Abstract
Upward trends in the incidence and mortality rates of colorectal cancer (CRC) in China over the past decade mean that it is critical to improve survival outcomes for patients with this malignancy. Analysis of genetic variants may identify biomarkers that have a role in CRC susceptibility and clinical outcomes in Chinese patients with CRC. RAD52 is a key mediator during DNA strand exchange and homologous recombination within mammalian cells. In this study, we explored the effects of RAD52 single nucleotide polymorphisms (SNPs) in the susceptibility and clinicopathological characteristics of Chinese Han patients with CRC. Five RAD52 SNPs (rs1051669, rs10774474, rs11571378, rs6489769, and rs7963551) were analyzed using TaqMan SNP genotyping in 281 patients with CRC and 309 healthy controls. Among those aged over 60 years in the total population, carriers of the variant C allele or at least one T allele of the rs1051669 SNP were at a lower risk of CRC than carriers of the wild-type CC variant of rs1051669, while in those carrying the rs7963551 SNP, the GT or GT+GG alleles were associated with an increased risk of CRC compared with patients carrying TT alleles. We indicated a significant correlation between RAD52 rs7963551 polymorphism and lymph node metastasis in CRC patients. In all patients, the T-T-T-T-T, C-T-T-T-T, and C-T-A-C-T haplotypes were associated with an increasing risk of CRC. Our findings suggest that 4 RAD52 SNPs (rs1051669, rs10774474, rs11571378, and rs6489769) might contribute to the prediction of CRC susceptibility. In conclusion, our study demonstrated that RAD52 polymorphisms were associated with CRC in a Chinese Han cohort.
Keywords: Chinese Han, colorectal cancer, polymorphisms, RAD52, susceptibility
1. Introduction
Over the last decade, incidence rates of colorectal cancer (CRC) have increased rapidly in China,[1] where it is now the third most common cancer after lung cancer and gastric cancer in the Chinese Han population.[2] It is therefore critical to improve survival outcomes for patients with this malignancy. Early diagnosis of CRC is challenging: patients often have no specific symptoms; lower abdominal symptoms are very common and mostly related to non-neoplastic diseases, not CRC; diagnosis involves several steps starting with when the patient detects the first symptoms, a consultation with a general practitioner, a referral to the specialist, and a waiting period for a diagnostic procedure, mainly involving colonoscopy, an invasive procedure; and endoscopic resources for diagnosis are usually limited.[3] Alterations in gene expression that lead to CRC are reflected by dysregulated levels of nucleic acids and proteins, which can act as potential CRC molecular markers, providing diagnostic, prognostic, and predictive treatment response information.[4] Genetic information in CRC tumors will help make the best treatment decisions for the patient, clarify their prognosis and their predicted response to treatment.[4]
Human RAD52 exists in an oligomeric form and acts as an important member involved in the homologous recombination pathway to repair double-strand DNA breaks.[5] The RAD52 gene is located on chromosome 12p12.2-13 and several single nucleotide polymorphisms (SNPs) have been identified in the RAD52 promoter and 3′-untranslated regions.[6] SNPs are often performed as genetic markers for the prediction of patient response to susceptibility and development of CRC.[7] A recent polymorphism study showed that the RAD52 rs1051669 AA genotype was associated with a significantly increased risk of developing CRC.[8] Interestingly, in another genetic study, the functional RAD52 rs7963551 polymorphism contributed to risk susceptibility of hepatitis B virus related hepatocellular carcinoma in Chinese populations.[9] An association between RAD52 genetic polymorphisms and CRC risk and prognosis remains unclear in Chinese Han individuals. Accordingly, we performed a case–control study to evaluate 5 SNPs in the RAD52 gene in Chinese Han patients with CRC.
2. Materials and methods
2.1. Patients and blood samples
We collected blood specimens from 281 patients (cases) diagnosed with CRC at Dongyang People's Hospital during 2015 and 2016. The control group consisted of 309 healthy participants without a history of cancer. All participants provided written informed consent for the study, which was approved by the Ethics Committee of Dongyang People's Hospital. CRC pathological stages were determined in all patients according to their medical records and the 6th edition of the American Joint Committee on Cancer (AJCC) tumor, node, metastasis (TNM) staging system. Standardized questionnaires completed by patients at hospital admission provided data on age, sex, smoking status, and alcohol consumption.
2.2. Selection of RAD52 polymorphisms
Two RAD52 SNPs were selected from a 2-kb region upstream of RAD52 (rs 10774474 and rs11571378), 2 (rs1051669 and rs7963551) were selected from the 3’un-translated region of RAD52, and 1 (rs6489769) was selected from the intron of RAD52; all SNPs had minor allele frequencies of greater than 5%. The rs7963551 SNP is known to contribute to susceptibility to hepatocellular carcinoma,[9] while patients with cervical cancer who carry ≥1 of 3 RAD52 variants (rs10774474T, rs11571378A, and rs1051669A) have demonstrated poorer progression-free survival and overall survival than those without any of the variant alleles.[10]
2.3. Genomic DNA extraction
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA blood kit (Qiagen, CA), according to the manufacturer's instructions. Extracted DNA was prepared for genotyping by polymerase chain reaction (PCR) and stored at -20°C until needed.
2.4. Genotyping by real-time PCR
Five RAD52 SNP probes were purchased from Thermo Fisher Scientific Inc. (Thermo Fisher, Applied Biosystems, Foster City), and assessment of allelic discrimination for RAD52 SNPs was conducted using a Roche LightCycler 480 Instrument II (Roche, Mannheim, Germany). Data were further analyzed with LightCycler 480 Software, Version 1.5 (Roche). PCR was carried out in a total volume of 10 μL containing 20 to 70 ng genomic DNA, 1 U Taqman Genotyping Master Mix (Applied Biosystems, Foster City, CA), and 0.25 μL probes. The protocol included an initial denaturation step at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
2.5. Statistical analysis
Differences between the 2 groups were considered significant if P values were less than .05. Hardy–Weinberg equilibrium was assessed using Chi-square goodness-of-fit tests for bi-allelic markers. Clinical and demographic characteristics were compared for the patients and controls using Mann–Whitney U tests and Fisher exact tests. The odds ratios (ORs) and 95% confidence intervals (CIs) for associations between genotype frequencies and the risk of CRC or clinicopathological characteristics were estimated by multiple logistic regression models, after controlling for other covariates. All data were analyzed using Statistical Analytic System software (version 9.1, 2005; SAS Institute, Cary, NC).
3. Results
All study participants were of Chinese Han ethnicity. Clinicopathological and demographic data are summarized in Table 1. Age and gender distributions were comparable between the 281 cases and 309 controls (P > .05). Patients were classified by the AJCC TNM (6th edition) as clinical stage I/II (n = 127; 45.2%), or clinical stage III/IV (n = 154; 54.8%).
Table 1.
Demographic and clinicopathological characteristics in healthy controls and patients with colorectal cancer.
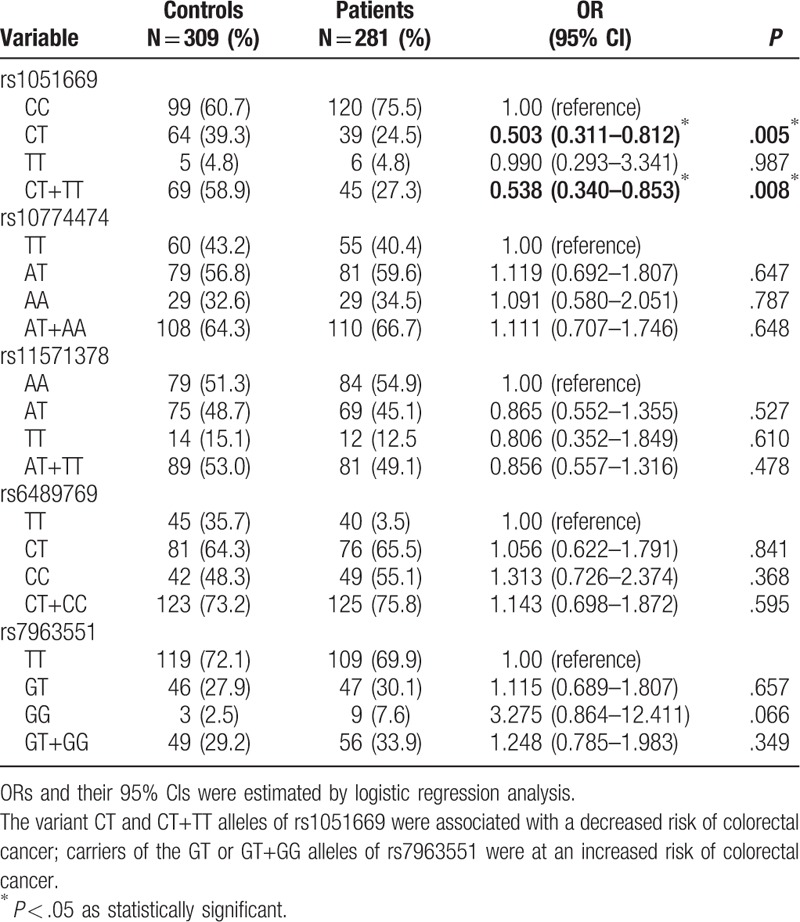
To evaluate whether the 5 RAD52 SNPs (rs1051669, rs10774474, rs11571378, rs6489769, and rs7963551) were associated with the risk of CRC, we genotyped all study participants. Genotyping distributions and associations between CRC and RAD52 gene polymorphisms are summarized in Table 2. In both CRC patients and controls, the alleles with the highest distribution frequency at rs1051669, rs10774474, rs11571378, rs6489769, and rs7963551 were homozygous C/C, heterozygous A/T, homozygous A/A, heterozygous C/T, and homozygous T/T, respectively. To our surprise, there were no significant between-group differences in distribution frequencies. However, further analysis of all study participants aged over 60 years revealed that individuals carrying CT or CT+TT genotypes at rs1051669 had half the risk of developing CRC compared with individuals carrying the wild-type CC polymorphic allele (CT allele carriers: rate ratio 0.503, 95% CI: 0.311–0.812, P < .05; CT+TT allele carriers: rate ratio 0.538, 95% CI: 0.340–0.853, P < .05) (Table 3). In contrast, carriers of the variant GT or GT+GG alleles of rs7963551 were at an increased risk of CRC.
Table 2.
Distribution frequency of RAD52 genotypes in 309 controls and 281 patients with colorectal cancer.
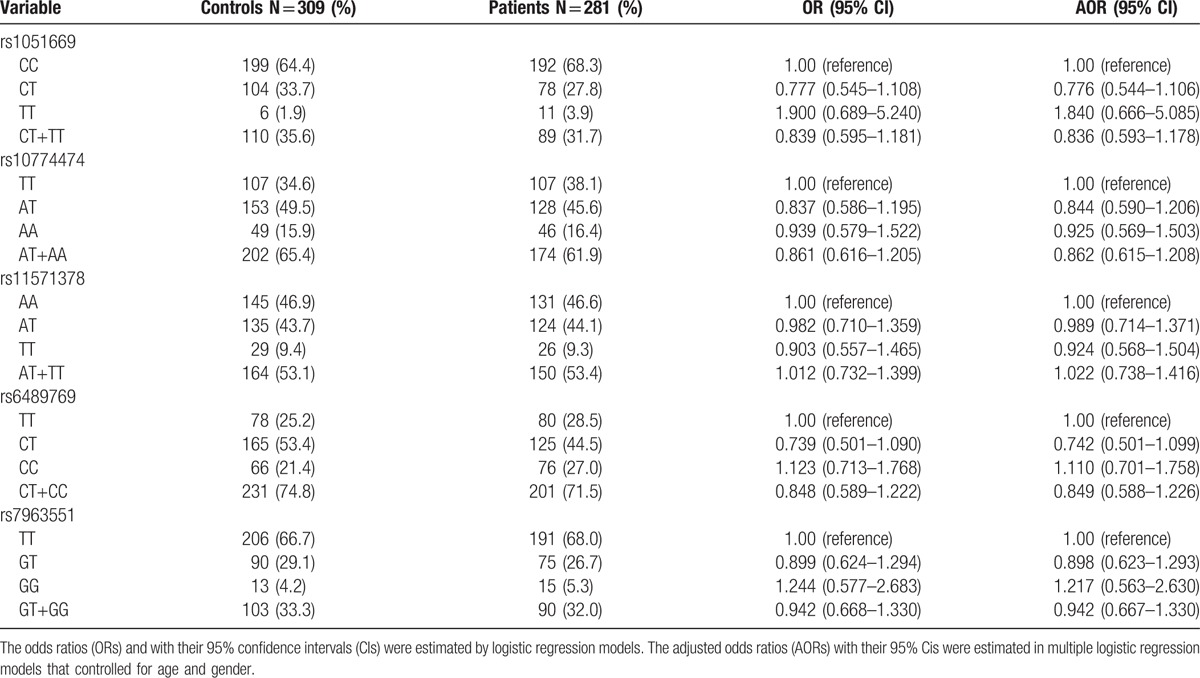
Table 3.
Odds ratios (ORs) and 95% confidence intervals (CIs) of distribution frequencies associated with RAD52 genotypes in study participants aged over 60 years.
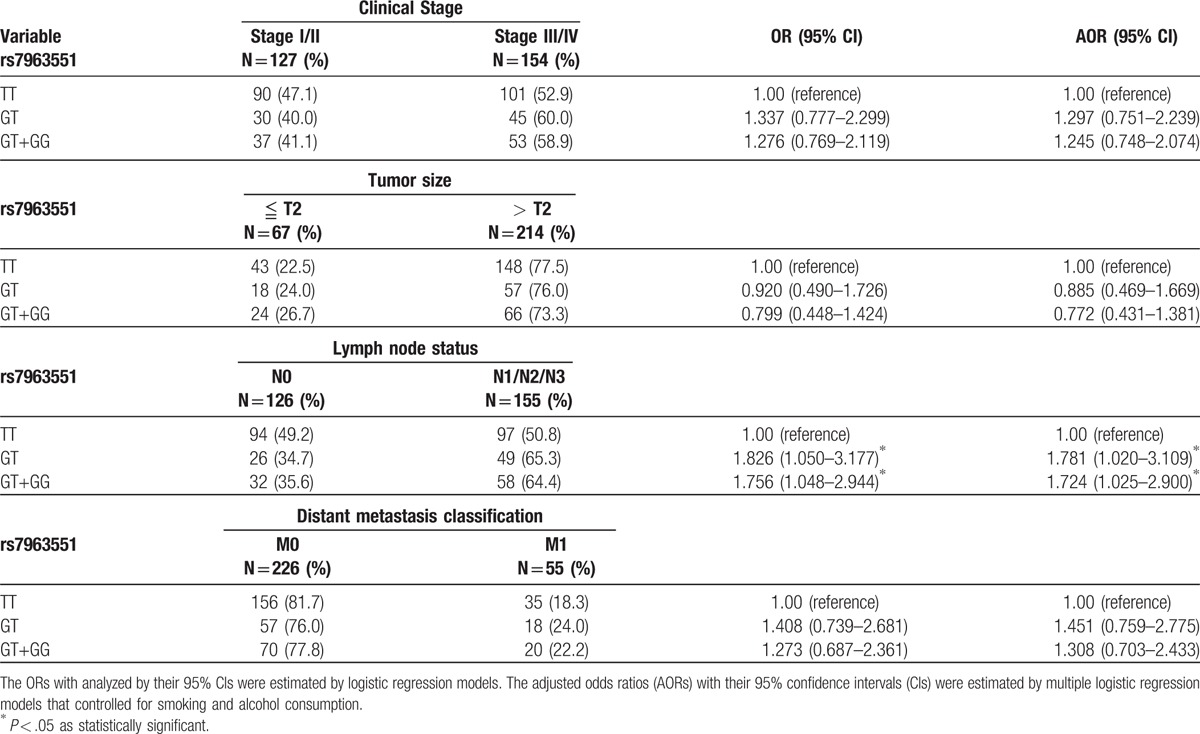
Next, RAD52 genotypes were evaluated in the CRC cohort to clarify the role of RAD52 polymorphisms in clinical TNM stage, primary tumor size, lymph node metastasis, and distant metastasis (Table 4). To reduce the possible interference of confounding variables, adjusted odds ratios (AORs) with 95% CIs were estimated by multiple logistic regression models controlling for smoking status and alcohol consumption. We observed a significant correlation between rs7963551 variants and lymph node metastasis (TT vs GT: AOR 1.781, 95% CI: 1.020–3.109, P < .05; TT vs GT+GG: AOR 1.724, 95% CI: 1.025–2.900, P < .05, respectively) (Table 4). No such significant findings were observed between other RAD52 genotypes and clinicopathological status (data not shown).
Table 4.
Odds ratio (OR) and 95% confidence interval (CI) of clinical status and RAD52 rs7963551 genotypic frequencies in 281 patients with colorectal cancer.
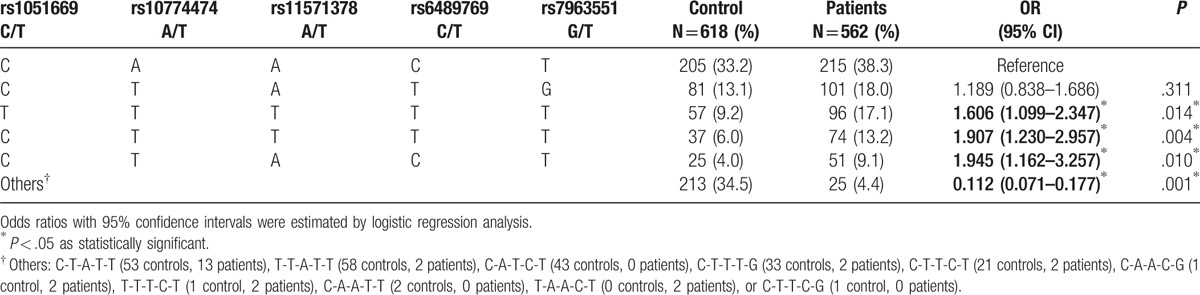
We used HaploView software and the PHASE program to explore common haplotypes in the RAD52 gene. Linkage disequilibrium (LD) patterns are depicted in Fig. 1. As summarized in Table 5, compared with the reference group C-A-A-C-T (RAD52 rs1051669/rs10774474/rs11571378/rs6489769/rs7963551), carriers of the T-T-T-T-T, C-T-T-T-T, or C-T-A-C-T haplotypes had approximately twice the risk of CRC (T-T-T-T-T: OR 1.606, 95% CI: 1.099–2.347; C-T-T-T-T: OR 1.907, 95% CI: 1.230–2.957; C-T-A-C-T: OR 1.945, 95% CI: 1.162–3.257). In contrast, carriers with other haplotypes, including C-T-A-T-T (53 controls, 13 patients), T-T-A-T-T (58 controls, 2 patients), C-A-T-C-T (43 controls, 0 patients), C-T-T-T-G (33 controls, 2 patients), C-T-T-C-T (21 controls, 2 patients), C-A-A-C-G (1 control, 2 patients), T-T-T-C-T (1 control, 2 patients), C-A-A-T-T (2 controls, 0 patients), T-A-A-C-T (0 controls, 2 patients), or C-T-T-C-G (1 control, 0 patients) were about 10% less likely to develop CRC (OR 0.112, 95% CI: 0.071–0.177) (Table 5).
Figure 1.
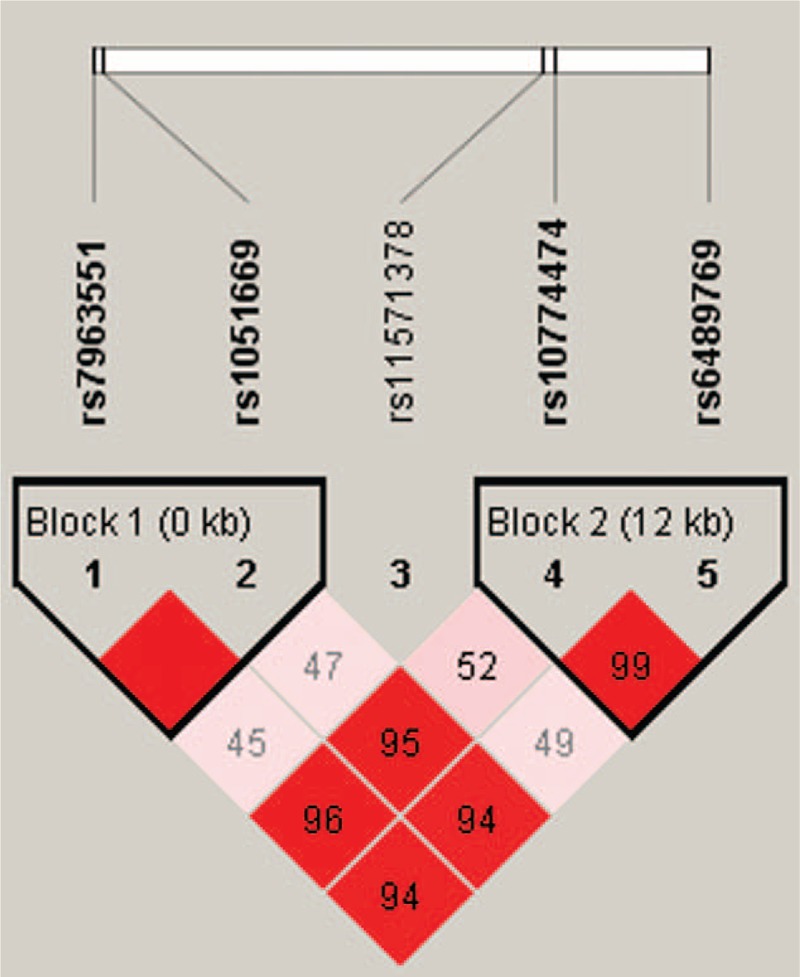
A linkage disequilibrium map of single nucleotide polymorphisms in the RAD52 gene in 309 healthy controls and 281 patients with colorectal cancer. Blocks 1 and 2 depict the pairwise D’ plots and haplotype blocks obtained from HaploView.
Table 5.
Distribution frequency of RAD52 haplotypes in healthy controls and patients with colorectal cancer.
4. Discussion
The etiology of CRC encompasses a combination of genetic and environmental factors. Some risk factors are related to lifestyle and can be reduced by implementing changes in terms of dietary and physical activity habits; a sedentary lifestyle is thought to increase the risk of developing CRC and is related to obesity, an important risk factor for CRC.[11,12] Moreover, unhealthy nutritional habits increase the chances of developing CRC by up to 70%; smoking and alcohol consumption are also linked to increased CRC risk.[12] In our study, age and gender were closely matched between cases and controls. We were unable to obtain information on smoking and alcohol consumption in controls, so were unable to determine any associations between CRC patients and controls in respect to these risk factors.
Having the ability to accurately predict the possibility of an individual developing CRC by considering identifiable variables, including genetic risk factors, would be extremely useful. Previous research has reported that the rs7963551C allele in RAD52 reduced breast cancer risk in Chinese Han women.[13] In this study, we hypothesized that genetic variants of RAD52 may influence clinical outcomes in CRC patients. Five SNPs were included in our case–control study.
A previous study reported that RAD52 variants appeared to predict prognosis in patients with cervical squamous cell carcinoma.[10] Although our results showed no such association with CRC in our overall study cohort, when we analyzed data from patients aged over 60 years, we found that 1 of the 5 RAD52 gene polymorphisms was associated with a low incidence of CRC. Interestingly, a previous report identified that the RAD52 rs7963551 polymorphism was significantly correlated with a decreased risk of small cell lung cancer among Chinese individuals.[14] We demonstrate in this study that the presence of GT and GT+GG alleles at rs7963551 was significantly associated with increased lymph node metastasis in CRC patients. In contrast, RAD52 polymorphisms at rs1051669, rs10774474, rs11571378, and rs6489769 were not significantly related to status. Notably, a recent study from the Czech Republic has reported that individuals carrying the AA genotype or the variant A allele in rs1051669 were at an increased risk of developing colon cancer (P ≤ .2); carriers of the heterozygous TC genotype of rs11571475 were at a decreased risk for colon cancer (P = .05), while carriers of the GT+GG genotype of rs7963551 also had a lower risk of developing CRC (P = .05).[8] Our results align with this finding and thus may provide insights into the development of targeted therapy for CRC in patients with the rs1051669 SNP.
In epigenetic DNA methylation, variant polymorphisms in the 3’ untranslated region can regulate gene expression and impact upon the risk of developing tumors.[15] The rs7963551 polymorphism is located in the 3’ untranslated region and may regulate RAD52 gene expression. LD and haplotype are generally used as genetic markers and predicted the susceptibility of diseases.[16] We evaluated the impact of different haplotype combinations of 5 RAD52 SNPs and found that the T-T-T-T-T, C-T-T-T-T, and C-T-A-C-T haplotypes contribute to susceptibility of developing CRC. These haplotypes suggest that RAD52 gene polymorphisms rs1051669, rs10774474, rs11571378, and rs6489769 are strongly associated with CRC susceptibility. In this study, we did not perform a detailed functional analysis of rs7963551. The issue of how these SNPs affect RAD52 gene expression in CRC cells is yet to be clarified. Moreover, some patient survival data were unavailable because patients had only recently enrolled in the study. Larger studies are needed to assess the correlations between RAD52 polymorphisms and CRC progression.
In summary, our findings demonstrate a significant association between RAD52 SNPs and distant CRC metastases in a cohort of Chinese Han individuals. We showed a significant correlation between rs7963551 variants and lymph node metastasis. Thus, RAD52 might serve as a genetic prognostic marker for CRC treatment.
Footnotes
Abbreviations: AJCC = American Joint Committee on Cancer, AOR = adjusted odds ratios, CI = confidence interval, CRC = colorectal cancer, GTEx = genotype-tissue expression, OR = odds ratio, PCR = polymerase chain reaction, SNP = single nucleotide polymorphisms, TNM = tumor, node, metastasis.
Funding/support: This work was supported by grants from the Jinhua Science and Technology Bureau (2015-3-088).
The authors have no conflicts of interest to declare.
References
- [1].Zhu J, Tan Z, Hollis-Hansen K, et al. Epidemiological trends in colorectal cancer in China: an ecological study. Dig Dis Sci 2017;62:235–43. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zheng R, Zeng H, et al. Annual report on status of cancer in China, 2011. Chin J Cancer Res 2015;27:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vega P, Valentin F, Cubiella J. Colorectal cancer diagnosis: pitfalls and opportunities. World J Gastrointest Oncol 2015;7:422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gonzalez-Pons M, Cruz-Correa M. Colorectal cancer biomarkers: where are we now? BioMed Res Int 2015;2015:149014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lok BH, Powell SN. Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin Cancer Res 2012;18:6400–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mortensen UH, Bendixen C, Sunjevaric I, et al. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci U S A 1996;93:10729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Canavese M, Ngo DT, Maddern GJ, et al. Biology and therapeutic implications of VEGF-A splice isoforms and single-nucleotide polymorphisms in colorectal cancer. Int J Cancer 2017;140:2183–91. [DOI] [PubMed] [Google Scholar]
- [8].Naccarati A, Rosa F, Vymetalkova V, et al. Double-strand break repair and colorectal cancer: gene variants within 3’ UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget 2016;7:23156–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li Z, Guo Y, Zhou L, et al. Association of a functional RAD52 genetic variant locating in a miRNA binding site with risk of HBV-related hepatocellular carcinoma. Mol Carcinog 2015;54:853–8. [DOI] [PubMed] [Google Scholar]
- [10].Shi TY, Yang G, Tu XY, et al. RAD52 variants predict platinum resistance and prognosis of cervical cancer. PLoS One 2012;7:e50461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Van Blarigan EL, Meyerhardt JA. Role of physical activity and diet after colorectal cancer diagnosis. J Clin Oncol 2015;33:1825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Odegaard AO, Koh WP, Yuan JM. Combined lifestyle factors and risk of incident colorectal cancer in a Chinese population. Cancer Prev Res (Phila) 2013;6:360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jiang Y, Qin Z, Hu Z, et al. Genetic variation in a hsa-let-7 binding site in RAD52 is associated with breast cancer susceptibility. Carcinogenesis 2013;34:689–93. [DOI] [PubMed] [Google Scholar]
- [14].Han S, Gao F, Yang W, et al. Identification of an SCLC susceptibility rs7963551 genetic polymorphism in a previously GWAS-identified 12p13.33 RAD52 lung cancer risk locus in the Chinese population. Int J Clin Exp Med 2015;8:16528–35. [PMC free article] [PubMed] [Google Scholar]
- [15].Vodicka P, Pardini B, Vymetalkova V, et al. Polymorphisms in non-coding RNA genes and their targets sites as risk factors of sporadic colorectal cancer. Adv Exp Med Biol 2016;937:123–49. [DOI] [PubMed] [Google Scholar]
- [16].Montazeri Z, Theodoratou E, Nyiraneza C, et al. Systematic meta-analyses and field synopsis of genetic association studies in colorectal adenomas. Int J Epidemiol 2016;45:186–205. [DOI] [PMC free article] [PubMed] [Google Scholar]


