Abstract
Machine Learning (ML) methods are now influencing major decisions about patient care, new medical methods, drug development and their use and importance are rapidly increasing in all areas. However, these ML methods are inherently complex and often difficult to understand and explain resulting in barriers to their adoption and validation. Our work (RFEX) focuses on enhancing Random Forest (RF) classifier explainability by developing easy to interpret explainability summary reports from trained RF classifiers as a way to improve the explainability for (often non-expert) users. RFEX is implemented and extensively tested on Stanford FEATURE data where RF is tasked with predicting functional sites in 3D molecules based on their electrochemical signatures (features). In developing RFEX method we apply user-centered approach driven by explainability questions and requirements collected by discussions with interested practitioners. We performed formal usability testing with 13 expert and non-expert users to verify RFEX usefulness. Analysis of RFEX explainability report and user feedback indicates its usefulness in significantly increasing explainability and user confidence in RF classification on FEATURE data. Notably, RFEX summary reports easily reveal that one needs very few (from 2–6 depending on a model) top ranked features to achieve 90% or better of the accuracy when all 480 features are used.
Keywords: Random Forest, Explainability, Interpretability, Stanford FEATURE
1. Introduction, Background and Motivation
Machine Learning (ML) methods applied on large amounts of biological, medical and life science data for use in academic, R&D and business environments are now influencing major decisions about patient care, new medical methods, drug development and their use and importance are rapidly increasing in all areas. However, algorithms and software implementing ML methods are inherently complex and often difficult to understand and explain both to non-experts as well as experts. In addition, ML training databases used to derive predictive models are often large and complex, with noisy data, and in many cases are imbalanced containing much fewer positive class samples than background samples making commonly used “average” classification accuracy measures inadequate. All this makes it very challenging to understand, evaluate and be confident about results of ML performance. The interest in explaining how ML systems work is lately also driven by general public and funding agencies given the penetration of ML in all aspects of our lives and not only in bio-science. This is indicated by articles in popular press, many recent blogs, new DARPA program on explainable AI [1], FDA requirements for future data mining [2]), as well as by recent workshops focused on this subject (e.g. 2016 ICML Workshop on Human Interpretability in Machine Learning; PSB 2018 Workshop on Machine Learning and Deep Analytics for Biocomputing: Call for Better Explainability). Problems arising with the lack of explainability are being increasingly documented and discussed [3]. However, review of the published scientific literature on explainability in ML shows that very few research efforts and methods focus specifically on ML explainability. In addition, there is practically no work following the tried-and-true best practice of “user centered design” in which one engages users who are the ultimate judges and beneficiaries of explainability. We believe that the importance and benefits of being able to explain why and how ML decisions models make their decisions to non-ML experts and experts alike (e.g. explainability) are critical and must be addressed. We can further define explainability in ML as model explainability - why and how the trained ML model works overall, and sample explainability - how ML made a decision for a specific data sample (e.g. sample under investigation or sample from the training database). ML training is outside of our scope since it is usually well explained. Note that the ML approach can be reproducible but it still may not be sufficiently explainable. The improved explainability of ML in biocomputing and other areas will result in the following benefits: a) increased confidence of application and domain experts who are key decision makers (and who are often non ML-experts) in adopting ML; b) better testing and prevention of cases where ML approach produces results based on fundamentally wrong reasons (e.g. based on features not available in real application, wrong data in training databases or imperfect algorithm); c) easier evaluation, audit and verification of ML results for granting agencies, government organizations like FDA, and editors/publishers who need to decide what is being published and with what level of detail; d) simplification and reduction of the cost of application of ML in practice (e.g. by knowing which smaller feature subsets produce adequate accuracy more cost effective systems can be built); e) improved “maintenance” where ML method has to be changed or tuned to new data or decision needs; and f) possible discovery of new knowledge and ideas (e.g. by discovering new patterns and factors that contribute to ML decisions)
1.1 Random Forest (RF) Classifiers
RF is a popular and powerful ensemble supervised classification method [4]. Due to its superior accuracy and robustness, and some ability to offer insights by ranking of its features, RF has effectively been applied to various machine learning applications, including many in bioinformatics and medical imaging. RF consists of a set of decision trees, each of which is generated by the bagging algorithm with no pruning, forming a “forest” of classifiers voting for a particular class. To train a RF, two parameters, the number of tress (ntree) in the forest and the number of randomly selected features/variables used to evaluate at each tree node (mtry), must be supplied, as well as a training database with ground-truth class labels. RF also allows adjustment of the voting threshold or cutoff (fraction of trees in the forest needed to vote for a given class), which is used to compute recall, precision and f-score. The accuracy estimate built into the RF algorithm and all its software implementations is called Out of Bag Error (OOB), which measures the average misclassification ratio of samples not used for RF training. One of the RF algorithm’s strengths, and reasons we chose it, is its ability to calculate various feature/variable importance measures which can form the basis for enhancing its explainability [4, 6]. For this work we chose MDA (mean decrease in accuracy) as our main feature importance (ranking) measure. MDA measures the average increase of the error rate (i.e. decrease of accuracy) against random permutation of feature values across OOB cases. With a trained RF, the values of OOB cases for a tree are first permuted along the m-th feature. Then error rate with and without this permutations are recorded and their difference computed. This is repeated for all decision trees and the average of these differences gives the m-th feature’s MDA. We leverage the fact that MDA can be computed for + and − class separately (e.g. MDA+ and MDA−), thus providing better explainability. For main measure of RF classification accuracy, given that in most cases we have unbalanced training data (as in our case study), instead of commonly used OOB we use f-score (f=2 * (precision*recall)/(precision + recall)) determined using K-fold (we use K=5) stratified cross validation (SCV) [5, 7] where we independently partition samples to K folds for positive and negative sample pools first, then merge positive and negative folds to form the K folds that preserves the class distribution of the original dataset. The SCV procedure is then repeated, with varying of the RF tree voting cutoff threshold to maximize f-score.
1.2 Related work on Explainability for Random Forest Classifiers
Basic RF classification results traditionally comprise: information on the training data; optimal RF parameters; and the set of estimated accuracy measures with description of evaluation methods being used. Current methods for RF explainability fall into two basic categories. Feature ranking uses RF-provided variable importance measures like e.g. RF-provided Gini, MDA (mean decrease in accuracy) or others, to present them in tables or horizontal bar charts sorted by chosen variable importance measure, as in [8, 9, 10, 22, 23]. Highly ranked features are then assumed to play important role RF predictions, which in turn may offer some insights into the observed process or can even be used to clean-up training databases [23]. However, this information is insufficient for more substantial explainability. In addition, feature ranking is seldom done for + and − class separately, thus posing problems for frequent case of imbalanced data sets. Enhanced ranked feature representation with more details for helping RF epxlainability has been reported by [11]. One innovative idea to look at pairs of highly ranked feature and extract positive and negative pair-wise feature interactions has been reported in [12]. The second basic approach is rule extraction from trained RF. This method consists of: a) performing standard RF training; b) defining rules by analyzing trained RF trees (resulting in very large set of rules, order of 100 K); and c) reducing the number and complexity of extracted rules by optimization e.g. minimizing some metrics (accuracy, coverage, rule complexity…) to reduce to 10s – 100s of rules, each with 1–10 or so conditions [13–17]. Common problem with this approach is still a large number of complex rules hard to interpret by humans and lack of tradeoffs between accuracy and number of rules used. Our prior work on explainability for RF was motivated by our original joint work with Stanford Helix team on applying Support Vector Machines (SVM) [18] and RF [5] to their FEATURE data [19] where we show very good classification results measured by high recall and precision. In [5] we made first attempts to improve explainability by using RF-provided variable importance measures but did not analyze positive vs. negative classes separately and achieved very limited explainability improvements. The published work on explainability for RF (and other ML methods) can be summarized as follows: a) in spite of the fact that explainability is geared toward non-expert and expert human users no design consideration and formal evaluations related to human usability of proposed explanations and representations have been attempted; b) proposed explainability representations do not offer easy to use and critically important tradeoffs between accuracy and complexity of ML; c) analysis of + vs. − class separately (critical for a common case of unbalanced training data) has seldom been done; and d) feature reduction is generally not applied before explainability steps, thus necessitating complex approaches using large numbers of features impeding the explainability.
1.3 User-Centered Approach in Enhancing Random Forest Explainability - RFEX
RFEX method starts with standard approach to RF classification, using training database and standard RF tools/algorithms producing base RF accuracy estimates. In a series of steps RFEX then produces a RFEX summary report which is to be used by human (often non-expert) users to improve the explainability of original trained RF classifier (approach advocated in [1]). In developing RFEX we took a user-centered-approach (which to the best of our knowledge has not been tried by others): we guide our RFEX method by user-centered explainability questions or requirements collected by discussions and observations with interested practitioners, and then we test usefulness of RFEX as it is applied to FEATURE data by formal usability experiments. Based on our experience and investigation (especially in common case of imbalanced data) most users will lack full understanding of how and why RF works based only on the traditionally provided information (e.g. info on training data, optimized RF parameters, accuracy evaluation methods and estimates) and would pose a number of explainability questions to gain more insights and confidence before adopting it:
Can the explainability analysis be done for + and − class separately (critical in frequent case of imbalanced training data)?
What are most important features contributing to ML prediction and how do they rank in importance? What is the relationship of most important features for + vs. − class, is there any overlap?
What is the loss/tradeoffs of accuracy if I use only certain subset of most important features?
What is “direction” of features? Abundance (“more of it” or “presence”) or deficiency (“less of it” or “absence”)? What thresholds I can use to determine this?
Which features interact together?
Can this analysis be presented in an easy to understand and simple summary for ML/domain experts and non-experts?
We then use these explainability questions as “user-driven requirements” for the design of RFEX resulting in one page RFEX explainability summary report (one page was a design goal). RFEX is implemented and extensively tested on Stanford FEATURE data. Most importantly (and to the best of our knowledge never done before) we also performed formal RFEX usability study with 13 users of various experience in RF and FEATURE to assess RFEX utility in increase of RF classification results’ explainability.
2. Case Study: RFEX Applied to Stanford FEATURE data
Stanford FEATURE [19] is a system for classifying protein functional sites from electrochemical signatures/properties around those functional sites. FEATURE data is organized as feature vectors each describing a site in a three dimensional protein structure, using 80 physicochemical properties (features) in 6 concentric spherical shells, each 1.25 Ångstroms thick, yielding 480 feature values per vector. Each feature is denoted by the physicochemical property name, followed by its shell location (Si). FEATURE data i.e. the training database used for RF training, contains feature vectors at known positive (functional site) and negative (background) class labels for each protein functional model [18]. FEATURE training data is highly imbalanced e.g. there are two to three orders of magnitude more negative (background) vs. positive (functional sites) samples. For the work in this paper we used the same 7 FEATURE models selected in experiments in [5], which are subset of models analyzed in [18], see Table 1.
Table 1.
Summary of RF basic accuracy using all 480 features (described by several accuracy measures) for 7 FEATURE models used in this study
| model | num.positive | num.negative | mtry | recall | precision | fscore | oob | positive.oob | negative.oob |
|---|---|---|---|---|---|---|---|---|---|
| ASP_PROTEASE.4.ASP.OD1 | 1585 | 48577 | 40 | 0.99180 | 0.99873 | 0.99525 | 0.00032 | 0.00883 | 0.00004 |
| EF_HAND_1.1.ASP.OD1 | 1811 | 48145 | 40 | 0.91275 | 1.00000 | 0.95439 | 0.00268 | 0.07289 | 0.00004 |
| EF_HAND_1.1.ASP.OD2 | 1811 | 50290 | 40 | 0.91496 | 0.99941 | 0.95532 | 0.00248 | 0.07013 | 0.00004 |
| EF_HAND_1.9.GLN.NE2 | 15 | 47325 | 10 | 0.13333 | 1.00000 | 0.23529 | 0.00027 | 0.86667 | 0.00000 |
| IG_MHC.3.CYS.SG | 2017 | 49081 | 40 | 0.98017 | 0.98266 | 0.98141 | 0.00123 | 0.01487 | 0.00067 |
| PROTEIN_KINASE_ST.5.ASP.OD1 | 1096 | 48924 | 40 | 0.94162 | 0.99901 | 0.96947 | 0.00112 | 0.05018 | 0.00002 |
| TRYPSIN_HIS.5.HIS.ND1 | 446 | 50007 | 40 | 0.94177 | 0.99767 | 0.96892 | 0.00050 | 0.05381 | 0.00002 |
2.1 Creation of RFEX Summary Reports
We first estimate “base RF classification accuracy” by training RF on FEATURE data using all 480 features and we estimate accuracy using f-score with 5 fold stratified cross validation (SCV). We use ntree = 500 and vary mtry as {10, 20, 40} to find the optimal combination maximizing f-score. This experiment confirmed high RF predictive power for all 7 models (as reported before in [5]). Table 1 shows 7 models, and for each model their training data and several base RF accuracy measures using all 480 features. For our analysis we use Open Source packages which implement RF and provide MDA measures as well as various methods for RF training, including SCV, namely R package [20 ] and caret tool kit [21], along with Python integration and application code. We then proceed in developing “explainable RF model/representation” using RFEX approach which involves a series of steps and strategies (including novel explainability measures) designed to explicitly answer all 6 explainability questions above. We show details of experiments for one FEATURE model, namely ASP_PROTEASE.4.ASP.OD1 and then present RFEX one page summaries for two FEATURE models (ASP_PROTEASE.4.ASP.OD1 and EF_HAND_1.1.ASP.OD1), shown in Fig 2 and Fig 3. Detailed experimental results and RFEX summary reports for all 7 models are presented in [7]. All our analysis is performed separately for positive (functional sites) and negative (background) class (explainability question 1).
Fig 2.
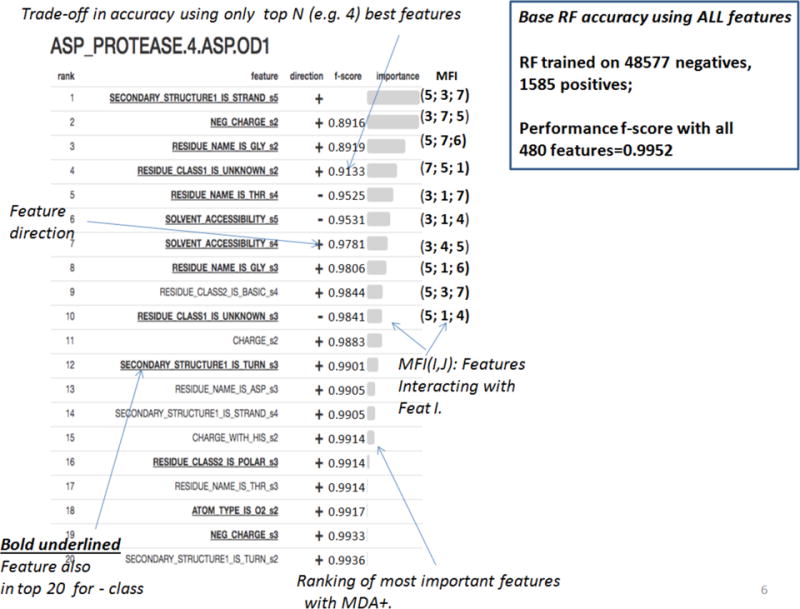
RFEX one page summary report for ASP_PROTEASE.4.ASP.OD1, with explanation of graphical elements as they relate to explainability questions
Fig 3.
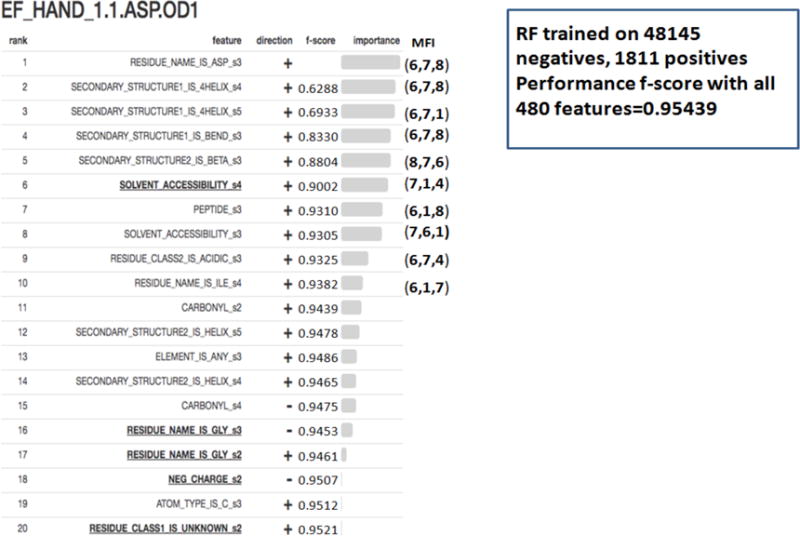
RFEX one page summary report for EF_HAND_1.1.ASP.OD1
To rank features by importance (explainability question 2) we use MDA + (ranking for positive class) and MDA − (ranking for negative class) provided from above trained RF classifiers using standard RF tools, as explained in Section 1.1. By leveraging feature rankings for positive and negative class separately (seldom done in published literature) we achieve more explainability given that FEATURE data is highly unbalanced. Indeed, this is justified by the fact that this method produces sets of differently ranked features for + and − class, as seen below in Table 2, showing 20 top ranked features for + and − class separately. Features appearing in both lists are bold.
Table 2.
Top 20 ranked features for ASP_PROTEASE.4.ASP.OD1) for positive class (MDA+ ranked) and negative class (MDA− ranked), with their feature direction (+/− columns). Features appearing in both lists are bold
| Top Features by +MDA | +/− | Top Features by −MDA | +/− |
|---|---|---|---|
| NEG_CHARGE_s2 | + (0.91) | RESIDUE_NAME_IS_GLY_s2 | − (0.99) |
| RESIDUE_CLASS1_IS_UNKNOWN_s2 | + (0.84) | RESIDUE_CLASS1_IS_UNKNOWN_s2 | − (0.99) |
| RESIDUE_NAME_IS_GLY_s2 | + (0.82) | RESIDUE_CLASS2_IS_POLAR_s2 | − (0.93) |
| SECONDARY_STRUCTURE_IS_STRAND_s5 | + (0.96) | RESIDUE_NAME_IS_LEU_s5 | − (0.96) |
| RESIDUE_NAME_IS_GLY_s3 | + (0.88) | SECONDARY_STRUCTURE1_IS_STRAND_s5 | − (0.88) |
| RESIDUE_CLASS1_IS_UNKNOWN_s3 | + (0.89) | PEPTIDE_s2 | − (0.85) |
| SOLVENT_ACCESSIBILITY_s5 | − (0.93) | SOLVENT_ACCESSIBILITY_s1 | + (0.83) |
| SOLVENT_ACCESSIBILITY_s4 | − (0.82) | RESIDUE_NAME_IS_GLY_s3 | − (0.96) |
| RESIDUE_NAME_IS_THR_s4 | + (0.86) | ATOM_TYPE_IS_O2_s2 | − (0.95) |
| ATOM_TYPE_IS_O2_s2 | + (0.86) | NEG_CHARGE_s2 | − (0.95) |
| SECONDARY_STRUCTURE1_IS_TURN_s3 | + (0.90) | RESIDUE_CLASS1_IS_UNKNOWN_s3 | − (0.96) |
| RESIDUE_CLASS2_IS_BASIC_s4 | − (0.99) | MOBILITY_s5 | + (0.92) |
| CHARGE_WITH_HIS_s2 | + (0.95) | SOLVENT_ACCESSIBILITY_s4 | + (0.92) |
| CHARGE_s2 | + (0.93) | SECONDARY_STRUCTURE1_IS_TURN_s3 | − (0.89) |
| NEG_CHARGE_S3 | + (0.88) | RESIDUE_NAME_IS_THR_S4 | − (0.94) |
| RESIDUE_NAME_IS_THR_s3 | + (0.77) | RESIDUE_CLASS2_IS_POLAR_s3 | − (0.95) |
| SECONDARY_STRUCTURE1_IS_TURN_s2 | + (0.84) | SOLVENT_ACCESSIBILITY_S5 | + (0.92) |
| RESIDUE_CLASS2_IS_POLAR_s3 | + (0.83) | RESIDUE_CLASS1_IS_HYDROPHOBIC_s5 | − (0.82) |
| SECONDARY_STRUCTURE1_IS_STRAND_s4 | + (0.94) | NEG_CHARGE_s3 | − (0.86) |
| RESIDUE_NAME_IS_ASP_s3 | + (0.93) | ELEMENT_IS_ANY_s4 | + (0.50) |
We then follow with critical (and seldom used by others) step of early complexity and dimensionality reduction where we aim to provide tradeoffs between using the subset of feature vs. loss of accuracy (explainability question 3). We focus on positive class and first re-train RF on top 2 ranked features from Table 2 using 5-fold SCV on original training data and record average f-score and its variation (measured by standard deviation). We then add next top ranked feature and retrain RF only on those 3 features. We repeat this adding top ranked features one by one until top 20th feature to obtain graph in Fig. 1 showing that by using very small subset of features (less than 20 from total of 480) one can achieve almost full base accuracy.
Fig. 1.
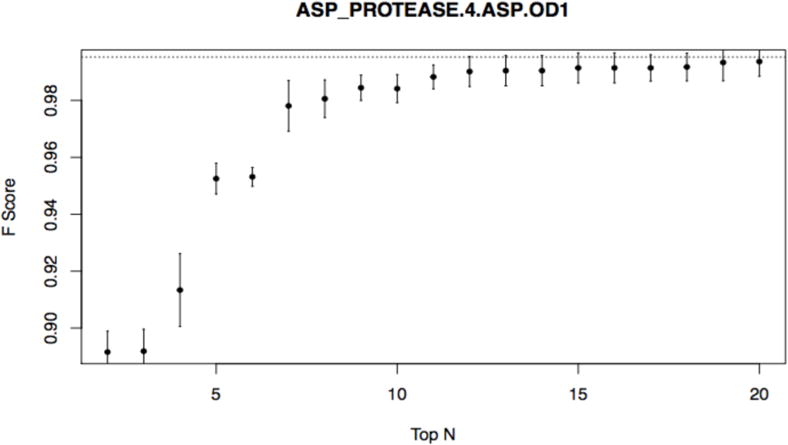
Trade-off of accuracy (average f-score and its variance from 5-fold CV) using Top N ranked subset of features (top 2, top 3 and so on), for positive class. Base accuracy using all 480 features is at dotted line
To understand the feature direction (+/− columns in Table 2) we introduce novel measure DIR(I) as + (n) or − (n) denoting fraction of times (n) when feature I was above (+) (abundance) or below (−) (deficiency) the threshold when making correct prediction, for all trees in the forest making a correct prediction, and for all test samples. We measure feature direction for top ranked 20 features, separately for positive and negative class (explainability question 4), shown in Table 2 as +/− columns. We also recorded histograms of threshold values used for top 5 ranked features but this information proved to be hard to use due to its variability. The table 2 also reveals some important confidence building explainability information: a) set of features best predicting positive vs. negative class is different and/or ranked differently; b) some of these features overlap (e.g. appear in both lists), and in those cases their direction is opposite; and c) all features are clearly either abundant or deficient (e.g. have high value of n). To measure which features “interact” or co-occur in making correct classifications (explainability question 5), we compute novel measure of Mutual Feature Interaction MFI(I,J) for features I and J as a count of times features I and J appear on the same tree path making a correct prediction, for all trees in RF ensemble, and for all test samples. We show top 3 co-occurring features for each of the top 10 ranked features (see Fig. 2). Note that MFI only measures statistical pair-wise feature co-occurrences and not necessarily causality.
Finally, we carefully designed a one page summary RFEX report (explainability question 6) intended to be easy to read and interpret for expert and non-experts alike, and to answer all 6 explainability questions above. It is provided for positive and negative class separately. We show two RFEX summary reports, first for ASP_PROTEASE.4.ASP.OD1 (positive class) in Fig. 2, annotated with explanations of what elements relate to 6 explainability questions (in italics), and the second one for EF_HAND_1.1.ASP.OD1 (positive class) in Fig. 3. One way is to use RFEX summary report is to verify whether it matches known intuition or biochemical patterns already known (e.g. by looking for “presence” or abundance (marked +) or “absence” or deficiency (marked −) of highly ranked features. This in turn would increase users’ confidence in RF predictions. Indeed, for the given two examples (ASP_PROTEASE, EF_HAND_1), there is evidence that the ranked features match our intuitive understanding of the active site structure, supported by the PROSITE [24] pattern matching. For ASP_PROTEASE, there is a required glycine residue that is one amino acid away from the active site; the ranked list indicates that atoms belonging to glycine residue(s) 2.5 to 3.75 Ångstroms (shells S2 and S3; RESIDUE_NAME_IS_GLY_s2, RESIDUE_NAME_IS_GLY_s3) are a positive predictor, and negative for background. For EF_HAND_1, alpha helix residues near the coordinating ASP residue is part of the motif definition. Reassuringly this rule contributes as two of the top 3 features for positive prediction (SECONDARY_STRUCTURE_IS_4HELIX_s4, SECONDARY_STRUCTURE_IS_4HELIX_s5). Another way of interpreting RFEX in general is to look at highly ranked features and use their presence or absence to indicate main predictive factors, which potentially can bring new insights (the approach we used in [22]). Finally, one can easily and efficiently use RFEX summary reports to explore tradeoffs between number of features used (with their names and direction) and classification accuracy by looking at f-score column for RFEX summary reports. This shows that for all 7 investigated FEATURE models (except for EF_HAND_1.9.GLN.NE2 which had only 15 training samples), it suffices to use only from 2–6 (depending on a model) top ranked features to achieve 90% or better of the accuracy (f-score) when all 480 features are used.
3. RFEX Usability Evaluation
The goals of RFEX usability evaluation were to assess: a) the increase of explainability e.g. users’ confidence and understanding of how and why RF works by using our RFEX approach compared to traditional methods of presenting RF classification results; b) obtain users’ feedback on the utility of each of the RFEX explainability summary report features. The usability evaluation was anonymous and was performed by users on their own time and place, based on the package of information and usability questionnaire sent to them with 11 questions. There were 13 users of varied experience in FEATURE and RF. User skill level in RF and FEATURE was assessed by users rating their expertise with 4 and 5 (e.g. “expert” level), or 1,2,3 (e.g. “non-expert”). Users in this study were grouped in 4 groups: a) FEATURE and RF NON-experts (4 users); b) FEATURE experts, RF non-experts (3 users); c) FEATURE non-experts, RF experts (2 users); and d) FEATURE experts, RF Experts (4 users). Group a) in some way corresponds to high level management or general public; group b) to bio scientists who are not versed in computational ML like RF; and group d) to computational bio scientists versed in biology (e.g. FEATURE) domain as well as ML (e.g. RF). Users were first asked to review Exhibit A (traditional ways of presenting RF results on FEATURE as in our prior work in [5]) for two models - ASP_PROTEASE.4.ASP.OD1 and EF_HAND_1.1.ASP.OD1) and assess their understanding of how and why RF works. Users were then given RFEX one page explainability summary reports for the above models (Exhibit B) and then asked to rate any gain in confidence and understanding of why and how RF works. Users were also asked to grade usefulness of particular RFEX summary report graphical and presentation features, as well as assess RFEX applicability to other RF and ML applications. In Table 3 below we show averages of answers to most important usability questions (4 out of 11), grouped by user expertise level as explained above (higher number indicates better rating).
Table 3.
Average of user responses to 4 most important usability questions for various user groups
| Question | ALL users (13) | FEATURE and RF NON-experts (4) | FEATURE experts and RF NON-experts (3) | FEATURE NON-experts and RF experts (2) | FEATURE and RF experts (4) |
|---|---|---|---|---|---|
| Estimate your increase in confidence of RF classification of FEATURE data after using RFEX summaries | 2.7 (SD 2.2) | 2.5 | 2 | 0.5 | 4.5 |
| Estimate your increase in understanding why and how RF works on FEATURE data (e.g. RF Explainability) using RFEX one page summaries | 3.3 (SD 1.7) | 3.25 | 3.7 | 1 | 4.25 |
| I believe RFEX approach will be useful for other applications of RF | 4.4 (SD 0.5) | 4.5 | 4.3 | 4 | 4.5 |
| I believe RFEX approach (or its modifications) will be useful for other machine learning methods | 4.0 (SD 0.8) | 4.5 | 3.3 | 3.5 | 4.25 |
For all 13 users, increase in understanding was significant (average of 3.3 for second question on a scale of 1…5) and increase in confidence in RF was good (average of 2.7 for first question). All users rated usefulness of RFEX method for other applications of RF and other ML approaches with 4 or 5, indicating strong RFEX promise at least at the conceptual level (averages for third and fourth questions were 4.4 and 4.0 respectively with small standard deviation). Analysis of usefulness of each feature of RFEX reporting where all users graded (not rated) them on a scale 1 to 5, indicated that most useful features were indeed those used to guide our design: one-page RFEX explainability summary design; feature ranking; presenting tradeoffs between number of features used and accuracy. In fact, all RFEX presentation features except thresholds used to test for abundance or deficiency of features were graded as above 3.7 in their usefulness. Positive user feedback on specific visualization format of RFEX summary points to importance of careful user-centered design for general ML explainability. Users also preferred to see up to top 3 Mutual Feature Interactions (MFI). The biggest increase both in confidence and understanding of how RF works on FEATURE data was achieved by user group d) (FEATURE and RF experts e.g. “Computational bio-scientists”), with averages of 4.5 and 4.25 on third and fourth questions. Users in group a) (FEATURE and RF NON-experts e.g. “Managers or general public”) showed significant increase in understanding (average of 3.25 on second question) and good increase in confidence (average of 2.5 on first question). Users in group b) (FEATURE experts and RF NON-experts e.g. “Bio scientists not versed in RF)” also showed strong increase in understanding of why and how RF works (average of 3.7 on second question) but moderate increase in overall confidence (average 2 on first question) which in part could be explained by their lack of RF expertise.
4. Conclusions and Future Work
Our RFEX method focuses on enhancing Random Forest (RF) classifier explainability by augmenting traditional information on RF classification results with one page RFEX summary report which is easy to interpret by users of various levels of expertise. RFEX method was designed and evaluated by never used before user-centered-approach driven by explainability questions and requirements collected from discussions with interested practitioners. It was implemented and extensively tested on Stanford FEATURE data. To assess usefulness of RFEX method for users, we performed formal usability testing with 13 expert and non-expert users which indicated its usefulness in increasing epxlainability and user confidence in RF classification on FEATURE data. Notably, RFEX summary reports easily reveal that one needs very few top ranked features (from 2-6 depending on a model) to achieve 90% or better of the accuracy achieved when all 480 features are used. Based on user feedback and our analysis we believe RFEX approach is directly applicable for other RF applications and to other ML methods where some form of feature ranking is available. Our future work includes applying RFEX on other RF applications and creation of a toolkit to automate RFEX creation.
Acknowledgments
We thank Prof. L. Kobzik, Dr. L. Buturovic and Prof. K. Okada for valuable feedback, J. Yang for helping with paper graphics and T. Murray and J. Schwartz for help in organizing our usability study. We also thank 13 anonymous usability reviewers. The work has partially been supported by NIH grant R01 LM005652 and by SFSU Center for Computing for Life Sciences.
References
- 1.DARPA program on Explainable AI. ( http://www.darpa.mil/program/explainable-artificial-intelligence), downloaded 07/04/17.
- 2.Duggirala HJ, et al. Use of data mining at the Food and Drug Administration. J Am Med Inform Assoc. 2016;23:428–434. doi: 10.1093/jamia/ocv063. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman S, Rosset S, Perlich C. Leakage in Data Mining: Formulation, Detection, and Avoidance. ACM Transactions on Knowledge Discovery from Data. 2012 Dec;6(4):1–21. [Google Scholar]
- 4.Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 5.Okada K, Flores L, Wong M, Petkovic D. Microenvironment-Based Protein Function Analysis by Random Forest. Proc ICPR (International Conference on Pattern Recognition); Stockholm. 2014. [Google Scholar]
- 6.Liaw, Wiener M. Classification and regression by randomforest. R News. 2002;2(3):18–22. [Online],: http://CRAN.R-project.org/doc/Rnews/ [Google Scholar]
- 7.Vigil A. MS Thesis. San Francisco State University, Computer Science Department; Dec, 2016. Building Explainable Random Forest Models with Applications in Protein Functional Analysis. [Google Scholar]
- 8.Malik H, Chowdhury I, Tsou H, Jiang Z, Hassan A. Understanding the rationale for updating a function’s comment. IEEE Int Conf on Software Maintenance. 2008 Oct; [Google Scholar]
- 9.Delen D. A comparative analysis of machine learning techniques for student retention management. Decision Support Systems. 2010 Nov;49(4):498–506. [Google Scholar]
- 10.Dale J, Popescu L, Karp P. Machine learning methods for metabolic pathway prediction. BMC Bioinformatics. 2010;11:15. doi: 10.1186/1471-2105-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng S. Unboxing the Random Forest Classifier: The Threshold Distributions. Airbnb Engineering and Data Science. https://medium.com/airbnb-engineering/unboxing-the-random-forest-classifier-the-threshold-distributions-22ea2bb58ea6, downloaded 07/04/17.
- 12.Kelly C, Okada K. IEEE Int Symposium on Biomedical Imaging. ISBI; 2012. Variable Interaction measures with Random Forest Classifiers. [Google Scholar]
- 13.Mashayekhi M, Gras R. Rule Extraction from Random Forest: the RF+HC Methods. Advances in Artificial Intelligence, Volume 9091 of the series Lecture Notes in Computer Science. 2015 Apr 29;:223–237. [Google Scholar]
- 14.Liu S, Dissanayake S, Patel S, Dang X, Milsna T, Chen Y, Wilkins D. Learning accurate and interpretable models based on regularized random forests regression. BMC Systems Biology. 2014;8(Suppl 3):S5. doi: 10.1186/1752-0509-8-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naphaporn S, Sinthupinyo S. 2011 International Conference on Information and Electronics Engineering. Vol. 6. IPCSIT; Singapore: 2011. Integration of Rules from a Random Forest. [Google Scholar]
- 16.Hara S, Hayashi K. ICML Workshop on Human Interpretability in Machine Learning. WHI; NY, USA: 2016. Making Tree Ensembles Interpretable. [Google Scholar]
- 17.Phung L, Chau V, Phung N. Extracting Rule RF in Educational Data Classification: From a Random Forest to Interpretable Refined Rules. Int Conf on Advanced Computing and Applications (ACOMP) 2015 [Google Scholar]
- 18.Buturovic L, Wong M, Tang G, Altman R, Petkovic D. High precision prediction of functional sites in protein structures. PLoS ONE. 9(3):e91240. doi: 10.1371/journal.pone.0091240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei L, Altman RB. Recognizing complex, asymmetric functional sites in protein structures using a Bayesian scoring function. J Bioinform Comput Biol. 2003;1(1):119–38. doi: 10.1142/s0219720003000150. [DOI] [PubMed] [Google Scholar]
- 20.R Core Team, R. A Language and Environment for Statistical Computing R Foundation for Statistical Computing. Vienna, Austria: 2013. http://www.R-project.org. [Google Scholar]
- 21.Kuhn M. The caret package. https://topepo.github.io/caret/, downloaded 07-04-17.
- 22.Petkovic D, Sosnick-Pérez M, Okada K, Todtenhoefer R, Huang S, Miglani N, Vigil A. Frontiers in Education FIE. Vol. 2016. Erie, PA: 2016. Using the Random Forest Classifier to Assess and Predict Student Learning of Software Engineering Teamwork. [Google Scholar]
- 23.Aeverman B, McCorison J, et al. PSB 2017. Hawaii: Jan, 2017. Production of Preliminary Quality Control Pipeline for Single Niclei RNA-SQ and its Application in the Analysis of Cell Type Diversity of Post-Mortem Human Brain Neocortex. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sigrist C, de Castro E, Cerutti L, Cuche B, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]


