Abstract
Two new silver(I) complexes, namely [Ag(qyAm)2](CF3SO3) (1) and [Ag(qyTAm)2](CF3SO3) (2), (qyAm = 2-(quinonyl)iminoadamantane, qyTAm = 2-(quinonyl)iminotriazaadamantane) have been synthesized and characterized by elemental analyses, 1H NMR, IR, electronic absorption spectroscopy, and X-ray diffraction. The coordination geometry of the silver center in both complexes is distorted tetrahedral where their respective qyAm and qyTAm ligand bind in a bidentate fashion using the imine and quinoline nitrogen atoms. Complex 2 is soluble in water and exhibits strong antimicrobial actions on both Gram-negative (E. coli, and P. aeruginosa) and Gram-positive (S. aureus) bacteria. The minimal inhibitory concentration (MIC) values for complex 2 (4, 4, and 8 μg for E. coli, P. aeruginosa, and S. aureus, respectively) are comparable to MIC values of silver nitrate and silver sulfadiazine.
Keywords: Antibacterial, Silver complex, Water soluble, Triazaadamantane moiety, Schiff base ligands
1. Introduction
Antimicrobial resistance has emerged as an increasing problem among developed nations in the hospital settings [1]. Antibacterial drugs that were once effective against different pathogens are no longer effective in eradication of bacterial infections. Pseudomonas aeruginosa, a Gram-negative bacteria, rarely causes infection in healthy people and yet is one of the major causes of death in hospitalized patients [2]. P. aeruginosa often becomes multi-drug resistant due to indiscriminate overuse of antimicrobial drugs and consequently has become more difficult to treat over the past few decades [3]. Similarly Staphylococcus aureus, an opportunistic Gram-positive pathogen often encountered in the medical settings, has acquired resistance to methicillin-derived antibiotics and is notoriously difficult to treat [3]. Both P. aeruginosa and S. aureus cause life-threatening infection in patients with severe burn-wounds which results from delay in the wound healing process due to bacterial colonization [4]. Bacterial colonization in wounds is often polymicrobial and hence broad-spectrum antibiotics are the best options for treatment. As a consequence, novel broad-spectrum antimicrobials for effective treatment of infections in wound burns are of paramount importance.
Silver has been known to possess antimicrobial properties for more than two thousand years and is considered a broad-spectrum antimicrobial agent [5]. Pharmaceutical application of silver was first recognized with the use of silver nitrate (AgNO3) in early 1800s for the treatment of ulcers [6]. The discovery of penicillin and its potent antibacterial properties led to a diminution of interest in silver and it was not until the 1960s when use of silver was reintroduced in the form of 0.5% AgNO3 solution to treat wounds on burn victims [6,7]. In addition, silver has been utilized in creams for topical treatment of burns and wounds in the form of silver sulfadiazine (AgSD). AgSD exhibits high efficacy in eradication of P. aeruginosa with minimum side effects in the wound healing process compared to AgNO3 [7].
It is well established that metallic silver causes minimum damage to living organism and cationic Ag+ is the biologically active species in applications of silver-containing compounds and formulations [5]. The antimicrobial properties of Ag+ are mainly attributed to its ability to bind thiols on proteins and enzymes [5], block cellular respiration [5], cause cell membrane damage [5], and bind to nucleic acids [8]. In addition to silver salts, silver complexes derived from various ligands have regained attention as antimicrobial agents [9]. Research in this area has indicated that the choice of ligand is of crucial importance in the development of antimicrobial silver complexes. Ideally, the ligands coordinated to silver should provide some desired physical, chemical, and/or biological properties that ultimately result in antimicrobial activity superior to that of pure silver salts. Although a number of silver complexes have been developed toward this goal, their biological use is often limited due to poor water solubility as in the case of AgSD [10].
Recently, we have been interested in the design and synthesis of ligands that could impart enhanced solubility of their silver complexes and rapid accumulation inside bacteria. In particular, we focused on the adamantyl moiety (Fig. 1a) which is lipophilic and has been shown to facilitate the process of cellular uptake of the compounds into which it is incorporated [11]. To date the adamantyl functionality has been widely utilized in the pharmaceutical industry including in many antiviral, antimicrobial and anticancer drugs used worldwide [11, 12]. 1-Adamantylamine, shown in Fig. 1c, is a derivative of adamantane that also possesses medicinal properties. Following this lead we first synthesized the Schiff base ligand 2-(quinonyl)iminoadamantane (qyAm) and the silver complex [Ag(qyAm)2](CF3SO3) (1) derived from it. The limited solubility of complex 1 in water however forced us to look for a better alternative. A scrutiny of the literature revealed that 1,3,5-triaza-7-phosphaadamantane (PTA, Fig. 1b) has been widely utilized in the synthesis of metal complexes with excellent cellular uptake properties [13,14]. In addition, PTA is water-soluble, and metal complexes derived from it also show high solubility in aqueous media. Such complexes have been reported to possess anticancer properties [15–19]. Therefore in our next attempt we incorporated 1,3,5-triazaadamant-7-amine (Fig. 1d), a derivative of PTA, in the Schiff base frame and isolated 2-(quinonyl)iminotriazaadamantane (qyTAm). This water-soluble ligand afforded complex [Ag(qyTAm)2](CF3SO3) (2) which exhibits significant solubility in aqueous media. Herein we report the syntheses, structures, and spectral characterization of the two complexes. In addition, the antibacterial properties of complex 2 have been assessed against both Gram-negative (Escherichia coli and P. aeruginosa) and Gram-positive (S. aureus) bacteria and their antimicrobial efficacies compared to that of silver nitrate and AgSD.
Fig. 1.
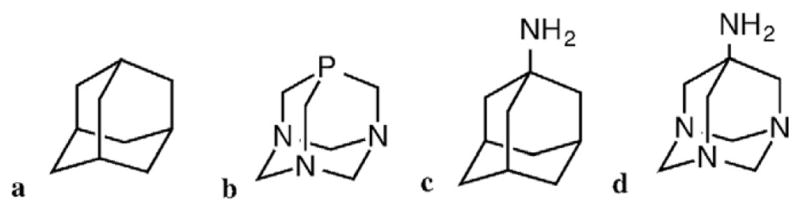
Structures of adamantane (a), 1,3,5-triaza-7-phosphaadamantane (PTA, b), 1-adamantylamine (c) and 1,3,5-triazaadamant-7-amine (d).
2. Experimental
2.1. Materials and methods
All reagents were of commercial grade and were used without further purification. The solvents were purified according to a standard procedure [20]. Quinoline-2-carboxaldehyde, silver trifluoromethanesulfonate and silver nitrate were purchased from Sigma Aldrich. 1-adamantylamine was purchased from Alpha Aesar. 1,3,5-triazaadamant-7-amine was purchased from MolPort. A PerkinElmer Spectrum-One FT-IR spectrometer was employed to obtain the IR spectra of the compounds. UV–vis spectra were recorded on a Varian Cary 50 UV–vis spectrophotometer. 1H NMR spectra at 298 K were monitored on a Varian Unity Inova 500 MHz instrument. Microanalyses (C, H, N) were performed using a Perkin-Elmer 2400 Series II elemental analyzer.
2.2. Synthesis of ligands
2.2.1. 2-(Quinonyl)iminoadamantane (qyAm)
Quinoline-2-carboxaldehyde 519.6 mg (3.3 mmol) and 1-adamantylamine 500 mg (3.3 mmol) were added to 35 mL of dry methanol and allowed to stir at room temperature for 18 h. The color of the solution changed from clear to pale-yellow within 30 min. After 18 h the volume of the solution was reduced to 5 mL and 15 mL of diethyl ether was added to it. The white solid thus separated was filtered and washed with cold methanol (yield: 777 mg, 81.2%). Anal. Calcd. for C20H22N2: C, 82.74; H, 7.67; N, 9.68. Found: C, 82.71; H, 7.65; N, 9.58. (KBr, cm−1): 2907 s, 2844 s, 1633 w, 1595 m, 1557 w, 1500 m, 1449 w, 1429 w, 1343 w, 1304 m, 1116 w, 1087 m, 890 w, 843 m, 753 s. 1H NMR (CDCl3, δ from TMS): 8.51 (s, 1H), 8.19 (d, 1H), 8.14 (d, 1H), 8.09 (d, 1H), 7.80 (d, 1H), 7.71 (t, 1H), 7.54 (t, 1H), 2.18 (s, 4H), 1.86 (s, 6H), 1.71(d, 5H). 13C NMR (CDCl3, δ from TMS): 156.62, 155.88, 147.68, 136.31, 129.56, 129.35, 128.64, 127.62, 127.02, 118.18, 58.38, 42.97, 36.47, 29.47.
2.2.2. 2-(Quinonyl)iminotriazaadamante (qyTAm)
2-(Quinonyl)iminotriazaadamante (qyTAm) was synthesized by following a procedure reported by us in a previous account [21]. The yield of the ligand (white solid) was 668 mg (68.8%). Anal. Calcd. for C17H19N5: C, 69.64; H, 6.58; N, 23.92. Found: C, 69.60; H, 6.55; N, 23.87. IR (KBr, cm−1): 2930 m, 2910 m, 2860 m, 1710 w, 1637 w, 1596 w, 1501 w, 1447 w, 1363 w, 1295 w, 1241 m, 1218 w, 1111 w, 1027 s, 993 s, 835 m, 821 s, 792 w, 751 m, 638 m. 1H NMR (CDCl3, δ from TMS): 8.43 (s, 1H), 8.15 (m, 2H), 8.09 (d, 1H), 7.82 (d, 1H), 7.73 (t, 1H), 4.52 (d, 3H), 4.15 (d, 3H), 3.54 (s, 6H). 13C NMR (CDCl3, δ from TMS): 160.27, 155.22, 137.04, 130.36, 129.94, 128.18, 128.00, 118.19, 74.06, 62.62.
2.3. Synthesis of complexes
2.3.1. [Ag(qyAM)2](CF3SO3) (1)
To a solution of 200 mg (0.779 mmol) of AgCF3SO3 in 15 mL in dry methanol, a batch of 263 mg (01.559 mmol) of qyAM in 10 mL of dry methanol was added and the mixture was allowed to stir for 18 h at room temperature. Next, the volume of the reaction mixture was reduced to 10 mL and 15 mL of diethyl ether was added. The white precipitate thus obtained was filtered, washed with cold methanol and dried in vacuo (yield: 470 mg, 72%). X-ray quality crystals were obtained by layering hexanes over solution of the complex in dichloromethane. Anal. Calcd. for C41H44N4O3F3SAg: C, 58.84; H, 5.32; N, 6.72. Found: C, 58.89; H, 5.29; N, 6.76. IR (KBr, cm−1): 2905 s, 2848 s, 645 w, 1592 w, 1508 m, 1453 w, 1431 w, 1343 w, 1307 m, 1263 s, 1223 m, 1154 s, 1031 s, 994 w, 934 w, 829 m, 757 m, 636 s, 572 w, 517 w. 1H NMR (CDCl3): 8.90 (s, 1H), 8.58 (d, 1H), 8.11 (d, 1H), 7.92 (d, 1H), 7.73 (d, 1H), 7.60 (t, 1H), 7.56 (t, 1H). 2.13 (s, 3H), 1.91 (s, 6H), 1.67 (d, 4H), 1.59 (d, 2H).
2.3.2. [Ag(qyTAm)2](CF3SO3) (2)
A batch of 460 mg 1.558 (mmol) of qyTAm in 10 mL of dry methanol was added to a solution of 200 mg (0.779 mmol) of AgCF3SO3 in 15 mL of dry methanol and the mixture was allowed to stir for 24 h at room temperature. Next, the volume of the pale yellow solution was reduced to 10 mL and 10 mL of diethyl ether was added. The white solid thus precipitated was filtered and dried in vacuo. X-ray quality crystals were obtained by slow evaporation of a solution of the complex in methanol. Yield: 468 mg (71%). Anal. Calcd. for C35H38N10O3F3SAg: C, 49.85; H, 4.58; N, 16.65. Found: C, 49.87; H, 4.53; N, 16.60. IR (KBr, cm−1) 2904 s, 2848 s, 1645 w, 1591 w, 1507 m, 1452 w, 1430 w, 1343 w, 1307 m, 1262 s, 1223 s, 1153 s, 1087 m, 1031 s, 994 w, 933 w, 828 m, 757 m 636 s, 571 w, 516 w. 1H NMR (CDCl3): 8.52 (s, 1H), 8.24 (d, 1H), 8.15 (d, 1H), 8.08 (d, 1H), 7.86 (d, 1H), 7.75 (t, 1H), 7.60 (t, 1H). 4.53 (s, 3H), 4.17 (s, 3H), 3.57 (d, 6H).
2.4. X-ray data collection and structure refinement
One suitable crystal from each sample vial was mounted on MitiGen loop with Paratone-N oil (Hampton Research) and transferred to the diffractometer. Data were collected on Bruker single crystal X-ray diffractometer equipped with PHOTON 100 and APEX II detector for complex 1 and 2 respectively. For complex 1 the data were collected with synchrotron radiation source by the ϕ and ω-scan technique in the range 3 ≤ 2θ ≤ 79 (Table 1) while for complex 2 sealed tube Mo source was employed and the data were collected by ω-scan technique in the range 3 ≤ 2θ ≤ 50 (Table 1). Data integration and reduction were performed with SAINT module [22]. The multi scan semi-empirical absorption correction was applied to the collected reflections using SADABS for both cases. The structures were generated using ShelXT (intrinsic phasing) [23] using Olex2 [24] graphical user interface and subsequently refined by full-matrix least squares procedure on F2 using ShelXL refinement package [25]. For both structures appropriate SHELX compatible constraints and restraints were applied to model some atoms with relatively higher anisotropic displacement parameters. All hydrogen atoms were included in calculated positions on the C atoms to which they are bonded, with C – H = 0.93 Å and Uiso(H) = 1.2Ueq(C). Calculations were also performed using the SHELXTL 2014 program package [26].
Table 1.
Crystal data and structure refinement parameters for 1 and 2.
| 1 | 2 | |
|---|---|---|
| Empirical formula | C41H44N4O3F3SAg | C35H38N10O3F3SAg |
| FW | 837.73 | 843.68 |
| Temp (K) | 150 | 298 |
| Crystal system | Monoclinic | Monoclinic |
| Space group | P2(1)/n | P2(1)/n |
| a (Å) | 18.9803(7) | 19.2289(7) |
| b (Å) | 18.6631(7) | 18.6472(6) |
| c (Å) | 22.8795(8) | 23.2634(8) |
| α (°) | 90 | 90 |
| β (°) | 112.823(2) | 112.7390(10) |
| γ (°) | 90 | 90 |
| V (Å3) | 7470.1(5) | 7693.1(5) |
| Z | 8 | 8 |
| Density (calcd) (g cm−3) | 1.490 | 1.457 |
| Abs coeff (mm−1) | 0.814 | 0.641 |
| No. of unique reflns | 34,412 (Rint = 0.0425) | 12,764 (Rint = 0.0193) |
| R1b (I > 2σ(I)) | 0.0397 | 0.0902 |
| wR2c | 0.1021 | 0.2651 |
| GOFa on F2 | 1.021 | 1.037 |
(No = number of observations, Nv = number of variables).
R1 = Σ||Fo| – |Fc|| / Σ|Fo|.
.
2.5. Antibacterial studies
The bacterial growth kinetics experiments were performed by following procedures reported by Chen and coworkers with a few modifications [27]. Briefly, the bacteria were first grown by spreading frozen liquid culture on a Luria Broth (LB) agar plate and incubating it at 37 °C for 18 h. Next, a single colony was isolated from this plate and used to inoculate 3 mL of LB. The bacterial culture was incubated for 18 h at 37 °C under constant shaking at 250 rpm. A 1 mL aliquot of this bacterial culture was then centrifuged at 5000 rpm for 5 min. The pellet thus obtained was re-suspended in sterile nano-pure water. This process was repeated once more and finally the pellet was added to a 5 mL volume of nano-pure water to obtain a new cell suspension having an absorbance of 0.1 at 600 nm. A 96-well plate was utilized to perform all growth experiments in triplicates, where the final volume of each well was 200 μL. The constituents consisted of 20 μL of the bacterial suspension (A600 = 0.1), 30 μl of sterile LB, varying amounts of solutions of the compounds (100, 80, 60, 40, 20, 10, 5, and 0 μL) and the corresponding amount of water required to bring the final volume to 200 μL. Upon inoculation and addition of the complexes, the plate was immediately placed in a plate reader at 37 °C where the optical density was measured at 600 nm for each independent well every minute for 24 h with 10 s mixing periods between reads.
3. Results and discussion
3.1. Synthesis and spectral properties of 1 and 2
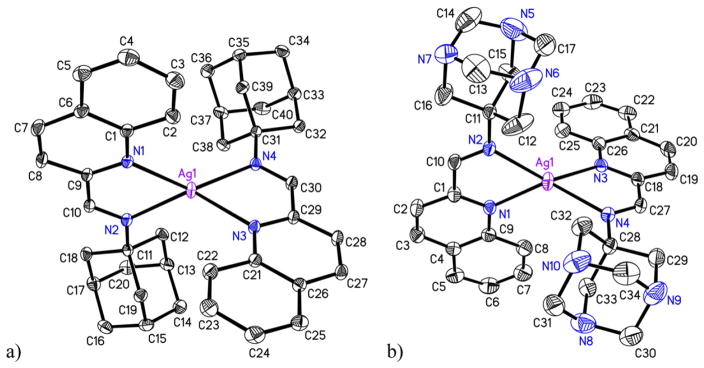
Reactions of qyAm and qyTAm with AgCF3SO3 in dry methanol afforded Ag(I) complexes 1 and 2 respectively as white crystalline solids. As evident from their structures (shown in Fig. 2a and b), both ligands bind to the Ag(I) center in a bidentate fashion utilizing the N donor atom of the quinoline ring and the imine N atom. Such coordination causes red shift of the absorption band maximum from 290 nm (in qyAm) to 325 nm in case of complex 1 in dichloromethane and from 303 nm (in qyTAm) to 330 nm in case of complex 2 in pure water. In addition, the 1H chemical shift values for complex 1 and 2 show notable differences in peak positions as compared to the free ligands further indicating coordination of the ligand. In case of complex 1 the proton from the imine functionality appears downfield (δ 8.90 ppm) as compared to the proton from the imine in the free ligand qyAm (δ 8.51 ppm). With complex 2, the proton from the imine functionality also appears down field (δ 8.52 ppm) as compared to the proton from the imine of the free ligand qyTAm (δ 8.43 ppm).
Fig. 2.
Molecular structures of the cations of (a) complex 1 and (b) complex 2. The thermal ellipsoids are drawn at 50% probability level. The hydrogen atoms and the counter anions are omitted for the sake of clarity.
3.2. Structures of 1 and 2
The molecular structures of complexes 1 and 2 were authenticated by single crystal X-ray crystallography and are shown in Fig. 2. Crystal data collection parameters are listed in Table 1 while selected metric parameters are presented in Table 2. Both complexes crystallize in monoclinic P2(1)/n space group with two crystallographically independent molecules in the asymmetric units. The coordination geometry around Ag is distorted tetrahedral. In 1 both chelate rings (constituted by Ag1, N1, C9, C10, N2 and Ag1, N3, C29, C30, N4 atoms) are reasonably planar with mean deviations of 0.033 and 0.039 Å respectively. The corresponding planes in 2 complex exhibit similar trend (mean deviations 0.027 and 0.028 Å). The dihedral angles between the two chelate planes for complexes 1 and 2 are 80.4 and 78.3° respectively. In each case, the average Ag–N distance (2.328 and 2.327 Å for 1 and 2 respectively) is quite similar for the two cations within the asymmetric unit. Distorted tetrahedron coordination geometry is not uncommon for Ag complexes with N donor ligands. A similar Ag complex with N donor namely, [Ag(bpy-OH)2]+ (where bpy-OH = bis(hydroxymethyl)-2,2′-bipyridne) exhibits distorted tetrahedral geometry with average Ag–N distance of 2.320 Å [28] comparable to that observed for the present complexes. No noticeable π-π stacking or other intermolecular interactions are realized upon careful examination of the crystal packing for both the structures.
Table 2.
Selected bond distances (Å) and angles (°) for complexes 1 and 2
| Complex 1
|
Complex 1
|
|||
|---|---|---|---|---|
| Molecule 1 | Molecule 2 | Molecule 1 | Molecule 2 | |
| Ag1–N1 | 2.3501(11) | – | 2.355(5) | – |
| Ag1–N2 | 2.3183(12) | – | 2.308(5) | – |
| Ag1–N3 | 2.3541(12) | – | 2.359(5) | – |
| Ag1–N4 | 2.3156(11) | – | 2.305(6) | – |
| Ag2–N5 | – | 2.3265(11) | – | 2.317(6) |
| Ag2–N6 | – | 2.3224(12) | – | 2.326(5) |
| Ag2–N7 | – | 2.3155(11) | – | 2.324(6) |
| Ag2–N8 | – | 2.3196(11) | – | 2.322(6) |
| N1–Ag1–N2 | 72.27(4) | – | 72.44(18) | – |
| N1–Ag1–N4 | 139.16(4) | – | 131.0(2) | – |
| N2–Ag1–N3 | 130.07(4) | – | 138.6(2) | – |
| N1–Ag1–N3 | 119.34(4) | – | 115.30(19) | – |
| N4–Ag1–N2 | 132.79(4) | – | 135.4(2) | – |
| N4–Ag1–N3 | 72.66(4) | – | 72.5(2) | – |
| N7–Ag2–N8 | – | 72.60(4) | 72.9(2) | |
| N7–Ag2–N5 | – | 130.73(4) | 131.3(2) | |
| N8–Ag2–N5 | – | 123.73(4) | 134.7(2) | |
| N7–Ag2–N6 | – | 136.75(4) | 126.2(2) | |
| N8–Ag2–N6 | – | 129.55(4) | 129.1(2) | |
| N5–Ag2–N6 | – | 72.82(4) | 72.5(2) | |
3.3. Solubility in aqueous media
Rapid precipitation (as AgCl) in biological media often poses as a problem in the treatment of burn wounds with AgNO3. Such precipitation renders the bioactive Ag+ ions unavailable at the wound bed. Because effective treatment of infections requires sustained release of Ag+, researchers have attempted use of different silver complexes to resist the rapid exit of the bioactive “drug” from the interaction zone [19, 28,29]. In the present study only 2 exhibits solubility in water (1 mg/mL) and we have carefully measured the stability of 2 in water at 37° C. As shown in Fig. S1 (Supporting information), complex 2 slowly releases Ag+ in such solution. Periodic checking of the absorption spectrum of a solution of 2 in water reveals that the spectrum slowly transitions to the spectrum of the ligand over a period of 2 h. This observation confirms the fact that 2 is an effective silver-donating complex that ensures slow release of Ag+ when applied to infected wounds. It is important to note that constant exposure of such solutions to visible light does not expedite the rate of silver release (as monitored by absorption spectroscopy), a fact that confirms the stability of the complex toward light.
3.4. Antibacterial studies
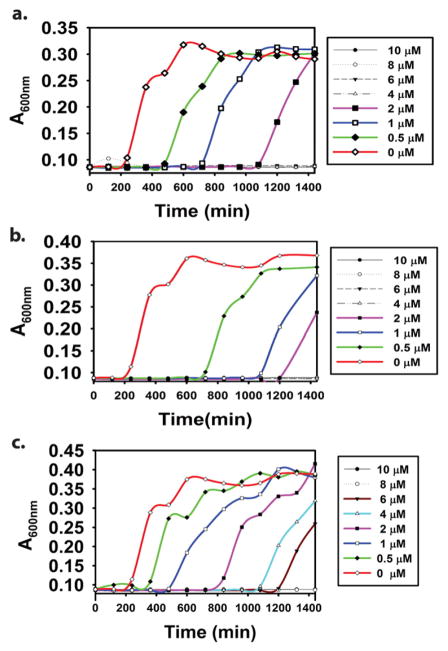
Antibacterial activities of both complexes have been assessed in the present study against two Gram-negative (E. coli and P. aeruginosa) and one Gram-positive (S. aureus) bacteria. The ligands by themselves display no antimicrobial activity against any of the three bacteria up to a concentration of 100 μM. While complex 1 exhibits limited solubility in water, complex 2 is readily soluble in water and several bacterial growth media. This allowed us to determine the minimal inhibitory concentration (MIC) of complex 2. MIC is defined as the lowest concentration of the compound able to inhibit the growth of the microorganism. In case of E. coli, 2 exhibited a MIC value of 2 μM, a value comparable to AgNO3 (2 μM) as well as AgSD (4 μM Table 3). As previously stated, both AgNO3 and AgSD are regularly utilized in Burn Victim Wards of hospitals for wound healing purposes. The bacterial growth curves for P. aeruginosa with increasing concentration of 2 are shown in Fig. 3. With this Gram-negative pathogen 2 exhibited a slightly higher MIC value of 4 μM. The corresponding MIC values for AgNO3 and AgSD were 4 μM and 8 μM, respectively. A somewhat higher MIC value (8 μM) was obtained for 2 against the Gram-positive bacterium S. aureus. Incidentally, higher MIC values were also noted for both AgNO3 (6 μM) and AgSD (8 μM) against this nosocomial pathogen (Table 3). These higher MIC values most possibly arise from differences in cell membrane structure and compositions of the Gram-positive and Gram-negative bacteria employed in this study [30]. Bacterial growth curves for both E. coli and S. aureus are included in Supplementary information.
Table 3.
MIC values of the different silver salts and complex 2
| E. coli | P. aeruginosa | S. aureus | |
|---|---|---|---|
| Compound | MIC (μM) | MIC (μM) | MIC (μM) |
| AgNO3 | 2 | 4 | 6 |
| AgSD | 4 | 8 | 8 |
| 2 | 4 | 4 | 8 |
Fig. 3.
Bacterial growth curves for P. aeruginosa in presence of complex 2 (a), AgNO3 (b), and AgSD (c). The y-axis absorbance is directly proportional to bacterial population.
To date the antimicrobial and antifungal effects of a few silver complexes have been reported. In a recent report, Smolenski et al. have described the antimicrobial activity of a highly light-stable and water-soluble silver complex derived from N-methyl-1,3,5-triaza-7-phosphaadamantane (mPTA) against both Gram-positive and Gram-negative bacteria [30]. Polymeric complexes with silver-organic network with bridging PTA ligands also exhibit solubility in water and significant antibacterial and antifungal activity [31]. Interesting antimicrobial behavior has been noted with a silver(I) nicotinate cluster [Ag(Hmna)6] and its anionic form (H2mna = 2-mercaptonicotinic acid). The neutral complex has shown activity against both Gram-positive and Gram-negative bacteria while the anionic complex exhibited activity only against Gram-negative bacteria [32]. The difference has been attributed to solubility, transport phenomena, and ligand exchange equilibria. Along the same line, a neutral and water-soluble complex of silver(I) with L-histidine namely [Ag(Hhis)], exhibits a wide spectrum of antibacterial and antifungal activity [33]. It is generally believed that facile ligand replacement by biological ligands in silver(I)—N bonding compounds is a key factor that leads to wide spectrum of antimicrobial activity [34]. The solubility properties and unusually strong antibacterial activity (Table 3) of the present complex 2 with silver(I)—N bonding appears to support this hypothesis. Although the ligand qyTAm itself does not exhibit significant antibacterial properties, the presence of triazaadamantyl moiety in the ligand frame facilitates increased cellular uptake of 2 and increases its solubility in aqueous media. Taken together, the strong antibacterial activity of complex 2 demonstrates the relationship between solubility and structure/activity of silver(I) complexes which can be further explored toward isolation of more effective silver-containing pharmaceuticals to thwart wound infections.
4. Conclusions
In conclusion two Ag(I) complexes 1 and 2, derived from designed Schiff base ligands, have been synthesized and structurally characterized. Complex 2 is readily soluble in water and releases Ag+ slowly in such solution. This property of 2 has been exploited to eradicate Gram-negative and Gram-positive bacteria. The MIC values of 2 against both classes of pathogen are very comparable to AgNO3 and AgSD. The slow release of Ag+ from 2 in water could be an advantage in keeping the silver donor more in circulation (compared to AgNO3) and avoid frequent dressing of infected burn wounds to remove the precipitated AgCl.
Supplementary Material
Acknowledgments
Financial support from the NSF grant DMR-1409335 is gratefully acknowledged. JJ was supported by the NIH grant 2R25GM058903. The Advanced Light Source is supported by the Director, Office of Basic Energy Sciences of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Abbreviations
- AgSD
Silver sulfadiazine
- PTA
1,3,5-triaza-7-phosphaadamantane
- mPTA
N-methyl-1,3,5-triaza-7-phosphaadamantane
- qyAm
2-(quinonyl)iminoadamantane
- qyTAm
2-(quinonyl)iminotriazaadamantane
- LB
Luria Broth
- MIC
Minimal inhibitory concentration
- H2mna
2-mercaptonicotinic acid
- Hhis
L-histidine
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jinorgbio.2016.12.009.
References
- 1.Jones RN. Am J Med. 1996;100:3S–12S. doi: 10.1016/s0002-9343(96)00102-7. [DOI] [PubMed] [Google Scholar]
- 2.Korvick JA, Yu VL. Antimicrob Agents Chemother. 1991;35:2167–2172. doi: 10.1128/aac.35.11.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones RN, Pfaller MA. Diagn Microbiol Infec Dis. 1998;31:379–388. doi: 10.1016/s0732-8893(98)00037-6. [DOI] [PubMed] [Google Scholar]
- 4.Bowler PG, Duerden BI, Armstrong DG. Clin Microbiol Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fong J, Wood F. Int J Nanomedicine. 2006;1:441–449. doi: 10.2147/nano.2006.1.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra I. J Antimicrob Chemother. 2007;59:587–590. doi: 10.1093/jac/dkm006. [DOI] [PubMed] [Google Scholar]
- 7.Klasen HJ. Burns. 2000;26:131–138. doi: 10.1016/s0305-4179(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 8.Jensen RH, Davidson N. Biopolymers. 1966;4:17–32. [Google Scholar]
- 9.Azocar IM, Gomez G, Levin P, Paez M, Muñoz H, Dinamarca N. J Coord Chem. 2014;67:3840–3853. [Google Scholar]
- 10.Rai M, Yadav A, Gade A. Biotechnol Adv. 2009;27:76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Lamoureux G, Artavia G. Curr Med Chem. 2010;17:2967–2978. doi: 10.2174/092986710792065027. [DOI] [PubMed] [Google Scholar]
- 12.Wanka L, Iqbal K, Schreiner PR. Chem Rev. 2013;113:3516–3604. doi: 10.1021/cr100264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babak MV, Meier SM, Huber KVM, Reynisson J, Legin AA, Jakupec MA, Roller A, Stukalov A, Gridling M, Bennett KL, Colinge J, Berger W, Dyson PJ, Superti-Furga G, Keppler BK, Hartinger CG. Chem Sci. 2015;6:2449–2456. doi: 10.1039/c4sc03905j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murraya BS, Babak MV, Hartinger CG, Dyson PJ. Coord Chem Rev. 2016;306:86–114. [Google Scholar]
- 15.Allardyce CS, Dyson PJ. Dalton Trans. 2016;45:3201–3209. doi: 10.1039/c5dt03919c. [DOI] [PubMed] [Google Scholar]
- 16.Lee RFS, Escrig S, Croisier M, Clerc-Rosset S, Knott GW, Meibom A, Davey CA, Johnsson K, Dyson PJ. Chem Commun. 2015;51:16486–16489. doi: 10.1039/c5cc06983a. [DOI] [PubMed] [Google Scholar]
- 17.Kilpin KJ, Crot S, Riedel T, Kitchen JA, Dyson PJ. Dalton Trans. 2014;43:1443–1448. doi: 10.1039/c3dt52584h. [DOI] [PubMed] [Google Scholar]
- 18.Atrian-Blasco E, Gascon S, Rodriguez-Yoldi MJ, Laguna M, Cerrada E. Eur J Inorg Chem. 2016;17:2791–2803. [Google Scholar]
- 19.Smolenski P, Jaros SW, Pettinari C, Lupidi G, Quassinti L, Bramucci M, Vitali LA, Petrelli D, Kochel A, Kirillov AM. Dalton Trans. 2013;42:6572–6581. doi: 10.1039/c3dt33026e. [DOI] [PubMed] [Google Scholar]
- 20.Armarego WLF, Chai CLL. Purification of Laboratory Chemicals. 7. Butterworth Heinmann; Oxford: 2003. [Google Scholar]
- 21.Jimenez J, Chakraborty I, Carrington SJ, Mascharak PK. Dalton Trans. 2016;45:13204–13213. doi: 10.1039/c6dt01358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.SAINT-plus (Version 7.03) Bruker AXS Inc; Madison, Wisconsin, USA: 2015. [Google Scholar]
- 23.Sheldrick GM. Acta Cryst. 2015;A71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolomanov OV, Bourhis LJ, Gildea RJ, Howard JAK, Puschmann H. J Appl Crystallogr. 2009;42:339–341. doi: 10.1107/S0021889811041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheldrick GM. Acta Cryst. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 26.Sheldrick GM. Acta Cryst. 2015;C71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas-Andrade M, Cho AT, Hu P, Lee SJ, Deming CP, Sweeney SW, Saltikov C, Chen S. J Mater Sci. 2015;50:2849–2858. [Google Scholar]
- 28.deBoer TR, Chakraborty I, Olmstead MM, Mascharak PK. Cryst Growth Des. 2014;14:4901–4905. doi: 10.1021/cg501175g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaros SW, Smolenski P, Guedes da Silva MFC, Florek M, Krol J, Staroniewicz Z, Pombeiro AJL, Kirillov AM. CrystEngComm. 2013;15:8060–8064. [Google Scholar]
- 30.Smolenski P, Pettinari C, Marchetti F, Guedes da Silva MFC, Lupidi G, Patzmay GVB, Petrelli D, Vitali LA, Pombeiro AJL. Inorg Chem. 2015;54:434–440. doi: 10.1021/ic501855k. [DOI] [PubMed] [Google Scholar]
- 31.Jaros SW, Guedes da Silva MFC, Florek M, Oliveira MC, Smolenski P, Pombeiro AJL, Kirillov AM. Cryst Growth Des. 2014;14:5408–5417. [Google Scholar]
- 32.Tsyba I, Bun-kit Mui B, Bau R, Noguchi R, Nomiya K. Inorg Chem. 2003;42:8028–8032. doi: 10.1021/ic030149m. [DOI] [PubMed] [Google Scholar]
- 33.Nomiya K, Takahashi S, Noguchi R, Nemoto S, Takayama T, Oda M. Inorg Chem. 2000;39:3301–3311. doi: 10.1021/ic990526o. [DOI] [PubMed] [Google Scholar]
- 34.Medici S, Peana M, Crisponi G, Nurchi VM, Lachowicz JI, Remelli M, Zoroddu MA. Coord Chem Rev. 2016;327–328:349–359. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.