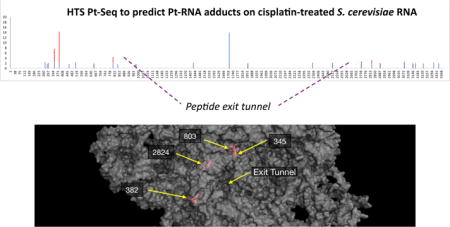
Abstract
Methods to map small-molecule binding sites on cellular RNAs are important for understanding interactions with both endogenous and exogenous compounds. Pt(II) reagents are well-known DNA and RNA crosslinking agents, but sequence-specific and genome-wide identification of Pt targets following in-cell treatment is challenging. Here we describe application of high-throughput ‘Pt-Seq’ to identify Pt-rRNA adducts following treatment of S. cerevisiae with cisplatin.
Graphical abstract
Noncoding RNA makes up a significant portion of the eukaryotic genome.1 Work has attributed regulatory roles to much of this noncoding RNA,2 highlighting RNA’s importance in the functional landscape of cells.3 Recent work has focused on the potential for RNA-targeting small molecules as potential drug candidates.4 Cisplatin (Fig. 1A), one of the most widely use anticancer drugs and the focus of this work, is known to bind RNA.5,6,7 Many RNA-based processes are disrupted by Pt compounds,8 but there is little information on global accumulation of Pt in cells.
Fig. 1.
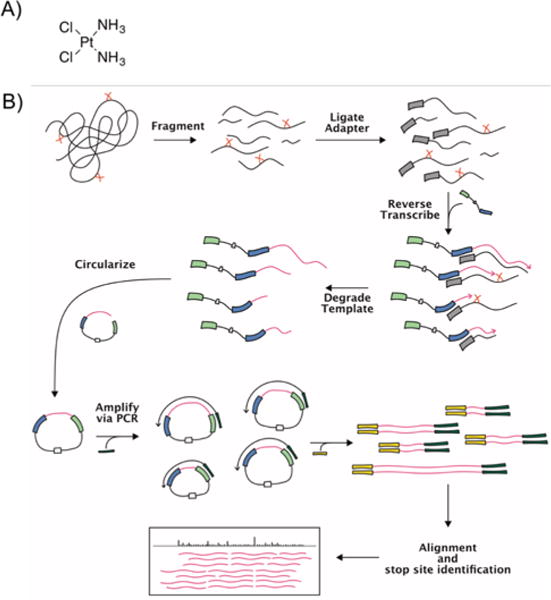
A) Cisplatin. B) Pt-Seq workflow. Pt-Seq uses a modified Mod-Seq workflow (15) to identify sites of platinum adducts on cellular RNA following cisplatin treatment. Briefly, cells are treated with cisplatin and extracted RNA is fragmented. An adapter containing an RT primer is 3′-end ligated and then extended using reverse transcription. The resulting cDNAs are stalled by platination sites, resulting in unique 3′-truncated fragments from treated samples. These fragments are circularized, PCR-amplified with primers compatible with Illumina high-throughput sequencing, and analyzed by the Pt-Seq pipeline. See †ESI for details.
Much work has supported the hypothesis that cellular RNA may be a significant target of cisplatin.5,6,7 Research has suggested that Pt adducts from cisplatin treatment accumulate to a greater degree on RNA than DNA,6,9 and that most Pt-RNA accumulation is on prevalent ribosomal RNA.6 Ribosomes present an attractive target for cisplatin as they are large RNA structures,10 crucial for cellular reproduction, and unlike DNA, present in the cytoplasm. Toxic agents such as sarcin and ricin act on the ribosome,11 as well as numerous ribosome targeting antibiotics, demonstrating its susceptibility to small molecule modification.12 Platinum adducts from cisplatin treatment have been observed on ribosomal RNA from E. coli and S. cerevisiae and have been mapped to single-nucleotide resolution using targeted primer extension.5,6,7,13 A recent crystallographic study has also identified Pt binding sites on purified T. thermophilus ribosomes complexed in vitro with mRNA and tRNA. In this study, the ribosomes were treated briefly in vitro with unactivated cisplatin.14 While an interesting molecular view of potential adduct types was unveiled, their relation to adducts formed during in-cell treatment is not established.
A comprehensive picture of platinum accumulation on cellular RNA resulting from drug treatment is still missing. Current methods of mapping platinum adducts on RNA are largely confined to reverse transcription (RT) primer extension analysis5,6,13 or mass spectrometry.7 Recent work has combined primer extension with high-throughput sequencing library generation in order to identify RNA modifications on a genome-wide scale. Notable examples are Mod-Seq and StructureFold, in which cellular RNA is treated with DMS to methylate cytosine and adenine residues,15,16 and methods based on SHAPE (selective 2′-hydroxyl acylation followed by primer extension) chemistry, which uses a library of alkylating reagents to modify the 2′ hydroxide of RNA.17,18 These methods use high levels of small molecule modifications to report on in-cell RNA structure based on accessibility. Here, towards predicting more specific small molecule binding sites, we have developed ‘Pt-Seq,’ which harnesses the power of high-throughput sequencing to map cisplatin-induced lesions on cellular RNA in a single sequencing experiment (Fig. 1B).
S. cerevisiae was chosen for this initial study based on our prior work using yeast to explore cisplatin accumulation on cellular RNA.6 Yeast share many conserved cellular processes with mammals and like all eukaryotic organisms express large amounts of noncoding RNAs.19 In prior studies, a few cisplatin adducts have been predicted on S. cerevisiae rRNA using targeted reverse transcription primer extension analysis.6,13 Because of this, S. cerevisiae is a good model system for developing our high-throughput platinum mapping technique.
The [Pt(NH3)2]-RNA adduct formed by cisplatin treatment is known to cause stalling of processive enzymes such as reverse transcriptases.5 Current methods of mapping Pt adducts that are based on extending radiolabeled RT primers rely on bands of increased intensity relative to background under treatment conditions on sequencing gels. While effective, these techniques are poorly suited for high-throughput analysis of large structured RNAs as the resolvable window is limited to approximately 100 nucleotides per experiment and each primer needs to be individually optimized and labelled. Pt-Seq is based on work by Talkish et al.15 and adapts RT primer extension for high-throughput sequencing (Fig. 1B). Briefly, RNA is extracted from treated and untreated cells, fragmented, and an adapter annealed to its 3′ end. This adapter serves as a priming site for the Mod-Seq primer based on Ingolia et al.20 This primer is extended using reverse transcriptase, the resulting cDNA fragments are size-selected and 5′ phosphorylated, and then the construct is circularized using Circligase (Epicentre). After circularization, PCR is performed using indexed primers containing Illumina adapters to generate sequencing libraries. After sequencing, reads are adapter trimmed, aligned to the yeast genome, and fed into the Mod-seq pipeline15 to identify platinum adduct sites.
We chose three treatments for this study: mock treatment control, 100 μM cisplatin, and 200 μM cisplatin. Two biological replicates were performed for each condition. Reads per condition vary from 3.8–7.3 million. Between 50–74% of reads aligned to the yeast genome with no mismatches, and 80–90% align with one mismatch. Estimated distribution across genes was generated using HTSeq-count21 which counts reads within gene features. Read distribution was predominantly on ribosomal RNA, as expected (Tables S1 and S2, †ESI).
We chose to use only reads with no mismatches to explore platinum accumulation on ribosomal RNA. We fed our trimmed and aligned reads into the Mod-Seq script15 to identify sites of RT stalling, setting a minumum 1.5-fold-enrichment threshold, and compared cisplatin treatments vs. control to derive the location of platinum adducts to single-nucleotide resolution. The results demonstrate a treatment effect by cisplatin. As drug concentration is increased 2-fold, the number of sites demonstrating >1.5-fold stop enrichment increase 2.2-fold for 25S rRNA (Fig. 2). Higher treatment concentration results in some overlapping and many new stop sites. Interestingly, for overlapping sites, increasing cisplatin treatment concentration from 100 to 200 μM did not always result in an increase in relative stop intensity, and occasionally higher concentrations of cisplatin result in decreased relative stop intensity (Fig. 2A and B). This could be due to changes in RNA structure at the higher Pt concentrations.
Fig. 2.
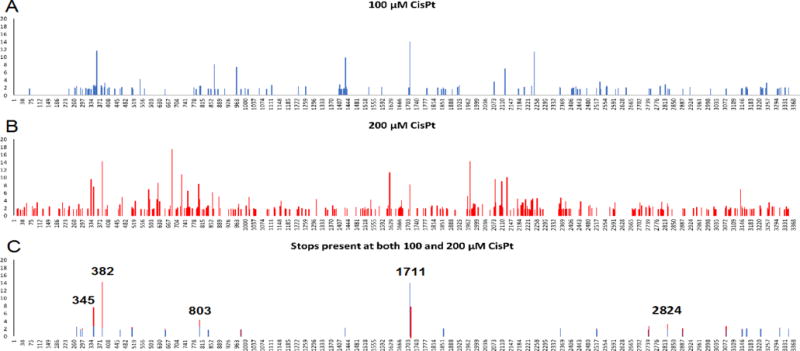
Enrichment of RT stop sites on 25S rRNA following cisplatin treatment of S. cerevisiae. A) 100 μM cisplatin treatment B) 200 μM cisplatin treatment. Increasing cisplatin concentration results in increased stop site observation. C) Stops present in both treatment concentrations, with a cutoff of 1.5-fold enrichment as described in text.
To further narrow our focus for closer analysis of the 25S large ribosomal subunit, we compared stops across 100 and 200 μM cisplatin treatment conditions at >1.5-fold enrichment over background, and then considered only those which were present in both treatments. Stops which were present in both conditions were then compared with a list of known modified nucleotides22 and any matches removed to ensure no overlap. Out of the 141 and 304 stop sites observed at 100 and 200 μM cisplatin, 26 are shared by both treatments (Fig. 2C). The median stop intensity for these paired stops was 1.8 at 100 μM cisplatin and 1.95 at 200 μM cisplatin, further exhibiting a treatment effect.
For this subset of 26 sites, the five sites with highest fold-enrichment are at positions G345, U382, C803, C1171, and G2824 (Fig. 2C). C1711 is located on helix 58, which is the binding site for ribosomal protein L30e, an indispensable ribosomal protein. Solvent accessible surface area calculations suggest that the C1711 N3 position is inaccessible (not shown), but that as a whole helix 58 is highly exposed. Helix 58′s interaction with protein L30e primarily relies on the unusual tertiary structure of helix 58, a kink-turn motif.23,24 C1711 is located just before this motif. Together, H58 and L30e form an intersubunit bridge, one of several contacts between the large and small ribosomal subunits.23,25
G2824 is located at the base of helix 89 (Fig. 3), in the “accommodation corridor,” a passageway located along the interface between H89 and the H90-92 structure of the large ribosomal subunit.26 It is immediately adjacent to the peptidyl transfer center,27 and close to the tRNA A-site. Interestingly, it is close to the binding sites of anisomycin and narciclasine, eukaryotic ribosome-targeting antibiotics that inhibit peptide bond formation.12 Furthermore, mutations to nearby residues (2820, 2829) result in non-viable ribosomes,27 suggesting a region particularly sensitive to modification. This implies a previously unexplored model for cisplatin inhibition of the ribosome. G345 and U382 are located near helix 23 and 24, respectively (Fig. 3). U382 is highly solvent exposed, located near the surface of the large ribosomal subunit opposite the small subunit. G345 is near the surface of the large ribosomal subunit and the 5.8S ribosomal RNA.
Fig. 3.
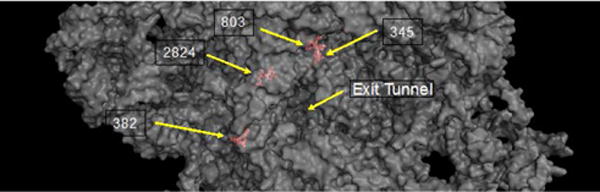
Three-dimensional views of the yeast ribosome (PDB 4V7R) from the solvent-exposed side of the large ribosomal subunit with major cisplatin adduct sites noted in red. 25S rRNA is shown as a partially transparent surface.
We utilized traditional targeted RT primer extension analysis to verify our high-throughput sequencing approach (Fig. 4) on RNA isolated from yeast cultures treated with 200 μM cisplatin (see Methods, ESI). We chose to analyze the region near helix 58 since it is highly solvent exposed, making it a good candidate for RT primer extension. Using a RT primer spanning 1761–1779 a number of stops are observed upstream of C1711 in the region spanning G1599 through G1646 (Fig. 4A and C) that broadly match regions identified by Pt-Seq.
Fig. 4.
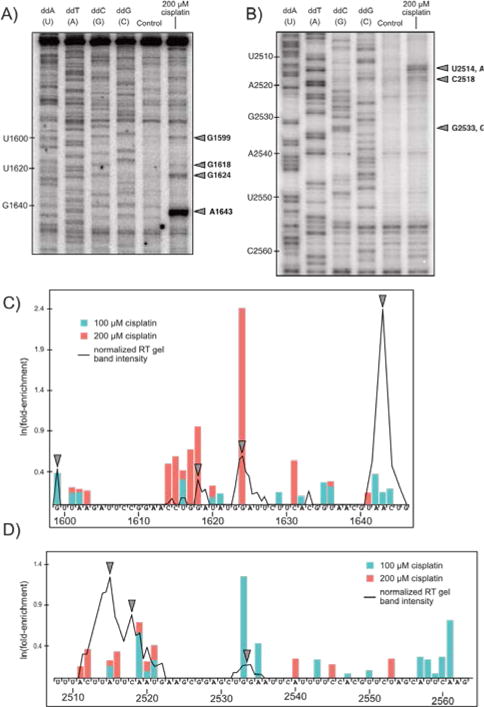
Comparison of gel-based RT primer extension analysis with results predicted by Pt-Seq. Two regions of the yeast 25S large ribosomal subunit (A: 1600–1650 and B: 2510–2560) were subject to traditional gel-based primer extension analysis. Lanes 1–4 are dideoxy sequencing ladders, lane 5 is 0 μM cisplatin control, and lane 6 is 200 μM cisplatin (6 hours treatment). Stop sites are marked with arrows. (C, D) Band intensity down each lane (ImageJ) overlaid on a bar plot of Pt-Seq results for both 100 and 200 μM cisplatin, shifted one nucleotide 3′ to illustrate RT stop sites.
A second primer annealed near helix 80 (3′ end 2605) reliably extended through U2508 and covered G2563 through U2510 (Fig. 4B), showing a strong stop at U2514 / A2515, a stop at C2518, and a weak stop at G2533/G2534 (Fig. 4B and D). Pt-Seq-determined stops from 2511–2521 reflect the broad set of stops from 2511–2522 observed via primer extension and suggest this region is particularly prone to modification. Together, these data support Pt-Seq as a high-throughput method of identifying cisplatin-derived adducts on cellular RNA.
Notably, not all strong Pt-Seq stop sites are represented in the gel-based RT assays and vice-versa. We used Circligase to create single-stranded circular vector-like products post-RT for PCR amplification as described by Talkish et al.15 Circularization avoids a second potentially low-yielding ligation step and Circligase itself is very efficient: the manufacturer reports near 100% conversion from linear to circular products of control sequences. Circligase does have a 3′ substrate bias toward primarily A and T residues in the 3′ position for circularization (Epicentre). Transforming our Pt-Seq results from adduct site to stop site (+1 nt) demonstrates a strong preference for A and T residues in the 3′ position at both treatment concentrations (not shown). This bias is present for both concentrations of cisplatin and also observed in control treatment (not shown), and suggests that results are biased towards those adjacent to a preferred Circligase substrate. This factor may explain the failure to recapitulate some strong stops seen by others by traditional RT analysis in the yeast peptidyl transferase center.6,13
In this study, we have performed a high-throughput sequencing-based survey of Pt-RNA adducts formed following treatment of S. cerevisiae with cisplatin. A majority of predicted Pt-RNA sites were located on ribosomal RNA, which also comprises the vast majority of yeast RNA (>80% by nucleotide).28 Platinum-induced stops are also observed on tRNAs and cytoplasmic RNAs such as SCR1 as well as transposable elements (not shown). On the 25S ribosomal subunit, a clear dose-dependent response is observed with increased cisplatin treatment concentration, with an increase in the number, median, and maximum stop intensity observed with doubling concentration of cisplatin (Fig 2). Results from Pt-Seq results compare well with gel-based RT mapping methods and infer regions highly prone to modification (Fig. 4).
Our strongest cisplatin-derived stops are located primarily in the highly conserved core of the ribosome (Fig. 3). G2824 is particularly interesting, as it is located in the core of the ribosome, near the peptidyl transferase center, where peptide bond formation occurs. Numerous eukaryotic-specific antibiotics bind in this region and mutations to this region results in nonviable ribosomes, suggesting this region is particularly sensitive to modification. Cisplatin-derived Pt adducts following in vitro treatment have been observed by crystallography to bind regions near ribosome-targeting antibiotics.14 Taken together, these results suggest a previously unexplored mechanism for cisplatin inhibition of the ribosome which warrants further investigation. Adapting Pt-Seq to mammalian cell lines, as has been done for high-throughput RNA structure mapping protocols using either RT truncation17 or mutational mapping,29,30 will be an important next step.
Supplementary Material
Acknowledgments
Financial support from the National Science Foundation (CHE1413677 to VJD) and the National Institutes of Health (GM007759-29 to KP) and technical assistance from E. Reister are gratefully acknowledged.
Footnotes
Electronic Supplementary Information (ESI) available Detailed materials and methods, Pt-induced stop counts on RNA from cisplatin-treated S. cerevisiae
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Mattick JS. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Hum Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Sharp PA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 5.Rijal K, Chow CS. Chem Commun. 2009;1:107–109. doi: 10.1039/b816633a. [DOI] [PubMed] [Google Scholar]
- 6.Hostetter AA, Osborn MF, DeRose VJ. ACS Chem Biol. 2012;7:218–225. doi: 10.1021/cb200279p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dedduwa-Mudalige GNP, Chow CS. Int J Mol Sci. 2015;16:21392–21409. doi: 10.3390/ijms160921392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman EG, Hostetter AA, Osborn MF, Miller AL, DeRose VJ. Met Ions Life Sci. 2011;9:347–77. doi: 10.1039/9781849732512-00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akaboshi M, Kawai K, Maki H, Akuta K, Ujeno Y, Miyahara T. Jpn. J Cancer Res. 1992;83:522–6. doi: 10.1111/j.1349-7006.1992.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore PB, Steitz TA. RNA. 2003;9:155–159. doi: 10.1261/rna.2127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tumer NE, Li XP. Curr Top Microbiol Immunol. 2012;357:1–18. doi: 10.1007/82_2011_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loubresse N Garreau de, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. Nature. 2014;513:517–524. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 13.Osborn MF, White JD, Haley MM, DeRose VJ. ACS Chem Biol. 2014;9:2404–2411. doi: 10.1021/cb500395z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melnikov SV, Dieter S, Steitz TA, Polikanov YS. Nucleic Acids Res. 2016;44:4978–4987. doi: 10.1093/nar/gkw246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talkish J, May GG, Lin Y, Woolford JL, Jr, McManus CJ. RNA. 2014;20:713–720. doi: 10.1261/rna.042218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Bouvier E, Kwok CK, Ding Y, Nekrutenko A, Bevilacqua PC, Assmann SM. Bioinformatics. 2015;31:2668–2675. doi: 10.1093/bioinformatics/btv213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Incarnato E, Neri F, Anselmi F, Oliviero S. Genome Biol. 2014;15:491. doi: 10.1186/s13059-014-0491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Delneri D, O’Keefe RT. Biochem Soc Trans. 2012;40:907–911. doi: 10.1042/BST20120042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Pyl PT, Huber W. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piekna-Przybylska D, Decatur WA, Fournier MJ. RNA. 2007;13:305–312. doi: 10.1261/rna.373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenner L, Melnikov S, Loubresse M Garreau de, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. Curr Op Struct Biol. 2012;22:759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Bifano AL, Atassi T, Ferrara T, Driscoll DM. BMC Mol Biol. 2013;14:12. doi: 10.1186/1471-2199-14-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halic M, Becker T, Frank J, Spahn CM, Beckmann R. Nat Struct Mol Biol. 2005;12:467–468. doi: 10.1038/nsmb933. [DOI] [PubMed] [Google Scholar]
- 26.Petrov AN, Meskauskas A, Roshwalb SC, Dinman JD. Nucleic Acids Res. 2008;36:6187–6198. doi: 10.1093/nar/gkn643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakauskaite R, Dinman JD. RNA. 2011;17:855–864. doi: 10.1261/rna.2593211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warner JR. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 29.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Nat Protoc. 2015;10:1643–1669. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. Nat Meth. 2017;14:75–82. doi: 10.1038/nmeth.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


