Abstract
This article provides a review of brain tissue alterations that may be detectable using diffusion magnetic resonance imaging MRI (dMRI) approaches and an overview and perspective on the modern dMRI toolkits for characterizing alterations that follow traumatic brain injury (TBI). Noninvasive imaging is a cornerstone of clinical treatment of TBI and has become increasingly used for preclinical and basic research studies. In particular, quantitative MRI methods have the potential to distinguish and evaluate the complex collection of neurobiological responses to TBI arising from pathology, neuroprotection, and recovery. dMRI provides unique information about the physical environment in tissue and can be used to probe physiological, architectural, and microstructural features. Although well‐established approaches such as diffusion tensor imaging are known to be highly sensitive to changes in the tissue environment, more advanced dMRI techniques have been developed that may offer increased specificity or new information for describing abnormalities. These tools are promising, but incompletely understood in the context of TBI. Furthermore, model dependencies and relative limitations may impact the implementation of these approaches and the interpretation of abnormalities in their metrics. The objective of this paper is to present a basic review and comparison across dMRI methods as they pertain to the detection of the most commonly observed tissue and cellular alterations following TBI.
Keywords: diffusion MRI, traumatic brain injury, axonal injury, neuroinflammation
1. INTRODUCTION
Despite the long history of traumatic brain injury (TBI) as a prevalent cause of death and disability in humans, defining the neurobiological underpinnings of damage and recovery following TBI remains a central challenge. The complex collection of physiological, cellular, and molecular changes that follow TBI can appear to be remarkably heterogeneous, but at the same time they are highly organized into coordinated responses such as neurodegeneration, inflammation, and regeneration. The corpus of histological studies spanning a variety of experimental animal models of TBI have provided crucial insights about the pathomechanisms and cellular alterations that accompany posttraumatic tissue change, but considerable work remains to determine the spatiotemporal evolution of abnormalities, interrelationships among different tissue responses, and their impact on health and behavioral outcomes. Noninvasive imaging in animal models has the potential to build on what is known from histology by providing longitudinal and whole‐brain information, but for this approach to be successful it is essential to first improve the understanding of how imaging abnormalities correspond to tissue and cellular changes.
Diffusion magnetic resonance imaging (dMRI) methods are particularly promising for the development of imaging markers of TBI pathology because they are sensitive to microscale water displacement as a proxy for tissue environment geometry and provide a range of quantitative scalar metrics across the whole brain. Furthermore, dMRI may be combined with other conventional or advanced magnetic resonance imaging (MRI) methods such as arterial spin labeling, susceptibility‐weighted imaging, or a variety of contrast agent MRI approaches to provide complementary and comprehensive outcome measures. Standard dMRI methods and especially diffusion tensor imaging (DTI) have already demonstrated sensitive detection of abnormalities in a number of experimental models of TBI. In the past decade, multiple advanced dMRI approaches have extended beyond the conventional models with the goals of improving the physical description of water diffusion (e.g., by modeling “non‐Gaussian” diffusion) or parameterizing dMRI with respect to the expected biological environment (e.g., by modeling cellular compartments and/or fiber geometry). These new tools will be valuable if they are able to improve the sensitivity or specificity of dMRI following TBI; however, we lack a systematic understanding of how dMRI methods differ from one another for detecting and describing tissue alterations.
A number of excellent reviews exist to describe the current understanding of cellular mechanisms of TBI in general (Bramlett & Dietrich, 2015; Pekna & Pekny, 2012) and within particular areas of neurobiology including neurodegeneration (Johnson, Stewart, & Smith, 2013; Stoica & Faden, 2010), inflammation (Burda, Bernstein, & Sofroniew, 2016; Ziebell & Morganti‐Kossmann, 2010), and myelin changes (Armstrong, Mierzwa, Marion, & Sullivan, 2016), among others. As well, several existing reviews have been published regarding MRI and DTI to study human TBI (Brody, Mac Donald, & Shimony, 2015; Duhaime et al., 2010; Hulkower, Poliak, Rosenbaum, Zimmerman, & Lipton, 2013), and recently a pertinent overview and summary of advanced dMRI tools and their relevance to clinical outcomes was published (Douglas et al., 2015). The focus of the present review is to combine what is known from work in experimental models of TBI about tissue and cellular alterations that may affect the physical tissue environment with a comparative description of the major methods for dMRI that may be differentially sensitive to TBI‐related tissue change alongside several important caveats for their use and interpretation. The first section provides a categorical summary of cellular response to trauma, emphasizing alterations with microstructural, architectural, or neuroanatomical manifestations that may give rise to detectable dMRI abnormalities, including a review of the existing dMRI studies in experimental TBI models. The second section contains a comparative overview of presently available dMRI methods from standard approaches to advanced techniques. The objective of this article is to provide a reference for the current understanding of these topics as well as a perspective to help guide selection of dMRI tools based on particular aspects of TBI questions.
2. THE PHYSICAL TISSUE ENVIRONMENT CHANGES FOLLOWING TRAUMA
dMRI is able to detect abnormalities only when the physical tissue environment changes in a way that substantially alters the movement of water within a voxel, such as the loss or gain of cellular boundaries or by organization or disorganization of oriented structures. Because dMRI measurements record signals from the motion of water molecules in small imaging volumes (voxels), their sensitivity is related to the magnitude of structural changes incurred by injury. This implies that dMRI is sensitive to abnormal cerebral physiology (e.g., edema, vascular injury, disruption of water homeostasis), changes in tissue composition (e.g., increased or decreased cellularity), and alterations in cellular morphology (e.g., glial reactivity or neurite density changes). On the other hand, functional and molecular changes are less likely to be detected by dMRI directly but may be associated with detectable changes indirectly. Table 1 lists the major categories of cellular changes arranged according to cell type that may give rise to altered tissue environment and thus affect dMRI measurements.
Table 1.
Cellular Alterations Known to Follow TBI and Associated dMRI Changes
| Cell type or compartment | TBI‐related alterations | Tissue environment | Expected diffusion changes | Major citations | dMRI evidence |
|---|---|---|---|---|---|
| Neurons | cell loss necrosis and apoptosis | atrophy, cavitation, unmasking | decreased diffusivity and anisotropy, increased anisotropy | Sato et al., 2001; Chen et al., 2003; Coleman, 2005; Stoica & Faden, 2010 | Assaf et al., 1997; Van Putten et al., 2005; Immonen et al., 2009; Laitinen et al., 2015 |
| axonal injury | axon morphology changes including beading and varicosities | reduction in anisotropy and reduction in diffusion, especially in the axial direction | Johnson et al., 2013 | Mac Donald, Dikranian, Bayly, et al., 2007; Budde et al., 2009; Jiang et al., 2011; Li et al., 2011; Bennett et al., 2012; van de Looij et al., 2012 | |
| neural plasticity sprouting, arborization | increased number of coherent processes and new collaterals | increased anisotropy and/or changed orientation | Bach‐y‐Rita, 2003; Yiu & He, 2006; Werner & Stevens, 2015; Meaney & Smith, 2015 | Kharatishvili et al., 2007; Hutchinson et al., 2012; Sierra et al., 2015 | |
| Oligodendrocytes | demyelination direct damage, chronic pathology | degenerating or lost | decreased anisotropy | Armstrong et al., 2016 | Jiang et al., 2011; Budde et al., 2011; Li et al., 2014; Mac Donald, Dikranian, Song, et al., 2007 |
| myelination repair remyelination | regenerating | normalized anisotropy | |||
| Astrocytes | Hypertrophy | increased number or thickness of glial processes, possibly organized or directional | increased or decreased anisotropy, decreased diffusivity | Norton et al., 1992; Sofroniew, 2005, 2009; Sofroniew & Vinters, 2010; Wilhelmsson et al., 2006; Suzuki et al., 2012; Sun & Jakobs, 2012; Pekny et al., 2014; Burda et al., 2016 | Budde et al., 2011; Zhuo et al., 2012; Mac Donald, Dikranian, Song, et al., 2007 |
| proliferation | increased cellularity | decreased diffusivity | |||
| glial scaring | dense glia, increased organization | decreased diffusivity, increased anisotropy | |||
| Microglia | phagocytosis | amoeboid stage microglia | increased diffusivity | Kreutzberg, 1996; Graeber, 2010; Wake & Fields, 2011; Ziebell et al., 2012; Roth et al., 2014 | |
| neural repair and support | rod‐microglia | possible increased anisotropy | |||
| Intracellular space | cytotoxic edema | cell swelling | decreased diffusivity | Moseley et al., 1990; Pierpaoli et al., 1993; Bramlett & Dietrich, 2004 | Hanstock et al., 1994; Smith et al., 1995; Alsop et al., 1996; Unterberg et al., 1997; Assaf et al., 1997; Stroop et al., 1998; Albensi et al., 2000; Van Putten et al., 2005; Immonen et al., 2009; Frey et al., 2014 |
| Extracellular space | vasogenic edema | excess extracellular fluid | increased diffusivity |
Note: For each major cell type of the brain (column 1), different types of abnormalities that have been observed to follow experimental brain injury are categorized (column 2). The resulting changes to the tissue environment (column 3) and water diffusivity and anisotropy (column 4) are given as well as relevant citations for the neurobiological phenomenon (column 5) and dMRI observations (column 6).
2.1. The physiologic response to TBI and alterations of diffusivity
Brain contusion is a common outcome of TBI and can be readily identified on T2‐weighted MRI scans once blood products have accumulated in the parenchyma or extracellular fluid has increased (vasogenic edema). In the hyperacute phase, dMRI has been shown to be uniquely sensitive to early pathophysiologies that are not detectable using conventional MRI (Smith et al., 1995). The early sensitivity of dMRI to TBI changes is similar to observations in ischemic stroke, for which diffusivity is known to decrease robustly during the first few hours in the absence of other MRI changes (Moseley et al., 1990). The first dMRI studies of TBI were performed to characterize diffusivity changes during this period in experimental models of focal contusion and provided evidence for both decreased (Alsop, Murai, Detre, McIntosh, & Smith, 1996; Stroop et al., 1998; Unterberg et al., 1997) and increased (Hanstock, Faden, Bendall, & Vink, 1994) diffusivity in the hours after injury. While these findings may seem contradictory, experimental differences such as injury severity can explain opposite changes in diffusivity (Smith et al., 1995) and also highlight the potential for dMRI to distinguish different pathophysiological features of tissue. In particular, mechanisms of cellular damage including metabolic disruption, beading, and cytotoxic edema have been proposed to explain acutely reduced diffusivity following stroke and injury, while vasogenic edema is generally accepted to underlie increased diffusivity. While cellular disruption generally precedes vasogenic edema in stroke, this may not always be the case in brain injury (Bramlett & Dietrich, 2004). For instance, observations in a model of focal cortical ischemia (Pierpaoli et al., 1993) have demonstrated increased diffusivity in nonischemic brain regions with edema adjacent to regions of decreased diffusivity where ischemic damage was later confirmed by histology. The implication of this for TBI research is that acutely increased diffusivity may indicate brain regions that undergo edema without cellular disruption, and possibly these areas will not progress to degenerative outcomes, while regions with acutely decreased diffusivity are more likely to have metabolic or other cellular disruption that will result in degeneration. It is important to also note that the detection of these changes is unlikely to be the same for ex vivo dMRI measurements as for in vivo measurements given the absence of physiologic mechanisms and water homeostasis as well as the reduction in extracellular space upon tissue fixation. Figure 1 demonstrates several of these points by showing DTI maps for in‐vivo and ex‐vivo mouse and ferret brains following experimental TBI.
Figure 1.
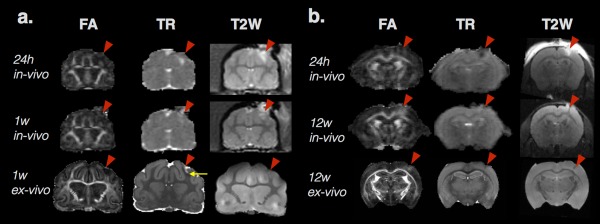
Two examples of DTI metric abnormalities following experimental TBI in ferret (a) and mouse (b) brains. For each species, the in vivo and ex vivo FA and trace (TR) maps and T2‐weighted images are shown from the same animal after controlled cortical impact (CCI site indicated by red arrowhead). Several key features of diffusion changes after TBI are demonstrated in this figure including heterogeneity of diffusivity abnormalities within regions of edema shown by values of TR that are increased (a), decreased (b), or normal (a and b) within tissue regions with T2 hyperintensity. Distinct profiles of TR and FA can also be found in this figure by comparing images in the middle row where TR is relatively normal for both the ferret and mouse brains, but FA is decreased in the ferret brain white matter at 1 week (a) and increased in the mouse brain cortex at 12 weeks (b). By comparing the middle and last rows of in vivo and ex vivo maps from the same animal at the same time point, a distinct pattern can be found for the ferret brain at 1 week (a) in which subdomains of increased TR (near the red arrowhead) and decreased TR (yellow arrow) can be found in regions of unremarkable in vivo TR. In contrast, the same region of increased FA can be found in both the in vivo and ex vivo mouse brain 12 weeks after CCI (b). The observations depicted in this figure demonstrate several of the key points described in the text
Diffusivity findings from these early studies have been replicated many times by more advanced work and extended to describe increased diffusivity at chronic time points corresponding to histological evidence of lesioned tissue in regions with abnormal initial diffusivity (Albensi et al., 2000; Assaf, Beit‐Yannai, Shohami, Berman, & Cohen, 1997; Van Putten, Bouwhuis, Muizelaar, Lyeth, & Berman, 2005). In addition, abnormalities of diffusivity following injury have been associated with meaningful outcomes including atrophy, seizure susceptibility (Frey et al., 2014), and learning impairment (Immonen et al., 2009).
2.2. Cellular alterations after TBI can affect dMRI scalar parameters
We have discussed above the ability of physiologic changes to influence the magnitude of water diffusion in ways that are detectable by basic dMRI methods. With the advent of DTI and other subsequent advanced dMRI approaches, it has become possible to probe changes in the tissue environment arising from the altered microstructure and local architecture of neurons and glia. This capability has compelled several dozen studies of experimental TBI intended to identify and characterize imaging markers that are associated with one or more of the robust cellular alterations that follow brain injury. Below are discussed the most evident neuronal and glial alterations that may contribute to changes detectable using DTI or other dMRI approaches.
2.2.1. Neuronal, axonal, and myelin alterations
Neuron cell bodies, dendrites, and axons encompass a wide range of spatial dimensions and morphologic features that may affect the characteristics of water diffusion in tissue. For example, hippocampal subfields can be distinguished on the basis of different dMRI values observed to be related to dendrite morphology (Shepherd, Özarslan, King, Mareci, & Blackband, 2006), and cortical anisotropy during development has been shown to reflect changes in neurite density and complexity (Huang, Yamamoto, Hossain, Younes, & Mori, 2008). While neuronal features in gray matter tissue regions may be detectable using dMRI, the far more common use is to characterize the myelinated axons of the brain's white matter pathways using diffusion anisotropy measures that are highly sensitive to cylindrical geometry. Following TBI, neuron loss, axonal damage, demyelination, and neural reorganization may drastically alter tissue composition as well as the local tissue geometry in ways that are detectable using dMRI.
Neuronal cell death
The irreversible loss of neurons defines neurodegenerative disorders resulting in the loss of brain function, and TBI has been associated with several mechanisms of neuronal death, each affecting the time course and tissue environment of the brain following injury (Chen, Pickard, & Harris, 2003; Sato, Chang, Igarashi, & Noble, 2001; Stoica & Faden, 2010). Neuronal death may result from direct immediate physical damage induced by biomechanical forces that disrupt cell membranes, causing primary cell loss, or may be initiated by a range of secondary cell loss processes that lead to active or programmed cell death. The loss of neurons in the tissue environment should ultimately lead to increased diffusivity from the loss of cell membranes and gross morphological alterations such as cavitation and atrophy, but cell death mechanisms that precede these changes may or may not encompass morphological features, such as cell swelling, that affect diffusivity (Stoica & Faden, 2010). When neuron death occurs, the axon and myelin sheath degenerate rapidly in a process known as Wallerian degeneration, which can lead to atrophy in the chronic phase. In the hours after injury, an accumulation of organelles in the axon causes swelling, and the myelin sheath is stretched. This is followed by disruption of the cytoskeleton and granular disintegration that completely degrades the axon within several days following injury. Morphological changes that arise from Wallerian degeneration and that may influence the tissue environment are in common with those described below for direct axonal damage and myelin pathology. In fact, there is evidence and strong support for a convergent set of degeneration mechanisms following a diverse range of initial neuron injury (Coleman, 2005).
Evidence for the contribution of cell loss to dMRI changes has been demonstrated in a number of studies by comparing dMRI metrics with cellularity (Van Putten et al., 2005) or by correlation of the MRI and histologically defined values for lesion volume (Assaf et al., 1997). For robust dMRI changes in the acute stage, numerous tissue alterations are likely present in these regions of primary damage and cellular loss near the injury site. The impact of secondary cell loss on the tissue environment during the chronic phase has been documented and associated with dMRI outcomes for several regions including the increased mean diffusivity (MD) in the hippocampus (Laitinen, Sierra, Bolkvadze, Pitkänen, & Gröhn, 2015) and increased fractional anisotropy (FA) in the thalamus (Immonen et al., 2009).
Traumatic and diffuse axonal injury
Biomechanical forces in the brain caused by trauma preferentially injure the long thin axonal processes of neurons that connect different neuroanatomical regions (Johnson et al., 2013). While some axonal loss may result from direct disruption of the cell membrane, the majority of axonal damage is secondary and follows a course of cellular changes that are most robust in the days after injury, but have been found to persist for much longer and may lead to axonal degeneration or functional impairment. While gross degeneration of axons may cause changes in the composition of tissue, typically a subset of axons within a tract are affected. The effects of axonal damage that are most consequential for detection by dMRI are changes in the axonal geometry to which diffusion techniques are remarkably sensitive. In particular, disruption of axonal transport from cytoskeletal stretching leads to the appearance of axonal varicosities and retraction bulbs along the axon, which can reduce diffusivity especially along the axon and increase the amount of restricted water contributing to the diffusion signal. Early DTI studies of mouse white matter identified decreases in anisotropy and diffusivity along the axon (axial diffusivity, Dax) 4 to 24 hr following controlled cortical impact (CCI) consistent with traumatic axonal injury pathology and confirmed by amyloid beta precursor protein and neurofilament staining (Mac Donald, Dikranian, Song, et al., 2007) in the absence of major changes in myelin basic protein expression. This is consistent with findings in a study of isolated axonal injury found to be related to decreased FA and axial diffusivity (Budde, Xie, Cross, & Song, 2009). In addition, numerous subsequent experimental TBI studies of CCI (Budde, Janes, Gold, Turtzo, & Frank, 2011; Davoodi‐Bojd et al., 2014; Jiang et al., 2011; Li et al., 2014; Wang et al., 2013; Zhuo et al., 2012), closed head injury (CHI) (Bennett, Mac Donald, & Brody, 2012; Li, Li, Feng, & Gu, 2011; van de Looij et al., 2012), and blast (Budde et al., 2013; Calabrese et al., 2014) have been able to detect histologically verified axonal pathology using dMRI approaches.
Myelin damage and loss
Neuronal degeneration and axonal damage are intricately related to pathophysiology and structural features of the myelin sheath (Armstrong et al., 2016), which has considerable influence on the dMRI signal (Beaulieu, 2002). If a neuron is damaged to the point of disconnection and degeneration, the myelin sheath collapses irreversibly and the remaining debris lingers in the interstitial space until it is eventually cleared. Demyelination is a different process by which direct damage causes apoptosis of the oligodendrocytes that form the myelin sheath, and often a subset of demyelinated axons are interspersed within a white matter tract. Unlike axonal degeneration, this process can be reversible with remyelination and restoration of neuronal function.
As described in the previous section, TBI‐related changes to myelinated axons are unlikely to present a single pathologic feature, but the relative contribution of axonal and myelin alterations may accessible to dMRI methods. For example, while reduced axial diffusivity was reported to acutely follow CCI in the presence of axonal damage without myelin changes, a second study in the same model (Mac Donald, Dikranian, Bayly, Holtzman, & Brody, 2007) found that weeks after injury, when demyelination became evident, reduced anisotropy was no longer accompanied by reduced axial diffusivity but, instead, by increased radial diffusivity. This is supported by similar findings of increased radial diffusivity in a model with demyelination without axonal damage (Sun et al., 2006) as well as subsequent studies demonstrating DTI changes primarily in anisotropy with abnormal myelin staining (Budde et al., 2011; Jiang et al., 2011; Li et al., 2014). Notably, changes in myelination and myelinated fibers may influence anisotropy differently depending on the tissue investigated. For example, loss of myelinated fibers in white matter was observed to reduce FA during the chronic period, while their loss in the thalamus resulted in increased FA (Laitinen et al., 2015).
Sprouting, remodeling, and regeneration
The neuronal response to brain injury also includes remodeling of neurocircuitry and neurite plasticity with the potential to promote recovery (Bach‐y‐Rita, 2003; Werner & Stevens, 2015). These processes appear to be more complex and variable than those related to neural damage and highly modulated by molecular signaling in the cell environment for both promotion and inhibition of axonal growth (Werner & Stevens, 2015; Yiu & He, 2006). Nevertheless, there is a clear behavioral time course for functional recovery that implies the presence of neural repair and/or circuit remodeling in the brain. At the cellular level, a time course has been proposed in which synaptic changes drive remodeling in the days and weeks following injury, and more widespread connections in neurocircuitry become evident during the chronic period following injury (Meaney & Smith, 2015).
The most evident relationship between dMRI metrics and neural reorganization following TBI has been found in the hippocampus following focal experimental TBI in rats. In the hippocampus, a pattern of acutely decreased and chronically increased diffusivity has been reported (Kharatishvili, Immonen, Gröhn, & Pitkänen, 2007) as well as changes in anisotropy and tissue orientation on the side of injury (Hutchinson, Rutecki, Alexander, & Sutula, 2012; Sierra, Laitinen, Gröhn, & Pitkänen, 2015). In several of these studies, the observed dMRI changes were associated with mossy fiber sprouting, which is a well‐established rewiring of hippocampal circuitry associated with epileptogenesis (Sutula et al., 1998) and known to result following TBI (Golarai, Greenwood, Feeney, & Connor, 2001). While it is appealing that increases in FA can be used as a marker of neuronal plasticity, great care must be taken in making this interpretation as other sources of anisotropy are present in the gray matter that may change with injury such as organized gliosis (Budde et al., 2011).
2.2.2. Glial alterations
As more becomes known about the function of glial cells in the central nervous system (CNS), it is clear that they are vital to a range of functional processes in the healthy brain including in homeostatic regulation, synapse structure and signaling, vascular coupling, and waste removal, among others. Astrocytes are at least as numerous as neurons in the brain (Herculano‐Houzel, 2014; Sofroniew & Vinters, 2010) with nonoverlapping, but functionally connected astrocyte domains of 55 microns in mice to 145 microns in humans (Oberheim et al., 2009; Sun & Jakobs, 2012), and each astrocyte exerts influence over most of the cellular elements in its domain, especially synapses. White matter astrocytes (fibrous) have smaller cell bodies, fewer and less branched processes, and elongated morphology along axons compared with gray matter (protoplasmic) astrocytes, and they also stain more readily for glial fibrillary acidic protein (GFAP) (Lundgaard, Osório, Kress, Sanggaard, & Nedergaard, 2014; Sofroniew & Vinters, 2010). Microglia are smaller than astrocytes, and unlike astrocytes, they are not directly connected to neural or vascular networks; rather, each microglial cell in healthy tissue surveys its own territory with mobile processes that are highly sensitive to changes in the microenvironment. While astrocytes, microglia, and other “supporting cells” of the CNS are far from quiescent in the healthy brain, their response to injury is by comparison robust, diverse, and coordinated across multiple cell types with major roles in the consequences of injury (Burda & Sofroniew, 2014). Furthermore, changes in glial cells are remarkably morphological in nature, demonstrating cytoarchitectural alterations that seem likely to influence the microstructural tissue environment in ways that are detectable by dMRI.
Astrocyte reactivity: Hypertrophy, proliferation, and glial scar formation
Astrocytes become “reactive” in response to a range of triggering events or environmental conditions including neuroinflammation, ischemia, and mechanical injury (Burda et al., 2016; Sun and Jakobs, 2012). Reactive astrocytosis plays a major role in neuroprotection, especially during the acute period, but can also hamper regenerative processes and recovery from injury at later time points (Pekny, Wilhelmsson, & Pekna, 2014; Sofroniew, 2005). Following TBI, astrocyte reactivity is dependent on the nature and severity of injury and marked by hypertrophy and proliferation during the acute period that may either subside or lead to the formation of glial scaring during the chronic period. Hypertrophy of astrocytes is a hallmark feature of reactivity driven by the upregulation of intermediate filament proteins (e.g., vimentin and GFAP) and likely to be consequential for dMRI measurements, but the interpretation of staining for GFAP, a common marker of glial reactivity, has several caveats. In particular, GFAP staining is specific to the cytoskeleton and labels only 15% of the astrocyte volume, so the staining pattern may not represent the true cellular morphometry (Sun and Jakobs, 2012). Nevertheless, GFAP immunohistochemistry (IHC) is an important indicator of changes in the number, length, and thickness of astrocyte processes to measure hypertrophic morphology and has been used successfully to describe quantitative changes in morphology that follow experimental TBI (Norton, Aquino, Hozumi, Chiu, & Bronson, 1992).
Proliferation or migration of astrocytes near injury would also potentially affect the environment by changing tissue composition. Whether astrocytes actively migrate toward the site of injury is not presently clear (Wilhelmsson et al., 2006), but the proliferation of new astrocytes has been demonstrated following TBI, although it is limited to severe injury and typically localized near the injury site and involved in the formation of a glial scar (Sofroniew, 2009; Suzuki, Sakata, Kato, Connor, & Morita, 2012). In milder injury, another caveat for the interpretation of GFAP staining has been observed when a greater number of GFAP‐positive cells are found after injury that are not from new astrocytes but, rather, from upregulation of GFAP in cells that would not normally stain positive (Sofroniew & Vinters, 2010).
Formation of a glial scar is the chronic outcome of reactive astrocytosis and provides a physical barrier to cordon off damaged or toxic tissue to protect viable tissue, and while the conventional view is that the glial scar also inhibits regenerative growth by cell signaling (Silver & Miller, 2004), this paradigm has recently been called into question by evidence that it may instead promote regeneration in some cases (Anderson et al., 2016). The glial scar itself is a very dense region of astrocytes absent of neuronal projections and vasculature. In such an environment, the diffusion of water is expected to be decreased compared with healthy tissue. Furthermore, astrocytes in the glial scar typically demonstrate a highly oriented and elongated morphology, which has been observed to increase diffusion measurements of anisotropy in experimental TBI (Budde et al., 2011).
Several dMRI studies have demonstrated a strong relationship between dMRI measures and astrocytic changes following trauma indicated by GFAP staining (Budde et al., 2011; Mac Donald, Dikranian, Bayly, et al., 2007; Zhuo et al., 2012). While early decreases in diffusivity have been shown to be colocalized with increased GFAP staining in cortical tissue near the injury site (Zhuo et al., 2012), a different profile of dMRI change has been observed during the chronic period in which anisotropy is increased and associated with increased anisotropy of GFAP staining in the same region of gray matter near the injury site (Budde et al., 2011). In the white matter, astrocyte reactivity was observed to accompany axonal damage and associated with decreases in FA during the chronic period, but it did not contribute to decreased FA in the acute period (Mac Donald, Dikranian, Bayly, et al., 2007). Taken together, these studies demonstrate the heterogeneity of DTI outcomes that can arise from the same pathomechanism depending on the time after injury and tissue type affected. Along with the notable lack of specificity of dMRI metrics—that is, multiple cellular alterations could lead to the same DTI abnormality—the development of advanced dMRI methods that are able to disentangle various features of tissue change would be consequential for improved identification and interpretation of pathology following TBI.
Microglial reactivity: Altered form and function of the immune cells of the CNS
Microglia are early responders to tissue environment change and the primary cell type for the brain's immune response (Graeber, 2010; Kreutzberg, 1996; Wake & Fields, 2011). Like astrocytes, microglia become “reactive” in response to injury in a graded manner, but with morphological features that are quite distinct. The classic pattern of microglial reactivity is defined by morphological stages (Ziebell, Adelson, & Lifshitz, 2015) whereby microglial processes retract and the cell body enlarges such that the resting state or “ramified” microglial morphology with extensive processes is quite distinct from the reactive or “amoeboid” state with morphology that is nearly spherical. In addition to these hallmark phenotypes of microglial activation, other observations of morphological alteration are known to follow injury including “honeycomb”‐ or “jellyfish”‐like arrangement of microglial processes (Roth et al., 2014) acutely and rod morphology that is most evident a week after injury (Ziebell, Taylor, Cao, Harrison, & Lifshitz, 2012). Clearly, the morphological response of microglia is remarkably diverse and appears to reflect information about the tissue state following injury, making this cell type a promising target for examination by dMRI, although the small size of microglial cells may limit their influence on the tissue environment.
3. THE MODERN dMRI TOOLKIT AND DETECTION OF POST‐TBI TISSUE CHANGE
It is evident from the previous section that a substantial and growing body of dMRI studies has emerged that demonstrates the sensitivity of diffusion weighted imaging (DWI)‐ and DTI‐derived diffusivity and anisotropy metrics for detecting TBI‐related brain abnormalities across a wide range of changes in the brain tissue environment. However, a fundamental limitation of these studies is the low specificity of DTI metrics for identifying particular cellular features of interest. In part, this can be addressed by studies that incorporate histological outcomes to substantiate the interpretation of DTI changes. Another promising avenue for improving the utility of dMRI in TBI research is application of more advanced modeling methods that may provide additional or more specific measures of tissue and cellular changes following TBI, and a number of potentially advantageous dMRI approaches have been developed in recent years (see Figure 2 for representative scalar maps across different diffusion models). To this end, this section presents a concise and systematic overview of conventional and advanced dMRI methods and a perspective on their relative strengths and limitations in the context of TBI research.
Figure 2.
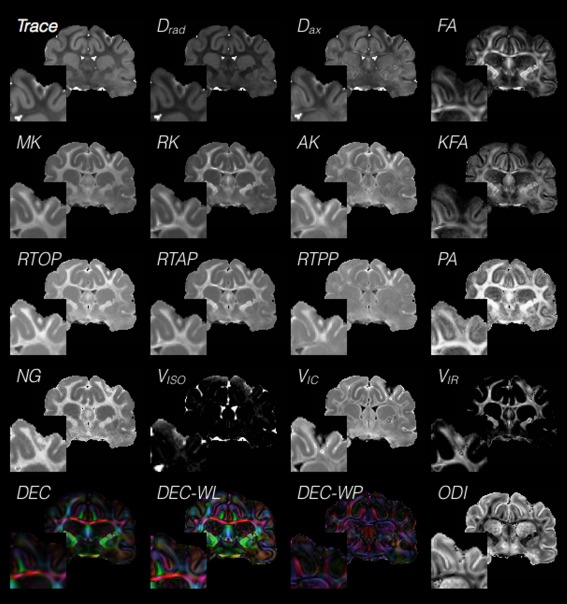
Cross‐model comparison of scalar maps in the injured brain. A range of tissue and injury‐related contrasts may be visually observed in this collage of 16 representative metrics in the same slice from different dMRI models. This cross‐model view of scalar maps demonstrates the potential for nonredundant information about regions of injury that may be gleaned from different models. DTI metrics of fractional anisotropy (FA), trace (TR), axial and radial diffusivity (Dax and Drad), directionally encoded color (DEC) map weighted by lattice index, DEC weighted by Westin linear anisotropy (WL) and DEC weighted by Westin planar anisotropy (WP), DKI metrics of mean kurtosis (MK), axial and radial kurtosis (AK and RK) and kurtosis FA (KFA), MAP‐MRI metrics of return to the origin, axis, and plane probabilities (RTOP, RTAP, and RTPP), propagator anisotropy (PA) and non‐Gaussianity (NG) and NODDI metrics of compartment volume fractions for isotropic free water (Viso), intracellular water (Vic) and intracellular restricted water (Vir), and orientation dispersion index (ODI). Insets of each map show tissue near the injury site where dMRI values are expected to be abnormal
The general pipeline that is used to perform most dMRI techniques is (1) acquisition of DWI volumes having particular diffusion weighting direction sets and b‐values, (2) correction of DWI volumes for artifacts and distortions, (3) modeling of the DWI data (or application of a mathematical transform) to generate scalar metric maps, and (4) analysis and interpretation of the scalar metric maps to answer an experimental question. While this pipeline appears to be sequential in nature, there is interdependence of each step on the others that requires careful planning of all steps prior to data collection. Table 2 outlines basic information related to these steps across different dMRI approaches.
Table 2.
Comparative Overview of Diffusion MRI Models
| Acquisition | ||||||
|---|---|---|---|---|---|---|
| Modeling approach | DWI sampling | DWI weighting | Metrics | Seminal references | ||
| ADC | S = S 0 e −bD | Few DWIs; one b=0 and 1 DWI | Low | ADC | Eccles et al., 1988 | |
| DTI | S = S 0 e −b:D | Few DWIs; one shell, minimum six directions | Low | Combinations of λ1, λ2, λ3. e.g. FA, TR, WL, WP | Basser et al., 1994 | |
| DKI |
|
Moderate number of DWIs; two shells | Low and moderate only | DTI metrics and mean kurtosis, axial and radial kurtosis and KFA | Jensen et al., 2005; Tabesh et al., 2011; Glenn et al., 2015 | |
| MAP | Asymmetric simple harmonic oscillator reconstruction and estimation | Moderate to many DWIs; multishell acquisition | Low, moderate, and high | DTI metrics and non‐Gaussianity, zero‐displacement probabilities, propagator anisotropy, ODFs | Özarslan et al., 2013; Avram et al., 2016 | |
| DSI | Model‐free | Many DWIs; Cartesian grid | Low, moderate, and high | ODFs possible to generate zero‐displacement probabilities | Tuch et al., 2003 | |
| Q‐ball | Model‐free | Moderate no. of DWIs; single‐shell HARDI acquisition | High | ODFs possible to generate zero‐displacement probabilities | Tuch, 2004 | |
| CHARMED | Intra/extra‐axonal compartments modeled by restricted/hindered sheets and cylinders | Multishell acquisition | Low, moderate, and high | Restricted and hindered component fractions; cone of uncertainty | Assaf & Basser, 2005 | |
| Axcaliber | Similar to CHARMED, but with additional modeling of axon diameter | Multishell acquisition | Low, moderate, and high flexible | CHARMED metrics and axon diameter | Assaf et al., 2008 | |
| NODDI | Watson distributed cylinders and sticks | Moderate no. of DWIs: multishell acquisition | Low, moderate, and high (flexible) | Cellular fractions, orientation dispersion index | Zhang et al., 2012; Tariq et al., 2016 | |
| WMTI | Intra/extra‐axonal compartments modeled with the Gaussian part of the DKI model | Moderate no. of DWIs; two shells (same as DKI) | Low and moderate only | Axonal water fraction, intra‐axonal diffusivity, extra‐axonal radial/axial diffusivity, extra‐axonal tortuosity | Fieremans et al., 2011 | |
| CSD tractography | Constrained spherical deconvolution | Single‐shell HARDI acquisition | High | ODFs and tractograms | Tournier et al., 2004, 2012 | |
Note: For physical and biophysical dMRI models (column 1, light and dark gray shading, respectively), a summary of the modeling approach (column 2) and acquisition strategy (columns 3 and 4) is given as well as the primary scalar metrics (column 5). Abbreviations: ADC ‐ Apparent Diffusion Coefficient; DWI ‐ diffusion weighted image; DTI ‐ Diffusion Tensor Imaging; TR ‐ Trace; WL ‐ Westin's Linear Anisotropy; WP ‐ Westin's Planar Anisotropy; DKI ‐ Diffusion Kurtosis Imaging; KFA ‐ Kurtosis FA; MAP ‐ Mean Apparent Propagator; ODF ‐ Orientation Distribution Function; DSI ‐ Diffusion Spectrum Imaging; CHARMED ‐ Composite Hindered And Restricted Model of Diffusion; NODDI ‐ Neurite Orientation Dispersion and Density Imaging; WMTI ‐ White Matter Tract Integrity; CSD ‐ Constrained Spherical Deconvolution; HARDI ‐ High Angular Resolution Diffusion Imaging.
Diffusion modeling frameworks may be roughly categorized into two main categories: (1) physical or “signal‐driven” diffusion models, which aim at measuring and characterizing of probabilistic water displacement profile (a.k.a. the diffusion propagator); and (2) biological or “microstructure‐driven” models, which aim to provide a more direct assessment of tissue compartments and their biological attributes by directly incorporating a priori biological information (assumptions) into the model. Each of these approaches provides a set of scalar outcome metrics that are based on the fitted parameters of the model, and the maps of these scalars are used to visualize and quantify differences across brain regions or to identify abnormalities in experimental models.
The first class of techniques—physical diffusion modeling—is the most prevalent and includes DTI (Basser, Mattiello, & LeBihan, 1994), diffusion kurtosis imaging (DKI) (Jensen, Helpern, Ramani, Lu, & Kaczynski, 2005; Tabesh, Jensen, Ardekani, & Helpern, 2011), and diffusion propagator representation models (Callaghan, Eccles, & Xia, 1988; Callaghan, MacGowan, & Packer, 1990). These models directly relate the MRI signal for each DWI to the diffusion weighting (i.e., b‐value in seconds per millimeter squared) and the orientation of the applied diffusion sensitizing gradients, which can be related to the q‐vector (1/mm) in reciprocal displacement space. The simplest dMRI approach uses as few as two DWI measurements to fit a single exponential decay to estimate the apparent diffusion coefficient (mm2/s) (Eccles, Callaghan, & Jenner, 1988) of water present in the tissue in the direction of the applied diffusion weighting. DTI, the most commonly used diffusion model, is an extension of this approach to model water diffusion in three dimensions by the diffusion tensor (Basser et al., 1994). The eigenvalues (λ1, λ2, and λ3) of the diffusion tensor can be used to represent the axes of a diffusion ellipsoid related to the preferred orientation and magnitude of water diffusion and to provide a set of scalar metrics based on mathematical arrangements of the eigenvalues that report a particular feature of the diffusion tensor. The trace of the diffusion tensor (TR = λ1 + λ2 + λ3) or mean diffusivity (MD = TR/3) and the fractional anisotropy (FA) (Pierpaoli & Basser, 1996) are the most commonly reported DTI scalar metrics, but others, including axial (Dax = λ1) and radial (Drad = [λ2+λ3]/2) diffusivity and Westin linear, planar, and spherical anisotropy (WL, WP, and WS) (Westin et al., 2002), can also be applied to provide more specific geometric information about water diffusion derived from the diffusion tensor.
While DTI already offers a quantitative and sensitive way to detect and evaluate changes in water diffusion that arise from alterations in the tissue environment, the diffusion tensor approximation is a simplified representation that cannot convey higher‐order signal behavior (i.e., non‐Gaussianity) or complex orientational information (i.e., multifiber geometry). To address these limitations, numerous models have been developed with the goal of quantifying the diffusion propagator, P(r)—the 3D probability distribution for water displacement (Callaghan et al., 1990). The earliest propagator measurement approaches relied directly on the Fourier relationship between P(r) and the MRI signal of DWI data collected in q‐space E(q) (Callaghan et al., 1988). Q‐space imaging methods of this type are model‐free and simply rely on taking a mathematical transform of E(q) at each voxel. These include diffusion spectrum imaging and Q‐ball imaging (Tuch, 2004; Tuch, Reese, Wiegell, & Wedeen, 2003), which produce orientation distribution functions (ODFs) that allow visualization of different fiber orientations in the same voxel.
More recently, model‐dependent approaches to propagator representation have been proposed that report information about water diffusion that is “non‐Gaussian.” For example, DKI (Glenn, Helpern, Tabesh, & Jensen, 2015; Jensen et al., 2005; Tabesh et al., 2011) expands the DTI model with an additional term (Table 2) that includes the kurtosis tensor, and several metrics may be calculated from this new tensor including the mean, radial, and axial kurtosis (MK, RK, and AK) as well as the kurtosis FA (KFA). In addition to new DKI metrics that probe non‐Gaussian information in the dMRI signal, the conventional DTI metrics that are calculated using this model are more stable than for DTI alone (Hutchinson et al., 2017; Veraart et al., 2011), and the DWI sampling for DKI is modest compared with higher‐order models and therefore requires relatively shorter scan times. However, DKI is highly dependent on the DWI sampling scheme, potentially leading to inconsistent DKI values across studies. Several studies of CCI in the rat have suggested that DKI markers, especially increases in MK, may be more sensitive to particular abnormalities such as the lesion boundary (Jiang et al., 2011) and the temporal evolution of abnormalities, which is distinct from the pattern of DTI changes (Zhuo et al., 2012). However, DKI has also been observed to be less sensitive to abnormalities in a repetitive mild TBI model (Yu et al., 2017), suggesting that in some cases the benefit of increased specificity is offset by reduced sensitivity.
While DKI assumes a particular signal behavior for all non‐Gaussian diffusion, other higher‐order models have been developed that are more flexible and extensive for describing non‐Gaussian diffusion. For example, mean apparent propagator (MAP)‐MRI (Avram et al., 2016; Özarslan et al., 2013) and ensemble average propagator approaches (Cheng, Jiang, & Deriche, 2012; Hosseinbor, Chung, Wu, & Alexander, 2013) use carefully selected functional series expansion to fit DWI data sampled over a wide range of b‐values with many orientations. The coefficients of these models provide an approximation of the diffusion propagator that may be visualized as ODFs or generate scalar maps of relevant propagator metrics. For MAP‐MRI, these include zero‐displacement probabilities (return to origin, axis, and plane probability, or RTOP, RTAP, and RTPP, respectively), non‐Gaussianity (NG), and propagator anisotropy (PA). While the greater complexity of techniques that measure the propagator provides more detailed information about water diffusion that may lead to improved specificity, these models require considerably more DWI data with greater diffusion weighting.
While physical diffusion models present scalar metrics that are related to the extent and preferred orientation of water diffusion, biophysical models incorporate a priori assumptions about the tissue environment to estimate features such as cellular fraction volumes, axon and tract dimensions, or neurite density and coherence. These approaches are appealing for identification of abnormalities in biological terms; however, caution is warranted not only to consider the validity of model assumptions across different healthy tissue types but also to consider how the biophysical model does or does not accommodate pathological processes. For example, tractography approaches (Tournier, Calamante, & Connelly, 2012; Tournier, Calamante, Gadian, & Connelly, 2004), which represent white matter “tracts” using DTI or ODF information, may misrepresent pathological changes that increase tissue organization, such as organized gliosis as a false increase in tract density or length, or other pathological changes such as gliosis may decrease white matter anisotropy even if the tracts are still present, which may lead to a misleading loss of tract representation (see Figure 3 and references; Budde et al., 2011). This said, tractography can be used effectively to illustrate white matter differences or to delineate known tract anatomy when interpretation is made carefully with reference to known neuroanatomy and/or accompanied by histological or functional validation.
Figure 3.
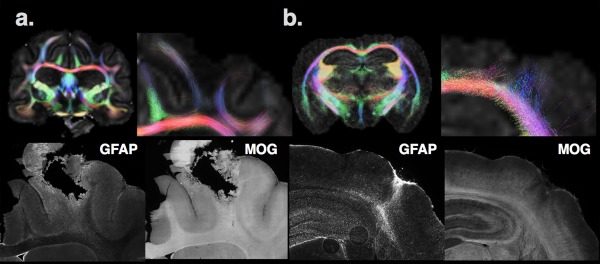
Two examples of caveats are shown for the biophysical representation of diffusion MRI information by tractography in the ferret (a) and mouse (b) brain using the same approach and parameters. In the ferret brain near the site of a penetrating injury, FA is low and few tracts can be found in the body of the white matter compared with the contralateral side. However, investigation of this region using IHC of the same brain reveals the presence of myelinated axons (indicated by MOG IHC—myelin oligodendrocyte glycoprotein) and upregulated staining of astrocytes (by GFAP IHC—glial fibrillary acidic protein). The interpretation of the tractography in this case could indicate a loss of white matter fibers, when in fact the underlying pathology appears to be more related to gliosis. In the mouse brain (b), a region of increased FA and aberrant “tracts” can be found in the cortex near the injury site; however, inspection by IHC reveals a disruption of MOG staining and upregulation and organized GFAP staining in this tissue region of the same animal. The interpretation of tractography in this case could suggest cortical plasticity, when in fact the underlying alteration is more related to glial changes. This is similar to a finding reported by Budde et al. (2011). Taken together, this figure emphasizes the need for careful interpretation of dMRI findings, especially for biophysical models such as tractography, which directly report neurobiological metrics based model assumptions
Other biophysical methods have been developed to provide scalar metrics that report local information about the tissue environment. For example, the composite hindered and restricted model of diffusion (CHARMED) (Assaf & Basser, 2005) and, later, AxCaliber (Assaf, Blumenfeld‐Katzir, Yovel, & Basser, 2008) were developed to estimate white matter fiber orientation and axon diameter in part to improve the sensitivity of dMRI to detect abnormalities in disorders affecting the white matter and may have particular relevance for evaluating axonal damage and myelination changes. Cellular compartment models based on multiexponential dMRI modeling or more advanced approaches have also been devised to estimate the intra‐ and extracellular contribution to the dMRI signal and to further model axonal dimensions (Jelescu, Veraart, Fieremans, & Novikov, 2016; Jelescu, Zurek, Winters, & Veraart, 2016; Kunz et al., 2014; Panagiotaki et al., 2012). More recently, several models have extended compartmental modeling to include features of “white matter tract integrity” (WMTI; Fieremans, Jensen, & Helpern, 2011) or “neurite orientation dispersion and density” (NODDI; Tariq, Schneider, Alexander, Gandini Wheeler‐Kingshott, & Zhang, 2016; Zhang, Schneider, Wheeler‐Kingshott, & Alexander, 2012), which may be advantageous for the detection of pathological or regenerative changes to specific neuroanatomical structures (i.e., white matter or neurites). While initial work using biophysical modeling has demonstrated correlations between histological and dMRI‐derived values for white matter axonal fraction and diameter following CCI in rats (Wang et al., 2013), much remains to be understood about the ability of biophysical models to accurately report tissue parameters. Furthermore, biophysical models have been shown to depend on specific fixed model parameters (e.g., compartmental diffusivity values), DWI sampling, and image quality (Jelescu, Veraart, et al., 2016), such that extreme care should be given to application of these models.
4. A PRACTICAL AND CONCEPTUAL PERSPECTIVE FOR THE USE OF ADVANCED dMRI METHODS IN TBI RESEARCH
The past several decades have seen considerable growth in our ability to characterize TBI‐related brain changes by incorporating dMRI into studies of experimental brain injury models. From early work to define diffusivity changes related to pathophysiology following TBI to more recent endeavors to relate DTI abnormalities with a range of cellular alterations, dMRI has emerged as a promising tool for providing microscale information about brain abnormalities that is quantitative, whole‐brain, and noninvasive.
The future of these efforts will be better enabled if dMRI approaches can be used in combination with one another and alongside other MRI modalities as a set of specialized tools to detect specific pathology, predict outcomes, or target treatments. However, for this to become possible we must first systematically understand the similarities and differences across dMRI models including their relative advantages and limitations. A recent study comparing the dependence of scalar metrics from DTI, DKI, MAP‐MRI, and NODDI in normal brain tissue identified several practical implications of moving to models with greater complexity or biological assumptions (Hutchinson et al., 2017). In particular, DKI metrics were found to be the most vulnerable to DWI sampling and image quality, while MAP‐MRI required the greatest DWI sampling and was dependent on the initial fitting of the diffusion tensor. The NODDI model was found to have stability over a range of DWI sampling and image quality, but to depend greatly on the selection of fixed model parameters (e.g., interstitial diffusivity). Greater understanding of the influence of experimental parameters on dMRI outcome measures as well as identifying redundancy and novelty of information across models is an active area of imaging research and will aid in the effective use of these methods.
Despite the challenges that accompany more complex diffusion modeling, there is also promise in their ability to better characterize posttraumatic brain abnormalities than existing methods. This was recently demonstrated in a study combining DTI and DKI metrics to evaluate markers of pathology after experimental TBI in rats (Zhuo et al., 2012). While cortical MD was decreased 2 hr after injury and then increased 1 week later, the MK was consistently elevated at both times and furthermore corresponded to astrocytosis. While more work remains to define whether and how DKI and other advanced methods provide additional or more specific information that corresponds to individual or organized pathogenesis, this study and others employing advanced dMRI methods to evaluate experimental TBI (Davoodi‐Bojd et al., 2014; Fozouni et al., 2013; Wang et al., 2013; Yu et al., 2017) have begun to build an important foundation to understand and apply advanced diffusion imaging in the context of TBI research.
The combination of a modern dMRI toolkit with other MRI modalities and advances in the understanding of neurobiological responses to TBI has the potential to improve the spatiotemporal characterization of TBI across various experimental models and provide outcome measures that may be directly translated as imaging markers in human studies or used in experimental models for the development of therapies.
CONFLICT OF INTEREST
The authors declare no relevant conflicts of interest.
AUTHOR CONTRIBUTIONS
E.H. wrote this article, reviewed the literature. and performed image acquisition and processing for the MRI figures; S.S. contributed insight about neurobiology of TBI and generated the histology figures; A.A. provided crucial knowledge and discussion about dMRI theory and methods; S.J. provided neurobiology expertise related to TBI and enabled histology contributions; C.P. provided diffusion MRI expertise, training, and discussion and enabled dMRI acquisition and processing for figures.
ACKNOWLEDGMENTS
The authors thank Miki Komlosh for her expertise and practical assistance in dMRI acquisition and Okan Irfanoglu for his expertise and guidance for dMRI data processing and modeling. The CNRM MRI core facility, especially Asamoah Bosomtwi and Alexandru Korotcov, provided expert support for in vivo MRI, and the NICHD intramural program supported the ex vivo MRI. The authors also thank the Congressionally Directed Medical Research Programs (CDMRP) for funding this work (award numbers W81XWH‐13‐2‐0019 and W81XWH‐13‐2‐0018) and the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF) for administration.
Hutchinson EB, Schwerin SC, Avram AV, Juliano SL, Pierpaoli C. Diffusion MRI and the detection of alterations following traumatic brain injury. J Neuro Res. 2018;96:612–625. https://doi.org/10.1002/jnr.24065
Funding Information Congressionally Directed Medical Research Programs, Award Numbers W81XWH‐13‐2‐0019 (C.P.) and W81XWH‐13‐2‐0018 (S.J.), and the Center for Neuroscience and Regenerative Medicine (S.J. and C.P.).
Significance Noninvasive magnetic resonance imaging may enable important observations about how the brain changes following injury. Diffusion magnetic resonance imaging (dMRI) is particularly promising for studying traumatic brain injury (TBI) because it quantitatively characterizes the physical tissue environment, which may change greatly after injury. While a number of conventional and advanced dMRI approaches have been developed that may benefit the detection of post‐TBI abnormalities, much remains to be known about the cellular underpinnings and relative value of these methods. This article reviews TBI‐related cellular alterations that influence the tissue environment and compares common dMRI tools for detecting such changes.
REFERENCES
- Albensi, B. C. , Knoblach, S. M. , Chew, B. G. , O'Reilly, M. P. , Faden, A. I. , & Pekar, J. J. (2000). Diffusion and high resolution MRI of traumatic brain injury in rats: Time course and correlation with histology. Experimental Neurology, 162, 61–72. [DOI] [PubMed] [Google Scholar]
- Alsop, D. C. , Murai, H. , Detre, J. A. , McIntosh, T. K. , & Smith, D. H. (1996). Detection of acute pathologic changes following experimental traumatic brain injury using diffusion‐weighted magnetic resonance imaging. Journal of Neurotrauma, 13, 515–521. [DOI] [PubMed] [Google Scholar]
- Anderson, M. A. , Burda, J. E. , Ren, Y. , Ao, Y. , O'Shea, T. M. , Kawaguchi, R. , … Sofroniew, M. V. (2016). Astrocyte scar formation aids central nervous system axon regeneration. Nature, 532, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, R. C. , Mierzwa, A. J. , Marion, C. M. , & Sullivan, G. M. (2016). White matter involvement after TBI: Clues to axon and myelin repair capacity. Experimental Neurology, 275(Pt. 3), 328–333. [DOI] [PubMed] [Google Scholar]
- Assaf, Y. , & Basser, P. J. (2005). Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. Neuroimage, 27, 48–58. [DOI] [PubMed] [Google Scholar]
- Assaf, Y. , Beit‐Yannai, E. , Shohami, E. , Berman, E. , & Cohen, Y. (1997). Diffusion‐ and T2‐weighted MRI of closed‐head injury in rats: A time course study and correlation with histology. Magnetic Resonance Imaging, 15, 77–85. [DOI] [PubMed] [Google Scholar]
- Assaf, Y. , Blumenfeld‐Katzir, T. , Yovel, Y. , & Basser, P. J. (2008). AxCaliber: A method for measuring axon diameter distribution from diffusion MRI. Magnetic Resonance in Medicine, 59, 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram, A. V. , Sarlls, J. E. , Barnett, A. S. , Özarslan, E. , Thomas, C. , Irfanoglu, M. O. , … Basser, P. J. (2016). Clinical feasibility of using mean apparent propagator (MAP) MRI to characterize brain tissue microstructure. NeuroImage, 127, 422–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach‐y‐Rita, P. (2003). Theoretical basis for brain plasticity after a TBI. Brain Injury, 17, 643–651. [DOI] [PubMed] [Google Scholar]
- Basser, P. J. , Mattiello, J. , & LeBihan, D. (1994). MR diffusion tensor spectroscopy and imaging. Biophysical Journal, 66, 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu, C. (2002). The basis of anisotropic water diffusion in the nervous system—A technical review. NMR in Biomedicine, 15, 435–455. [DOI] [PubMed] [Google Scholar]
- Bennett, R. E. , Mac Donald, C. L. , & Brody, D. L. (2012). Diffusion tensor imaging detects axonal injury in a mouse model of repetitive closed‐skull traumatic brain injury. Neuroscience Letters, 513, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramlett, H. , & Dietrich, W. D. (2015). Long‐term consequences of traumatic brain injury: Current status of potential mechanisms of injury and neurologic outcomes. Journal of Neurotrauma, 32, 1834–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramlett, H. M. , & Dietrich, D. W. (2004). Pathophysiology of cerebral ischemia and brain trauma: Similarities and differences. Journal of Cerebral Blood Flow & #Metabolism, 24, 133–150. [DOI] [PubMed] [Google Scholar]
- Brody, D. L. , Mac Donald, C. L. , & Shimony, J. S. (2015). Current and future diagnostic tools for traumatic brain injury: CT, conventional MRI, and diffusion tensor imaging. Handbook of Clinical Neurology, 127, 267–275. [DOI] [PubMed] [Google Scholar]
- Budde, M. D. , Janes, L. , Gold, E. , Turtzo, L. , & Frank, J. A. (2011). The contribution of gliosis to diffusion tensor anisotropy and tractography following traumatic brain injury: Validation in the rat using Fourier analysis of stained tissue sections. Brain, 134(Pt. 8), 2248–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde, M. D. , Shah, A. , McCrea, M. , Cullinan, W. E. , Pintar, F. A. , & Stemper, B. D. (2013). Primary blast traumatic brain injury in the rat: Relating diffusion tensor imaging and behavior. Frontiers in Neurology, 4, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde, M. D. , Xie, M. , Cross, A. H. , & Song, S.‐K. K. (2009). Axial diffusivity is the primary correlate of axonal injury in the experimental autoimmune encephalomyelitis spinal cord: A quantitative pixelwise analysis. Journal of Neuroscience, 29, 2805–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda, J. E. , Bernstein, A. M. , & Sofroniew, M. V. (2016). Astrocyte roles in traumatic brain injury. Experimental Neurology, 275(Pt. 3), 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda, J. E. , & Sofroniew, M. V. (2014). Reactive gliosis and the multicellular response to CNS damage and disease. Neuron, 81, 229–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, E. , Du, F. , Garman, R. H. , Johnson, G. A. , Riccio, C. , Tong, L. C. , & Long, J. B. (2014). Diffusion tensor imaging reveals white matter injury in a rat model of repetitive blast‐induced traumatic brain injury. Journal of Neurotrauma, 31, 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan, P. T. , Eccles, C. D. , & Xia, Y. (1988). NMR microscopy of dynamic displacements: k‐space and q‐space imaging. Journal of Physics E: Scientific Instruments, 21, 820. [Google Scholar]
- Callaghan, P. T. , MacGowan, D. , & Packer, K. J. (1990). High‐resolution q‐space imaging in porous structures. Journal of Magnetic Resonance, Series B, 90, 177–182. [Google Scholar]
- Chen, S. , Pickard, J. D. , & Harris, N. G. (2003). Time course of cellular pathology after controlled cortical impact injury. Experimental Neurology, 182, 87–102. [DOI] [PubMed] [Google Scholar]
- Cheng, J. , Jiang, T. , & Deriche, R. (2012). Nonnegative definite EAP and ODF estimation via a unified multi‐shell HARDI reconstruction. Medical Image Computing and Computer‐Assisted Intervention: MICCAI International Conference on Medical Image Computing and Computer‐Assisted Intervention, 15(Pt. 2), 313–321. [DOI] [PubMed] [Google Scholar]
- Coleman, M. (2005). Axon degeneration mechanisms: Commonality amid diversity. Nature Reviews Neuroscience, 6, 889–898. [DOI] [PubMed] [Google Scholar]
- Davoodi‐Bojd, E. , Chopp, M. , Soltanian‐Zadeh, H. , Wang, S. , Ding, G. , & Jiang, Q. (2014). An analytical model for estimating water exchange rate in white matter using diffusion MRI. PloS One, 9(5), e95921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, D. B. , Iv, M. , Douglas, P. K. , Anderson, A. , Vos, S. B. , Bammer, R. , … Wintermark, M. (2015). Diffusion tensor imaging of TBI: Potentials and challenges. Topics in Magnetic Resonance Imaging, 24, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhaime, A.‐C. C. , Gean, A. D. , Haacke, E. M. , Hicks, R. , Wintermark, M. , Mukherjee, P. , … Riedy, G. ; Common Data Elements Neuroimaging Working Group Members, Pediatric Working Group Members . (2010). Common data elements in radiologic imaging of traumatic brain injury. Archives of Physical Medicine and Rehabilitation, 91, 1661–1666. [DOI] [PubMed] [Google Scholar]
- Eccles, C. D. , Callaghan, P. T. , & Jenner, C. F. (1988). Measurement of the self‐diffusion coefficient of water as a function of position in wheat grain using nuclear magnetic resonance imaging. Biophysical Journal, 53, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieremans, E. , Jensen, J. H. , & Helpern, J. A. (2011). White matter characterization with diffusional kurtosis imaging. NeuroImage, 58, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni, N. , Chopp, M. , Nejad‐Davarani, S. P. , Zhang, Z. G. , Lehman, N. L. , Gu, S. , … Jiang, Q. (2013). Characterizing brain structures and remodeling after TBI based on information content, diffusion entropy. PloS One, 8(10), e76343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, L. , Lepkin, A. , Schickedanz, A. , Huber, K. , Brown, M. S. , & Serkova, N. (2014). ADC mapping and T1‐weighted signal changes on post‐injury MRI predict seizure susceptibility after experimental traumatic brain injury. Neurological Research, 36, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn, G. R. , Helpern, J. A. , Tabesh, A. , & Jensen, J. H. (2015). Quantitative assessment of diffusional kurtosis anisotropy. NMR in Biomedicine, 28, 448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golarai, G. , Greenwood, A. C. , Feeney, D. M. , & Connor, J. A. (2001). Physiological and structural evidence for hippocampal involvement in persistent seizure susceptibility after traumatic brain injury. Journal of Neuroscience, 21, 8523–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber, M. B. (2010). Changing face of microglia. Science, 330, 783–788. [DOI] [PubMed] [Google Scholar]
- Hanstock, C. C. , Faden, A. I. , Bendall, M. R. , & Vink, R. (1994). Diffusion‐weighted imaging differentiates ischemic tissue from traumatized tissue. Stroke, 25, 843–848. [DOI] [PubMed] [Google Scholar]
- Herculano‐Houzel, S. (2014). The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia, 62, 1377–1391. [DOI] [PubMed] [Google Scholar]
- Hosseinbor, A. P. , Chung, M. K. , Wu, Y.‐C. C. , & Alexander, A. L. (2013). Bessel Fourier orientation reconstruction (BFOR): An analytical diffusion propagator reconstruction for hybrid diffusion imaging and computation of q‐space indices. NeuroImage, 64, 650–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Yamamoto, A. , Hossain, M. , Younes, L. , & Mori, S. (2008). Quantitative cortical mapping of fractional anisotropy in developing rat brains. Journal of Neuroscience, 28, 1427–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulkower, M. B. , Poliak, D. B. , Rosenbaum, S. B. , Zimmerman, M. E. , & Lipton, M. L. (2013). A decade of DTI in traumatic brain injury: 10 years and 100 articles later. AJNR American Journal of Neuroradiology, 34, 2064–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, E. B. , Avram, A. V. , Irfanoglu, M. O. , Koay, C. G. , Barnett, A. S. , Komlosh, M. E. , … Pierpaoli, C. (2017). Analysis of the effects of noise, DWI sampling, and value of assumed parameters in diffusion MRI models. Magnetic Resonance in Medicine, 78(5), 1767‐1780. doi: 10.1002/mrm.26575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, E. B. , Rutecki, P. A. , Alexander, A. L. , & Sutula, T. P. (2012). Fisher statistics for analysis of diffusion tensor directional information. Journal of Neuroscience Methods, 206, 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen, R. J. , Kharatishvili, I. , Niskanen, J.‐P. P. , Gröhn, H. , Pitkänen, A. , & Gröhn, O. H. (2009). Distinct MRI pattern in lesional and perilesional area after traumatic brain injury in rat—11 months follow‐up. Experimental Neurology, 215, 29–40. [DOI] [PubMed] [Google Scholar]
- Jelescu, I. O. , Veraart, J. , Fieremans, E. , & Novikov, D. S. (2016). Degeneracy in model parameter estimation for multi‐compartmental diffusion in neuronal tissue. NMR in Biomedicine, 29, 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelescu, I. O. , Zurek, M. , Winters, K. V. , & Veraart, J. (2016). In vivo quantification of demyelination and recovery using compartment‐specific diffusion MRI metrics validated by electron microscopy. Neuroimage, 132, 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J. H. , Helpern, J. A. , Ramani, A. , Lu, H. , & Kaczynski, K. (2005). Diffusional kurtosis imaging: The quantification of non‐gaussian water diffusion by means of magnetic resonance imaging. Magnetic Resonance in Medicine, 53, 1432–1440. [DOI] [PubMed] [Google Scholar]
- Jiang, Q. , Qu, C. , Chopp, M. , Ding, G. L. , Davarani, S. P. , Helpern, J. A. , … Mahmood, A. (2011). MRI evaluation of axonal reorganization after bone marrow stromal cell treatment of traumatic brain injury. NMR in Biomedicine, 24, 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, V. E. , Stewart, W. , & Smith, D. H. (2013). Axonal pathology in traumatic brain injury. Experimental Neurology, 246, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharatishvili, I. , Immonen, R. , Gröhn, O. , & Pitkänen, A. (2007). Quantitative diffusion MRI of hippocampus as a surrogate marker for post‐traumatic epileptogenesis. Brain, 130(Pt. 12), 3155–3168. [DOI] [PubMed] [Google Scholar]
- Kreutzberg, G. W. (1996). Microglia: A sensor for pathological events in the CNS. Trends in Neurosciences, 19, 312–318. [DOI] [PubMed] [Google Scholar]
- Kunz, N. , Zhang, H. , Vasung, L. , O'Brien, K. R. , Assaf, Y. , Lazeyras, F. , … Hüppi, P. S. (2014). Assessing white matter microstructure of the newborn with multi‐shell diffusion MRI and biophysical compartment models. NeuroImage, 96, 288–299. [DOI] [PubMed] [Google Scholar]
- Laitinen, T. , Sierra, A. , Bolkvadze, T. , Pitkänen, A. , & Gröhn, O. (2015). Diffusion tensor imaging detects chronic microstructural changes in white and gray matter after traumatic brain injury in rat. Frontiers in Neuroscience, 9, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Li, X.‐Y. Y. , Feng, D.‐F. F. , & Gu, L. (2011). Quantitative evaluation of microscopic injury with diffusion tensor imaging in a rat model of diffuse axonal injury. European Journal of Neuroscience, 33, 933–945. [DOI] [PubMed] [Google Scholar]
- Li, N. , Yang, Y. , Glover, D. P. , Zhang, J. , Saraswati, M. , Robertson, C. , & Pelled, G. (2014). Evidence for impaired plasticity after traumatic brain injury in the developing brain. Journal of Neurotrauma, 31, 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgaard, I. , Osório, M. J. , Kress, B. T. , Sanggaard, S. , & Nedergaard, M. (2014). White matter astrocytes in health and disease. Neuroscience, 276, 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Donald, C. L. , Dikranian, K. , Bayly, P. , Holtzman, D. , & Brody, D. (2007). Diffusion tensor imaging reliably detects experimental traumatic axonal injury and indicates approximate time of injury. Journal of Neuroscience, 27, 11869–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Donald, C. L. , Dikranian, K. , Song, S. K. , Bayly, P. V. , Holtzman, D. M. , & Brody, D. L. (2007). Detection of traumatic axonal injury with diffusion tensor imaging in a mouse model of traumatic brain injury. Experimental Neurology, 205, 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney, D. F. , & Smith, D. H. (2015). Cellular biomechanics of central nervous system injury. Handbook of Clinical Neurology, 127, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, M. E. , Cohen, Y. , Mintorovitch, J. , Chileuitt, L. , Shimizu, H. , Kucharczyk, J. , … Weinstein, P. R. (1990). Early detection of regional cerebral ischemia in cats: Comparison of diffusion‐ and T2‐weighted MRI and spectroscopy. Magnetic Resonance in Medicine, 14, 330–346. [DOI] [PubMed] [Google Scholar]
- Norton, W. T. , Aquino, D. A. , Hozumi, I. , Chiu, F. C. , & Brosnan, C. F. (1992). Quantitative aspects of reactive gliosis: A review. Neurochemical Research, 17, 877–885. [DOI] [PubMed] [Google Scholar]
- Oberheim, N. A. , Takano, T. , Han, X. , He, W. , Lin, J. H. , Wang, F. , … Nedergaard, M. (2009). Uniquely hominid features of adult human astrocytes. Journal of Neuroscience, 29, 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özarslan, E. , Koay, C. G. , Shepherd, T. M. , Komlosh, M. E. , İrfanoğlu, M. O. , Pierpaoli, C , & Basser, P. J. (2013). Mean apparent propagator (MAP) MRI: A novel diffusion imaging method for mapping tissue microstructure. NeuroImage, 78, 16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotaki, E. , Schneider, T. , Siow, B. , Hall, M. G. , Lythgoe, M. F. , & Alexander, D. C. (2012). Compartment models of the diffusion MR signal in brain white matter: A taxonomy and comparison. NeuroImage, 59, 2241–2254. [DOI] [PubMed] [Google Scholar]
- Pekna, M. , & Pekny, M. (2012). The neurobiology of brain injury. Cerebrum: The Dana Forum on Brain Science, 2012, 9. [PMC free article] [PubMed] [Google Scholar]
- Pekny, M. , Wilhelmsson, U. , & Pekna, M. (2014). The dual role of astrocyte activation and reactive gliosis. Neuroscience Letters, 565, 30–38. [DOI] [PubMed] [Google Scholar]
- Pierpaoli, C. , & Basser, P. J. (1996). Toward a quantitative assessment of diffusion anisotropy. Magnetic Resonance in Medicine, 36, 893–906. [DOI] [PubMed] [Google Scholar]
- Pierpaoli, C. , Righini, A. , Linfante, I. , Tao‐Cheng, J. H. , Alger, J. R. , & Di Chiro, G. (1993). Histopathologic correlates of abnormal water diffusion in cerebral ischemia: Diffusion‐weighted MR imaging and light and electron microscopic study. Radiology, 189, 439–448. [DOI] [PubMed] [Google Scholar]
- Roth, T. L. , Nayak, D. , Atanasijevic, T. , Koretsky, A. P. , Latour, L. L. , & McGavern, D. B. (2014). Transcranial amelioration of inflammation and cell death after brain injury. Nature, 505, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M. , Chang, E. , Igarashi, T. , & Noble, L. J. (2001). Neuronal injury and loss after traumatic brain injury: Time course and regional variability. Brain Research, 917, 4554. [DOI] [PubMed] [Google Scholar]
- Shepherd, T. M. , Özarslan, E. , King, M. A. , Mareci, T. H. , & Blackband, S. J. (2006). Structural insights from high‐resolution diffusion tensor imaging and tractography of the isolated rat hippocampus. NeuroImage, 32, 14991509. [DOI] [PubMed] [Google Scholar]
- Sierra, A. , Laitinen, T. , Gröhn, O. , & Pitkänen, A. (2015). Diffusion tensor imaging of hippocampal network plasticity. Brain Structure & #Function, 220, 781–801. [DOI] [PubMed] [Google Scholar]
- Silver, J. , & Miller, J. H. (2004). Regeneration beyond the glial scar. Nature Reviews Neuroscience, 5, 146–156. [DOI] [PubMed] [Google Scholar]
- Smith, D. H. , Meaney, D. F. , Lenkinski, R. E. , Alsop, D. C. , Grossman, R. , Kimura, H. , … Gennarelli, T. A. (1995). New magnetic resonance imaging techniques for the evaluation of traumatic brain injury. Journal of Neurotrauma, 12, 573–577. [DOI] [PubMed] [Google Scholar]
- Sofroniew, M. V. (2005). Reactive astrocytes in neural repair and protection. Neuroscientist, 11, 400–407. [DOI] [PubMed] [Google Scholar]
- Sofroniew, M. V. (2009). Molecular dissection of reactive astrogliosis and glial scar formation. Trends in Neurosciences, 32, 638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew, M. V. , & Vinters, H. V. (2010). Astrocytes: Biology and pathology. Acta Neuropathologica, 119, 7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoica B. A., & Faden, A. I. (2010). Cell death mechanisms and modulation in traumatic brain injury. Neurotherapeutics, 7, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroop, R. , Thomale, U. W. , Päuser, S. , Bernarding, J. , Vollmann, W. , Wolf, K. J. , … Unterberg, A. W. (1998). Magnetic resonance imaging studies with cluster algorithm for characterization of brain edema after controlled cortical impact injury (CCII). Acta Neurochirurgica Supplement, 71, 303–305. [DOI] [PubMed] [Google Scholar]
- Sun, D. , & Jakobs, T. C. (2012). Structural remodeling of astrocytes in the injured CNS. Neuroscientist, 18, 567–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S.‐W. W. , Liang, H.‐F. F. , Trinkaus, K. , Cross, A. H. , Armstrong, R. C. , & Song, S.‐K. K. (2006). Noninvasive detection of cuprizone induced axonal damage and demyelination in the mouse corpus callosum. Magnetic Resonance in Medicine, 55, 302–308. [DOI] [PubMed] [Google Scholar]
- Sutula, T. , Zhang, P. , Lynch, M. , Sayin, U. , Golarai, G. , & Rod, R. (1998). Synaptic and axonal remodeling of mossy fibers in the hilus and supragranular region of the dentate gyrus in kainate‐treated rats. Journal of Comparative Neurology, 390, 578–594. [DOI] [PubMed] [Google Scholar]
- Suzuki, T. , Sakata, H. , Kato, C. , Connor, J. A. , & Morita, M. (2012). Astrocyte activation and wound healing in intact‐skull mouse after focal brain injury. European Journal of Neuroscience, 36, 3653–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabesh, A. , Jensen, J. H. , Ardekani, B. A. , & Helpern, J. A. (2011). Estimation of tensors and tensor‐derived measures in diffusional kurtosis imaging. Magnetic Resonance in Medicine, 65, 823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, M. , Schneider, T. , Alexander, D. C. , Gandini Wheeler‐Kingshott, C. A. , & Zhang, H. (2016). Bingham‐NODDI: Mapping anisotropic orientation dispersion of neurites using diffusion MRI. NeuroImage, 133, 207–223. [DOI] [PubMed] [Google Scholar]
- Tournier, J. , Calamante, F. , & Connelly, A. (2012). MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology, 22, 53–66. [Google Scholar]
- Tournier, J. D. , Calamante, F. , Gadian, D. G. , & Connelly, A. (2004). Direct estimation of the fiber orientation density function from diffusion‐weighted MRI data using spherical deconvolution. NeuroImage, 23, 1176–1185. [DOI] [PubMed] [Google Scholar]
- Tuch, D. S. (2004). Q‐ball imaging. Magnetic Resonance in Medicine, 52, 1358–1372. [DOI] [PubMed] [Google Scholar]
- Tuch, D. S. , Reese, T. G. , Wiegell, M. R. , & Wedeen, V. J. (2003). Diffusion MRI of complex neural architecture. Neuron, 40, 885–895. [DOI] [PubMed] [Google Scholar]
- Unterberg, A. W. , Stroop, R. , Thomale, U. W. , Kiening, K. L. , Päuser, S. , & Vollmann, W. (1997). Characterisation of brain edema following "controlled cortical impact injury" in rats. Acta Neurochirurgica Supplement, 70, 106–108. [DOI] [PubMed] [Google Scholar]
- van de Looij, Y. , Mauconduit, F. , Beaumont, M. , Valable, S. , Farion, R. , Francony, G. , … Lahrech, H. (2012). Diffusion tensor imaging of diffuse axonal injury in a rat brain trauma model. NMR in Biomedicine, 25, 93–103. [DOI] [PubMed] [Google Scholar]
- Van Putten, H. P. , Bouwhuis, M. G. , Muizelaar, J. P. , Lyeth, B. G. , & Berman, R. F. (2005). Diffusion‐weighted imaging of edema following traumatic brain injury in rats: Effects of secondary hypoxia. Journal of Neurotrauma, 22, 857–872. [DOI] [PubMed] [Google Scholar]
- Veraart, J. , Poot, D. H. , Van Hecke, W. , Blockx, I. , Van der Linden, A. , Verhoye, M. , & Sijbers, J. (2011). More accurate estimation of diffusion tensor parameters using diffusion Kurtosis imaging. Magnetic Resonance in Medicine, 65, 138–145. [DOI] [PubMed] [Google Scholar]
- Wake, H. , & Fields, R. D. (2011). Physiological function of microglia. Neuron Glia Biology, 7, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Chopp, M. , Nazem‐Zadeh, M.‐R. R. , Ding, G. , Nejad‐Davarani, S. P. , Qu, C. , … Jiang, Q. (2013). Comparison of neurite density measured by MRI and histology after TBI. PloS One, 8(5), e63511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J. K. , & Stevens, R. D. (2015). Traumatic brain injury: Recent advances in plasticity and regeneration. Current Opinion in Neurology, 28, 565–573. [DOI] [PubMed] [Google Scholar]
- Westin, C. F. F. , Maier, S. E. , Mamata, H. , Nabavi, A. , Jolesz, F. A. , & Kikinis, R. (2002). Processing and visualization for diffusion tensor MRI. Medical Image Analysis, 6, 93–108. [DOI] [PubMed] [Google Scholar]
- Wilhelmsson, U. , Bushong, E. A. , Price, D. L. , Smarr, B. L. , Phung, V. , Terada, M. , … Pekny, M. (2006). Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proceedings of the National Academy of Sciences, 103, 17513–17518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu, G. , & He, Z. (2006). Glial inhibition of CNS axon regeneration. Nature Reviews Neuroscience, 7, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Shukla, D. K. , Armstrong, R. C. , Marion, C. M. , Radomski, K. L. , Selwyn, R. G. , & Dardzinski, B. J. (2017). Repetitive model of mild traumatic brain injury produces cortical abnormalities detectable by magnetic resonance diffusion imaging, histopathology, and behavior. Journal of Neurotrauma, 34, 1364–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Schneider, T. , Wheeler‐Kingshott, C. A. , & Alexander, D. C. (2012). NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage, 61, 1000–1016. [DOI] [PubMed] [Google Scholar]
- Zhuo, J. , Xu, S. , Proctor, J. L. , Mullins, R. J. , Simon, J. Z. , Fiskum, G. , & Gullapalli, R. P. (2012). Diffusion kurtosis as an in vivo imaging marker for reactive astrogliosis in traumatic brain injury. NeuroImage, 59, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell, J. M. , Adelson, P. D. , & Lifshitz, J. (2015). Microglia: Dismantling and rebuilding circuits after acute neurological injury. Metabolic Brain Disease, 30, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell, J. M. , & Morganti‐Kossmann, M. (2010). Involvement of pro‐ and anti‐inflammatory cytokines and chemokines in the pathophysiology of traumatic brain injury. Neurotherapeutics, 7, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell, J. M. , Taylor, S. E. , Cao, T. , Harrison, J. L. , & Lifshitz, J. (2012). Rod microglia: Elongation, alignment, and coupling to form trains across the somatosensory cortex after experimental diffuse brain injury. Journal of Neuroinflammation, 9, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]


