Abstract
An estimated 50% of depressed patients are inadequately treated by available interventions. Even with an eventual recovery, many patients require a trial and error approach, as there are no reliable guidelines to match patients to optimal treatments and many patients develop treatment resistance over time. This situation derives from the heterogeneity of depression and the lack of biomarkers for stratification by distinct depression subtypes. There is thus a dire need for novel therapies. To address these known challenges, we propose a multi-scale framework for fundamental research on depression, aimed at identifying the brain circuits that are dysfunctional in several animal models of depression as well the changes in gene expression that are associated with these models. When combined with human genetic and imaging studies, our preclinical studies are starting to identify candidate circuits and molecules that are altered both in models of disease and in patient populations. Targeting these circuits and mechanisms can lead to novel generations of antidepressants tailored to specific patient populations with distinctive types of molecular and circuit dysfunction.
Introduction
Depression, or Major Depressive Disorder (MDD), is characterized by the persistence of negative thoughts and emotions that disrupt mood, cognition, motivation and behavior. Depression is the leading cause of disability worldwide affecting over 300 million people (World Health Organization, 2017). This burden has been rising steadily, with an 18% increase in prevalence between 2005 and 2015. The illness occurs throughout the lifespan, from childhood through old age, is ~two-fold more common in women than men, and has higher incidence during puberty, peripartum periods, and menopause. Depression in mothers has detrimental effects on the fetus and young children (O’Donnell & Meaney, 2016). Depression is chronic: half of those who experience one episode of depression have recurrent episodes, with increasing frequency and severity of episodes over time. Depression is a leading cause of suicide and is associated with several common medical conditions, such as obesity, diabetes, stroke, Parkinson’s disease and multiple sclerosis as well as a greater risk for Alzheimer’s disease and sudden cardiac death. The impact of depression on humanity cannot be overstated
While depression is diagnosed as a single entity, MDD, by the Diagnostic Statistical Manual (DSM) (2013), there are 681 combinations of symptoms that could meet the DSM criteria, reflecting the heterogeneity of symptoms, etiologies and pathophysiologies. Several described subtypes of depression, notably melancholic, psychotic, or atypical depression, are distinguished solely by self-report criteria with no objective biological indicators. Depression is moderately heritable with as much as ~35% of the risk associated with genetic predisposition (Geschwind & Flint, 2015), but is also highly influenced by adverse life experiences (Otte et al., 2016).
Multiple modalities of treatment are effective for depression, including antidepressant medications, psychotherapies, and various brain stimulation techniques. Nonetheless, fewer than half of MDD patients achieve full remission with a first treatment (Rush, 2007). Further, matching a patient to his/her optimal treatment generally requires multiple trials of different treatments, with the sobering observation that the more treatments tried without success, the less likely a successful outcome. In sum, there remains a huge unmet need for a “precision medicine” approach to depression, with an important next step requiring development of treatments designed selectively for biologically-defined subtypes of this broad, heterogeneous syndrome (Drysdale et al., 2017; Williams 2016).
A significant percentage of all MDD patients exhibit resistance to all available standard treatments. The evolution of resistance can develop in patients previously responsive to treatment or as a progressive, deteriorating illness course over time (Thase & Schwartz, 2015). Resistance can manifest as the presence of residual depressive symptoms following treatment as well as loss of effectiveness with ongoing treatment. Treatment options with increasing resistance are limited and generally involve continued use of the same modalities, including combination, augmentation or switching medications, introduction of electroconvulsive therapy (ECT) or trials of other neurostimulation strategies. These approaches risk complications, including increased toxicity with higher medication dosages and combination regimens.
Treatment Resistant Depression (TRD) represents a heterogeneous state with likely multiple causal mechanisms. TRD patients exhibit the same diversity of symptoms, course, history and co-occurring conditions as for treatment-responsive MDD. However, very little is known about what distinguishes patients who do or do not respond to treatment. The extent to which individuals with TRD versus treatment-responsive MDD differ in etiology or pathophysiology remains mostly obscure, although there are several reports that a history of early life stress increases treatment-resistance (Bernet & Stein, 1999; Nanni et al., 2012; Williams et al., 2016) and that individuals with TRD exhibit differences in brain circuit function (McGrath et al., 2016; Dunlop et al., 2017). Nevertheless, the underlying mechanisms are not known. Consequently, TRD remains an operational definition—and with several different definitions suggested, for example, referring to failure to respond to treatment within a depressive episode or failure to respond to a previously effective treatment in a subsequent episode (Fava, 2003; Conway et al., 2017). Thus, a major goal of current research is to establish more precise, biologically-based definitions of TRD as well as new antidepressant treatments targeting that underlying pathophysiology. Characterization of the biological heterogeneity of the TRD patient population is therefore both a necessity and a challenge. Strategies that consider biological subtypes rather than merely number and type of past treatments are needed. Yet, despite the compelling need for new treatments, especially for TRD, most pharmaceutical companies no longer prioritize depression given recent failures in drug discovery and the view that not enough is known about the underlying biology of depression to provide a rational path forward.
Animal models play an essential role in drug discovery in virtually all fields of medicine, but are particularly challenging in the case of depression (Nestler & Hyman, 2010). As with MDD itself, animal models must equally consider strategies that address symptom and etiological heterogeneity, while being mindful that only some human behaviors are amenable to study in non-human model systems: e.g., motivation, anhedonia, negative affect, hypothalamic dysregulation and homeostasis can be addressed, but not sadness, guilt, ruminations or suicidality. Several acute and chronic stress models have been used, but until recently it has been difficult to distinguish between adaptive vs. maladaptive responses to the stress. While stress is a risk factor for human depression, most individuals exposed to chronic stress do not develop depressive disorders. The issue of susceptibility is thus of paramount importance for animal studies of the biological basis for the relationship between stress and specific symptoms of depression. Additionally, many studies validate the models based on antidepressant response, thereby skewing away from identifying novel mechanisms of therapeutic actions. Indeed, there has not yet been a concerted effort to model the emergence of treatment resistance in rodents. This would require using animals with some genetic or developmental liability, exposing them to multiple bouts of stress and antidepressant treatment, and characterizing a worsening course and the emergence to treatment resistance.
This review focuses on novel strategies in antidepressant drug discovery, particularly for patients with TRD. The authors came together four years ago to create the Depression Task Force sponsored by the Hope for Depression Research Foundation (http://www.hopefordepression.org/). Our goal is to use a reverse translation strategy to model the key features of depression, including its emergence, course and treatment response or resistance. We developed an interactive platform to test and integrate a set of complementary animal models of depression. These models are used to shed light on the pathophysiology of depression, including the relevant neural circuitry and the underlying genetic and molecular mechanisms. Anchored by an interactive “big data” analytic platform, we use a multi-scale, systems biology approach that leverages advances in genomics and neural circuitry, and integrates the discoveries in these animal models with findings from MDD patients and high-risk cohorts. Our goal is to identify biomarkers that will advance patient subtyping and treatment stratification, and facilitate the development of novel targeted interventions.
Brain Plasticity and Vulnerability in the Context of Brain-Body Interactions: Historical Overview
Studies of the neurobiology of depression, including TRD, focus largely on the association between stress and depression, with the hope that an understanding of the biological pathways that link stress to depression would inform on the pathophysiology of the disorder. The hypothalamic-pituitary-adrenal (HPA) axis, which controls secretion of both corticotrophin-releasing factor (CRF) and glucocorticoids, is central to the stress response. Mutations of several HPA genes have been used as genetic models of depression. For example, glucocorticoid receptor (GR) overexpression in the forebrain acts during development to increase anxiety and affective lability (Wei et al., 2004, 2012). Likewise, CRF or CRF1 receptor overexpression in mouse associates with enhanced depressive-life features (De Kloet et al., 2005). Key to understanding the neurobiology and pathophysiology of depression was the discovery of steroid hormone receptors, including GR, in brain regions that moderate virtually every aspect of brain function, which has broadened the definition of “neuroendocrinology” to include the reciprocal communication between the brain and body via hormonal and neural pathways (McEwen et al., 2015c). Discovery of GRs in hippocampus became the “gateway” for other discoveries and a better understanding of the meaning of “stress” in terms of the concepts of allostasis and allostatic load and overload, where life style and health behaviors are key factors along with stressful experiences themselves (McEwen, 1998; McEwen et al., 2015c). This research led to the discovery of structural and functional plasticity in the brain, mediated in part by hormones, which facilitated the emergence of the science of “epigenetics” by revealing effects of the social and physical environment on adult as well as developing brain structure and function (Meaney & Ferguson-Smith, 2010; Bagot et al., 2014; McEwen et al., 2015c; Box 1). Cellular and molecular mechanisms for plasticity emerged and revealed other mechanisms of steroid hormone action than direct genomic stimulation, including actions on mitochondria (McEwen et al., 2015a). Besides steroid hormones, metabolic hormones enter and affect the brain and their relationship to brain metabolism and mitochondrial function has become important for understanding disorders like diabetes, depression and dementia (Rasgon & McEwen, 2016). Finally, hormone actions via epigenetic mechanisms, beginning preconception, during gestation and in infancy and childhood and operating over the life course. are changing the way we look at the development of disorders and the possibilities for intervention (Meaney & Ferguson-Smith, 2010; Bagot et al., 2014; Halfon et al., 2014, Rasgon & McEwen, 2016).
BOX 1. Core Processes Involved in Mental and Physical Health Over the Life Course.
It must be emphasized that the following mechanisms and etiological processes have not yet been compared sufficiently between TRD versus treatment-responsive MDD. Establishing areas of similarities and differences between the two is a high priority of current research.
Allostasis and allostatic load/overload
Stressful experiences can precipitate depression. The brain is the central organ of stress adaptations because it perceives and determines what is threatening, and activates the behavioral and physiological responses to the stressor, which promote adaptation (“allostasis”) but also contribute to pathophysiology (“allostatic load/overload”) when dysregulated (McEwen, 1998). Health-promoting behaviors are also an essential component of successful allostasis, along with adequate sleep and normal circadian function as well as efficient energy metabolism. Health-damaging behaviors contribute to allostatic load/overload. The mediators of allostasis include not only cortisol and epinephrine, but also the parasympathetic nervous system, pro- and anti-inflammatory cytokines, and metabolic hormones (McEwen, 2007). Moreover, the brain uses a host of interacting mediators to alter neural circuitry and function (McEwen et al., 2015a).
Epigenetics
Epigenetics now refers to the ongoing regulation of gene expression via an array of molecular processes involving post-translational histone modifications, methylation of cytosine bases on DNA, the actions of transcription factors and numerous chromatin-regulatory proteins, translational regulation of RNAs by microRNAs and RNA splicing and editing (Meaney & Ferguson-Smith, 2010; Bagot et al., 2014; Mehler, 2008). Transposable elements including retrotransposons and DNA transposons comprise around 40% of the human genome and also play an emerging regulatory role in stress and aging in the brain (McEwen et al., 2015a).
Structural and functional plasticity
The adult, as well as developing, brain possesses a remarkable ability to adapt by showing structural and functional plasticity in response to stressful and other experiences, including neuronal replacement in VHIP and dendritic remodeling and synapse plasticity and turnover throughout the nervous system. Structural and functional allostatic plasticity is particularly evident in the hippocampus, a key structure for episodic and spatial memory and mood regulation, where structural plasticity has been investigated using a combination of morphological, molecular, pharmacological, electrophysiological and behavioral approaches (McEwen, 2007). The hippocampus was the first brain structure outside of the hypothalamus found to possess stress and sex steroid hormone receptors and it provided a gateway into the hormone sensitivity of the rest of the brain (McEwen et al., 2015b). The amygdala, involved in fear, anxiety, and aggression, and the prefrontal cortex, important for working memory, executive function, and sell-regulation, both show structural plasticity. In amygdala, basolateral neurons expand dendrites after chronic stress (Chattarji et al., 2015), while medial PFC neurons, as well as hippocampal neurons, show dendritic shrinkage from the same stress (McEwen & Morrison, 2013). The NAc also shows altered spine density with stress (Christoffel et al., 2011; Warren et al., 2014).
Circadian disruption affects the brain as well as systemic physiology, leading to shrinkage of PFC dendrites and cognitive rigidity as well as insulin and leptin resistance (McEwen & Karatsoreos, 2015). Likewise, poor sleep impairs parasympathetic/sympathetic balance, increases systemic inflammation and Impairs glucose regulation (McEwen & Karatsoreos, 2015). Diabetes and insulin resistance are risk factors for depression, which in turn increases risk for dementia (Rasgon & McEwen, 2016).
Mitochondria, which have their own DNA and are inherited from the mother, are glucocorticoid and estrogen sensitive for regulating calcium sequestration and free radical balance, and make their own contributions to allostasis and allostatic load and overload (Picard et al., 2014).
Peripheral organ function
Recent work has emphasized the profound interplay between the brain and peripheral organs in controlling normal health, and there are an increasing number of examples where such interactions have been demonstrated to influence depression in humans and stress responses in animals. Examples include the cardiovascular system, metabolism and immunity and inflammation, to name a few (Hodes et al., 2015b; Finnell and Wood, 2016; Wohleb et al., 2016).
Sex differences
There are important sex differences in how the brain responds to stressors, as well as structural and functional plasticity differences (McEwen and Morrison, 2013; Hodes et al., 2015a; Labonté et al., 2017), which contribute to the important concept that males and females do most of the same things equally well cognitively and emotionally but differ in the “strategies” that they use. The entire brain has non-genomic as well as genomic receptors for sex hormones in both sexes and many neural processes are affected (McEwen & Milner, 2017). Further work is needed to determine the role of such hormonal factors, as opposed to chromosomal and other mechanisms, that are responsible for the dramatic sex differences seen in stress responses and depression.
Poverty and early life adversity, interacting with alleles of certain genes, produce lasting effects on brain and body via epigenetic mechanisms (Hackman et al., 2010), leading to multimorbidity of mental and physical disorders (Hyde et al., 2016). Preconception epigenetic factors (Rasgon & McEwen, 2016) and stressful experiences before conception (Rodgers & Bale, 2015) and during gestation (O’Donnell & Meaney, 2017) also have important influences.
The plasticity and vulnerability of the brain are the keys to understanding and treating MDD. Acute and chronic stress alter cortico-limbic circuitry, which sub-serves executive functions essential for decision making, as well as the regulation of affective states. This altered circuit functioning also affects systemic physiology via neuroendocrine, autonomic, immune, and metabolic mediators, which in turn can modify behavioral outcomes, thus underscoring a bidirectional alignment of behavioral states with peripheral organ function (McEwen et al 2015c). The immune system provides a useful example: recent studies have revealed the dramatic effect of chronic stress on levels of pro-inflammatory cytokines in the periphery and, in turn, the potent influence of systemic cytokines on the susceptibility versus resilience of individuals to subsequent stress (Hodes et al., 2015b). Most changes induced by acute or chronic stress are likely adaptive in most individuals and in most circumstances and promote resilience (Feder et al., 2009; McEwen et al., 2015b). However, in susceptible individuals, stress-related responses become maladaptive, and sometimes irreversible (McEwen et al., 2015a). In this latter situation, the goal would be to induce additional changes that compensate for this maladaptive state of plasticity, perhaps in part by inducing mechanisms of natural resilience that promote active recovery, as will be discussed in this review. An overarching goal for future research is to better understand what differentiates TRD and treatment-responsive MDD at the levels of neural circuits and peripheral physiology.
Convergence and Divergence Across Multiple Rodent Models of Depression and Treatment Resistance
The heterogeneity of depression argues for the use of multiple animal models to capture both the diversity of the causes and symptoms as well as the common mechanisms that might underlie certain symptoms that are shared across all models. Each animal model likely recapitulates abnormalities seen in only a subset of patients or a subset of features of the broad syndrome, with many molecular-cellular mechanisms being unique to a given animal model. At the same time, other molecular-cellular mechanisms might be shared across multiple models, with such convergence representing final common pathways that contribute to core symptoms of depression seen in most patients. It is essential, as noted earlier, for the field to turn its attention to using animal models that capture features of TRD, namely, that all individual animals do not respond fully to available antidepressants.
Animal models of depression can be broadly divided into those that manipulate the environment using different types of stressors, as stress is a common trigger of clinical depression, and those that manipulate or exploit the genetics of vulnerability to depression. More recently, several models have focused on the interplay of genetic, environmental and developmental factors.
Chronic stress models
Several chronic stress paradigms have been used as models of depression (see Kollack-Walker et al., 1999; Akil, 2005; Berton & Nestler, 2006). Several involve subjecting adult rodents to repeated or persistent stress, such as chronic social defeat stress, chronic variable stress, chronic isolation stress, or chronic restraint stress. Chronic stress models, including isolation stress, are also used during juvenile periods and often compared to enriched environmental conditions (Isgor et al., 2004). Other chronic stress paradigms subject rodents to stress early in life, such as daily periods of maternal separation (e.g., Francis et al., 2002; Peña et al., 2017). In prenatal stress models, pregnant dams are subjected to various forms of acute or chronic stress (Weinstock, 2008). Chronic administration of corticosterone has been used as a pharmacological means of recapitulating the effects of excessive glucocorticoid secretion seen in many of the behavioral models (David et al., 2009; Olausson et al., 2013). In each of these paradigms, behavioral abnormalities are assessed with a series of acute assays to test an animal’s behavioral and physiological state. Examples include measures of social behavior (e.g., social approach-avoidance, vocalizations), reward-based tests (e.g., sucrose preference, novelty-seeking, intracranial self-stimulation, sexual behavior), acute stress tests (e.g., forced swimming, tail suspension, learned helplessness), exploration-based tests (e.g., open field, dark-light, elevated plus maze), neuroendocrine tests (e.g., plasma corticosterone), fearfulness/anxiety (e.g., novelty-induced suppression of feeding, fear conditioning), body weight and so on. Most chronic stress paradigms induce behavioral abnormalities interpreted as depression- and/or anxiety-like, although a clear distinction is difficult to discern in animals and indeed the two conditions are highly comorbid in humans. Antidepressant-like actions are characterized by the prevention or reversal of these behavioral abnormalities, with responses to standard antidepressants sometimes requiring repeated (weeks) of administration whereas responses to ketamine (an experimental, rapidly acting antidepressant in humans) are seen acutely.
These various chronic stress paradigms have distinct features. Chronic social defeat stress and maternal separation induce long-lasting behavioral abnormalities, which make it possible to study reversal of symptoms with repeated antidepressant administration, while the other assays—characterized typically by shorter-lived symptoms—usually measure prevention of the abnormalities with prior or concurrent treatment. Chronic social defeat stress has the additional advantage of revealing resilience in that, while roughly two-thirds of the mice succumb to the stress, the remaining third avoid the depression-like behavioral abnormalities (Krishnan et al., 2007). All of the aforementioned chronic stress models have been validated in both male and female rodents, although chronic variable stress uniquely reveals a greater vulnerability in females that matches the clinical situation (Hodes et al., 2015a).
Genetic mutant mouse models
These models typically focus on altering the expression of genes implicated either in stress responsiveness, such as the HPA genes noted above, or mechanisms of action of antidepressants. For example, mutations of serotonin receptors implicated in antidepressant responses have served as useful models. Complete knockout of the 5-HT1A receptor produces an increase in anxiety-related behavior and resistance to serotonin-selective reuptake inhibitor (SSRI) antidepressants (but not tricyclic antidepressants) (Santarelli et al., 2003). These effects are mediated by distinct circuits. The anxiety phenotype is recapitulated by knocking down the 5-HT1A receptor specifically from serotonergic neurons in the midbrain raphe nuclei during an early postnatal period, while this manipulation has no effect on the antidepressant response. In contrast, knocking out the receptor in adulthood from the hippocampal dentate gyrus (DG) does not alter anxiety- or depression-like phenotypes, but instead blocks the effects of SSRIs on these phenotypes (Donaldson et al., 2014; Nautiyal, 2017; Samuels et al., 2015). This double dissociation illustrates the complexity of the brain circuitry underlying affective states and antidepressant responses, but also provides opportunities for targeting circuits that are distinct from those engaged by SSRIs.
Selective breeding of rodent lines
An alternative to manipulating the expression of specific target genes is the use of selective breeding for behavioral traits that capture key facets of affective behavior. Common genetic factors underlie multiple stress-related psychiatric diseases that fall in the broad categories of either “internalizing disorders” or “externalizing disorders” (Kendler et al., 1992; Cerdá et al., 2010). Two personality traits emerge as key predictors of psychopathology: neuroticism (which includes high trait anxiety) is a strong predictor of internalizing disorders (Khan et al., 2005), and high sensation seeking is a predictor of externalizing disorders (Zuckerman M & Kuhlman, 2000). Our High Responder-Low Responder (HR-LR) breeding model in the rat aims at capturing differences in the genetics of environmental reactivity by focusing on the traits of sensation-novelty seeking vs. spontaneous anxiety (Flagel et al., 2014). Over the course of 50 generations, the bred HR and LR lines have provided strong evidence of differences in genetic predisposition to anxiety-, depressive- and addictive-like behaviors, as well as insight into the neurobiological and molecular genetic underpinnings. These genetic differences express early in life, and can be modified by early interventions to reset temperament into adulthood (e.g., Turner et al., 2011). These selectively bred lines serve as a model of differential susceptibility or resilience to depression. As well, stress manipulations across the lifespan can be layered upon this background to study the interplay of genes and environment on the behavioral phenotype and response to antidepressant treatments.
Another rodent model that has been useful to study depression-related behavior and to screen for antidepressants are the Flinders Sensitive (FSL) and Resistant (FRL) lines (Overstreet, 1993). The Flinders line was originally selectively bred for increased responses to an anticholinesterase agent and, ultimately, led to a line that exhibits behavioral, neurochemical and pharmacological features that have been reported in depressed individuals, including reduced appetite, abnormal REM sleep patterns and psychomotor retardation. Multiple classes of antidepressants, but not psychomotor stimulants, reverse these depression-related phenotypes. The FSL rats with depressive-like traits have a deficiency of acetyl-L-carnitine (LAC), accompanied by metabolic dysfunction (high insulin, glucose and triglycerides), suggesting a state of insulin resistance, a recognized risk factor for depression in humans. Pharmacological supplementation of LAC corrects the depressive traits as well as the metabolic dysfunction, reinforcing the notion of a bidirectional interaction between brain and peripheral organ functions (Bigio et al., 2016). Interestingly, the antidepressant-like effects of LAC can be observed after only 3 days of administration (Nasca et al., 2013).
Gaps in animal models
Despite the use of these many animal models, key facets of the human depression syndrome have not been optimally modeled. Very few studies utilize multiple hit models, where for example an animal is subjected to several bouts of stress throughout life. Recent studies of multiple hits demonstrate that rodents with a history of chronic stress exposure exhibit a different reaction to a novel stressor or corticosterone elevation than a stress-naïve animal (Gray et al., 2014; Datson et al., 2013), and that exposure to stress early in life increases an animal’s susceptibility to different forms of stress in adulthood (Peña et al., 2017). Likewise, genetic variation moderates the impact of chronic stress and our existing animal studies are poor at modeling such gene x environment interactions.
Even less well studied is the trajectory of animals exposed to multiple bouts of stress plus multiple courses of antidepressant treatments, which is common in TRD. Both childhood adversity and stress in adulthood predict increased TRD following antidepressant treatments (Thase, 2011; Nanni et al., 2012; Williams et al., 2016). These findings form the basis for novel and clinically-relevant animal models examining the mechanisms by which early life adversity or repeated stressful events in adulthood might influence responses to antidepressants. There is a clear need to match animal model development to central challenges seen in human depression, with a focus on capturing features unique to TRD.
The availability of this range of animal models is a unique opportunity to study both convergence and divergence of biological changes associated with depression or treatment response across them. It informs research at multiple levels of analyses, from behavior to neural circuitry to underlying cellular, molecular and genetic mechanisms. Below, we summarize our current understanding at the level of neural circuits and genetics/genomics, and point to strategies for new discoveries, greater integration and possible translation to human clinical applications.
Neural Circuitry of Depression
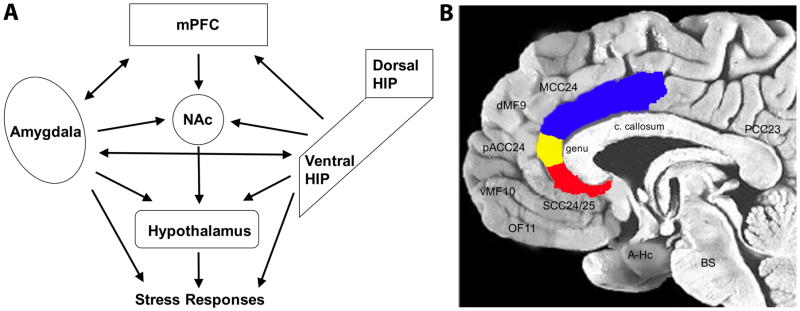
Parallel but complementary studies using animal models, and both in vivo imaging and post-mortem studies in humans, suggest that depression does not arise through pathology in a single brain region or cell type, but instead is mediated by altered functioning across an integrated cortico-limbic circuit in the forebrain (FIG 1) (Harris & Gordon, 2015; Heshmati & Russo, 2015). Key network nodes include regions of prefrontal cortex (PFC), connected with numerous subcortical structures including the hippocampus (particularly its most ventral part; VHIP), amygdala nuclei, and nucleus accumbens (NAc), among numerous other brain regions. Each of these structures is altered by stress in animals and depression in humans, both at the functional (i.e., glucose metabolism, blood flow), the cytoarchitectural (i.e., morphological changes to neurons and glia within these regions) and molecular levels (i.e., altered gene expression). Moreover, manipulations of each of these structures in animals and in patients alter emotion-related behaviors, including many that are responsive to both classical and novel antidepressants as well as behavioral treatments (cognitive therapy in patients; enrichment and exercise in animals) (Goldapple et al., 2004; Perez-Sepulveda et al., 2013), neurostimulation in animals and patients (e.g., deep brain stimulation, ECT and repetitive transcranial magnetic stimulation [rTMS]) (Mayberg et al.,, 2005; Fox et al., 2012; Riva-Posse et al., 2014, 2017) and optogenetic and Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) manipulations in animals (e.g., Covington et al., 2010; Chaudhury et al., 2013; Gunaydin et al., 2014; Hamani et al., 2014; Ferenczi et al., 2015; Insel et al., 2015; Hultman et al., 2016).
Figure 1.
Cortico-limbic circuitry implicated in mood regulation and depression. (A) Simplified schematic diagram of the cortico-limbic circuitry and the many interactions across the various brain regions. Not all known connections are depicted. Likewise, not all outputs of each region are depicted. mPFC, medial prefrontal cortex; HIP, hippocampus; NAc, nucleus accumbens. (B) Midline sagittal view of the human brain illustrating the location of major PFC regions, with the anterior cingulate cortex highlighted: blue, MCC24, mid-cingulate cortex; yellow, pACC24, pre-genual anterior cingulate cortex; red, SCC24/25, subcallosal cingulate cortex. Other brain regions noted include: dMF9, dorsomedial frontal cortex; vMF10, ventromedial frontal; OF11, orbitofrontal; A-Hc, amygdala-hippocampus in the temporal lobe; BS, brainstem; PCC23, posterior cingulate cortex; c. callosum, corpus callosum.
Studies in humans
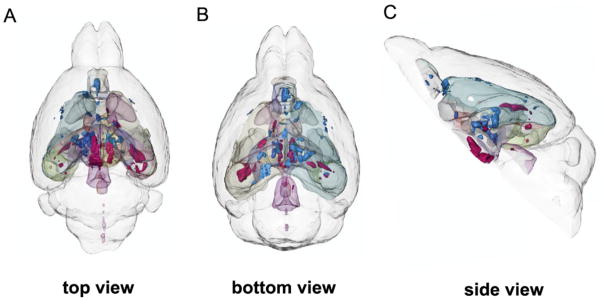
Neuroimaging studies using magnetic resonance imagining (MRI) and positron emission tomography (PET) have successfully characterized brain states of depressed patients (Mayberg, 2009). As the technology and analytic techniques have matured, there is growing emphasis not only on properties of individual regions, but also on their organization within integrated pathways and distributed neural networks (Craddock et al., 2012; Hyett et al., 2015; Drysdale et al., 2016; Williams, 2016). Differential modulation of these defined networks by various treatments can be evaluated, providing added perspective to understanding mechanisms mediating clinical response and remission. Variability in baseline patterns can be further evaluated against known clinical phenotypes (Mayberg, 2003; McGrath et al., 2014; Williams, 2016; Dunlop et al., 2017). Advances in small animal in vivo MRI technologies can identify the effects of chronic stress or novel antidepressant treatments on connectivity within depression-related neural circuits (FIG 2) (e.g., Anacker et al., 2016), thus allowing for convergence of animal and human studies at the level of brain circuitry.
Figure 2.
Three-dimensional rendering of all significant volume changes after chronic social defeat stress in mice. Red clusters indicate positive correlations with social avoidance, and blue clusters indicate negative correlations with social avoidance. (A) Top view. (B) Bottom view. (C) Side view. From Anacker et al. (2016).
Studies comparing MDD patients to healthy controls report relative hyperactivity of limbic regions, including the amygdala, insula and subcallosal cingulate cortex (SCC), as well as atrophy of the hippocampus and hypoactivity in the dorsolateral prefrontal cortex (Ressler & Mayberg, 2007). However, average differences between groups may mask important heterogeneity between individuals (Williams, 2016; Mayberg, 2003), with some patients failing to show these changes (variable hippocampal atrophy), or even demonstrating opposite patterns (e.g., increased metabolism in the dorsolateral prefrontal cortex) (Goldapple et al., 2004). This variability in brain states across patients likely has important implications for clinical subtyping relevant to treatment response and resistance studied using specific animal models.
As one example, studies using resting state metabolic activity assessed by 18F-fluorodeoxyglucose PET found six brain regions differentially associated with the outcomes among MDD randomized to treatment with either an SSRI or cognitive behavioral therapy (McGrath et al., 2013; Dunlop et al., 2015). Activity in the right anterior insula emerged as the optimal candidate for treatment selection with relative hyperactivity in the subcallosal cingulate cortex, further differentiating those patients who went on to be resistant to both interventions (McGrath et al., 2014). A parallel strategy using resting state functional connectivity and resting state fMRI has similarly demonstrated treatment-specific prediction patterns. Functional connectivity patterns between the subcallosal cingulate (thought to be the homologue of the infralimbic PFC in rodents) and the ventromedial and ventrolateral PFC and midbrain similarly predicted differential outcomes to both CBT and medication (Dunlop et al., 2017). Studies such as these are beginning to define biological differences between treatment response and non-response in human MDD.
To optimally leverage these findings from human studies towards the development of valid animal models, it is important to consider the anatomical localization of frontal lobe findings in both imaging and post-mortem studies. Abnormalities have been reported for the dorsolateral and ventrolateral PFC (Brodmann Areas BA 9,46,10,47), as well as orbitofrontal and ventromedial PFC (BA 11, 10, 9). Cingulate changes involve multiple ventral, dorsal and posterior sectors (BA 25/32, 24a,24c, 23) (FIG 1). While there is clear homology for the some of the reported subcortical and limbic regions between humans and animals (e.g., amygdala, hippocampus, insula, NAc, midbrain and brainstem), such homologies are less certain for frontal and cingulate regions (Heilbronner et al., 2016). Despite these limitations, there is solid foundation for complementary and mutually informative studies of core behaviors across humans and animal models of depression based on these patient observations.
Developmental studies of emotion-related neural circuitry in humans emphasize the involvement of the same cortico-limbic regions in stress vulnerability throughout life. Perinatal environmental conditions that increase the risk for depression, such as birth outcomes, childhood socio-economic status and the quality of parental mental health and childcare, reveal sustained effects on the structure and connectivity of amygdala, hippocampus and PFC as well as on individual differences in stress reactivity that associate with adversity-related amygdala hyperactivity to threat as measured by fMRI (Buss et al., 2007; Luby et al., 2012, 2016; Noble et al., 2015; Gee et al., 2013; Rifkin-Graboi et al., 2013; Teicher & Samson, 2016) as well as dopaminergic responses to stress in the NAc (Pruessner et al., 2004). Importantly, many of these reports reveal gender-dependent influences, with increased effects of childhood adversity on girls that are not unique to mental health outcomes (Buss et al., 2007, 2012; O’Donnell & Meaney, 2017). Importantly, studies of unaffected youth with a familiar history of depression show alterations in cortico-limbic anatomy that parallel those observed in depression. Alterations in these targeted neural circuits thus emerge early in life and predate the onset of depression. These findings are consistent with those of the GR and 5HT1A mouse models noted above, where the relevant effects were established in early development. Therefore, studies of the developmental origins of the risk for depression emphasize neural circuits similar to those emerging from studies of adult humans, as well as of animal models.
Studies in animal models
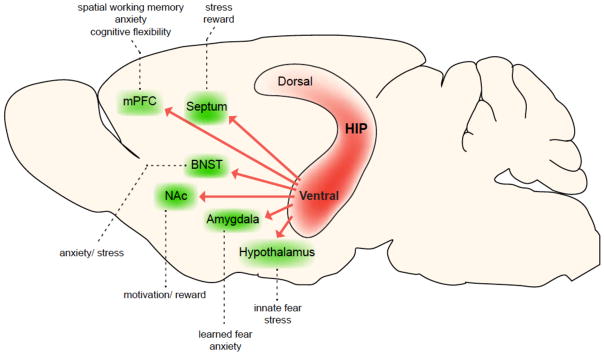
The dysregulated circuitry seen in human subjects is reflected in many of the rodent animal models of depression. Of course, animal work can add greater granularity and inform subsequent human analyses. One of the most highly implicated structures in stress responses in animals is the VHIP (Fanselow & Dong, 2010; Strange et al., 2014). Several forms of acute and chronic stress alter VHIP function and its manipulation likewise controls responses to stress (Kheirbek et al., 2013). For example, the 5-HT1A receptor within the ventral DG (VDG) is necessary for the antidepressant-like effects of SSRIs, as noted earlier (Samuels et al., 2015). The DG is the only part of the hippocampus where new neurons are continuously produced throughout adulthood in all mammalian species including humans (Spalding et al., 2013). We and others showed that hippocampal neurogenesis is necessary (Santarelli et al., 2003) and sufficient (Hill et al., 2015) for some but not all of the behavioral effects of several classes of antidepressants (David et al., 2009; Bessa et al., 2009). Interestingly, 5-HT1A receptors are expressed in mature neurons (but not young neurons) in the DG. These studies suggest therefore a combined influence of young and mature neurons within this brain structure. Indeed, there is evidence that young neurons modulate the activity of mature neurons within the DG and that the resulting output influences the downstream circuitry (CA3, CA1) to control mood and cognition (Denny et al., 2014; Redondo et al., 2014). There is also mounting evidence that these regions of VHIP play important roles in affective state, with outputs to distinct limbic regions producing different, and in some cases opposite, effects on depression- and anxiety-related outcomes (FIG 3) (Bagot et al., 2015; Kheirbek et al., 2013; Padilla-Coreano et al., 2016).
Figure 3.
Projections from the ventral HIP modulate distinct emotional behaviors. The ventral HIP sends direct projections to numerous structures that directly influence emotional behaviors (green structures indicate ventral HIP projection targets and dotted lines describe their putative behavioral consequences upon activation). These projections contribute to distinct aspects of behavior, such as spatial working memory through the mPFC, fear, anxiety and stress responses through the mPFC, amygdala, hypothalamus, septum and BNST, and reward-seeking behaviors through the NAc and septum. Moreover, there is evidence that many of these projections arise from largely non-overlapping cell populations within the CA1 pyramidal layer, including projections to the amygdala, mPFC, NAc, septum and lateral hypothalamus (Jin and Maren, 2015; Okuyama et al., 2016; Parfitt et al., 2017; Xu et al., 2016; Jimenez JC and Hen R, personal communication). Abbreviations: HIP, hippocampus; mPFC, medial prefrontal cortex; BNST, bed nucleus of stria terminalis; NAc, nucleus accumbens.
Animal models implicate other components of this cortico-limbic circuitry in emotional regulation under normal conditions and in stress-induced pathological states. In particular, the NAc, and its dopaminergic inputs from the ventral tegmental area, are crucial for the long-lasting anhedonic-like and social avoidance responses to chronic social defeat stress (Chaudhury et al., 2013; Russo and Nestler, 2013). Optogenetic and biochemical manipulations that mimic the effects of social defeat increase anhedonia and social avoidance, with the opposite manipulations reducing these depression-related behaviors and promoting resilience. Likewise, several regions of PFC and amygdala regulate responses to acute and chronic stress (see Covington et al., 2010; Duman, 2014; Gunaydin et al., 2016; Sharma et al., 2016; Lau et al., 2016). Neuroimaging studies of animals resilient or susceptible to social defeat reflect the importance of both structure and connectivity of the NAc, VHIP and basolateral amygdala (BLA), together with PFC regions in predicting behavioral responses to chronic stress (Anacker et al., 2015). It will be important in future studies to directly address the functioning of this circuitry in models that recapitulate aspects of TRD.
Depression Genetics and Genomics
Genetics and genomics influence the risk for depression. What is unclear is: 1) What is the nature of genetic vs. environmental influences, and their interactions, on the propensity for depression? And 2) What are the genes and gene networks involved in defining that heritable propensity for depression, and what biological systems do they affect? Strands of evidence are beginning to shed light on the answers. A third question is the extent to which genetic factors that contribute to MDD overall differ between those individuals with TRD versus treatment-responsive illness. As will be seen, answering this question will be an extremely challenging task given the requirement for literally tens and hundreds of thousands of subjects in order to achieve genetic findings of genome-wide statistical significance. Nevertheless, the expectation is that parsing genetic data by treatment response will over time help us define the biological basis of TRD.
What is the role of genetics in depression?
There is clear evidence that vulnerability to depression and other affective disorders is attributable to heritable, genetic factors. The evidence derives from several approaches, including twin studies showing that first-degree relatives of depressed patients exhibit a 3-fold increase in risk for depression. Such analyses place the heritability of depression at ~35% (Geschwind & Flint, 2015), although such estimates include gene x environment interaction effects. This degree of heritability is significantly lower than for many other psychiatric disorders. For example, heritability estimates for bipolar disorder are between 60–85%, and relatives of bipolar patients exhibit a 10-fold increase in the risk of developing the illness (Smoller & Finn, 2003). Finally, as with other psychiatric illnesses, there is a lack of specificity in the inherited risk (Dean et al., 2010; Zhu et al., 2012), suggesting that part of the genetic predisposition determines global vulnerability rather than specific outcomes.
What genes are involved?
Given the complexity of the depression syndrome, it would be extremely valuable to identify at least one set of determinants—i.e., the genetic factors—to increase prediction, allow prevention and guide early diagnosis and treatment. Intensive efforts to pinpoint depression risk genes were disappointing until recently. In 2013, a “mega-analysis” conducted by the MDD Working Group and the Psychiatric GWAS (gene-wide association studies) Consortium examined 1.2 million single-nucleotide polymorphisms (SNPs) in 18,759 subjects of European ancestry (9,240 MDD and 9,519 controls), followed by a replication phase with 6,783 MDD cases and 50,695 controls (MDD Working Group, 2013). Despite the analysis of over 76,000 individuals, the authors concluded that they were “unable to identify robust and replicable findings,” as no locus achieved genome-wide significance. This finding is consistent with previous attempts that also showed weak or unreplicable effects. This outcome stands in sharp contrast with successful GWAS of individuals with bipolar disorder, schizophrenia, or autism, which uncovered both unique and shared loci that confer vulnerability to these disorders (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; De Rubeis et al., 2014; Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011; Sklar et al., 2011). The authors of the mega-analysis for depression speculated that the syndrome may be particularly heterogeneous and therefore requires much larger samples to uncover the relevant genetic factors. The inability to pinpoint the genes does not mean they do not exist, but rather that there are so many of them, with each playing a minor role in the overall risk for the illness in the population.
A more recent study analyzed samples acquired by the genetic testing company, 23andMe, which deals directly with consumers (Hyde et al., 2016). It should be noted that the 23andMe samples relied solely on self-report in response to queries about being diagnosed with and treated for past depressive episodes. In the initial phase, over ~76,000 individuals who self-reported a diagnosis of depression were compared to ~232,000 individuals with no self-reported diagnosis of the illness. A meta-analysis was conducted combining the identified loci with publically available MDD genomic data. Genetic loci identified through this initial meta-analysis were verified through an independent replication study with over 45,000 cases and 106,000 controls also from 23andMe. In all, ~460,000 subjects were included in the various components of this study in addition to the pre-existing GWAS data. The joint analysis across all three sets of data resulted in a total of 17 independent SNPs from 15 different chromosomal regions that reached genome-wide significance. The identified genes were enriched for those involved in transcriptional regulation during neural development. A second analysis from the same 23andMe population focused on SNPs associated with response to various classes of antidepressants. The clearest findings were related to buproprion and implicated pathways associated with circadian rhythm and growth factor-associated neuroplasticity (Li et al., 2016).
The 23andMe analyses provided the enormous scale of study needed for capturing some of the genetic variability that contributes to depression vulnerability in the general population. However, there are concerns about methodological aspects of this approach, especially the self-report of diagnosis (Abbasi, 2017). Indeed, recent findings show that most adults with depression are not appropriately diagnosed and treated (which would contaminate the control group) and most diagnoses and treatments of depression do not appropriately match the illness severity (Olfson et al., 2016). Additionally, the identified loci still accounted for only ~1–2% of the variance in the risk for depression. Nevertheless, the 23andMe analyses provide new leads in the search for genetic factors that broadly contribute to the illness and that are worthy of further scientific study, as will be discussed below.
An alternative approach to finding depression vulnerability genes is to focus on a more homogeneous genetic background. The CONVERGE study, which focused on Han Chinese females, succeeded in pinpointing candidate genes for further analyses (Peterson et al., 2015). Whether these genes provide a basis for understanding depression in other populations remains to be determined. Nevertheless, this approach complements the study of extremely large and diverse populations to gradually and progressively compile a complete list of genetic factors that underlie the ~35% heritability of depression.
Finally, it is important to note that psychiatric disorders including depression, bipolar disorder, schizophrenia and autism share significant genetic etiology as stated earlier (Cross-Disorder Group of the PGC, 2013), and that many of the genes discovered across these illnesses are regulatory and developmental in nature, often pointing to epigenetic pathways (Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015). These findings suggest that a portion of the genetic architecture of psychiatric illnesses is not specific for a certain illness, but rather influences a global vulnerability perhaps through effects on responses to adversity. We note that increased stress reactivity is a risk factor for virtually all forms of psychiatric illness. The nature of the specific illness may then emerge based on other, unique genetic factors, life history and stochastic events during development (Kendler et al., 2003).
What next?
The picture emerging confirms the view that genetic studies will not, on their own, yield sufficient information to understand the biological bases of depression. The interaction between genetics and environmental in defining development, and neuroplasticity throughout life, confound any efforts to isolate any single factor, such as genetic vulnerability. We suggest that it is critical to use a convergent approach, coupling the hard-won genetic information with our knowledge of brain organization, the underlying developmental program and the mechanisms of neuroplasticity that mediate the impact of the environmental, as well as the impact of the illness itself, on the brain (Akil et al., 2010). This approach requires the use of a range of animal models that capture various facets of human depression and the use of state-of-the-art genomic and neurobiological strategies to gain new insights into the function of vulnerability genes and their role in the development, expression and treatment responsiveness of human depression.
Post-mortem human studies
Genome-wide methods have been used to map gene expression changes in post-mortem brains of humans with a history of severe clinical depression. Earlier studies utilized microarray technology, with recent efforts moving to RNA-sequencing (RNA-seq) (e.g., Bernard et al., 2011; Choudary et al., 2005; Duric et al., 2013; Evans et al., 2004; Gaiteri and Sibille, 2011; Iwamoto et al., 2004; Kang et al., 2007; Klempan et al., 2009; Labonte et al., 2017; Lalovic et al., 2010; Li et al., 2013; Mehta et al., 2010; Sequeira et al., 2007, 2012; Togichi et al., 2008; Wang et al., 2008). There is a broad transcriptional dysregulation throughout the depressed brain, especially pronounced in the cortico-limbic circuitry implicated in human depression. Functional pathway analyses reveal alterations in gene families related to neuroplasticity, including growth factors (Turner et al., 2012; Duman & Duman, 2015), as well as broad scale disruptions in gene regulation, for example, manifested in altered circadian rhythms (Bunney et al., 2015). Beyond changes in expression levels, there is mounting evidence of other classes of regulatory changes, including alterations in microRNAs (Dwivedi et al., 2014), and of epigenetic signatures (Bagot et al., 2014; Sun et al., 2015) associated with depression.
Establishing the significance of these post-mortem observations requires parallel studies in animal models to ascertain changes functionally related to the pathophysiology of depression, and to antidepressant response or treatment resistance. As multiple brain regions from larger numbers of patients and control subjects are analyzed, it will become increasingly possible to carry out such comparisons with animal data. Moreover, imaging genetics consortia such as the ENIGMA project (Thompson et al., 2014) permit exploration of GWAS examining variation in brain structure in both healthy controls and patients. These datasets are accessible on-line (http://enigma.ini.usc.edu/enigma-vis/) and allow researchers to query the association of specific genetic variants with MRI-based measures of brain structure. The integration of genetic findings, gene expression changes and neural phenotypes associated with depression is a critical starting place for reverse translation—i.e., the use of animal models to reproduce these changes, understand their significance and test their causal relation to specific facets of depressive symptoms.
Transcriptomic and Epigenomic Data Across Multiple Animal Models
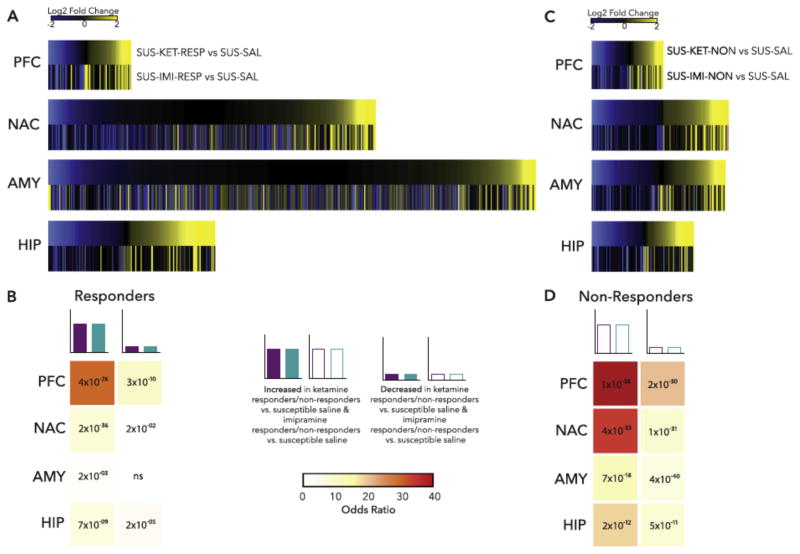
Initial work in animal models of depression utilized DNA microarray technology, with RNA-seq increasingly used in recent years. The latter has the marked advantage of quantifying expression changes in individual splice variants of a gene as well as in several types of non-coding RNAs (e.g., microRNAs, long non-coding RNAs), which we now know play important roles in cell regulation. Moreover, transcripts revealed by RNA-seq can be better aligned with GWAS results, which often identify variants in regions of the genome that do not directly code for proteins. A recent study, for example, performed RNA-seq on four brain regions—NAc, mPFC, vHIP, and BLA—at three time points after chronic social defeat stress, separately examining susceptible and resilient subgroups of mice (Bagot et al., 2016a). A follow up study carried out RNA-seq on the same four brain regions after repeated imipramine or acute ketamine treatment of susceptible mice, examining mice that responded behaviorally or not to these treatments (Bagot et al., 2016b), one of the first efforts to study the molecular basis of treatment resistance in an animal model. These large datasets provide new insight into stress vulnerability and treatment responses. The data suggest that resilience is the far more plastic state, with many more genes showing altered expression in most brain regions in resilient mice compared with susceptible mice. From this perspective, susceptibility appears to represent, in part, a failure of this plasticity. Moreover, treatment response is complex, associated with the reversal of some of the changes in gene expression seen in susceptibility, with the induction of a subset of changes seen in resilience, and with the induction of many additional changes in gene expression not related to natural susceptibility or resilience (FIG 4). Even though repeated imipramine and acute ketamine reverse behavioral abnormalities in roughly an equal fraction of susceptible mice, largely distinct gene changes in distinct brain regions are associated with treatment response to the two drugs. Non-response, in contrast, is characterized by a lack of the gene expression changes seen in animals that respond to treatment as well as the induction of unique gene expression changes that might oppose treatment response. Each of these findings now defines numerous follow up studies to establish causal relationships among specific genes and behavioral outcomes, and to perform similar analyses on more sophisticated rodent models of treatment resistance involving multiple bouts of stress and multiple courses of antidepressant treatment.
Figure 4.
Example of transcriptomic analysis of treatment response vs. non-response in a mouse model. Mice were subjected to chronic social defeat stress. Susceptible mice were treated with repeated imipramine or single dose ketamine and responders and non-responders were identified. Four brain regions were then subjected to RNA-seq. (A) Heatmaps show the union of ketamine response (SUS-KET-RESP vs. SUS-SAL) and imipramine response (SUS-IMI-RESP vs. SUS-SAL) differentially expressed genes (DEGs) rank ordered by log2 fold change of ketamine response and scaled by relative number of DEGs. (B) Table of p value (text) and odds ratio (warmer colors indicating increasing odds ratio) for Fisher’s exact test for enrichment of ketamine response DEGs in imipramine response DEGs. (C) Heatmaps show the union of ketamine nonresponse (SUS-KET-NON vs. SUS-SAL) and imipramine nonresponse (SUS-IMI-NON vs. SUS-SAL) DEGs rank ordered by log2 fold change of ketamine nonresponse and scaled by relative number of DEGs. (D) Table of p value (text) and odds ratio (warmer colors indicating increasing odds ratio) for Fisher’s exact test for enrichment of ketamine nonresponse DEGs in imipramine nonresponse DEGs. *p<0.05. AMY, amygdala; HIP, hippocampus; NAC, nucleus accumbens; ns, nonsignificant; PFC, prefrontal cortex. From Bagot et al., 2016b.
RNA-seq analysis has also been used to define the developmental origins of individual differences in stress reactivity. Peripubertal environmental enrichment dampens behavioral responses to stress and associates with transcriptional changes in the VDG that are enriched for genes implicated in neurogenesis (Zhang et al., submitted). These findings are consistent with the proposed role for neurogenesis in defining individual differences in stress reactivity. Also, taken together with the studies of chronic social defeat stress, the results are consistent with the idea that a relative absence of plasticity renders individuals susceptible to chronic stress.
It is also now possible to use a variety of genetic reporter mice to study changes in gene expression in individual cell populations (e.g., Heiman et al., 2014). Using a mouse line targeting the highly stress-sensitive pyramidal neurons of the CA3 region of hippocampus, transcriptome-wide changes in response to acute and chronic stress were identified using RNA-seq (Gray et al., 2017). Further, this reporter line was crossed to mice harboring the BDNF Val66Met variation, which revealed highly distinct translational profiles of CA neurons to chronic stress as a function of BDNF activity that are associated with stress susceptibility (Gray et al. 2017). Interestingly, BDNF Met allele carriers display many of the same expression changes at baseline (prior to stress exposure) that are exhibited by normal (BDNF Val) mice after stress exposure, suggesting that the genetic polymorphism confers a stress-like phenotype at the molecular level (Gray et al., 2017).
RNA-seq studies of the VDG have begun to identify candidate genes that confer response or non-response to LAC, which as noted above exerts fast-acting antidepressant-like effects in rodent models (Bigio et al., 2016). Heatmap representation shows wide differences in global RNA expression profiles in VDG between FSL rats and their controls, FRL rats, that are partially reversed with LAC treatment, indicating that LAC corrected many of the gene expression changes associated with depressive-like phenotypes of FSL rats. Interestingly, the transcriptome of FSL rats resistant to a low dose of LAC after an acute stress spanned the RNA profile of both FSL and FRL rats. Although a large number of gene expression changes were rescued by LAC in the treatment-resistant FSL group, many genes remained unaltered and other genes changed as a result of the acute stress, consistent with findings in the chronic social defeat stress model noted above (Bagot et al., 2016b) that treatment resistance is an active process in which new clusters of genes are altered rather than solely a lack of correcting global RNA expression profiles in resistant individuals (Bigio et al., 2016).
In parallel with RNA-seq, our groups and others are beginning to employ several genome-wide approaches (e.g., ChIP-seq, ATAC-seq, whole-genome and TET-assisted bisulfite sequencing) to map the global epigenome in animal depression models (e.g., Hunter et al., 2012; Dias et al., 2014; Sun et al., 2015; Zhang et al., submitted). The utility of mapping the epigenome is based on the notion that long-lasting changes in gene expression will be reflected by chromatin modifications that provide novel insight into the mechanisms underlying the transcriptional regulation. Studies of chromatin modifications have the additional advantage of moving beyond measures of steady-state RNA levels by revealing genes that are “primed” or “desensitized” by one stress exposure to respond differently to a subsequent exposure to stress or antidepressant treatment. Studies of DNA methylation can identify particularly long-lasting epigenetic modifications of relevance for transcription that are candidate mechanisms for the sustained effects of environmental conditions (Meaney & Ferguson-Smith, 2010). Epigenetic investigations will therefore prove to be particularly helpful in characterizing the multiple hit models outlined above. As these genome-wide epigenome maps are generated, their overlay with RNA-seq data will provide an increasingly complete view of how chronic stress and antidepressant treatments control the transcriptional output of specific brain regions and of specific cell types (both neuronal and non-neuronal) within a given region. It will also be possible to integrate multiple platforms of gene and chromatin data with brain circuit and brain imaging data, for example, using the developmental trajectory of expression of given genes in the Allen Brain Atlas and studying the degree to which the same genes are regulated across the cortico-limbic circuitry.
Most RNA-seq datasets are analyzed for transcripts that show different levels of expression after stress or antidepressant treatment, while genome-wide epigenomic mapping data are analyzed for genomic regions that display significant differences in enrichment of a given chromatin modification. However, the very large size of these datasets (often involving many terabytes) allows more complex bioinformatics approaches such as weighted gene co-expression network analysis (WGCNA) (Parikshak et al., 2013; Song and Zhang, 2015). WGCNA identifies key driver or “hub” genes, which are inferred to control the expression of larger gene networks. This includes identifying genes that display a hub role selectively in a stress- or drug-treated state in regions comprising depression circuits (Gaiteri et al., 2014; Bagot et al., 2016a; Labonté et al., 2017).
Convergence Across Animal Models and Human Depression
Given the inherent limitations of animal models of depression, it is imperative to correlate the exploration of transcriptional and epigenetic mechanisms in animals with abnormalities seen in the depressed human brain. The latter approach is fraught with its own limitations, such as the difficulty in obtaining high quality tissue with short post-mortem intervals, and the complications of comorbid conditions and variable, complex personal histories of stress and treatment exposures. Thus, overlaying transcriptional and epigenetic data with human genetic findings (e.g., Ding et al., 2015; Fromer et al., 2016) has the unique advantage of drawing upon the strengths of both approaches. Such overlays offer the additional possibility of validating a given animal model, not only by recapitulating certain symptoms (referred to as face validity), but also by determining the degree to which transcriptional abnormalities in the depressed human brain are recapitulated in a given animal study in relation to human brain structure. For example, some of the same genes that are implicated as genetic risk factors for depression are also highly regulated in animal models (Box 2). This convergence provides important validation of the role of these specific genes in contributing to depression. At the same time, such analyses provide an important layer of validation of the human genetic findings, where numerous genes approach genome-wide significance, but appear to exert very small effects on risk. Convergence between these genes with transcriptomic regulation in animal models and depressed human brain would guide geneticists and neuroscientists alike on where to place the focus moving forward. However, it is important to emphasize that studies of genes in animal models should not be restricted to genes that also display known genetic variations in depression. Only a small subset of all such genes have yet been identified, each such gene contributes a minute fraction to overall heritability of depression and there is no reason to assume a priori that genes that are most robustly regulated by stress are the same that underlie genetic predisposition to stress susceptibility. One example is provided by Sdk1 (encoding the cell adhesion molecule sidekick 1), which serves as a hub gene in mouse depression models (Bagot et al., 2016a) and for which a genetic variant in humans moderates the relation between antenatal maternal mood and childhood behavioral problems (Gupta et al., submitted), even though there is no evidence that it is a risk gene for depression per se.
Box 2. Examples of Convergence Between Transcriptional Abnormalities in Rodent Depression Models and Genetic Risk Factors for Human Depression.
Although at very early stages, there is already an impressive convergence between transcriptional changes seen in animal models and human genetic findings and several of these convergent genes offer promising targets for antidepressant drug discovery. An important caveat of these analyses is the lack of information available about the genetic underpinnings of TRD as opposed to overall MDD. OLFM4 is one of the genetic loci associated with depression identified in the recent 23andMe study (Hyde et al., 2016). OLFM4 (encoding olfactomedin-4) is highly expressed in human amygdala and cortex and regulated by chronic social defeat stress primarily in the amygdala (Bagot et al., 2016a). Likewise, expression of OLFM4 is induced in VTA and NAc by early life stress and in the hippocampus by environmental enrichment. While OLFM4 does not appear to be abnormally expressed in the depressed human brain, it is part of a significant depression-related gene module in depressed human brain identified by WGCNA (Labonté et al., 2017). Olfactomedins are implicated in neurodevelopment through effects on cell adhesion and known to interact with AMPA glutamate receptors. Another olfactomedin gene, OLFM3, emerged as a significant target in a GWAS of working memory (Heck et al., 2014).
SLC6A15—also implicated in the 23andMe study (Hyde et al., 2016), encodes BOAT2, a sodium-dependent neutral amino acid transporter that is increased significantly in PFC in human depression and in early life stress models in rodents (Labonté et al., 2017). Slc6a15 is also part of a significant gene module associated with stress susceptibility vs. resilience in the mouse social defeat paradigm (Bagot et al., 2016a). An earlier GWAS revealed an association between an SLC6A15 variant and the risk for depression that was confirmed in a meta-analysis across additional independent samples (Kohli et al., 2011). The presence of a risk allele was associated with down-regulation of Scl6a15 expression in hippocampus, with alterations in hippocampal volume, HPA axis activity and performance on cognitive tasks (Kohli et al., 2011; Schuhmacher et al., 2013). Manipulation of SLC6A15 in mice alters hippocampal glutamate levels, and loss of the gene protected against negative effects of chronic stress, whereas overexpression increased stress vulnerability (Santarelli et al., 2015, 2016). Rare coding variants in SLC6A15 that increase proline uptake have been identified in a sample of major depression (Quast et al., 2013).
The PENK gene, identified in the 23andMe study (Hyde et al., 2016), encodes proenkephalin, and is part of a highly-ranked depression-related gene module in depressed human brain by WGCNA (Labonté et al., 2017). It is also a highly-ranked gene associated with stress susceptibility vs. resilience in mice (Bagot et al., 2016a) and is one of the genes in the DG most highly affected by environmental enrichment and SSRIs (Sillaber et al., 2014; Samuels et al., 2014; Zhang et al., submitted). Multiple opioid receptor systems have been implicated in depression (Lutz & Kieffer, 2013). which may relate to the unique antidepressant properties of tianeptine (see text).
This convergence analysis identifies several additional genes not previously associated with depression, including HACE1, MEF2C, MLF1, VRK1, and BAZ1A, and two long non-coding RNAs that are highly regulated in human depression datasets. BAZ1A (which encodes a chromatin remodeling protein) is of particular interest: polymorphisms at this locus correlate with treatment response to bupropion (Li et al., 2016), while Baz1a is consistently induced in the NAc of several chronic stress models in mice and in human depression, where its induction has been shown to mediate stress susceptibility, and its suppression antidepressant-like effects, in both males and females (Sun et al., 2015).
Another recent study (Dass et al., submitted) underscores the potential for convergence between human and animal datasets. The researchers used a publicly available GWAS dataset that examined genetic variants associated with addiction (Study of Addiction: Genetics and Environment or SAGE; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1) that includes measures of childhood adversity. Focusing only on subjects with a history of adversity permitted analysis of genetic variants associated with resilience (adversity/no addiction) and susceptibility (adversity/addiction). The resulting variants nominally (p<0.05) distinguishing resilient and susceptible individuals were clustered to form a polygenic risk score for susceptibility or “PRSsus” (Purcell et al. 2009), validated using a completely independent dataset. The validation showed that the PRSsus predicted the association between childhood adversity and externalizing symptoms in adulthood. In parallel, an RNA-seq study of VDG from mice in the chronic social defeat stress paradigm revealed transcripts that mapped to 275 genes that distinguished resilient and susceptible animals, of which 116 were also on the PRSsus derived from the human GWAS data, including the SDK1 gene noted above (Bagot et al., 2016a). These findings reflect the value of convergence between human and animal genomic data.
It is necessary to complement the transcriptomic and epigenetic analyses of brain tissue on which we focus in this review with several other “omic” analyses in both humans and animal models. Proteomic, metabolomic, and lipidomic studies are needed to examine other forms of regulation that contribute to the depressed state and its treatment. Likewise, it is necessary to study various measures not only in brain, but in blood or saliva as well in efforts to identify robust peripheral biomarkers for depression and treatment response. The fact that very different transcriptional changes are seen across several brain regions in stressed animals and depressed humans indicates the likelihood that different changes will predominate in peripheral tissues. However, in certain cases epigenetic or transcriptional signals associated with clinically-relevant environmental conditions are apparent in both blood and brain (Kundakovica et al., 2015). For example, epigenetic modifications of a GR gene promoter associated with early life adversity are apparent both in human hippocampus (McGowan et al., 2009) and human peripheral cells (Turecki & Meaney, 2016). Moreover, certain blood changes—even if different from those seen in brain—might reflect some aspect of brain function that could be used to parse distinct subtypes of the human syndrome or predict the responsiveness of individuals to one treatment vs. another. The profound co-morbidity of depression with immunologic, metabolic and cardiovascular function encourages such thinking.
Gene x Environment Interactions
Environmental influences such as developmental history, early life stress and repeated exposure to trauma, as well as perhaps stochastic events during development, contribute almost two thirds of the variance to depression (Otte et al., 2016). Moreover, the illness has a dynamic course, whereby exposure of a vulnerable individual to negative or traumatic events can trigger of initial episodes of depression. However, the association between a triggering event and a depressive episode becomes less evident in more severe recurrent cases (Cai et al., 2015), where the illness takes on a progressively worsening “life of its own” with relapse in the absence of environmental triggers. This could reflect a loss of adaptive plasticity (see above) in response to stress that results from the illness itself, leading to an increased propensity for relapse. In this scheme, genetic vulnerability, environmental factors, and maladaptive neuroplasticity each play an important and highly interactive role in the expression and dynamic course of the illness.
Childhood adversity strongly predicts the risk for depression and associates with increased “neuroticism” (Roy, 2002). Neuroticism refers to emotional instability and increased stress sensitivity, which influence the interaction between stressful life events and negative affect, including symptoms of depression and anxiety (Kendler et al., 2004). There is dramatic individual variation in the mental health outcomes subsequent to severe childhood adversity; at least part of this variation is explained by genetics (Zhang & Meaney, 2010; Klengel & Binder, 2013). For example, some studies have identified an interaction between childhood adversity and genetic variants in or near genes associated with serotonergic signaling, such as a length variation in the promoter region of the SLC6A4 gene, which encodes the serotonin transporter (Caspi et al., 2003; Karg et al., 2011). In these studies, the short ‘s’ allele, which reduces transcriptional efficiency and transporter expression (Lesch et al. 1996), associates weakly with increased sensitivity to childhood adversity (Caspi et al., 2003) and reduced treatment response (Keers & Uher, 2012). However, the latest and largest meta-analysis (Culverhouse et al, 2017) found no interaction between stress and the ‘s’ allele, which suggests that the interaction between stress and the serotonin transporter polymorphism is not broadly generalizable and may only be observed in limited situations such as after childhood trauma. Nevertheless, one consistent finding in all these studies is the profound effect of stress on increasing the risk for depression (Donaldson et al, 2016; Culverhouse et al, 2017). There is also evidence for enhanced amygdala sensitivity to threat, as assessed by fMRI, amongst s-allele carriers (Pezawas et al., 2005). Rhesus macaques bear an analogous length variation of the gene, and infants with the less transcriptionally active s-variant show higher emotional reactivity, greater sensitivity to stress and increased stress-related health outcomes. These effects are, in turn, highly dependent upon the early social rearing context, thus reflecting a gene x environment interaction. Consistent with this model, mice lacking Scl6a4 throughout life display increased depressive-like behaviors as adults and pharmacological blockade of the transporter in mice exclusively during early postnatal development increases adult depressive-like behavior in contrast to the antidepressant-like effects seen with adult treatment (Ansorge et al., 2004).
As with the serotonin transporter, genetic variations in certain serotonin receptors are implicated in depression or antidepressant response. A polymorphism in the promoter of the HTR1A gene (encoding the 5-HT1A receptor) associates with depression and treatment resistance. This HTR1A polymorphism influences 5-HT1A receptor levels during early postnatal development and the impact of the polymorphism is stronger when combined with early life stressors (Donaldson et al., 2016). Interestingly, the polymorphism influences the binding of a family of transcription factors to the promoter of the HTR1A gene, and the expression of one family member, Hes5, is modified in several depression models (Albert, 2012; Lemonde et al., 2003; and our unpublished data).
Meta-analyses of studies of candidate gene x environment interactions commonly report small effect sizes and suffer from many of the same limitations as those afflicting GWAS. Genes operate in networks and thus genetic influences are likely to be better reflected in analyses that capture a broad range of genetic vulnerability. One such approach is that of polygenic risk scores derived from large GWAS. A polygenic score based on GWASs of depression reflects the composite genetic burden for interactions with childhood adversity in predicting depression (Peyrot et al., 2014) and provides opportunities for novel genomic approaches. Qiu et al. (2017) used two independent cohorts and found that the polygenic risk scores for depression of the child, but not the mother, moderated the relation between antenatal maternal depression and fetal cortico-limbic development. Mining the offspring polygenic risk scores for those SNPs that significantly accounted for this G (genetic vulnerability for depression) x E (antenatal maternal depressive symptoms) interaction, mapped the SNPs to genes and performed gene pathway analyses with FDR correction. The results identified glutamate receptor signaling and the SNARE complex as candidate biological systems mediating the impact of antenatal maternal depression on fetal amygdala development. This study reflects the opportunities for novel, genomic approaches based on existing GWAS datasets that can then be aligned to animal models to directly explore manipulations of resulting candidate pathways.
How Can We Use This Multi-Scale Information to Generate Novel Antidepressants?
Integration of multi-scale analyses are already providing a compelling algorithm to help guide antidepressant drug discovery efforts. We are proposing a reverse translation approach that starts by recognizing the complexity of the human syndrome with a focus on TRD, employs a broad range of animal models, overlays gene regulatory data from these models with studies of the post-mortem depressed human brain and GWAS and integrates studies using human and animal neuroimaging together with genetic risk factors for the syndrome. We argue that the animal models should be made more complex by characterizing the molecular, cellular, circuit and behavioral consequences of multiple bouts of chronic stress interspersed with repeated antidepressant treatments, to more accurately reflect the life course of TRD in humans. Studies focusing on stress resilience vs. susceptibility represent a major advance that maps onto the human condition. To increase our understanding of treatment resistance, the focus should move away from relying on standard antidepressants (e.g., SSRIs, tricyclics, etc.). to validate our models, towards developing models where the efficacy of these antidepressants is either low to begin with or wanes with repeated episodes. Greater focus should be placed on molecular, cellular and circuit mechanisms of depression in our models that are not addressed by today’s antidepressants. This experimental focus is required to identify mechanisms of antidepressant action that apply to individuals who fail today’s standard treatments. Particular emphasis should be expended in the development of clinically-relevant models for TRD, which is perhaps the most pressing challenge.
Already, our initial analyses point to several convergent genes linked to depression in human genetic studies and showing interesting and consistent patterns of regulation in our combined animal datasets. While a great deal more work is needed, as outlined in this review, the data are beginning to suggest novel, tangible approaches to new therapeutics. Examples of such early “hits” are given in Box 2. Antagonists of the neutral amino acid transporter, SCL6A15 (BOAT2), strategies to restore the activity of proteins encoded by genes suppressed in the depressed state by the chromatin remodeling factor, BAZ1A, or direct manipulation of olfactomedins, as just three of many examples, would all be predicted to exert antidepressant activity. In addition, genome-wide analyses of transcriptional and epigenetic regulation across several rodent depression models are driving novel clinical trials. Examples include ezogabine, a potentiator of the KCNQ family of K+ channels, which is approved for use in the U.S. and abroad for the treatment of epilepsy although it is not used widely, as well as LAC, a naturally occurring metabolite discussed earlier. Early clinical trials with these agents in TRD are promising (e.g., Van Dam et al., 2016).
Another recent area of focus of the Depression Task Force is the atypical antidepressant tianeptine (McEwen et al., 2010), which, although not available in the US, has been used clinically in Europe, Asia, and South America for 30 years. In comparison to tricyclic antidepressants and SSRIs, tianeptine displays improved tolerability and reduced incidence and severity of side effects. There is also evidence that tianeptine acts more rapidly than typical antidepressants, with initial therapeutic benefits observed after only one week. Two recent small clinical trials have suggested that tianeptine may be efficacious in TRD, for example, patients refractory to SSRI monotherapy. Tianeptine’s direct molecular target has been unclear (McEwen et al., 2010), but it was recently shown to bind selectively to the human mu-opioid receptor (MOR) and to act as a full agonist at this receptor without inducing tolerance or dependence (Gassaway et al, 2014). In mice, genetic deletion or pharmacological blockade of MOR blocks the behavioral actions of tianeptine, including its antidepressant-like effects (Samuels et al., 2017). Work is now underway in our laboratories to study the antidepressant mechanisms of tianeptine as well as of novel, potentially non-addicting MOR agonists.
Conclusion and Future Directions
Recent advances in methodologies to study genetic and epigenetic mechanisms, as well as the functioning of precise brain microcircuits, prompt new optimism for our ability to parse the broad, heterogeneous syndrome of human depression into biologically-defined subtypes and to generate more effective and rapidly-acting treatments based on a knowledge of disease etiology and pathophysiology and circuit dynamics. We outline here a multi-dimensional systems biology approach to depression and TRD specifically that leverages diverse datasets spanning animals and humans across multiple brain regions and time points in the life cycle to identify novel targets for antidepressant drug discovery efforts.
Despite recent criticism of animal models of psychiatric disorders, some of it quite valid, there is simply no path forward without the judicious use of animals (Nestler and Hyman, 2010). In the absence of known depression-causing genes of very strong effect and high penetrance, it is simply impossible for studies of molecular mechanisms in cell-based models to be the major driving force of antidepressant drug discovery efforts. Rather, we articulate a highly coordinated effort that makes use of multiple animal models, with each model recognized for its utility but also its limitations. By combining multiple models across species it is possible to identify several convergent mechanisms that are crucial in mediating susceptibility vs. resilience as well as subtyping for treatment selection at all stages of illness. By incorporating multiple bouts of stress and exposure to various antidepressant treatments, across the lifespan of the individual, these models provide an ideal platform to test causal mechanisms mediating the development of treatment resistance.
Animal models are essential for several reasons. Animals are required to demonstrate the causal involvement of individual genes and molecules in depression-related outcomes. Animal models also allow for the control and manipulation of the environment, and for understanding the mechanisms by which a given environmental conditions (e.g., early life stress) promote widely divergent outcomes. Delineating precise gene x environment interactions will be an important step in moving the field forward. Finally, animal models are required to understand how a collection of genes, combined with environmental exposures, during a lifetime control the functioning of individual neuronal and non-neuronal cell types and ultimately the functioning of the neural circuits in which those neurons operate. The ability to navigate from gene to cell to circuit to behavior and back is revolutionizing neuroscience and promises fundamental advances in understanding and treating disease at long last.
This accumulating evidence from animal models must be overlaid with human datasets. As animal and human studies converge on candidate mechanisms for treating TRD, it will be essential to recapture the largely lost art of experimental human pharmacology. Psychopharmacological studies with humans are an essential link in translating research from preclinical models to the clinic, and yet have largely disappeared from our current landscape. Rather than relying solely on large scale clinical trials, the field needs to study the neural and behavioral effects of numerous novel mechanisms in humans. Such small clinical studies must be carefully designed to ensure that a bona fide answer is obtained: there must be evidence that the mechanism is being engaged in the human brain and the study must be sufficiently powered to detect meaningful outcomes. Such approaches will be augmented dramatically by the degree to which such trials can be targeted to subsets of TRD patients that display the deficits associated with the mechanism under study. We summarize in this review a small number of examples of how a multi-scale approach can identify novel, putative antidepressant mechanisms, some of which are now being studied in human populations. The hope and expectation is that this uniquely broad effort will overcome obstacles encountered over the past 60 years and provide fundamentally new paths forward to antidepressant treatments.
Acknowledgments
Preparation of this review was supported by the Hope for Depression Research Foundation (HDRF). The authors thank Dr. Steven Roose (Columbia University) for helpful discussions.
References
- Abbasi J. 23andMe, big data, and the genetics of depression. JAMA. 2016;317:14–16. doi: 10.1001/jama.2016.14136. [DOI] [PubMed] [Google Scholar]
- Akil H. Stressed and depressed. Nat Med. 2005;1:116–118. doi: 10.1038/nm0205-116. [DOI] [PubMed] [Google Scholar]
- Akil H, Brenner S, Kandel E, Kendler KS, King MC, Scolnick E, Watson JD, Zoghbi HY. Medicine. The future of psychiatric research: genomes and neural circuits. Science. 2010;327:1580–1581. doi: 10.1126/science.1188654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert PR. Transcriptional regulation of the 5-HT1A receptor: implications for mental illness. Phil Trans R Soc London Series B, Biol Sci. 2012;367:2402–2415. doi: 10.1098/rstb.2011.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anacker C, Hen R. Adult hippocampal neurogenesis and cognitive flexibility - linking memory and mood. Nat Rev Neurosci. 2017;18:335–346. doi: 10.1038/nrn.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anacker C, Scholz J, O’Donnell KJ, Allemagne-Grand R, Diorio J, Bagot R, Nestler EJ, Hen R, Lerch J, Meaney MJ. Neuroanatomical differences associated with stress susceptibility and resilience. Biol Psychiatry. 2015;79:840–849. doi: 10.1016/j.biopsych.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge MS, Zhou M, Lira A, Hen R, Gingrich JA. Early-life blockade of the 5-HT transporter alters emotional behavior in adult mice. Science. 2004;306:879–881. doi: 10.1126/science.1101678. [DOI] [PubMed] [Google Scholar]
- Bagot RC, Labonte B, Peña CJ, Nestler EJ. Epigenetic signaling in psychiatric disorders: stress and depression. Dialogues Clin Neurosci. 2014;16:281–295. doi: 10.31887/DCNS.2014.16.3/rbagot. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot RC, Parise EM, Peña CJ, Zhang HX, Maze I, Chaudhury D, Persaud B, Cachope R, Bolaños-Guzmán CA, Cheer JF, Deisseroth K, Han MH, Nestler EJ. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nat Commun. 2015;6:7062. doi: 10.1038/ncomms8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang JS, Huang XJ, Schlüter OM, Maze I, Peña CJ, Heller EA, Issler O, Wang MH, Song WM, Stein JL, Liu SC, Doyle MA, Scobie KN, Sun HS, Neve RL, Geschwind D, Dong Y, Shen L, Zhang B, Nestler EJ. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron. 2016a;90:969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot RC, Cates HM, Purushothaman I, Vialou V, Heler EA, Labonté B, Peña CJ, Shen L, Nestler EJ. Ketamine and imipramine reverse transcriptional signatures of susceptibility and induce resilience-specific gene expression profiles. Biol Psychiatry. 2016b;81:285–295. doi: 10.1016/j.biopsych.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa JM, Ferreira D, Melo I, Marques F, Cerqueira JJ, et al. The mood-improving actions of antidepressants do not depend on neurogenesis but are associated with neuronal remodeling. Mol Psychiatry. 2009;14:764–773. doi: 10.1038/mp.2008.119. [DOI] [PubMed] [Google Scholar]
- Bunney B, Li J, Walsh D, Stein R, Vawter M, Cartagena P, Barchas J, Schatzberg A, Myers R, Watson S, Akil H, Bunney W. Circadian dysregulation of clock genes: clues to rapid treatments in major depressive disorder. Molecular Psychiatry. 2015;20(1):48–55. doi: 10.1038/mp.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss C, Meaney MJ, Lupien S, Pruessner J. Maternal care modulates the relationship between prenatal risk and hippocampal volume. Journal of Neuroscience. 2007;27:2592–2595. doi: 10.1523/JNEUROSCI.3252-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss C, Davis EP, Shahbaba B, Pruessner JC, Head K, Sandman CA. Maternal cortisol over the course of pregnancy and subsequent child amygdala and hippocampus volumes and affective problems. Proc Natl Acad Sci USA. 2012;109(20):E1312–E1319. doi: 10.1073/pnas.1201295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard R, Kerman IA, Thompson RC, Jones EG, Bunney WE, Barchas JD, Schatzberg AF, Myers FM, Akil H, Watson SJ. Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol Psychiatry. 2011;16:634–646. doi: 10.1038/mp.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet CZ, Stein MB. Relationship of childhood maltreatment to the onset and course of major depression in adulthood. Depress Anxiety. 1999;9:169–174. [PubMed] [Google Scholar]
- Berton O, Nestler EJ. New approaches to antidepressant drug discovery: beyond monoamines. Nature Rev Neurosci. 2006;7:137–151. doi: 10.1038/nrn1846. [DOI] [PubMed] [Google Scholar]
- Bigio B, Mathe AA, Sousa VC, Svenningsson P, McEwen BS, Nasca C. Epigenetics and energetics in ventral hippocampus mediate rapid antidepressant action: Implications for treatment resistance. Proc Natl Acad Sci USA. 2016;113:7906–7911. doi: 10.1073/pnas.1603111113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N, Chang S, Li Y, Li Q, Hu J, Liang J, Song L, Kretzschmar W, Gan X, Nicod J, Rivera M, Deng H, Du B, Li K, Sang W, Gao J, Gao S, Ha B, Ho HY, Hu C, Hu J, Hu Z, Huang G, Jiang G, Jiang T, Jin W, Li G, Li K, Li Y, Li Y, Li Y, Lin YT, Liu L, Liu T, Liu Y, Liu Y, Lu Y, Lv L, Meng H, Qian P, Sang H, Shen J, Shi J, Sun J, Tao M, Wang G, Wang G, Wang J, Wang L, Wang X, Wang X, Yang H, Yang L, Yin Y, Zhang J, Zhang K, Sun N, Zhang W, Zhang X, Zhang Z, Zhong H, Breen G, Wang J, Marchini J, Chen Y, Xu Q, Xu X, Mott R, Huang GJ, Kendler K, Flint J. Molecular signatures of major depression. Curr Biol. 2015;25:1146–1156. doi: 10.1016/j.cub.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Cerdá M, Sagdeo A, Johnson J, Galea S. Genetic and environmental influences on psychiatric comorbidity: a systematic review. J Affect Disord. 2010;126(1–2):14–38. doi: 10.1016/j.jad.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattarji S, Tomar A, Suvrathan A, Ghosh S, Rahman MM. Neighborhood matters: divergent patterns of stress-induced plasticity across the brain. Nat Neurosci. 2015;18:1364–1375. doi: 10.1038/nn.4115. [DOI] [PubMed] [Google Scholar]
- Chaudhury D, Walsh JJ, Friedman AK, Juarez B, Ku SM, Koo JW, Ferguson D, Tsai HC, Pomeranz L, Christoffel DJ, Nectow AR, Ekstrand M, Domingos A, Mazei-Robison MS, Mouzon E, Lobo MK, Neve RL, Friedman JM, Russo SJ, Deisseroth K, Nestler EJ, Han MH. Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature. 2013;493:532–536. doi: 10.1038/nature11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, Myers RM, Bunney WE, Jr, Akil H, Watson SJ, Jones EG. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci USA. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffel DJ, Golden SA, Dumitriu D, Robison AJ, Janssen WG, Ahn HF, Krishnan V, Reyes CM, Han MH, Ables JL, Eisch AJ, Dietz DM, Ferguson D, Neve RL, Greengard P, Kim Y, Morrison JH, Russo SJ. IκB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J Neurosci. 2011;31:314–321. doi: 10.1523/JNEUROSCI.4763-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway CR, George MS, Sackheim HA. Toward an evidence-based, operational definition of treatment-resistant depression. When enough is enough. JAMA Psychiatry. 2017;74:9–10. doi: 10.1001/jamapsychiatry.2016.2586. [DOI] [PubMed] [Google Scholar]
- Covington HE, III, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, LaPlant Q, Mouzon E, Ghose S, Tamminga CA, Neve RL, Deisseroth K, Nestler EJ. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci. 2010;30:16082–16090. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock RC, Holtzheimer PE, Hu XP, Mayberg HS. Disease state prediction from resting state functional connectivity. Mag Resonance Me. 2009;62:1619–28. doi: 10.1002/mrm.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse RC, Saccone NL, Horton AC, Ma Y, Anstey KJ, Banaschewski T, et al. Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.44. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson NA, van den Oever JM, Korobko OB, Magarinos AM, de Kloet ER, McEwen B. Prior history of chronic stress changes the transcriptional response to glucocorticoid challenge in the dentate gyrus region of the male rat hippocampus. Endocrinology. 2013;154:3261–3272. doi: 10.1210/en.2012-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DJ, Samuels BA, Rainer Q, Wang JW, Marsteller D, Mendez I, Drew M, Craig DA, Guiard BP, Guilloux JP, et al. Neurogenesis-dependent and -independent effects of fluoxetine in an animal model of anxiety/depression. Neuron. 2009;62:479–493. doi: 10.1016/j.neuron.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean K, Stevens H, Mortensen PB, Murray RM, Walsh E, Pedersen PB. Full Spectrum of Psychiatric Outcomes Among Offspring With Parental History of Mental Disorder. Arch Gen Psychiatry. 2010;67:822–829. doi: 10.1001/archgenpsychiatry.2010.86. [DOI] [PubMed] [Google Scholar]
- De Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimäki T, Lin CF, Ma'ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnström K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Rüther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, Tomm NK, Turi GF, Losonczy A, Hen R. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagnostic Statistical Manual (DSM) American Psychiatric Association; Washington DC: 2013. [Google Scholar]
- Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, Scobie K, Bagot R, LaBonté B, Ribeiro E, Liu X, Kennedy P, Vialou V, Ferguson D, Peña C, Calipari ES, Koo JW, Mouzon E, Ghose S, Tamminga C, Neve R, Shen L, Nestler EJ. β-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. 2014;516:51–55. doi: 10.1038/nature13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H, French BJ, Lewis DA, Tseng GC, Sibille E. Molecular and genetic characterization of depression: overlap with other psychiatric disorders and aging. Mol Neuropsychiatry. 2015;1:1–12. doi: 10.1159/000369974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Piel DA, Santos TL, Richardson-Jones J, Leonardo ED, Beck SG, Champagne FA, Hen R. Developmental effects of serotonin 1A autoreceptors on anxiety and social behavior. Neuropsychopharmacology. 2014;39:291–302. doi: 10.1038/npp.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, le Francois B, Santos TL, Almli LM, Boldrini M, Champagne FA, Arango V, Mann JJ, Stockmeier CA, Galfalvy H, et al. The functional serotonin 1a receptor promoter polymorphism, rs6295, is associated with psychiatric illness and differences in transcription. Transl Psychiatry. 2016;6:e746. doi: 10.1038/tp.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A, Schatzberg AF, Sudheimer K, Keller J, Mayberg HS, Gunning FM, Alexopoulos GS, Fox MD, Pascual-Leone A, Voss HU, Casey BJ, Dubin MJ, Liston C. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2017;23(1):28–38. doi: 10.1038/nm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS. Pathophysiology of depression and innovative treatments: remodeling glutamatergic synaptic connections. Dialogues Clin Neurosci. 2014;16:11–27. doi: 10.31887/DCNS.2014.16.1/rduman. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman CH, Duman RS. Spine synapse remodeling in the pathophysiology and treatment of depression. Neurosci Lett. 2015;5(601):20–29. doi: 10.1016/j.neulet.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop BW, Kelley ME, McGrath CL, Craighead WE, Mayberg HS. Preliminary Findings Supporting Insula Metabolic Activity as a Predictor of Outcome to Psychotherapy and Medication Treatments for Depression. J Neuropsychiatry Clin Neurosci. 2015;27:237. doi: 10.1176/appi.neuropsych.14030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop B, Rajendra J, Craighead E, Kelley M, McGrath C, Choi K, Kinkead B, Nemeroff C, Mayberg H. Functional Connectivity of the Subcallosal Cingulate Identifies Differential Outcomes to Treatment with Cognitive Behavioral Therapy or Antidepressant Medicaiton for Major Depressive Disorder. Am J Psych. 2017;174:533–545. doi: 10.1176/appi.ajp.2016.16050518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, Jurjus GJ, Dieter L, Duman RS. Altered expression of synapse and glutamate related genes in postmortem hippocampus of depressed subjects. Int J Neuropsychopharmacol. 2013;16:69–82. doi: 10.1017/S1461145712000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci. 2014;16(1):43–61. doi: 10.31887/DCNS.2014.16.1/ydwivedi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entringer S, Buss C, Wadhwa PD. Prenatal stress, development, health and disease risk: A psychobiological perspective-2015 Curt Richter Award Paper. Psychoneuroendocrinology. 2015;62:366–375. doi: 10.1016/j.psyneuen.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, Lopez JF, Thompson RC, Meng F, Stead JD, Walsh DM, Myers RM, Bunney WE, Watson SJ, Jones EG, Akil H. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci USA. 2004;101:15506–15511. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS, Dong HW. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava GA. Can long-term treatment with antidepressant drugs worsen the course of depression? J Clin Psychiatry. 2003;64:123–133. doi: 10.4088/jcp.v64n0204. [DOI] [PubMed] [Google Scholar]
- Feder A, Nestler EJ, Charney DS. Psychobiology and molecular genetics of resilience. Nat Rev Neurosci. 2009;10:446–457. doi: 10.1038/nrn2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi EA, Zalocusky KA, Liston C, Grosenick L, Warden MR, Amatya D, Katovich K, Mehta H, Patenaude B, Ramakrishnan C, Kalanithi P, Etkin A, Knutson B, Glover GH, Deisseroth K. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351(6268):aac9698. doi: 10.1126/science.aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel SB, Waselus M, Clinton SM, Watson SJ, Akil H. Antecedents and consequences of drug abuse in rats selectively bred for high and low response to novelty. Neuropharmacology. 2014;76:425–436. doi: 10.1016/j.neuropharm.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Buckner RL, White MP, Greicius MD, Pascual-Leone A. Efficacy of transcranial magnetic stimulation targets for depression is related to intrinsic functional connectivity with the subgenual cingulate. Biol Psychiatry. 2012;72(7):595–603. doi: 10.1016/j.biopsych.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DD, Diorio J, Plotsky PM, Meaney MJ. Environmental enrichment reverses the effects of maternal separation on stress reactivity. J Neuroscience. 2002;22:7840–7843. doi: 10.1523/JNEUROSCI.22-18-07840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AK, Juarez B, Ku SM, Zhang H, Calizo RC, Walsh JJ, Chaudhury D, Zhang S, Hawkins A, Dietz DM, Murrough JW, Ribadeneira M, Wong EH, Neve RL, Han MH. KCNQ channel openers reverse depressive symptoms via an active resilience mechanism. Nat Commun. 2016;7:11671. doi: 10.1038/ncomms11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, Klei LL, Kramer R, Pinto D, Gümüş ZH, Cicek AE, Dang KK, Browne A, Lu C, Xie L, Readhead B, Stahl EA, Xiao J, Parvizi M, Hamamsy T, Fullard JF, Wang YC, Mahajan MC, Derry JM, Dudley JT, Hemby SE, Logsdon BA, Talbot K, Raj T, Bennett DA, De Jager PL, Zhu J, Zhang B, Sullivan PF, Chess A, Purcell SM, Shinobu LA, Mangravite LM, Toyoshiba H, Gur RE, Hahn CG, Lewis DA, Haroutunian V, Peters MA, Lipska BK, Buxbaum JD, Schadt EE, Hirai K, Roeder K, Brennand KJ, Katsanis N, Domenici E, Devlin B, Sklar P. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Sibille E. Differentially expressed genes in major depression reside on the periphery of resilient gene coexpression networks. Front Neurosci. 2011;5:95. doi: 10.3389/fnins.2011.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassaway MM, Rives ML, Kruegel AC, Javitch JA, Sames D. The atypical antidepressant and neurorestorative agent tianeptine is a μ-opioid receptor agonist. Transl Psychiatry. 2014;4:e411. doi: 10.1038/tp.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee DG, Gabard-Durnam LJ, Flannery J, Humphreys KL, Telzer EH, Hare TA, Bookheimer SY, Tottenham N. Early developmental emergence of human amygdala–prefrontal connectivity after maternal deprivation. Proc Natl Acad Sci USA. 2013;110:15638–15643. doi: 10.1073/pnas.1307893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349:1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldapple K, Segal Z, Garson C, Lau M, Bieling P, Kennedy S, Mayberg H. Modulation of cortical-limbic pathways in major depression: treatment specific effects of Cognitive Behavior Therapy compared to paroxetine. Arch Gen Psych. 2004;61:34–41. doi: 10.1001/archpsyc.61.1.34. [DOI] [PubMed] [Google Scholar]
- Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2014;19:1171–1178. doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JG, Rubin TG, Kogan JF, Marrocco J, Weidmann J, Lindkvist S, Lee FS, Schmidt EF, McEwen BS. Translational profiling of stress-induced neuroplasticity in the CA3 pyramidal neurons of BDNF Val66Met mice. Mol Psychiatry. 2017 doi: 10.1038/mp.2016.219. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, Tye KM, Anikeeva P, Malenka RC, Deisseroth K. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackman DA, Farah MJ, Meaney MJ. Socioeconomic status and the brain: mechanistic insights from human and animal research. Nat Rev Neurosci. 2010 Sep;11(9):651–659. doi: 10.1038/nrn2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon N, Larson K, Lu M, Tullis E, Russ S. Lifecourse health development: past, present and future. Maternal Child Health J. 2014;18:344–365. doi: 10.1007/s10995-013-1346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamani C, Amorim BO, Wheeler AL, Diwan M, Driesslein K, Covolan L, Butson CR, Nobrega JN. Deep brain stimulation in rats: different targets induce similar antidepressant-like effects but influence different circuits. Neurobiol Dis. 2014;71:205–214. doi: 10.1016/j.nbd.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AZ, Gordon JA. Long-range neural synchrony in behavior. Annu Rev Neurosci. 2015;38:171–194. doi: 10.1146/annurev-neuro-071714-034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck A, Fastenrath M, Ackermann S, Auschra B, Bickel H, Coynel D, Gschwind L, Jessen F, Kaduszkiewicz H, Maier W, Milnik A, Pentzek M, Riedel-Heller SG, Ripke S, Spalek K, Sullivan P, Vogler C, Wagner M, Weyerer S, Wolfsgruber S, de Quervain DJ, Papassotiropoulos A. Converging genetic and functional brain imaging evidence links neuronal excitability to working memory, psychiatric disease, and brain activity. Neuron. 2014;81(5):1203–1213. doi: 10.1016/j.neuron.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronner SR, Rodriguez-Romaguera J, Quirk GJ, Groenewegen HJ, Haber SN. Circuit-Based Corticostriatal Homologies Between Rat and Primate. Biol Psychiatry. 2016;80(7):509–521. doi: 10.1016/j.biopsych.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9(6):1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heshmati M, Russo SJ. Anhedonia and the brain reward circuitry in depression. Curr Behav Neurosci Rep. 2015;2(3):146–153. doi: 10.1007/s40473-015-0044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AS, Sahay A, Hen R. Increasing Adult Hippocampal Neurogenesis is Sufficient to Reduce Anxiety and Depression-Like Behaviors. Neuropsychopharmacology. 2015;40:2368–2378. doi: 10.1038/npp.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, Magida J, Brancato A, Takahashi A, Flanigan ME, Ménard C, Aleyasin H, Koo JW, Lorsch ZS, Feng J, Heshmati M, Wang M, Turecki G, Neve R, Zhang B, Shen L, Nestler EJ, Russo SJ. Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. J Neurosci. 2015a;35:16362–16376. doi: 10.1523/JNEUROSCI.1392-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes GE, Kana V, Menard C, Merad M, Russo SJ. Neuroimmune mechanisms of depression. Nat Neurosci. 2015b;18:1386–1393. doi: 10.1038/nn.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman R, Mague SD, Li Q, Katz BM, Michel N, Lin L, Wang J, David LK, Blount C, Chandy R, Carlson D, Ulrich K, Carin L3, Dunson D, Kumar S, Deisseroth K, Moore SD1, Dzirasa K. Dysregulation of Prefrontal Cortex-Mediated Slow-Evolving Limbic Dynamics Drives Stress-Induced Emotional Pathology. Neuron. 2016;91(2):439–452. doi: 10.1016/j.neuron.2016.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RG, Murakami G, Dewell S, Seligsohn M, Baker ME, Datson NA, McEwen BS, Pfaff DW. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA. 2012;109:17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, Tung JY, Hinds DA, Perlis RH, Winslow AR. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyett MP, Breakspear MJ, Friston KJ, Guo CC, Parker GB. Disrupted effective connectivity of cortical systems supporting attention and interoception in melancholia. JAMA Psychiatry. 2015;72(4):350–358. doi: 10.1001/jamapsychiatry.2014.2490. [DOI] [PubMed] [Google Scholar]
- Insel N, Pilkiw M, Nobrega JN, Hutchison WD, Takehara-Nishiuchi K, Hamani C. Chronic deep brain stimulation of the rat ventral medial prefrontal cortex disrupts hippocampal-prefrontal coherence. Exp Neurol. 2015;269:1–7. doi: 10.1016/j.expneurol.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgor C, Kabbaj M, Akil H, Watson SJ. Delayed effects of chronic, variable stress during peripubertal-juvenile period on hippocampal morphology, cognitive and stress axis functions in rats. Hippocampus. 2004;14:636–648. doi: 10.1002/hipo.10207. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Jin J, Maren S. Fear renewal preferentially activates ventral hippocampal neurons projecting to both amygdala and prefrontal cortex in rats. Sci Rep. 2015;5:8388. doi: 10.1038/srep08388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Adams DH, Simen A, Simen BB, Rajkowska G, Stockmeier CA, Overholser JC, Meltzer HY, Jurjus GJ, Konick LC, Newton SS, Duman RS. Gene expression profiling in postmortem prefrontal cortex of major depressive disorder. J Neurosci. 2007;27:13329–13340. doi: 10.1523/JNEUROSCI.4083-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keers R, Uher R. Gene–environment interaction in major depression and antidepressant treatment response. Curr Psychiatry Rep. 2012;14(2):129–137. doi: 10.1007/s11920-011-0251-x. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, et al. Major depression and generalized anxiety disorder. Same genes, (partly) different environments? Arch Gen Psychiatry. 1992;49:716–722. doi: 10.1001/archpsyc.1992.01820090044008. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Kuhn J, Prescott CA. The interrelationship of neuroticism, sex, and stressful life events in the prediction of episodes of major depression. Am J Psychiat. 2004;161:631–636. doi: 10.1176/appi.ajp.161.4.631. [DOI] [PubMed] [Google Scholar]
- Khan AA, Jacobson KC, Gardner CO, Prescott CA, Kendler KS. Personality and comorbidity of common psychiatric disorders. Br J Psychiatry. 2005;186:190–196. doi: 10.1192/bjp.186.3.190. [DOI] [PubMed] [Google Scholar]
- Kheirbek MA, Drew LJ, Burghardt NS, Costantini DO, Tannenholz L, Ahmari SE, Zeng H, Fenton AA, Hen R. Differential Control of Learning and Anxiety along the Dorsoventral Axis of the Dentate Gyrus. Neuron. 2013;77:955–968. doi: 10.1016/j.neuron.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempan TA, Sequeira A, Canetti L, Lalovic A, Ernst C, Ffrench-Mullen J, Turecki G. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol Psychiatry. 2009;14:175–189. doi: 10.1038/sj.mp.4002110. [DOI] [PubMed] [Google Scholar]
- Klengel T, Binder EB. Gene-environment interactions in major depressive disorder. Can J Psychiatry. 2013;58:76–83. doi: 10.1177/070674371305800203. [DOI] [PubMed] [Google Scholar]
- Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K, Czamara D, Alexander M, Salyakina D, Ripke S, Hoehn D, Specht M, Menke A, Hennings J, Heck A, Wolf C, Ising M, Schreiber S, Czisch M, Müller MB, Uhr M, Bettecken T, Becker A, Schramm J, Rietschel M, Maier W, Bradley B, Ressler KJ, Nöthen MM, Cichon S, Craig IW, Breen G, Lewis CM, Hofman A, Tiemeier H, van Duijn CM, Holsboer F, Müller-Myhsok B, Binder EB. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70:252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollack-Walker S, Don C, Watson SJ, Akil H. Differential expression of c-fos mRNA within neurocircuits of male hamsters exposed to acute or chronic defeat. J Neuroendocrinol. 1999;11(7):547–559. doi: 10.1046/j.1365-2826.1999.00354.x. [DOI] [PubMed] [Google Scholar]
- Kozak MJ, Cuthbert BN. The NIMH research domain criteria initiative: background, issues, and pragmatics. Psychophysiology. 2016;53:286–97. doi: 10.1111/psyp.12518. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ, Laplant Q, Graham A, Lutter M, Lagace DC, Ghose S, Reister R, Tannous P, Green TA, Neve RL, Chakravarty S, Kumar A, Eisch AJ, Self DW, Lee FS, Tamminga CA, Cooper DC, Gershenfeld HK, Nestler EJ. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, Champagne FA. DNA methylation of BDNF as a biomarker of early-life adversity. Proc Nat Acad Sci USA. 2015;112:6807–6813. doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B, Engmann O, Purushothaman I, Hodes GE, Lorsch ZS, Hamilton PJ, Calipari ES, Scarpa JR, Loh YHE, Issler O, Kronmna H, Walker D, Pfau M, Doyle M, Neve R, Russo S, Kazarskis A, Mechawar N, Turecki G, Zhang B, Shen L, Tamminga C, Nestler EJ. Sex-specific transcriptional signatures in human depression. Nat Med. 2016 doi: 10.1038/nm.4386. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalovic A, Klempan T, Sequeira A, Luheshi G, Turecki G. Altered expression of lipid metabolism and immune response genes in the frontal cortex of suicide completers. J Affect Disord. 2010;120:24–31. doi: 10.1016/j.jad.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Lau T, Bigio B, Zelli D, McEwen BS, Nasca C. Stress-induced structural plasticity of medial amygdala stellate neurons and rapid prevention by a candidate antidepressant. Mol Psychiatry. 2017;22:227–234. doi: 10.1038/mp.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonde S, Turecki G, Bakish D, Du L, Hrdina PD, Bown CD, Sequeira A, Kushwaha N, Morris SJ, Basak A, et al. Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J Neurosci. 2003;23:8788–8799. doi: 10.1523/JNEUROSCI.23-25-08788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas JD, Schatzberg AF, Jones EG, Myers RM, Watson SJ, Jr, Akil H, Bunney WE. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci USA. 2013;110:9950–9955. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Tian C, Seabrook GR, Drevets WC, Narayan VA. Analysis of 23andMe antidepressant efficacy survey data: implication of circadian rhythm and neuroplasticity in bupropion response. Transl Psychiatry. 2016;6:e889. doi: 10.1038/tp.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby JL, Barch DM, Belden A, Gaffrey MS, Tilman R, Babb C, Nishino T, Suzuki H, Botteron KN. Maternal support in early childhood predicts larger hippocampal volumes at school age. Proc Natl Acad Sci USA. 2012;109(8):2854–2859. doi: 10.1073/pnas.1118003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby JL, Belden A, Harms MP, Tillman R, Barch DM. Preschool is a sensitive period for the influence of maternal support on the trajectory of hippocampal development. Proc Natl Acad Sci USA. 2016 doi: 10.1073/pnas.1601443113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz PE, Kieffer BL. Opioid receptors: distinct roles in mood disorders. Trends Neurosci. 2013;36:195–206. doi: 10.1016/j.tins.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, Heath AC, Holsboer F, Lucae S, Madden PA, Martin NG, McGuffin P, Muglia P, Noethen MM, Penninx BP, Pergadia ML, Potash JB, Rietschel M, Lin D, Müller-Myhsok B, Shi J, Steinberg S, Grabe HJ, Lichtenstein P, Magnusson P, Perlis RH, Preisig M, Smoller JW, Stefansson K, Uher R, Kutalik Z, Tansey KE, Teumer A, Viktorin A, Barnes MR, Bettecken T, Binder EB, Breuer R, Castro VM, Churchill SE, Coryell WH, Craddock N, Craig IW, Czamara D, De Geus EJ, Degenhardt F, Farmer AE, Fava M, Frank J, Gainer VS, Gallagher PJ, Gordon SD, Goryachev S, Gross M, Guipponi M, Henders AK, Herms S, Hickie IB, Hoefels S, Hoogendijk W, Hottenga JJ, Iosifescu DV, Ising M, Jones I, Jones L, Jung-Ying T, Knowles JA, Kohane IS, Kohli MA, Korszun A, Landen M, Lawson WB, Lewis G, Macintyre D, Maier W, Mattheisen M, McGrath PJ, McIntosh A, McLean A, Middeldorp CM, Middleton L, Montgomery GM, Murphy SN, Nauck M, Nolen WA, Nyholt DR, O'Donovan M, Oskarsson H, Pedersen N, Scheftner WA, Schulz A, Schulze TG, Shyn SI, Sigurdsson E, Slager SL, Smit JH, Stefansson H, Steffens M, Thorgeirsson T, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Völzke H, Weilburg JB, Willemsen G, Zitman FG, Neale B, Daly M, Levinson DF, Sullivan PF. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS. Modulating Dysfunctional Limbic-Cortical Circuits in Depression: Towards Development of Brain-based Algorithms for Diagnosis and Optimised Treatment. Br Med Bull. 2003;65:193–207. doi: 10.1093/bmb/65.1.193. [DOI] [PubMed] [Google Scholar]
- Mayberg HS. Targeted Modulation of Neural Circuits: A New Treatment Strategy for Depression. J Clin Inv. 2009;119(4):717–25. doi: 10.1172/JCI38454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS, Lozano A, Voon V, Kennedy S, McNeely H, Hamani C, Schwalb J, Seminowicz D. Deep Brain Stimulation for Treatment Resistant Depression. Neuron. 2005;45:651–660. doi: 10.1016/j.neuron.2005.02.014. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Protective and Damaging Effects of Stress Mediators. New England J Med. 1998;338:171–179. doi: 10.1056/NEJM199801153380307. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Karatsoreos IN. Sleep Deprivation and Circadian Disruption: Stress, Allostasis, and Allostatic Load. Sleep medicine clinics. 2015;10:1–10. doi: 10.1016/j.jsmc.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Milner TA. Understanding the broad influence of sex hormones and sex differences in the brain. J Neurosci Res. 2017;95:24–39. doi: 10.1002/jnr.23809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Chattarji S, Diamond DM, Jay TM, Reagan LP, Svenningsson P, Fuchs E. The neurobiological properties of tianeptine (Stablon): from monoamine hypothesis to glutamatergic modulation. Mol Psychiatry. 2010;15(3):237–249. doi: 10.1038/mp.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Morrison JH. The brain on stress: Vulnerability and plasticity of the prefrontal cortex over the life course. Neuron. 2013;79:16–29. doi: 10.1016/j.neuron.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Bowles NP, Gray JD, Hill MN, Hunter RG, Karatsoreos IN, Nasca C. Mechanisms of stress in the brain. Nat Neurosci. 2015a;18:1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Gray JD, Nasca C. 60 years of neuroendocrinology: Redefining neuroendocrinology: stress, sex and cognitive and emotional regulation. J Endocrinol. 2015b;226:T67–83. doi: 10.1530/JOE-15-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Gray J, Nasca C. Recognizing Resilience: Learning from the Effects of Stress on the Brain. Neurobiology of Stress. 2015c;1:1–11. doi: 10.1016/j.ynstr.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO1, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CL, Kelley ME, Holtzheimer PE, Dunlop BW, Craighead WE, Franco AR, Craddock RC, Mayberg HS. Toward a Neuroimaging Treatment Selection Biomarker for Major Depressive Disorder. JAMA Psychiatry. 2013;70(8):821–829. doi: 10.1001/jamapsychiatry.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CL, Kelley ME, Dunlop BW, Holtzheimer PE, Craighead WE, Mayberg HS. Pretreatment Brain States Identify Likely Failures to Standard Treatments for Depression. Biol Psychiatry. 2014;76(7):527–535. doi: 10.1016/j.biopsych.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Ferguson-Smith A. Epigenomic regulation of the neural transcriptome: The meaning of the marks. Nature Neuroscience. 2010;13:1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Menke A, Binder EB. Gene expression studies in major depression. Curr Psychiatry Rep. 2010;12:135–144. doi: 10.1007/s11920-010-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM, Noland RC, Kovalik JP, Seiler SE, Davies MN, DeBalsi KL, Ilkayeva OR, Stevens RD, Kheterpal I, Zhang J, Covington JD, Bajpeyi S, Ravussin E, Kraus W, Koves TR, Mynatt RL. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012;15:764–777. doi: 10.1016/j.cmet.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasca C, Xenos D, Barone Y, Caruso A, Scaccianoce S, Matrisciano F, Battaglia G, Mathé AA, Pittaluga A, Lionetto L, Simmaco M, Nicoletti F. L-acetylcarnitine causes rapid antidepressant effects through the epigenetic induction of mGlu2 receptors. Proc Natl Acad Sci USA. 2013;110:4804–4809. doi: 10.1073/pnas.1216100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasca C, Zelli D, Bigio B, Piccinin S, scaccianoce S, Nistico R, McEwen BS. Stress dynamically regulates behavior and glutamatergic gene expression in hippocampus by opening a window of epigenetic plasticity. Proc Natl Acad Sci USA. 2015;112:14960–14965. doi: 10.1073/pnas.1516016112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni V, Uher R, Danese A. Childhood maltreatment predicts unfavorable course of illness and treatment outcome in depression: A met-analysis. Am J Psychiatry. 2012;169:141–151. doi: 10.1176/appi.ajp.2011.11020335. [DOI] [PubMed] [Google Scholar]
- Nautiyal KMH, Hen R. Serotonin receptors in depression: from A to B. F1000Research. 2017;6:123. doi: 10.12688/f1000research.9736.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nature Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble KG, Houston SM, Brito NH, Bartsch H, Kan E, Kuperman JM, et al. Family income, parental education and brain structure in children and adolescents. Nature neuroscience. 2015;2:773–8. doi: 10.1038/nn.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KJ, Meaney MJ. Fetal origins of mental health: The Developmental Origins of Health and Disease Hypothesis. Am J Psychiatry. 2017;174:319–328. doi: 10.1176/appi.ajp.2016.16020138. [DOI] [PubMed] [Google Scholar]
- Okuyama T, Kitamura T, Roy DS, Itohara S, Tonegawa S. Ventral CA1 neurons store social memory. Science. 2016;353:1536–1541. doi: 10.1126/science.aaf7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olfson M, Blanco C, Marcus SC. Treatment of Adult Depression in the United States. JAMA Intern Med. 2016;176:1482–1491. doi: 10.1001/jamainternmed.2016.5057. [DOI] [PubMed] [Google Scholar]
- Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, Mohr DC, Schatzberg AF. Major depressive disorder. Nat Rev Dis Primers. 2016;2:16065. doi: 10.1038/nrdp.2016.65. [DOI] [PubMed] [Google Scholar]
- Padilla-Coreano N, Bolkan SS, Pierce GM, Blackman DR, Hardin WD, Garcia-Garcia AL, Spellman TJ, Gordon JA. Direct Ventral Hippocampal-Prefrontal Input Is Required for Anxiety-Related Neural Activity and Behavior. Neuron. 2016;89:857–866. doi: 10.1016/j.neuron.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt GM, Nguyen R, Bang JY, Aqrabawi A, Tran MM, Seo DK, Richards BA, Kim JC. Bidirectional Control of Anxiety-Related Behaviours in Mice: Role of Inputs Arising from the Ventral Hippocampus to the Lateral Septum and Medial Prefrontal Cortex. Neuropsychopharmacology. 2017;42:1715–1728. doi: 10.1038/npp.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña CJ, Kronman HG, Walker DM, Cates HM, Bagot RC, Purushothaman I, Issler O, Loh YE, Leong T, Kiraly DD, Goodman E, Neve RL, Shen L, Nestler EJ. Early life stress confers lifelong susceptibility in mice via ventral tegmental area OTX2. Science. 2017;356:1185–1188. doi: 10.1126/science.aan4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Sepulveda JA, Flagel SB, Garcia-Fuster MJ, Slusky RJ, Aldridge JW, Watson SJ, Akil H. Differential impact of a complex environment on positive affect in an animal model of individual differences in emotionality. Neuroscience. 2013;248:436–447. doi: 10.1016/j.neuroscience.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Cai N, Bigdeli TB, Li Y, Reimers M, Nikulova A, Webb BT, Bacanu SA, Riley BP, Flint J, Kendler KS. The genetic architecture of major depressive disorder in Han Chinese women. JAMA Psychiatry. 2016;74:162–168. doi: 10.1001/jamapsychiatry.2016.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrot WJ, Milaneschi Y, Abdellaoui A, Sullivan PF, Hottenga JJ, Boomsma DI, et al. Effect of polygenic risk scores on depression in childhood trauma. Br J Psychiatry. 2014;205:113–119. doi: 10.1192/bjp.bp.113.143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Meyer-Lindenberg A, Drabant EM, Verchinski BA, Munoz KE, Kolachana BS, Egan MF, Mattay VS, Hariri AR, Weinberger DR. 5-HTTLPR polymorphism impacts human cingulate-amygdala interactions: a genetic susceptibility mechanism for depression. Nat Neurosci. 2005;8:828–834. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- Picard M, Juster RP, McEwen BS. Mitochondrial allostatic load puts the 'gluc' back in glucocorticoids. Nat Rev Endocrinol. 2014;10:303–310. doi: 10.1038/nrendo.2014.22. [DOI] [PubMed] [Google Scholar]
- Pruessner JL, Champagne FA, Meaney MJ, Dagher A. Parental care and neuroendocrine and dopamine responses to stress in humans: A PET imaging study. J Neuroscience. 2004;24:2825–2831. doi: 10.1523/JNEUROSCI.3422-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olausson P, Kiraly DD, Gourley SL, Taylor JR. Persistent effects of prior chronic exposure to corticosterone on reward-related learning and motivation in rodents. Psychopharmacology (Berl) 2013;225(3):569–577. doi: 10.1007/s00213-012-2844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Cuboni S, Bader D, Altmann A, Weber P, Arloth J, Röh S, Brückl T, Ising M, Kopczak A, Erhardt A, Hausch F, Lucae S, Binder EB. Functional coding variants in SLC6A15, a possible risk gene for major depression. PLoS One. 2013;8:e68645. doi: 10.1371/journal.pone.0068645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Shen M, Buss C, Kwek K, Saw S-M, Chong Y-S, Gluckman PD, Wadhwa PD, Entringer S, Stner M, Gilmore JH, Karani N, Heim CM, O’Donnell KJ, Holbrook JD, Fortier MV, Meaney MJ the GUSTO study Group. Antenatal Maternal Depressive Symptoms and Neonatal Cortico-Limbic Morphology: Candidate Biological Mechanisms Revealed by Genetic Vulnerability. Cerebral Cortex. 2017 doi: 10.1093/cercor/bhx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet DH. The Flinders sensitive line rats: a genetic animal model of depression. Neurosci Biobehav Rev. 1993;17:51–68. doi: 10.1016/s0149-7634(05)80230-1. [DOI] [PubMed] [Google Scholar]
- Rasgon NL, McEwen BS. Insulin resistance-a missing link no more. Mol Psychiatry. 2016;21:1648–1652. doi: 10.1038/mp.2016.162. [DOI] [PubMed] [Google Scholar]
- Redondo RL, Kim J, Arons AL, Ramirez S, Liu X, Tonegawa S. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 2014;513:426–430. doi: 10.1038/nature13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Mayberg HS. Targeting abnormal neural circuits in mood and anxiety disorders: from the laboratory to the clinic. Nat Neurosci. 2007;10:1116–1124. doi: 10.1038/nn1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin-Graboi A, Bai J, Chen H, Hameed WB, Sim LW, Tint MT, Leutscher-Broekman B, Chong YS, Gluckman PD, Fortier MV, Meaney MJ, Qiu A. Prenatal maternal depression associates with microstructure of right amygdala in neonates at birth. Biological Psychiatry. 2013;74:837–844. doi: 10.1016/j.biopsych.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva-Posse, Choi KS, Holtzheimer PE, McIntyre CC, Gross RE, Chaturvedi A, Crowell AL, Garlow S, Mayberg HS, et al. Defining Critical White Matter Matter Pathways Mediating Successful Subcallosal Cingulate Deep Brain Stimulation for Treatment-Resistant Depression. Biol Psychiatry. 2014;76(12):963–969. doi: 10.1016/j.biopsych.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva-Posse P, Choi KS, Holtzheimer PE, Crowell AL, Garlow SJ, Rajendra JK, McIntyre CC, Gross RE, Mayberg HS. A connectomic approach forsubcallosal cingulate DBS surgery: prospective targeting in treatment resistant depression. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.59. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Bale TL. Germ cell origins of posttraumatic stress disorder risk: The transgenerational impact of parental stress experience. Biol Psychiatry. 2015;78:307–314. doi: 10.1016/j.biopsych.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A. Childhood trauma and neuroticism as an adult: possible implication for the development of the common psychiatric disorders and suicidal behaviour. Psychological Medicine. 2002;32(8):1471–1474. doi: 10.1017/s0033291702006566. [DOI] [PubMed] [Google Scholar]
- Rush AJ. STAR*D: what have we learned? Am J Psychiatry. 2007;164:201–204. doi: 10.1176/ajp.2007.164.2.201. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Charney DS. Next generation antidepressants. Proc Natl Acad Sci USA. 2103;110:4441–4442. doi: 10.1073/pnas.1301593110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14:609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels BA, Leonardo ED, Dranovsky A, Williams A, Wong E, Nesbitt AM, McCurdy RD, Hen R, Alter M. Global state measures of the dentate gyrus gene expression system predict antidepressant-sensitive behaviors. PLoS One. 2014;9:e85136. doi: 10.1371/journal.pone.0085136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels BA, Anacker C, Hu A, Levinstein MR, Pickenhagen A, Tsetsenis T, Madroñal N, Donaldson ZR, Drew LJ, Dranovsky A, et al. 5-HT1A receptors on mature dentate gyrus granule cells are critical for the antidepressant response. Nat Neurosci. 2015;18:1606–1611. doi: 10.1038/nn.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels BA, Nautiyal KM, Kruegel AC, Levinstein MR, Magalong VM, Gassaway MM, Grinnell SG, Han J, Ansonoff MA, Pintar JE, Javitch JA, Sames D, Hen R. The Behavioral Effects of the Antidepressant Tianeptine Require the Mu-Opioid Receptor. Neuropsychopharmacology. 2017 doi: 10.1038/npp.2017.60. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Santarelli S, Wagner KV, Labermaier C, Uribe A, Dournes C, Balsevich G, Hartmann J, Masana M, Holsboer F, Chen A, Müller MB, Schmidt MV. SLC6A15, a novel stress vulnerability candidate, modulates anxiety and depressive-like behavior: involvement of the glutamatergic system. Stress. 2016;19(1):83–90. doi: 10.3109/10253890.2015.1105211. [DOI] [PubMed] [Google Scholar]
- Santarelli S, Namendorf C, Anderzhanova E, Gerlach T, Bedenk B, Kaltwasser S, Wagner K, Labermaier C, Reichel J, Drgonova J, Czisch M, Uhr M, Schmidt MV. The amino acid transporter SLC6A15 is a regulator of hippocampal neurochemistry and behavior. J Psychiatr Res. 2015;68:261–269. doi: 10.1016/j.jpsychires.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Schuhmacher A, Lennertz L, Wagner M, Höfels S, Pfeiffer U, Guttenthaler V, Maier W, Zobel A, Mössner R. A variant of the neuronal amino acid transporter SLC6A15 is associated with ACTH and cortisol responses and cognitive performance in unipolar depression. Int J Neuropsychopharmacol. 2013;16:83–90. doi: 10.1017/S1461145712000223. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study of schizophrenia identifies five novel loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Klempan T, Canetti L, Ffrench-Mullen J, Benkelfat C, Rouleau A, Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Morgan L, Walsh DM, Cartagena PM, Choudary P, Li J, Schataberg AF, Watson SJ, Akil H, Myers RM, Jones EG, Bunney WE, Vawter MP. Gene expression changes in the prefrontal cortex, anterior cingulate cortex and nucleus accumbens of mood disorders subjects that committed suicide. PLoS One. 2012;7:e35367. doi: 10.1371/journal.pone.0035367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler SE, Koves TR, Gooding JR, Wong KE, Stevens RD, Ilkayeva OR, Wittmann AH, DeBalsi KL, Davies MN, Lindeboom L, Schrauwen P, Schrauwen-Hinderling VB, Muolo DM. Carnitine Acetyltransferase Mitigates Metabolic Inertia and Muscle Fatigue during Exercise. Cell Metab. 2015;22:65–76. doi: 10.1016/j.cmet.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Powers A, Bradley B, Ressler KJ. Gene × environment determinants of stress- and anxiety-related disorders. Annu Rev Psychol. 2016;67:239–261. doi: 10.1146/annurev-psych-122414-033408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillaber I, Panhuysen M, Henniger MS, Ohl F, Kuhne C, Putz B, Pohl T, Deussing JM, Paez-Pereda M, Holsboer F. Profiling of behavioral changes and hippocampal gene expression in mice chronically treated with the SSRI paroxetine. Psychopharmacology. 2008;200:557–572. doi: 10.1007/s00213-008-1232-6. [DOI] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N and the Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- Song WM, Zhang B. Multiscale embedded gene co-expression network analysis. PLoS Comput Biol. 2015;11:e1004574. doi: 10.1371/journal.pcbi.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, Boström E, Westerlund I, Vial C, Buchholz BA, et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, Moser EI. Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci. 2014;15:655–669. doi: 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- Sun H, Damez-Werno DM, Scobie KN, Shao NY, Dias C, Rabkin J, Koo JW, Korb E, Bagot RC, Ahn FH, Cahill ME, Labonté B, Mouzon E, Heller EA, Cates H, Golden SA, Gleason K, Russo SJ, Andrews S, Neve R, Kennedy PJ, Maze I, Dietz DM, Allis CD, Turecki G, Varga-Weisz P, Tamminga C, Shen L, Nestler EJ. ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat Med. 2015;21:1146–1153. doi: 10.1038/nm.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Sasaki T, Kato N, Kato T. Gene expression profiling of major depression and suicide in the prefrontal cortex of postmortem brains. Neurosci Res. 2008;60:184–191. doi: 10.1016/j.neures.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Teicher MH, Samosn JA. Annual Research Review: Enduring neurobiological effects of childhood abuse and neglect. Journal of Child Psychology and Psychiatry. 2016;57(3):241–266. doi: 10.1111/jcpp.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thase ME. Treatment-resistant depression: prevalence, risk factors, and treatment strategies. J Clin Psychiatry. 2011;72(5):e18. doi: 10.4088/JCP.8133tx4c. [DOI] [PubMed] [Google Scholar]
- Thase ME, Schwartz TL. Choosing medications for treatment-resistant depression based on mechanism of action. J Clin Psychiatry. 2015;76:720–727. doi: 10.4088/JCP.14052ah2c. [DOI] [PubMed] [Google Scholar]
- Thompson PM and the Alzheimer’s Disease Neuroimaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study (SYS) Group. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 8(2):153–182. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Meaney MJ. Effects of the social environment and stress on glucocorticoid receptor gene methylation: A systematic review. Biological Psychiatry. 2016;79:87–96. doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Clinton SM, Thompson RC, Watson SJ, Akil H. Fibroblast growth factor-2 (FGF-2) augmentation early in life alters hippocampal development and rescues the anxiety phenotype in vulnerable animals. Proc Natl Acad Sci USA. 2011;108(19):8021–8025. doi: 10.1073/pnas.1103732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Watson SJ, Akil H. The fibroblast growth factor family: neuromodulation of affective behavior. Neuron. 2012;76(1):160–174. doi: 10.1016/j.neuron.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam NT, Kautz M, Friedman AK, Han MH, Nestler EJ, Charney DS, Iosifescu DV, Murrough JW. The potassium channel modulator ezogabine decreases symptomatology and increases reward response in depression. SOBP Abs 2016 [Google Scholar]
- Wang SS, Kamphuis W, Huitinga I, Zhou JN, Swaab DF. Gene expression analysis in the human hypothalamus in depression by laser microdissection and real-time PCR: the presence of multiple receptor imbalances. Mol Psychiatry. 2008;13:786–799. doi: 10.1038/mp.2008.38. [DOI] [PubMed] [Google Scholar]
- Warren BL, Sial OK, Alcantara LF, Greenwood MA, Brewer JS, Rozofsky JP, Parise EM, Bolaños-Guzman CA. Altered gene expression and spine density in nucleus accumbens of adolescent and adult male mice exposed to emotional and physical stress. Dev Neurosci. 2014;36:250–260. doi: 10.1159/000362875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Lu X-Y, Liu L, Schafer G, Shieh K-R, Burke S, Robinson TE, Watson SJ, Seasholtz AF, Akil H. Glucocorticoid receptor overexpression in forebrain: a mouse model of increased emotional lability. Proceedings of the National Academy of Sciences. 2004;101(32):11851–11856. doi: 10.1073/pnas.0402208101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Fentress HM, Hoversten MT, Zhang L, Hebda-Bauer EK, Watson SJ, Seasholtz AF, Akil H. Early-life forebrain glucocorticoid receptor overexpression increases anxiety behavior and cocaine sensitization. Biol Psychiatry. 2012;71(3):224–31. doi: 10.1016/j.biopsych.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock M. The long-term behavioural consequences of prenatal stress. Neurosci Biobehav Rev. 2008;32(6):1073–1086. doi: 10.1016/j.neubiorev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Williams LM. Precision psychiatry: a neural circuit taxonomy for depression and anxiety. Lancet Psychiatry. 2016;3:472–480. doi: 10.1016/S2215-0366(15)00579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Debattista C, Duchemin AM, Schatzberg AF, Nemeroff CB. Childhood trauma predicts antidepressant response in adults with major depression: data from the randomized international study to predict optimized treatment for depression. Transl Psychiatry. 2016;6:e799. doi: 10.1038/tp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb ES, Franklin T, Iwata M, Duman RS. Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci. 2016;17:497–511. doi: 10.1038/nrn.2016.69. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) WHO reference number: WHO/MSD/MER/2017.2. 2017. Depression and Other Common Mental Disorders. [Google Scholar]
- Xu C, Krabbe S, Grundemann J, Botta P, Fadok JP, Osakada F, Saur D, Grewe BF, Schnitzer MJ, Callaway EM, et al. Distinct Hippocampal Pathways Mediate Dissociable Roles of Context in Memory Retrieval. Cell. 2016;167:961–972. e91. doi: 10.1016/j.cell.2016.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Meaney MJ. Epigenetics and the environmental regulation of the genome and its function. Annual Reviews of Psychology. 2010;61:439–466. doi: 10.1146/annurev.psych.60.110707.163625. [DOI] [PubMed] [Google Scholar]
- Zhu X, Need AC, Petrovski S, Goldstein DN. One gene, many neuropsychiatric disorders: lessons from Mendelian diseases. Nature Neuroscience. 2014;7:773–781. doi: 10.1038/nn.3713. [DOI] [PubMed] [Google Scholar]
- Zuckerman M, Kuhlman DM. Personality and risk-taking: common biosocial factors. J Pers. 2000;68(6):999–1029. doi: 10.1111/1467-6494.00124. [DOI] [PubMed] [Google Scholar]