Abstract
This study was aimed to evaluate the immunogenicity of recombinant plasmid deoxyribonucleic acid (DNA), pBud-H5-green fluorescent protein (GFP)-interferon-regulatory factor (IRF)3 following delivery using polyamidoamine (PAMAM) dendrimer and transactivator of transcription (TAT)-conjugated PAMAM dendrimer as well as the effect of IRF3 as the genetic adjuvant. BALB/c mice were vaccinated transdermally with pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3. The expression analysis of H5 gene from the blood by using quantitative real-time reverse transcriptase polymerase chain reaction confirmed the ability of PAMAM dendrimer as a carrier for gene delivery, as well as the ability of TAT peptide to enhance the delivery efficiency of PAMAM dendrimer. Mice immunized with modified PAMAM by TAT peptide showed higher hemagglutination inhibition titer, and larger CD3+/CD4+ T cells and CD3+/CD8+ T cells population, as well as the production of cytokines, namely, interferon (IFN)-γ, interleukin (IL)-2, IL-15, IL-12, IL-6, and tumor necrosis factor-α compared with those immunized with native PAMAM. These results suggest that the function of TAT peptide as a cell-penetrating peptide is able to enhance the gene delivery, which results in rapid distribution of H5 in the tissues of the immunized mice. Furthermore, pBud-H5-GFP co-expressing IRF3 as a genetic adjuvant demonstrated the highest hemagglutination inhibition titer besides larger CD3+/CD4+ and CD3+/CD8+ T cells population, and strong Th1-like cytokine responses among all the systems tested. In conclusion, TAT-PAMAM dendrimer-based delivery system with IRF3 as a genetic adjuvant is an attractive transdermal DNA vaccine delivery system utilized to evaluate the efficacy of the developed DNA vaccine in inducing protection during challenge with virulent H5N1 virus.
Keywords: influenza virus, DNA vaccine, vaccine delivery, dendrimer, TAT peptide
Introduction
Influenza virus as an enveloped virus belonging to the Orthomyxoviridae family is a major pathogen that represents an ongoing threat to several species as diverse as poultry, swine, and mammals, including humans, principally through their ability to cause respiratory morbidity and mortality. When an outbreak of avian influenza (AI) occurs in a high population density area, vaccination is one of the most effective and cost-beneficial interventions to prevent mortality and reduce morbidity from the infectious pathogen.1 The outcome of the implementation of a vaccination policy on the dynamics of infection is mainly to reduce the susceptibility to infection while at the same time reducing the amount of virus shed into the environment.2
Various kinds of experimental AI vaccines against H5N1 have been described. Categories of vaccines include the following: inactivated whole virus vaccines, live attenuated vaccines,3 recombinant subunit vaccines, and reverse genetic vaccines.4 While many of these vaccines have been shown to induce protective immunity in the laboratory under optimal conditions, the final proof of protection and efficacy is still anticipated. The overwhelming majority of AI vaccines produced and sold were of oil-emulsion-inactivated whole AI virus vaccines delivered via the parenteral route (subcutaneous or intramuscular), and less frequently, recombinant-vectored vaccines. With the difficulties in controlling the current influenza virus epizootic disease using existing strategies, including vaccination, there is a clear need to examine alternative vaccine strategies.5,6
With the advances in modern vaccine technologies, deoxyribonucleic acid (DNA) vaccines have been proposed as the potential solution in the development of effective vaccines since they are safer than traditional whole-killed viral vaccines, relatively inexpensive to manufacture and store, and have the potential to simultaneously stimulate humoral and cellular immune responses.7 Despite the advantages of DNA vaccine, there are only 4 licensed DNA vaccines being used in the veterinary field until now, including vaccines against the West Nile virus in horses,8 infectious hematopoietic necrosis in schooled salmon,9 canine melanoma,10 and growth hormone-releasing hormone product in swine.11 One of the key challenges in producing DNA vaccine is poor immunogenicity of the vaccine primarily due to poor tissue distribution and expressions following transfection of the transgene. However, generally it is believed that the development in DNA vaccine technology will reveal appropriate and customized vaccines to various disorders in the future, representing efficient and economically accessible prophylactic and therapeutic vaccines.7 Furthermore, the immunogenic potency of DNA-based vaccines can be significantly increased if the delivery of DNA is improved, and additional immunologic stimulant is provided. Hence, various carriers such as cationic liposomes, polysaccharides, and cationic polymers (ie, polyethyleneimine, dendrimers, and chitosan) have been used to enhance the uptake of DNA vaccine, and eventually its ability to induce immune responses.12,13 Generally, dendrimers have a high ionic interaction with DNA, hence are able to produce very stable DNA complexes.14 Polyamidoamine (PAMAM) dendrimers have also been studied as an effective transdermal carrier for different drugs and transgenes. For example, transdermal delivery of 8-methoxypsoralen (8-MOP) by using G3 and G4 PAMAM dendrimers behind the ear of mice showed better skin permeation and higher concentration of 8-MOP in the epidermis and dermis.15 Transdermal delivery of the plasmid DNA by collagen membrane-supported dendrimer/DNA were also associated with better gene expression in the mouse skin compared with uncomplexed plasmid DNA.16
Furthermore, the efficiency of PAMAM dendrimer in the delivery of DNA can be further enhanced by linking to cell-specific ligands, cell-penetrating peptide (CPP) and oligopeptide.17 One of the well-described CPP is the human immunodeficiency virus-1 (HIV-1) transactivator of transcription (TAT), where it has the ability to cross the plasma membrane of neighboring cells.18 Studies have shown dramatically higher and longer gene expression in animals administered with a combination of modified TAT peptide and cationic lipids or polycations.19,20 Moreover, genetic adjuvants are characterized as expression vectors of cytokines that are able to modulate immune responses when incorporated in a vaccine model. The immune modulatory effects of cytokines such as interferon-regulatory factors (IRFs), interleukin (IL)-1, IL-12, interferon (IFN)-α, IFN-γ, IL-6, IL-8, and granulocyte-macrophage colony-stimulating factor on DNA vaccine potency have been investigated by several researchers.21–23
Our in vitro study showed successful permeation of plasmid encoding the H5-green fluorescent protein (GFP) genes (pIRES-H5-GFP) as DNA vaccine with TAT-conjugated PAMAM dendrimer as the delivery platform across the artificial skin membrane, and in vitro transfection expression.24 The aim of this study was to investigate the potential of PAMAM dendrimer as a DNA vaccine carrier, and the effect of TAT peptide on the delivery efficiency of PAMAM while facilitating permeation of DNA vaccine through the skin of experimental laboratory animals. Moreover, the effects of IRF3 as a genetic adjuvant on the humoral and cellular immunity following delivery of the developed DNA plasmid were also evaluated in this study.
Materials and methods
Construction of recombinant DNA plasmids
The expression vector pBudCE4.1 was used to construct recombinant DNA plasmids expressing the H5-GFP and IRF3 genes. The H5-GFP genes were first cloned into the pBud expression vector downstream of the elongation factor (EF)-1α promoter to generate pBud-H5-GFP. The H5-GFP gene was amplified from the recombinant plasmid pIRES as a template using a pair of H5 specific primers listed in Table 1. The polymerase chain reaction (PCR) product of pIRES-H5-GFP was purified using PCR clean up Kit (Qiagen, Germantown, MD, USA), prior to double digestion with restriction enzymes. The purified PCR product and the expression vector pBudCE4.1 were digested with NotI and XhoI (Fermentas, Burlington, Canada). Thereafter, the DNA plasmid pBud-IRF3-H5-GFP was constructed by insertion of the IRF3 into pBud downstream of the cytomegalovirus (CMV) promoter in the pBud-H5-GFP to construct the pBud-IRF3-H5-GFP. The IRF3 gene was amplified from recombinant plasmid pBoost2 as a template using a pair of IRF3 specific primers listed in Table 1. The gene for IRF3 was inserted into the PstI and XbaI (Fermentas) sites downstream of the CMV promoter. Heat shock method was used to transform the constructed plasmids, pBud-IRF3-H5-GFP into the Escherichia coli Top 10 competent cells. The pBud-IRF3-H5-GFP construct was purified using an endotoxin-free plasmid purification kit (Qiagen, Valencia, CA, USA). The quantity and quality of the purified plasmids were assessed by optical density at 260 and 280 nm and by electrophoresis in 1% agarose gel (Promega, Madison, WI, USA).
Table 1.
Primers for amplification of the H5 and IRF3 genes
| Gene | Primer | Nucleotide sequence (5′ to 3′) |
|---|---|---|
| H5 | Forward | ATTGGCGGCCGCACCATGGAGAAAATAGTGC |
| Reverse | AGCTCTCGAGTTACTTGTACAGCTCGTCCATG | |
| IRF3 | Forward | ATTGCTGCAGACCATGGAAACCCCGAAACC |
| Reverse | AGCTTCTAGATCAGATATTTCCAGTGGCCTGG |
Abbreviation: IRF, interferon-regulatory factor.
Screening of the recombinant plasmids
To confirm the presence of the inserts, the extracted plasmids were subjected to single and double digestion by using appropriate restriction enzymes. Single digestion at the unique restriction enzyme sites within the recombinant plasmids was carried out to linearize and analyze the total size of the DNA, while double digestion was performed to release the inserted genes. The digestion reaction mixture was set up as follows: 1 μL of appropriate 10× reaction buffer, 1 μL restriction enzymes 5U/μL (Fermentas), 0.5 μg plasmid DNA and dH2O to a final volume of 10 μL, and then incubated at 37°C for 3 h. Finally, the digested plasmids were analyzed on 1% (w/v) agarose gel. Following confirmation of the inserts, the plasmid DNA was subjected to DNA sequencing (Medigene, Seoul, South Korea) to determine the sequences and the reading frames of the inserts. Nucleotide sequences of the inserts were evaluated by homology search against a database with Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast/) from the National Center for Biotechnology Information (NCBI), USA.
Preparation of PAMAM dendrimer-TAT-plasmid complex (polyplex)
The TAT-PAMAM/plasmid DNA polyplexes were prepared by mixing TAT-PAMAM and 2 μg of plasmid DNA, at N/P (TAT-PAMAM/DNA) ratio of 6:1 (+/−) based on our previous study in serum-free DMEM medium followed by 20 min incubation at room temperature.24
DNA immunization of BALB/c mice
Sixty 4-week-old BALB/c mice were divided into 6 groups. Mice in the treatment groups were vaccinated transdermally behind the ear twice at day 1 (start of the trial) and at day 14 post-immunization as a booster. For transdermal vaccination, the animals were anesthetized with ketamine (100 mg/kg):xylazine (10 mg/kg), and vaccine preparations in a volume of 50 μL were applied to the skin behind the ear, and spread over the entire area using the side of a pipette tip. Group 1 was left unimmunized and served as the negative control. Group 2 was vaccinated with pBud-PAMAM complex as plasmid and dendrimer control. Group 3 and 4 were vaccinated with the pBud-H5-GFP and PAMAM/pBud-H5-GFP complex, respectively. Group 5 was vaccinated with TAT-PAMAM/pBud-H5-GFP and finally, group 6 was vaccinated with TAT-PAMAM/pBud-H5-GFP-IRF3.
Blood samples were drawn from the tail and subjected to RNA extraction to quantify the H5 gene expression prior to vaccination, at days 3 and 7 after the first immunization, and finally, at days 14 and 28. The collected serum samples were subjected to cytokine evaluation and detection of antibody titer against H5. Three weeks after the booster immunization, mice were anesthetized intraperitoneally using ketamine (100 mg/kg):xylazine (10 mg/kg), and blood samples were collected using terminal heart puncture. The mice were sacrificed and their spleen were removed and kept at −80°C until further analysis. All animal procedures were performed according to Malaysian Law Act 772, Animal Welfare Act 2015 and were approved by the Animal Care and Use Committee of the Faculty of Veterinary Medicine (UPM/IACUC/AUP-R016/2015), Universiti Putra Malaysia (UPM).
Expression analysis of the H5 gene in the vaccinated mice
Total RNA was extracted from the blood of the vaccinated mice using QIAamp RNA blood mini Kit (Qiagen, USA) based on the manufacturer’s instructions. The quantitative real-time reverse transcriptase-PCR assay based on TaqMan probe was developed for quantification of H5 RNA from the blood of different vaccinated groups. The extracted RNA was transcribed into complementary DNA (cDNA) by reverse transcriptase using high capacity RNA to cDNA Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. A MyTaq TM HS mix kit (Bioline, Taunton, MA, USA) was mixed to a total volume of 20 μL with DNase-free water for the assay. The data was analyzed using the CFX manager version 3.1 software (Bio-Rad, Hercules, CA, USA). Absolute quantification methods were performed via the standard curves. The absolute copy numbers of H5 gene were determined based on a standard curve.
Determination of antibody titers against H5 by hemagglutination inhibition (HI) test
Hemagglutination (HA) assay was carried out using an inactivated AIV, H5N1 subtype (Harbin Weike Biotechnology Development Company, Harbin, China). Calculation of the HA endpoint is based on the highest dilution at which complete hemagglutination occurred, and was considered as 1 HA unit, while the HI titer is the reciprocal of the highest serum dilution that fully inhibit hemagglutination. The HA and HI assays were performed as per the World Organisation for Animal Health (OIE) procedure.
Determination of T cell responses using flow cytometry
Splenocytes were obtained by meshing the spleen using a 70 μL cell strainer (Sigma, St Louis, MO, USA) followed by centrifugation at 1,800× g for 5 min. The pellet was washed with PBS (pH =7.4), then resuspended in 1 mL lysis buffer, and was kept on ice for 5 min followed by another centrifugation step at 2,000× g for 5 min. After washing the pellet with PBS, 2.5×106 cells were resuspended in 100 μL PBS. Finally, the splenocytes were stained with mouse monoclonal antibodies specific for CD3 (fluorescein isothiocyanate [FITC]), CD4 (Phycoerythrin), and CD8 alpha (Allophycocyanin) followed by incubation on ice for 1 h. Then, the samples were washed with PBS and centrifuged at 400× g for 10 min and analyzed using the FACSCalibur™ flow cytometer (BD Biosciences, San Jose, CA, USA) and CellQuestPro™ software (BD Biosciences).
Quantification of cytokines expression levels using multiplex bead-based immunoassay
Expression levels of 9 cytokines, namely, pro-inflammatory (IL-1β, tumor necrosis factor [TNF]-α, and IL-6), Th1-like (IFN-γ, IL-2, IL-15, and IL-12), and Th2-like (IL-4 and IL-10) cytokines were determined from the serum samples using Mouse Cytokine/Chemokine Magnetic Bead Panel, MCYTOMAG-70K (Milliplex MAP Kit, Millipore Corporation, Bedford, MA, USA). The test was performed by using standard procedures according to the instructions provided by the manufacturer.
Statistical analysis
The mean values of the results were statistically analyzed by 1-way analysis of variance to determine the differences between treatment means at the 5% (P<0.05) significance level. Data was statistically analyzed using the Minitab software (Minitab, State College, PA, USA).
Results
Construction of pBud-H5-GFP-IRF3
In order to construct the DNA plasmid encoding IRF3 and H5-GFP genes, the H5-GFP genes were first cloned into pBud expression vector downstream of the EF-1α promoter to generate pBud-H5-GFP. Thereafter, the DNA plasmid of pBud-IRF3-H5-GFP was constructed by insertion of the IRF3 gene into pBud downstream of the CMV promoter in the pBud-H5-GFP. In order to verify the presence of the putative H5-GFP recombinants, the extracted plasmid from the randomly selected transformants was subjected to restriction enzyme analysis (Figure 1). Single digestion of pBud-H5-GFP with XhoI produced a single fragment of about 7,672 bp, which correlated with the expected size of the recombinant DNA plasmid. Double digestion of the plasmid with NotI and XhoI resulted in 2 fragments, 4,546 and 3,126 bp corresponding to the pBud CE4.1 and the H5-GFP gene, respectively.
Figure 1.
Analysis of the inserted gene (H5-GFP) by using restriction enzyme digestion and polymerase chain reaction.
Notes: M: GeneRulerTM DNA ladder Kit, 1 Kbp (Fermentas, Burlington, Canada); lane 1, 2, 3: single digestion with XhoI; lane 4, 5: double digestion with NotI and XhoI. Single digestion of pBud-H5-GFP with XhoI produced a single fragment of about 7,672 bp, which correlate with the expected size of the recombinant DNA plasmid. Double digestion of the plasmid with NotI and XhoI, resulted in two fragments, 4,546 and 3,126 bp related to pBud CE4.1 and the H5-GFP gene, respectively.
Abbreviations: DNA, deoxyribonucleic acid; GFP, green fluorescent protein.
Single digestion of pBud-IRF3-H5-GFP plasmid with PstI produced a fragment of about 8,935 bp, which corresponds to the expected size of the recombinant plasmid. Double digestion of the plasmid with PstI and XbaI resulted in two fragments of about 1,263 and 7,672 bp corresponding to IRF3 and pBud-H5-internal ribosome entry site (IRES)-GFP, respectively (Figure 2). The final product of the constructed plasmid is shown in Figure 3.
Figure 2.
Analysis of the inserted gene in pBud IRF3-H5-GFP by using restriction enzyme digestion and polymerase chain reaction.
Notes: M: GeneRulerTM DNA ladder Kit, 1 Kbp (Fermentas); lane 1, 2, 3: single digestion with PstI; lane 4, 5: double digestion with PstI and XbaI. Single digestion of pBud IRF3-H5-GFP plasmid with PstI produced a fragment of about 8,935 bp which correspond to the expected size of the recombinant plasmid. Double digestion of the plasmid with PstI and XbaI resulted in two fragments of about 1,263 and 7,672 bp corresponding to IRF3 and pBud-H5-IRES-GFP, respectively.
Abbreviations: GFP, green fluorescent protein; IRF, interferon-regulatory factor.
Figure 3.
Map of final construct of recombinant plasmid DNA, pBud-H5-IRES-GFP/IRF3 encoding H5-GFP and IRF3 genes at sites of NotI and XohI downstream the EF-1α promoter and XbaI and PstI downstream the CMV promoter, respectively.
Abbreviations: GFP, green fluorescent protein; IRF, interferon-regulatory factor; PCMV, cytomegalovirus promoter.
The identical sequence of the inserted genes was confirmed by double-stranded sequencing. The sequences of the H5 and IRF3 genes in the recombinant DNA were compared with published H5 and IRF3 gene sequence using the Mega software. The data revealed that the inserted H5 and IRF3 in pBud have 100% homology with the published H5 and IRF3 genes, respectively.
Expression analysis of the H5 gene in the vaccinated mice
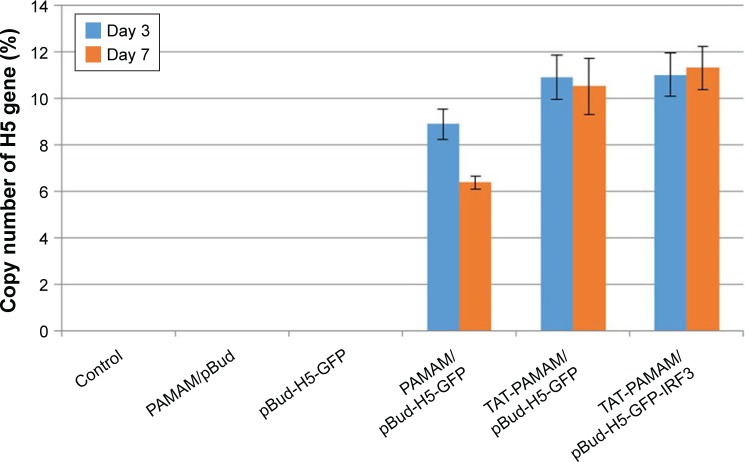
The H5 gene expression at 3 and 7 days after initial immunization through transdermal route showed significantly (P<0.05) higher expression of H5 gene in mice vaccinated with PAMAM/pBud-H5-GFP complex in comparison with pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP complex compared with PAMAM/pBud-H5-GFP. However, the differences in the expression of H5 gene in the group vaccinated with TAT-PAMAM/pBud-H5-GFP-IRF3 complex in comparison with TAT-PAMAM/pBud-H5-GFP complex were not significant (P>0.05). The mice inoculated with pBud-H5-GFP, PAMAM/pBud, and those unimmunized failed to demonstrate detectable H5 gene (Figure 4).
Figure 4.
The H5 gene expression study in the mice vaccinated transdermal with PAMAM/pBud, pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3 after 3 and 7 days.
Notes: The significant (P<0.05) higher expression of H5 gene observed in mice vaccinated with PAMAM/pBud-H5-GFP complex in comparison to pBud-H5-GFP and TAT-PAMAM/pBud-H5-GFP complex compared to PAMAM/pBud-H5-GFP.
Abbreviations: GFP, green fluorescent protein; IRF, interferon-regulatory factor; PAMAM, polyamidoamine; TAT, transactivator of transcription.
H5 antibody response following DNA vaccination by using different delivery systems
As shown in Table 2, the mice inoculated with PAMAM/pBud, pBud-H5-GFP, and those unimmunized failed to demonstrate detectable HI titer. In mice vaccinated with PAMAM/pBud-H5-GFP, no measurable HI antibody titer was detected at day 14 post-vaccination. Two weeks after transdermal booster immunization, the mice showed an average HI titer of 3.3±0.4. The HI antibody titer kept increasing with time and reached 4±0.9 at 3 weeks after booster immunization. Two weeks after the first immunization, mice immunized with TAT-PAMAM/pBud-H5-GFP showed the HI antibody titer of 2.3±0.4. The immunized mice showed increasing antibody responses to HA, where the HI antibody titer increased with time and reached 4.6±0.8 after 2 weeks of booster immunization. The highest HI titer of 6.3±0.4 was detected at the third week after booster vaccination. The antibody responses in mice vaccinated with TAT-PAMAM/pBud-H5-GFP-IRF3 showed HI titer of 4.9±0.8 at day 14 post-vaccination. Rapidly increasing antibody responses to HA were observed at the second week after booster immunization, where the HI antibody titer increased with time and reached 6.64±0.4. At 3 weeks after booster vaccination, the HI titer reached 8.9±0.8.
Table 2.
Mean HI results of serum samples from immunized mice
| Group | HI titers at different days post immunization
|
||
|---|---|---|---|
| Day 14 | Day 28 | Day 35 | |
| pBud-H5-GFP | ND | ND | ND |
| PAMAM/pBud-H5-GFP | ND | 3.3±0.4 | 4±0.9 |
| TAT-PAMAM/pBud-H5-GFP | 2.3±0.4 | 4.6±0.8 | 6.3±0.4 |
| TAT-PAMAM/pBud-H5-GFP-IRF3 | 4.9±0 | 6.64±0.4 | 8.9±0.8 |
| PAMAM/pBud | ND | ND | ND |
| Negative control | ND | ND | ND |
Abbreviations: GFP, green fluorescent protein; HI, hemagglutinin inhibition; IRF, interferon-regulatory factor; PAMAM, polyamidoamine; TAT, transactivator of transcription; ND, not detected.
Immunophenotyping of T cell responses following DNA vaccination by using different delivery systems
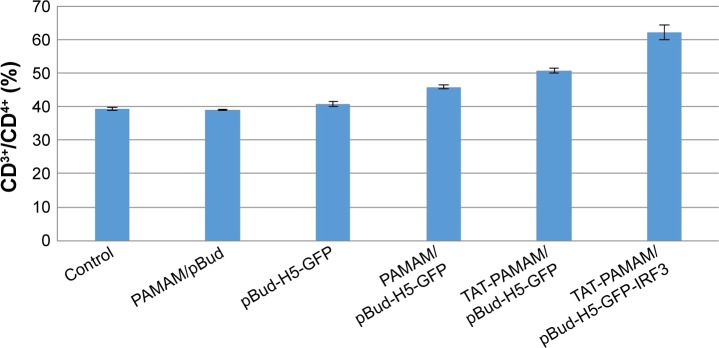
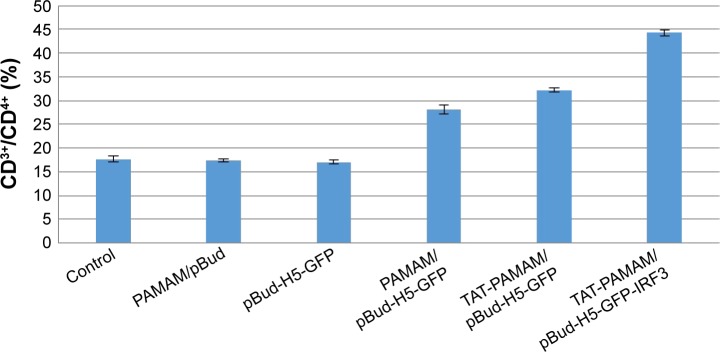
As shown in Figure 5, transdermal-vaccinated mice with pBud-H5-GFP showed no significant increase in the CD4+ T lymphocytes populations compared with the control group. Mice vaccinated with H5-GFP plasmid DNA using TAT-conjugated PAMAM dendrimer showed significant increase in the population of CD4+ T cells compared with native PAMAM dendrimer. The population of CD4 cells in mice vaccinated with H5-GFP-IRF3 plasmid DNA using TAT-conjugated PAMAM dendrimer was significantly higher (P<0.05) compared with the other treatment group of mice.
Figure 5.
Population of CD3+/CD4+ T cells in inoculated mice with PAMAM/pBud, pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3.
Notes: Vaccination with H5-GFP plasmid DNA using TAT-conjugated PAMAM dendrimer showed the significant population of CD4+ T cells compared with native PAMAM dendrimer. The population of CD4 in vaccination with H5-GFP-IRF3 plasmid DNA using TAT-conjugated PAMAM dendrimer was significant compared to the other resultant groups.
Abbreviations: GFP, green fluorescent protein; IRF, interferon-regulatory factor; PAMAM, polyamidoamine; TAT, transactivator of transcription.
As summarized in Figure 6, the percentage of the T cells population in transdermal-immunized mice with TAT-PAMAM/pBud-H5-GFP was significantly higher (P<0.05) than that of mice immunized with PAMAM/pBud-H5-GFP alone, while the highest population of CD8+ T lymphocytes was detected in mice vaccinated with TAT-PAMAM/pBud-H5-GFP-IRF3.
Figure 6.
Population of CD3+/CD8+ T cells in inoculated mice with pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3.
Notes: The percentage of the T cells population in mice transdermal immunized with TAT-PAMAM/pBud-H5-GFP was significantly higher (P<0.05) than that in mice immunized with PAMAM/pBud-H5-GFP. Significantly, the highest population of CD8+ T lymphocytes was detected in mice vaccinated with TAT-PAMAM/pBud-H5-GFP-IRF3.
Abbreviations: GFP, green fluorescent protein; IRF, interferon-regulatory factor; PAMAM, polyamidoamine; TAT, transactivator of transcription.
Quantification of cytokines expression levels following DNA vaccination by using different delivery systems
Overall, as shown in Tables 3–5, the levels of all cytokines in the control group were similar to those vaccinated with PAMAM/pBud and pBud-H5-GFP-IRF3 group through transdermal administration (P>0.05). Most of the differences in the expression levels between the inoculated mice and control were detected at 2 and 3 weeks after the booster immunization.
Table 3.
Serum IL-1β, IL-6, and TNF-α levels in mice immunized with PAMAM/pBud, pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3
| Cytokine | Groups | Level of cytokines (pg/mL) at different days post immunization
|
||
|---|---|---|---|---|
| Day 14 | Day 28 | Day 35 | ||
| IL-1β | Control | 18.91±0.48 | 19.04±0.52 | 18.02±0.42 |
| PAMAM/pBud | 17.89±0.52 | 17.1±0.25 | 18.11±0.62 | |
| pBud-H5-GFP | 17.53±0.89 | 16.98±0.52 | 17.88±0.63 | |
| PAMAM/pBud-H5-GFP | 18.01±0.77 | 22.06±0.74 | 28.27±0.95 | |
| TAT-PAMAM/pBud-H5-GFP | 25.5±0.98 | 5.66±1.25 | 22.06±0.84 | |
| TAT-PAMAM/pBud-H5-GFP-IRF3 | 28.73±0.98 | 4.5±1.25 | 23.81±2.32 | |
| IL-6 | Control | 33.39±0.42 | 34.52±0.45 | 38.85±0.45 |
| PAMAM/pBud | 39.33±0.21 | 34.61±0.52 | 33.99±0.45 | |
| pBud-H5-GFP | 40.68±0.54 | 44.39±0.52 | 45.87±0.61 | |
| PAMAM/pBud-H5-GFP | 50.21±0.52 | 40.68±0.68 | 86.2±0.75 | |
| TAT-PAMAM/pBud-H5-GFP | 102.23±0.87* | 102.18±0.44 | 133.11±0.87 | |
| TAT-PAMAM/pBud-H5-GFP-IRF3 | 105.74±0.98* | 70.4±0.58 | 423±0.98* | |
| TNF-α | Control | 9.67±0.42 | 10.11±0.45 | 10.25±0.45 |
| PAMAM/pBud | 5.29±0.36 | 5.88±0.54 | 6.03±0.45 | |
| pBud-H5-GFP | 6.23±0.45 | 6.41±0.35 | 9.67±0.45 | |
| PAMAM/pBud-H5-GFP | 10.42±0.45 | 10.48±0.52 | 139.64±0.74* | |
| TAT-PAMAM/pBud-H5-GFP | 15.03±0.58 | 14.74±0.92 | 203.23±0.87* | |
| TAT-PAMAM/pBud-H5-GFP-IRF3 | 15.34±0.68 | 16.23±0.68 | 271.52±0.75* | |
Note:
P<0.05.
Abbreviations: GFP, green fluorescent protein; IL, interleukin; IRF, interferon-regulatory factor; PAMAM, polyamidoamine; TAT, transactivator of transcription; TNF, tumor necrosis factor.
Table 4.
Serum IFN-γ, IL-2, IL-12, and IL-15 levels in mice immunized with PAMAM/pBud, pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-conjugated PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3
| Cytokine | Groups | Level of cytokines (pg/mL) at different days post immunization
|
||
|---|---|---|---|---|
| Day 14 | Day 28 | Day 35 | ||
| INF-γ | Control | 12.44±0.42 | 12.52±0.45 | 8.34±0.45 |
| PAMAM/pBud | 12.31±0.51 | 11.9±0.52 | 12.17±0.45 | |
| pBud-H5-GFP | 12.84±0.41 | 13.89±0.87 | 13.2±0.62 | |
| PAMAM/pBud-H5-GFP | 12.44±0.52 | 11.59±0.92 | 24.38±0.72* | |
| TAT-PAMAM/pBud-H5-GFP | 12.48±0.41 | 12.31±0.82 | 52.96±0.84* | |
| TAT-PAMAM/pBUD-H5-GFP-IRF3 | 37.73±0.74* | 71.21±0.98* | 126.82±0.84* | |
| IL-2 | Control | 13.19±0.52 | 12.81±0.52 | 11.1±0.45 |
| PAMAM/pBud | 16.9±0.72 | 11.7±0.82 | 13.46±0.65 | |
| pBud-H5-GFP | 12.78±0.54 | 14.27±0.82 | 14.23±0.45 | |
| PAMAM/pBud-H5-GFP | 12.83±0.51 | 32.6±0.92 | 31.54±0.81* | |
| TAT-PAMAM/pBud-H5-GFP | 14.66±0.65 | 14.71±0.66 | 51.71±0.78* | |
| TAT-PAMAM/pBUD-H5-GFP-IRF3 | 17.69±0.65 | 26.95±0.98 | 153.05±1.25* | |
| IL-12 | Control | 96±0.45 | 94.3±0.65 | 79.2±0.52 |
| PAMAM/pBud | 85±0.45 | 88±0.45 | 85±0.45 | |
| pBud-H5-GFP | 103.03±0.95 | 90.44±0.45 | 99.56±0.45 | |
| PAMAM/pBud-H5-GFP | 115±0.92 | 90.63±0.96 | 162.09±0.65* | |
| TAT-PAMAM/pBud-H5-GFP | 120.99±0.58 | 175.84±0.89 | 253.23±0.85* | |
| TAT-PAMAM/pBUD-H5-GFP-IRF3 | 139.64±0.82 | 162±0.85 | 580±6.21* | |
| IL-15 | Control | 26.33±0.52 | 27.44±0.65 | 26.89±0.54 |
| PAMAM/pBud | 12.85±0.48 | 13.52±0.52 | 14.25±0.42 | |
| pBud-H5-GFP | 9.07±1.25 | 2.44±0.96 | 11.84±0.52 | |
| PAMAM/pBud-H5-GFP | 40.73±0.98 | 39.01±2.54 | 72.67±1.25* | |
| TAT-PAMAM/pBud-H5-GFP | 52.96±0.96 | 40.73±2.25 | 65.08±3.25* | |
| TAT-PAMAM/pBUD-H5-GFP-IRF3 | 61.86±1.25 | 69.05±4.25 | 163.64±1.58* | |
Note:
P<0.05.
Abbreviations: GFP, green fluorescent protein; IFN, interferon; IL, interleukin; IRF, interferon-regulatory factor; PAMAM, polyamidoamine; TAT, transactivator of transcription.
Table 5.
Serum IL-4 and IL-10 levels in mice immunized with PAMAM/pBud, pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-conjugated PAMAM/pBud-H5-GFP and TAT-PAMAM/pBud-H5-GFP-IRF3
| Cytokine | Groups | Level of cytokines (pg/mL) at different days post immunization
|
||
|---|---|---|---|---|
| Day 14 | Day 28 | Day 35 | ||
| IL-4 | Control | 1.79±0.45 | 3.47±0.45 | 4.28±0.51 |
| PAMAM/pBud | 1.81±0.45 | 3.47±0.52 | 4.07±0.65 | |
| pBud-H5-GFP | 4.16±0.42 | 5.44±0.45 | 5.68±0.45 | |
| PAMAM/pBud-H5-GFP | 4.12±0.68 | 5.45±0.45 | 5.85±0.45 | |
| TAT-PAMAM/pBud-H5-GFP | 6.02±0.85* | 5.47±0.52 | 5.85±0.52 | |
| TAT-PAMAM/pBUD-H5-GFP-IRF3 | 6.14±0.74* | 5.55±0.87 | 5.48±0.75 | |
| IL-10 | Control | 31.97±0.32 | 30.25±0.25 | 32.45±0.25 |
| PAMAM/pBud | 30.23±0.54 | 30.2±0.45 | 30.21±0.45 | |
| pBud-H5-GFP | 31.62±0.95 | 31.26±0.45 | 32.14±0.45 | |
| PAMAM/pBud-H5-GFP | 31.85±0.54 | 26.64±0.45 | 32.52±0.52 | |
| TAT-PAMAM/pBud-H5-GFP | 32.59±0.78 | 32.69±0.65 | 32.54±0.87 | |
| TAT-PAMAM/pBUD-H5-GFP-IRF3 | 33.7±0.62 | 25.62±0.78 | 33.49±0.84 | |
Note:
P<0.05.
Abbreviations: GFP, green fluorescent protein; IL, interleukin; IRF, interferon-regulatory factor; PAMAM, polyamidoamine; TAT, transactivator of transcription.
No significant difference (P>0.05) in IL-1β expression was observed in mice vaccinated with PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3. Meanwhile, mice vaccinated with TAT-PAMAM/pBud-H5-GFP and TAT-PAMAM/pBud-H5-GFP-IRF3 showed a significant increase (P<0.05) in the serum IL-6 level at day 14 compared with control. The highest expression of IL-6 was detected at day 35 in the mice inoculated with TAT-PAMAM/pBud-H5-GFP-IRF3. Significant upregulation in the expression levels of TNF-α was detected at day of 35 for mice vaccinated with PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3 compared with control (Table 3).
Mice immunized with PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3 showed the highest expression of IFN-γ, IL-2, IL-12, and IL-15 at day 35 compared with control. Vaccination with TAT-PAMAM/pBud-H5-GFP showed higher expression of IFN-γ and IL-12 compared with PAMAM/pBud-H5-GFP at day 35. Mice vaccinated with TAT-PA-MAM/pBud-H5-GFP-IRF3 showed a significant (P<0.05) level of IFN-γ in all the studied time points (days 14, 28 and 35) compared with PAMAM/pBud-H5-GFP and TAT-PAMAM/pBud-H5-GFP (Table 4).
The immunized mice vaccinated with TAT-PA-MAM/pBud-H5-GFP and TAT-PAMAM/pBud-H5-GFP-IRF3 showed a significant (P<0.05) increase in IL-4 at day 14 compared with control. Significantly (P<0.05) higher level of IL-4 was detected in groups vaccinated with TAT-conjugated PAMAM/pBud-H5-GFP compared with PAMAM/pBud-H5-GFP, while no significant results were observed for this group compared with TAT-PAMAM/pBud-H5-GFP-IRF3. No significant results were observed for IL-10 in all resultant groups (Table 5).
Discussion
Our previous study showed that PAMAM and TAT-PAMAM formed compact nanometer-sized polyplexes with positive charges that are considered to be important for their adsorption to negatively charged cellular membranes prior to its cellular uptake through internalization mechanisms. Additionally, in vitro transfection and transient gene expression study in Vero cells using different delivery systems incorporating PAMAM dendrimer and TAT-conjugated PAMAM dendrimer revealed the potency of these nanoparticles for gene delivery. Permeability value of DNA plasmid using PAMAM and TAT-conjugated PAMAM dendrimers through the artificial membrane, Pion’s skin PAMPA addressed the cellular uptake facilitation of the plasmid DNA polyplexes after the insertion of the TAT peptide into the polymeric vector, and offered it as a non-invasive method of penetrating DNA plasmid through the skin.24
In this study, the potential of PAMAM dendrimer and TAT-PAMAM complex facilitating permeability of DNA vaccine through the skin of experimental animal, and the effect of the IRF3 as a genetic adjuvant on the immunogenicity of recombinant DNA plasmid were investigated. For this purpose, transdermal inoculation of pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3 was carried out in BALB/c mice. Transcutaneous immunization is a useful strategy for genetic vaccine immunization to induce detectable immune responses. Moreover, vaccination through the skin offers potential for painless, needle-free, and self-applicable immunization that can be useful for annual influenza vaccination of large populations.25 Different compounds have been used as carrier for DNA vaccine delivery through skin vaccination. One study showed that plasmid expressing hepatitis B surface antigen encapsulated in liposome glued on bare skin of mice ear induced humoral immunization responses.26 Transdermal delivery of the pCF1CAT DNA by collagen membrane-supported dendrimer/DNA induced greater CAT expression in the mouse skin compared with uncomplexed DNA.16 The transdermal delivery with cationic liposomes of envelope protein genes of Japanese encephalitis virus induced effective and protective antibodies against the infection.26
In order to evaluate the ability of PAMAM and TAT- PAMAM dendrimers for gene delivery, H5 gene expression of blood was determined at days 3 and 7 after immunization in BALB/c mice. Significantly (P<0.05) higher expression of H5 gene in mice vaccinated with DNA plasmid using TAT-conjugated dendrimer compared with native PAMAM dendrimer indicated the advantage of the TAT-PAMAM dendrimer preparation over the native PAMAM dendrimer and comparable with pBud-H5-GFP. The skin is known to be a site rich in antigen-presenting cells, some of which are specific to the skin, including epidermal Langerhans cells and dermal dendritic cells. In addition, antigen may be taken up directly by lymphatic vessels in the skin for transport to antigen-presenting cells in the lymph nodes. Numerous studies have investigated the delivery of DNA vaccines through transdermal administration and analyzing the transgene expression. In order to ensure successful diffusion of large vaccine molecules into the skin, different techniques have been employed such as the use of chemical enhancers, ultrasound, electroporation, gene gun, and microneedles.27 For example, intradermal injection of the polymer/DNA complexes into mice using 3 structurally distinct cationic polymers (branched polyethylenimine [PEI], linear poly [2-aminoethyl methacrylate] [PAEM], and diblock copolymer [PEG-b-PAEM]), and analysis of the transgene expression revealed that the polyplexes appeared to result in more transfected skin cells than naked plasmid. Additionally, it has been shown that naked plasmid level dropped to below detection limit after 24 h, whereas polyplexes persisted for up to 2 weeks.28 Several in vivo studies have used a combination of modified HIV-1 TAT peptide with cationic lipids and PEI in the delivery of plasmid DNA encoding a luciferase gene. The results showed that animals administered with TAT/cationic lipid/DNA intramuscularly developed significantly higher and longer luciferase expression than those with TAT/DNA, cationic lipid/DNA or DNA alone.19,20
Significantly higher HI titer in mice vaccinated with TAT-PAMAM/pBud-H5-GFP than those vaccinated with PAMAM/pBud-H5-GFP exhibited an efficient gene delivery system where the TAT peptide was conjugated to the PAMAM dendrimer. Significantly (P<0.05) high HI titer in mice vaccinated with TAT-PAMAM/pBud-H5-GFP-IRF3 than those with TAT-PAMAM/pBud-H5-GFP showed the effect of IRF3 as a genetic adjuvant in humoral immunity induction. IRFs play a critical role in the activation of type I interferon, IFN-α and IFN-β as well as interferon-stimulated genes and other cytokines. IFN-α and IFN-β, in turn, stimulate both innate and acquired immune responses. IRF3 serves as direct transducers of virus-mediated signaling pathways. The expression of IRF3 is sufficient for the induction of IFN-β in infected cells. Downregulation or null mutations of IRF3 in the IFN-α/β pathway inhibit the expression of IFN-α/β, while defects in IRF3 and IRF7 completely abolish IFN-α/β expression.29 From the present study, the immunodulatory activity of IRF3 as genetic adjuvant in the induction of humoral responses was observed. Thus it is suggested that the mechanism of IRF3 function is through the IFN-α/β pathway.
It has been shown that cellular immune responses play a key role in eliminating viral infections as well as in vaccine-induced protective immunity. Therefore, quantification of specific T cell responses has become a focus of interest in order to understand protective immunity in many infectious diseases. In the present study, proliferation of CD3+/CD4+ and CD3+/CD8+ T cells in vaccinated mice with pBud-H5-GFP, PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP and TAT-PAMAM/pBud-H5-GFP-IRF3 were measured using flow cytometry immunophenotyping. The modification of PAMAM dendrimer with TAT peptide showed enhanced activity in T cell responses. The use of TAT peptide resulted in <2-fold increases in the number of CD8+ T lymphocytes. Previous study has shown that the addition of TAT is associated with a much higher activation of CD8+ T lymphocytes and in vivo protective efficacy compared with the control group.30 The effect of TAT peptide on CD4+ T cells was not as significant as those on CD8+ T cells. H5 DNA vaccine co-delivered with IRF3 showed good adjuvant activities for T-cell responses against HA. These results reveal that IRF3 serves as genetic adjuvant for DNA-induced CD8+ and CD4+ responses. Previous study showed that the incorporation of IRF1, IRF3, and IRF7 into influenza virus DNA vaccines induced humoral and/or cellular immune responses against the viral infection.29 However, IRF1 and IRF3 are in favor of inducing strong humoral and cellular immune responses, while IRF7 enhanced both humoral and cellular immune responses. In another study, DNA vaccines encoding IRF3 and IRF7 stimulated humoral and cellular immune responses to protect against vaccinia virus infection.31 As mentioned previously, the IRF3 has a critical role in the induction of IFN-α/β as they are the most important mammalian cytokines in the innate and adaptive immunity. IFN-α/β also play a role in the development of other immune cell types, such as CD4 and CD8 T cells.32
Cytokines can be quantitated at various levels, including mRNA detection by real-time PCR, measurements of intracellular proteins by fluorescence-activated cell sorter analysis of stained permeabilized cells, quantification secreted cytokines via bioassays, enzyme-linked immunosorbent assays, radioactive immunosorbent assays, and microarrays. Recent advances in the simultaneous detection of proteins have resulted in various particle-based flow cytometric assays. The advantages of multiplexed bead-based immunoassays, which make it an attractive alternative to other bioassays, are speed and high-throughput, sample size, accuracy and reproducibility, versatility, and flexibility.33,34
In this study, mouse cytokines, including pro-inflammatory cytokines, (IL-1β, TNF-α, and IL-6), Th1 (IFN-γ, IL-2, IL-15, and IL-12) and Th2 (IL-4 and IL-10), were simultaneously measured in the serum samples collected at the initial day before vaccination, 14 days after immunization, and 2 and 3 weeks after booster immunization of inoculated mice by using the Luminex assay. The multiplex analysis of cytokines in mice inoculated with PAMAM/pBud-H5-GFP, TAT-PAMAM/pBud-H5-GFP, and TAT-PAMAM/pBud-H5-GFP-IRF3 revealed significant up-regulation in the expression of TNF-α, IL-6, IFN-γ, IL-2, IL-15, and IL-12, thus showing an activation of a mixed pro-inflammatory and Th1-like cytokines, which may be of importance for viral clearance. The induction of Th1 cytokines is valuable for the elimination of influenza infection since it has been shown that Th1 responses are essential for viral clearance during the late phases of influenza A virus infection in mice.35 The higher expression of cytokines such as TNF-α, IL-6, IFN-γ, IL-12, and IL-2 in mice immunized with TAT-PAMAM/pBud-H5-GFP compared with PAMAM/pBud-H5-GFP could probably be associated with rapid distribution of H5 in the tissue of the immunized mice where PAMAM dendrimer was modified with TAT peptide.
Determination of serum cytokines profile in the mice vaccinated with TAT-PAMAM/pBud-H5-GFP-IRF3 revealed that IRF3 enhances IFN-γ secretion. Transcription factors IRF3 activates toll-like receptor-independent type I IFN production, which are very important in inducing IFN-γ production.32 IFN-α/β is one of the most abundant cytokines released by macrophages during viral infections. It has been shown that IFN-α/β produced by virus-infected macrophages directly stimulates IFN-γ production in T cells. IFN-γ is one of the key cytokines directing T cell immune responses toward Th1-type and cell-mediated immunity. Additionally, TAT-PAMAM/pBud-H5-GFP-IRF3 vaccination increased the level of IL-2, IL-15, IL-12, IL-6, and TNF-α cytokines, which are also able to enhance the IFN-γ production. Studies conducted in IL-12 knockout mice have shown that IL-12 is essential in enhancing IFN-γ gene expression following endotoxin administration. Moreover, it was shown that IRF3 was involved in IL-6 expression during murine encephalomyelitis virus infection of macrophages.36 Additionally, it was found that IRF3 is required for maximum induction of nitric oxide (NO) produced by macrophages in response to exogenous IL-6. But further studies are required to define the mechanism of IRF3 involvement in the induction of IL-6.37 These results led us to assume that IRF3 has the good activity as a genetic adjuvant in the induction of mixed pro-inflammatory reactions as well as Th1 responses. The effect of IRF3 in the induction of pro-inflammatory and Th1 responses was supported by flow cytometry T cell immunophenotyping obtained from mice inoculated with TAT-PAMAM/pBud-H5-GFP-IRF3 that showed an increase in the CD4+ T cell population. This population of CD4+ T cell probably associates with the stimulation of Th1 and pro-inflammatory type cytokines, IL-2, IL-15, IL-12, IL-6, and TNF-α. Hence, this study revealed the advantage of the conjugation between PAMAM dendrimer and TAT to improve the efficacy of H5 DNA vaccines which allows better distribution and higher expression levels of the encoded antigen following vaccination via the skin. Additionally, the expression of IRF3 incorporated in the DNA vaccine with TAT-conjugated dendrimer platform showed the ability to enhance the immune stimulatory potency, therefore, enhancing the vaccine-induced immune response.
Conclusion
This is the first study to report on the testing of TAT/PAMAM dendrimer system for transdermal delivery of DNA plasmids encoding the IRF3 in mice. The overall results showed better skin permeation and higher cellular uptake of the pBud-H5-GFP-IRF3 when nanoparticles were formulated using TAT/PAMAM. Incorporation of the IRF3 gene as the genetic adjuvant in this delivery system resulted in the induction of strong antibody and cell-mediated immune responses as well as enhanced Th1-like cytokine production against AI virus in mice. These results lead us to conclude that the outstanding transfection efficiency with relatively low cytotoxicity and ease of preparation would make TAT-PAMAM dendrimer as a promising non-viral vector along with IRF3 as a genetic adjuvant for the induction of appropriate immune responses. Further studies are required to determine whether the improved immune responses can be translated into protection in mice following a lethal dose of the H5N1 virus. In addition, although the TAT-PAMAM system synthesized has the ability to deliver the plasmid DNA transdermally, the suitability of the delivery system for topical administration of DNA vaccine requires further study.
Acknowledgments
The study was supported by HICOE Grant no: 6369101, from the Ministry of Higher Education, Government of Malaysia.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lee YT, Kim KH, Ko EJ, et al. New vaccines against influenza virus. Clin Exp Vaccine Res. 2014;3(1):12–28. doi: 10.7774/cevr.2014.3.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Organisation for Animal Health . Manual of diagnostic tests and vaccines for terrestrial animals. Paris, France: Office International Des Epizooties; 2008. pp. 1092–1106. [Google Scholar]
- 3.Yazdanbakhsh M, Kremsner PG. Influenza in Africa. PLoS Med. 2009;6(12):e1000182. doi: 10.1371/journal.pmed.1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sedova E, Shcherbinin D, Migunov A, et al. Recombinant influenza vaccines. Acta Naturae. 2012;4(4):17–27. [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention Vaccine Adjuvants. 2016. Available from: https://www.cdc.gov/vaccinesafety/concerns/adjuvants.html.
- 6.Luke CJ, Subbarao K. Vaccines for pandemic influenza. Emerg Infect Dis. 2006;12(1):66–72. doi: 10.3201/eid1201.051147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira VB, Zurita-Turk M, Saraiva TDL, et al. DNA vaccines approach: from concepts to applications. World J Vaccines. 2014;4(2):50–71. [Google Scholar]
- 8.Davidson AH, Traub-Dargatz JL, Rodeheaver RM, et al. Immunologic responses to West Nile virus in vaccinated and clinically affected horses. J Am Vet Med Assoc. 2005;226(2):240–245. doi: 10.2460/javma.2005.226.240. [DOI] [PubMed] [Google Scholar]
- 9.Garver KA, LaPatra SE, Kurath G. Efficacy of an infectious hematopoietic necrosis (IHN) virus DNA vaccine in Chinook Oncorhynchus tshawytscha and sockeye O. nerka salmon. Dis Aquat Organ. 2005;64(1):13–22. doi: 10.3354/dao064013. [DOI] [PubMed] [Google Scholar]
- 10.Bergman PJ, Camps-Palau MA, McKnight JA, et al. Development of a xenogeneic DNA vaccine program for canine malignant melanoma at the Animal Medical Center. Vaccine. 2006;24(21):4582–4585. doi: 10.1016/j.vaccine.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 11.Khan AS, Bodles-Brakhop AM, Fiorotto ML, Draghia-Akli R. Effects of maternal plasmid GHRH treatment on offspring growth. Vaccine. 2010;28(8):1905–1910. doi: 10.1016/j.vaccine.2009.10.093. [DOI] [PubMed] [Google Scholar]
- 12.Bolhassani A, Safaiyan S, Rafati S. Improvement of different vaccine delivery systems for cancer therapy. Mol Cancer. 2011;10:3. doi: 10.1186/1476-4598-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun K, Li X, Jiang J, et al. Distribution characteristics of DNA vaccine encoded with glycoprotein C from Anatid herpesvirus 1 with chitosan and liposome as deliver carrier in ducks. Virol J. 2013;10:89. doi: 10.1186/1743-422X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro G, Tros de Ilarduya C. Activated and non-activated PAMAM dendrimers for gene delivery in vitro and in vivo. Nanomedicine. 2009;5(3):287–297. doi: 10.1016/j.nano.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Borowska K, Wołowiec S, Rubaj A, Głowniak K, Sieniawska E, Radej S. Effect of polyamidoamine dendrimer G3 and G4 on skin permeation of 8-methoxypsoralene – in vivo study. Int J Pharm. 2012;426(1–2):280–283. doi: 10.1016/j.ijpharm.2012.01.041. [DOI] [PubMed] [Google Scholar]
- 16.Bielinska AU, Yen A, Wu HL, et al. Application of membrane-based dendrimer/DNA complexes for solid phase transfection in vitro and in vivo. Biomaterials. 2000;21(9):877–887. doi: 10.1016/s0142-9612(99)00229-x. [DOI] [PubMed] [Google Scholar]
- 17.Choi JS, Nam K, Park JY, Kim JB, Lee JK, Park JS. Enhanced transfection efficiency of PAMAM dendrimer by surface modification with L-arginine. J Control Release. 2004;99(3):445–456. doi: 10.1016/j.jconrel.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 18.Yi WJ, Yang J, Li C, et al. Enhanced nuclear import and transfection efficiency of TAT peptide-based gene delivery systems modified by additional nuclear localization signals. Bioconjug Chem. 2011;23(1):125–134. doi: 10.1021/bc2005472. [DOI] [PubMed] [Google Scholar]
- 19.Yamano S, Dai J, Hanatani S, et al. Efficient in vivo gene delivery using modified Tat peptide with cationic lipids. Biotechnol Lett. 2014;36(7):1447–1452. doi: 10.1007/s10529-014-1497-2. [DOI] [PubMed] [Google Scholar]
- 20.Yamano S, Dai J, Hanatani S, et al. Long-term efficient gene delivery using polyethylenimine with modified Tat peptide. Biomaterials. 2014;35(5):1705–1715. doi: 10.1016/j.biomaterials.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Grunwald T, Ulbert S. Improvement of DNA vaccination by adjuvants and sophisticated delivery devices: vaccine-platforms for the battle against infectious diseases. Clin Exp Vaccine Res. 2015;4(1):1–10. doi: 10.7774/cevr.2015.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda K, Wada Y, Shimada M. Recent developments in preclinical DNA vaccination. Vaccines (Basel) 2014;2(1):89–106. doi: 10.3390/vaccines2010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki S, Takeshita F, Xin KQ, Ishii N, Okuda K. Adjuvant formulations and delivery systems for DNA vaccines. Methods. 2003;31(3):243–254. doi: 10.1016/s1046-2023(03)00140-3. [DOI] [PubMed] [Google Scholar]
- 24.Bahadoran A, Moeini H, Bejo MH, Hussein MZ, Omar AR. Development of Tat-conjugated dendrimer for transdermal DNA vaccine delivery. J Pharm Pharm Sci. 2016;19(3):325–338. doi: 10.18433/J3G31Q. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Li B, Wu MX. Effective and lesion-free cutaneous influenza vaccination. Proc Natl Acad Sci U S A. 2015;112(16):5005–5010. doi: 10.1073/pnas.1500408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang R, Zhuang F, Meng Z, Deng M, Zheng C, Duan M. A new potent route of DNA vaccine inoculation: DNA-liposome complexes on bare skin induce antigen-special antibody responses. Molecules. 2003;8(1):120–126. [Google Scholar]
- 27.Gill HS, Kang SM, Quan FS, Compans RW. Cutaneous immunization: an evolving paradigm in influenza vaccines. Expert Opin Drug Deliv. 2014;11(4):615–627. doi: 10.1517/17425247.2014.885947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palumbo RN, Zhong X, Panus D, Han W, Ji W, Wang C. Transgene expression and local tissue distribution of naked and polymer-condensed plasmid DNA after intradermal administration in mice. J Control Release. 2012;159(2):232–239. doi: 10.1016/j.jconrel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki S, Amara RR, Yeow WS, Pitha PM, Robinson HL. Regulation of DNA-raised immune responses by cotransfected interferon regulatory factors. J Virol. 2002;76(13):6652–6659. doi: 10.1128/JVI.76.13.6652-6659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leifert JA, Lindencrona JA, Charo J, Whitton JL. Enhancing T cell activation and antiviral protection by introducing the HIV-1 protein transduction domain into a DNA vaccine. Hum Gene Ther. 2001;12(15):1881–1892. doi: 10.1089/104303401753153938. [DOI] [PubMed] [Google Scholar]
- 31.Bramson JL, Dayball K, Hall JR, et al. Super-activated interferon-regulatory factors can enhance plasmid immunization. Vaccine. 2003;21(13):1363–1370. doi: 10.1016/s0264-410x(02)00694-1. [DOI] [PubMed] [Google Scholar]
- 32.Gustafsson T.The Role of Dendritic Cells in Adjuvant-Induced Immune Responses [PhD thesis]. Göteborgs universitet; 2013 [Google Scholar]
- 33.Elshal MF, McCoy JP. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods. 2006;38(4):317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan SS, Smith MS, Reda D, Suffredini AF, McCoy JP., Jr Multiplex bead array assays for detection of soluble cytokines: comparisons of sensitivity and quantitative values among kits from multiple manufacturers. Cytometry B Clin Cytom. 2004;61(1):35–39. doi: 10.1002/cyto.b.20021. [DOI] [PubMed] [Google Scholar]
- 35.Van der Goot JA, Koch G, de Jong MC, Van Boven M. Quantification of the effect of vaccination on transmission of avian influenza (H7N7) in chickens. Proc Natl Acad Sci U S A. 2005;102(50):18141–18146. doi: 10.1073/pnas.0505098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore TC, Cody L, Kumm PM, Brown DM, Petro TM. IRF3 helps control acute TMEV infection through IL-6 expression but contributes to acute hippocampus damage following TMEV infection. Virus Res. 2013;178(2):226–233. doi: 10.1016/j.virusres.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore TC, Petro TM. IRF3 and ERK MAP-kinases control nitric oxide production from macrophages in response to poly-I: C. FEBS Lett. 2013;587(18):3014–3020. doi: 10.1016/j.febslet.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]