Analysis of Pathogens In Vivo Applied to the Fungus Candida albicans. MBio. 2015;6(5). ARTN e00942-15 doi: 10.1128/mBio.00942-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostuni MA, Lamanuzzi LB, Bizouarn T, Dagher MC, Baciou L. Expression of functional mammal flavocytochrome b(558) in yeast: comparison with improved insect cell system. Biochim Biophys Acta. 2010;1798(6):1179–88. Epub 2010/02/23. doi: 10.1016/j.bbamem.2010.02.016 . [DOI] [PubMed] [Google Scholar]
- 42.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–49. Epub 1997/09/23. . [DOI] [PubMed] [Google Scholar]
- 43.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148(1–2):126–38. Epub 2012/01/24. doi: 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317(5839):815–9. doi: 10.1126/science.1140748 . [DOI] [PubMed] [Google Scholar]
- 45.Jenull S, Tscherner M, Gulati M, Nobile CJ, Chauhan N, Kuchler K. The Candida albicans HIR histone chaperone regulates the yeast-to-hyphae transition by controlling the sensitivity to morphogenesis signals. Sci Rep. 2017;7(1):8308 Epub 2017/08/18. doi: 10.1038/s41598-017-08239-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chi MH, Craven KD. RacA-Mediated ROS Signaling Is Required for Polarized Cell Differentiation in Conidiogenesis of Aspergillus fumigatus. PLoS One. 2016;11(2):e0149548 Epub 2016/02/20. doi: 10.1371/journal.pone.0149548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin S, Gao Z, Wang C, Huang L, Kang Z, Zhang H. Nitric Oxide and Reactive Oxygen Species Coordinately Regulate the Germination of Puccinia striiformis f. sp. tritici Urediniospores. Front Microbiol. 2016;7:178 Epub 2016/03/05. doi: 10.3389/fmicb.2016.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67(7):2982–92. doi: 10.1128/AEM.67.7.2982-2992.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller AF, Teixeira M, et al. Superoxide dismutases and superoxide reductases. Chem Rev. 2014;114(7):3854–918. Epub 2014/04/02. doi: 10.1021/cr4005296 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.da Silva Dantas A, Patterson MJ, Smith DA, Maccallum DM, Erwig LP, Morgan BA, et al. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol Cell Biol. 2010;30(19):4550–63. Epub 2010/08/04. doi: 10.1128/MCB.00313-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dantas Ada S, Day A, Ikeh M, Kos I, Achan B, Quinn J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules. 2015;5(1):142–65. Epub 2015/02/28. doi: 10.3390/biom5010142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasa K, Kim J, Yee S, Kim W, Choi W. A MAP kinase pathway is implicated in the pseudohyphal induction by hydrogen peroxide in Candica albicans. Mol Cells. 2012;33(2):183–93. Epub 2012/02/24. doi: 10.1007/s10059-012-2244-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasution O, Srinivasa K, Kim M, Kim YJ, Kim W, Jeong W, et al. Hydrogen peroxide induces hyphal differentiation in Candida albicans. Eukaryot Cell. 2008;7(11):2008–11. Epub 2008/09/16. doi: 10.1128/EC.00105-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu W, Solis NV, Ehrlich RL, Woolford CA, Filler SG, Mitchell AP. Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol. 2015;13(2):e1002076 Epub 2015/02/19. doi: 10.1371/journal.pbio.1002076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacCallum DM. Massive induction of innate immune response to Candida albicans in the kidney in a murine intravenous challenge model. FEMS Yeast Res. 2009;9(7):1111–22. Epub 2009/10/22. doi: 10.1111/j.1567-1364.2009.00576.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nett JE, Andes DR. Fungal Biofilms: In Vivo Models for Discovery of Anti-Biofilm Drugs. Microbiol Spectr. 2015;3(3). doi: 10.1128/microbiolspec.MB-0008-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nett JE, Zarnowski R, Cabezas-Olcoz J, Brooks EG, Bernhardt J, Marchillo K, et al. Host Contributions to Construction of Three Device-Associated Candida albicans Biofilms. Infect Immun. 2015;83(12):4630–8. Epub 2015/09/16. doi: 10.1128/IAI.00931-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao YL, Zhou TT, Guo HS. Hyphopodium-Specific VdNoxB/VdPls1-Dependent ROS-Ca2+ Signaling Is Required for Plant Infection by Verticillium dahliae. PLoS Pathog. 2016;12(7):e1005793 Epub 2016/07/28. doi: 10.1371/journal.ppat.1005793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei W, Zhu W, Cheng J, Xie J, Jiang D, Li G, et al. Nox Complex signal and MAPK cascade pathway are cross-linked and essential for pathogenicity and conidiation of mycoparasite Coniothyrium minitans. Sci Rep. 2016;6:24325 Epub 2016/04/14. doi: 10.1038/srep24325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryder LS, Dagdas YF, Mentlak TA, Kershaw MJ, Thornton CR, Schuster M, et al. NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc Natl Acad Sci U S A. 2013;110(8):3179–84. Epub 2013/02/06. doi: 10.1073/pnas.1217470110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valdivia A, Duran C, San Martin A. The role of Nox-mediated oxidation in the regulation of cytoskeletal dynamics. Curr Pharm Des. 2015;21(41):6009–22. Epub 2015/10/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reddi AR, Culotta VC. SOD1 Integrates Signals from Oxygen and Glucose to Repress Respiration. Cell. 2013;152(1–2):224–35. Epub 2013/01/22. doi: 10.1016/j.cell.2012.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komalapriya C, Kaloriti D, Tillmann AT, Yin Z, Herrero-de-Dios C, Jacobsen MD, et al. Integrative Model of Oxidative Stress Adaptation in the Fungal Pathogen Candida albicans. PLoS One. 2015;10(9):e0137750 Epub 2015/09/15. doi: 10.1371/journal.pone.0137750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3(5):1076–87. Epub 2004/10/08. doi: 10.1128/EC.3.5.1076-1087.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, et al. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20(10):1451–8. Epub 2010/09/03. doi: 10.1101/gr.109553.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jimenez-Lopez C, Collette JR, Brothers KM, Shepardson KM, Cramer RA, Wheeler RT, et al. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot Cell. 2013;12(1):91–100. Epub 2012/11/13. doi: 10.1128/EC.00290-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–27. Epub 2004/10/12 doi: 10.1016/j.gene.2004.06.021 . [DOI] [PubMed] [Google Scholar]
- 68.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. Epub 1991/01/01. . [DOI] [PubMed] [Google Scholar]
- 69.Li CX, Gleason JE, Zhang SX, Bruno VM, Cormack BP, Culotta VC. Candida albicans adapts to host copper during infection by swapping metal cofactors for superoxide dismutase. Proc Natl Acad Sci U S A. 2015;112(38):E5336–42. Epub 2015/09/10. doi: 10.1073/pnas.1513447112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nett JE, Cain MT, Crawford K, Andes DR. Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. J Clin Microbiol. 2011;49(4):1426–33. Epub 2011/01/14. doi: 10.1128/JCM.02273-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson CJ, Cabezas-Olcoz J, Kernien JF, Wang SX, Beebe DJ, Huttenlocher A, et al. The Extracellular Matrix of Candida albicans Biofilms Impairs Formation of Neutrophil Extracellular Traps. PLoS Pathog. 2016;12(9):e1005884 Epub 2016/09/14. doi: 10.1371/journal.ppat.1005884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pierce CG, Uppuluri P, Tristan AR, Wormley FL Jr., Mowat E, Ramage G, et al. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc. 2008;3(9):1494–500. Epub 2008/09/06. doi: 10.1038/nport.2008.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72(10):6023–31. Epub 2004/09/24. doi: 10.1128/IAI.72.10.6023-6031.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Expression of the various members of the FRE family listed by ORF designation were examined by qRT-PCR as described in Materials and Methods. Shown is the fold change in expression after 1 hour stimulation of hyphal morphogenesis by IMDM compared to yeast-form cells (cultured as in Fig 1). Results represent the averages of triplicate cultures. Genes that were previously characterized as cupric or ferric metalloreductases [37–39] or genes induced during fungal invasion of the kidney [40] are indicated by + marks. The C. albicans orthologue to S. cerevisiae Yno1 [18] is indicated.
(TIF)
(A,B) WT SC5314 and isogenic fre8Δ/Δ cells were induced to form hyphae by culturing cells seeded at 4 x 106 cells/ml at 37°C with 5% serum in the presence of the indicated levels of farnesol or methanol vehicle. Following four hours, cells were either (A) photographed or (B) assayed for viability by XTT as described in Materials and Methods where results represent the averages of biological triplicates. The decrease in cell mass/viability with 300 and 400 μM farnesol is statistically significant as determined by ANOVA with Tukey post-test; ****p<0.0001. (C) SC5314 cells seeded at 4 x 106 cells/ml (low density) were cultured for four hours at 37°C in YPD supplemented with or without 5% serum or with conditioned 5% serum media derived from high density WT or fre8Δ/Δ cultures (6 x 107 cells/ml). The conditioned medium was obtained by removing cells from the high density cultures through centrifugation. Results show that low density SC5314 forms hyphae at 37°C even in the absence of serum (-serum), but hyphal formation is blocked by conditioned medium from high density WT and fre8Δ/Δ cultures, indicative of quorum sensing [48]. Photographs are representative of 5–10 images over 2 experimental trials.
(TIF)
Mice were infected with either C. albicans WT SC5314 or the isogenic fre8Δ/Δ or the FRE8 complemented fre8Δ/Δ (fre8Δ/Δ Res) strain by lateral tail vein injection. Following 48 hours of infection, kidney and spleen were harvested and examined for (A) RNA markers of inflammation in the kidney by qRT-PCR as described in Materials and Methods, and (B) CFUs. Results are from 7–8 mice from each group. (A) The mRNA levels of the indicated inflammatory markers is shown as a fold change over uninfected controls. In all three infected strains, the increases in TNF- α, IL-6 and IL-17A are statistically significant compared to uninfected controls (****p<0.0001; ***p<0.0007). There is no statistically significant difference between WT and fre8Δ/Δ for any samples as determined by a one-way ANOVA with a Tukey post-test. There was a small (<2 fold) increase in expression of IL-17 and IL6 in the fre8Δ/Δ RES compared to WT, but the significance of this small variation is uncertain. (B) CFUs are shown as a function of tissue wet weight. The difference between fre8Δ/Δ and the fre8Δ/Δ strain complemented with FRE8 (fre8Δ/Δ RES) is significant (*p = 0.039). There is no statistically significant difference in CFUs obtained from spleen.
(TIF)
The indicated yeast strains were seeded at OD600 = 0.001 and grown at 30°C in YPD where growth by OD600 was either monitored continuously (TOP) or following a 16 hour period (BOTTOM). Results represent the averages of triplicate cultures (TOP) or of two to five experimental trials of hyphal morphogenesis (BOTTOM).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.