Abstract
Bruch’s membrane opening-minimum rim width (BMO-MRW) is a recently proposed structural parameter which estimates the remaining nerve fiber bundles in the retina and is superior to other conventional structural parameters for diagnosing glaucoma. Measuring this structural parameter requires identification of BMO locations within spectral domain-optical coherence tomography (SD-OCT) volumes. While most automated approaches for segmentation of the BMO either segment the 2D projection of BMO points or identify BMO points in individual B-scans, in this work, we propose a machine-learning graph-based approach for true 3D segmentation of BMO from glaucomatous SD-OCT volumes. The problem is formulated as an optimization problem for finding a 3D path within the SD-OCT volume. In particular, the SD-OCT volumes are transferred to the radial domain where the closed loop BMO points in the original volume form a path within the radial volume. The estimated location of BMO points in 3D are identified by finding the projected location of BMO points using a graph-theoretic approach and mapping the projected locations onto the Bruch’s membrane (BM) surface. Dynamic programming is employed in order to find the 3D BMO locations as the minimum-cost path within the volume. In order to compute the cost function needed for finding the minimum-cost path, a random forest classifier is utilized to learn a BMO model, obtained by extracting intensity features from the volumes in the training set, and computing the required 3D cost function. The proposed method is tested on 44 glaucoma patients and evaluated using manual delineations. Results show that the proposed method successfully identifies the 3D BMO locations and has significantly smaller errors compared to the existing 3D BMO identification approaches.
Keywords: SD-OCT, ophthalmology, retina, segmentation, optic disc, Bruch’s membrane opening
Graphical Abstract

1. Introduction
Glaucoma is one of the major causes of blindness and it is estimated that the number of people with glaucoma will increase to 79.6 million in 2020 (Quigley and Broman, 2006). There are different procedures to assess the functional and structural deficits caused by glaucoma. Visual field tests are the clinical standard for monitoring functional change. Color (stereo) fundus photography and, more recently, spectraldomain optical coherence tomography (SD-OCT) are two structural imaging modalities for monitoring the structural changes due to this progressive disease (Abràmoff et al., 2010). In order to be able to monitor the glaucomatous structural changes occurring within the optic nerve head region, the cup-to-disc ratio (CDR), defined as the cup area over the optic disc area, is a common parameter to compute which becomes larger as glaucoma progresses due to the increase in the loss of nerve fiber bundles in the neuroretinal rim. More recently, Bruch’s membrane opening-minimum rim width (BMO-MRW), defined as the minimum Euclidean distance from Bruch’s membrane opening to the internal limiting membrane (ILM) surface (Fig. 1), was introduced which measures the remaining neuroretinal rim tissue (Reis et al., 2012). Recent studies showed that BMO-MRW is superior to other structural parameters for diagnosing open-angle glaucoma (Chauhan et al., 2013), although note that the retinal nerve fiber layer thickness (RNFLT) may still be preferable for monitoring longitudinal change (Gardiner et al., 2015). Furthermore, the BMO is used to help calculate the depth of anterior lamina cribrosa surface, which is also utilized for monitoring glaucomatous damage (Sredar et al., 2013; Ren et al., 2014; Lee et al., 2014).
Figure 1.

Illustration of (a) retinal structures including ILM and BM surfaces, BMO points and BMO-MRW parameter on a single SD-OCT slice and (b) BMO points in 3D.
Beside the necessity of identifying BMO points for computing the BMO-MRW, the BM surface ending points also define the true optic disc boundary. The 3D imaging ability of SDOCT machines showed that the disc margin (DM) from fundus photographs does not always coincide with the outer border of rim tissue, however, BMO, also referred to as the neural canal opening (NCO), is the true outer border of rim tissue (optic disc boundary) which remains unaltered during intraocular pressure changes due to glaucoma (Chauhan and Burgoyne, 2013; Chauhan et al., 2013; Reis et al., 2012). Hence, considering the inherent subjectivity and required time for manual delineations, in order to be able to identify the true optic disc boundary and compute the structural parameter BMO-MRW automatically, having a reliable automated approach for segmentation of BMO points is of great desire.
Various approaches have been employed for segmenting the optic disc from SD-OCT volumes including pixel-based classification methods (Lee et al., 2010; Abràmoff et al., 2009; Miri et al., 2013), model-based approaches (Fu et al., 2015), and graph-based approaches (Hu et al., 2010; Miri et al., 2015), among which some techniques are also utilized for segmenting the optic disc from fundus photographs (Abràmoff et al., 2007; Cheng et al., 2013; Xu et al., 2007; Yin et al., 2011; Lowell et al., 2008; Chràsteka et al., 2005; Merickel Jr et al., 2006, 2007; Zheng et al., 2013). The approaches that attempted to segment the BMO from SD-OCT volumes mostly focused on 2D segmentation of BMO points. For instance, the works in (Hu et al., 2010) and (Miri et al., 2015) focused on segmenting the 2D projection of BMO points, while Fu et al. found the BMO points from a number of individual 2D B-scans and fitted an ellipse to the points to obtain the complete ring-shape BMO segmentation (Fu et al., 2015). The method proposed by Belghith et al. also segments BM surface and BMO points from each radial slices independently and then an elliptical shape is utilized as a prior to estimate the curve that best represent the BMO points (Belghith et al., 2014, 2016).
Our proposed method in (Antony et al., 2014) was the first step towards directly obtaining a 3D segmentation, where we presented an automated iterative graph-theoretic approach for segmenting multiple surfaces with a shared hole. The method was applied to segment the junction of the inner and outer segments (IS/OS) of the photoreceptors and the Bruch’s membrane (BM) surfaces and their shared hole (i.e. BM opening). This method needs an initial 2D segmentation of BMO points, which is obtained from a 2D projection image, using a method similar to the existing 2D approaches (Hu et al., 2010; Miri et al., 2015). The corresponding z-values are identified by projecting the 2D segmentations onto the BM surface. Since the layer segmentation around the ONH region is not precise, the computed z-values are not always accurate. In order to allow for correcting the z-values, an iteration phase was added to the method within which the z-values along with new layer segmentations are identified as part of the proposed surface+hole method. The updated layers produces a new 2D projection image from which an updated 2D BMO segmentation is obtained and the iterations continue. This surface + hole approach was shown to be more accurate than existing 2D approaches; however, the presence of externally oblique border tissue (Reis et al., 2012), which attaches to the end of BM surface and appears very similar to the ending point of BM surface sometimes confuses the algorithm and causes continuing of its iterative search on the border tissue such that instead of the end of the BM surface, the BMO is identified on the border tissue.
Correspondingly, the purpose of this work is to address the limitation of our previous approach by eliminating the iteration phase and presenting an automated machine-learning graph-theoretic approach that segments the BMO points as a 3D ring in radially resampled SD-OCT volumes. More specifically, similar to our previous approach, a 2D initial segmentation is obtained using a graph-theoretic approach. The volume in the z-direction is downsampled to achieve an isotopic grid and an estimated 3D location of BMO points is computed by projecting the (r, θ) pairs onto the BM surface. Instead of defining a mathematical model for BMO points, which is not feasible as they appear differently even in slices corresponding to a single subject, we learn the intensity-based attributes of BMO points a priori. In particular, based on a random-forest classifier trained using BMO intensity models, the likelihood of each voxel being a BMO point is generated. The inverted likelihood map will serve as the cost function for finding the 3D BMO path using a shortest-path approach. The final BMO segmentation in the original image resolution is obtained by refining the z-coordinates using a similar approach as used to find the BMO path within the downsampled volume.
2. Methods
The overall flowchart of the proposed method is shown in Fig. 2. The four major components of the proposed method are: 1) a preprocessing step including transferring the SD-OCT volumes to the radial domain and segmenting intraretinal surfaces (Section 2.1), 2) identifying the 2D projected locations of the BMO points using a graph-theoretic approach (Section 2.2), 3) computing a cost function for identifying the BMO 3D path using a machine-learning approach (Section 2.3), and 4) identifying the 3D location of BMO points (as a 3D path within the SD-OCT volumes) using a shortest path method (Section 2.4) and refining the path in the z-direction (Section 2.5).
Figure 2.
Flowchart of overall method.
2.1. Preprocessing
In the preprocessing step, the SD-OCT volume is transfered to the radial domain and the intraretinal surfaces that are needed for identifying the BMO points are segmented. If the original SD-OCT volume in the Cartesian domain is represented by I(x, y, z), the radial volume IR(r, θ, z) is obtained using bilinear interpolation of the original volume with angular precision of 1° degree. The radial transformation is performed because the BMO points are more obvious in the radial volume than in the original SD-OCT volume where BMO points are less obvious in the slices close to the upper/lower part of disc boundary and are not present in the slices that do not intersect with optic disc (Fig. 3). With such a transformation, there are two BMO points in each radial slice and unlike the original domain, Bruch’s membrane opening has a stiffer shape especially in the r-direction (i.e. displacement of the BMO points in-between slices in the r-direction is minimal). This property will be used later as a constraint for the graph-theoretic approach in Appendix A.0.2 and Section 2.4.
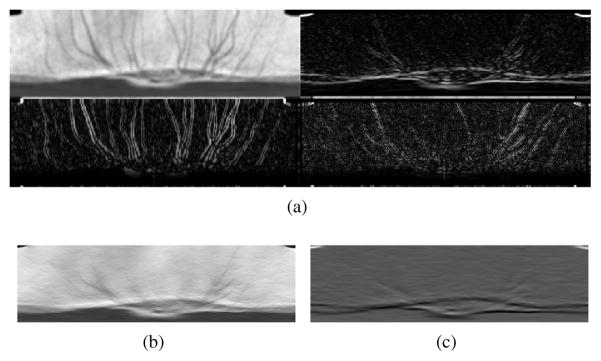
Figure 3.

(a) Illustration of lines and their corresponding B-scans in the original domain and (b) sampling the original volume for creating the radial volume. The shape of the opening is more consistent in (b) than in (a) as the BMO points (orange circles) in (a) become close to each other which causes identification of the points to be more difficult.
Once the radial volume is created, intraretinal surfaces are segmented from the radial volume using a theoretical multi-resolution graph-based approach (Garvin et al., 2009; Lee et al., 2010). Specifically, the intraretinal surface segmentation problem is transformed to an optimization problem with a number of specific constraints and the goal is finding a set of feasible surfaces with the minimum cost, simultaneously (Garvin et al., 2009). Lee et al. (Lee et al., 2010) proposed a multi-resolution approach for executing the algorithm which speeds up the segmentation. We employed this method for segmenting three intraretinal surfaces from the radial volumes: the first surface is called the internal limiting membrane (ILM), surface two is located at the junction of the inner and outer segments of photoreceptors (IS/OS), and surface three, called Bruch’s membrane (BM) surface, is the outer boundary of the retinal pigment epithelium (RPE). These surfaces are segmented because (1) the ILM surface will be used as a constraint for finding the 2D and the 3D BMO path as well as computing BMO-MRW, (2) IS/OS and BM surfaces will be used for creating the radial projection image, and (3) the BM surface also will be used for identifying the estimated 3D location of the BMO points. Since the RPE-complex, bounded by IS/OS and BM surfaces, does not exist inside the ONH, the surface segmentations corresponding to the second and third surfaces are not meaningful inside the opening. Hence, the second and the third surfaces are interpolated inside the ONH opening which is approximated by a circle larger than the typical size of the ONH opening (1.73 mm radius) and centered at the center of the ONH (Miri et al., 2015). Use of interpolated surfaces within this circle is beneficial for computing a projection image (discussed in the next paragraph) with a visible interface at the projected BMO locations (Hu et al., 2010). The lowest point of the ILM surface is considered as the approximated center of the ONH (Fig. 4a).
Figure 4.

Radial surface segmentation and projection image creation. (a) Example radial scan with segmented surfaces where red, green, and yellow are the ILM, IS/OS, and BM surfaces, respectively. Note that the IS/OS and BM surfaces are interpolated inside the ONH. (b) The projection image obtained as described in Section 2.1. (c) The reformatted radial projection image in which the 2D BMO projection locations appear as a horizontal path.
The radial projection image is created by averaging the intensities of the RPE-complex (the sub-volume between the blue and yellow surfaces in Fig. 4a) in the z-direction (Fig. 4b). In particular, the intensities of the sub-volume bounded by 15 voxels (29.3 μm) above the second surface and 15 voxels below the third surface are averaged in the z-direction and the resulting projection image is reformatted such that radial and θ values appear on vertical and horizontal axes, respectively and the BMO projection path appears as an approximately horizontal boundary (Fig. 4c).
2.2. Identifying 2D Projected Location of BMO Endpoints
In this section, an initial 2D segmentation of BMO points is computed using a graph-theoretic approach. Since the method has similarities with our previous work reported in (Miri et al., 2015) and for the sake of clarity and space, only a brief summary of the approach is presented here. The differences between the approach utilized in this work and our work reported in (Miri et al., 2015) is that the 2D cost function for segmenting the projected BMO points in this work does not include region terms and in addition to the hard boundary smoothness constraint, a shape prior constraint is also enforced when BMO projection points are identified. More details of this step are mentioned in Appendix A.
The 2D BMO points are identified in the form of a path within the radial projection image (Fig. 4c). The cost function associated with the graph-theoretic approach includes an edge-based (aka on-boundary) cost function which contains the information regarding the unlikelihood of pixels locating on the BMO 2D path. This cost function is computed by applying an asymmetric 2D Gaussian derivative filter to a vessel-free projection image. The vessel-free image is obtained by applying Haar stationary wavelet transform (SWT) to the radial projection image and removing blood vessels using modification of SWT coefficients.
Since the BMO 2D path appears as a very smooth horizontal boundary in the radial projection image, we utilize the graph-theoretic approach in (Song et al., 2013) to incorporate the shape prior information in the graph-theoretic approach where deviations from the expected shape of the BMO 2D path are penalized using a convex function (here a quadratic function is utilized). Therefore, the total cost of the BMO 2D path consists of an edge term and a shape term.
Finally, the 2D BMO locations are projected onto the BM surface to obtain an approximation of the 3D BMO to be used in subsequent steps.
2.3. Computing Machine-Learning-Based 3D Cost Function
We propose to formulate the problem of identifying the BMO points as a finding a minimum-cost path within an SDOCT volume (Section 2.4). The cost of the path is minimized with respect to a cost function which consists of two terms: the node cost, CV, includes information regarding the similarity of each node to a BMO point and the edge term, CE, controls the smoothness of the path. In order to compute the 3D cost function (CV) for more precisely identifying the 3D BMO locations, a machine-learning based approach is utilized. First, to obtain an isotropic grid, the radial OCT volume is downsampled in the z-direction to reflect its physical resolution such that the size of each pixel in the r-z plane changes from 30μm × 2μm to 30μm × 30μm. For each subject in the training set, a set of intensity-based features are extracted from each BMO location. An intensity model of BMO (referred to as the low-resolution PCA model as the features are computed from a downsampled volume) is created from the extracted features using principal component analysis (PCA). In order to train a random forest (Breiman, 2001) classifier, the same set of intensity-based features are extracted from BMO points (positive) as well as non-BMO points (negative) and projected to the low-resolution PCA model. The classifier computes the probability of being a BMO point for each point in the vicinity of estimated 3D BMO locations (explained in Section 2.3.2) for subjects in the test set. The inverted probability map will serve as the 3D cost function of the shortest path method for identifying the 3D BMO locations (Section 2.4).
2.3.1. Computing 3D PCA-Intensity BMO Model
In order to compute the 3D low-resolution intensity model of the BMO, three types of intensity features are computed at each BMO point: 1) neighborhood intensity profile (NIP), 2) steerable Gaussian derivatives (SGD), and 3) Gabor features. Since the radial OCT volumes are created by sampling the original OCT volume in a circular pattern (Fig. 3b), the first and last slices are considered as neighbors during feature extraction. The low-resolution PCA intensity model consists of a number of PCA-based models created from each feature category. Before feature extraction, the intensities of the radial OCT volume are linearly normalized to the [0, 255] (i.e., mapping the minimum intensity to 0, the maximum intensity to 255, and scaling linearly between) interval such that the intensity scale throughout the dataset is consistent.
The NIP feature provides information regarding the 3D intensity profile of the query point’s neighborhood. NIP features are computed using a mask (Fig. 5) of size 9 × 9 × 3 within which 48 neighbors around the query point in each of current, previous, and next slices are marked (a total of 144 features). The 48 neighbors are inspired by the positions of offsets used in the FAST-ER corner detector (Rosten et al., 2010). The differences of the query point’s intensity and the intensities of its 144 neighbors are computed to learn the 3D intensity profile around a BMO point.
Figure 5.

Illustration of NIP feature locations for a query point p.
A set of 3D steerable Gaussian derivative filters with σ = {0.5: 0.5: 3} and ϕ = {0°: 30°: 359°} are used to reflect the intensity change around a BMO point (a total of 72 features). Assume is a symmetric 3D Gaussian filter and (···)ϕ represents the rotation operator in the r-z plane such that is the rotated version of ℱσ(r, z, θ) at angle in the r-z plane. Thus, the 3D steerable Gaussian derivative filter, , can be expressed as follows:
| (1) |
As depicted in Eq. 1, the derivative is taken only in the r-z plane, however, in order to incorporate the 3D contextual information, the SGD filters are designed in 3D to integrate information from neighboring slices as well.
A set of Gabor filters (Grigorescu et al., 2002) extract localized frequency information (i.e. textural information) from the region of interest. The Gabor filters are represented as:
| (2) |
where σ, γ, λ, ϕ, and ψ are the scale, spatial aspect ratio, wavelength, orientation, and the phase offset, respectively. Since the Gabor filters are symmetric, the filter bank includes 6 scales and 6 orientations of σ = {0.5: 0.5: 3} and ϕ = {0°, 30°: 179°}. The wavelength (λ) was set to σ/0.56 as this corresponds to a half-response spatial frequency bandwidth of one octave. The spatial aspect ratio and the phase offset were set to γ = 1 and ψ = 0°, respectively.
Once the intensity features are extracted from the BMO points, in order to reduce the dimensionality of the feature set (144 NIP, 72 SGD, and 36 Gabor features), PCA was utilized to create 13 PCA-based intensity models by retaining more than 90% of the variation in each category. The PCA-based intensity models consisted of 1 NIP model, 6 SGD models corresponding to the 6 different scales used for creating SGD filters, and 6 Gabor models corresponding to 6 scales of Gabor filters.
2.3.2. Classification
As mentioned earlier, a random forest classifier is employed for computing the 3D cost function (CV). For training, the search region is limited to a small vicinity of each BMO point which is the region inside a donut defined by two ellipses centered at each BMO location. It is known that BMO always locates below the ILM surface; hence, if any parts of the search region fall above the ILM surface, they are excluded for feature extraction (e.g. the shaded region in Fig. 6a). The area inside the smaller ellipse represents the inherent subjectivity of manual delineation for identifying a BMO point and the positive class members are selected from this region. Since the training set is highly skewed (i.e. only one BMO point exists in each search region) and to reflect the inter-observer variability, we consider each BMO point along with its 4-neighborhood pixels as positive class (a total of 5) and randomly sampled 20 points from the search region (the area between the outer and inner ellipses) to represent the negative class (Fig. 6a). The purpose of the inner ellipse is to prevent sampling the negative class from inside this region. Note that the inner ellipse is drawn a bit larger than its actual size in Fig. 6a for illustration purposes. The appropriate size of the search region was computed in the training set such that in addition to the manual delineation (true BMO locations), an estimated 3D location for each BMO in the training set was computed by projecting the 2D location (i.e. (r, θ)-values) of each BMO point (that obtained in Appendix A.0.2) to the BM surface. The estimated and true locations of BMO points were compared to obtain the range of estimation error. The radii of outer ellipse were computed such that more than 99% of the estimated BMO locations were included in the search region (rbig = 300μm, rsmall = 120μm). The radii of the inner ellipse were set to rbig = 90μm, rsmall = 45μm. The same set of intensity features that were used for creating the low-resolution PCA model were extracted for both classes. The features were projected to the low-resolution PCA model using their corresponding eigenvectors. The number of trees was set to 500 and the number of variables available for splitting at each tree node was set to square root of total number of predictors (28 NIP, 12 SGD, and ).
Figure 6.

(a) Illustration of the donut of search region for training set. The negative class (cyan dots) are randomly sampled from the area between ellipses and the positive class (yellow crosses) are taken from the area inside the small purple ellipse. (b) Illustration of search region for testing set which is an ellipse with the same size as the outer ellipse (dark blue) of the donut around the estimated 3D BMO location (yellow cross). The green shaded areas in (a) and (b) are excluded from the search region due to the fact that BMO never locates above the ILM surface. (c) A slice (corresponding to the B-scan shown in (b)) of the 3D cost function utilized for identifying the BMO 3D path which is obtained by inverting the output of the RF classifier.
For testing, in order to locate the search region, which is an ellipse with the same size as the outer ellipse of the donut used for training, we need an estimation of the BMO location. Similar to obtaining the estimated location of BMO points in the training set, the 2D location (i.e. (r, θ)-values) of each BMO point computed in Section 2.2 were projected to the BM surface (the yellow surface in Fig. 6b) to estimate the corresponding z-values. All voxels inside the search region in the testing set are potential candidates for being a BMO point (Fig. 6b). Hence, the intensity features are extracted for all points inside the search region and projected to the low-resolution PCA model using their corresponding eigenvectors.
The RF classifier computed the probability of being a BMO point for all voxels inside the search region to generate the 3D likelihood map. The inverse (1 − p) of the likelihood map (Fig. 6c) was utilized as the node cost, CV, in the total cost function of finding the 3D BMO path. This means assigning lower costs to nodes with higher likelihood of being a BMO point.
2.4. Identifying the 3D BMO Path Using Dynamic Programming
The problem of finding the BMO 3D path is formulated as a shortest-path problem. Here, the goal is to find a path with minimum cost that satisfies a set of constraints. Note that for constructing the graph, we rearrange the radial OCT volumes of size 200 × 180 × 1024 to have size of 100 × 360 × 1024 such that there is one BMO point in each slice. The purpose of rearrangement is that in the rearranged radial volume the BMO points appear as a closed loop path in 3D and it allows us to model the problem as finding a shortest path within the rearranged radial OCT volume. The 3D node cost (CV) also undergoes the same rearrangement to match its corresponding OCT volume. Assume that a directed acyclic graph (DAG) 𝒢(V, E) is constructed from the radial volume ℛ of size R × Z × Θ with non-negative edge and node costs and we want to find the BMO locations as a minimum-cost 3D path ℘ = {V℘, E℘} within 𝒢. Each voxel in the volume is represented by a node in the graph; hence, the number of nodes in the graph, |V|, is equal to the number of voxels in the volume. Each node is connected only to its neighbors (i.e., the feasible nodes in the subsequent slice). As there is only one BMO endpoint in each slice, the nodes in the same slice are not connected to each other. The neighboring constraint in the r-direction and z-direction are Δr = 2 and Δz = 1, respectively which means the neighborhood of each node is a 3 × 5 rectangle in the next slice such that there is an edge between the node and its neighbors (Fig 7). The neighbors of each node in the subsequent slice is specified according to the maximum variations of BMO paths in-between slices in the training set. Additionally, since the BMO path is a closed circular path in the Cartesian domain, we need to ensure that the computed shortest path is a closed loop (i.e., the first and last radial slices are considered as adjacent slices).
Figure 7.

Graph construction. There are weighted edges between each node (green) and its neighboring nodes (red). The neighboring constraint in r-direction (Δr) and in z-direction (Δz) determine the amount of allowed variation from slice to slice.
There are two types of costs in the graph: edge costs and node costs. The edge costs are responsible for smoothness of the path and they penalize the deviations from the BMO location in the previous slice. The penalizing function in r-direction is fr = (rθ1 − rθ2)2 and in the z-direction it is fz = |zθ1 − zθ2| which generates the following edge weights for all neighborhoods:
| (3) |
Since the BMO path has a smaller variation in the r direction than in the z direction, the penalty for variation in the r direction is heavier (quadratic function) than in the z direction (linear function). The cost of the edge (u, v), Cuv, connecting nodes u = (zu, ru, θu) and v = (zv, rv, θv) is obtained by adding the weights (assuming Wr and Wz elements are expanded at −2: 2 and −1: 1 in r and z directions, respectively) as follows:
| (4) |
Hence, the total cost of a feasible BMO path is as follows:
| (5) |
where CV is the 3D node cost computed by the random forest classifier in Section 2.3.2 and 0 < β < 1 (set to 0.15 empirically) determines the importance of edge cost with respect to the node cost. In order to reflect the anatomical information that the BMO can touch the ILM surface but never pass it, the nodes in the search region that fall above the ILM surface are not considered as neighbors of any node in the graph (i.e., there is no edge connecting them to any other nodes).
In order to compute the minimum-cost shortest path through the constructed graph, a dynamic programming (DP) approach is utilized. In order to solve the problem using DP we need to define the subproblem and the recursive formulation to solve the problem as well as the base case. Suppose we add a dummy node, s, and we connect all nodes in the first slice to s using identical zero-cost edges. Now the problem becomes to find the minimum-cost path from s to any node located in the last slice (the enforcement of circularity constraint will be explained later). We know that if s ⇝ u ⇝ v is a shortest path from s to v, this implies that s ⇝ u is a shortest path from s to u, as well; otherwise if there were a shorter path between s and u we would obtain a better path between s and v by replacing the s ⇝ u with the shorter path. But we assumed that s ⇝ u ⇝ v is a shortest path between s and v, so we have a contradiction. Based on this idea, we define the subproblem. Let us assume OPT(v, k) is the minimum-cost path from s to node v in slice k, the recursive formulation for OPT(v, k) can be written as follows:
| (6) |
and the base case is OPT(v, 1) = CV(rv, zv, θv). The optimal solution can be efficiently computed using dynamic programming by computing the values in order of increasing k.
Here, the circularity constraint means that, assuming the first and last slices are adjacent slices, the path ends at one of the allowed neighbors of the starting node. In order to ensure that this condition is satisfied, we found all possible shortest paths that satisfied the circularity condition and picked the path with the minimum cost. More specifically, in order to enforce the circularity constraint, the following steps need to be performed:
Force the path to start at a specific node in the first slice and specify the allowed ending points (its neighbors) in the last slice (according to the smoothness constraints).
Find the minimum-cost path for the specified configuration of starting-ending points.
Repeat steps 1 and 2 until all candidates in the first slice are examined.
Choose the path with the minimum cost among all the shortest paths as the 3D BMO path.
The enforcement in step 1 was performed by deliberately increasing the cost of all nodes in the search region of the first slice except for the starting node and repeating the same action for all nodes in the search region of the last slice except for the neighboring nodes.
2.5. Refinement of BMO Path in the z-Direction
The BMO path computed in Section 2.4 needs further refinement as the path was found in the downsampled (in the z-direction with 30 × 30μm voxel size in r-z plane) volume. Processing the data initially in the isotropic grid was beneficial for easily setting up the smoothness/neighborhood constraints for graph construction, saving time and space for extracting the features, training the RF classifier, and finding the shortest path (especially for computing all possible shortest paths satisfying the circularity constraint). In order to obtain the BMO path in the original image resolution (30×2μm voxel size in r-z plane), a similar method as used to find the BMO path in the downsampled volume was utilized. However, the search region for refinement was restricted such that it only consisted of those 15 voxels in the original resolution that corresponded to the BMO point found in the lower resolution. Therefore, as r-values were fixed during this step, the refined z-values were computed as a shortest path with the circularity constraint within a small 2D image of size 15 × 360 in Z × Θ.
The node cost computation procedure is similar to that of Section 2.3. From the volumes in the training set, a new intensity model reflecting the intensity profile around the BMO in the original resolution was created (referred to as the high-resolution PCA model as the features are computed from the original resolution). The model was obtained using the same set of features as in Section 2.3.1 and applying PCA such that 90% of the variation in the feature set was retained. For training the RF classifier a 15-voxel length array around each BMO point in the z-direction was involved. The positive class (BMO) included the BMO point itself and the voxels right above and below it (in order to resemble the inter-observer variability) and the negative class (non-BMO) included the rest of the array (a total of 12). The intensity-based features of all points in positive and negative classes were computed and projected to the high-resolution PCA model and a separate random forest classifier, RFz, was trained utilizing the projected intensity features. The number of trees and variables to be randomly extracted at each decision split were set to 500 and the square root of number of features, respectively. Note that while the refinement of the BMO path occurs after computing the initial BMO estimate from the downsampled data, training this high-resolution classifier does not depend on the output of the first low-resolution classifier in any way and thus is trained independently.
As mentioned above, each BMO point (found in the lower resolution) contributes 15 voxels from the original resolution to the test set. The intensity-based features were computed for all points in the test set and projected to the high-resolution PCA model. The trained RFz classifier produced the likelihood map of each node being BMO. The inverted likelihood map served as the node cost in the graph construction. The graph 𝒢Z = (VZ, EZ) consisted of 15 × 360 nodes and edges existed only between each node and its neighbors in the subsequent slice. The furthest neighbor of each node has at most Δd distance in the z-direction for which the Δd is computed from the training set. The edge weight increases (Fig. 8) as the distance of neighbors becomes larger via the penalizing function fd = |zd1θ1 − zd2θ2|. The final 3D BMO path was obtained by finding the shortest path with circularity constraint using the dynamic programming approach similar to that in Section 2.4.
Figure 8.

Refinement Graph, 𝒢Z, construction. The red box indicates one voxel on the BMO path in the downsampled volume which corresponds to 15 voxels in the original resolution. The edges are color coded with warmer colors corresponding to higher weights. For a particular node (green) in slice θ1, all nodes in the subsequent slice, θ1 + 1, with a distance less than Δd are considered as neighbors. The yellow nodes indicate those nodes with a distance larger than Δd.
3. Experimental Methods
3.1. Data
The training dataset includes 25 SD-OCT volumes centered at the optic nerve head that were acquired using a Cirrus HD-OCT device (Carl Zeiss Meditec, Inc., Dublin, CA) at the University of Iowa from the dataset in (Abràmoff et al., 2009). In this dataset, patients that met the definition of suspected glaucoma, open-angle glaucoma, angle-closure glaucoma, or combined-mechanism glaucoma were included. The size of each scan was 200×200×1024 voxels with a voxel size of 30×30×2 μm in the x-y-z direction. Similarly, the testing set includes 44 patients diagnosed with glaucoma suspect or open-angle glaucoma and optic nerve head (ONH)-centered SD-OCT volumes were obtained in one eye of each patient.
3.2. Reference Standard
For the training set, the BMO points were marked on all radial B-scans such that one expert first traced the BMO points on the 3D SD-OCT volume with two additional experts providing corrections resulting in a final tracing that was the result of the consensus of three experts through a discussion. For the testing set, the BMO points were identified on 20 evenly spaced randomly-picked radial slices by consensus of manual delineations from three experts (a total of 40 BMO points for each subject). More specifically, the first slice was randomly chosen from the first 9 slices in the cube and the remaining 19 slices were evenly distributed with respect to the first slice. For instance, if the random number was 4 then the set of selected slices was {4, 13, 22, …, 157, 166, 175}.
3.3. Experiments
As two independent datasets were available for training and testing purposes, the proposed method used the entire training set for creating the BMO models and training the classifiers and it was tested on the entire test set. The performance of the proposed BMO identification method (BMOproposed) as well as our previous iterative method (Antony et al., 2014) (BMOiterative) were evaluated using the reference standard obtained on the 20 slices of each subject with manual delineation (BMOmanual). The signed and unsigned distances of automated BMO points with manual BMO points in the r-direction and z-direction, were measured separately. If the automated methods identified the BMO closer to the optic disc center, the sign of distance in the r-direction was positive. Similarly, if the automated BMO (BMOproposed/BMOiterative) located below the manual BMO (BMOmanual), the sign of distance in the z-direction was positive. In addition, the distance of automated and manual BMO points in the r-z plane was measured. A paired t-test was utilized to compare the error measurements (a p-value < 0.05 was considered significant).
Furthermore, as enabling automated computuation of the BMO-MRW measure was one of the major motives of our proposed method, this structural parameter was computed using the manual (BMOmanual-MRW), the proposed method (BMOproposed-MRW) and the iterative approach (BMOiterative-MRW). The signed and unsigned differences of BMOproposed-MRW and BMOiterative-MRW with BMOmanual-MRW (called the BMO-MRW error) were calculated and compared using paired t-tests with p-values < 0.05 being considered significant. The Pearson correlation as well as root mean square error (RMSE) of automated BMO-MRW measures with respect to the manual BMO-MRW were also computed. Zou’s method for comparing two overlapping correlations based on two dependent groups (Zou, 2007) was utilized to compare the Pearson correlations of BMOmanual-MRW with BMOproposed-MRW and BMOiterative-MRW (p-values < 0.05 were considered significant).
4. Results
Fig. 9 shows example results of BMO identification and BMO-MRW computation using the automated methods and the reference standard. For better visualization, only the central part of each B-scan is shown. The first row in Fig. 9 depicts an example that both automated methods successfully identified the BMO points. The second and third examples demonstrate how presence of the externally oblique border tissue causes erroneous BMO identification for the automated approaches; however, the proposed method is less affected than the iterative approach. The externally oblique border tissue attaches to the end of BM surface and resembles the BMO points which influences the automated methods. The last example demonstrates a case that the results of both iterative approach and the proposed method are affected by the presence of an ambiguous border tissue. In such cases, without using the 3D contextual information, it is very difficult (even for humans) to mark the exact location of BMO. Note that when there is no border tissue present, the iterative and our proposed methods have comparable performances (Fig. 9).
Figure 9.
Example results, left column is the original B-scan along with the ILM surface and the right column demonstrates the segmentation results. The blue, yellow, and green circles indicate the BMOproposed, BMOiterative, and BMOmanual, respectively. The lines connecting the BMO points to the ILM surface, indicate the corresponding BMO-MRW measures.
The unsigned and signed BMO identification errors in the r-direction, z-direction, and r-z plane are reported in Table 1. Note that the standard deviation values are computed over the averages of the subjects. Meaning that for each subject in the testing set the average of errors in the 20 slices (with BMO points manually marked on them) are computed and the standard deviation is computed over the averages of the subjects. Based on BMO identification errors, the proposed method outperformed our previous iterative approach (Antony et al., 2014) in the z-direction, r-direction, and r-z plane (p < 0.05). Similarly, the signed BMO identification error showed that the proposed method has significantly lower errors in the r- and z-directions than the iterative approach (p < 0.05). The maximum unsigned BMO identification error in the r- and z-directions and in the r-z plane for the proposed (iterative) method were 76.5 (148.5), 42.8 (92.4), and 85.1 (166.17) μm, respectively.
Table 1.
Unsigned and signed BMO identification error in the r-direction, z-direction, and r-z plane in μm (Mean± SD).
| Error | Unsigned
|
Signed
|
||
|---|---|---|---|---|
| (Antony et al., 2014) | Proposed Method | (Antony et al., 2014) | Proposed Method | |
| r-direction | 49.53±30.41 | 37.98±14.91 | 26.49±40.22 | −9.49±24.58 |
| z-direction | 31.58±21.06 | 22.28± 8.58 | 25.45±14.37 | 8.33±17.72 |
| r-z plane | 63.03±36.28 | 49.28±16.78 | – | – |
Table 2 shows the measurements associated with the structural parameter BMO-MRW including unsigned and signed BMO-MRW error and RMSE. Based on the unsigned BMOMRW error, the proposed method had a significantly lower BMO-MRW error than the iterative approach (p < 0.05). Similarly, the signed BMO-MRW error showed that BMOproposed-MRW has significantly smaller bias than BMOiterative-MRW (p < 0.05). The mean±standard deviation of manual and automated BMO-MRW measures were 182.41±86.07 μm (BMOmanual-MRW), 184.46±81.61 μm (BMOproposed-MRW), and 187.33±88.51 μm (BMOiterative-MRW), respectively. The maximum unsigned BMO-MRW error of the proposed (iterative) method was 38.08 (75.10) μm. Furthermore, the proposed method had a smaller RMSE than the iterative approach for computing the BMO-MRW (p < 0.05).
Table 2.
BMO-MRW error measurements in μm (Mean± SD).
| Error | (Antony et al., 2014) | Proposed Method |
|---|---|---|
| Unsigned error | 26.65±13.27 | 22.22± 5.99 |
| Signed error | 6.61±18.59 | − 0.30±12.44 |
| RMSE | 17.99± 8.15 | 11.62± 4.63 |
The comparison of Pearson correlations of BMOmanual-MRW with BMOproposed-MRW (0.992) and BMOiterative-MRW (0.980) using the method in (Zou, 2007) showed that the proposed method had significantly higher correlation with the reference standard than the iterative approach (p < 0.05). Additionally, Fig. 10 depicts the Bland-Altman plots of BMOproposed-MRW and BMOiterative-MRW with respect to BMOmanual-MRW where the tighter fit and smaller error of the proposed method in computing the BMO-MRW are observable.
Figure 10.

The Bland-Altman plots of (a) BMOiterative-MRW and (b) BMOproposed-MRW in comparison with BMOmanual-MRW. The proposed method has a tighter fit and lower error than the iterative approach.
5. Discussion and Conclusion
In this paper, we presented a machine-learning graph-based approach for automated segmentation of Bruch’s membrane opening from SD-OCT volumes. Our results showed that the proposed method successfully identifies the opening points of the BM surface such that it enables computing the structural parameter Bruch’s membrane opening-minimum rim width (BMO-MRW) automatically. The proposed method first identifies the estimated location of BMO points by finding the BMO projection locations using a similar graph-theoretic approach reported in (Miri et al., 2015) with incorporation of shape prior and mapping them onto the BM surface. The estimated 3D locations were then utilized to more precisely find the 3D BMO loop by formulating the problem as computing the minimum-cost path within the 3D volume using dynamic programming. We learned the intensity-based attributes of BMO points (in the form of PCA models) using a random forest classifier which computed the cost function needed for identifying the minimum-cost path.
To the best of our knowledge, our previous iterative graph-theoretic approach (Antony et al., 2014) was one of the first non-commercialized approaches reported for identifying the 3D BMO points automatically that included incorporation of 3D context. A graph-theoretic approach was utilized for identifying the initial 2D segmentation of BMO points, and while the (r, θ) pairs kept unchanged, we looked for the corresponding z-values as the hole within a 3D surface. Upon finding the 3D BMO locations as part of the surface + hole method, a new radial projection image was created from which the updated 2D locations were identified. This iteration was repeated until convergence. While use of 3D context has advantages over other strategies that independently search for the BMO in each slice, in this paper, we aimed to improve our previous method by removing the iterations and eliminating the two-step identification of the (r, θ) pairs and z values and instead, finding the (r, θ, z) coordinates as part of a 3D loop in a single run.
The other limitation of our previous work was that in the presence of externally oblique border tissue (Reis et al., 2012), the iterations would continue to search for the BMO point along the anterior surface of the border tissue. The presence of externally oblique border tissue confused the algorithm and caused the algorithm to find the BMO point on the border tissue instead of at the end of the BM surface. The presence of externally oblique border tissue was definitely one of the main reasons for the large positive signed border positioning errors of the prior iterative method reported in Table 1. For instance, the three outliers in Fig. 10a are illustrating errors in measuring BMO-MRW caused by large errors in identifying the exact location of BMO points due to the presence of externally oblique border tissue (EOBT). With these three subjects removed, our proposed approach still has significantly better results (compared to our previous iterative method) when considering the BMO identification error; however, the BMO-MRW error measurements, while still better (i.e., smaller) using our proposed approach, were no longer significantly better. To better understand these results, it is important to remember that BMO-MRW errors (with the BMO-MRW being a measure that reflects the minimum distance from the BMO to the ILM surface) is only an indirect measure of the BMO segmentation quality, whereas the BMO position error is a much more direct measure. The issue of the presence of border tissue was addressed in the proposed method to a certain extent by preventing the deviation from the expected shape of path using the penalizing edge weights.
The penalizing weights not only helped with avoiding the border tissues, but they were also effective in dealing with the vessels shadows. Unlike a typical BMO point locating at the end of a bright band, co-localizing a BMO point with a blood vessel causes the BMO point to be located inside a dark region (Fig. 11). Presenting the penalizing weights helped the BMO path to keep the natural trend of the BMO trajectories in the volume and not deviate from the expected shape of the BMO path. However, it must be noted that setting up high penalizing weights is not the solution for the issues of presence of border tissue and blood vessel shadows. The reason is that even though higher penalizing weights leads to a stiffer BMO path with minimal variation in the r- and z-directions; there are cases that the BMO path must be capable of following the “true” natural translation of the BMO points throughout the volume. Hence, there is a trade off between being capable of dealing with the presence of border tissue and blood vessel shadows and following the true displacements of the BMO path throughout the volume.
Figure 11.

An example of identifying the BMO point using the proposed method within the shadow of a large blood vessel.
Additionally, the issue of the presence of border tissue and the blood vessels were also dealt with in identifying the 2D location of the BMO points. This was performed by enforcing the prior shape constraint in the r-direction during applying the graph-theoretic approach. Identifying an accurate 2D location of BMO points is important as it leads to a better estimated 3D location which requires smaller search region to find the actual BMO point. The size of the search region was computed based on the accuracy of estimated 3D BMO points in the training set. Shrinking the searching area decreases the BMO point candidates and the reduces the size of the graph to be solved.
Furthermore, having 180 radial slices (because of dense sampling with angular resolution of θ = 1°) enables us to rely on 3D contextual information with more certainty. Meaning that the BMO path has smaller variations in between slices in the r and z directions; hence, smoothness constraints (Δr and Δz in Fig. 7), which determine the allowed neighborhood of each node in the graph, become tighter. This is beneficial in dealing with situations where a BMO point colocalizes with a blood vessel shadow or in the presence of EOBT as the BMO path has a small allowed variation and this prevents from being deviated from the actual path and producing large errors.
In contrast, the standard radial slices that are available from using protocols with 24 slices (angular resolution of θ = 15°), such as those used in (Reis et al., 2012; Chauhan and Burgoyne, 2013), would require looser smoothness constraints and smaller weights for penalizing functions (Eq. 4) to be capable of following possibly larger variations of BMO points in the r and z directions. The looser smoothness constraints and smaller penalizing weights might be less effective in dealing with blood vessel shadows and/or the presence of EOBT. When considering other protocols, if there is enough information in the y direction (i.e. enough number of B-scans) in the Cartesian domain such that extracting a radial volume with sufficient number of radial slices (i.e. sufficiently small angular resolution) becomes feasible, then our proposed method can be considered for identifying the BMO points from the SD-OCT volume to best take advantage of 3D contextual information.
Our proposed method was mostly implemented in (nonoptimized) MATLAB with the exception of the radial surface segmentation and 2D projected BMO identification steps that were performed in C++. We utilized a 64-bit Operating System with Intel(R) Xeon(R) CPU @ 3.70GHz and 64.0 GB memory. The method takes approximately 30 to 40 minutes to complete. The most time consuming part of the proposed method is enforcing the circularity constraint on the BMO path using dynamic programming as explained in Section 2.4. Since the runs that are needed to be performed in order to find the minimum-cost path with circularity constraint are completely independent from each other, they could be performed in parallel. Since this was not done in our current implementation, the running time of the algorithm could be considerably decreased by implementing the circularity constraint enforcement in a parallel fashion. In addition, implementing the algorithm in a different programming language such as C++ and optimizing the code would also be expected to dramatically improve the speed of the proposed method.
In summary, we proposed a machine-learning graph-based approach for segmentation of BMO points from SD-OCT volumes. After transferring the volumes to the radial domain, radial projection images were computed by segmenting the intraretinal surfaces. The projection images were processed using SWT in order to create the vessel-free images in which the blood vessels were significantly suppressed. An edge-based cost function was obtained from the gradient of vessel-free images to incorporate in a graph-theoretic approach for finding the 2D location of BMO points (r, θ). The volumes were downsampled in the z-direction to achieve an isotropic grid (with the same size as the physical resolution) and an estimated 3D location of BMO points, (r, z, θ), were obtained by projecting the 2D coordinations onto the BM surface. An elliptical search region around each estimated location was considered to form a 3D tube from which the 3D BMO points were identified by looking for the minimum-cost path within the searching tube using dynamic programming. The cost function for finding the minimum-cost path was computed by inversing the likelihood map generated by a RF classifier based on the resemblance of each point inside the searching tube to the BMO intensity models. Once the BMO points were identified, a similar approach as used to find the BMO path within the downsampled volume was utilized to refine the z-values such that the 3D coordinates of BMO points in image resolution were obtained.
This genuine 3D segmentation method enabled more accurate identification of BMO points which results in more precise computation of BMO-MRW measure and consequently, more accurate diagnosis of glaucoma.
Acknowledgments
This work was supported, in part, by the National Institutes of Health grants R01 EY018853 and R01 EY023279; the Department of Veterans Affairs Rehabilitation Research and Development Division (Iowa City Center for the Prevention and Treatment of Visual Loss and Career Development Award 1IK2RX000728); and the Marlene S. and Leonard A. Hadley Glaucoma Research Fund.
Appendix A. Identifying 2D BMO Path
The projected locations of BMO points are identified from the radial projection image in the form of a 2D path utilizing a graph-theoretic approach and an edge-based cost function computed from the radial projection image. The approach is similar to our previous work in (Miri et al., 2015), except that here, the total cost function does not include the in-region cost term and shape prior information is also enforced.
Appendix A.0.1. Computing the Edge-Based Cost Function
An edge-based cost function is designed to have information regarding the location of the boundary of interest in the image (here, the 2D BMO trajectory). More specifically, the edge-based cost function is a probability map that contains the unlikelihood of pixels locating on the boundary. Therefore, we would like to have a cost function with low values on the expected location of the boundary and high values everywhere else.
The BMO boundary and the retinal blood vessels are the two major structures of the reformatted radial projection images. With the help of the Haar stationary wavelet transform (SWT) (Fowler, 2005), we eliminate the effect of retinal blood vessels in computing the edge-based cost function. The directionality of the SWT enables us separate the blood vessels (mostly appear vertically) from the BMO boundary (located horizontally). The blood vessels are mostly present in the diagonal and vertical coefficients, whereas the BMO boundary appears in the horizontal coefficients (Fig. A.12a). Furthermore, we can catch blood vessels with different sizes as SWT is a multiscale transformation. In order to compensate for the presence of the blood vessels, the diagonal and vertical coefficients are set to zero in all 4 scales while retaining the horizontal and approximation coefficients. The inverse SWT transformation using the modified coefficients generates a 2D radial projection image in which the blood vessels are significantly suppressed which is called vessel-free radial projection image (Fig. A.12b).
Figure A.12.
An example of edge-based cost function computation. (a) The 2nd scale of SWT decomposition. Note that BMO boundary appears in the horizontal coefficients while the blood vessels mostly appear in vertical and diagonal coefficients. (b) The vessel-free radial projection image. (c) The edge-based cost function computed by applying the Gaussian derivative filter, ℱσrσθ (r, θ), to the vessel-free radial projection image.
In order to compute the edge-based cost function, an asymmetric 2D Gaussian derivative filter ℱσrσθ (r, θ) is applied to the vessel-free projection image Iv f (r, θ) with the aim of capturing the dark-to-bright transitions. The filter has larger scale in the θ-direction (σ = 4) than the r-direction (σ = 2) and computes the derivatives only in the r-direction (vertical axis). If a general 2D Gaussian filter in the radial domain can be written as:
| (A.1) |
the asymmetric Gaussian derivative filter applied to the vessel-free image can be expressed as ℱσr,σθ (r, θ) = ∂𝒢σr,σθ/∂r, where σr and σθ are the standard deviations in the r and θ directions, respectively. The edge-based cost function is shown in Fig. A.12c. Therefore, the total edge-cost representing the cost of pixels on a 2D BMO path, ℬ(θ), can be expressed as
| (A.2) |
and the goal is to select ℬ(θ) that minimizes the cost function Cedge.
Appendix A.0.2. Segmenting BMO 2D Path
In order to identify the minimum-cost 2D BMO path, a graph-theoretic approach proposed by Song et al. (Song et al., 2013) is utilized. This approach, is an extension to the method proposed by Garvin et al. (Garvin et al., 2009) that enables us to constrain the geometry of the boundary of interest based on prior shape information of the boundary. For instance, as mentioned in Section 2.1, it is known that in the radial projection image, the BMO boundary is very smooth across slices with minimal changes in the r-direction. This shape-prior knowledge was enforced as a soft smoothness constraint along with the hard smoothness constraints (Song et al., 2013).
More specifically, consider a 2D image I(r, θ) of size R×Θ in the radial domain and the 2D BMO boundary ℬ(θ) as a function of θ that maps each θ-value to its corresponding r-value. In addition, assume that the function intersects with each column (each θ) once (at r) and the function uses a two-neighbor relationship. Two types of constraints are applied to the neighboring columns, the hard smoothness constraint and the shape-prior (soft smoothness) constraints. The hard smoothness constraint for a pair of neighboring columns (θ1, θ2) in the θ-direction can be written as below
| (A.3) |
where Δr is the maximum allowed change of r between two neighboring columns. In order to incorporate the shape prior information, in addition to hard smoothness constraints, the deviation from the expected shape inside the allowed constraint is penalized as well (Song et al., 2013). A convex function f (h) penalizes the cost of the boundary if the change of the boundary is deviated from its expected shape. Specifically, for any pair of neighboring columns (identified by Nc) such as (θ1, θ2) on boundary ℬ, if the expected shape change of boundary B between (θ1, θ2) is m(θ1,θ2) the cost of the shape term can be written as:
| (A.4) |
Here, m(θ1,θ2) = 0 as we expect to have very smooth BMO boundary with minimal change between neighboring columns. The penalizing function f needs to be a convex function for which a quadratic function is chosen. The total cost of finding the initial 2D BMO boundary ℬ consists of both edge-based and shape prior cost functions which can be expressed as follows:
| (A.5) |
The coefficient α determines the impact of shape prior cost function with respect to the edge-based cost function. Using a training set, the parameter α was set to 0.7 in this study. The optimal boundary can be found by computing the maxflow/mincut in the arc-weighted graph as in (Song et al., 2013).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abràmoff MD, Alward WLM, Greenlee EC, Shuba L, Kim CY, Fingert JH, Kwon YH. Automated segmentation of the optic disc from stereo color photographs using physiologically plausible features. Invest Ophthalmol Vis Sci. 2007;48:1665–1673. doi: 10.1167/iovs.06-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abràmoff MD, Garvin MK, Sonka M. Retinal imaging and image analysis. IEEE Rev Biomed Eng. 2010;3:169–208. doi: 10.1109/RBME.2010.2084567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abràmoff MD, Lee K, Niemeijer M, Alward WLM, Greenlee EC, Garvin MK, Sonka M, Kwon YH. Automated segmentation of the cup and rim from spectral domain OCT of the optic nerve head. Invest Ophthalmol Vis Sci. 2009;50:5778–5784. doi: 10.1167/iovs.09-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony BJ, Miri MS, Abràmoff MD, Kwon YH, Garvin MK. Automated 3D segmentation of multiple surfaces with a shared hole: segmentation of the neural canal opening in SD-OCT volumes. Proc. MICCAI 2014, LNCS, Part I; Boston, USA. 2014. pp. 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belghith A, Bowd C, Medeiros FA, Hammel N, Yang Z, Weinreb RN, Zangwill LM. Does the location of Bruch’s membrane opening change over time? Longitudinal analysis using San Diego Automated Layer Segmentation Algorithm (SALSA) Glaucoma. 2016;57:675–682. doi: 10.1167/iovs.15-17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belghith A, Bowd C, Weinreb RN, Zangwill LM. A hierarchical framework for estimating neuroretinal rim area using 3D spectral domain optical coherence tomography (SD-OCT) optic nerve head (ONH) images of healthy and glaucoma eyes. Proc. IEEE Eng. Med. Bio. Soc. (EMBS); Chicago, USA. 2014. pp. 3869–3872. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Chauhan BC, Burgoyne CF. From clinical examination of the optic disc to clinical assessment of the optic nerve head: a paradigm change. Am J Ophthalmol. 2013;156:218–227. doi: 10.1016/j.ajo.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BC, O’Leary N, AlMobarak FA, Reis AS, Yang H, Sharpe GP, Hutchison DM, Nicolela MT, Burgoyne CF. Enhanced detection of open-angle glaucoma with an anatomically accurate optical coherence tomography-derived neuroretinal rim parameter. Ophth. 2013;120:535–543. doi: 10.1016/j.ophtha.2012.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Liu J, Xu Y, Yin F, Wong DWK, Tan NM, Tao D, Cheng CY, Aung T, Wong TY. Superpixel classification based optic disc and optic cup segmentation for glaucoma screening. IEEE Trans Med Imag. 2013;32:1019–1032. doi: 10.1109/TMI.2013.2247770. [DOI] [PubMed] [Google Scholar]
- Chràsteka R, Wolfa M, Donatha K, Niemanna H, Paulusb D, Hothornc T, Lausenc B, Lmmerd R, Mardind C, Michelsond G. Automated segmentation of the optic nerve head for diagnosis of glaucoma. Med Imag Anal. 2005;9:297–314. doi: 10.1016/j.media.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Fowler JE. The redundant discrete wavelet transform and additive noise. IEEE Sig Proc Let. 2005;12:629–632. [Google Scholar]
- Fu H, Xu D, Lin S, Wong DWK, Liu J. Automatic optic disc detection in OCT slices via low-rank reconstruction. IEEE Trans Biomed Eng. 2015;62:1151–1158. doi: 10.1109/TBME.2014.2375184. [DOI] [PubMed] [Google Scholar]
- Gardiner SK, Boey PY, Yang H, Fortune B, Burgoyne CF, Demirel S. Structural measurements for monitoring change in glaucoma: comparing retinal nerve fiber layer thickness with minimum rim width and area. Invest Ophthalmol Vis Sci. 2015;56:6886–6891. doi: 10.1167/iovs.15-16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin MK, Abràmoff MD, Wu X, Russell SR, Burns TL, Sonka M. Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans Med Imag. 2009;28:1436–1447. doi: 10.1109/TMI.2009.2016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorescu SE, Petkov N, Kruizinga P. Comparison of texture features based on gabor filters. IEEE Trans Image Process. 2002;11:1160–1167. doi: 10.1109/TIP.2002.804262. [DOI] [PubMed] [Google Scholar]
- Hu Z, Abràmoff MD, Kwon YH, Lee K, Garvin MK. Automated segmentation of neural canal opening and optic cup in 3D spectral optical coherence tomography volumes of the optic nerve head. Invest Ophthalmol Vis Sci. 2010;51:5708–5717. doi: 10.1167/iovs.09-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Niemeijer M, Garvin MK, Kwon YH, Sonka M, Abràmoff MD. Segmentation of the optic disc in 3-D OCT scans of the optic nerve head. IEEE Trans Med Imag. 2010;29:159–168. doi: 10.1109/TMI.2009.2031324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Kim TW, Weinreb RN, Lee EJ, Girard MJA, Mari JM. Anterior lamina cribrosa insertion in primary open-angle glaucoma patients and healthy subjects. PLOS ONE. 2014;9:e114935. doi: 10.1371/journal.pone.0114935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell J, Hunter A, Steel D, Basu A, Ryder R, Fletcher E, Kennedy L. Optic nerve head segmentation. IEEE Trans Med Imag. 2008;23:256–264. doi: 10.1109/TMI.2003.823261. [DOI] [PubMed] [Google Scholar]
- Merickel MB, Jr, Abràmoff MD, Sonka M, Wu X. Proc SPIE, Med Imag 2007: Imag Proc. 2007. Segmentation of the optic nerve head combining pixel classification and graph search; p. 651215.p. 10. [Google Scholar]
- Merickel MB, Jr, Wu X, Sonka M, Abràmoff MD. Optimal segmentation of the optic nerve head from stereo retinal images. Proc SPIE, Med Imag 2006: Phys Func Struc Med Imag. 2006:61433B, 8. [Google Scholar]
- Miri MS, Abràmoff MD, Lee K, Niemeijer M, Wang JK, Kwon YH, Garvin MK. Multimodal segmentation of optic disc and cup from SD-OCT and color fundus photographs using a machine-learning graph-based approach. IEEE Trans Med Imag. 2015;34:1854–1866. doi: 10.1109/TMI.2015.2412881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri MS, Lee K, Niemeijer M, Abràmoff MD, Kwon YH, Garvin MK. Multimodal segmentation of optic disc and cup from stereo fundus and SD-OCT images. Proc. SPIE, Med. Imag. 2013: Imag. Proc; Orlando, Florida. 2013. p. 86690O.p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis AS, Sharpe GP, Yang H, Nicolela MT, Burgoyne CF, Chauhan BC. Optic disc margin anatomy in patients with glaucoma and normal controls with spectral domain optical coherence tomography. Ophth. 2012;119:738–747. doi: 10.1016/j.ophtha.2011.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Yang H, Gardiner SK, Fortune B, Hardin C, Demirel S, Burgoyne CF. Anterior lamina cribrosa surface depth, age, and visual field sensitivity in the portland progression project. Invest Ophthalmol Vis Sci. 2014;55:1531–1539. doi: 10.1167/iovs.13-13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosten E, Porter R, Drummond T. Faster and better: a machine learning approach to corner detection. IEEE Trans Pattern Anal Mach Intell. 2010;32:105–119. doi: 10.1109/TPAMI.2008.275. [DOI] [PubMed] [Google Scholar]
- Song Q, Bai J, Garvin MK, Sonka M, Buatti JM, Wu X. Optimal multiple surface segmentation with shape and context priors. IEEE Trans Med Imag. 2013;32:376–386. doi: 10.1109/TMI.2012.2227120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sredar N, Ivers KM, Queener HM, Zouridakis G, Porter J. 3D modeling to characterize lamina cribrosa surface and pore geometries using in vivo images from normal and glaucomatous eyes. Biomed Opt Express. 2013;4:1153–1165. doi: 10.1364/BOE.4.001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chutatape O, Sung E, Zheng C, Kuan PCT. Optic disk feature extraction via modified deformable model technique for glaucoma analysis. Patt Rec. 2007;40:2063–2076. [Google Scholar]
- Yin F, Liu J, Ong SH, Sun Y, Wong DWK, Tan NM, Cheung C, Baskaran M, Aung T, Wong TY. Model-based optic nerve head segmentation on retinal fundus images. 33rd Ann. Int. Conf. IEEE EMBS; Boston, Massachusetts USA. 2011. pp. 2626–2629. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Stambolian D, O’Brien J, Gee JC. Optic disc and cup segmentation from color fundus photograph using graph cut with priors. Proc. MICCAI 2013, LNCS, Part II; Nagoya, Japan. 2013. pp. 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou GY. Toward using confidence interval to compare correlations. Psychol Methods. 2007;12:399–413. doi: 10.1037/1082-989X.12.4.399. [DOI] [PubMed] [Google Scholar]