Abstract
Thymidylate is synthesized de novo in all living organisms for replication of genomes. The chemical transformation is reductive methylation of deoxyuridylate at C5 to form deoxythymidylate. All eukaryotes including humans complete this well-understood transformation with thymidylate synthase utilizing 6R-N5-N10-methylene-5,6,7,8-tetrahydrofolate as both a source of methylene and a reducing hydride. In 2002, flavin-dependent thymidylate synthase was discovered as a new pathway for de novo thymidylate synthesis. The flavin-dependent catalytic mechanism is different than thymidylate synthase because it requires flavin as a reducing agent and methylene transporter. This catalytic mechanism is not well-understood, but since it is known to be very different from thymidylate synthase, there is potential for mechanism-based inhibitors that can selectively inhibit the flavin-dependent enzyme to target many human pathogens with low host toxicity.
Keywords: Thymidylate, Flavin, Thymidylate synthase, Enzyme, Mechanisms, N5-flavin adduct
1. Introduction
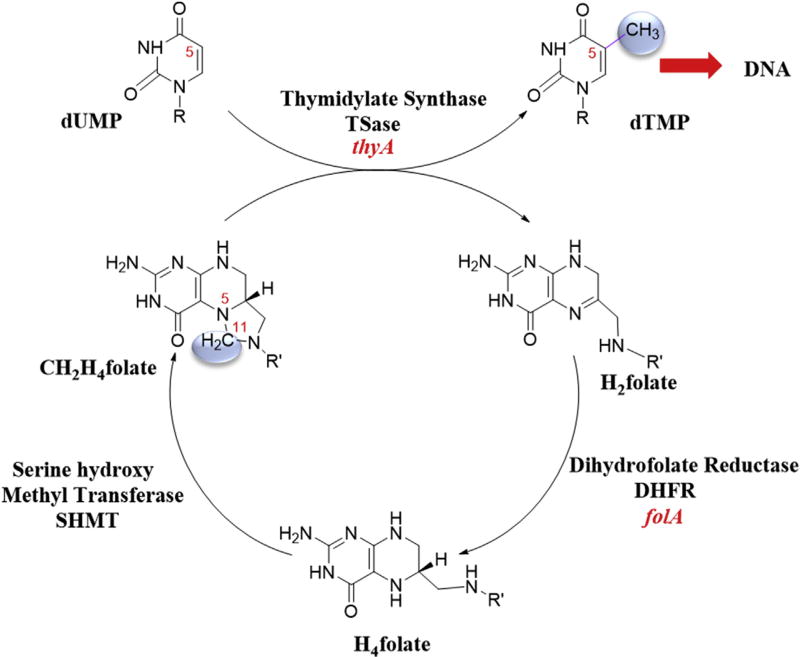
Thymidylate (2′-deoxythymidine-5′-monophosphate; dTMP) cannot be directly synthesized from its RNA counterpart by ribonucleotide reductase. Thus, it is synthesized de novo in all living organisms. The last step of the thymidylate synthesis is catalyzed by the enzyme Thymidylate Synthase (TSase; classical TSase EC 2.1.1.45) encoded by thyA. TSase catalyzes the reductive methylation at C5 of the uracil moiety of dUMP (2′-deoxyuridine-5′-monophosphate). The source of the methylene carbon and reducing hydride for the conversion is 6R-N5-N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4folate; MTHF) [1]. At the end of each catalytic cycle of TSase, CH2H4folate is converted to dihydrofolate (H2folate; DHF) and the cellular CH2H4folate pool is replenished by the activity of Dihydrofolate Reductase (DHFR) (coded by folA) and Serine hydroxymethyltransferase (SHMT) consecutively (Scheme 1).
Scheme 1.
Enzymes involved in reductive methylation of dUMP and the coupled cycle for CH2H4folate regeneration. Highlighted in blue is the methylene being transferred from N5 of CH2H4folate to C5 of dUMP.
Thymidylate synthesis is crucial in cell proliferation. Hence, the enzymes involved in this biosynthetic pathway are targets for chemotherapeutic and antibiotic drug design. Chemotherapeutic agents such as 5-fluorouracil (5-F-dUMP) and Raltitrexed target classical TSase for skin, colon, and ovarian cancers [2,3]. Methotrexate, used clinically for treating cancer and autoimmune disorders, targets DHFR and trimethoprim is specific to bacterial DHFR inhibition [4,5].
Until 2002 thymidylate was thought to be synthesized either de novo by TSase or by scavenging exogenous thymidine compounds by the action of thymidine kinase (Tdk). Genomic analysis on archaea and bacteria revealed many organisms lacking thyA do not carry folA as well as the gene coding for thymidine kinase, yet they survived in thymidine deficient media [6]. This observation led to the discovery of a new gene thyX, which codes for a different class of thymidylate synthases: Flavin-Dependent Thymidylate Synthase (FDTS, EC 2.1.1.148) [6].
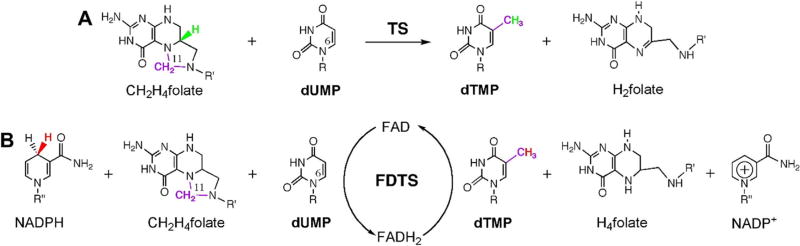
Besides flavin (flavin adenine dinucleotide; FAD) dependence, a striking difference between classical TSase and FDTS is the role of CH2H4folate. In TSase, CH2H4folate is both the methylene and hydride source to form dTMP (Scheme 2A) [7], however in FDTS it serves only as the carbon source. FDTS catalyzed dTMP formation can be dissected into two half-reactions with respect to the oxidation state of flavin. In the reductive half-reaction (FAD → FADH2), flavin is reduced by NADPH (nicotinamide adenine dinucleotide phosphate) and during the oxidative half-reaction flavin is oxidized to form the product dTMP. Unlike TSase, at the end of each catalytic cycle of FDTS, CH2H4folate is converted to H4folate (Scheme 2B) instead of H2folate.
Scheme 2.
Thymidylate Synthase reactions. (A) The reaction catalyzed by classical Thymidylate Synthase (TSase; TS). (B) Reaction catalyzed by FDTS enzyme. R = 2′-deoxyribose-5′-phosphate; R′ = (p-aminobenzoyl)-glutamate; R″ = adenosine-5′-pyrophosphate-ribityl. Reprinted with permission from Ref. [7].
It was postulated that FDTS might be a bifunctional enzyme with both TSase and DHFR activity [8,9]. Several experimental observations refuted this claim: (i) when R–[6-3H]-CH2H4folate is used, the product dTMP was not labeled with tritium and the tritium was exclusively present on H4folate [10]; (ii) in D2O, FDTS formed singly deuterated dTMP, suggesting proton exchange occurs between reduced flavin and the solvent before it transfers the hydride to the nucleotide [10,11]; (iii) when tritiated NADPH (R- and S-[4-3H] NADPH) was used in the reaction, all of the tritium was in water rather than in H4folate or dTMP [10]. Observation (iii) can be attributed to hydride transfer from NADPH to N5 of FAD, forming FADH2, at which stage exchange with water happens. These observations ruled out the possibility of FDTS serving as a bifunctional enzyme in organisms lacking thyA/folA genes.
The gene thyX is present in ~30% of microorganisms including severe pathogens (e.g. Mycobacterium tuberculosis, Rickettsia prowazekii, and Helicobacter pylori) [6]. Some of these pathogens rely solely on FDTS for thymidylate synthesis (e.g. Rickettsia sp. and H. pylori). TSase and FDTS are dissimilar in both primary sequence and structure (Fig. 1), and their catalytic mechanisms differ significantly. Thus, FDTS is identified as an excellent antibiotic target. Flavin-dependent thymidylate biosynthesis encourages investigation into a separate family of flavoprotein and thymidylate synthase folds. This mechanism is complicated with flavin capabilities, but provides an opportunity for inhibitor design targeting pathogen propagation.
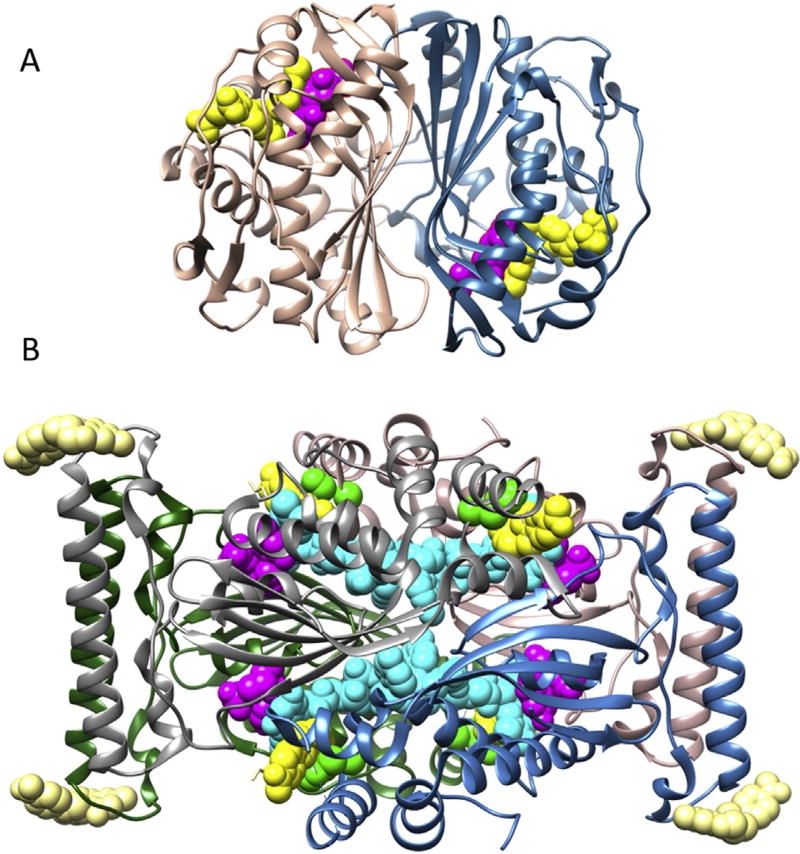
Fig. 1.
Thymidylate synthase structures. (A) Classical Thymidylate Synthase (TSase) is a homodimer with two identical active sites each binding dUMP and CH2H4folate. (B) Flavin-Dependent Thymidylate Synthase (FDTS) is a homotetramer with four identical active sites each binding dUMP, CH2H4folate, and FAD with possible allosteric sites for CH2H4folate. Each subunit is colored differently and ligand coloring is as follows: His53 (green), folate (yellow), FAD (cyan), and dUMP (magenta). PDB codes are 2KCE (TSase) and 4GT9 (FDTS).
2. Structural peculiarities of a flavoprotein
Since 2002, genome comparison and forty-two crystallographic structures of oxidized Thermatoga maritima (Tm), M. tuberculosis (Mtb), Paramecium bursaria Chlorella virus 1 (PBCV-1), Streptomyces cacaoi (Sc), H. pylori (Hp), and Corynebacterium glutamicum (Cg) FDTS indicate an enzyme unrelated to TSase, DHFR, and other flavoproteins [6,11–24]. The gene thyX produces an assembly with less than 30% sequence identity to TSase, DHFR, and all known proteins while producing a unique fold (Fig. 1) [12,13].
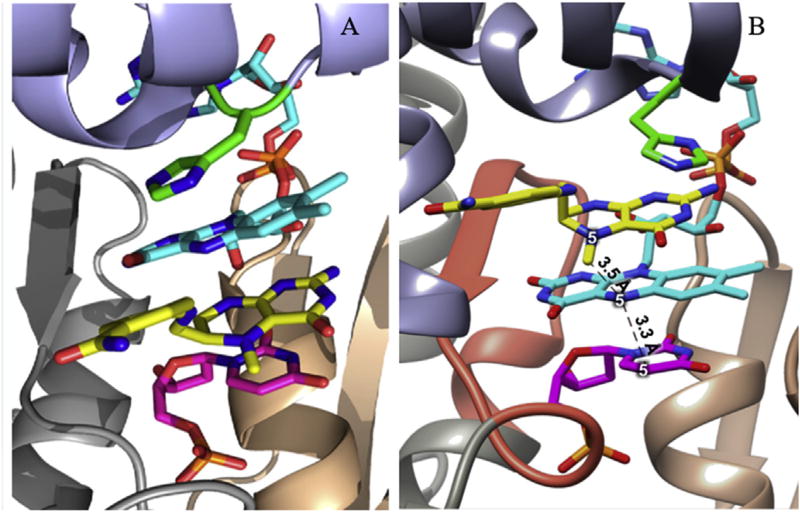
The center of the FDTS homotetramer is a large void intersecting four identical active sites each created at interfaces of three subunits and each binding dUMP, CH2H4folate, and FAD (Fig. 1B) [12–14,16,17,25]. The reactive configuration of ligands was first inspired by TSase where dUMP and folate are neighboring (Fig. 2A) [17], however this is false and the known reactive configuration of ligands is shown in Fig. 2B [17,26]. Crystallographic structures have agreed on <4 Å gaps between the central ring of FAD's isoalloxazine system and the neighbors it is π-stacked between: the pyrimidine ring of dUMP and the pterin ring of folate (Fig. 2B) [7,11,13,17,18,27–29]. Notably although not catalytically required, His53 (TmFDTS) stacks with folate (Fig. 2B).
Fig. 2.
Active site view of TmFDTS-dUMP-FAD-folate complex. The folate derivative is folinic acid, an analogue of the reactive form of CH2H4folate. The carbonyl oxygen of folinic acid has been omitted for clarity. (A) Folinic acid and FAD modeled in opposite locations with His53 rotated and C2′–C3′ and C1′-N10 of FAD rotated to allow for binding. (B) Crystallographic structure of FDTS active site with known reactive locations of all substrates and the FAD binding motif shown as a red ribbon. Color scheme is the same as Fig. 1. PDB code is 4GTA. Figure 2A reproduced with permission from Ref. [17].
Active site nuances of FDTS are intriguing because FDTS performs the same net catalysis as TSase and DHFR, yet neither TSase nor DHFR use an FAD cofactor. A rigid, preformed helix-loop-strand FAD binding motif, a novel feature for redox-active flavoenzymes, has been characterized for FDTS through studies with apoenzyme, apoenzyme with dUMP or dUMP analogues, holoenzyme, and holoenzyme with dUMP or dUMP analogues [13]. Additionally, Dym and Eisenberg [30] examined relationships between sequence and structure of thirty-two families of FAD binding proteins, determining four families of FAD folds. Each of the four families was identified with a specific conserved sequence motif characteristic of the pyrophosphate binding site [13]. The FAD fold of FDTS is not contained within these four FAD fold families, thus indicating a unique FAD binding domain for redox flavoenzymes corresponding to amino acids residues 75–91 for TmFDTS (shown red in Fig. 2B) [13]. Part of this unique peptide segment was characterized for T. maritima and PBCV-1 and has become known as the substrate binding loop (residues 86–97 TmFDTS, partially shown in Fig. 2B) and plays an integral role in dUMP binding [13,17,18]. The sequence of residues 75–97 for TmFDTS is QWFRHRIASYNELSGRYSKLSYE [17]. This flexible region becomes ordered when dUMP binds and its relevance to inhibitor design is discussed later in this review. Additionally, the FAD binding motif of FDTS is unlike other flavoproteins in that FAD ligands are paired among two subunits where two FAD ligands interact through the ribose of AMP leaving the AMP moieties near one another, but having no direct interactions [13,19,31,32]. Also, pyrophosphate hydrogen bonding interactions which hold the tail of the flavin tight to FDTS are known to be specific to FDTS [13,30]. Tight tail binding allows for active site plasticity because it needs to accommodate FAD and FADH2 which leaves the isoalloxazine ring of the flavin solvent accessible [13]. This differs from many flavoproteins which tightly bind the isoalloxazine ring [19].
Structural features of the FDTS active site and possible allosteric sites complicate experiments related to kinetics and inhibitor design. A series of crystal structures published in 2012 have evidence for CH2H4folate allosteric sites at the periphery (light yellow MTHF in Fig. 1B) of TmFDTS [17]. These allosteric sites include three hydrogen bonds with amino acid residues and one with a water molecule [17]. These sites have not been shown to be catalytically important and may continue to be a hurdle by complicating the kinetic pathway by allosteric cooperativity [17]. The folate binding site has been proposed to be compatible with NADPH through docking experiments showing no steric clashes with multiple NADPH orientations, an important observation relevant to the reductive half-reaction for FDTS and in accordance with known flavin and folate dependent enzymes [17,18,27]. This has been corroborated with kinetic data [33]. The only successful attempt at obtaining crystallographic structures with NADP+ or NADPH bound showed for MtbFDTS that NADP+ replaced 5-Br-dUMP and FAD from all active sites, the adenine moieties were deeply buried, which left the nicotinamide ring exposed with some conformational freedom, and the binding mode among three subunits was analogous to FAD [20]. Collectively, structural observations indicate a complex, well-ordered active site with three ligands (reductive half-reaction), which posits rare flavin chemistry and new avenues for mechanism-based inhibitors.
3. Kinetic considerations
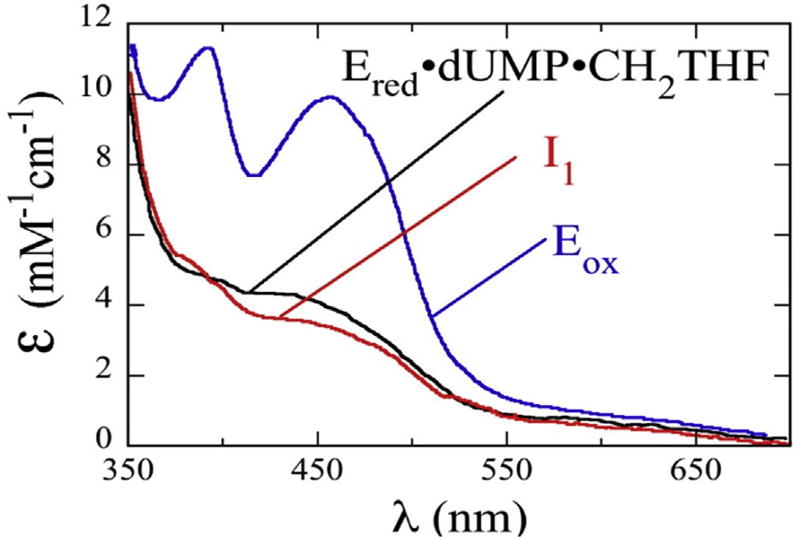
As isolated, FDTSs from many but not all organisms have flavin bound and appear yellow, which indicates flavin is oxidized, however upon reduction (e.g. dithionite, NADPH, ferrodoxin) the yellow color vanishes, which indicates reduced flavin. Spectrophotometric studies have shown noticeable changes in the ultraviolet–visible (UV–vis) absorption spectra and the presence of reaction intermediates for the TmFDTS catalyzed formation of dTMP (oxidative half-reaction; Fig. 3) [15,34,35]. Under anaerobic conditions, addition of either substrate, dUMP or CH2H4folate, to reduced enzyme first leaves the UV–vis spectrum relatively unchanged, however addition of the second substrate significantly changes the absorbance of the FDTS complex (black trace in Fig. 3). Once the reaction progresses (red trace in Fig. 3), an intermediate involving a slight change in flavin hydroquinone electronic environment without a change in redox state of the flavin is observed [34]. After the reaction is complete, the absorbance spectrum indicates oxidation of the flavin [6,34,35].
Fig. 3.
Absorbance spectra of FDTS catalysis. These are deconvoluted spectra generated from using singular-value decomposition of raw absorption data fit to a two-step reaction. The blue trace is oxidized enzyme, the black trace is the reduced enzyme complex, and the red trace is suggested to be a bound flavin intermediate. Adapted from Ref. [34].
Initial velocity studies with NADPH as the reducing agent showed substrate inhibition by dUMP and NADPH [10,16]. Rates of FAD reduction and FADH2 oxidation depend on the presence of dUMP since dUMP is known to be an activator of the reductive half-reaction of FDTS [36]. Binding of dUMP stabilizes a conformation of FAD in which FAD can interact with NADPH more favorably to accelerate FAD reduction [15,36].
Common for flavoproteins, FDTS has oxidase functionality, as it catalyzes the reduction of molecular oxygen (O2) to hydrogen peroxide (H2O2) with NADPH, among other reducing agents [37,38]. Examining oxidase activity with respect to activation kinetics of dUMP and CH2H4folate has proven useful for elucidating binding constants, and the order and release of cofactors and products for TmFDTS [37]. FDTS exhibits oxidase activity with normal Michaelis-Menten kinetics and an apparent Km, O2 = 29 ± 2 µM at 65 °C [37]. This is similar to flavoproteins which have oxidase activity as the primary function (ex: vanillyl-alcohol oxidase Km, O2 = 28 µM and dimethylglycine oxidase Km, O2 = 41 ± 3 µM) [39,40], which suggests FDTS has an O2 binding site or at very high O2 concentration O2 dependent steps become kinetically favored [7,37,38]. CH2H4folate and O2 compete for the FDTS-dUMP-FADH2-NADP+ complex and at 2 µM CH2H4folate concentration the oxidase activity gets inhibited [7,37,41].
4. Chemical mechanisms of FDTS
Throughout the years, classical TSase has been studied extensively, and its mechanism is well established (Scheme 3a). First, protonation of N10 of CH2H4folate forms a reactive iminium ion (step 1). Next, the pyrimidine ring of dUMP is activated via Michael addition at C6 by a conserved active site cysteine (C146 in Escherichia coli) (step 2). This makes the C5 nucleophilic and it attacks the Schiff base iminium ion of CH2H4folate to form an enzyme-bound covalent bridged ternary complex (step 3). Proton abstraction from C5 of dUMP (Step 4) followed by hydride transfer from H4folate to the enzyme bound exocyclic methylene complex (step 5) yields the products, dTMP and H2folate. The mechanism based chemotherapeutic drug 5-F-dUMP inhibits classical TSase by forming a “dead-end” complex (step 4) obstructing the formation of the exocyclic methylene intermediate [1,42].
Scheme 3.
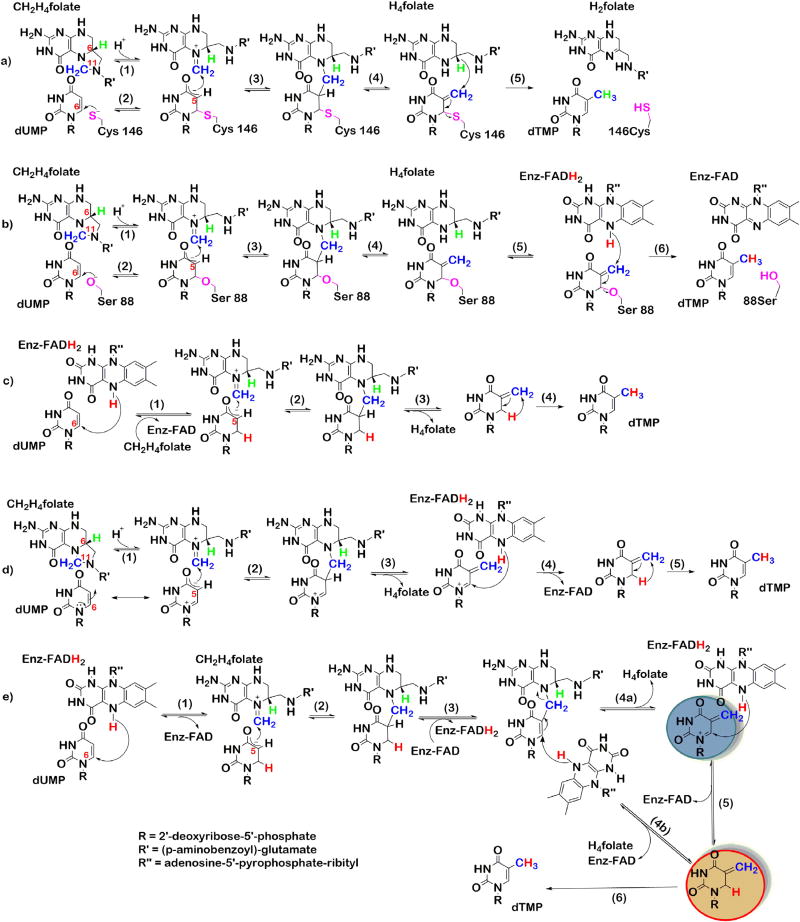
TSase mechanisms; a). classical TSase. b). FDTS where serine acts as the nucleophile. c). mechanism for FDTS where hydride of flavin acts as the nucleophile. d). substrate polarization in the active-site of FDTS. e). mechanism that combines c and d to explain the proton exchange at C5 of dUMP. Adapted with permission from Ref. [29].
Mechanisms b, c, d, and e in Scheme 3 [29] demonstrate proposed mechanisms (up until 2016) for the oxidative half-reaction of FDTS catalyzed thymidylate synthesis. Unlike TSase, FDTS does not have a cysteine in its active site. However, sequence alignment between different thyX genes suggested the only nucleophilic candidate in close proximity (~4 Å) to the C6 of dUMP is a conserved serine residue [10,13]. Hence, an analogous mechanism (Scheme 3b) to classical TSase was proposed for FDTS. In this mechanism, serine (S88 in TmFDTS) acts as the enzymatic nucleophile to activate the dUMP ring for the subsequent Mannich-type condensation with the iminium ion on CH2H4folate. Instead of H4folate, FADH2 reduces the exocyclic methylene intermediate to form the final product dTMP [9].
Mutational studies on TmFDTS revealed both S88A and S88C mutations resulted in active proteins (apparent kcat S88A was 17-fold slower and for S88C it was 400-fold slower than the WT), which suggests that serine is not the enzymatic nucleophile responsible for the activation of the dUMP ring [11]. Activity of the mutant proteins and the absence of contamination from classical TSase suggested serine might not be the active site nucleophile. Since there are no other nucleophilic residues in proximity to C6 of dUMP, either water (hydroxide), the hydride of reduced flavin, or N5 of the reduced flavin was suspected to be the nucleophile. However, there are no basic residues to deprotonate water in the active site [13]. In addition, 5-carba-5-deaza-FAD substituted FDTS was still active, which rules out the possibility of N5 of reduced flavin being the nucleophile. In an attempt to follow the flow of hydrides during FDTS catalysis, the reaction was carried out in D2O (>99.6% D) at 37 °C and at 65 °C (which is closer to the physiological temperature of T. maritima). Both temperatures lead to a singly deuterated dTMP product. 1H NMR and 2H NMR data indicated that at the lower temperature, 60% of the dTMP had a deuterium at C6, while the remaining 40% of the dTMP had a deuterium at C7. At 65 °C, the deuteriumwas solely present at C7 [11]. Formation of 6D-dTMP by deuteride transfer from N5 of flavin is supported by the crystal structure (4GTA), where the distance between C6 of dUMP and N5 of flavin is 3.4 Å [13]. All this information suggested a novel mechanism for FDTS (Scheme 3c), where the hydride of the reduced flavin acts as the nucleophilic activator (step 1). Methylene transfer from CH2H4folate (step 3) is followed by 1,3-hydride shift to yield the product dTMP (step 4) [11].
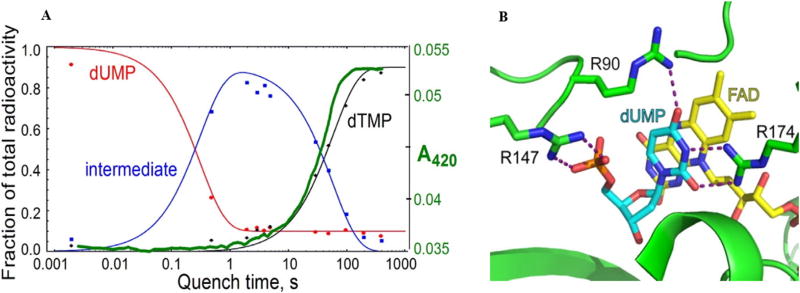
If the activation of dUMP is the first step in the oxidative half-reaction (Scheme 3c, step 1), flavin needs to get oxidized when incubated with dUMP. However, spectroscopic studies on reduced FDTS revealed flavin does not become oxidized when incubated with 5-F-dUMP (an analogue of dUMP that does not hinder the initial Michael addition step) (Fig. 3) [34]. Oxidation of flavin requires both dUMP and CH2H4folate to be bound to the protein. In addition, the oxidation of flavin takes place slightly before the product starts to accumulate (Fig. 4A, green trace) [43].
Fig. 4.
A) Single-turnover FDTS reaction kinetics overlaid with stopped-flow flavin absorbance trace (green). Reduced flavin (FADH2) has no 420 nm absorbance, while oxidized flavin (FAD) does. B) the arrangement of R90, R174, and the phosphate of dUMP (cyan) is proposed to stabilize a resonance form of dUMP with enhanced nucleophilicity at C5. Also, note the proximity of the phosphate oxygens to C5 of the uracil. PDB code 1O26. Figures reprinted with permission from Refs. [43 and 21], respectively.
These observations prompted further investigation on what the nucleophilic activator might be. In the FDTS active site, dUMP binds in a curled conformation where the phosphate group is pointing toward the C6 and the oxygen atoms of the phosphate moiety are in close enough proximity to C6 within 3.5 and 4 Å of it to perform a nucleophilic attack (Fig. 4B) [34]. To test this possibility, 5-dUMPS (phosphothioate analogue of dUMP) was used, where, a terminal oxygen atom is replaced with a more nucleophilic sulfur atom. This modified substrate did not increase the rate of product formation, which is inconsistent with the phosphate moiety serving as the nucleophile.
Efforts toward the identification of the potential nucleophile for the activation of the dUMP ring were unsuccessful. Thus, it was postulated that upon binding, dUMP is electronically polarized in the active site of FDTS and does not require any activation by nucleophilic attack at C6. Negative charge on the phosphate group near N1 and C6 of dUMP and positive charges from R90 and R174 (TmFDTS) near carbonyl oxygen of C4 could stabilize and induce the polarization of dUMP, resulting in higher electron density at C5 (Fig. 4B) [34]. Additionally, recently published 13C-NMR data on TmFDTS-dUMP complex revealed N3 of dUMP is deprotonated. This causes C4 and C2 carbonyl carbons to shift ~8 ppm downfield (with 13C and 15N labeled dUMP) with respect to free dUMP in solution at pH 8 [21]. This observation further supports the role of R174 in stabilizing the negative charges on N3 and C4 carbonyl oxygen. The authors argue that deprotonation of N3 makes any negative charge at C5 more localized. As a result, C5 is a better nucleophile for the condensation with the CH2H4folate. With these observations in hand, mechanism d was proposed. In this mechanism upon binding, polarized dUMP nucleophilically attacks the iminium ion (step 2, Scheme 3d) of CH2H4folate without the aid of any nucleophilic activation as previously proposed. This mechanism is consistent with oxidation of flavin taking place at a later stage of the mechanism (step 4, Scheme 3d). Furthermore, studies by Frederick et al. showed the phosphate moiety of dUMP might be the base that abstracts the proton from C5 of dUMP to yield the exocyclic methylene intermediate [21].
Mishanina et al. isolated and characterized an acid-trapped intermediate of TmFDTS (intermediate kinetics are shown in Fig. 4A; dUMP trace is in red and product dTMP in black) [44]. Formation of the trapped species 5-hydroxymethyl-dUMP (5-HM-dUMP, blue trace Fig. 4A) can be explained via either of the mechanisms c or d (Scheme 3). In order to distinguish between mechanisms c and d, acid quenching was repeated in D2O (>99.9%) [43] with the expectation of trapping singly deuterated 5-HM-dUMP (one mass unit heavier) if mechanism c was taking place. On the other hand, if mechanism d is operative, then the intermediate trapped in D2O would not be heavier than the intermediate trapped in H2O. 5-HM-dUMP isolated from acid quenching in D2O did not contain deuterium. Nevertheless, 100% of the dTMP formed was singly deuterated, corroborating previously reported observations [11]. These observations support mechanism c (Scheme 3). Surprisingly, more than 60% of the unreacted dUMP (incubated with reduced flavin) was singly deuterated. Longer incubation of dUMP with the oxidized enzyme did not label the substrate, and the labeling did not depend on the availability of CH4H2folate. Control experiments revealed the position of the labeling to be at C5 of dUMP. Neither mechanism c nor d could solely explain the observations of the acid quenching in D2O. Hence, a new mechanism (Scheme 3e) was put forward to reconcile both mechanisms and account for all the experimental evidence obtained. In this mechanism, the hydride of reduced flavin activates the dUMP (step 1). Once the methylene is transferred to C6 of dUMP (step 2), oxidized flavin abstracts the hydride back from C6 (step 3). Steps 1–3 are fast and occur within the dead time of the stopped-flow instrument, therefore, flavin appears to be reduced (Fig. 4A green trace) until steps 4b and 5. The exocyclic methylene intermediate (highlighted in blue) formed in step 4a will not be deuterated during the acid quench and is thus responsible for the formation of the non-deuterated intermediate. The intermediate from step 4b (highlighted in red) explains the previously reported formation of 6D-dTMP.
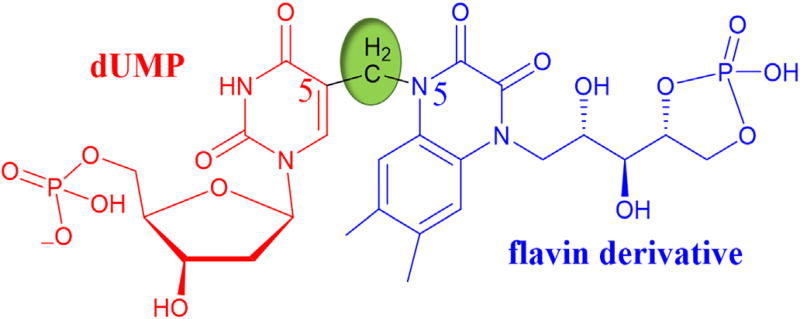
Until 2016 all the mechanisms proposed for FDTS (Scheme 3b, c, d, and e) suggested that the methylene is transferred directly from CH2H4folate to C5 of dUMP. This was proposed in analogy to TSase mechanism (Scheme 3a) where methylene is directly transferred from N5 of CH2H4folate to C5 of dUMP. Isolation and characterization of a base-trapped intermediate of TmFDTS shed light on how the methylene is transferred to C5 of dUMP [26]. This trapped derivative of the intermediate had a covalent methylene bridge between dUMP and N5 of flavin ([M − H]− 705.1212 Da, Fig. 5) [26].
Fig. 5.
Base-trapped dUMP-CH2-FAD adduct. Methylene transferred from CH2H4folate is highlighted in green. Figure adapted with permission from Ref. [26].
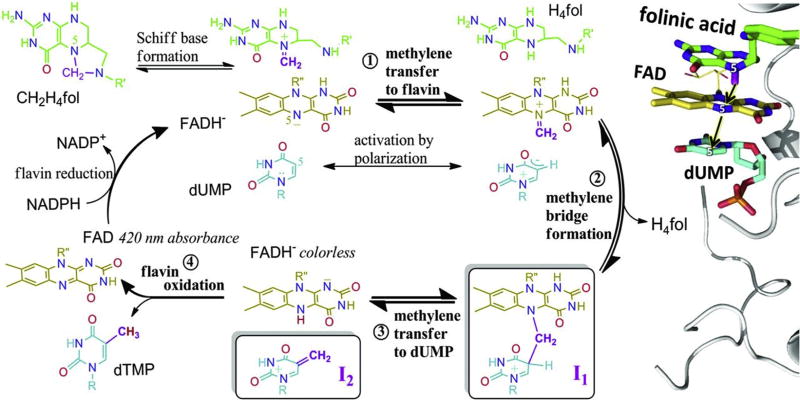
The structure of the trapped intermediate led us to propose a new mechanism for FDTS (Fig. 6) where N5 of reduced flavin acts as the methylene carrier. Unlike TSase, the methylene from CH2H4folate is not transferred directly to C5 of dUMP; rather, it is facilitated by N5 of reduced flavin. The folate-bound crystal structure of TmFDTS solved in 2012 (Fig. 2A) supports this claim. In 2016, the inactivity of 5-deaza-FAD reconstituted FDTS studied after denaturation to remove the residual FAD provided a strong piece of evidence for this flavin-mediated methylene transfer. With all this new information the mechanism in Fig. 6 was proposed [26].
Fig. 6.
Proposed chemical mechanism for FDTS. (Left) Methylene to be transferred to N5 of flavin is in purple. Hydride on N5 reduced flavin is in red. (Right) Active site of TmFDTS (PDB ID 4GTA) with bound dUMP, FAD and folinic acid, a stable analogue of CH2H4folate. For clarity, the carbonyl oxygen of folinic acid is omitted. The structure is inverted to match the orientation of ligands in the left mechanism. Reprinted with permission from Ref. [26].
The Schiff base formed on N5 of CH2H4folate is being attacked by nucleophilic N5 of reduced flavin (step 1). Then, C5 of the activated dUMP performs a nucleophilic attack on the iminium-ion on flavin (step 2) to form a dUMP-CH2-FADH intermediate (I1) which gets trapped in base (the structure of the authentic intermediate is deduced from the chemically modified compound (Fig. 5). Exocyclic methylene intermediate (I2) formed after the methylene transfer to dUMP (step 3) is trapped in acid as 5-HM-dUMP. Finally, the reduced flavin gets oxidized after transferring the N5 hydride to C7 of the exocyclic methylene intermediate.
Besides FDTS, there are several reports of CH2-N5 FADH imine formation [45]. The enzyme TrmFO methylates a uracil nucleotide in tRNA where N5 of flavin facilitates the methylene transfer from CH2H4folate to C5 of the uracil moiety [46]. In the absence of the substrate, an enzyme bound N5 flavin adduct (Cys53-CH2-FADH, Bacillus subtilis) was observed. This supports the formation of a CH2-flavin Schiff base during it's catalysis. Enzymes like alkyl-dihydroxyacetone phosphate synthase (ADPS) and UDP-galactopyranose form N5-imine adducts during catalysis [47,48]. An early work of Blankenhorn et al. showed the formation of a CH2- FADH Schiff base during flavin-dependent carbonyl activation [49]. The impact of this chemistry on rational inhibitor design will be discussed in the following section.
5. Inhibitor design
Inhibitors with high specificity for FDTS over TSase are sought, especially in low nanomolar Ki range, which is something that may be aided by the fact that FDTS is the only known uracil-methylating enzyme without a catalytic nucleophile [28,43]. Substrate analogues have been examined for this endeavor, and crystal structures of TmFDTS with a variety of possible inhibitors have been obtained. Of particular interest are the structures of Raltitrexed and folinic acid because the active site of TmFDTS appears to accommodate these substrates with similar interactions compared to CH2H4folate with folinic acid inhibiting FDTS [17]. Additionally, folinic acid is analogous to the reactive form of CH2H4folate, further supporting the stacking of bound substrates and separation of the original methylene donor and the final methylene acceptor by FAD [17]. Inhibitor design based on an FAD scaffold or binding site has not been explored and has limited potential. Since FAD is a common enzyme cofactor, FAD-based inhibitors may target FDTS, but will likely inhibit necessary, innocent proteins. However, two avenues exist for inhibitor design. There is potential to design an inhibitor to lock open the substrate binding loop, or FAD binding motif, since its closed conformation is essential to keep substrates bound [19]. It may be possible to design a scaffold with high affinity to the FAD ribosyl and adenine moiety binding locations since they have very high affinity while the isoalloxazine ring has more freedom. Efforts to design and search for dUMP analogues that inhibit FDTS often have relied on maintaining substrate conformation in the active site. Along with conserving pyrimidine ring π-stacking interactions, the phosphoryl moiety of dUMP analogues shows crucial interactions with catalytic triad residues, together directing a recent study by Kogler et al. to synthesize possible inhibitors, leading to specific modifications at C5 [50]. The most promising drug leads found from this study were dUMP analogues containing long carbon chains at C5 with a propargylamide near the pyrimidine ring which showed IC50 values as low as 0.91 µM(compared to 5-F-dUMP, 0.29 µM) and good selectivity over TSase [50].
Another avenue of inhibitor design for specificity of FDTS over TSase will take advantage of active site accessibility, where TSase has a much more cumbersome, deeply buried active site compared to FDTS [28]. This route would allow more chemical space to be explored for FDTS inhibitors due to the availability of space, as large inhibitors would face a steric barrier eliminating them from binding to TSase [28]. One of the few known FDTS-specific inhibitors possibly exploiting this functionality is an amide derivative of 5-propargyl-dUMP that showed 100-fold specificity towards FDTS [28,50].
A route of inhibition for FDTS yet to be implemented is forming flavin-folate adducts by exploiting isoalloxazine N5 nucleophilicity towards folate by functionalizing folate derivatives into more electrophilic species, then reacting them with reduced flavin to generate an adduct that would serve as the inhibitor [26]. Similarly, forming flavin-nucleotide adducts may have the same outcome, but would rely on electrophilic N5 of oxidized flavin. In either case, the bridged, “dead-end” species could serve as an irreversible inhibitor of FDTS.
Inhibitor screening has become high-throughput with the advent of modern docking algorithms and large compound databases. Virtual screening of the dUMP binding site for MtbFDTS (superimposable active site with TmFDTS) found 2000 potential inhibitor candidates with thirteen able to make native substratelike interactions with local arginines and substrate π-stacking interactions [26]. Some of the thirteen selected compounds exhibited no inhibition, while those containing imidazole-pyidazine scaffolds exhibited up to 29% inhibition [19]. The most effective inhibitors in this study have the opportunity to π-stack, as is the case with the native substrate, and the addition of bulkier groups than methyl at position 1 of this moiety improved inhibition [27]. A similar study used high-throughput screening to experimentally test 40,000 compounds against MtbFDTS, and yielded seven compounds with low IC50 and no TSase activity with the best lead having an IC50 of 710 ± 20 nM [51]. This study observed 1,4-benzoaxaine ring-containing compounds as having an important role in slow, tight binding inhibitors.
The future of FDTS inhibition will likely provide new therapeutics to quench the resistance of many pathogens. However, it is likely that potent drug candidates will arise from knowledge gained from virtual screening studies and mechanism-based scaffolds which will be aided by a complete FDTS mechanism. A lucid description of the flavin environment in FDTS catalysis will not only add to the flavin toolbox, but help prevent unwanted thymidylate synthase inhibition.
6. Conclusions
Although classical thymidylate synthase and flavin-dependent thymidylate synthase catalyze the same net chemical reaction to form thymidylate, peculiarities between structure and function, as well as presence in many pathogenic organisms, mark the latter enzyme as an effective drug target. Deconvolution of flavin's involvement in the catalytic mechanism and targeting the idiosyncrasies of the flavin fold and coupled substrate stacking will lead to new understanding of nature's chemistry tools and improve rational drug design. Progress of drug design through screening and mechanism-based design has afforded few compounds selective to the pathogenic enzyme with low inhibitory concentration. Elucidating the enigmatic flavin-dependent mechanism will require crystallographic structures of the enzyme in a reactive, reduced state and further observation of reaction intermediates.
Acknowledgments
The work has been funded by NIH RO1 GM110775 to AK. KK was supported by a training fellowship from the University of Iowa Center for Biocatalysis and Bioprocessing and the NIH-funded Predoctoral Training Program in Biotechnology (2 T32 GM008365).
Abbreviations
The following abbreviations are used in this manuscript
- dTMP
2′-deoxythymidine-5′-monophosphate
- dUMP
2′-deoxyuridine-5′-monophosphate
- TSase
Thymidylate synthase
- DHFR
Dihydrofolate reductase
- FDTS
Flavin dependent thymidylate synthase
- MTHF, CH2H4folate
N5,N10-methylene-5,6,7,8,-tetrahydrofolate
- DHF, H2folate
Dihydrofolate
- THF, H4folate
Tetrahydrofolate
- FAD
Flavin adenine dinucleotide
- NADPH
Nicotinamide adenine dinucleotide phosphate
- AMP
Adenosine 5′-monophosphate
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 2.Kelly C, et al. Use of raltitrexed as an alternative to 5-fluorouracil and capecitabine in cancer patients with cardiac history. Eur. J. Cancer. 2013;49(10):2303–2310. doi: 10.1016/j.ejca.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 4.Cronstein BN. Low-dose methotrexate: a mainstay in the treatment of rheumatoid arthritis. Pharmacol. Rev. 2005;57(2):163–172. doi: 10.1124/pr.57.2.3. [DOI] [PubMed] [Google Scholar]
- 5.Huovinen P, et al. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 1995;39(2):279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myllykallio H, et al. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297(5578):105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 7.Koehn EM, Kohen A. Flavin-dependent thymidylate synthase: a novel pathway towards thymine. Arch. Biochem. Biophys. 2010;493(1):96–102. doi: 10.1016/j.abb.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson EF, et al. Mechanistic characterization of Toxoplasma gondii thymidylate synthase (TS-DHFR)-dihydrofolate reductase. Evidence for a TS intermediate and TS half-sites reactivity. J. Biol. Chem. 2002;277(45):43126–43136. doi: 10.1074/jbc.M206523200. [DOI] [PubMed] [Google Scholar]
- 9.Atreya CE, Anderson KS. Kinetic characterization of bifunctional thymidylate synthase-dihydrofolate reductase (TS-DHFR) from Cryptosporidium hominis: a paradigm shift for ts activity and channeling behavior. J. Biol. Chem. 2004;279(18):18314–18322. doi: 10.1074/jbc.M400009200. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal N, et al. Mechanistic studies of a flavin-dependent thymidylate synthase. Biochemistry. 2004;43(32):10295–10301. doi: 10.1021/bi0490439. [DOI] [PubMed] [Google Scholar]
- 11.Koehn EM, et al. An unusual mechanism of thymidylate biosynthesis in organisms containing the thyX gene. Nature. 2009;458(7240):919–923. doi: 10.1038/nature07973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn P, et al. Crystal structure of thy1, a thymidylate synthase complementing protein from Thermotoga maritima at 2.25 A resolution. Proteins. 2002;49(1):142–145. doi: 10.1002/prot.10202. [DOI] [PubMed] [Google Scholar]
- 13.Mathews II, et al. Functional analysis of substrate and cofactor complex structures of a thymidylate synthase-complementing protein. Structure. 2003;11(6):677–690. doi: 10.1016/s0969-2126(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 14.Sampathkumar P, et al. Structure of the Mycobacterium tuberculosis flavin dependent thymidylate synthase (MtbThyX) at 2.0A resolution. J. Mol. Biol. 2005;352(5):1091–1104. doi: 10.1016/j.jmb.2005.07.071. [DOI] [PubMed] [Google Scholar]
- 15.Graziani S, et al. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria Chlorella virus-1. J. Biol. Chem. 2004;279(52):54340–54347. doi: 10.1074/jbc.M409121200. [DOI] [PubMed] [Google Scholar]
- 16.Leduc D, et al. Functional evidence for active site location of tetrameric thymidylate synthase X at the interphase of three monomers. Proc. Natl. Acad. Sci. U. S. A. 2004;101(19):7252–7257. doi: 10.1073/pnas.0401365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koehn EM, et al. Folate binding site of flavin-dependent thymidylate synthase. Proc. Natl. Acad. Sci. U. S. A. 2012;109(39):15722–15727. doi: 10.1073/pnas.1206077109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graziani S, et al. Catalytic mechanism and structure of viral flavin-dependent thymidylate synthase ThyX. J. Biol. Chem. 2006;281(33):24048–24057. doi: 10.1074/jbc.M600745200. [DOI] [PubMed] [Google Scholar]
- 19.Mathews II. Flavin-dependent thymidylate synthase as a drug target for deadly microbes: mutational study and a strategy for inhibitor design. J. Bioterror Biodef. 2013;S12:004. doi: 10.4172/2157-2526.S12-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampathkumar P, et al. NADP+ expels both the co-factor and a substrate analog from the Mycobacterium tuberculosis ThyX active site: opportunities for anti-bacterial drug design. J. Mol. Biol. 2006;360(1):1–6. doi: 10.1016/j.jmb.2006.04.061. [DOI] [PubMed] [Google Scholar]
- 21.Stull FW, et al. Deprotonations in the reaction of flavin-dependent thymidylate synthase. Biochemistry. 2016;55(23):3261–3269. doi: 10.1021/acs.biochem.6b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baugh L, et al. Increasing the structural coverage of tuberculosis drug targets. Tuberc. (Edinb) 2015;95(2):142–148. doi: 10.1016/j.tube.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basta T, et al. Mechanistic and structural basis for inhibition of thymidylate synthase ThyX. Open Biol. 2012;2(10):120120. doi: 10.1098/rsob.120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesley SA, et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. U. S. A. 2002;99(18):11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason A, et al. A lag-phase in the reduction of flavin dependent thymidylate synthase (FDTS) revealed a mechanistic missing link. Chem. Commun. (Camb) 2006;16:1781–1783. doi: 10.1039/b517881a. [DOI] [PubMed] [Google Scholar]
- 26.Mishanina TV, et al. An unprecedented mechanism of nucleotide methylation in organisms containing thyX. Science. 2016;351(6272):507–510. doi: 10.1126/science.aad0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luciani R, et al. Virtual screening and X-ray crystallography identify nonsubstrate analog inhibitors of flavin-dependent thymidylate synthase. J. Med. Chem. 2016;59(19):9269–9275. doi: 10.1021/acs.jmedchem.6b00977. [DOI] [PubMed] [Google Scholar]
- 28.Mishanina TV, Koehn EM, Kohen A. Mechanisms and inhibition of uracil methylating enzymes. Bioorg Chem. 2012;43:37–43. doi: 10.1016/j.bioorg.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi M, Karunaratne K, Kohen A. Flavin-dependent thymidylate synthase as a new antibiotic target. Molecules. 2016;21(5) doi: 10.3390/molecules21050654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10(9):1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster CE, et al. Crystal structure of human quinone reductase type 2, a metalloflavoprotein. Biochemistry. 1999;38(31):9881–9886. doi: 10.1021/bi990799v. [DOI] [PubMed] [Google Scholar]
- 32.Park HW, et al. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268(5219):1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 33.Becker HF, et al. Substrate interaction dynamics and oxygen control in the active site of thymidylate synthase ThyX. Biochem. J. 2014;459(1):37–45. doi: 10.1042/BJ20131567. [DOI] [PubMed] [Google Scholar]
- 34.Conrad JA, et al. Detection of intermediates in the oxidative half-reaction of the FAD-dependent thymidylate synthase from Thermotoga maritima: carbon transfer without covalent pyrimidine activation. Biochemistry. 2014;53(32):5199–5207. doi: 10.1021/bi500648n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattis SG, Palfey BA. Direct observation of the participation of flavin in product formation by thyX-encoded thymidylate synthase. J. Am. Chem. Soc. 2005;127(3):832–833. doi: 10.1021/ja0432214. [DOI] [PubMed] [Google Scholar]
- 36.Laptenok SP, et al. Ultrafast real-time visualization of active site flexibility of flavoenzyme thymidylate synthase ThyX. Proc. Natl. Acad. Sci. U. S. A. 2013;110(22):8924–8929. doi: 10.1073/pnas.1218729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, et al. Oxidase activity of a flavin-dependent thymidylate synthase. FEBS J. 2009;276(10):2801–2810. doi: 10.1111/j.1742-4658.2009.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattevi A. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 2006;31(5):276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Fraaije MW, van Berkel WJ. Catalytic mechanism of the oxidative demethylation of 4-(methoxymethyl)phenol by vanillyl-alcohol oxidase. Evidence for formation of a p-quinone methide intermediate. J. Biol. Chem. 1997;272(29):18111–18116. doi: 10.1074/jbc.272.29.18111. [DOI] [PubMed] [Google Scholar]
- 40.Basran J, et al. Mechanistic aspects of the covalent flavoprotein dimethylglycine oxidase of Arthrobacter globiformis studied by stopped-flow spectrophotometry. Biochemistry. 2002;41(14):4733–4743. doi: 10.1021/bi025519h. [DOI] [PubMed] [Google Scholar]
- 41.Chernyshev A, et al. The relationships between oxidase and synthase activities of flavin dependent thymidylate synthase (FDTS) Chem. Commun. (Camb) 2007;(27):2861–2863. doi: 10.1039/b700977a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finer-Moore JS, Santi DV, Stroud RM. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme: thymidylate synthase mutagenesis, function, and structure. Biochemistry. 2003;42(2):248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 43.Mishanina TV, Corcoran JM, Kohen A. Substrate activation in flavin-dependent thymidylate synthase. J. Am. Chem. Soc. 2014;136(30):10597–10600. doi: 10.1021/ja506108b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishanina TV, et al. Trapping of an intermediate in the reaction catalyzed by flavin-dependent thymidylate synthase. J. Am. Chem. Soc. 2012;134(9):4442–4448. doi: 10.1021/ja2120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piano V, Palfey BA, Mattevi A. Flavins as covalent catalysts: new mechanisms emerge. Trends Biochem. Sci. 2017 doi: 10.1016/j.tibs.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Hamdane D, et al. FAD/folate-dependent tRNA methyltransferase: flavin as a new methyl-transfer agent. J. Am. Chem. Soc. 2012;134(48):19739–19745. doi: 10.1021/ja308145p. [DOI] [PubMed] [Google Scholar]
- 47.Razeto A, et al. The crucial step in ether phospholipid biosynthesis: structural basis of a noncanonical reaction associated with a peroxisomal disorder. Structure. 2007;15(6):683–692. doi: 10.1016/j.str.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 48.Soltero-Higgin M, et al. A unique catalytic mechanism for UDP-galactopyranose mutase. Nat. Struct. Mol. Biol. 2004;11(6):539–543. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 49.Blankenhorn G, Ghisla S, Hemmerich P. Model studies on flavin-dependent carbonyl-activation: reduction of carbonyl compounds by flavohydroquinone. Z Naturforsch B. 1972;27(9):1038–1040. doi: 10.1515/znb-1972-0909. [DOI] [PubMed] [Google Scholar]
- 50.Kogler M, et al. Synthesis and evaluation of 5-substituted 2'-deoxyuridine monophosphate analogues as inhibitors of flavin-dependent thymidylate synthase in Mycobacterium tuberculosis. J. Med. Chem. 2011;54(13):4847–4862. doi: 10.1021/jm2004688. [DOI] [PubMed] [Google Scholar]
- 51.Asrar RA, et al. Discovery of a new Mycobacterium tuberculosis thymidylate synthase X inhibitor with a unique inhibition profile. Biochem. Pharmacol. 2017;135:69–78. doi: 10.1016/j.bcp.2017.03.017. [DOI] [PubMed] [Google Scholar]