SUMMARY
Although apoptotic cells (ACs) contain nucleic acids that can be recognized by Toll-like receptors (TLRs), engulfment of ACs does not initiate inflammation in healthy organisms. Here we identified macrophage populations that continually engulf ACs in distinct tissues and found that these macrophages shared characteristics compatible with immunologically silent clearance of ACs, including high expression of AC recognition receptors, low expression of TLR9, and reduced TLR responsiveness to nucleic acids. Removal from tissues resulted in loss of many of these characteristics and the ability to generate inflammatory responses to AC-derived nucleic acids, suggesting that cues from the tissue microenvironment program macrophages for silent AC clearance. The transcription factors KLF2 and KLF4 controlled the expression of many genes within this AC clearance program. The coordinated expression of AC receptors with genes that limit responses to nucleic acids may thus represent a central feature of tissue macrophages that ensures maintenance of homeostasis.
INTRODUCTION
Nucleic acids are recognized by multiple pattern recognition receptors (PRRs), including endosomal Toll-like receptors (TLRs) specific for DNA (TLR9) and various forms of RNA (TLR3, TLR7, TLR8, and TLR13)(Barbalat et al., 2011). This strategy affords broad recognition of multiple pathogen classes, and its failure can render the host susceptible to infection by a variety of pathogens (Barrat et al., 2016). However, the cost of this broad recognition is the potential for inappropriate responses to self-derived nucleic acids, which can lead to autoimmunity or autoinflammatory diseases (Sharma et al., 2015).
Multiple mechanisms limit recognition of self nucleic acids by TLRs. For instance, TLR9 preferentially recognizes DNA that contains unmethylated CpG dinucleotides, motifs that are more frequent in microbial DNA than mammalian DNA (Coch et al., 2009; Krieg et al., 1995; Yasuda et al., 2009). In addition, endosomal localization of nucleic-acid sensing TLRs limits access to extracellular self DNA and RNA (Barton et al., 2006; Mouchess et al., 2011). Bypassing this compartmentalization can disrupt homeostasis. For example, the generation of immune complexes containing nucleic acids can lead to Fc receptor-mediated uptake of endogenous nucleic acids, activation of endosomal TLRs, and subsequent autoimmune responses (Boulé et al., 2004; Leadbetter et al., 2002; Means et al., 2005).
Avoidance of self nucleic acid recognition during clearance of apoptotic cells (ACs) presents additional challenges. First, the volume of cargo that must be cleared is immense; it has been estimated that millions of cells die by apoptosis in the human body every day (Fond and Ravichandran, 2016). If clearance is disrupted, accumulation of ACs can lead to immune stimulation and, eventually, autoimmune disease (Asano et al., 2004; Baumann et al., 2002; Hanayama et al., 2004). Second, professional phagocytes that engulf ACs, such as macrophages and dendritic cells, express TLRs capable of nucleic acid recognition. Third, after recognition by a variety of phagocytic receptors (Miyanishi et al., 2007; Park et al., 2008; Scott et al., 2001), ACs traffic to phagosomes, the same organelles that house nucleic-acid sensing TLRs. Nevertheless, AC-derived nucleic acids do not typically initiate inflammatory responses. This avoidance is generally attributed to AC-induced expression of anti-inflammatory mediators. ACs can induce anti-inflammatory cytokine production as well as cell autonomous anti-inflammatory signaling pathways in phagocytes, as shown mainly through in vitro studies (A-Gonzalez et al., 2009; Freire-de-Lima et al., 2006; McDonald et al., 1999; Rothlin et al., 2007). However, in vivo AC clearance is a constant process, and it remains unclear how the innate immune system balances induction of anti-inflammatory responses while maintaining the ability to respond to pathogens.
Tissue-resident macrophages are proposed to be important mediators of AC clearance (Fond and Ravichandran, 2016). Several macrophage populations engulf ACs injected into mice (Baratin et al., 2017; McGaha et al., 2011; Miyake et al., 2007; Uderhardt et al., 2012; Wang et al., 2008), and apoptotic intestinal epithelial cells are engulfed by a dendritic cell (DC) subset and two macrophage populations in the intestine (Cummings et al., 2016). However, the identities of the cells that clear ACs from most tissues in the steady state remain unclear. This issue is particularly interesting in light of evidence that macrophages from different tissues are quite heterogeneous (Gautier et al., 2012b). Much of this diversity is controlled by local signals from tissues that induce gene expression and dictate the phenotype and function of resident macrophages (Gosselin et al., 2014; Lavin et al., 2014; Okabe and Medzhitov, 2014). It remains unclear whether this environmental programming induces heterogeneity in the ability of different resident macrophage populations to clear ACs, or influences responses to ACs.
To investigate how self-tolerance is maintained during AC-clearance, we identified macrophage populations in several tissues that efficiently engulf ACs in vivo at steady state. We found that the tissue microenvironment programmed these macrophages to limit responses to low doses of nucleic acid TLR ligands and to lack expression of TLR9. The transcription factors KLF2 and KLF4 were critical regulators of this AC-clearance program: KLF2 and KLF4 induced expression of genes that facilitated AC uptake as well as negatively regulated TLR signaling. Our findings suggest that coordinated expression of receptors that promote AC clearance with genes that limit innate responses to the nucleic acids within ACs may represent a central feature of tissue macrophages that ensures maintenance of homeostasis in the face of constant AC clearance.
RESULTS
Identification of macrophage populations that clear apoptotic cells in vivo
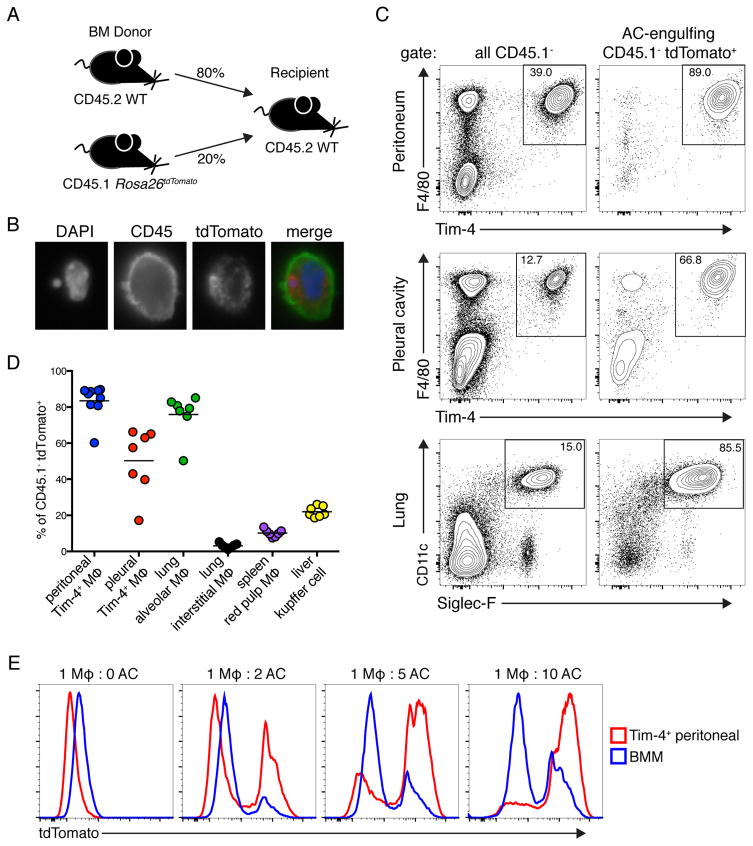
To investigate how tolerance to AC-derived ligands is maintained we sought to identify cells that clear ACs under homeostatic conditions in vivo. We generated mixed bone marrow chimeric mice: C57BL/6 (WT) bone marrow expressing the congenic marker CD45.2 was mixed with bone marrow expressing the congenic marker CD45.1 and the fluorescent protein tdTomato and injected into irradiated WT recipients (Figure 1A). In these mice the only cells that genetically encode for tdTomato also express CD45.1. Therefore, we reasoned that these mice could be used to identify cells involved in AC clearance, as tdTomato+CD45.2+CD45.1− cells must have acquired tdTomato by engulfing a CD45.1+ cell. The use of congenic markers allowed us to exclude any tdTomato+CD45.2+CD45.1+ cells resulting from cell-cell fusion. It is possible that cells acquired tdTomato by engulfing necrotic cells or cell debris, but one would not expect significant necrotic death under homeostatic conditions. A recent study used a similar system to track engulfment of GFP+ cells and observed low level constitutive engulfment, which they also attributed to steady-state clearance of ACs (Cummings et al., 2016).
Figure 1. Identification of tissue resident macrophage populations that clear apoptotic cells in vivo.
(A) Diagram of bone marrow chimeras used to identify AC-engulfing cells at steady state. (B) Representative immunofluorescence image of AC-engulfing cells isolated from chimeras. Cells were stained with DAPI and antibody against CD45. (C) Representative flow cytometric analyses of tissues from bone marrow chimeras. Cells were gated on CD45.1− cells or AC-engulfing cells (CD45.1− tdTomato+), as indicated. (D) Quantification of AC engulfment by the indicated macrophage populations in tissues. (E) CD45.2+ Tim-4+ pMacs and CD45.2+ BMMs were incubated with CD45.1+ tdTomato+ ACs at indicated ratios and analyzed by flow cytometry. All data are representative of at least three independent experiments. See also Figure S1.
Analysis of tissues after ten weeks of reconstitution identified a population of CD45.1−tdTomato+ cells that was not present in control chimeras generated with CD45.1+ bone marrow lacking tdTomato (Figure S1A). Immunofluorescence microscopy confirmed that these cells contained tdTomato+ cell corpses, as shown by the colocalization of tomato and DNA within WT cells (Figure 1B).
We identified three tissue-resident macrophage populations as the primary engulfers of dead hematopoietic cells at steady state in their respective tissues: peritoneal macrophages (pMacs) that express the AC recognition receptor Tim-4, pleural cavity macrophages that express Tim-4, and lung alveolar macrophages, which do not express Tim-4 (Figures 1C, 1D, and S1B). To confirm the results of the bone marrow chimera system we independently established that these populations efficiently engulf labeled ACs in vivo and in vitro (Figures 1E, S1C, and S1D). In other tissues analyzed, we did not identify a distinct macrophage population that was the primary engulfer of dead hematopoietic cells. However, we did identify several macrophage populations that engulf dead cells to a lesser extent: CD11b+ interstitial macrophages in the lung, red pulp macrophages in the spleen, and Kupffer cells in the liver (Figures 1D, S1B and S1E). We noted that CD11cmid migratory dendritic cells represented about 25% of CD45.1−tdTomato+ cells in the skin draining lymph node, suggesting that they may acquire dead cells in the skin and then migrate (Figures S1B and S1E). In several organs we observed a B220+ population that engulfed dead hematopoietic cells. These cells are likely B lymphocytes that have engulfed apoptotic bodies as has previously been described for B-1 cells (Rodriguez-Manzanet et al., 2010). We also identified a dead cell engulfing CD45− population in the liver, which is likely either hepatocytes or endothelial cells that have previously been described to engulf ACs (Dini et al., 2002). The differences in CD45.1−tdTomato+ cells between organs were not due to large differences in the number of available ACs, as the percentage of activated Caspase-3/7+tdTomato+ cells was similar in all organs we examined, except the liver which had more ACs (Figure S1F).
AC-engulfing macrophage populations do not express TLR9
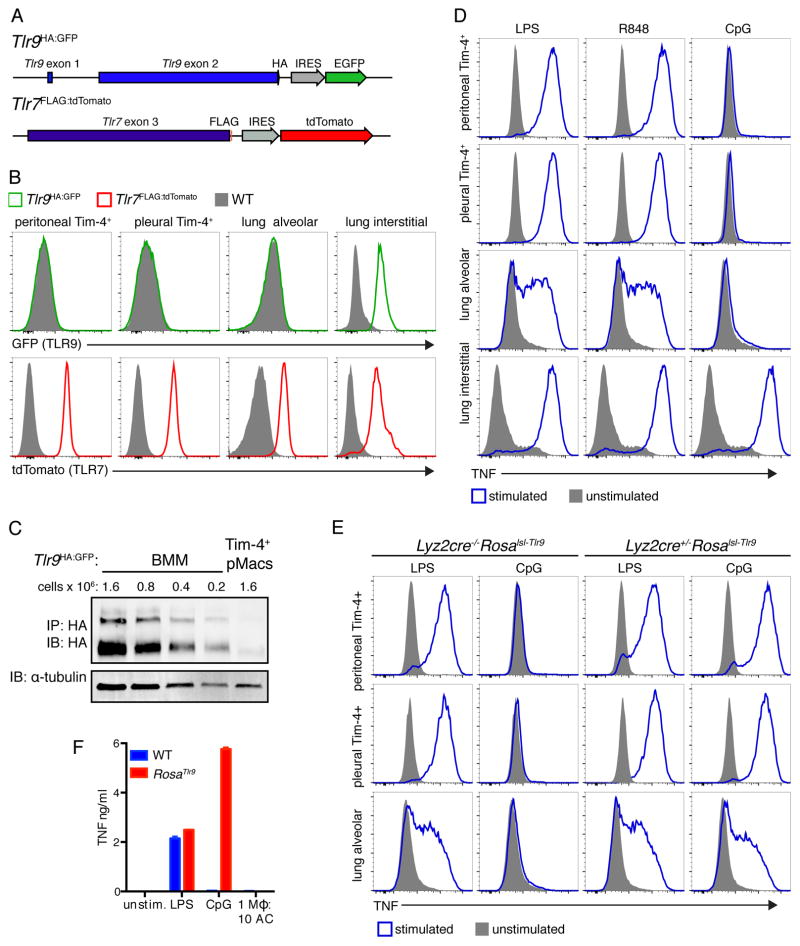
We chose to further characterize the peritoneal, pleural, and alveolar macrophages because these populations represented the clearest examples of cells dedicated to AC clearance in our system. We also noted that these three macrophage populations are localized to body cavities and wondered whether they share features that enable silent clearance of dead cells at these sites. As a first step we analyzed expression of TLR9 and TLR7 using reporter mice (Figure 2A). Examining endogenous TLR expression is notoriously challenging due to a lack of reliable antibodies, so we generated reporter mice to enable analysis of endogenous TLR expression levels with C-terminal epitope tags and fluorescent proteins driven by internal ribosome entry sequences downstream of the TLR coding sequence (Tlr9HA:GFP and Tlr7FLAG:tdTomato). Intriguingly, the three AC-engulfing macrophage populations described above did not express TLR9 (Figure 2B and 2C). Other macrophage populations, such as red pulp macrophages, Kupffer cells, lung CD11b+ interstitial macrophages, and F4/80mid peritoneal and pleural macrophages did express TLR9 (Figures 2B and S2A). TLR7 was expressed in most macrophages studied.
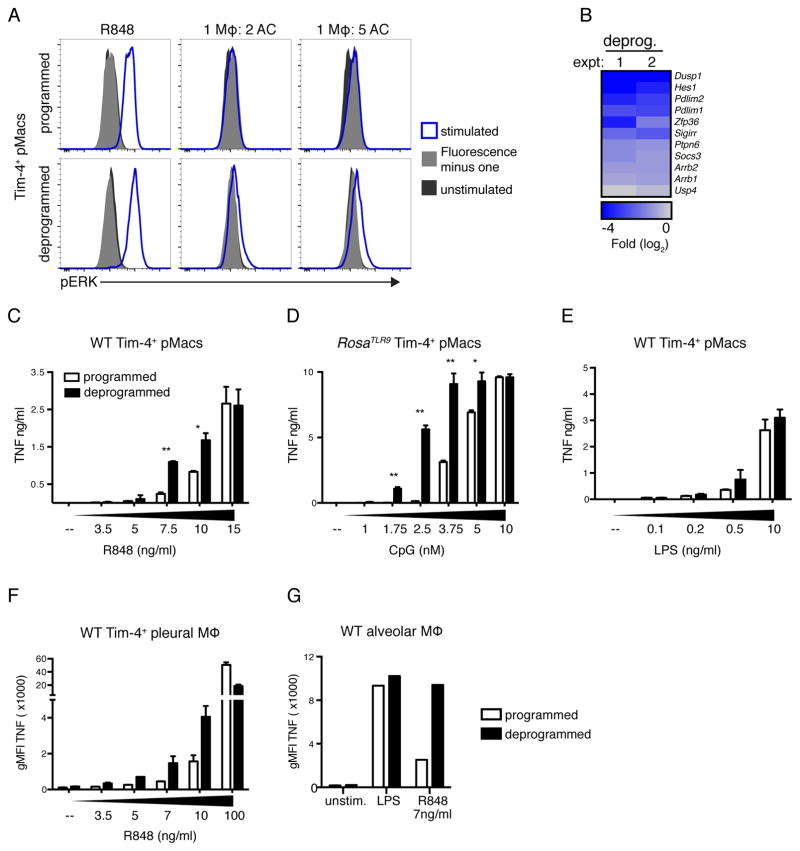
Figure 2. AC-engulfing macrophages do not express TLR9.
(A) Diagram of reporter mice generated to examine TLR expression. (B) Representative histograms of fluorescent protein expression by gated macrophages harvested from Tlr9HA:GFP, Tlr7FLAG:tdTomato, or WT mice. Peritoneal and pleural Tim-4+ macrophages are live F4/80+Tim-4+ cells. Alveolar macrophages are live SiglecF+CD11c+CD64+CD11b− cells. Lung interstitial macrophages are live CD11b+F4/80+MHCII+ cells. (C) Cell lysates from Tim-4+ pMacs and BMMs from Tlr9HA:GFP mice were immunoprecipitated with anti-HA resin and eluted protein was analyzed by anti-HA western. The two bands represent ER localized TLR9 (larger band) and endosomal cleaved TLR9 (smaller band). An anti-tubulin immunoblot was performed on lysates. (D) TNF production by macrophages from WT mice stimulated ex vivo with TLR ligands. (E) TNF production by macrophages from Lyz2cre+/−Rosalsl-Tlr or littermate control Lyz2cre−/−Rosalsl-Tlr9 mice stimulated ex vivo with the indicated TLR ligands (F). TNF production by RosaTlr9 Tim-4+ pMacs stimulated with TLR ligands or ACs. All data are representative of at least three independent experiments. See also Figure S2.
We confirmed that the three AC-engulfing macrophage populations lack TLR9 by stimulating macrophages ex vivo with TLR ligands. These macrophages produced TNF in response to stimulation with LPS (TLR4 ligand) or R848 (TLR7 ligand) but not CpG oligonucleotides (ODN), a synthetic TLR9 ligand (Figures 2D and S2B). CD11b+ interstitial macrophages and red pulp macrophages generated robust responses to TLR9 stimulation. Interestingly, TLR9 responses generated by Kupffer cells were muted compared to responses to other TLR ligands (Figure S2B). Kupffer cells were also intermediate in engulfment of ACs (Figure 1D), suggesting that there may be an inverse correlation between AC engulfment ability and expression of TLR9. Altogether, these data led us to hypothesize that tight regulation of TLR9 expression is one mechanism used to avoid responses to self-derived DNA.
Forcing expression of TLR9 in AC-engulfing macrophages only slightly enhances inflammation during pristane-induced lupus
To investigate whether lack of TLR9 expression in AC-engulfing macrophages is needed to maintain self-tolerance we forced these macrophages to express TLR9. We generated mice that ectopically express TLR9 in all cells (RosaTlr9) or in LysM+ cells (Lyz2cre+/−Rosalsl-Tlr9) by crossing mice in which TLR9 is inserted behind a floxed stop cassette in the Rosa26 locus to EIIA-cre or Lyz2-cre mice, respectively. Tim4+ pMacs, pleural cavity macrophages, and alveolar macrophages from Lyz2cre+/−Rosalsl-Tlr9 mice responded to CpG ODN (Figure 2E). This result confirms that AC-engulfing macrophages control responses to TLR9 ligands through TLR9 expression. However, despite this forced TLR9 responsiveness, Tim-4+ pMacs from RosaTlr9 mice did not respond to ACs in vitro (Figure 2F). This lack of response was not due to reduced AC engulfment (Figure S2C).
When housed unperturbed in our specific pathogen-free mouse facility, Lyz2cre+/−Rosalsl-Tlr9 mice showed no evidence of inflammatory disease over twelve months (data not shown), demonstrating that forced expression of TLR9 in AC-engulfing macrophages is not sufficient to disrupt homeostasis. To investigate whether increased cell death could promote disease, we employed the pristane-induced model of lupus, in which intraperitoneal injection of pristane oil induces substantial apoptosis followed by recruitment of inflammatory cells and development of a lupus-like disease (Calvani et al., 2005). TLR7 and TLR9 have been implicated in the pathogenesis of this disease model (Lee et al., 2008; Summers et al., 2010). Compared to Lyz2cre−/−Rosalsl-Tlr9 littermate controls, Lyz2cre+/−Rosalsl-Tlr9 mice had small increases in the number of cells recruited into the peritoneum at two weeks (Figures S2D and S2E). After nine months Lyz2cre+/−Rosalsl-Tlr9 mice still demonstrated a small increase in peritoneal cell numbers (Figures S2D and S2F). Sera from these mice also contained increased anti-dsDNA IgG (Figure S2G). Altogether, the overexpression of TLR9 in LysM+ cells slightly enhanced inflammation in the pristane model of lupus; however, the subtlety of these differences indicated that additional mechanisms must be limiting tissue macrophage responses to nucleic acids within ACs.
Tissue environments program resident macrophages for silent clearance of ACs
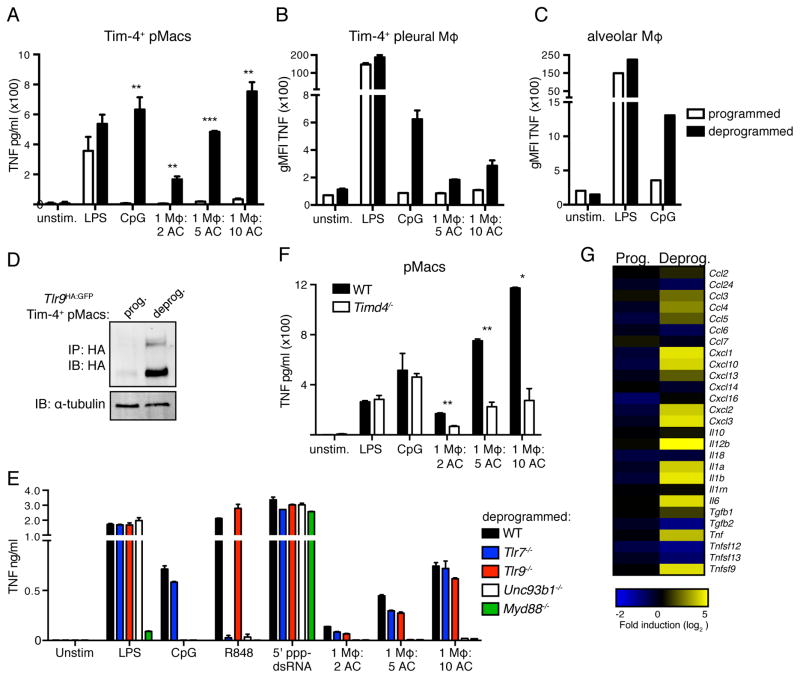
Tissue-resident macrophages are programmed by cues from their environment to express specific transcription factors that shape their identity and function (Gautier et al., 2012b; Gosselin et al., 2014; Lavin et al., 2014; Okabe and Medzhitov, 2014). We hypothesized that AC-engulfing macrophages may be subject to similar programming of which inhibition of TLR9 expression is only one facet. To investigate this possibility, we compared isolated Tim-4+ pMacs that had been cultured ex vivo overnight to macrophages that had been cultured ex vivo for three days. The macrophages cultured for three days not only gained responsiveness to CpG ODN but also began responding to ACs (Figures 3A and S3A). Similar results were obtained with alveolar macrophages and Tim-4+ pleural macrophages (Figures 3B and 3C). This altered response corresponded with increased expression of TLR9 protein (Figure 3D) and was not due to changes in AC uptake (Figure S3B). Tim-4+ pMacs from mice lacking the TLR signaling adaptor MyD88 or Unc93b1, a trafficking chaperone required for endosomal TLR function, failed to respond to ACs (Lee et al., 2013; Tabeta et al., 2006)(Figures 3E and S3B). Tlr7−/− and Tlr9−/− pMacs still responded to ACs, suggesting that a combination of RNA and DNA ligands are responsible for the response to ACs. In contrast, bone marrow-derived macrophages (BMMs) failed to engulf ACs efficiently and did not produce TNF (Figures S3C and 1E). We confirmed that the observed responses were to nucleic acids within ACs rather than free nucleic acids by using pMacs lacking Tim-4. Tim-4 is required for efficient uptake of ACs by pMacs (Miyanishi et al., 2007; Wong et al., 2010) (Figure S3B). Reduced AC uptake by Timd4−/− pMacs correlated with reduced production of TNF (Figure 3F).
Figure 3. AC-engulfing macrophages removed from their tissue environment generate inflammatory responses to ACs.
(A) TNF production by Tim-4+ pMacs cultured overnight (programmed) or for 60h (deprogrammed) before stimulation with TLR ligands or ACs. Data are representative of at least three independent experiments. (B) TNF production by programmed or deprogrammed pleural cells from WT mice stimulated with TLR ligands or ACs. Data are representative of at least three independent experiments. (C) TNF production by programmed or deprogrammed lung cells from WT mice stimulated with TLR ligands. Data are representative of at least three independent experiments. (D) Lysates from programmed and deprogrammed Tlr9HA:GFP Tim-4+ pMacs were immunoprecipitated with anti-HA resin and eluted protein was analyzed by anti-HA western. An anti-tubulin immunoblot was performed on lysates. Data are representative of three independent experiments (E) TNF production by deprogrammed Tim-4+ pMacs from WT, Tlr7−/−, Tlr9−/−,Unc93b1−/−, or Myd88−/− mice stimulated with TLR ligands, RIG-I ligand, or ACs. Data are representative of at least two independent experiments. (F) TNF production by deprogrammed pMacs from WT or Timd4−/− mice stimulated with TLR ligands and ACs. Data are representative of two independent experiments. (G) RNA sequencing analysis of programmed and deprogrammed Tim-4+ pMacs stimulated with ACs. Data are presented as fold stimulated over unstimulated and are results from one experiment with technical duplicates averaged so only one value is shown. See also Figure S3 and S4.
To obtain a more complete picture of the responses to ACs made by ex vivo tissue macrophages we used RNAseq. Tim-4+ pMacs generated a limited transcriptional response to ACs that did not include the production of pro- or anti-inflammatory cytokine mRNA (Figures 3G and S3D). However, ex vivo cultured Tim-4+ pMacs generated a robust response to ACs, including increased expression of inflammatory cytokines such as Tnf, Il1a, Ilb1, and Cxcl2 (Figure 3G). Altogether, these results demonstrate that macrophages removed from their tissue of residence can no longer avoid recognition of nucleic acids within ACs. We refer to this change in macrophage responsiveness to ACs as “deprogramming”.
Resident pMacs are influenced by signals from the omentum that induce expression of pMac signature genes (i.e., genes more highly expressed in pMacs than in other tissue macrophages). Retinoic acid generated in the omentum induces a number of genes in pMacs, including the transcription factor Gata6, which in turn induces the expression of many other pMac signature genes (Gautier et al., 2014; Okabe and Medzhitov, 2014; Rosas et al., 2014). To test whether these previously described environmental cues were responsible for the AC-clearance program in Tim-4+ pMacs, we supplied macrophages undergoing deprogramming with omentum culture supernatant, as described by others (Okabe and Medzhitov, 2014). While this treatment improved macrophage survival, it did not prevent responses to CpG ODN or ACs after a three-day culture (Figure S3E, and data not shown). Therefore, we continued culturing pMacs in omentum culture supernatant during deprogramming for the rest of these studies. Gata6 did not induce the AC-clearance program, as increasing Gata6 expressing using lentiviral transduction did not prevent responses to CpG ODN or AC stimulation (Figures S3F and S3G). Therefore, the signals that program pMac identity are not sufficient to maintain the AC-clearance program in ex vivo tissue macrophages.
To more thoroughly compare programmed and deprogrammed Tim-4+ pMacs, we analyzed gene expression by RNA sequencing and confirmed the results by nCounter analysis (Figure S4E). The expression of many previously identified pMac signature genes decreased during the deprogramming period, further demonstrating that deprogrammed macrophages were losing aspects of their tissue-programmed identity even in the presence of omentum culture supernatant (Figure S4A, (Gautier et al., 2012b)). However, expression of macrophage core genes (i.e., genes expressed by tissue-resident macrophage populations but not dendritic cells) did not show a similar reduction (Figure S4B). In addition, there was no coordinated change in gene expression indicative of a shift from M2 to M1 or vice versa ((Jablonski et al., 2015), Figure S4C).
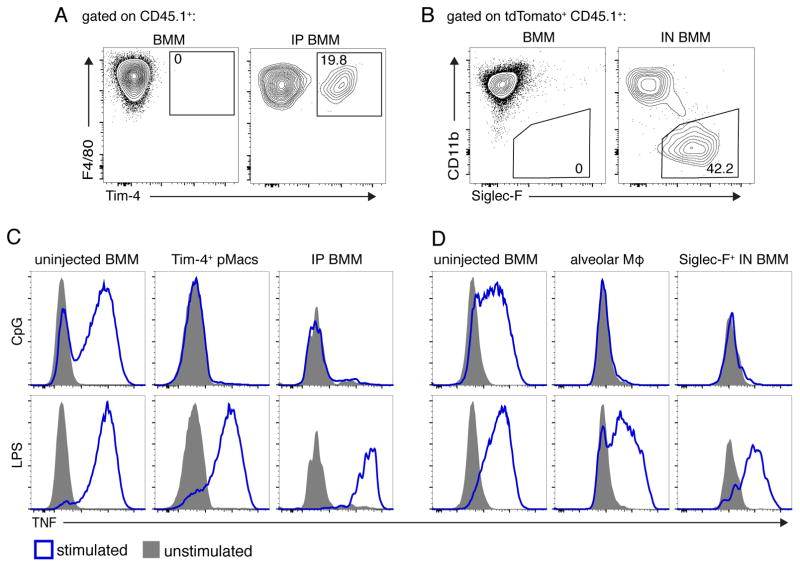
We next tested directly whether signals from the tissue environment can confer the AC-clearance program. Congenically marked BMMs differentiated with M-CSF for seven days in vitro were injected into the peritoneum or administered intra-nasally. After several weeks in the peritoneum or lungs, a proportion of the transplanted BMMs began to express Tim-4 or Siglec-F (a marker of alveolar macrophages), respectively (Figures 4A and 4B). Furthermore, when harvested and stimulated with TLR ligands, both populations of transplanted BMMs failed to respond to CpG ODN (Figures 4C and 4D). We excluded the possibility that newly programmed transplanted BMMs arose from a small population of contaminating progenitor cells, since we did not observe significant proliferation of BMMs after IP injection (Figure S4F). These results suggest that environmental cues induce a program in certain populations of tissue macrophages that facilitates silent clearance of ACs. This program induces high expression of AC recognition receptors and low expression of TLR9.
Figure 4. BMMs gain AC-clearance programming in tissue environments.
(A) Flow cytometric analyses of uninjected CD45.1+ WT BMMs (left) or BMMs transferred into the peritoneum for three weeks (right). (B) Flow cytometric analyses of uninjected CD45.1+ tdTomato+ cells (left) or BMMs transferred intranasally (IN) and analyzed after 39d (right). (C) TNF production by BMMs transferred as described in (A), harvested, and stimulated with TLR ligands. (D) TNF production by BMMs transferred as described in (B), harvested, and stimulated with TLR ligands. SiglecF+ cells are gated as shown in panel B. All data are representative of three independent experiments. See also Figure S4.
Programmed AC-engulfing macrophages have a higher activation threshold for endosomal TLR responses
Next, we more carefully analyzed endosomal TLR signaling in programmed and deprogrammed macrophages. Interestingly, while deprogrammed Tim-4+ pMacs generated a small but consistent phospho-ERK signal in response to ACs, programmed macrophages did not (Figure 5A). Thus, TLR signaling in programmed macrophages was impaired at a step prior to phosphorylation of ERK. We also noted the decreased expression of several inhibitors of TLR signaling in deprogrammed macrophages (Figure 5B). Some of these inhibitors, such as SOCS3, DUSP1, and SHP-1, inhibit TLR signaling prior to ERK phosphorylation (Kondo et al., 2012).
Figure 5. Programmed macrophages have a higher activation threshold for TLR7 and TLR9 responses.
(A) Tim-4+ pMacs were stimulated with R848 or ACs for 30min, and phospho-ERK1/2 signal was measured by flow cytometry. Data are representative of three independent experiments. (B) Negative regulators of TLR signaling that are significantly (p < 0.05) reduced in deprogrammed Tim-4+ pMacs relative to programmed Tim-4+ pMacs as measured by RNA sequencing. Results of two independent experiments are shown with replicates averaged. (C–E) TNF production by programmed and deprogrammed Tim-4+ pMacs stimulated with increasing doses of TLR ligands. Data are representative of at least three independent experiments. (D) To compare CpG responses, Tim-4+ pMacs were isolated from RosaTlr9 mice. (F and G) Pleural (F) or lung (G) cells from WT mice were harvested and incubated overnight or for 60h before stimulation. Data are representative of two independent experiments. See also Figure S5.
We hypothesized that tissue macrophages involved in AC clearance may require a high threshold for initiation of endosomal TLR signaling. This high threshold could prevent responses to weak ligands. Several aspects of AC-derived nucleic acids make them weak ligands for endosomal TLRs. For instance, mammalian DNA contains very few unmethylated CpG dinucleotides, and upon induction of apoptosis, DNA and RNA are actively degraded (Coch et al., 2009; Enari et al., 1998; King et al., 2000; Krieg et al., 1995). To compare the activation threshold for TLR signaling between programmed and deprogrammed macrophages, we measured responses to low doses of TLR ligands. Deprogrammed macrophages generated a more robust response to low levels of TLR7 ligands, TLR13 ligands, and TLR9 ligands (Figures 5C–G and S5A–F). Importantly, Tlr7 and Tlr13 expression was not affected by deprogramming (Figures S4D). Altogether, these data are consistent with a model wherein programmed macrophages maintain a higher threshold for endosomal TLR activation and cannot generate responses to the less stimulatory ligands derived from ACs.
Inflammatory cues induce TLR9 expression without enabling responses to ACs
Although the absence of TLR9 in peritoneal, pleural, and alveolar macrophages may be beneficial when clearing ACs, this lack of expression could be detrimental during infection. We therefore examined how inflammatory signals affect the AC-clearance program. As a first step, we incubated Tim-4+ pMacs overnight with various cytokines followed by stimulation with TLR ligands or ACs. Tim-4+ pMacs pretreated with interferon (IFN)γ or IFNβ increased TLR9 expression and gained responsiveness to TLR9 stimulation (Figures 6A, 6B, and S6A). However, IFNγ or IFNβ treatment did not enable responses to ACs, indicating that the cytokines did not reverse the entire AC-clearance program. This lack of response was not due to an inability to engulf ACs (Figure S6C). IFNγ pretreatment also induced TLR9 responses in pleural and alveolar macrophages (Figure 6C).
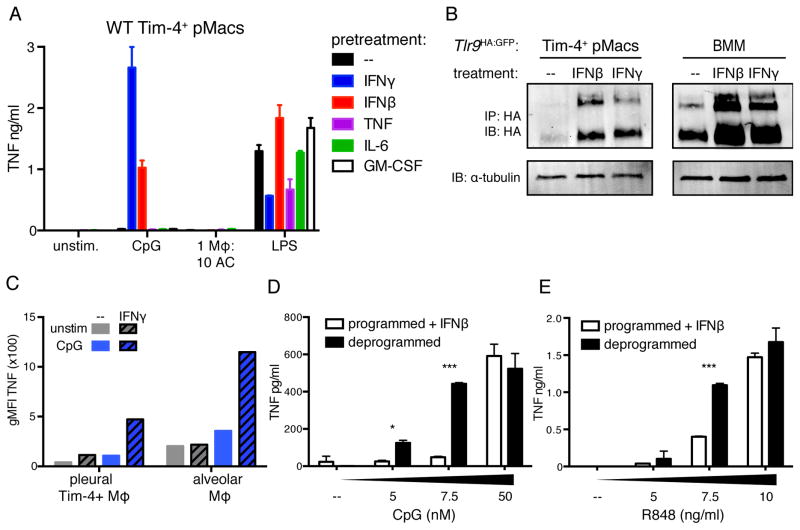
Figure 6. Inflammatory cues induce TLR9 expression in AC-engulfing macrophages.
(A) TNF production by Tim-4+ pMacs treated overnight with the indicated cytokines before stimulation with TLR ligands or ACs. Data are representative of at least three independent experiments. (B) Tim-4+ pMacs and BMMs from Tlr9HA:GFP mice were cultured overnight ± IFNγ or IFNβ, and TLR9 levels in lysates were measured by anti-HA immunoprecipitation and immunoblot. An anti-tubulin immunoblot was performed on lysates. Wells of Tim-4+ pMacs correspond to lysate of 1.6e6 cell equivalents, and wells of BMMs correspond to lysate of 0.4e6 cell equivalents. Data are representative of at least three independent experiments. (C) TNF production by lung and pleural cells harvested from WT mice and incubated overnight ± 100ng/ml IFNγ before stimulation with CpG ODN. Data are representative of two independent experiments. (D–E) TNF production by programmed Tim-4+ pMacs incubated overnight with IFNβ or by untreated deprogrammed Tim-4+ pMacs stimulated with increasing doses of CpG (D) or R848 (E). Data are representative of three independent experiments. See also Figure S6.
The lack of response to ACs in macrophages exposed to inflammatory signals suggested that these cells maintain a high threshold for endosomal TLR signaling. To test this possibility, we compared programmed Tim-4+ pMacs that had been pretreated with IFNβ to deprogrammed Tim-4+ pMacs. Despite expressing similar amounts of TLR9, IFNβ treated Tim-4+ pMacs generated weaker responses to low doses of CpG ODN and R848 (Figures 6D, 6E, S6B, S6D, and S6E). Although prolonged expression of TLR9 in AC-engulfing macrophages may be detrimental, the inability of these macrophages to respond to low-level nucleic acid stimulation may prevent responses to self-derived nucleic acids during acute inflammation.
KLF2 and KLF4 control an AC-clearance program in macrophages
We next attempted to identify transcription factors that control the AC-clearance program. Our RNAseq analysis identified many transcription factors whose expression was lower in deprogrammed Tim-4+ pMacs (Figure S7A). These transcription factors included Gata6 and RARβ that influence peritoneal macrophage identity. In addition, expression of the transcription factors Kruppel-like factor 2 (KLF2) and KLF4 was significantly decreased after deprogramming. These transcription factors contribute to gene regulation in a variety of biological contexts including macrophage polarization and repression of myeloid cell activation (Liao et al., 2011; Mahabeleshwar et al., 2011).
To examine whether these transcription factors regulate the AC-clearance program we first assessed their importance for inhibition of TLR9 expression in the peritoneal environment. To this end, we used Cas9 genome editing to generate BMMs lacking each transcription factor and injected these cells into the peritoneum of congenically marked mice. BMMs transduced with empty vector or with guides for Gata6 or Rarb lost the ability to respond to TLR9 stimulation (Figures 7A and 7B). In contrast, BMMs transduced with guides specific for Klf2 continued to respond to TLR9 stimulation, and Klf4 targeted BMMs had an intermediate response (Figures 7A and 7B). These data suggest that KLF2, potentially with contributions from KLF4, inhibits TLR9 responses in resident pMacs.
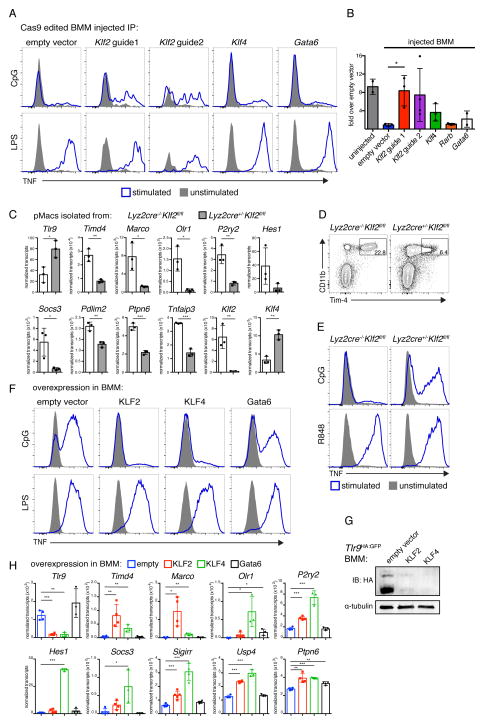
Figure 7. KLF2 and KLF4 imprint an AC-clearance program on macrophages.
(A and B) BMMs from Cas9 expressing mice were transduced with vectors encoding guide RNAs targeting the indicated transcription factors and injected IP into congenically marked mice. 14d after injection peritoneal cells were harvested and stimulated with TLR ligands. Uninjected BMMs transduced with empty vector were stimulated as a control. (B) Quantification of IP injected BMMs responses to CpG displayed relative to injected empty vector response. Data are the combined results from two independent experiments with n = 2–4 per group. (C) F4/80+ pMacs were isolated from Lyz2cre+/−Klf2fl/fl mice and littermate control Lyz2cre−/−Klf2fl/fl mice. Levels of mRNA were quantified using a Nanostring nCounter. n = 3 per group from independent experiments. (D and E) Peritoneal cells from Lyz2cre+/−Klf2fl/fl mice and littermate control Lyz2cre−/−Klf2fl/fl mice were harvested and (D) analyzed by flow cytometry for expression of the indicated markers or (E) stimulated with TLR ligands. Data are representative of three independent experiments. (F) TNF production by WT BMMs transduced with retroviral vectors encoding KLF2, KLF4, Gata6 or with empty vector and stimulated with the indicated TLR ligands. Data are representative of at least two independent experiments. (G) Tlr9HA:GFP BMMs were transduced with retroviral vectors encoding KLF2, KLF4, or empty vector. Lysates were subjected to immunoprecipitation with anti-HA resin and eluted TLR9 protein was visualized by anti-HA immunoblot. An anti-tubulin immunoblot was performed on lysates. Data are representative of at least two independent experiments. (H) WT BMMs were transduced with retroviral vectors encoding KLF2, KLF4, Gata6, or with empty vector. Levels of mRNA were quantified using a Nanostring nCounter. n = 3–4 per group from independent experiments. See also Figure S7.
To determine whether KLF2 controls expression of other aspects of the AC-clearance program, we analyzed gene expression in pMacs isolated from Lyz2cre+/−Klf2fl/fl mice and Lyz2cre−/−Klf2fl/fl littermate control mice. Expression of Timd4 was reduced in KLF2-deficient pMacs, as was expression of the AC recognition receptors Marco and Olr1 and P2ry2, an ATP receptor demonstrated to aid in recruitment of phagocytes to ACs (Figure 7C) (Elliott et al., 2009; Oka et al., 1998; Wermeling et al., 2007). We confirmed reduced expression of Tim-4 on the surface Lyz2cre+/−Klf2fl/fl pMacs by flow cytometry (Figure 7D). High expression of negative regulators of TLR signaling also required KLF2; mRNA levels of Hes1, Socs3, Pdlim2, Ptpn6, and Tnfaip3 were lower in KLF2-deficient pMacs. Lyz2cre+/−Klf2fl/fl pMacs also expressed higher levels of TLR9 than littermate controls and responded to CpG ODN (Figure 7E). These results indicate that KLF2 is required for all aspects of the AC-clearance program.
We also tested whether KLF2 and KLF4 are sufficient to drive expression of the AC-clearance program in BMMs. Ectopic expression of KLF2- and KLF4 in BMMs reduced TLR9 responses but not TLR4 responses, while Gata6 overexpression had little effect (Figures 7F, S7C, and S7D). KLF2 and KLF4 inhibited TLR9 expression and drove expression of many of the AC-clearance program genes (Figures 7G and 7H).
These data demonstrate that the transcription factors KLF2 and KLF4 induce a series of coordinated changes, including the reduced expression of TLR9 and the increased expression of inhibitors of TLR signaling and receptors that recognize ACs. Notably, while alveolar macrophages express low levels of Klf2, their expression of Klf4 is high (data from the Immunological Genome Project), suggesting that KLF4 may play a more prominent role in programming alveolar macrophages.
DISCUSSION
We characterized three macrophage populations adept at AC clearance in the peritoneum, pleural cavity, and lungs. We demonstrated that these AC-engulfing macrophages are programmed by their tissue environments to silently clear ACs. This programming included inhibition of TLR9 expression as well as increased expression of AC recognition receptors and negative regulators of TLR signaling. We hypothesize that the resulting increased threshold of activation in these cells enables silent clearance of large numbers of ACs.
Several mechanisms that limit inflammatory signaling in cells that engulf ACs have been previously described, many involving induction of inhibitors upon AC recognition, but our work identified distinct features intrinsic to certain tissue macrophages that enable silent AC clearance. A recent study identified phagocyte populations that engulf apoptotic epithelial cells in the intestine (Cummings et al., 2016). After AC engulfment these populations demonstrate reduced expression of genes involved in inflammatory responses and increased expression of negative regulators of the immune response. In contrast, the macrophages we characterized appeared to be preprogrammed for silent AC clearance and did not demonstrate large changes in gene expression after engulfment of ACs. We also did not observe increased expression of anti-inflammatory cytokines in pMacs in response to ACs. The Tyro3, Axl, and Mer (TAM) receptors recognize ACs and inhibit TLR signaling by increasing expression of SOCS genes (Rothlin et al., 2007). However, expression of TAM receptors did not decrease in deprogrammed macrophages, and SOCS genes were not increased in programmed macrophages in response to ACs. Therefore, we do not believe that the AC-clearance program involves this receptor family. We propose that macrophages that engulf fewer ACs have fewer requirements for endosomal TLR regulation and therefore can use these alternative mechanisms to prevent responses to ACs. We identified several macrophage populations, including the splenic red pulp macrophages and lung interstitial macrophages, which were not the primary AC engulfers in their tissue and seemed to lack AC clearance programming. Since programmed macrophages may be vulnerable to infection, it is likely beneficial for some tissue macrophages to retain high responsiveness to nucleic acid ligands.
Interestingly, the three AC-engulfing macrophage populations we identified reside in cavities and are motile. Many other tissue-resident macrophage populations are immobilized (Stamatiades et al., 2016). Peritoneal macrophages can be recruited directly from the peritoneal cavity into the liver to repair damage after liver injury (Wang and Kubes, 2016). ACs release find-me signals that recruit phagocytes to dying cells (Elliott et al., 2009; Truman et al., 2008). Perhaps cavity dwelling macrophages can be recruited to dying cells in their cavities as well as neighboring organs. As cavity resident cells these macrophage populations may also have limited exposure to circulating proteins, such as DNASE1L3, which digests DNA in microparticles released from ACs (Sisirak et al., 2016), One interesting possibility is that cavity resident macrophages utilize additional mechanisms to limit responses to ACs because of reduced DNase activity in these compartments.
We demonstrated that the transcription factors KLF2 and KLF4 induced expression of AC recognition receptors as well as negative regulators of TLR signaling. In this way these transcription factors link enhanced AC-engulfment ability with reduced TLR signaling. This coordination may ensure that macrophages with enhanced abilities to engulf ACs do not generate inappropriate responses to self nucleic acids. Mice lacking KLF2 in LysM+ cells display increased levels of proinflammatory cytokines at steady state in their sera and show enhanced inflammation during infection (Mahabeleshwar et al., 2011). Perhaps dysregulation of macrophage AC engulfment ability and endosomal TLR responses contributes to this inflammatory phenotype.
Klf2 and Klf4 expression is not restricted to AC-engulfing macrophages (data from the Immunological Genome Project). Therefore, it is likely that other tissue-induced transcription factors and differences in chromatin structure are involved in induction of the AC-clearance program. Although some transcription factors involved in environmental programming are restricted to a single macrophage population others have more complex expression profiles. For example, the transcription factor PPARγ is required for the development and identity of alveolar macrophages as well as splenic red pulp macrophages (Gautier et al., 2012a; Schneider et al., 2014).
It remains to be determined how tissue environments impart an AC-clearance program. KLF4 expression is increased after macrophage precursors colonize the lung and begin differentiation into alveolar macrophages, suggesting that KLF4 expression is induced by the lung microenvironment (Mass et al., 2016). We were unable to demonstrate a role for the recognition of ACs as the initiating signal (data not shown). However, it may be difficult to recapitulate in vitro the continual engulfment of ACs that occurs in vivo. It is interesting to note that KLF2 and KLF4 expression is induced in endothelial cells following exposure to extracellular ATP released in response to shear stress (Sathanoori et al., 2015). ATP is also a known find-me signal released by ACs that induces phagocyte recruitment (Elliott et al., 2009). Continual exposure of AC-engulfing macrophages to ATP may induce expression of KLF2 and KLF4. We demonstrated that KLF2 and KLF4 increased expression of P2ry2, the receptor that recognizes this find-me signal, suggesting there could be feed forward regulation of ATP detection and expression of KLF2 and KLF4.
Programmed AC-engulfing macrophages had a higher activation threshold for endosomal TLR signaling. This feature likely prevents responses to less stimulatory self ligands, while allowing responses to microbial nucleic acids. Endosomally localized negative regulators of TLR signaling may regulate this threshold. It is also possible that trafficking of endosomal cargo is different in programmed versus deprogrammed macrophages. LC3-associated phagocytosis (LAP) has previously been demonstrated to facilitate phagosome maturation and degradation of internalized pathogens and ACs (Martinez et al., 2011; Sanjuan et al., 2007). Mice with deficiencies in LAP components demonstrate defective clearance of ACs and develop an autoinflammatory disorder (Martinez et al., 2016). Differences in the ability of deprogrammed macrophages to induce LAP may enable responses to AC-derived cargo.
The extent to which regulation of endosomal TLRs in human macrophages parallels that of mice is unclear. Expression of certain endosomal TLRs in human myeloid cells is thought to be more limited than their expression in mice. In human peripheral blood mononuclear cells, TLR7 and TLR9 are exclusively expressed in pDCs, while TLR8 is more broadly expressed in macrophages and conventional DCs (Kadowaki et al., 2001; Krug et al., 2001). These data are often cited to conclude that no human macrophages express these endosomal TLRs. However, given our growing appreciation of tissue macrophage heterogeneity, this concept is worth revisiting. For instance, human decidual macrophages have been shown to express several endosomal TLRs (Duriez et al., 2014). It will be important to expand our understanding of the regulation of nucleic acid sensing TLR expression and function in different human tissue-resident macrophages populations, especially in light of the AC clearance program that we described here. Dysregulated responses by human macrophages that clear ACs could contribute to autoimmune or inflammatory disorders.
Finally, our work suggests that there may be therapeutic value in understanding how the expression of TLRs and regulators of TLR signaling is controlled in specialized cell types. We demonstrated that expression of TLR9 in AC-engulfing macrophages can be modulated by inflammatory signals such as IFNβ and IFNγ. While this induction may enable pathogen detection, we also observed that forced expression of TLR9 could subtly exacerbate chronic inflammation. An IFN-signature has been described in subsets of patients suffering from autoimmune disorders, most notably lupus (Bennett et al., 2003). The link between type I IFN and disease in these patients is clearly complex, but one mechanism may be increased expression of nucleic acid sensing TLRs in macrophages that clear ACs. This continual expression may promote and prolong inflammation. If human AC-engulfing macrophages also utilize KLF2 and KLF4 to limit expression of endosomal TLRs and promote expression of negative regulators of TLR signaling, then increasing KLF2 or KLF4 expression may reduce inflammatory signaling. In this regard, it is notable that statin treatment can increase expression of KLF2 in human peripheral blood monocyte-derived macrophages (Tuomisto et al., 2008). Defining the gene expression programs and unique functional characteristics of distinct tissue macrophage populations will likely reveal additional therapeutic targets and open new avenues for treatment of immune disorders.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gregory Barton (barton@berkeley.edu).
Experimental Model and Subject Details
Mice
Mice were housed under specific pathogen-free conditions at the University of California, Berkeley. All mouse experiments were performed in accordance with the guidelines of the Animal Care and Use Committee at UC Berkeley. Unless noted mice (females and males) were analyzed at 6–12 weeks of age. Timd4−/− mice were obtained from Genentech (Wong et al., 2010). Lyz2cre+/−Klf2fl/fl mice were obtained from Jerry Lingrel (University of Cincinnati) (Weinreich et al., 2009).
Tlr7FLAG:tdTomato mice were generated by constructing a targeting vector encoding a FLAG tag on the 3′ end of the Tlr7 gene followed by IRES-tdTomato sequence. This vector was electroporated into C57BL/6-derived embryonic stem cells by the Mouse Biology Program at UC Davis. The vector also introduced a loxP-flanked neomycin resistance cassette. Targeting was assessed by southern blot and correctly targeted ES cells were injected into ICR/CD1 blastocysts. Chimeric males were mated with C57BL/6 background EIIA-cre females to remove the neomycin resistance cassette. The Tlr9HA:GFP mice were generated in a similar manner except the construct encoded an HA tag on the 3′ end of the Tlr9 gene followed by an IRES-EGFP sequence. To generate Rosalsl-Tlr9 a floxed region (containing eGFP, a Neo cassette, and a transcriptional stop sequence) followed by an HA tagged Tlr9 was inserted into the Rosa26 locus. The construct was generated by cloning cDNA encoding Tlr9 with a C-terminal HA tag into the pBigT vector to introduce a loxP-flanked cassette with a transcriptional stop sequence. The resulting construct was cloned into the ROSA26 eGFP DTA plasmid, thereby replacing DTA with HA tagged Tlr9. pBigT invloxP (pBigT) and ROSA26 eGFP DTA were a kind gift from Martinez-Barbera (Ivanova et al., 2005). The linearized targeting construct was electroporated into C57BL/6 ES cells. Correctly targeted ES clones were identified by southern blot and correctly targeted clones were injected into blastocysts. Chimeric mice were mated to achieve germline transmission.
Primary cell culture
BMMs were differentiated for seven days in RPMI complete media (RPMI-1640 supplemented with 10% (vol/vol) fetal calf serum, L-glutamine, penicillin-streptomycin, sodium pyruvate, and HEPES pH 7.2) supplemented with M-CSF containing supernatant from 3T3-CSF cells.
Unless noted peritoneal macrophages ex vivo for 60 hours, as well as overnight controls, were cultured in 25% (v/v) omentum supernatant in RPMI complete media. Omentum supernatant was generated by harvesting omenta from C57BL/6 mice and culturing the omenta in RPMI complete media at 1ml/omenta overnight. Supernatant was collected, filtered through a 0.22um filter, and frozen at −80°C for future use. Lung and pleural cells were harvested as described above and plated on non-tissue culture treated plates in RPMI complete media.
Method Details
Chimeric mice
Chimeric mice were generated by irradiation of C57BL/6 with 1000rads. Mice were reconstituted with 1x107 bone marrow cells (80% C57BL/6 and 20% CD45.1+ tdTomato+). Mice were analyzed 10–12 weeks after cell transfer.
Tissue harvest
Cells from the peritoneal and pleural cavities were recovered by lavage with ice cold PBS. Spleens and lymph nodes were digested with collagenase XI with DNAse I for 30min and single cell suspensions were generated by mechanical disruption. Perfused lungs were digested with collagenase XI with DNAse I for 45min, single cell suspensions were generated by mechanical disruption through a 100um filter, cells were resuspended in 44% isotonic percoll, underlayed with 67% percoll, and spun at 1550xg without brake. Cells from the interface were collected for analysis. Perfused livers were digested with collagenase VIII with DNase I for 45min, cells were resuspended in Hank’s buffered salt solution and centrifuged at 30xg for 3min. After low speed centrifugation cells in suspension were collected and underlayed with a solution of 25% isotonic Percoll and 50% Percoll, then spun for 15min at 1800xg without brake. Cell counts were obtained using Count Bright absolute counting beads.
Isolation of peritoneal Macrophages
Peritoneal cells were recovered by lavage with 5ml of ice cold PBS. For RNAseq and western blot experiments B cells were first depleted using anti-CD19-biotin antibody and biotin binder dynabeads. Tim-4+ cells were isolated using anti-TIM-4 antibody and anti-rat IgG microbeads. F4/80+ cells were isolated using anti-F4/80 antibody (BM8) and anti-rat IgG microbeads.
Apoptotic cell generation and engulfment
Thymi were harvest from WT or CD45.1+ RosatdTomato mice and single cell suspensions were generated by mechanical disruption through a 70um filter. Cells were irradiated with 600rad and incubated in RPMI complete media at 37°C for 4 hours. Apoptotic cells were then incubated with macrophages at the indicated ratios. For experiments examining AC engulfment capabilities macrophages were allowed to engulf ACs for 60min. For AC injection experiments 0.5e6 CD45.1+ RosatdTomato ACs were injected intraperitoneally per mouse; one hour after injection peritoneal cells were harvested as above.
Cell stimulations
Cells were plated in RPMI complete media. Tissue macrophages were stimulated directly ex vivo, after overnight culture, or after 60 hours in culture. Cells were stimulated with TLR ligands or ACs. For analysis of secreted cytokines, supernatant was collected 7 hours after stimulation. For intracellular cytokine staining, 30 minutes after stimulation brefeldin A (GolgiPlug) was added to cells before incubation for another 4 hours.
For overnight cytokine treatment, cytokines were added to RPMI complete media containing 25% omentum culture supernatant and incubated with macrophages for 16 hours. Concentrations of cytokines used were 10ng/ml IFNγ, 20U/ml IFNβ, 5ng/ml TNF, 50ng/ml IL-6, or 5ng/ml GM-CSF. For dose curve comparison and corresponding western blot IFNβ was used at 100U/ml.
Flow cytometry
Dead cells were excluded using a fixable live/dead stain or DAPI and all stains were carried out in PBS containing 2% FBS (v/v) and 2mM EDTA including anti-CD16/32 Fc blocking antibody and normal mouse serum. Cells were stained for 20min at 4°C with antibodies.
For intracellular TNF or Gata6 staining cells were permeabilized with Fix/Perm buffer (BD or eBioscience respectively) for 20min at 4°C. Cells were then stained with antibodies. For pERK staining cells were immediately fixed after stimulation with 1.6% PFA at room temperature, then washed and resuspended in ice-cold methanol. After overnight incubation at −20°C, cells were stained with p-ERK1/2 biotin followed by Steptavidin-BV42. All cells were analyzed on an LSRII or LSR Fortessa (BD Biosciences), and data was analyzed with FlowJo.
Activated Caspase-3/7 was measured using CellEvent Caspase-3/7 Green Flow Cytometry Assay, according to the manufacturer’s instructions.
BMM transfers
For peritoneal transfer experiments 1e6 differentiated CD45.1+ BMMs were injected into the peritoneum of CD45.2+ mice. Peritoneal cells were harvested 2 or 3 weeks after cell transfer and stimulated with TLR ligands as described above. For lung transfer experiments differentiated 2e6 CD45.1+ tdTomato+ BMMs were intranasally administered on three consecutive days. Lungs were harvested, as described above, 5.5 weeks after cell transfer. For CFSE proliferation experiments CD45.1+ BMMs were labeled with 5μM CFSE in PBS for 10min at 37°C prior to transfer.
Transcription factor overexpression
For retroviral transduction of BMMs a mouse stem cell virus (MSCV)-based retroviral vector was used that expressed the gene of interested followed by IRES- puromycin resistance gene-T2A-EGFP. VSV-G-pseudotyped retrovirus was made in GP2-293 packaging cells. GP2-293 cells were transfected with retroviral vectors and pVSV-G using Lipofectamine LTX. 24 hr post-transfection, cells were incubated at 32°C. 48 hr post-transfection viral supernatant was used to infect bone marrow cells in RPMI complete media containing M-CSF. Bone marrow cells and viral supernatant were spun at 1250xg for 90 min at 32°C then incubated overnight at 32°C. Bone marrow cells were re-transduced with fresh viral supernatant the following day. Two days after last transduction puromycin selection was added for three days.
For lentiviral overexpression a vector with a pHAGE backbone was used that expressed Gata6 followed by IRES-mCherry. Lentiviral particles were produced in 293T cells by transfecting with the lentiviral plasmid and the helper constructs psPAX2 and pCMV-VSV-G. Lentivirus was concentrated by filtering cell supernatant with 0.45μm filter then centrifuging at 47,850×g. Viral pellets were resupended in PBS. 0.5e6 transduction units were injected per mouse. One week after injection peritoneal cells were harvested.
Cas9 genome editing of BMMs
The lentiviral gRNA plasmid pKLV-U6gRNA(BbsI)-PGKpuro2ABFP was used. The sequences of the guide RNA target sites are as follows with the protospacer adjacent motif underlined: Gata6 - ACCGCCTCGGCGTCGAGCTGCGG, Klf2 guide 1 – TTCGCCAGCCCGTGCGAGCGCGG, Klf2 guide 2 - CTGGCCGCGAAATGAACCCGAGG, Klf4 – CTCCACGTTCGCGTCCGGCCCGG, Rarb - TATGGCGTCAGTGCCTGCGAGGG. BMMs from Rosacas9 mice (Jackson Laboratory) were transduced with lentivirus and selected with puromycin.
Pristane
Mice were intraperitoneally injected with 0.5ml of pristane that had been filtered through a 0.22um filter. Mice were analyzed after two weeks or after nine months.
Western Blot Analysis
Cells were lysed in RIPA buffer (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 0.5mM EGTA, 1% NP-40, 1% DOC, 0.1% SDS) containing protease inhibitors. Cell lysates were immunoprecipitated with anti-HA matrix. After washing in RIPA buffer, matrix beads were boiled in SDS-PAGE loading buffer. Protein was run on a 4–15% gel (and transferred to Immobilon-FL membrane. After blocking, membranes were probed with anti-HA antibody, followed by anti-rat-680 secondary or probed with anti-tubulin, followed by anti-mouse-800 secondary. Images were scanned using a Licor Odyssey. Images were quantified using ImageJ.
Enzyme-linked immunosorbent assay (ELISA) and CBA assay
Cytokines were measured using a Cytometric Bead Array assay according to the manufacturer’s instructions.
For anti-dsDNA ELISAs NUNC Maxisorp plates were coated first with poly-l-lysine at 50μg/ml overnight at −20°C, washed, then coated with 1 00μg/ml Calf thymus DNA overnight at 4°C. Plates were then blocked with PBS + 1% BSA (w/v) + 5% goat serum (v/v) at room temperature for four hours before serum samples diluted 1:500 in PBS + 1% BSA (w/v) + 5% goat serum (v/v) were added and incubated overnight at 4°C. Secondary anti-IgG all-biotin was used at 1:10 000 followed by Streptavidin-HRP. Plates were developed with 1mg/ml OPD in Citrate Buffer (PBS with 0.05M NaH2 PO4 and 0.02M Citric acid) with HCl acid stop.
RNA sequencing
Total RNA was prepared from directly ex vivo or deprogrammed (60 hour cultured) Tim-4+ pMacs using RNAzol, Turbo DNAse treated and purified using RNA clean and concentrator columns. 1 ug of total RNA was subsequently used for rRNA depletion using the RiboZero depletion kit to prepare RNA-Seq library by using the TruSeq RNA sample prep kit (Illumina) according to manufacturer’s instructions. 50-cycle single-end RNA sequencing was performed on a HiSeq 4000 (Illumina). Sequenced reads were quality-trimmed (cut-off of 30) with cutadapt and aligned to the mouse transcriptome (GRCm38, release 79) with Kallisto (Bray et al., 2016). Using R and Bioconductor packages (see list in table), counts for each transcript were extracted using Sleuth, collapsed at the gene level, and analyzed for differential expression using DESeq2. Q-values (Benjamini-Hochberg correction) lower than 0.05 were considered significant. Heat maps were generated using MeV (Multiple Experiment Viewer) software.
For the AC stimulation experiment AC derived RNA was analyzed as well as RNA from macrophages ± ACs. Peritoneal macrophages were stimulated with ACs immediately ex vivo (programmed) or after 60 hours in culture (deprogrammed). Two hours after addition of ACs RNA was prepared as above. Genes whose expression was greater than two fold higher in RNA isolated from ACs alone relative to macrophage RNA were considered AC-derived and excluded.
Quantitative PCR
Cells were lysed in RNAzol and RNA was purified according to manufacturer’s instructions and concentrated using a column. cDNA was prepared with iScript cDNA synthesis kit, and quantitative PCR was performed with SYBR Green on a StepOnePlus thermocycler (Applied Biosystems). Primer sequences for Gata6 and Klf4 were obtained from PrimerBank and synthesized by IDT.
For analysis of Cas9 genome editing genomic DNA from BMMs was prepared using a genomic DNA isolation kit. Primers were designed to bind the cut site and normalized using control primers that bind an unrelated site.
mRNA transcript count using Ncounter
The expression levels of a set of genes associated with tissue macrophages, AC clearance, or TLR signaling were analyzed using the NanoString nCounter Analysis System (NanoString Technologies). Raw counts of samples were normalized according to the manufacturer’s recommendations using reference genes as internal controls (Cltc, Gapdh, Gusb, Hprt, Pg1, and Tubb5). Normalization was performed using nSolver Analysis Software v3.0.
Quantification and Statistical Analysis
Statistical analysis was performed with the Prism software (GraphPad software). P-values were determined using unpaired two-tailed Students’s t-test. Where noted t-tests were performed on log transformed data to account for the non-normal distribution of the data. *p < 0.05, **p < 0.01, ***p < 0.001.
Data and Software Availability
The RNA sequencing data have been deposited in the Geo database at https://www.ncbi.nlm.nih.gov/geo/with the accession number GSE93820.
Supplementary Material
Acknowledgments
We thank Dr. Jerry Lingrel for providing Lyz2cre+/−Klf2fl/fl mice. We thank K. Ching for assistance with cloning and R. Vance and members of the Barton and Vance labs for constructive discussions and critical reading of the manuscript. This work was supported by the NIH (AI072429, AI105184 and AI063302 to G.M.B. and AI118841 and AR050256 to M.J.S) and by the Lupus Research Institute (Distinguished Innovator Award to G.M.B.). J.D. was supported by a Long-Term Human Frontier Science Program Fellowship (LT-000081-2013L).
Footnotes
Author Contributions
A.W.R. and G.M.B designed experiments. A.W.R. performed experiments. B.L.L. generated the Tlr9HA:GFP mouse. J.D. analyzed RNA sequencing data. S.J. and M.J.S. generated the Rosalsl-Tlr9 mouse. A.W.R. and G.M.B. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- A-Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, Deniz J, Ramirez C, Diaz M, Gallardo G, et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 2009;31:245–258. doi: 10.1016/j.immuni.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Miwa M, Miwa K, Hanayama R, Nagase H, Nagata S, Tanaka M. Masking of phosphatidylserine inhibits apoptotic cell engulfment and induces autoantibody production in mice. J Exp Med. 2004;200:459–467. doi: 10.1084/jem.20040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratin M, Simon L, Jorquera A, Ghigo C, Dembele D, Nowak J, Gentek R, Wienert S, Klauschen F, Malissen B, et al. T Cell Zone Resident Macrophages Silently Dispose of Apoptotic Cells in the Lymph Node. Immunity. 2017;47:349–362. e5. doi: 10.1016/j.immuni.2017.07.019. [DOI] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Barrat FJ, Elkon KB, Fitzgerald KA. Importance of Nucleic Acid Recognition in Inflammation and Autoimmunity. Annu Rev Med. 2016;67:323–336. doi: 10.1146/annurev-med-052814-023338. [DOI] [PubMed] [Google Scholar]
- Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- Baumann I, Kolowos W, Voll RE, Manger B, Gaipl U, Neuhuber WL, Kirchner T, Kalden JR, Herrmann M. Impaired uptake of apoptotic cells into tingible body macrophages in germinal centers of patients with systemic lupus erythematosus. Arthritis & Rheumatism. 2002;46:191–201. doi: 10.1002/1529-0131(200201)46:1<191::AID-ART10027>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulé MW, Broughton C, Mackay F, Akira S, Marshak-Rothstein A, Rifkin IR. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med. 2004;199:1631–1640. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Calvani N, Caricchio R, Tucci M, Sobel ES, Silvestris F, Tartaglia P, Richards HB. Induction of apoptosis by the hydrocarbon oil pristane: implications for pristane-induced lupus. J Immunol. 2005;175:4777–4782. doi: 10.4049/jimmunol.175.7.4777. [DOI] [PubMed] [Google Scholar]
- Coch C, Busch N, Wimmenauer V, Hartmann E, Janke M, Abdel-Mottaleb MMA, Lamprecht A, Ludwig J, Barchet W, Schlee M, et al. Higher activation of TLR9 in plasmacytoid dendritic cells by microbial DNA compared with self-DNA based on CpG-specific recognition of phosphodiester DNA. Journal of Leukocyte Biology. 2009;86:663–670. doi: 10.1189/jlb.0509314. [DOI] [PubMed] [Google Scholar]
- Cummings RJ, Barbet G, Bongers G, Hartmann BM, Gettler K, Muniz L, Furtado GC, Cho J, Lira SA, Blander JM. Different tissue phagocytes sample apoptotic cells to direct distinct homeostasis programs. Nature. 2016:1–21. doi: 10.1038/nature20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini L, Pagliara P, Carlà EC. Phagocytosis of apoptotic cells by liver: a morphological study. Microsc Res Tech. 2002;57:530–540. doi: 10.1002/jemt.10107. [DOI] [PubMed] [Google Scholar]
- Duriez M, Quillay H, Madec Y, Costa El H, Cannou C, Marlin R, de Truchis C, Rahmati M, Barré-Sinoussi F, Nugeyre M-T, et al. Human decidual macrophages and NK cells differentially express Toll-like receptors and display distinct cytokine profiles upon TLR stimulation. Front Microbiol. 2014;5:316–316. doi: 10.3389/fmicb.2014.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF, Park D, Woodson RI, Ostankovich M, Sharma P, et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature. 2009;461:282–286. doi: 10.1038/nature08296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Fond AM, Ravichandran KS. Advances in Experimental Medicine and Biology. Cham: Springer International Publishing; 2016. Clearance of Dying Cells by Phagocytes: Mechanisms and Implications for Disease Pathogenesis; pp. 25–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-de-Lima CG, Xiao YQ, Gardai SJ, Bratton DL, Schiemann WP, Henson PM. Apoptotic cells, through transforming growth factor-beta, coordinately induce anti-inflammatory and suppress pro-inflammatory eicosanoid and NO synthesis in murine macrophages. J Biol Chem. 2006;281:38376–38384. doi: 10.1074/jbc.M605146200. [DOI] [PubMed] [Google Scholar]
- Gautier EL, Chow A, Spanbroek R, Marcelin G, Greter M, Jakubzick C, Bogunovic M, Leboeuf M, van Rooijen N, Habenicht AJ, et al. Systemic analysis of PPARγ in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. The Journal of Immunology. 2012a;189:2614–2624. doi: 10.4049/jimmunol.1200495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Ivanov S, Williams JW, Huang SCC, Marcelin G, Fairfax K, Wang PL, Francis JS, Leone P, Wilson DB, et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. Journal of Experimental Medicine. 2014;211:1525–1531. doi: 10.1084/jem.20140570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012b;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miyasaka K, Aozasa K, Koike M, Uchiyama Y, Nagata S. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- Ivanova A, Signore M, Caro N, Greene NDE, Copp AJ, Martinez-Barbera JP. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis. 2005;43:129–135. doi: 10.1002/gene.20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski KA, Amici SA, Webb LM, de Ruiz-Rosado JD, Popovich PG, Partida-Sanchez S, Guerau-de-Arellano M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE. 2015;10:e0145342. doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J Exp Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KL, Jewell CM, Bortner CD, Cidlowski JA. 28S ribosome degradation in lymphoid cell apoptosis: evidence for caspase and Bcl-2-dependent and -independent pathways. Cell Death Differ. 2000;7:994–1001. doi: 10.1038/sj.cdd.4400731. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33:449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- Krug A, Towarowski A, Britsch S, Rothenfusser S, Hornung V, Bals R, Giese T, Engelmann H, Endres S, Krieg AM, et al. Toll-like receptor expression reveals CpG DNA as a unique microbial stimulus for plasmacytoid dendritic cells which synergizes with CD40 ligand to induce high amounts of IL-12. Eur J Immunol. 2001;31:3026–3037. doi: 10.1002/1521-4141(2001010)31:10<3026::aid-immu3026>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter EAE, Rifkin IRI, Hohlbaum AMA, Beaudette BCB, Shlomchik MJM, Marshak-Rothstein AA. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, Barton GM. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PY, Kumagai Y, Li Y, Takeuchi O, Yoshida H, Weinstein J, Kellner ES, Nacionales D, Barker T, Kelly-Scumpia K, et al. TLR7-dependent and FcgammaR-independent production of type I interferon in experimental mouse lupus. Journal of Experimental Medicine. 2008;205:2995–3006. doi: 10.1084/jem.20080462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahabeleshwar GH, Kawanami D, Sharma N, Takami Y, Zhou G, Shi H, Nayak L, Jeyaraj D, Grealy R, White M, et al. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity. 2011;34:715–728. doi: 10.1016/j.immuni.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, Jacome-Galarza CE, Händler K, Klughammer J, Kobayashi Y, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016:353. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PP, Fadok VA, Bratton D, Henson PM. Transcriptional and translational regulation of inflammatory mediator production by endogenous TGF-beta in macrophages that have ingested apoptotic cells. J Immunol. 1999;163:6164–6172. [PubMed] [Google Scholar]
- McGaha TL, Chen Y, Ravishankar B, Rooijen NV, Karlsson MCI. Marginal zone macrophages suppress innate and adaptive immunity to apoptotic cells in the spleen. Blood. 2011;117:5403–5412. doi: 10.1182/blood-2010-11-320028. [DOI] [PubMed] [Google Scholar]
- Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Asano K, Kaise H, Uemura M, Nakayama M, Tanaka M. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J Clin Invest. 2007;117:2268–2278. doi: 10.1172/JCI31990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- Mouchess ML, Arpaia N, Souza G, Barbalat R, Ewald SE, Lau L, Barton GM. Transmembrane Mutations in Toll-like Receptor 9 Bypass the Requirement for Ectodomain Proteolysis and Induce Fatal Inflammation. Immunity. 2011;35:721–732. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka KK, Sawamura TT, Kikuta KK, Itokawa SS, Kume NN, Kita TT, Masaki TT. Lectin-like oxidized low-density lipoprotein receptor 1 mediates phagocytosis of aged/apoptotic cells in endothelial cells. Proc Natl Acad Sci USA. 1998;95:9535–9540. doi: 10.1073/pnas.95.16.9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Jung MY, Kim HJ, Lee SJ, Kim SY, Lee BH, Kwon TH, Park RW, Kim IS. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 2008;15:192–201. doi: 10.1038/sj.cdd.4402242. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzanet R, Sanjuan MA, Wu HY, Quintana FJ, Xiao S, Anderson AC, Weiner HL, Green DR, Kuchroo VK. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proc Natl Acad Sci USA. 2010;107:8706–8711. doi: 10.1073/pnas.0910359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas M, Davies LC, Giles PJ, Liao CT, Kharfan B, Stone TC, O’Donnell VB, Fraser DJ, Jones SA, Taylor PR. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin CV, Ghosh S, Zuniga EI, Oldstone MBA, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Sathanoori R, Rosi F, Gu BJ, Wiley JS, Müller CE, Olde B, Erlinge D. Shear stress modulates endothelial KLF2 through activation of P2X4. Purinergic Signal. 2015;11:139–153. doi: 10.1007/s11302-014-9442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- Scott RS, McMahon EJ, Pop SM, Reap EA, Caricchio R, Cohen PL, Earp HS, Matsushima GK. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 2001;411:207–211. doi: 10.1038/35075603. [DOI] [PubMed] [Google Scholar]
- Sharma S, Fitzgerald KA, Cancro MP, Marshak-Rothstein A. Nucleic Acid-Sensing Receptors: Rheostats of Autoimmunity and Autoinflammation. J Immunol. 2015;195:3507–3512. doi: 10.4049/jimmunol.1500964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisirak V, Sally B, D’Agati V, Martinez-Ortiz W, Özçakar ZB, David J, Rashidfarrokhi A, Yeste A, Panea C, Chida AS, et al. Digestion of Chromatin in Apoptotic Cell Microparticles Prevents Autoimmunity. Cell. 2016;166:88–101. doi: 10.1016/j.cell.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatiades EG, Tremblay ME, Bohm M, Crozet L, Bisht K, Kao D, Coelho C, Fan X, Yewdell WT, Davidson A, et al. Immune Monitoring of Trans-endothelial Transport by Kidney-Resident Macrophages. Cell. 2016;166:991–1003. doi: 10.1016/j.cell.2016.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers SA, Hoi A, Steinmetz OM, O’Sullivan KM, Ooi JD, Odobasic D, Akira S, Kitching AR, Holdsworth SR. TLR9 and TLR4 are required for the development of autoimmunity and lupus nephritis in pristane nephropathy. Journal of Autoimmunity. 2010;35:291–298. doi: 10.1016/j.jaut.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7 doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Truman LA, Ford CA, Pasikowska M, Pound JD, Wilkinson SJ, Dumitriu IE, Melville L, Melrose LA, Ogden CA, Nibbs R, et al. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood. 2008;112:5026–5036. doi: 10.1182/blood-2008-06-162404. [DOI] [PubMed] [Google Scholar]
- Tuomisto TT, Lumivuori H, Kansanen E, Häkkinen SK, Turunen MP, van Thienen JV, Horrevoets AJ, Levonen AL, Ylä-Herttuala S. Simvastatin has an anti-inflammatory effect on macrophages via upregulation of an atheroprotective transcription factor, Kruppel-like factor 2. Cardiovasc Res. 2008;78:175–184. doi: 10.1093/cvr/cvn007. [DOI] [PubMed] [Google Scholar]
- Uderhardt S, Herrmann M, Oskolkova OV, Aschermann S, Bicker W, Ipseiz N, Sarter K, Frey B, Rothe T, Voll R, et al. 12/15-Lipoxygenase Orchestrates the Clearance of Apoptotic Cells and Maintains Immunologic Tolerance. Immunity –. 2012 doi: 10.1016/j.immuni.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Wang J, Kubes P. A Reservoir of Mature Cavity Macrophages that Can Rapidly Invade Visceral Organs to Affect Tissue Repair. Cell. 2016;165:668–678. doi: 10.1016/j.cell.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao Y, Yuan X, Xia W, Luo Y, Sun E, Chen ZK. The liver mediates apoptotic cell-induced immune regulation. Scand J Immunol. 2008;68:297–305. doi: 10.1111/j.1365-3083.2008.02141.x. [DOI] [PubMed] [Google Scholar]
- Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermeling F, Chen Y, Karlsson MCI. Class A scavenger receptors regulate tolerance against apoptotic cells, and autoantibodies against these receptors are predictive of systemic lupus. J Exp Med. 2007;204:2259–2265. doi: 10.1084/jem.20070600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Valdez PA, Tan C, Yeh S, Hongo JA, Ouyang W. Phosphatidylserine receptor Tim-4 is essential for the maintenance of the homeostatic state of resident peritoneal macrophages. Proc Natl Acad Sci USA. 2010;107:8712–8717. doi: 10.1073/pnas.0910929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Richez C, Uccellini MB, Richards RJ, Bonegio RG, Akira S, Monestier M, Corley RB, Viglianti GA, Marshak-Rothstein A, et al. Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. The Journal of Immunology. 2009;183:3109–3117. doi: 10.4049/jimmunol.0900399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.