Abstract
Muscle regeneration is a dynamic process during which cell state and identity change over time. A major roadblock has been a lack of tools to resolve a myogenic progression in vivo. Here we capitalize on a transformative technology, single-cell mass cytometry (CyTOF), to identify in vivo skeletal muscle stem cell and previously unrecognized progenitor populations that precede differentiation. We discovered two cell surface markers, CD9 and CD104, whose combined expression enabled in vivo identification and prospective isolation of stem and progenitor cells. Data analysis using the X-shift algorithm paired with single-cell force directed layout visualization, defined a molecular signature of the activated stem cell state (CD44+/CD98+/MyoD+) and delineated a myogenic trajectory during recovery from acute muscle injury. Our studies uncover the dynamics of skeletal muscle regeneration in vivo and pave the way for the elucidation of the regulatory networks that underlie cell-state transitions in muscle diseases and aging.
Adult muscle stem cells (MuSCs) drive skeletal muscle repair and regeneration1. Normally quiescent during homeostasis, MuSCs become activated upon muscle injury to replenish the stem cell pool, and simultaneously give rise to progeny that will differentiate and repair the damage2. While the role and phenotype of stem cells in muscle regeneration has been extensively studied, little is known about the myogenic progenitor stage, due to the lack of tools to capture this transient cell population in vivo.
In hematopoiesis, the identification and isolation of hematopoietic stem and progenitor cells by standard cell sorting methods3 and single-cell mass cytometry4 have enabled the discovery of the key pathways that go awry at specific stages of development5,6, profoundly impacting the treatment of a multitude of blood diseases3,7. Similarly, the elucidation of myogenic progenitors in vivo has the potential to define the key molecular events that govern cell-state transitions during the course of regeneration, and drive the development of new therapeutic strategies for muscle diseases.
To address the cellular and molecular complexity of the myogenic compartment, a major challenge in the muscle field, we applied a high-dimensional single-cell platform called Mass Cytometry, also known as Cytometry by Time of Flight (CyTOF). CyTOF enables the simultaneous measurements of up to 50 parameters per single cell using antibodies conjugated to metal isotopes4,8. The multidimensional attribute of CyTOF allowed us to identify previously unrecognized progenitor cell populations in vivo, based on surface marker combinations and to determine their role during muscle regeneration. This study defines an in vivo developmental progression from stem to progenitor cells in skeletal muscle, providing the foundation for future studies of cellular signaling dysfunction within these myogenic populations in the context of aging, dystrophy and other disease states. Moreover, it paves the way for future investigations of such cell populations in other systems.
RESULTS
Identification of a myogenic progression in vivo by single-cell mass cytometry
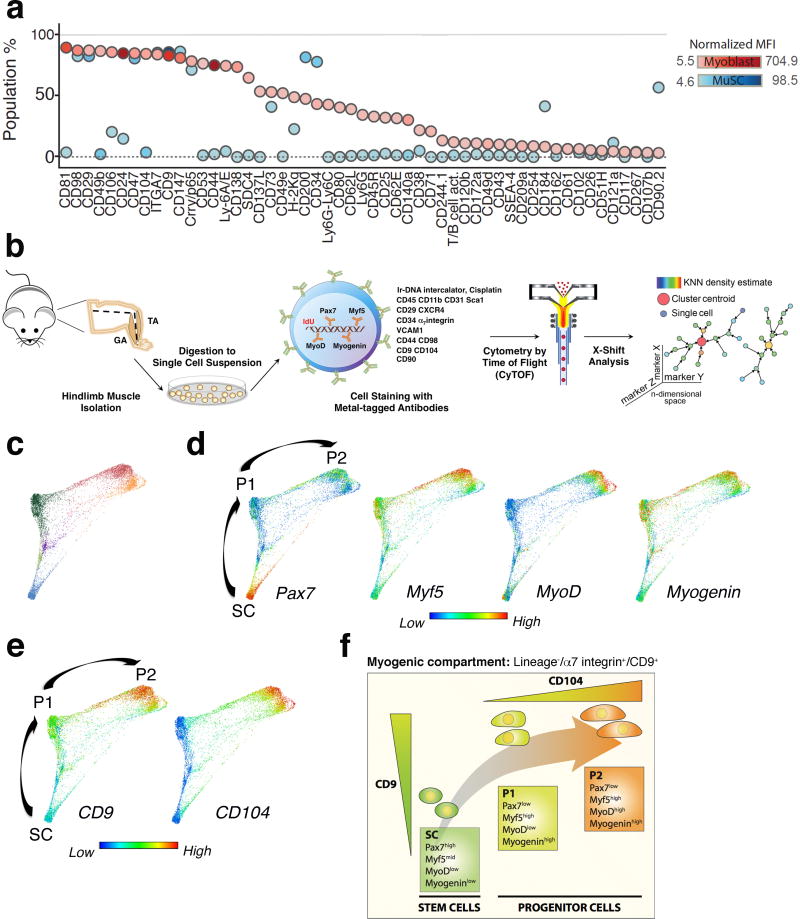
To discover surface markers that could uniquely distinguish between myogenic stem and progenitor cells in vivo in skeletal muscle, we performed a high-throughput fluorescence-based flow cytometry screen with 176 antibodies to integral membrane proteins in both MuSCs, isolated from Pax7-ZsGreen reporter mice9, and myoblasts, a primary culture system used to study the late stages of myogenic differentiation and fusion. Flow cytometry data analysis identified several surface markers (Fig.1a), for which antibodies were then included in our CyTOF panel based on two criteria: (i) presence of the markers on either MuSCs or myoblasts, (ii) differential expression levels on MuSCs versus myoblasts. In addition, the screen confirmed the expression on Pax7-ZsGreen MuSCs of known markers previously used to isolate MuSCs, such as α7 integrin and CD3410, β1 integrin/CD29 and CXCR4/CD18411, and VCAM/CD10612 (Fig.1a).
Figure 1. Identification of distinct cell surface markers that delineate a myogenic progression in vivo.
(a) Cell surface marker screening panel analysis of muscle stem cells (MuSCs) and myoblasts. A single-cell suspension of hindlimb muscle isolated from Pax7-ZsGreen reporter mice, and cultured myoblasts were stained with 176 cell surface antibodies and analyzed by fluorescence-based flow cytometry. MuSCs were identified as ZsGreen+ cells. The plot shows the fraction of cells expressing each cell surface marker (y axis) and the level of protein expression, indicated as intensity of blue (MuSCs) and red (Myoblasts). (b) CyTOF mass cytometry workflow. Tibialis Anterior (TA) and Gastrocnemius (GA) muscles were triturated, digested to a single-cell suspension, stained with isotope-chelated antibodies and run through the CyTOF instrument. Stained cells were passed through an inductively-coupled plasma, atomized, ionized, and the elemental composition was mass measured. Signals corresponding to each elemental tag were correlated to the presence of the respective isotopic marker. Data were analyzed using standard flow cytometry software and the clustering algorithm X-shift. (c) Live/Lineage−/α7integrin+/CD9+ cells gated from murine hindlimb muscles (TA and GA) were analyzed with the X-shift algorithm (K=30 was auto-selected by the switch-point finding algorithm) yielding six clusters (color-coded in blue, purple, light green, dark green, red and orange). These clusters were visualized using single-cell force-directed layout. Up to 2000 cells were randomly selected from each X-shift cluster, each cell was connected to 30 nearest neighbors in the phenotypic space and the graph layout was generated using the ForceAltas2 algorithm13–15 (representative experiment, n= 3 mice; 4 independent experiments). (d) Expression level of the myogenic transcription factors Pax7, Myf5, MyoD and Myogenin was visualized in the X-shift clusters shown in (c). Developmental time was inferred and three distinct populations were identified as SC, P1 and P2 (representative experiment, n= 3 mice). (e) Expression level of CD9 and CD104 was visualized in the X-shift clusters shown in (c) (representative experiment, n= 3 mice). (f) Model summarizing the expression pattern of the newly identified surface markers, CD9 and CD104, during the proposed transition from the stem cell (SC) to the progenitor (P1, P2) state.
To identify myogenic progenitors in vivo within skeletal muscle, we capitalized on the CyTOF technology and assembled a panel of 23 isotope-conjugated antibodies. These reagents were used to simultaneously measure, at the single-cell level, the expression of previously unrecognized surface markers revealed in our screen, known surface proteins used to isolate MuSCs by us and others, and myogenic transcription factors known to define distinct stages of myogenesis1 (Supplementary Table 1, Supplementary Fig.1a–d). Single-cell suspensions of hindlimb muscles, Gastrocnemius (GA) and Tibialis Anterior (TA) were prepared from 8-week old mice as described previously10 and processed for CyTOF analysis (Fig.1b). To distinguish stem and progenitor cell populations, we analyzed the CyTOF dataset with a recently developed K-nearest neighbor density-based clustering algorithm called X-shift13, which performed unsupervised clustering analysis of cells within the myogenic compartment, defined as Lineage−/α7integrin+ cells and subsequently refined to Lineage−/α7integrin+/CD9+ cells. With this new multivariate algorithm six clusters were generated, based on the expression of known and previously unrecognized surface markers, and key myogenic transcription factors. In order to visualize the spatial relationships between the cell types within these X-shift clusters, 2000 randomly sampled cells from each cluster were subjected to a force-directed layout13–15. The resultant map revealed that cells fell densely into three prominent clusters and were linked by sparsely populated paths (Fig.1c). Cells within the densely populated regions were characterized by differential expression of the myogenic transcription factors Pax7, Myf5, MyoD and Myogenin (Fig.1d), which are known to mark muscle stem cells, activated stem cells, committed and differentiated muscle cells, respectively1. The unique expression pattern of the myogenic transcription factors allowed us to infer developmental stage for these clusters, which we named stem cell population SC, based on high Pax7 expression, and progenitor cell populations P1 and P2, based on Myf5, MyoD and Myogenin expression (Fig.1d). The co-expression patterns of two surface markers identified in the screen, CD9, a tetraspannin16,17 and CD104, an integrin18–22, distinctively defined these populations. SC and P1 were distinguished by differential expression levels of CD9 (intermediate in SC and high in P1), whereas P2 expressed high levels of both CD9 and CD104 (Fig.1e). These in vivo data were supported by the results of the flow cytometry screen in which CD9 was expressed at higher levels in myoblasts than in MuSCs (Supplementary Fig.1e, f) and CD104 was expressed exclusively by myoblasts (Fig.1a, Supplementary Fig.1e). Importantly, the combination of these two surface markers identified myogenic progenitors in vivo, a critical missing link between stem cells and differentiated muscle cells.
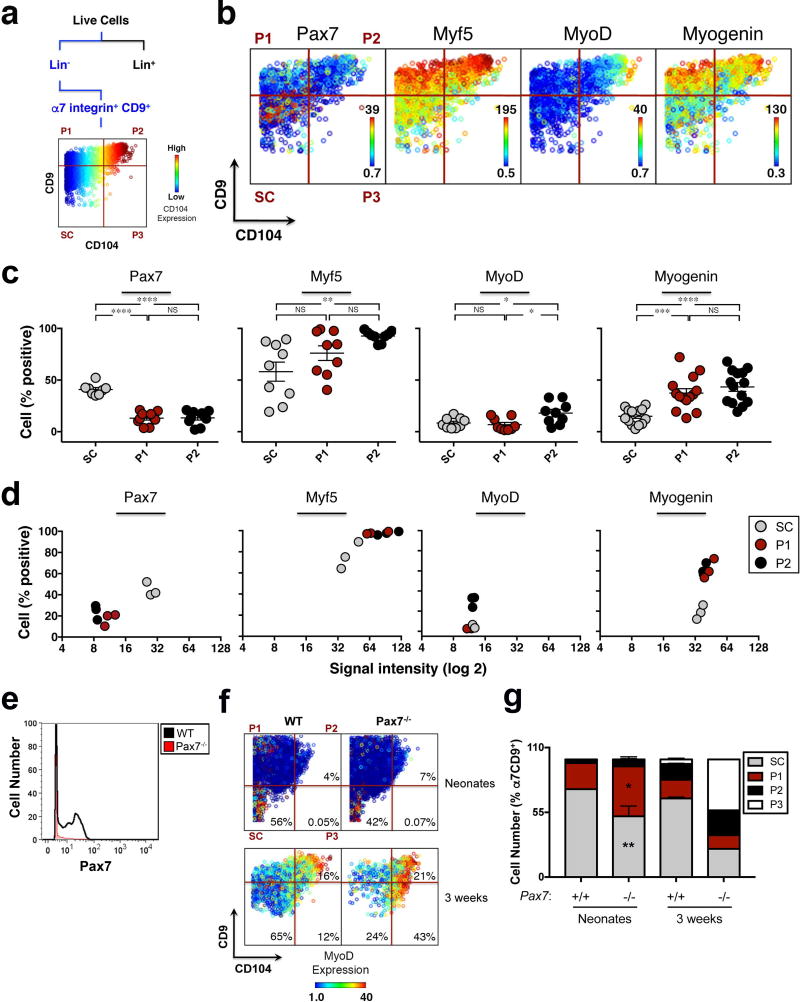
Data analysis by principal component analysis (PCA), a dimensionality reduction algorithm, revealed that the progenitor populations (P1 and P2) differ from the stem cell population (SC) based on their protein co-expression patterns (Supplementary Fig.1g). This corroborated the X-shift clustering analysis and led us to hypothesize that staining with antibodies against CD9 and CD104 could serve as a strategy to prospectively isolate stem (Pax7high/Myf5mid/MyoDlow/Myogeninlow) and progenitor (Pax7low/Myf5high/MyoDlow/high/Myogeninhigh) cells by Fluorescence Activated Cell Sorting (FACS) (Fig.1f). Importantly, the three X-shift clusters could be identified by manual gating, the gold-standard of flow cytometry analysis, solely based on the expression of CD9 and CD104 (Fig.2a, Supplementary Fig.1d). Manual gating also revealed a rare population, P3 (Fig.2a), whose frequency was too low for functional assays. Therefore, these two markers now provide a sorting strategy to prospectively isolate stem and progenitor cell populations that represent landmarks of myogenesis, as confirmed by the differential expression of the myogenic transcription factors Pax7, Myf5, MyoD and Myogenin, shown in the third dimension (Fig.2b).
Figure 2. Unique strategy for prospective isolation of stem and progenitor cells in vivo in skeletal muscle.
(a) Scheme of gating strategy for CyTOF data. Live cells are identified based on lack of cisplatin binding. Live/Lineage−/α7integrin+/CD9+ cells are selected and a biaxial dot plot of CD9 (y axis) by CD104 (x axis), colored by channel (CD104 expression), is shown. (b) Representative biaxial dot plot of CD9 by CD104 colored by channel, showing expression of Pax7, Myf5, MyoD and Myogenin in the stem and progenitor cells populations. (c) Data summary shows the fraction of cells (y axis) within populations SC, P1 and P2, expressing Pax7, Myf5, MyoD and Myogenin ((n=10 mice (Pax7), n= 9 (Myf5 and MyoD), 3 independent experiments; n=14 (Myogenin), 4 independent experiments). Line represents mean ± SEM. ANOVA test was performed with significance determined by Bonferroni's multiple comparisons test. (d) Data summary of myogenic transcription factor expression levels within populations SC, P1 and P2. Each graph shows the relationship between the percentage of positive cells (y axis) and the signal intensity within the positive population (x axis, log2) for the expression of Pax7, Myf5, MyoD and Myogenin (representative experiment, n=3 mice). (e) Histogram overlay of Pax7 expression in muscle isolated from Pax7 knock-out (Pax7−/−) and wild type (WT) mice and stained with an isotope-chelated antibody against Pax7. (f) Representative biaxial dot plots of CD9 by CD104 colored by channel, showing MyoD expression within populations SC, P1 and P2, in Pax7−/− and WT muscle isolated from neonates and 3 weeks old mice. (g) Stacked columns indicate the relative proportion of each population within the Live/Lineage−/α7 integrin+/CD9+ myogenic compartment in Pax7−/− and WT muscle isolated from neonates (mean ± SEM from n=3 mice, 2 independent experiments) and 3 weeks old mice (n=1 Pax7−/−; mean ± SEM from n=10 WT, 2 independent experiments). Multiple t-tests analysis with Bonferroni correction was used to determine difference between Pax7−/− and WT neonates within populations SC, P1, P2. *, **, *** and **** represent statistical significance at p<0.05, p<0.01, p<0.001 and p<0.0001 respectively. NS represents statistically non-significant.
Quantification of each population, identified in the biaxial dot plot of CD9 by CD104, showed that SC comprised the highest fraction of Pax7+ cells (Fig.2c), with the highest expression levels (Fig.2d). In contrast, P1 and P2 comprised the highest fraction of Myf5+ and Myogenin+ cells (Fig.2c), with the highest expression levels (Fig.2d). P2 also comprised a small fraction of MyoD+ cells (Fig.2b–d). These data are consistent with the identification of SC as the MuSC population and P1 and P2 as previously unrecognized progenitor populations (Fig.1c–e). Further, they suggest a model in which as cells transition from the stem cell state (SC) to the differentiated myogenic state, they sequentially upregulate the expression of CD9 and CD104, giving rise to P1 and then P2 (Fig.1f). In support of this model Pax7 knock-out (Pax7−/−) mice demonstrated a shift in the distribution of cell populations SC, P1, P2 and P3 (Fig.2f). Compared to wild type controls, the fraction of cells in SC in Pax7−/− mice was markedly diminished at the neonatal stage (Fig.2f, g) and progressively declined by 3 weeks of age (Fig.2f, g). At birth, the decrease in Pax7−/− cells within the SC population was paralleled by an increase in the fraction of cells in P1 (Fig.2g), and was accompanied by alteration of a key signaling pathway involved in the regulation of myogenic commitment23–26, as shown by increased phosphorylation of MAPKAPK2 (Supplementary Fig.2a), a direct target of p38α/β MAPK27. In one rare Pax7−/− mouse that survived to three weeks of age, a majority of α7integrin+/CD9+ muscle cells were in P2 and P3. The cells expressed high levels of MyoD (Fig.2f) and Myogenin (Supplementary Fig.2b), and exhibited increased phosphorylation of MAPKAPK2 (Supplementary Fig.2a), indicative of premature differentiation. Collectively, our data suggest that in Pax7−/− muscle, stem cells are progressively lost, the stem cell state is not maintained, and committed cells are the prevalent population by three weeks of age, when the role of Pax7 is required and mutant mice start to die28,29.
The newly identified progenitor populations originate from muscle stem cells and exhibit myogenic potential in vitro and in vivo
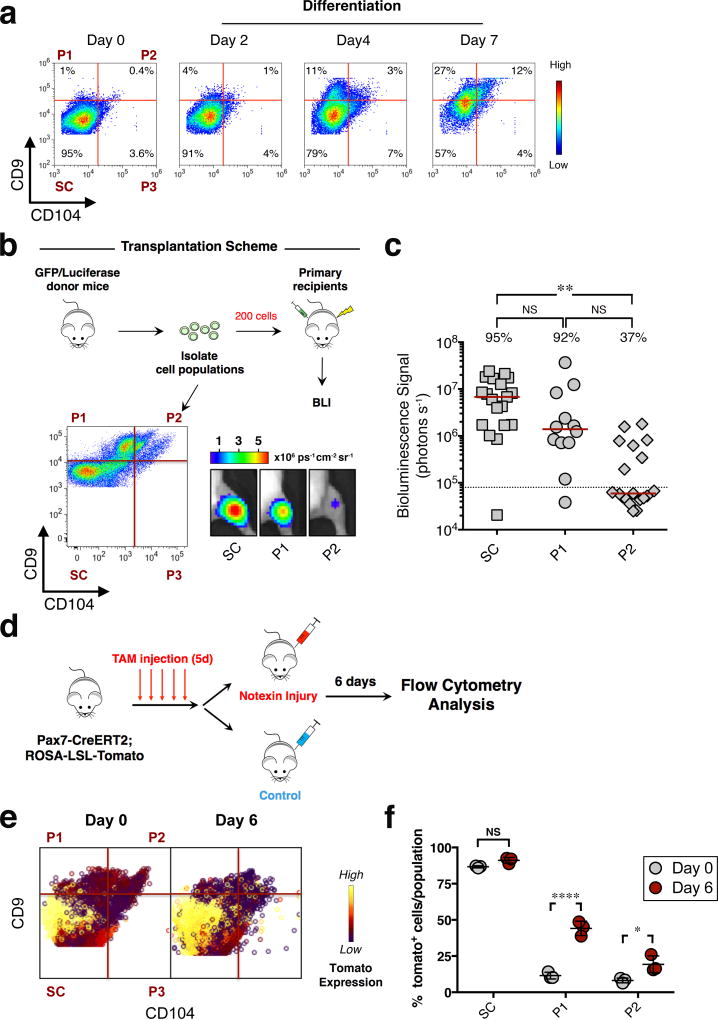
To determine if the progenitor populations were derived from MuSCs, we cultured α7integrin+/CD34+ MuSCs isolated from murine hindlimbs, and measured changes in the expression of CD9 and CD104 during commitment to differentiation. Sorted MuSCs were expanded on hydrogels for one week and then treated with differentiation media for 2, 4, and 7 days respectively (Fig.3a). On day 2 of the assay, the majority of cells comprised stem cells, SC. Over the course of one week the fraction of cells bearing the P1 phenotype progressively increased from 1% to 27%, whereas cells bearing the P2 phenotype increased from 0.4% to 12% (Fig.3a). These results are consistent with a muscle stem cell of origin giving rise to sequential populations of myogenic progenitor cells that could now be distinguished by differential expression of CD9 and CD104.
Figure 3. Progenitor cell populations originate from muscle stem cells and exhibit distinct regenerative capacity in vivo.
(a) Flow cytometry analysis of sorted α7integrin+/CD34+ cells, cultured in growth media for 1 week on biomimetic hydrogels and then in differentiation media on collagen-coated plates for 2, 4, 7 days respectively. Representative biaxial plots of CD9 by CD104 show the fraction of populations SC, P1, P2 and P3 at the indicated time points (n=5, 2 independent experiments). (b) Scheme depicting the in vivo assay of regenerative capacity. Hindlimb muscles isolated from GFP/Luciferase mice were digested to a single-cell suspension. Cell populations SC, P1 and P2, were sorted based on expression of CD9 and CD104 (dot plot, lower left panel) and transplanted (200 cells/injection) into the irradiated TA muscle of NOD/SCID mice. Representative BLI images at 5 weeks post transplant are shown (lower right panel). (c) Scatter plot shows the percentage of transplants from each condition that engrafted above threshold (dashed line, 80,000 photons/s) into recipient tissue and the BLI signal intensity (y axis). Line represents median BLI signal (n= 19 mice (SC and P2), 3 independent experiments; n= 12 (P1), 2 independent experiments). ANOVA test was performed with significance determined by Bonferroni’s multiple comparisons test. (d) Scheme depicting lineage-tracing experiment to track progenitor populations in vivo upon injury. (e) TA muscle isolated from uninjured and injured mice (day 6 post-injury) was digested to a single-cell suspension and stained using fluorescently-conjugated antibodies against lineage markers, α7integrin, CD9 and CD104. Representative biaxial dot plots of CD9 by CD104 colored by channel, show the expression of tdTomato in populations SC, P1 and P2 at day 0 and day 6-post injury (n=3 mice per condition). (f) The scatter plot shows the fraction of tomato+ cells in populations SC, P1 and P2 at day 0 and day 6-post injury (mean ± SD from n=3 mice per condition). Multiple t-tests analysis with Bonferroni correction was used to determine the difference between the two groups (uninjured and injured) within population SC, P1 and P2. *, **, and **** represents statistical significance at p<0.05, p<0.01 and p<0.0001. NS represents statistically non-significant.
To determine the functional capabilities of the myogenic progenitors in vitro and in vivo, two independent assays were designed. In the first assay, individual stem and progenitor cell populations were sorted, placed into culture for a week on hydrogel and then differentiated to induce fusion. Cells from both the stem cell population (SC) and progenitor cell populations (P1 and P2) retained proliferative capacity and successfully differentiated and fused in vitro (Supplementary Fig.2d). In the second assay, the regenerative potential of the stem cell population (SC) and the progenitor cell populations (P1, P2) were each assessed in vivo using standard assays10,30. Individual cell populations were isolated from GFP/Luciferase mice and transplanted into the irradiated TA muscles of NOD/SCID mice (Fig.3b). At 5 weeks post-transplant, the contribution of the donor cells to regenerated damaged tissue was determined by bioluminescence imaging (BLI) (Fig.3b, c). Based on engraftment frequencies and BLI signal intensity of the engrafted transplants, the regenerative potential of the populations was highest for SC and progressively lower for P1 and P2 (Fig.3b, c), consistent with the functional attributes of stem and two progressively more differentiated progenitor cell populations.
To visualize the MuSC (SC) to progenitor (P1, P2) transition in vivo we performed lineage tracing in response to acute injury, induced by notexin injection. In this scenario, Pax7CREERT2ROSA-LSL-tomato mice31 were injected with tamoxifen for 5 consecutive days, and injected with notexin in the TA and GA muscles 10 days later. Tissue was harvested and cells were analyzed by flow cytometry at day 6 post-injury (Fig.3d). Cell populations SC to P2 were identified based on differential expression of CD9 and CD104 and analyzed for the expression of tomato, indicative of prior Pax7 expression. In the pre-injury state, most tomato+ cells were in SC (Fig.3e, f; Supplementary Fig.2e). However, in response to injury, while the fraction of tomato+ cells in SC remained elevated, there was a marked increase in the fraction of tomato+ cells in P1 and P2 (Fig.3e, f; Supplementary Fig.2e). Together, these in vitro and in vivo functional studies demonstrate that the newly identified progenitor populations originate from MuSCs, have myogenic potential, and exhibit distinct regenerative capacity in vivo.
Trajectory of activated stem cells to progenitor cells revealed by CyTOF analysis after acute muscle injury
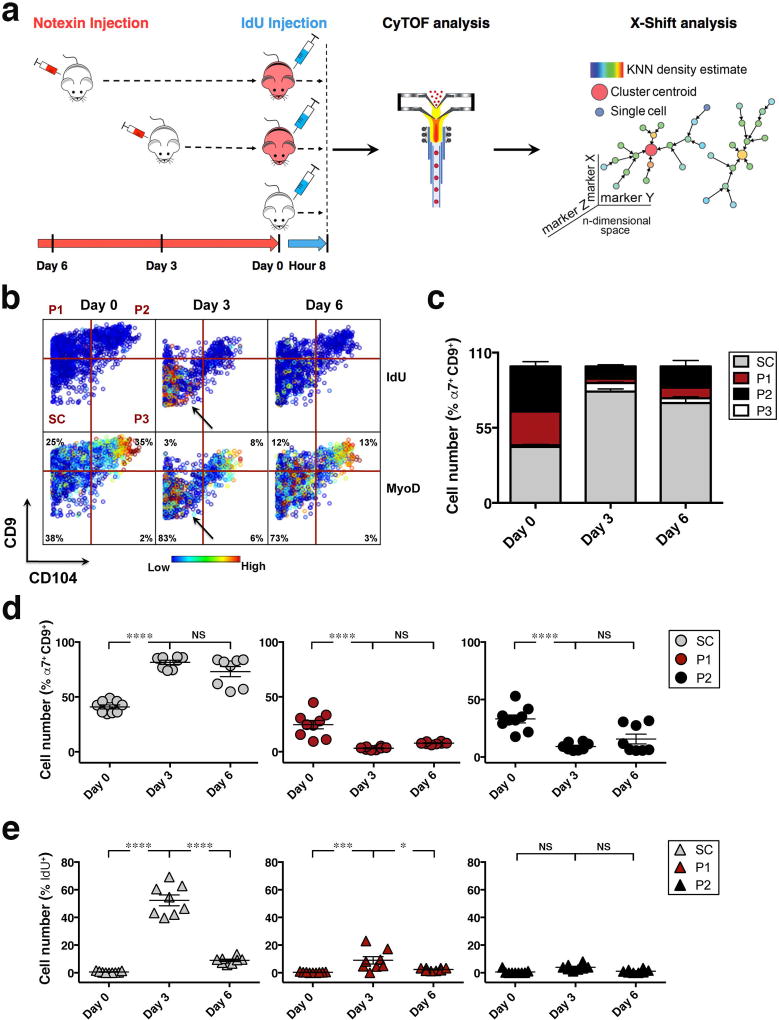
To capture the progression of myogenesis in vivo and define the cellular and molecular dynamics of regeneration, we employed CyTOF analysis of skeletal muscle during an injury time course by notexin injection. To mark cells in S-phase and follow changes in proliferation, IdU was injected 8 hours before tissue processing32 (Fig.4a). Muscle tissue was digested to a single-cell suspension and stained with the CyTOF antibody panel described above (Fig.1b and Supplementary Table 1)). Data analysis revealed that at day 3 post-injury the cells in population SC incorporated IdU, with a fraction exhibiting a concomitant increase in the expression of MyoD (Fig.4b). Consistent with the increased IdU incorporation, a marker of S-phase, there was a striking increase in the proportion of SC cells at day 3 post-injury. This was accompanied by a reduction in the proportion of P1 and P2 cells (Fig.4c, d). The sharp decline in the fraction of SC cells that incorporated IdU, from ~ 52% ± 4% at day 3 post-injury to ~ 9% ± 1% by day 6, and the minimal IdU incorporation into cells within P1 and P2 suggested that early after muscle injury, cells within SC are the most proliferative (Fig.4b, e).
Figure 4. CyTOF analysis reveals the cellular and molecular dynamics within stem and progenitor cell populations during recovery from acute injury.
(a) Experimental scheme depicts acute injury time course. Mice were acutely injured by notexin injection in the TA and GA muscles, 6 or 3 days prior to tissue collection and injected with IdU 8 hours prior to being sacrificed. Muscle tissues of 3 indicated groups were simultaneously collected at day 0, stained with isotope-chelated antibodies, run through the CyTOF instrument and analyzed using the X-shift clustering algorithm. (b) Representative biaxial dot plot of CD9 by CD104 colored by channel shows IdU incorporation (upper panels) and MyoD expression (lower panels) within individual populations during the injury time course (Day 0, Day 3, Day 6). Arrows indicate increased IdU incorporation and MyoD expression in the SC population at day 3. (c) The cells within each population in (b) are quantified as a fraction of the α7integrin+/CD9+ myogenic compartment during the injury time course. Stacked columns indicate the relative proportion of each population within the Live/Lineage−/α7 integrin+/CD9+ population at each time point (mean ± SEM from n= 8 mice, 3 independent experiments). (d) Scatter plots show cell number (%) for each population in (b) as a function of time (day) post-injury. Line represents mean ± SEM. (e) Scatter plot shows the fraction of IdU+ cells within each population in (b) during the time course of muscle injury. Line represents mean ± SEM. Statistical analyses were performed using two-way ANOVA test with significance level determined by Bonferroni’s multiple comparisons test. *, *** and **** represent statistical significance at p<0.05, p<0.001 and p<0.0001 respectively. NS represents statistically non-significant.
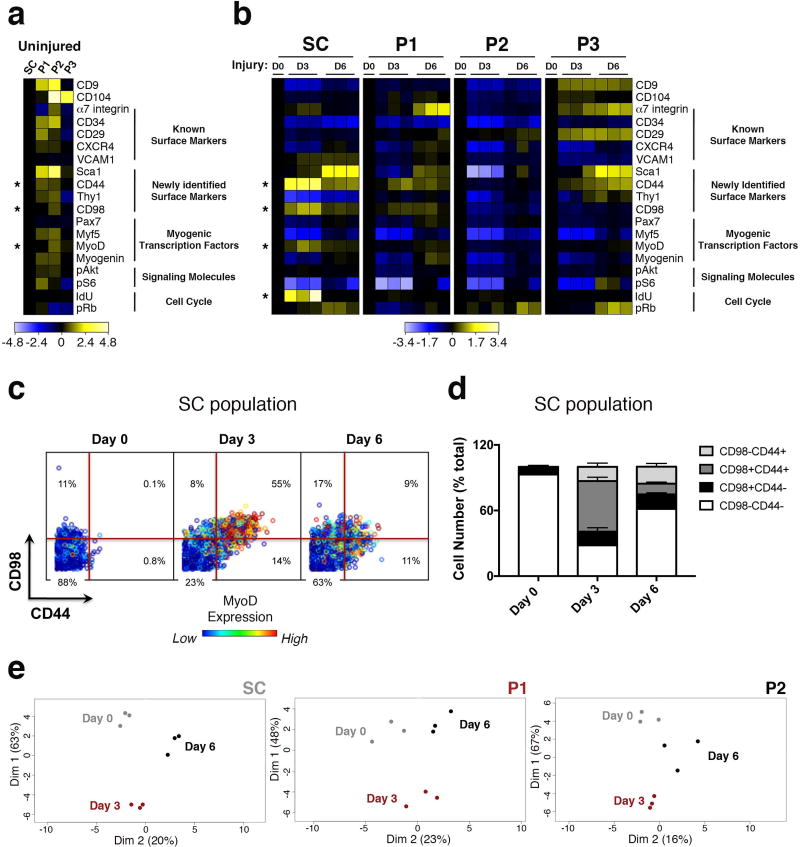
The CyTOF technology enabled high-resolution molecular analysis of muscle stem and progenitor cell populations during the functional response to injury. A heatmap showing protein expression levels for each population revealed that at the steady state, cell surface markers CD44, a glycoprotein implicated in myoblast migration and differentiation33, and CD98, an amino acid transporter upregulated in human skeletal muscle in response to increased availability of essential amino acids34, together with myogenic transcription factors Myf5, MyoD and Myogenin were expressed at higher levels in P1 and P2 compared to SC (Fig.5a). Upon injury, the most striking increases for IdU incorporation were seen on day 3 within population SC. Additionally, CD44 and CD98 were transiently co-expressed on day 3, concomitant with the transcription factor MyoD (Fig.5b–d), in about 50% of the SC population. All of these proteins returned to pre-injury levels by day 6 (Fig.5b), indicating that they identify a molecular signature of the activated stem cell state. PCA analysis (Fig.5e), corroborated by a supervised clustering analysis enforcing a two cluster solution (Supplementary Fig.3a), showed that at day 3 individual populations cluster away from day 0 and day 6, confirming that significant cellular and molecular alterations occur by day 3 and subside to a level approximating the uninjured state by day 6.
Figure 5. High dimensional analysis of acute muscle injury identifies a molecular signature of the activated stem cell state.
(a) Heatmap of protein expression in uninjured populations (transformed ratios compared to population SC, n=3 mice per condition). (b) Heatmap of protein expression in individual populations during the time course of injury (transformed ratios compared to day 0, n=3 mice per condition). (c) Representative biaxial dot plots of CD98 (y axis) by CD44 (x axis) colored by channel, showing MyoD expression in subsets of the SC population defined by CD98 and CD44 expression, during the injury time course (Day 0, Day 3, Day 6) (n=5, 2 independent experiments). (d) Cells within each CD98 by CD44 subset are quantified as a fraction of the SC population during the injury time course. Stacked columns indicate the relative proportion of each subset within the SC population at each time point (mean ± SEM from n=5, 2 independent experiments). Statistical analyses were performed using two-way ANOVA test for multiple comparisons with significance level determined by Bonferroni’s multiple comparisons test. The relative increase in the fraction of cells within the CD98+/CD44+ subset from Day 0 to Day 3 post-injury is highly significant (p<0.0001). (e) PCA plot of individual populations in (b) during the time course of injury. Proteins were clustered by their log2 median intensities (n=3 mice per condition)
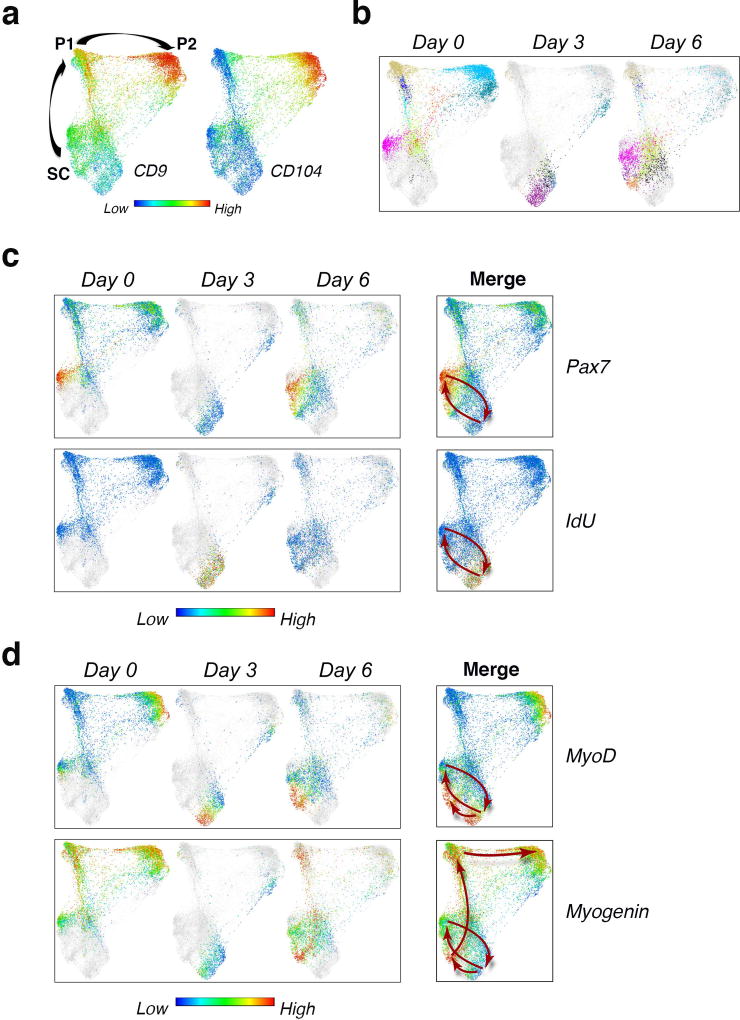
To gain increased resolution into the dynamic changes that occur during the 6 day course of recovery from injury, we applied the unsupervised X-shift analysis approach described above (Fig.1c–e). High-dimensional X-shift analysis of the mass cytometry dataset over time allowed us to construct a molecular map of regeneration as a continuum. A force-directed layout visualization combining cells from X-shift clusters at all time points revealed three dense regions that could be distinguished by the expression pattern of the surface markers CD9 and CD104 (Fig.6a). Analysis of the regeneration map over time revealed that the occupancy of cells in the dense regions differed at each time point (Fig.6b), which is consistent with the findings shown in Fig.4c, d. Moreover, the single-cell spatial resolution enabled by X-shift analysis paired with single-cell force directed layout data visualization, uncovered dynamic changes in the expression of Pax7, MyoD and Myogenin as well as IdU incorporation, revealing distinct patterns and marking a differentiation trajectory (Fig.6c, d). At the pre-injury state (day 0) there was no IdU incorporation into any of the stem or progenitor cell populations, consistent with the literature35,36. During the early phase of recovery from injury (day 3) cells in population SC, transited to a different location on the map, characterized by high IdU incorporation (Fig.6c). MyoD upregulation clearly marked a portion of these cells and its elevated expression persisted during the late phase of regeneration (day 6), when MyoD expression partially coincided with Myogenin upregulation (Fig.6d). Myogenin expression, during the late phase of recovery from injury extended in a unique pattern along the path that progresses from SC to P2, delineating a trajectory of myogenic differentiation in vivo, which is highlighted in the merged panel for days 0, 3, 6 (Fig.6d, lower right panel, arrows). The MyoD negative fraction of SC showed a distinctive upregulation of Pax7, indicating a return to quiescence (Fig.6c, d, upper right panels, arrows). In summary, unsupervised X-shift analysis of the myogenic compartment during the time-course of recovery from acute injury enabled us to visualize the spatial relationships between MuSCs (SC) and the newly identified progenitor cells (P1, P2), and uncover a dynamic myogenic trajectory that delineated cell-state transitions. Together, these data suggest an in vivo myogenic regeneration model in which activated MuSCs (SC) give rise to progeny, some of which express high levels of MyoD, commit and differentiate into progenitors (P1, P2), while others upregulate Pax7 expression and go back to quiescence to replenish the stem cell pool. Importantly, we have identified two surface markers, CD9 and CD104, whose differential expression allowed us to capture these myogenic progenitor populations in vivo.
Figure 6. High dimensional analysis of acute muscle injury uncovers cell state transitions in vivo.
(a) Live/Lineage−/α7 integrin+/CD9+ cells gated from hindlimb muscles isolated during the course of notexin injury (Day 0, Day 3, Day 6) were clustered with X-shift algorithm and cells within the resultant clusters were visualized using single-cell force-directed layout as described in Fig. 1c. The color code shows the expression level of CD9 and CD104 (representative analysis, n=9 mice). (b) Visualization of cells from the X-shift clusters at each time point (Day 0, Day 3, Day 6) using single-cell force directed layout. The color code shows X-shift clusters (n=3 mice per condition). (c) Visualization of cells from the X-shift clusters as in (b). The color code shows the expression level of Pax7 (upper panels), and IdU incorporation (lower panels). Arrows indicate the trajectory of SC over time. (d) Visualization of X-shift clusters as in (b). The color code shows the expression level of MyoD (upper panels) and Myogenin (lower panels). Arrows indicate the trajectory of SC as in (c) and the progression at day 6 from SC to P1 and P2 (highlighted by Myogenin expression).
DISCUSSION
Prior work by Nolan and colleagues has highlighted the potential of single-cell mass cytometry to resolve the heterogeneity in the hematopoietic compartment, based on cell surface markers4,5. Here we pioneered the application of CyTOF to simultaneously analyze cell surface markers and key myogenic transcription factors. This enabled us to resolve the heterogeneity of the myogenic compartment, yielding previously unknown progenitor cell populations, and delineating a myogenic progression from stem to progenitor cells in vivo in skeletal muscle. We showed that these progenitor cell populations originate from Pax7+ stem cells and exhibit myogenic function in vitro and in vivo, by a combination of lineage tracing and functional assays of myogenesis.
High-dimensional CyTOF analysis of skeletal muscle resolved the intermediate stages of myogenesis at an unprecedented level of detail and revealed the complex relationship between stem and progenitor states during regeneration, suggesting that these states are in constant flux. For example, during recovery from acute injury, SC, the most stem-like population, is activated to give rise to subsets that are MyoD+ and MyoD−, the latter returning to occupy a ground state identified by high Pax7. Indeed X-shift clustering analysis combined with single-cell force directed layout visualization of the myogenic compartment provided a high-resolution lens that revealed the cellular and molecular dynamics defining the response to acute muscle injury. It uncovered unanticipated heterogeneity within the SC compartment, leading to the identification of a molecular signature of the activated stem cell state, in addition to a differentiation trajectory from MuSCs (SC) to progenitor cells (P1 and P2) that could not have previously been envisioned.
Previous studies suggest that CD9, the myogenic progenitor marker identified here, has a functional role in the activation of the myogenic program. A member of the tetraspanin family, CD9 plays a key role in signal transduction and has been implicated in various biological processes, such as cell migration, homing, adhesion and fusion37. In systems other than muscle, CD9 has been shown to negatively regulate ADAM10 and ADAM17, two metalloproteases involved in activation of Notch signaling38–40, an essential pathway for MuSC maintainance41. Therefore, we speculate that upregulation of CD9 at the transition from MuSC (SC) to progenitor cell (P1, P2) could lead to indirect inhibition of Notch signaling via ADAMs, initiating commitment and differentiation. Another tetraspannin, CD82, has been recently identified on the surface of human muscle stem and progenitor cells42,43. Due to its low expression range in murine muscle cells (Supplementary Fig.3b), CD82 cannot faithfully discriminate the cell populations described here (SC, P1 and P2).
CD104, which marks progenitor population P2, is also known as integrin beta 4 and is a receptor for laminins18. It plays a key structural role in the hemidesmosomes of epithelial cells44. Like other integrins it mediates cell-matrix, cell-cell adhesion, and activates signaling pathways that regulate cell growth. CD104 has been implicated in the epithelial to mesenchymal transition20, tumor progression21 and cellular reprogramming to pluripotency15. In skeletal muscle, it has been shown to mark a heterogeneous cell population, which CD104 alone could not resolve to functional homogeneous subsets22.
The approach described here provides a new tool for identifying and isolating myogenic progenitors in vivo and therefore offers the opportunity to uncover key factors and the downstream signaling pathways that govern cell fate decisions and regulate the balance between cell states, during muscle homeostasis and regeneration. Understanding the fundamental biology and manipulating these cell states ex vivo will be instrumental for overcoming barriers to successful transplantation and for optimizing muscle cell therapeutic strategies. Furthermore, high-resolution single-cell studies of the signaling pathways that modulate the transition from MuSC to committed and differentiated muscle cell may provide insight into the mechanisms underlying the development of rhabdomyosarcoma, a highly heterogeneous tumor that affects children45. While here we present the simultaneous measurements of transcription factors and cell surface proteins, the complexity can be extended further. Single-cell mass cytometry has the unique attribute, over other available single-cell approaches, to study post-translational modifications and cell signaling in complex populations as they change over time.
In summary, the work presented here sets the stage for addressing questions that were previously beyond reach, such as uncovering how stem and progenitor cell interaction regulates tissue regeneration46,47, and establishes a foundation for investigating the cellular and molecular defects that characterize aging and muscle diseases. Such studies may reveal alterations in population distribution or cell signaling of fundamental interest and potential utility in the identification of novel therapeutic targets. These results provide a paradigm for other studies of solid tissues and their characteristic kinetic changes in the course of development. Finally, our work paves the way for high-resolution analysis of other regenerating tissues, with the potential to discover as yet unknown stem and progenitor cell populations with a role in development and disease.
Supplementary Material
Acknowledgments
We thank David Burns and Federico Gherardini for valuable discussion; Guojun Han for help with graphics; Dr. Kyba for Pax7-ZsGreen transgenic mice and Dr. Rudnicki for Pax7 knock-out mice; Kassie Koleckar, Peggy Kraft and Matthew Blake for technical assistance; the Stanford Shared FACS Facility for technical support. This study was supported by BD Biosciences Stem Cell grant (E.P.); US National Institutes of Health (NIH) grant K99AG042491 (B.D.C.); Muscular Dystrophy Association (MDA) development grant 217821 (A.T.V.H.), NIH grants NS089533 and AG020961, California Institute for Regenerative Medicine grant RB5-07469 and the Baxter Foundation (H.M.B.).
Footnotes
AUTHOR CONTRIBUTIONS
E.P. and H.M.B. conceived the study. E.P. designed and performed experiments, analyzed and interpreted data and wrote the manuscript. N.S. developed analysis algorithm, analyzed and interpreted data. H.M.B, W.J.F. and A.H. designed experiments, analyzed and interpreted data and wrote the manuscript. T.M., K.L.D, S.C.B, B.D.C and G.P.N analyzed and interpreted data. A.J. provided technical support with antibody conjugation and CyTOF data acquisition.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Chang NC, Rudnicki MA. Satellite cells: the architects of skeletal muscle. Curr. Top. Dev. Biol. 2014;107:161–181. doi: 10.1016/B978-0-12-416022-4.00006-8. [DOI] [PubMed] [Google Scholar]
- 2.Blau HM, Cosgrove BD, Ho ATV. The central role of muscle stem cells in regenerative failure with aging. Nature Medicine. 2015;21:854–862. doi: 10.1038/nm.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissman IL. Translating stem and progenitor cell biology to the clinic: barriers and opportunities. Science. 2000;287:1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 4.Bendall SC, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 8.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosnakovski D, et al. Prospective Isolation of Skeletal Muscle Stem Cells with a Pax7 Reporter. STEM CELLS. 2008;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerletti M, et al. Highly Efficient, Functional Engraftment of Skeletal Muscle Stem Cells in Dystrophic Muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Cheung TH, Charville GW, Rando TA. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samusik N, Good Z, Spitzer MH, Davis KL, Nolan GP. Automated mapping of phenotype space with single-cell data. Nature Methods. 2016:1–7. doi: 10.1038/nmeth.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacomy M, Venturini T, Heymann S, Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson G, et al. The tetraspanin CD9 affords high-purity capture of all murine hematopoietic stem cells. Cell Rep. 2013;4:642–648. doi: 10.1016/j.celrep.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Tachibana I, Hemler ME. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. The Journal of Cell Biology. 1999;146:893–904. doi: 10.1083/jcb.146.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke AS, Lotz MM, Mercurio AM. A novel structural variant of the human beta 4 integrin cDNA. Cell Adhes. Commun. 1994;2:1–6. doi: 10.3109/15419069409014197. [DOI] [PubMed] [Google Scholar]
- 19.Su L, Lv X, Miao J. Integrin beta 4 in neural cells. Neuromolecular Med. 2008;10:316–321. doi: 10.1007/s12017-008-8042-1. [DOI] [PubMed] [Google Scholar]
- 20.Masugi Y, et al. Upregulation of integrin β4 promotes epithelial-mesenchymal transition and is a novel prognostic marker in pancreatic ductal adenocarcinoma. Lab. Invest. 2015;95:308–319. doi: 10.1038/labinvest.2014.166. [DOI] [PubMed] [Google Scholar]
- 21.Guo W, et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 22.Liadaki K, et al. β4 integrin marks interstitial myogenic progenitor cells in adult murine skeletal muscle. J. Histochem. Cytochem. 2012;60:31–44. doi: 10.1369/0022155411428991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones NC, et al. The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell. The Journal of Cell Biology. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Troy A, et al. Coordination of Satellite Cell Activation and Self-Renewal by Par-Complex-Dependent Asymmetric Activation of p38α/β MAPK. Stem Cell. 2012;11:541–553. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgrove BD, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nature Medicine. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segalés J, Perdiguero E, Muñoz-Cánoves P. Regulation of Muscle Stem Cell Functions: A Focus on the p38 MAPK Signaling Pathway. Front Cell Dev Biol. 2016;4:91. doi: 10.3389/fcell.2016.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hausburg MA, et al. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. Elife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seale P, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 29.Maltzahn von J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- 32.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry. 2012;81A:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mylona E, Jones KA, Mills ST, Pavlath GK. CD44 regulates myoblast migration and differentiation. J. Cell. Physiol. 2006;209:314–321. doi: 10.1002/jcp.20724. [DOI] [PubMed] [Google Scholar]
- 34.Drummond MJ, et al. An increase in essential amino acid availability upregulates amino acid transporter expression in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2010;298:E1011–8. doi: 10.1152/ajpendo.00690.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 37.Leung KT, et al. The tetraspanin CD9 regulates migration, adhesion, and homing of human cord blood CD34+ hematopoietic stem and progenitor cells. Blood. 2011;117:1840–1850. doi: 10.1182/blood-2010-04-281329. [DOI] [PubMed] [Google Scholar]
- 38.Gutiérrez-López MD, et al. The sheddase activity of ADAM17/TACE is regulated by the tetraspanin CD9. Cell. Mol. Life Sci. 2011;68:3275–3292. doi: 10.1007/s00018-011-0639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arduise C, et al. Tetraspanins regulate ADAM10-mediated cleavage of TNF-alpha and epidermal growth factor. J. Immunol. 2008;181:7002–7013. doi: 10.4049/jimmunol.181.10.7002. [DOI] [PubMed] [Google Scholar]
- 40.Mizuno S, et al. A Disintegrin and Metalloprotease 10 is Indispensable for Maintenance of the Muscle Satellite Cell Pool. J. Biol. Chem. 2015 doi: 10.1074/jbc.M115.653477. jbc.M115.653477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 42.Uezumi A, et al. Cell-Surface Protein Profiling Identifies Distinctive Markers of Progenitor Cells in Human Skeletal Muscle. Stem Cell Reports. 2016;7:263–278. doi: 10.1016/j.stemcr.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander MS, et al. CD82 Is a Marker for Prospective Isolation of Human Muscle Satellite Cells and Is Linked to Muscular Dystrophies. Cell Stem Cell. 2016;19:800–807. doi: 10.1016/j.stem.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Neut R, Krimpenfort P, Calafat J, Niessen CM, Sonnenberg A. Epithelial detachment due to absence of hemidesmosomes in integrin beta 4 null mice. Nat. Genet. 1996;13:366–369. doi: 10.1038/ng0796-366. [DOI] [PubMed] [Google Scholar]
- 45.Saab R, Spunt SL, Skapek SX. Myogenesis and rhabdomyosarcoma the Jekyll and Hyde of skeletal muscle. Curr. Top. Dev. Biol. 2011;94:197–234. doi: 10.1016/B978-0-12-380916-2.00007-3. [DOI] [PubMed] [Google Scholar]
- 46.Hsu Y-C, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu Y-C, Li L, Fuchs E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell. 2014;157:935–949. doi: 10.1016/j.cell.2014.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.