SUMMARY
Ubiquitously expressed genes have been implicated in a variety of specific behaviors, including responses to ethanol. However, the mechanisms that confer this behavioral specificity have remained elusive. Previously, we showed that the ubiquitously expressed small GTPase Arf6 is required for normal ethanol-induced sedation in adult Drosophila. Here, we show that this behavioral response also requires Efa6, one of (at least) three Drosophila Arf6 guanine exchange factors. Ethanol-naïve Arf6 and Efa6 mutants were sensitive to ethanol-induced sedation and lacked rapid tolerance upon re-exposure to ethanol, when compared to wild-type flies. In contrast to wild type-flies, both Arf6 and Efa6 mutants preferred alcohol-containing food without prior ethanol experience. An analysis of the human ortholog of Arf6 and orthologs of Efa6 (PSD1-4) revealed that the minor G-allele of SNP rs13265422 in PSD3, as well as a haplotype containing rs13265422, were associated with increased frequency of drinking and binge drinking episodes in adolescents. The same haplotype was also associated with increased alcohol dependence in an independent European cohort. Unlike the ubiquitously expressed human Arf6 GTPase, PSD3 localization is restricted to the brain, particularly the prefrontal cortex (PFC). Functional magnetic resonance imaging (fMRI) revealed that the same PSD3 haplotype was also associated with a differential fMRI signal in the PFC during a Go/No-Go task, which engages PFC-mediated executive control. Our translational analysis therefore suggests that PSD3 confers regional specificity to ubiquitous Arf6 in the PFC to modulate human alcohol-drinking behaviors.
INTRODUCTION
Alcohol is one of the most used and abused drugs in the world.1,2 Excessive alcohol consumption can lead to alcohol use disorders (AUD) and addiction,1 and the behavioral changes that associate with these include tolerance and increased consumption and preference.3–5 Genetic predisposition has been estimated to contribute 40–60% to the development of AUDs,6–10 but the molecular mechanisms involved in this process remain poorly understood.
Numerous model organisms have been established to investigate genes and mechanisms that underlie the development of AUDs. Upon acute alcohol exposure, the vinegar fly, Drosophila melanogaster, exhibits behaviors similar to mammals,11,12 such as disinhibition13 and locomotor hyperactivity,14 followed by sedation.14 Repeat exposures can lead to rapid or chronic tolerance, i.e. reduced sensitivity to ethanol-induced sedation upon re-exposure.15 Flies also develop preference for alcohol consumption in a two choice paradigm, both within a day of being able to choose,16 or after a defined ethanol pre-exposure.17
Mechanistically, there is striking conservation, from flies to humans, of genes that regulate ethanol-responses,18,19 including the Rho-family of small GTPases. Rho GTPases are ubiquitously expressed signaling molecules that regulate actin dynamics and affect many neuronal processes, including addiction-related behaviors, like cocaine-induced place preference and sensitivity to alcohol.20 The Rho-family GTPase Rac1 acts in conjunction with the ubiquitously expressed GTPase Arf6 to regulate ethanol-induced behaviors.19,21 Arf6 regulates receptor trafficking and actin dynamics at the plasma membrane22–24 and acts in a signaling pathway that includes the insulin receptor, mTor, and S6 kinase.25 This pathway is activated by alcohol self-administration in rodents,26 and inhibition of the direct S6k activator mTor reduces drinking relapse.27 One potential problem for therapeutic drugs targeting members in this Arf6 pathway is toxicity, caused by the fact that the proteins involved are expressed in nearly every tissue in the body.28,29
Here, we use Drosophila to isolate conserved genes associated with human alcohol behaviors. We show that Drosophila Efa6 (also known as dPSD), a guanine exchange factor (GEF) and activator of Arf6, is required for normal alcohol-induced behaviors. We show that a single nucleotide polymorphism (SNP) in one of the four human orthologs, PSD3, and one haplotype phase containing that SNP are associated with the frequency of drinking in adolescents, and the same haplotype phase is also associated with alcohol dependence in an independent sample. PSD3 is specifically expressed in the brain, especially the prefrontal cortex (PFC).29 Functional brain imaging revealed an association of the same haplotype phase with differential activation in the right inferior frontal gyrus of adolescents during a Go/No-Go executive control task. The specific expression of PSD3 in the PFC, coupled with its association with specific phenotypes in prefrontal activation and in alcohol consumption suggests a general mechanism by which a ubiquitous signaling pathway can be controlled by a spatially restricted regulator. It also suggests a strategy for the development of specific therapeutic intervention with fewer side effects.
MATERIALS AND METHODS
Fly Methods
Flies were kept on standard cornmeal/molasses food at 25°C on a 12 hr light : 12 hr dark cycle at constant humidity (76 %) and were grown to regular density (~200–400 F1 flies per bottle). All fly lines were outcrossed (for at least 5 generations) to w− Berlin prior to behavioral experiments. Fly strains not described in21 were obtained from the Bloomington Drosophila Stock Center (Efa6PB, Stock #10314), and Dr. Yang Hong (Efa6KO, a knock-out line generated by homologous recombination).30 Adult males (unless otherwise noted) were collected 1–5 days after eclosion and used for experiments after at least one day of recovery within the next one to three days. Sedation and tolerance were determined by observing the flies’ loss-of-righting reflex (LORR).31 To do so, flies were exposed to a mixture of two air flows at predetermined ratios, one water-saturated (air), and the other saturated with ethanol (EtOH). The combined (EtOH/air) flowrate was kept at 150 units. Flies were visually inspected every 5 min for LORR upon gentle tapping during the exposure. The time when 50% of the flies sedated (ST-50) was determined for each tube of 20 flies, for an n = 1. For rapid tolerance, flies were exposed to ethanol for 30 min, and then re-exposed 4 hr later. Preference assays were done in an abbreviated, 16 hr, “2-bottle choice” capillary feeder (CAFÉ) assay,16 following a 20 min mock, or preference-inducing ethanol pre-exposure 24 hr prior.17 Sample sizes of at least 6 for sedation and tolerance and 12 groups of flies for CAFÉ preference were chosen. Genotypes were randomly assigned to the different exposure tubes/chambers, numerically coded, and the experimenter was blind to genotype during the assay. All groups of flies were included in the analyses. Data were analyzed using Prism (GraphPad 6 Software, La Jolla, CA). Chi-square test was used for the discontinuous data in Figure 1a, and ANOVAS for the rest of Figures 1–3 where values were normally distributed (D’Agostino & Pearson normality test with Bonferroni correction). No differences in variances were detected (Brown-Forsythe test). Only in Figure 2D was one variable not normally distributed (due to one “outlier”), but as all data points were included, a non-parametric test was performed in that case (Kruskal-Wallis, with Dunn’s post-hoc test).
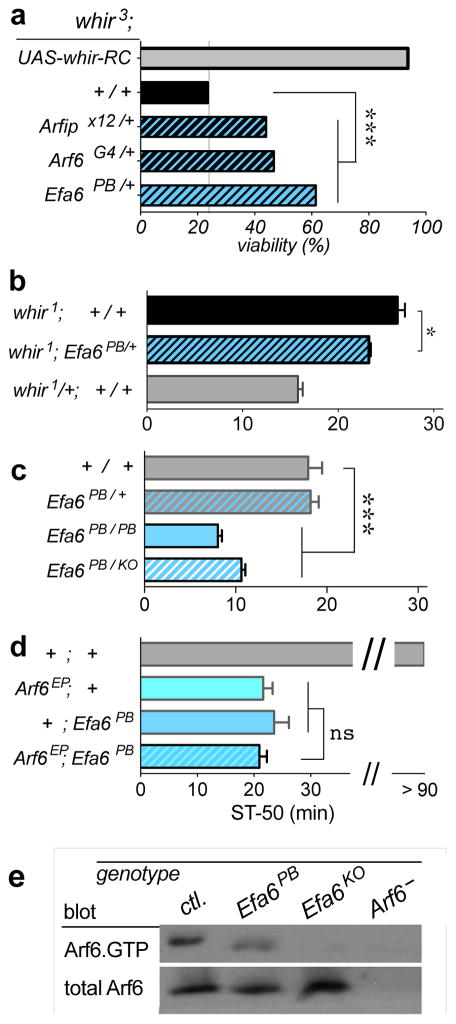
Figure 1.
Efa6 mutant phenotypes. (a) Viability phenotypes of various whir3 genotypes. Displayed are the ratios of surviving whir3/Y;genotype males to whir3/+;genotype females. All the genotypes are in the context of the whir3 mutation, indicated by the whir3; atop. The whir3 allele causes semi-lethality (whir3;+/+, black bar), which can be rescued by re-introducing the RhoGAP18B-PC isoform (whir3;UAS-whir-RC, where RC is the RNA transcript encoding the PC protein isoform, grey bar) driven by the Gal4-driver inserted in the whir3 mutant. (Chi-square test with Bonferroni correction, z = 12.4, df = 153.6,1, P < 0.001, n = 1776 flies). This isoform also rescues the whir3-mediated ethanol resistance.31 Heterozygous Arfip/+, Arf6/+, and Efa6/+ mutations partially suppress the whir3 semi-lethality phenotype (df = 113.3,3, ***P < 0.001, n > 329 flies per genotype). (b) Heterozygous Efa6 mutation partially suppresses the ethanol-resistance phenotype of whir1 mutants (One-way ANOVA with Dunnett’s multiple comparison vs. whir1, F(2,16) = 70.3, *P = 0.011, n = 8,5,6; top to bottom). In all of the Figures in this manuscript, error bars depicted are the standard error of the mean. Here, and in the following Figures, flies were exposed to 130/20 Ethanol/Air vapor (unless otherwise noted). Male flies were used in all behavioral experiments with the exception of whir1/+ females here (as the whir gene is on the X chromosome) and the females indicated in Supplementary Figures 2 and 3. (c) Homozygous Efa6PB mutants show significantly enhanced sensitivity to ethanol-induced sedation, as do Efa6PB/KO trans-heterozygous mutants (One-way ANOVA, with Dunnett’s multiple comparison vs. +/+ w− Berlin, F(3,33) = 48.0, ***P < 0.001, n = 6,7,11,13; top to bottom). The Efa6KO allele is a molecularly-targeted knock out, previously described.30 (d) Arf6;Efa6 double mutants are no more sensitive than either single mutant alone (F(2,33) = 4.6, ns P > 0.54, n = 12 per genotype). Flies were exposed to 30/120 Ethanol/Air, a low dose that did not sedate wild-type flies after 90 min. (e) Activated Arf6.GTP pull-down from head extracts, followed by anti-Arf6 Western blot. Both Efa6PB, and Efa6KO mutants show reduced Arf6 activation (and GTP-loading) compared to w− Berlin control (ctl.). Lysates from Arf6− flies contain undetectable levels of Arf6. A representative blot from 2 replicates is shown.
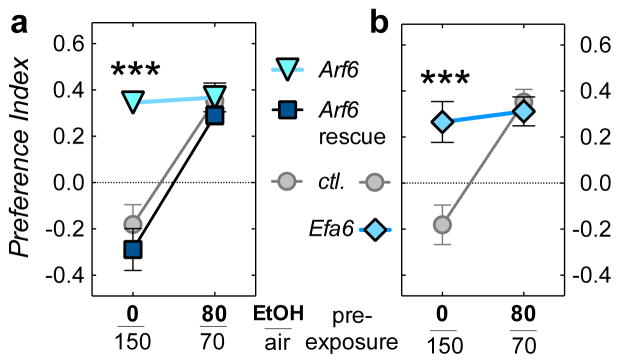
Figure 3.
Arf6 and Efa6 mutants show increased alcohol consumption preference. (a) Flies were offered a choice between liquid food, and liquid food containing 15% ethanol. Naïve wild-type flies (mock exposed to 0/150 Ethanol/Air) avoid ethanol (Preference Index < 0; w− Berlin ctl. flies). This changes to preference (PI > 0) after a 20 min 80/70 Ethanol/Air pre-exposure the day before (Two-way ANOVA, F(1,80)exposure = 44, P < 0.001, n =13,18,12,17,12,14, left to right, top to bottom). Arf6 mutants show high, naïve ethanol preference, independent of a pre-exposure (Dunnett’s post-hoc test, ***P < 0.001). This phenotype is rescued by UAS-Arf6-cDNA expression (Arf6 rescue, genotypes as in Figure 3A). (b) Efa6PB homozygotes display the same naïve preference phenotype (***P < 0.001, n = 13,18,12,12, left to right, top to bottom).
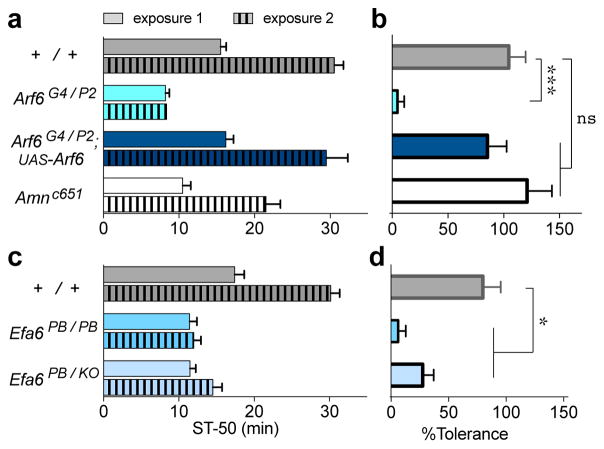
Figure 2.
Efa6 and Arf6 are required for rapid ethanol tolerance. (a) Flies were exposed for 30 min to 130/20 Ethanol/Air (exposure 1, plain bars), and after a 4 hr recovery they were re-exposed (exposure 2, striped). Two-way ANOVA reveals significant effects for exposure (F(1,96) = 89), genotype (F(3,96) = 42), and interaction (F(3,96) = 9, n = 14,11,13,14; top to bottom). Both Arf6 mutants (light blue, P < 0.01, Dunnett’s post-hoc test) and Amn (white, P < 0.05) are sensitive to ethanol-induced sedation during the first exposure when compared to the +/+ control, as previously reported.21,42 (b) Wild-type and Amn mutant flies develop tolerance (expressed as the % increase in ST-50 from first to second exposure), but Arf6 mutants do not (One-way ANOVA with Dunnett’s post-hoc test, ***P < 0.001). Note that expression of UAS-Arf6-cDNA driven by the endogenous Arf6 promoter (Gal4-expressing Arf6G4 allele) rescues both Arf6− sedation (a) and tolerance (b) phenotypes. (c) Mutants of Efa6 are sensitive, compared to +/+ control, to ethanol-induced sedation during the first exposure (P < 0.01, n = 8,6,9; top to bottom) and also develop significantly less tolerance (d) than wild-type flies. This is true for Efa6PB homozygous, and Efa6PB/KO trans-heterozygous flies (*P < 0.05, Kruskal-Wallis non-parametric test with Dunn’s post-hoc test. Non-normality of the data was due to one +/+ “outlier”, which was nevertheless included in the analysis). All +/+ controls in these experiments are w− Berlin flies.
Lethality Screen
To determine viability of the whir3 allele, we crossed whir3 virgins to wild type males. Because the whir gene is on the X-chromosone, this resulted in F1 progeny consisting of whir3 mutant males, and whir3/+ heterozygous, phenotypically normal females. At 25°C, only ~25% as many males survived, compared to females, hence the designation of whir3 as semi-lethal (note that individual flies were only scored as alive when reaching the adult stage, and the designation “semi-lethal” applied to the genotype, not individual flies of that genotype). At 28°C, only ~1% of whir3 males survived, and the suppressor screen was performed at that temperature. Unbiased mutations on the third chromosome (mut(III); limited to chromosome 3 only for the crossing scheme that follows) were crossed to UAS-hid virgins (to get rid of 3rd chromosome balancer chromosomes), resulting in mut(III)/UAS-hid progeny. Virgins of whir3 were then crossed to mut(III)/UAS-hid males. The whir3 mutant also contains a Gal4-driver that causes all whir3;+/UAS-hid males and whir3/+;+/UAS-hid females to die (data not shown), therefore the surviving flies were all of the desired whir3;mut(III)/+ and whir3/+;mut(III)/+ genotype. Vials with >5 surviving males were scored as putative suppressors, and the cross was repeated with the same 3rd chromosome mut(III), to confirm the suppression. All mutations had been mapped, and the affected genes determined (Exelixis, San Francisco, CA). We confirmed the insertion sites by PCR (see Supplementary Figure 1).
Biochemical Assays
Arf6.GTP levels were determined using a specific GG3A-PBD (protein binding domain) conjugated to GST, which binds to activated Arf6 only. Arf6.GTP was then pulled-down using glutathione-agarose beads and compared to total levels of Arf6 in 3% lysate in two independent experiments (Active Arf6 Pull-Down and Detection Kit, Thermo Scientific). Western blots were performed using anti-Arf6 antibody (1:1000, Sigma-Aldrich, #A5230) and visualized using enhanced chemiluminescence (Amersham).
Human Cohort and Analysis
The IMAGEN Study was approved by local ethics research committees at each research site: King’s College London, University of Nottingham, Trinity College Dublin, University of Heidelberg, Technische Universität Dresden, Commissariat à l’Energie Atomique et aux Energies Alternatives, and Universitätsklinikum Hamburg Eppendorf. Informed consent was sought from all participants and a parent/guardian of each participant. 1363 IMAGEN adolescent individuals (712 females) were included in the association analyses between frequency of drinking and binge drinking in the last 30 days (age mean = 16.46, range =13.81–18.80, s.d. = 0.51) and SNPs/haplotypes of PSD genes. Alcohol usage behaviors were assessed with the European School Survey Project on Alcohol and Other Drugs (ESPAD) questionnaire,32 and substantial alcohol use was observed in these 16-year old IMAGEN participants, of which 1004 (74%) individuals had experience of drinking in the last 30 days, while 528 (39%) individuals had experience of binge drinking, which is in line with previous findings.33 For the functional MRI experiments, 1771 IMAGEN individuals (914 females) were assessed for their blood oxygen-level dependent (BOLD) response contrast between ‘stop success vs. go success’ during the Stop Signal Task (Go/No-Go) at 14-years old (mean age: 14.43; range: 12.56–18.67; s.d = 0.42). See34 for description of the IMAGEN cohort and analysis, and the Supplementary Materials and Methods for additional detail. The Study of Addiction: Genetics and Environment (SAGE) dataset has been described by Bierut and colleagues35 which integrates alcohol dependent individuals and controls from different sub-datasets. Specifically, 2544 subjects of European descent were included in our analyses to match the genetic background of IMAGEN sample. Descriptive statistics for both IMAGEN and SAGE drinking behaviors are summarized in Supplementary Table 1 and 2.
Genetic and Haplotype Analyses
For SNP data, the linear regression/partial correlation was adopted to conduct univariate analysis between SNP and phenotypes. Given a sequence of SNPs with high linkage disequilibrium, e.g. D′, a haplotype analysis could help to investigate the underlying hidden haplotype structure through estimating most probable haplotype phases for each individual. Hence established haplotype phases represent allele combinations of a chromosomal region (i.e. determined haplotype blocks) that are inheritable without recombination and could help to capture information from non-genotyped SNPs.36,37 In the IMAGEN sample, the LD/haplotype blocks of the PSD3 gene were generated and illustrated through the Haploview software.38 The exact haplotype phases of both IMAGEN and SAGE samples were estimated with the PLINK software.39 To investigate sexual dimorphism, the gender-specific correlations were compared by using Fisher r-to-Z transformation. See Supplementary Materials and Methods for more details. The IMAGEN data are available from a dedicated database: https://imagen2.cea.fr.
RESULTS
Efa6 Genetically Interacts with Mutations of RhoGAP18B
The Drosophila Rho-family GTPase activating protein RhoGap18B acts on the ubiquitous small GTPase Rac1,40 which is downstream of the integrin cell adhesion molecule,19 and upstream of the actin-severing protein cofilin.40 Activity of this pathway in neurons modulates sensitivity to ethanol-induced sedation,19 and loss-of-function mutations in the white rabbit (whir) gene, encoding RhoGAP18B, cause resistance to ethanol-induced sedation.31 The two RhoGAP18B alleles whir1 and whir3 (caused by a transposable element inserted in two different locations in the gene) show similar extents of ethanol-induced resistance, but the stronger whir3 allele also causes reduced viability.31 In order to isolate additional genes contributing to the signaling cascade that includes RhoGAP18B, we performed a genetic modifier screen. We hypothesized that mutations that reduce the lethality of the whir3 allele would also reduce the ethanol-resistance of whir1 and would therefore be in a genetic network with RhoGAP18B regulating ethanol-induced behaviors. We performed a screen of 300 transposable elements on the third chromosome and isolated two suppressors of whir3 semi-lethality, Arfip and Efa6 (Figure 1a). Mutation in Arfaptin (Arfipx12) suppressed whir3 semi-lethality, and we have previously shown that the Arf6 and Rac1 GTPase-binding protein Arfaptin is involved in regulating ethanol-induced behaviors in the adult nervous system.21 The other suppressor we isolated was a mutation in Efa6 (Figure 1a), an activator of Arf6.41 Given that both Arfip and Efa6 are associated with Arf6, we tested if a mutation in Arf6 would also affect whir3 semi-lethality. As hypothesized, Arf6G4 loss of function mutants21 also suppressed whir3 semi-lethality (Figure 1a).
Mutations in Arf6 and Arfip suppressed the ethanol-resistance of whir1 flies.21 Because Efa6 is an activator of Arf6, we next tested whether mutation in Efa6 would also suppress whir1-mediated ethanol-resistance. Unlike experiments with the whir3 allele, this whir1 genetic interaction experiment was not confounded by developmental semi-lethality. Double mutant whir1;Efa6PB/+ flies showed a significant suppression of the whir1 ethanol-resistance phenotype (Figure 1b), while in a wild-type background, Efa6PB/+ heterozygotes showed no phenotype (Figure 1c). These data suggest that Efa6 acts in concert with RhoGAP18B to regulate ethanol-induced behaviors, as do Arfip and Arf6.21
Efa6 Activates Arf6 and Regulates Ethanol-Induced Sedation
We then tested whether Efa6 mutants alone would have an ethanol-induced sedation phenotype. The Efa6PB allele we isolated is a transposable element insertion in the open reading frame of Efa6, disrupting the Sec7 GEF domain essential for GTP loading (Supplementary Figure 1). Homozygous Efa6PB mutants were viable, but male sterile. This is the same phenotype that was previously reported for an Efa6KO knock out allele generated by targeted, homologous recombination at the Efa6 locus.30 Homozygous Efa6PB males showed significantly enhanced sensitivity to ethanol-induced sedation, as did Efa6PB/Efa6KO trans-heterozygotes (Figure 1c). Efa6 is therefore required to counteract ethanol-induced sedation.
Loss of either Efa6 or Arf6 caused enhanced sensitivity to ethanol sedation (Figure 1c).21 Therefore, we hypothesized that Efa6 acts as an activator of Arf6 in the specific physiological context of ethanol-induced sedation. We first tested this hypothesis genetically by generating Efa6;Arf6 double mutants. If loss of Efa6 function causes loss of Arf6 activity, then introducing a loss-of-function Arf6 mutation should not enhance the Efa6 phenotype any further. As predicted, Arf6, Efa6, and the Arf6;Efa6 double mutants all had indistinguishable ethanol-sensitivity phenotypes, and the introduction of the Arf6 loss-of-function mutation did not make the Efa6 phenotype any more severe (Figure 1d). This lack of enhanced sensitivity was not due to a floor effect, because we used a very low dose of ethanol during exposure, which was unable to sedate wild-type flies, and we have previously shown that the Arf6 phenotype can be enhanced with an unrelated, ethanol-sensitive Amn mutation.21
If Efa6 acts as an activator of Arf6, we next hypothesized that Efa6PB mutants should show reduced Arf6 activation as measured by GTP loading. We tested this prediction by specifically pulling down activated Arf6.GTP from head extracts in our set of mutants (Efa6PB, Efa6KO and Arf6). Western blots revealed a reduction of activated Arf6 in the mutants compared to wild-type controls (Figure 1e). Together, these data show that Efa6 acts upstream of Arf6 to regulate Arf6 activation and together, they counteract ethanol-induced sedation.
Efa6 and Arf6 Regulate Ethanol-Induced Tolerance and Consumption Preference
One hallmark of addiction is altered behavioral responses after prior alcohol exposure, including reduced sedation, increased tolerance, and heightened preference and consumption. We therefore tested if Efa6 and Arf6 regulate ethanol-induced behavioral plasticity in addition to naïve ethanol-induced sedation. Flies develop rapid functional tolerance after being exposed to a sedating dose of ethanol, a behavioral change that can be measured upon a second exposure.15 For example, it takes wild-type flies twice as long to sedate during the second exposure, indicating that they developed 100% tolerance relative to the sedation time during the first exposure (Figure 2a,b). Arf6 mutant flies were as sensitive to sedation during the second exposure as they were during the first exposure, indicating that they did not develop tolerance (Figure 2a,b). We were able to rescue that phenotype when expressing Arf6-cDNA driven by the endogenous Arf6 promoter (using the Arf6G4 mutant allele which includes a Gal4-driver expressed from endogenous Arf6 promoter/enhancers). Because a tolerance phenotype might be confounded by the initial sedation-sensitivity of Arf6 mutants, we also tested the ethanol-sensitive Amnc651 mutant for their tolerance. We found that while these mutants showed initial sensitivity to sedation (as expected),42 they developed tolerance similar to that of the wild type (Figure 2a,b). These results indicate that initial sensitivity to alcohol and rapid tolerance phenotypes can be separated, as found by Devineni and colleagues.43 Similar to Arf6 mutants, Efa6 mutants also failed to develop tolerance to repeat ethanol exposures (Figure 2c, d), further strengthening the functional connection between the two genes.
We then tested our Arf6 and Efa6 mutant flies for their ethanol preference in an assay similar to a two-bottle choice paradigm. We used the capillary feeder (CAFÉ) assay, where flies choose between two capillaries, one containing liquid food only, and the other liquid food with 15% ethanol. Wild-type flies showed initial indifference to ethanol on the first day and then acquire ethanol preference over the course of three to four days.16,44,45 Pre-exposing naïve flies to ethanol vapor, 24 hr before the choice assay, also results in acquisition of preference.17 In an abbreviated 16-hr CAFÉ, naïve flies showed acute aversion to alcohol (as found in),17 and a pre-exposure to vaporized ethanol (20 min, 80/70 ethanol/air mix, 24 hr prior to test) induced alcohol preference (Figure 3). Arf6 mutants showed high initial preference, which did not change upon pre-exposure, and this phenotype was rescued by expressing Arf6-cDNA (driven by the Arf6Gal4 mutant/driver; Figure 3a). Similarly, Efa6 mutants also showed high initial, and unchanging preference for ethanol (Figure 3b). Arf6 and Efa6 are therefore required for normal alcohol-induced tolerance and preference.
Because variants in the human ortholog of Efa6 showed a trend towards gender-specific phenotypes (albeit non-significant; see below), and the above experiments were performed in male flies (see also legend to Figure 1), we also tested Drosophila Arf6 and Ef6 mutant females. We found that the mutations equally affected acute aversion to alcohol in naïve males and females (Supplementary Figure 3), but that some of the tolerance and sedation phenotypes were more pronounced in males, compared to females (Supplementary Figure 2). Overall, however, the sexual dimorphism of these Drosophila alcohol phenotypes was subtle, and just as in human adolescents (see below), not pervasive.
Variants in Human PSD3 Associate with Frequency of Alcohol Consumption and with PFC Activation
Previous genes identified in Drosophila that function in alcohol-induced behaviors have orthologs associated with human alcohol behaviors.19 We therefore decided to examined the human orthologs of Efa6 and Arf6. The human genome encodes for one Arf6 ortholog, and four Efa6 orthologs, PSD1-4 (for Pleckstrin and Sec7 GEF domain-containing proteins – the same domains as found in Drosophila EFA6; see Supplementary Figure 4 for gene structures). We first studied association of PSD1-4 and human Arf6 with drinking behavior in adolescents, including frequency of drinking and binge drinking (Supplementary Table 1 and 2). In the IMAGEN sample of 16-year old European adolescents genotype information was available for 252 SNPs in PSD3, as well as one SNP in PSD1, nine SNPS in PSD2, two SNPs in PSD4, and two SNPs in the Arf6 gene region (Supplementary Table 3). In PSD1, PSD2, PSD4, and Arf6, there was no significant association (Supplementary Table 3). However, in PSD3 we found overall significant association of the minor G allele of rs13265422 (Figure 4, Supplementary Figure 5; minor allele frequency = 0.3) with frequency of drinking in the last 30 days (r = 0.11, t = 3.97, P = 7.3×10−5; P corrected for 266 SNPs of all five genes, as well as two phenotypes, based on 100,000 permutations, Pcorrected = 0.031). While this result was mainly observed in girls (r = 0.14, t = 3.63, P = 2.8×10−4), but less so in boys (r = 0.08, t = 1.99, P = 0.047), the directionality of association was similar in both genders, and there was no significant difference in the effect between boys and girls (z = 1.06, P = 0.29). We also found a nominally significant association of rs13265422 with frequency of binge drinking in the last 30 days, a measure of alcohol abuse (r = 0.06, t = 2.06, P = 0.040; girls: r = 0.09, t = 2.48, P = 0.013; boys: r = 0.02, t = 0.52, P = 0.60), and again there was no significant difference between boys and girls (z = 1.33, P = 0.18).
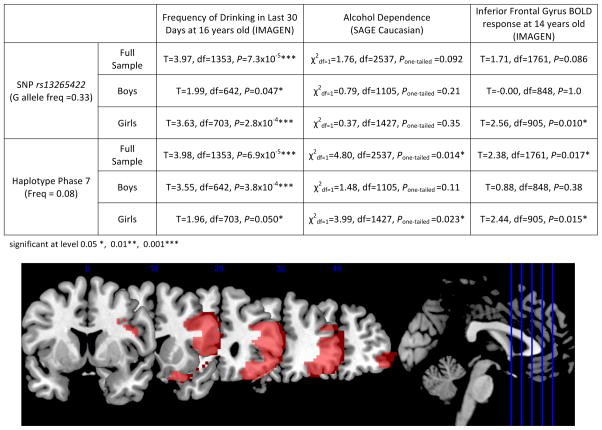
Figure 4.
Human PSD3 phenotypes. (Top) Associations of PSD3 with frequency of drinking (IMAGEN), alcohol dependence (SAGE Caucasian) and BOLD response during Go/No-Go task (IMAGEN), a measure of executive control. The minor G allele of SNP rs13265422 is associated with increased frequency of drinking in the last 30 days in the IMAGEN cohort of 16-year old European adolescents (GG-genotype average score = 1.77, n = 146; GT-genotype average score = 1.49 n = 620; TT-genotype average score = 1.34, n = 597). The haplotype phase 7 shows association not only with increased frequency of drinking in the last 30 days (IMAGEN), but also shows association with increased alcohol dependence in an independent sample of Caucasian adults (SAGE), as well as increased activation contrast between ‘No-Go success vs. Go success’ in the right inferior frontal gyrus (IMAGEN). Gender and sites were controlled for all analyses where applicable, and handedness was controlled for fMRI phenotypes. (Bottom) Illustration of right inferior frontal gyrus (rIFG) activation as red area in coronal view. The slides were acquired based on MNI sagittal coordinates 8, 18, 28, 38, 48 (from left to right).
The SNP rs13265422 is localized 40k nucleotides downstream of the stop codon (Supplementary Figure 5) and shows strong/moderate linkage disequilibrium with SNPs within the 3′ UTR of PSD3, i.e. rs3739398 (r2 = 0.50), rs3739396 (r2 = 0.47), rs901933 (r2 = 0.52), rs901934 (r2 = 0.35); rs13265422 might therefore be a marker of functional SNPs within the 3′ UTR that can potentially affect the stability and translation of mRNA.46
To further investigate the genetic structure of PSD3 we carried out a haplotype analysis of this gene (see Supplementary Materials and Methods), and detected 29 haplotype blocks (Supplementary Figure 6a). The haplotype block that contains SNP rs13265422 (Supplementary Figure 6b) was significantly associated with frequency of drinking (η2partial = 0.022, F(9,1345) = 3.37, P = 4.3×10−4, omnibus test; girls: η2partial = 0.042, F(9,695) = 3.40, P = 4.2×10−4; boys: η2partial = 0.039, F(9,634) = 2.85, P = 0.0026). This haplotype block contains 12 SNPs that give rise to ten individual haplotype phases with frequency > 1% (Supplementary Table 4). Within this block, haplotype phase 7 (Figure 4, Supplementary Table 5; frequency 8%), containing the G allele of rs13265422, showed the strongest association with frequency of drinking (r = 0.11, t = 3.98, P = 6.9×10−5; girls: r = 0.074, t = 1.96, P = 0.050; boys: r = 0.14, t = 3.55, P = 3.8×10−4), as well as frequency of binge drinking (r = 0.095, t = 3.50, P = 4.7×10−4; girls: r = 0.09, t = 2.27, P = 0.023; boys: r = 0.10, t = 2.57, P = 0.010); there was no statistically significant gender difference (z = −1.21, P = 0.23 for frequency of drinking; z = −0.29, P = 0.77 for frequency of binge drinking). We then investigated rs13265422 and the haplotype block containing rs13265422 in the European sample of the Study of Addiction: Genetics and Environment (SAGE) cohort (n = 2544, Supplementary Table 1), an independent adult alcohol-dependence sample. We did not find significant association with rs13265422 (OR = 1.09, χ2df=1 = 1.76, P = 0.18) (Figure 4), but we observed a very similar haplotype structure of ten individual haplotype phases (Supplementary Table 4). While there was no omnibus significance, we again found significant association of haplotype phase 7 (Figure 4, Supplementary Table 5) with increased alcohol dependence (OR = 1.28, χ2df=1 = 4.80, Pone-tailed = 0.014; girls: OR = 1.36, χ2df=1 = 3.99, Pone-tailed = 0.023, n = 1433; boys: OR = 1.22, χ2df=1 = 1.48, Pone-tailed = 0.11, n = 1111), and again no significant gender difference was observed (z = 0.41, P = 0.69).
Drosophila Arf6 is a ubiquitous protein23,47 involved in plasma membrane trafficking of numerous cell surface receptors,48 and Drosophila Efa6 is also expressed in all tissues.47 Human Arf6 is also expressed ubiquitously.29,49 In contrast, PSD3 is specifically expressed in the human brain, especially the PFC (Supplementary Figure 7).29,49 The PFC is required for behaviors involving executive control,50 including alcohol use disorders.51,52 We therefore analyzed activation of the PFC using functional MRI during a Stop Signal Task, which engages executive control in the PFC. In this task, probands are asked to press one of two buttons most of the time (Go), and not press either button (No-Go) one out of six times (on average). We selected a region of interest, right inferior frontal gyrus (rIFG; see Supplementary Materials and Methods), in the PFC and measured the association of ‘No-Go success vs. Go success’ during this task with the PSD3 SNP rs13265422 and haplotype phase 7. There was a trend towards association of the BOLD response of rIFG with rs13265422 (r = 0.04, t = 1.71, P = 0.086; girls r = 0.08, t = 2.56, P = 0.010; boys r = −0.00, t = −0.00, P = 1.00), and a significant association with haplotype phase 7 (r = 0.06, t = 2.38, P = 0.017; girls: r = 0.08, t = 2.44, P = 0.015; boys: r = 0.03, t = 0.88, P = 0.38; Figure 4) – indicating that in carriers of the risk genotype/haplotype a greater effort was required to carry out a successful inhibition. This might lead to a greater risk for increased frequency of alcohol drinking/binge drinking, which was also observed with this genotype/haplotype. While the differences were again driven by girls, the two genders were not significantly different (SNP: z = 1.78, P = 0.075; haplotype phase 7: z = 1.06, P = 0.29). In a whole brain survey (without region of interest hypothesis) of the same functional contrast we found no significant association after correction for multiple testing, neither for rs13265422 nor for PSD3 haplotype phase 7, however, both SNP and haplotype phase had their highest peak level significance in the right inferior frontal gyrus region (Supplementary Figure 8). Overall, our data suggest a modulating role for Efa6/PSD3 in brain activity during behavioral inhibition, and in the regulation of alcohol drinking.
DISCUSSION
We have previously shown that Arf6 acts in the adult nervous system to regulate ethanol-induced sedation in flies.21 Here, we expand on this genetic network regulating alcohol-induced behaviors in Drosophila (see Figure 5) by showing that one of its activators, Efa6, is also required for normal behavioral sensitivity to ethanol. In addition, both Arf6 and Efa6 are required for ethanol-induced tolerance, a behavioral change upon repeat alcohol exposure that is thought to be a precursor to alcohol addiction.53 Indeed, flies lacking either of these genes showed abnormal alcohol consumption preference. In humans, the minor G allele of SNP rs13265422 and a haplotype phase containing this SNP of PSD3, one of the four human Efa6 orthologs, were associated with increased drinking frequency and binge drinking frequency in adolescents. The same haplotype phase was also associated with alcohol dependence in the independent SAGE sample. Arf6 and PSD3 are involved in neurite and dendrite outgrowth54–56 and might therefore also be involved in neuronal plasticity underlying the behavioral changes seen during addiction. Arf6 is a ubiquitously-expressed protein in flies57,58 and humans.23,29,49 Drosophila Efa6 is also found in most tissues,57,58 with an enrichment in the nervous system.30 Human PSD3, on the other hand, is highly specific for the human brain and shows a particular enrichment in the PFC,29,49 a region known to be involved in addiction51,52 and critical for executive control.50 We show that in a Go/No-Go task, which engages prefrontal cortical areas, the same PSD3 haplotype phase also associated with differential fMRI activation, specifically in the rIFG. This PSD3 haplotype phase thus affected both alcohol-drinking behavior, as well as prefrontal activity during an executive control task. Insufficient executive control is well known to be a risk factor for externalizing disorders, including alcohol abuse and addiction.59
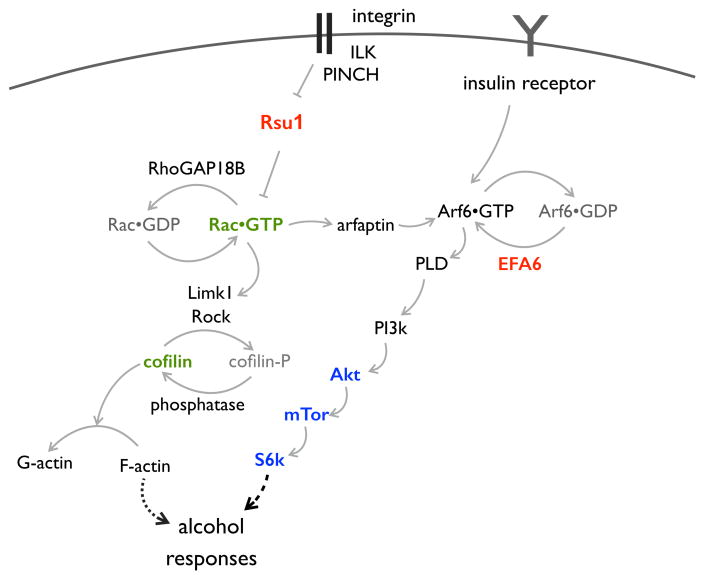
Figure 5.
Model of molecular mechanisms involved in Drosophila alcohol responses. Interactions (arrows) are based on our genetic and biochemical data19,21,25,31,40 (as well as other published data cited therein). Of particular relevance to this report: RhoGAP18B (encoded by the whir gene) binds to, and acts on Rac1.11,40 Rac1 is linked to Arf6 via Arfaptin, which binds to either activated GTPase.21 Here, we show that Efa6 is required for Arf6 activation and behavioral ethanol responses, and together, these biochemical data support our initial finding of a genetic interaction between whir and Efa6, placing them in the same network. Note that all molecules depicted here have mammalian orthologs, with the exception of RhoGAP18B, which contains a GTPase activating GAP domain and long stretches without any other characterized domains. Proteins whose genes are associated with human alcohol drinking are depicted in red (Rsu1,40 Efa6, this report), ones involved in rodent alcohol drinking in blue,27,60 and ones linked to rodent cocaine-induced behaviors in green (Rac1,cofilin).61,62 Abbreviations: ILK: integrin-linked kinase, PINCH: particularly interesting Cys-His rich protein, Rsu1: Ras-suppressor 1, Rac: Ras-related C3 botulinum toxin substrate, Limk1: LIM domain kinase 1, Rock: Rho-associated kinase, Arf6: ADP-ribosylation factor 6, PLD: phospholipase D, Akt: Thymona-associated kinase from the Ak strain, mTor: mechanistic target of rapamycin, S6k: S6 kinase.
Mutations in the Efa6 activator of ubiquitous Arf6 GTPase were identified in our genetic interaction screen with mutants for whir, encoding the RhoGAP18B inactivator of ubiquitous Rac1 GTPase. In this expanding genetic network regulating Drosophila behavioral ethanol responses (Figure 5), neuronal Arf6 acts in the signaling pathway linking the insulin receptor to S6 kinase to regulate ethanol-induced sedation in Drosophila.25 In rodents, inhibition of the mTor kinase in the homologous pathway can reduce alcohol drinking26 and relapse behavior.27 The mTor inhibitor rapamycin is a clinically used, FDA-approved drug for human use, but it causes a large number of side effects, likely due to the fact that the pathway (insulin receptor/Arf6/mTor/S6 kinase) is found in most every cell and tissue and controls cell growth and survival. Furthermore, both Arf6 and the Rho-family of small GTPases are ubiquitous proteins that are involved in many cellular and neuronal processes.20,47 In contrast, our study suggests a mechanism by which the restricted expression of a regulator, in this case PSD3, can confer highly specific regulation of this ubiquitous pathway: by activating Arf6 in specific anatomical regions and thereby also achieving regulation of specific behaviors. This has implications for the targeted treatment of specific behavioral disorders, in this case alcohol use disorders. By targeting the restricted PSD3, as opposed to the ubiquitous insulin receptor/Arf6/mTor/S6 kinase pathway, one might improve on therapeutic efficacy while decreasing side effects.
Supplementary Material
Acknowledgments
The authors thank the Bloomington stock center, and Yang Hong (Univ. Pittsburgh) for fly strains, and Michael Buszczak, Helmut Krämer, and Rothenfluh lab members for helpful discussions. This work was supported by the NIH (T32 fellowships DA007290 to D.A.G. and J.H.P., F32 AA021340 to S.A.O., K08 DK091316 to A.R.R., R01 AA019526 and R21 AA022404 to A.R.), the European Union-funded FP6 Integrated Project IMAGEN (Reinforcement-related behavior in normal brain function and psychopathology; LSHM-CT-2007-037286), the FP7 projects IMAGEMEND (602450) and MATRICS (603016), the Innovative Medicine Initiative Project EU-AIMS (115300-2), the European Research Council Award ‘STRATIFY’ as well as the Medical Research Council Programme Grant ‘Developmental pathways into adolescent substance abuse’ (93558). Further support was provided by the Swedish Funding Agency FORMAS, the MRC-ICMR Newton project ‘Consortium on Vulnerability to Externalizing Disorders and Addictions’ [c-VEDA] (MR/N000390/1), the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, the German Bundesministerium für Bildung und Forschung (BMBF grants 01GS08152; 01EV0711; eMED SysAlc 01ZX1311A; Forschungsnetz AERIAL), the NIH (R01 MH085772-01A1), as well as the NIH-BD2K (Big Data to Knowledge) grant U54 EB020403 – ENIGMA Center for Worldwide Medicine, Imaging and Genomics. A.R. was also supported by an Effie Marie Cain Scholarship in Biomedical Research from UT Southwestern Medical Center Dallas.
Footnotes
Conflict of interest: The authors declare no conflict of interest
AUTHOR CONTRIBUTIONS
D.A.G., J.H.P., S.A.O., S.F.A., and A.R. conceived, performed, and analyzed the Drosophila experiments. S.D., T.B., C.B., A.L.W.B., P.J.C., H.F., B.I., M.L., J.M., T.P., M.S., G.S., and the IMAGEN consortium acquired the human data. T.J., B.X., and G.S. analyzed the human data. D.A.G., T.J., J.H.P., J.L.H., A.R.R., G.S., and A.R. wrote the paper.
Supplementary information is available at Molecular Psychiatry’s website.
CONFLICT OF INTEREST
T.B. served in an advisory or consultancy role for Hexal Pharma, Lilly, Medice, Novartis, Otsuka, Oxford Outcomes, PCM Scientific, Shire, and Viforpharma. T.B. received conference attendance support and conference support or speaker’s fees from Lilly, Medice, Novartis, and Shire. T.B. is/has been involved in clinical trials conducted by Shire and Viforpharma. J.G. has received research funding from AstraZeneca, Eli Lilly & Co., Janssen- Cilag, and Bristol-Myers Squibb and speaker’s fees from AstraZeneca, Janssen-Cilag, and Bristol-Myers Squibb. This work is unrelated to above grants and relationships.
References
- 1.Edenberg HJ, Foroud T. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol. 2013;10:487–494. doi: 10.1038/nrgastro.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topper SM, Aguilar SC, Topper VY, Elbel E, Pierce-Shimomura JT. Alcohol disinhibition of behaviors in C. elegans. PLoS ONE. 2014;9:e92965. doi: 10.1371/journal.pone.0092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- 4.Newlin DB, Thomson JB. Alcohol challenge with sons of alcoholics: A critical review and analysis. Psychol Bull. 1990;108:383. doi: 10.1037/0033-2909.108.3.383. [DOI] [PubMed] [Google Scholar]
- 5.King AC, de Wit H, McNamara PJ, Cao D. Rewarding, stimulant, and sedative alcohol responses and relationship to future binge drinking. Arch Gen Psychiatry. 2011;68:389–399. doi: 10.1001/archgenpsychiatry.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick DM, Jones K, Saccone N, Hinrichs A, Wang JC, Goate A, et al. Endophenotypes successfully lead to gene identification: results from the collaborative study on the genetics of alcoholism. Behav Genet. 2006;36:112–126. doi: 10.1007/s10519-005-9001-3. [DOI] [PubMed] [Google Scholar]
- 7.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 8.True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- 9.Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut L, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 10.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 11.Narayanan AS, Rothenfluh A. I believe I can fly!: Use of Drosophila as a model organism in neuropsychopharmacology research. Neuropsychopharmacology. 2016;41:1439–1446. doi: 10.1038/npp.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaun KR, Devineni AV, Heberlein U. Drosophila melanogaster as a model to study drug addiction. Hum Genet. 2012;131:959–975. doi: 10.1007/s00439-012-1146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H-G, Kim Y-C, Dunning JS, Han K-A. Recurring ethanol exposure induces disinhibited courtship in Drosophila. PLoS ONE. 2008;3:e1391. doi: 10.1371/journal.pone.0001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf FW, Rodan AR, Tsai LTY, Heberlein U. High-Resolution Analysis of Ethanol-Induced Locomotor Stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. doi: 10.1016/s0896-6273(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 16.Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19:2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peru y Colón de Portugal RL, Ojelade SA, Penninti PS, Dove RJ, Nye MJ, Acevedo SF, et al. Long-lasting, experience-dependent alcohol preference in Drosophila. Addic Biol. 2014;19:392–401. doi: 10.1111/adb.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grotewiel M, Bettinger JC. Drosophila and Caenorhabditis elegans as Discovery Platforms for Genes Involved in Human Alcohol Use Disorder. Alcoholism Clin Exp Res. 2015;39:1292–1311. doi: 10.1111/acer.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ojelade SA, Jia T, Rodan AR, Chenyang T, Kadrmas JL, Cattrell A, et al. Rsu1 regulates ethanol consumption in Drosophila and humans. Proc Natl Acad Sci USA. 2015;112:E4085–93. doi: 10.1073/pnas.1417222112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothenfluh A, Cowan CW. Emerging roles of actin cytoskeleton regulating enzymes in drug addiction: actin or reactin’? Curr Opin Neurobiol. 2013;23:507–512. doi: 10.1016/j.conb.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peru y Colón de Portugal RL, Acevedo SF, Rodan AR, Chang LY, Eaton BA, Rothenfluh A. Adult neuronal Arf6 controls ethanol-induced behavior with Arfaptin downstream of Rac1 and RhoGAP18B. J Neurosci. 2012;32:17706–17713. doi: 10.1523/JNEUROSCI.1944-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song J, Khachikian Z, Radhakrishna H, Donaldson JG. Localization of endogenous Arf6 to sites of cortical actin rearrangement and involvement of Arf6 in cell spreading. J Cell Sci. 1998;111:2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- 23.Lebeda RA, Johnson SK, Stewart MI, Haun RS. Sequence, genomic organization, and expression of the human ADP-Ribosylation Factor 6 (ARF6) gene: a class III ARF. DNA and Cell Biology. 2004;22:737–741. doi: 10.1089/104454903770946719. [DOI] [PubMed] [Google Scholar]
- 24.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nature Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 25.Acevedo SF, Peru y Colón de Portugal RL, Gonzalez DA, Rodan AR, Rothenfluh A. S6 Kinase reflects and regulates ethanol-induced sedation. J Neurosci. 2015;35:15396–15402. doi: 10.1523/JNEUROSCI.1880-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neasta J, Ben Hamida S, Yowell Q, Carnicella S, Ron D. Role for mammalian target of rapamycin complex 1 signaling in neuroadaptations underlying alcohol-related disorders. Proc Natl Acad Sci USA. 2010;107:20093–20098. doi: 10.1073/pnas.1005554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barak S, Liu F, Ben Hamida S, Yowell QV, Neasta J, Kharazia V, et al. Disruption of alcohol-related memories by mTORC1 inhibition prevents relapse. Nat Neurosci. 2013;16:1111–1117. doi: 10.1038/nn.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos dos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, et al. FlyBase: Introduction of the Drosophila melanogaster release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–7. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016;44:D313–6. doi: 10.1093/nar/gkv1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Zhou W, Dong W, Watson AM, Hong Y. Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci USA. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LTY, Lasek AW, Heberlein U. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127:199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Morgan M, Hibell B, Andersson B, Bjarnason T, Kokkevi A, Narusk A. The ESPAD Study: implications for prevention. Drugs: Education, Prevention and Policy. 2009;6:243–256. [Google Scholar]
- 33.Viner RM, Taylor B. Adult outcomes of binge drinking in adolescence: findings from a UK national birth cohort. Journal of Epidemiology & Community Health. 2007;61:902–907. doi: 10.1136/jech.2005.038117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumann G, Loth E, Banaschewski T, Barbot A, Barker G, Büchel C, et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15:1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 35.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 37.Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, et al. Human genome sequence variation and the influence of gene history, mutation and recombination. Nature Genetics. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- 38.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojelade SA, Acevedo SF, Kalahasti G, Rodan AR, Rothenfluh A. RhoGAP18B isoforms act on distinct Rho-Family GTPases and regulate behavioral responses to alcohol via cofilin. PLoS ONE. 2015;10:e0137465. doi: 10.1371/journal.pone.0137465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naassila M, Ledent C, Daoust M. Low ethanol sensitivity and increased ethanol consumption in mice lacking adenosine A2A receptors. J Neurosci. 2002;22:10487–10493. doi: 10.1523/JNEUROSCI.22-23-10487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 43.Devineni AV, McClure K, Guarnieri D, Corl A, Wolf F. The genetic relationships between ethanol preference, acute ethanol sensitivity, and ethanol tolerance in Drosophila melanogaster. Fly. 2011 doi: 10.4161/fly.5.3.16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pohl JB, Baldwin BA, Dinh BL, Rahman P, Smerek D, Prado FJ, III, et al. Ethanol preference in Drosophila melanogaster is driven by its caloric value. Alcoholism Clin Exp Res. 2012;36:1903–1912. doi: 10.1111/j.1530-0277.2012.01817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu S, Chan T, Shah V, Zhang S, Pletcher SD, Roman G. The propensity for consuming ethanol in Drosophila requires rutabaga adenylyl cyclase expression within mushroom body neurons. Genes, Brain and Behav. 2012;11:727–739. doi: 10.1111/j.1601-183X.2012.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skeeles LE, Fleming JL, Mahler KL, Toland AE. The Impact of 3′UTR Variants on Differential Expression of Candidate Cancer Susceptibility Genes. PLoS ONE. 2013;8:e58609. doi: 10.1371/journal.pone.0058609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nature Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbins TW, Arnsten A. The neuropsychopharmacology of fronto-executive function: monoaminergic modulation. Annu Rev Neurosci. 2009;32:267–287. doi: 10.1146/annurev.neuro.051508.135535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu YL, Richardson HN. Alcohol, stress hormones, and the prefrontal cortex: A proposed pathway to the dark side of addiction. Neuroscience. 2014;277:139–151. doi: 10.1016/j.neuroscience.2014.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jasinska AJ, Chen BT, Bonci A, Stein EA. Dorsal medial prefrontal cortex (MPFC) circuitry in rodent models of cocaine use: implications for drug addiction therapies. Addic Biol. 2015;20:215–226. doi: 10.1111/adb.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volkow ND, Koob GF, McLellan AT. Neurobiologic advances from the brain disease model of addiction. N Engl J Med. 2016;374:363–371. doi: 10.1056/NEJMra1511480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanda M, Kamata A, Katsumata O, Fukunaga K, Watanabe M, Kondo H, et al. The postsynaptic density protein, IQ-ArfGEF/BRAG1, can interact with IRSp53 through its proline-rich sequence. Brain Research. 2009;1251:7–15. doi: 10.1016/j.brainres.2008.11.061. [DOI] [PubMed] [Google Scholar]
- 55.Hernández-Deviez DJ, Roth MG, Casanova JE, Wilson JM. ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol Biol Cell. 2004;15:111–120. doi: 10.1091/mbc.E03-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hernández-Deviez DJ, Casanova JE, Wilson JM. Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci. 2002;5:623–624. doi: 10.1038/nn865. [DOI] [PubMed] [Google Scholar]
- 57.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The modENCODE Consortium. Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whelan R, Conrod PJ, Poline J-B, Lourdusamy A, Banaschewski T, Barker GJ, et al. Adolescent impulsivity phenotypes characterized by distinct brain networks. Nat Neurosci. 2012;15:920–925. doi: 10.1038/nn.3092. [DOI] [PubMed] [Google Scholar]
- 60.Neasta J, Ben Hamida S, Yowell Q, Carnicella S, Ron D. Role for mammalian target of rapamycin complex 1 signaling in neuroadaptations underlying alcohol-related disorders. Proc Natl Acad Sci USA. 2010;107:20093–20098. doi: 10.1073/pnas.1005554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dietz DM, Sun H, Lobo MK, Cahill ME, Chadwick B, Gao V, et al. Rac1 is essential in cocaine-induced structural plasticity of nucleus accumbens neurons. Nat Neurosci. 2012;15:891–896. doi: 10.1038/nn.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toda S, Shen H-W, Peters J, Cagle S, Kalivas PW. Cocaine Increases Actin Cycling: Effects in the Reinstatement Model of Drug Seeking. J Neurosci. 2006;26:1579–1587. doi: 10.1523/JNEUROSCI.4132-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.