Abstract
Zoonotic A(H7N9) avian influenza viruses emerged in China in 2013 and continue to be a threat to human public health, having infected over 800 individuals with a mortality rate approaching 40%. Treatment options for people infected with A(H7N9) include the use of neuraminidase (NA) inhibitors. However, like other influenza viruses, A(H7N9) can become resistant to these drugs. The use of monoclonal antibodies is a rapidly developing strategy for controlling influenza virus infection. Here we generated a murine monoclonal antibody (3c10-3) directed against the NA of A(H7N9) and show that prophylactic systemic administration of 3c10-3 fully protected mice from lethal challenge with wild-type A/Anhui/1/2013 (H7N9). Further, post-infection treatment with a single systemic dose of 3c10-3 at either 24, 48 or 72 h post A(H7N9) challenge resulted in both dose- and time-dependent protection of up to 100% of mice, demonstrating therapeutic potential for 3c10-3. Epitope mapping revealed that 3c10-3 binds near the enzyme active site of NA, and functional characterization showed that 3c10-3 inhibits the enzyme activity of NA and restricts the cell-to-cell spread of the virus in cultured cells. Affinity analysis also revealed that 3c10-3 binds equally well to recombinant NA of wild-type A/Anhui/1/2013 and to a variant NA carrying a R289K mutation known to infer NAI resistance. These results suggest that 3c10-3 has the potential to be used as a therapeutic to treat A(H7N9) infections either as an alternative to, or in combination with, current NA antiviral inhibitors.
Keywords: Influenza, H7N9, Monoclonal antibody, Antiviral, Neuraminidase, NA epitope
1. Introduction
Influenza viruses pose a significant threat to global public health, affecting human as well as agricultural and wild animal species. Occasionally viruses cross the species barrier and pose the potential for a global pandemic. Avian influenza A viruses of the H7N9 subtype first infected humans in China in 2013 and have since resulted in 786 confirmed cases and 307 deaths (case-fatality risk 39%) as of May 2016 (Food and Agriculture Organization of the United Nations, 2016). Most infections are believed to result from direct exposure to poultry, but there is evidence that limited person-to-person spread has occurred (Farooqui et al., 2016; Qi et al., 2013). Although A(H7N9) is not yet capable of establishing sustained person-to-person transmission, these viruses are considered to have pandemic potential due to their ability to bind both avian and human-like receptors (α2,3 and α2,6-linked sialic acid, respectively) (Shi et al., 2013; Yang et al., 2013; Zhou et al., 2013).
Currently, there is no commercially-available vaccine for the prevention of A(H7N9) infection and the main treatment strategy for avian influenza infection consists of supportive medical care and the use of neuraminidase inhibitors (NAIs). The World Health Organization recommends treatment with NAIs as quickly as possible, for patients with suspected or confirmed A(H7N9) virus infection but their clinical effectiveness against this virus remains unknown. For seasonal viruses, the US Centers for Disease Control and Prevention (CDC) recommends the use of NAIs for patients with suspected or confirmed influenza who are hospitalized or at high risk for complications due to influenza, including patients less than 2 and more than 65 years of age, those with underlying medical conditions or compromised immunity, and pregnant women. However, NAIs only consistently reduce the severity of disease if taken within 48 h of the onset of clinical symptoms (Dobson et al., 2015). Further, there is evidence that A(H7N9) can gain NAI resistance while maintaining in vivo virulence in animal models (Hai et al., 2013; Itoh et al., 2015). Supporting this, virus with reduced susceptibility to the commonly-used NAI oseltamivir has been isolated from A(H7N9) patients treated with NAI (Hu et al., 2013). Thus, it is important to establish alternative antiviral options to mitigate the severity of disease caused by A(H7N9) infection.
The two major influenza envelope glycoproteins, hemagglutinin (HA) and NA, are the primary viral components known to induce protective humoral immune responses upon influenza vaccination or viral infection (Tosh et al., 2010). Antibodies targeting the dominant surface protein, HA, tend to be directly neutralizing while those targeting NA have the potential to reduce viral replication efficiency by blocking NA sialidase activity and thereby impairing viral budding and spread (Eichelberger and Wan, 2015; Johansson et al., 1989). Monoclonal antibodies (mAbs) are a rapidly advancing class of therapeutic proteins that possess wide clinical utility owing to their biocompatibility, high antigen specificity, and targeted immune stimulation dictated by the Fc subtype used. Here we describe a mAb that targets the NA of A(H7N9) and protects mice from lethal A(H7N9) challenge when administered in either a prophylactic or therapeutic setting.
2. Materials and methods
2.1. Animals, cells, viruses, and proteins
All research studies involving the use of animals were reviewed and approved by the Institutional Animal Care and Use Committee at the CDC in an Association for the Assessment and Accreditation of Laboratory Animal Care accredited facility. Six to eight week old female BALB/c mice (The Jackson Laboratory) were used for all experiments. Mice were anesthetized by isoflurane inhalation for all intranasal (i.n.) instillations.
Immortalized B cells were maintained in IMDM supplemented with 10% ultra-low IgG FBS (HyClone). Influenza viruses were propagated in embryonated chicken eggs and titers were determined on MDCK cells by plaque assay. RG32a is a reverse-genetics reassortant virus containing the HA and NA from A/Shanghai/2/2013(H7N9) (“Sh/2”) with the 6 internal genes from A/Puerto Rico/8/1934(H1N1) (“PR8”). H6N9 is a reverse-genetics reassortant virus containing the HA from A/Turkey/CA/BENN/1973(H6N1), NA from Sh/2 (H7N9), and the remaining genes from PR8. The reverse-genetic reassortants were constructed by standard molecular biology techniques (Dong et al., 2009). A/Anhui/1/2013(H7N9) (“Anhui/1”), A/California/07/2009(H1N1pdm09) (“Ca/07”) and A/Perth/16/2009 (H3N2) (“Perth/16”) are wild-type viruses isolated from human infections. All work with RG32a was performed at Biosafety Level 2 enhanced containment, while Anhui/1 challenge studies were performed at Animal Biosafety Level 3-enhanced containment. The remaining viruses were handled under BSL2 containment.
The recombinant his-tagged NA (recNA) from Anhui/1, Ca/07, and Perth/16 were produced in a baculovirus expression system and purified as previously described (Wan et al., 2015; Yang et al., 2010). Monoclonal antibody to N3 NA of A/Canada/44/2004 (H7N3) (FR-969), N2 NA of A/Perth/16/2009 (H3N2) (FR-1155), and N1 NA of A/Brisbane/59/2007 (H1N1) (FR-933) were obtained from the Influenza Reagent Resource (IRR). mAb 0-1h6 to H7 HA of Sh/2 was produced by our laboratory using hybridoma technology (manuscript in preparation).
To generate a panel of recNA with a single mutation at solvent-exposed residues, a codon optimized cDNA encoding the ectodomain (residues 36-465) of the NA gene of Sh/2 was synthesized (GenScript USA Inc., NJ) and sub-cloned into pIEx-4 vector (EMD Millipore, MA) using the In-Fusion HD cloning system (Clontech, CA). All subsequent recNA mutants were generated from this wild type pIEx-4-NA clone using the QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene, CA). Primer sequences used in mutagenesis PCR are available upon request. The resulting constructs were transiently transfected into sf9 cells (EMD Millipore, MA) using the Cellfectin II transfection reagent (Life Technologies, NY). All procedures were performed following protocols provided by manufacturers. The transfected cells were transferred into flasks and grown on a shaker at 27 °C for five days. All recNA proteins contained a His-Tag at the N-terminus followed by a tetramerization domain, from human vasodilator-stimulated phosphoprotein, and a thrombin cleavage site (Xu et al., 2008). The recNAs secreted in the culture supernatant were analyzed for their expression levels by western blot analysis. Supernatants were assessed using the MUNANA assay as described below to confirm correct conformation and functional activity of the mutated NA proteins (Yang et al., 2015). The recNA supernatants were used in biolayer interferometry assays without further purification.
2.2. Generation of mouse hybridoma secreting NA-specific mAb
Six to eight week old female BALB/c mice (The Jackson Laboratory) were immunized and boosted by intraperitoneal (ip) injection of 15 μg of a beta-propiolactone (BPL)-inactivated and purified RG32a virus. Three days post-boost, splenocytes were fused with SP2/IL-6 myeloma cells (American Type Culture Collection) using the ECM 2001 electrofusion apparatus (BTX Harvard Apparatus/Boston MA). After two brief washes with Cytofusion Medium C (BTX) the isolated lymphocytes were mixed with myeloma cells at a 1:1 ratio, resuspended at 10 × 106 cells/ml in Cytofusion Medium C and electrofusion was performed according to the manufacturer’s protocol. Fused cells were cultured in ClonaCell-HY Medium C (StemCell Technologies) for 48 h at 37 °C in a 5% CO2 incubator. Fused cells were centrifuged and re-suspended in 10 ml of ClonaCell-HY Medium C and then gently mixed with 90 ml of methylcellulose-based ClonaCell-HY Medium D (StemCell Technologies) containing HAT components, CloneDetect (Molecular Devices) and IL-6 (Roche). The fused cells were plated into 100-mm petri dishes (BD Biosciences) and allowed to grow at 37 °C in a 5% CO2 incubator. After 10 days, single hybridoma clones were picked by ClonePix FL (Molecular Devices, Sunnyvale CA) based on anti-IgG FITC intensities from CloneDetect reagent and transferred to 96-well cell culture plates (BD Biosciences) with 200 μl/well IMDM and 20% serum. Hybridoma culture media was changed prior to ELISA screening. 3c10-3 was retrieved after 2 rounds of limiting dilution and clonality was confirmed by sequence analysis.
2.3. Production and purification of mAb
3c10-3 secreting hybridoma cells were expanded in CELLine™ Disposable Bioreactor (Corning) according to the manufacturers’ instructions. mAb 3c10-3 was subsequently purified from tissue culture supernatant using a protein G column (GE Healthcare).
2.4. Enzyme linked immunosorbent assay
For detection of NA reactivity, ELISA assays were performed as previously described (Wilson et al., 2014). Briefly, Costar Hi Bind plates (Corning Inc., Tewksbury, MA) were coated overnight with BPL-inactivated virus (25 HA units/well) or recNA (100 ng/well) at 4 °C. Plates were blocked for 1 h with PBS/0.1% Tween-20 (PBST) containing 1.5% BSA (blocking buffer) at room temperature. All mAbs were serially titrated in blocking buffer and allowed to incubate with antigen-coated plates for 1 h at room temperature. After three PBST washes, wells were probed with goat anti-mouse (H&L)-HRP (Thermo Scientific) for 1 h at room temperature. Plates were washed three times with PBST and signal was developed with 1 step™ Turbo TMB-ELISA reagent (Thermo Scientific). Reactions were stopped with 1 N sulfuric acid and absorbance was read at 450 nm.
2.5. Neuraminidase antibody inhibition assays
Antibodies with neuraminidase inhibition activity were detected using Enzyme-Linked Lectin assay (ELLA) as described previously (Couzens et al., 2014). Briefly, serial two-fold diluted mAb were incubated with an H6N9 reassortant virus, bearing the NA of Sh/2 but an unrelated HA, in 96 well plates coated with fetuin for 16–18 h. Following incubation, horse radish peroxidase-labeled peanut agglutinin (lectin) were added to the reaction and incubated for 2 h, followed by TMB substrate to reveal enzymatic cleavage of fetuin by viral NA. The percent inhibition of NA enzymatic activity at each serum dilution was calculated by comparison with values from virus control wells (virus but no serum). End-point NAI titers were calculated as the reciprocal of the highest dilution with at least 50% inhibition.
For MUNANA-based neuraminidase inhibition assays, a series of 2-fold diluted purified recNA (Anhui/1) were mixed with 200 μM of MUNANA (Sigma-Aldrich) and a sigmoidal curve of NA generated. The recNA concentration (1.22 × 10−5 mg/ml) corresponding to the midpoint of the linear section of the curve was chosen for the inhibition assay. A series of 2-fold diluted mAb were mixed with recNA at the fixed concentration of recNA and incubated at room temperature for 1 h, followed by standard procedures of MUNANA assay. The mAb IC50 was calculated by plotting the results on a semi-log graph in Excel.
2.6. Plaque reduction neutralization assay
For pre-infection treatment studies, 300–500 pfu of RG32a was incubated for 30 min with mAb at the indicated concentration followed by infection of a monolayer of Madin-Darby canine kidney (MDCK) cells in six-well plates. After 1 h incubation, cells were washed 4 times with PBS and overlaid with agar supplemented with 1 μg/ml of trypsin without antibody. On day 3 post-infection, the agar overlay was removed, cells were fixed with 70% ethanol for 10 min, and plaques were visualized by staining with crystal violet. For post-infection treatment studies, MDCK cells were first incubated with 300–500 pfu of RG32a in the absence of mAb for 1 h and washed accordingly. The cells were then overlaid with agar supplemented with 1 μg/ml trypsin and the indicated concentration of mAb. Plaques were visualized as indicated above on day 3 post-infection. mAb 0–1h6, targeting the HA of Sh/2, was generated by our laboratory (manuscript in preparation) and served as positive control while mouse-IgG1, a kappa isotype-matched mAb (Crown Bioscience Inc.), served as negative control. To determine plaque size, images were imported into ImageJ2 and 10 random plaques were measured for each condition using the drawing tool, converted to millimeters2, and compared to the control plaques, respectively.
2.7. Prophylactic and therapeutic efficacy studies
To examine the prophylactic efficacy of 3c10-3, 6–8 week old female BALB/c mice (The Jackson Laboratory) (N = 5 per group) were passively immunized by intraperitoneal (i.p.) injection with the indicated dose of mAb in a final volume of 200 μl. Control mice received mouse IgG1, kappa isotype-matched mAb (Crown Bioscience Inc.) or saline. 24 h post transfer, mice were challenged with 5 MLD50 of Anhui/1 H7N9 by intranasal instillation. Mice were anesthetized by isoflurane inhalation for all intranasal (i.n.) instillations and the total volume did not exceed 50 μl.
To examine the therapeutic efficacy of 3c10-3, 6–8 week old female BALB/c mice (N = 5 per group) were challenged with 5 MLD50 of Anhui/1 and treated 1, 2 or 3 days later with a single instillation of mAb (i.p.) at the indicated dose. Control mice received a single instillation of 2 mg/kg mouse-IgG1, kappa isotype-matched mAb (Crown Bioscience Inc.) 1 day post infection. Mice were monitored daily for body weight and mortality for 14 days post-challenge. Mice reaching ≥25% weight loss were euthanized.
2.8. Binding kinetics and epitope mapping
Using biolayer interferometry (BLI) on the Octet Red96 System (ForteBio, Inc., Menlo Park, CA) the binding kinetics for mAb 3c10-3 binding to Anhui/1 recNA containing either R289 (wild type) or K289 was assessed, as previously described (Sleeman et al., 2013; Wilson et al., 2014). For epitope mapping BLI was used to determine 3c10-3 binding to a panel of recNAs with single surface exposed point mutations (Fig. 2a), as compared to wild-type Anhui/1 as previously described (Sleeman et al., 2013; Wilson et al., 2015). Briefly, expression levels of each mutated recNA were determined by western blot and the amount of recNA used in biosensor loading was normalized for equal loading. Anti-HIS biosensors (ForteBio, Inc.) were loaded with a constant amount of each recNA, followed by analysis of antigen-antibody interaction using protocols provided by the manufacturer. Unless otherwise specified, all amino acid residue numbers are in N9 numbering. Data are presented as percentage of antibody binding to mutant recNA compared to WT recNA (100%). A reduction of ≥50% in binding response for each mutant compared to the WT recNA was considered significant (Throsby et al., 2008).
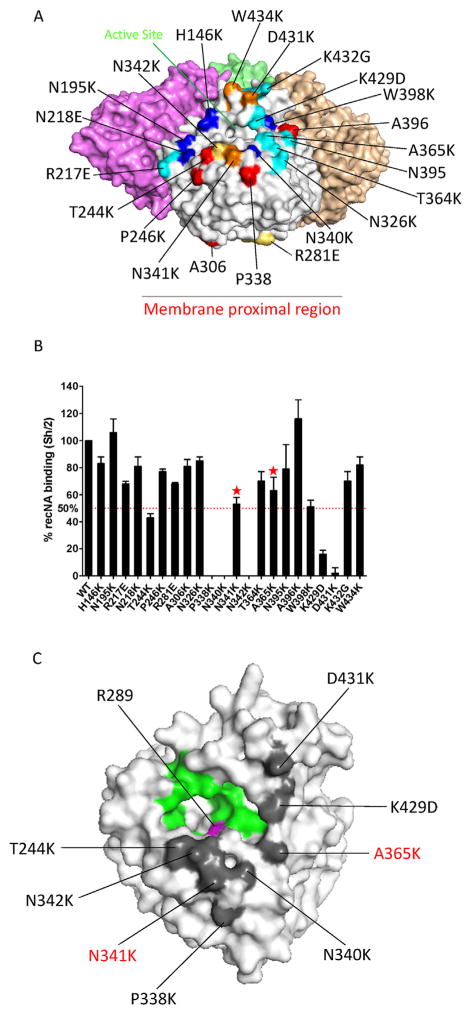
Fig. 2. Epitope map of 3c10-3 as determined by Biolayer Interferometry (BLI).
(A) Location of the amino acid position of 22 individual mutant recNA developed on a NA monomer of the 3-dimensional structure of a N9 tetramer (PDB accession code 4MWJ). The mutant panel is primarily based on previously identified essential amino acids of mAb binding to NA. Red; N1 (Wan et al., 2013), blue; N2 (Gulati et al., 2002; Lentz et al., 1984; Webster et al., 1984), cyan; N9 (Air et al., 1990; Tulip et al., 1992a; Tulip et al., 1992b), yellow; N8 (Saito et al.,1994) and orange; unique selected by us. (B) 3c10-3 binding affinity to each mutant recNA and the percent response compared to wild-type Sh/2 recNA was determined. A greater than 50% reduction in binding activity was the cutoff for significance. “
 ” signifies a mutation that resulted in an increased off rate. (C) The location of residues changes that reduce binding affinity, as identified by BLI, on the 3-dimensional structure of a NA monomer (PDB accession code 4MWJ). Gray; 3c10-3 epitope, green; enzyme active site (Air, 2012) and magenta; NAI resistance mutation (Sleeman et al., 2013). Red text indicates residue changes that significantly increased the 3c10-3 off rate. The structural figures were generated with PyMol software (Schrodinger).
” signifies a mutation that resulted in an increased off rate. (C) The location of residues changes that reduce binding affinity, as identified by BLI, on the 3-dimensional structure of a NA monomer (PDB accession code 4MWJ). Gray; 3c10-3 epitope, green; enzyme active site (Air, 2012) and magenta; NAI resistance mutation (Sleeman et al., 2013). Red text indicates residue changes that significantly increased the 3c10-3 off rate. The structural figures were generated with PyMol software (Schrodinger).
3. Results
3.1. 3c10-3 inhibits A/H7N9 virus NA activity
Splenocytes from a mouse immunized with BPL-inactivated A(H7N9) (RG32a) were fused with Sp2/IL6 myeloma cells. A hybridoma secreting mAb 3c10-3 was identified by screening for recN9 reactivity by ELISA. IgH and IgL sequence analysis confirmed that the hybridoma secreting 3c10-3 is clonal and of the isotype IgG1/Kappa (data not shown). 3c10-3 binds the recNA of Anhui/1 detectably at very low concentration (<5 ng/ml) and did not detectably bind the recNA of Ca/07 (N1) or Perth/16 (N2) (Table 1).
Table 1.
In vitro activity of 3c10-3. mAb were screened by ELISA for reactivity to the indicated recNA and further screened for NA enzyme inhibition by ELLA and MUNANA.
| ELISA (ng/ml)a | ELLA (ng/ml)b | MUNANA (ng/ml)b | |||
|---|---|---|---|---|---|
|
|
|
|
|||
| Anhui/1 recN9 | CA/07 recN1 | Perth/16 recN2 | Sh/2 H6N9 | Anhui/1 recN9 | |
| 3c10-3 | 4.88 | >5000 | >5000 | 3.125 | 6051 |
| FR-969 (Anti-N3) | >5000 | >5000 | >5000 | >2500 | >50,000 |
| FR-1155 (Anti-N2) | >5000 | >5000 | 19.53 | >2500 | >50,000 |
| FR-933 (Anti-N1) | >5000 | 1250 | >5000 | >2500 | >50,000 |
Data are shown as the minimum mAb concentration required to produce a 2-fold signal above negative control..
Data are shown as the minimum concentration of antibody required to inhibit 50% of enzyme activity.
To determine if 3c10-3 is capable of inhibiting the enzymatic activity of N9 in vitro, an enzyme-linked lectin assay (ELLA) and MUNANA assay was employed. 3c10-3 decreased the cleavage of the large substrate fetuin by Sh/2 N9 by 50% at a very low concentration (<4 ng/ml) and also demonstrated the ability to decrease Anhui/1 recN9 cleavage of the small substrate MUNANA at 6051 ng/ml (Table 1).
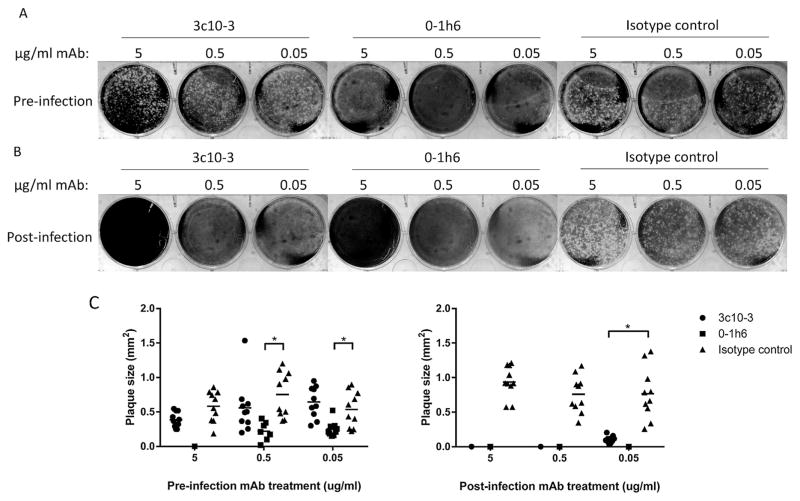
To further characterize 3c10-3 inhibition of NA enzymatic activity on the virus life cycle, plaque reduction assays were performed. The presence of 3c10-3 was shown to reduce A(H7N9) (RG32a) plaque size when supplemented into the agar overlay (Fig. 1b and Fig. 1c, right panel) while having minimal effect on plaque size when virus was only treated with 3c10-3 prior to infection (Fig. 1a and c, left panel) indicating that 3c10-3 inhibits cell-to-cell virus spread. As a control, anti-H7 mAb 0–1h6, which targets the HA1 domain of A(H7N9) (manuscript in preparation), directly neutralized virus infection and inhibited virus spread under the respective conditions.
Fig. 1. 3c10-3 reduces A(H7N9) plaque size when supplemented in media post-infection but does not prevent infection.
(A) Virus was incubated with the indicated concentration of mAb prior to infection of the MDCK monolayer. The infection overlay did not contain any mAb. (B) The MDCK monolayer was infected with virus and the indicated concentration of mAb was incorporated into the infection overlay. Cells were infected for 72 h prior to visualization. mAb 0–1h6 binds the HA1 domain of H7 HA and served as a positive control. (C) Diameters of RG32a plaque size were determined for both panel A (left) and B (right) by randomly selecting and measuring 10 plaques in ImageJ2. Diameters significantly different compared to those of the isotype control mAb (P < 0.05) are indicated by lines and asterisks, respectively.
3.2. 3c10-3 binds near the enzyme active site of NA
To determine critical residues involved in the binding footprint of the NA/3c10-3 complex, we developed a panel of 22 recNA, each containing a single point mutation of a surface exposed residue relative to that of the NA of Sh/2 (Fig. 2a), and tested their binding affinity as determined by BLI. A decrease of ≥50% binding activity was our cutoff for significance. Compared to wild type NA, a significant reduction in binding affinity was noted with NA encoding a change at residues T244, P338, N340, N342, K429 and D431 (Fig. 2b). A significant increase in off rate was also observed with changes at N341 and A365, a strong indication that these residues also contribute to the binding activity of 3c10-3 to NA. These mutated NAs with altered binding patterns to 3c10-3 were confirmed to have correct conformations as they were found to maintain similar neuraminidase activities in MUNANA assays and could be recognized by other N9 monoclonal antibodies (data not shown). These data imply that 3c10-3 directly inhibits the enzymatic activity of N9 NA by binding across the enzymatic active site (Fig. 2c).
3.3. 3c10-3 binds neuraminidase inhibitor resistant NA
The NA of A/Shanghai/1/2013(H7N9) has been shown to be resistant to the NA inhibitor, oseltamivir, due to a R289K substitution in the enzyme active site (Wu et al., 2013).
To determine if 3c10-3 maintains the ability to bind NA carrying the R289K substitution, recNA encoding this mutation was constructed and binding kinetics were determined using BLI. 3c10-3 bound both R289 and K289-containing NA with similar affinity, suggesting that 3c10-3 may be an alternative treatment option for NAI resistant viruses (Table 2).
Table 2.
3c10-3 binding kinetics toward wild type and recN9 with NAI resistance.
| KD (M) | KD error | kon(ms−1) | kon error | kdis(s−1) | kdis error | |
|---|---|---|---|---|---|---|
| A/Anhui/1 recN9 | 2.34 × 10−09 | 7.06 × 10−11 | 1.72 × 1005 | 1.05 × 1003 | 4.01 × 10−04 | 1.19 × 10−05 |
| A/Anhui/1 recN9 (R289K) | 3.11 × 10−09 | 7.42 × 10−11 | 1.65 × 1005 | 1.00 × 1003 | 5.14 × 10−04 | 1.19 × 10−05 |
3.4. Prophylactic administration of 3c10-3 protects mice against lethal challenge infection
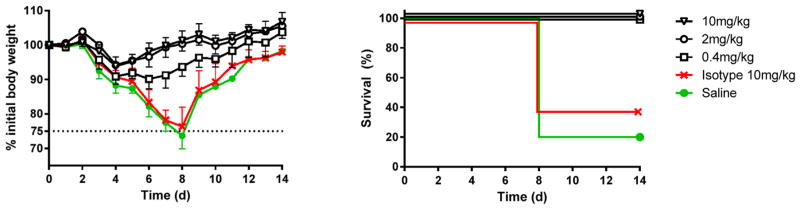
Prophylactic administration of anti-influenza mAb to individuals that are in close contact with an A(H7N9) infected human or poultry could be of great value. To evaluate the protective efficacy of 3c10-3 in vivo, mice were administered 10, 2 or 0.4 mg/kg of 3c10-3 i.p. 24 h prior to an i.n. challenge with 5 times the 50% mouse lethal dose (5MLD50) of Anhui/1 virus. Control mice received 10 mg/kg of isotype-matched mAb or saline, as indicated. Systemic administration of a single dose of 3c10-3 protected all mice from death for each experimental dose (Fig. 3) and resulted in ≤10% weight loss for the lowest dose tested (0.4 mg/kg) (Fig. 3). In contrast, day 8 post challenge, 60% of the isotype control and 80% of the saline control mice were sacrificed due to a >25% loss of weight (Fig. 3). Overall, these results indicate that 3c10-3 protects mice from A(H7N9) lethality and weight loss in a dose-dependent manner when administered prophylactically.
Fig. 3. Intraperitoneal delivery of mAb 3c10-3 affords prophylactic protection against A(H7N9) lethal challenge.
Six to eight week-old female BALB/c mice (n = 5 per group) were injected i.p. with 0.4, 2, or 10 mg/kg of mAb 3c10-3 and 24 h later infected with 5MLD50 of Anhui/1 virus. The percentage of initial body weight (mean ± std) and survival are shown. Control groups received 10 mg/kg of isotype-matched control antibody (IGg1/Kappa) or saline.
3.5. Therapeutic administration of 3c10-3 up to 72 h post lethal challenge protects mice from mortality
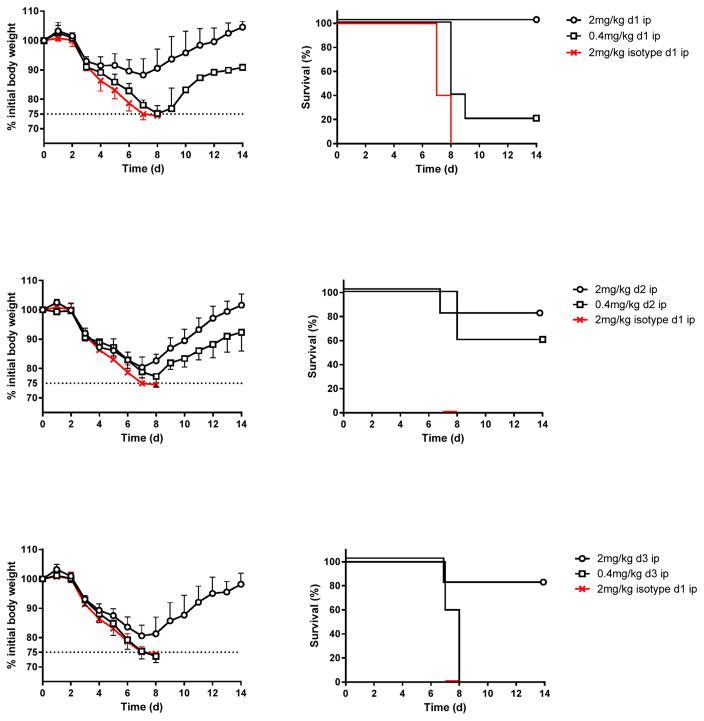
The ability of 3c10-3 to protect mice from a lethal A(H7N9) virus challenge when administered in the period after infection was tested to simulate a clinical situation. Mice were challenged with 5 MLD50 Anhui/1 and subsequently treated with a single dose of either 2 mg/kg or 0.4 mg/kg of 3c10-3 (i.p.) on day 1, 2 or 3 post-infection. Mice that received 2 mg/kg of isotype-matched mAb on day 1 post-infection served as control for all groups. Systemic administration of a single dose of 2 mg/kg on day 1 post-challenge protected all mice against death, while 80% were protected when the same dose was given on day 2 or 3 post-challenge. A single dose of 0.4 mg/kg conferred 20 and 60% protection against death when administered day 1 or 2 post-challenge respectively. None of the mice survived after receiving isotype matched mAb, or when 0.4 mg/kg of 3c10-3 was administered on day 3 post-challenge (Fig. 4).
Fig. 4. Intraperitoneal delivery of 3c10-3 affords therapeutic protection following A(H7N9) lethal challenge.
Six to eight week-old female BALB/c mice (n = 5 per group) were infected with 5MLD50 of Anhui/1 and later injected i.p. with 2 or 0.4 mg/kg of mAb 3c10-3 on d 1, 2 or 3 post-challenge. The percentage of initial body weight (mean ± std) and survival are shown. Control mice received 2 mg/kg of isotype-matched antibody (IGg1/Kappa) on d 1 post-challenge served as control.
4. Discussion
Avian A(H7N9) viruses continue to infect humans, especially in Asia, and pose a pandemic threat. NAI’s are currently the only clinically available class of antiviral drugs for treatment of A(H7N9) infections. However, the emergence of A(H7N9) viruses with resistance to NAI’s remains a concern. Here we show that the anti-NA mAb 3c10-3 protects mice from lethal challenge with A(H7N9) virus, and reduces morbidity and mortality when used therapeutically in already-infected mice. Although a number of anti-NA mAbs with protective efficacy in vivo have been described recently (Doyle et al., 2013; Jiang et al., 2013; Shoji et al., 2011; Wan et al., 2013; Wohlbold et al., 2016), this is the first report of an anti-N9 mAb that has therapeutic activity.
We performed antigenic mapping of 3c10-3 to identify the region of NA with which it interacts. In comparison to influenza hemagglutinin, antigenic sites on NA are poorly understood. Epitope mapping of the N1, N2, N8 and N9 subtypes, using mAb to select escape mutants, has revealed roughly 30 amino acids critical for antibody-NA binding (reviewed; (Air, 2012; Eichelberger and Wan, 2015)). The most thoroughly studied anti-N9 mAb include NC10 and NC41 that were raised against A/tern/Australia/G70C/1975 (H11/N9). The three-dimensional structure of these mAb in association with their substrate, revealing their binding footprint, has been determined (Air et al., 1990; Malby et al., 1994; Tulip et al., 1992a; Webster et al., 1987). Of the eight amino acids identified as essential for their binding to N9 (Air et al., 1990; Tulip et al., 1992b), one (365) was also identified in our epitope mapping, indicating that 3c10-3 has an overlapping footprint with these two N9-specific mAb and suggesting that this may be an immunodominant region. Crystallography study on the 3c10-3 and N9 complex are currently under way and will provide further information on how this mAb binds to its target.
The finding that 3c10-3 spans the enzyme active site of N9 is consistent with the profound functional inhibition of NA activity, including the cleavage of both small and large substrate, this antibody causes. The N9 amino acids identified in the binding of 3c10-3 to the NA of A/H7N9 are highly conserved amongst all known N9 isolates, ranging from 97.4% to 99.7% identity, based on sequence analysis of 649 unique N9 sequences deposited in GISAID (as of 9-16-2016; Data not shown). Variation in these residues may be of particular importance for antigenic drift of A(H7N9) viruses, and should be monitored as part of the ongoing surveillance of A(H7N9) viruses in human and animal reservoirs. More importantly, epi-topes of 3c10-3 are not overlapping R289, a residue known to confer NAI resistance in Group 2 NA when mutated (R292 in N2 numbering (Sleeman et al., 2013)), making it a better option for control of A(H7N9) NAI-resistant viruses.
Apart from the NA active site, a second receptor binding site with hemagglutinating activity has been reported for the N9 subtype of NA (Laver et al., 1984). The residues that reacted with sialic acid (Varghese et al., 1997) are highly conserved within currently circulating H7N9 isolates, indicating these recent isolates may also have this ability. Interestingly, two (A365 and K429) of the N9 amino acids we identified as part of the 3c10-3 footprint are also part of this sialic acid binding site. This suggests that, if the hemadsorbing activity of N9 is maintained in contemporary A/H7N9 viruses, 3c10-3 would also inhibit this activity.
Influenza vaccines are standardized by the amount of HA contained in each dose and traditionally, influenza vaccine studies have focused on measurement of anti-HA antibody responses as correlates of protection. However, it is well known that anti-NA antibodies also can confer some protection (Monto and Kendal, 1973; Murphy et al., 1972; Schild, 1969), although the level of NA in split vaccines vary and currently are not standardized. The growing number of reports detailing the protective efficacy of anti-NA mAb in vivo (Doyle et al., 2013; Jiang et al., 2013; Shoji et al., 2011; Wan et al., 2013; Wohlbold et al., 2016), further support a re-emphasis on including anti-NA antibody responses in the analysis of protection against influenza infection and provides additional support for standardizing the amount of NA contained in each vaccine dose.
Our findings demonstrate that 3c10-3 can effectively prevent A(H7N9) lethality on its own. There have also been reports on the efficacy of mAb(s) against the HA of A(H7N9) virus infections in the mouse model (Chen et al., 2015; Dunand et al., 2015; Tan et al., 2016; Tharakaraman et al., 2015). Since treatment with 3c10-3, or the above mentioned HA-targeting mAb, alone both prevents and treats influenza virus infection, it seems plausible that an antibody cocktail targeting both influenza HA and NA may be a viable therapeutic approach, or could be administered to at-risk individuals (e.g. those in close contact with infected humans or poultry) as a prophylactic measure. Such a combination therapy approach may reduce the potential risk of the development of A(H7N9) escape mutants induced by monotherapy. Since 3c10-3 is of murine origin, humanization of this mAb could potentially increase its therapeutic value in humans by avoiding a human anti-mouse antibody response; this work is currently in progress.
Although we demonstrate that 3c10-3 is capable of direct inhibition of NA enzymatic activity and of virus spread in vitro, the mechanism of action affording protection in vivo is likely to be a combination of inhibition of NA enzyme activity coupled with Fc receptor-guided effector functions such as complement-mediated and/or antibody-dependent cell-mediated cytotoxicity (ADCC). Studies are currently under way to better understand the mechanisms of 3c10-3 protective efficacy in vivo.
Acknowledgments
We thank Lee Pitts, Joo Lee and Jason Goldstein for performing the fusion and subsequent ClonePix selection of hybridoma clones. This work was supported by the Centers for Disease Control and Prevention.
References
- Air GM. Influenza neuraminidase. Influenza Other Respir Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Air GM, Laver WG, Webster RG. Mechanism of antigenic variation in an individual epitope on influenza virus N9 neuraminidase. J Virol. 1990;64:5797–5803. doi: 10.1128/jvi.64.12.5797-5803.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang J, Bao L, Guo L, Zhang W, Xue Y, Zhou H, Xiao Y, Wang J, Wu F, Deng Y, Qin C, Jin Q. Human monoclonal antibodies targeting the haemagglutinin glycoprotein can neutralize H7N9 influenza virus. Nat Commun. 2015;6:6714. doi: 10.1038/ncomms7714. [DOI] [PubMed] [Google Scholar]
- Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, Eichelberger M. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods. 2014;210c:7–14. doi: 10.1016/j.jviromet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Dobson J, Whitley RJ, Pocock S, Monto AS. Oseltamivir treatment for influenza in adults: a meta-analysis of randomised controlled trials. Lancet (London, England) 2015;385:1729–1737. doi: 10.1016/S0140-6736(14)62449-1. [DOI] [PubMed] [Google Scholar]
- Dong J, Matsuoka Y, Maines TR, Swayne DE, O’Neill E, Davis CT, Van-Hoven N, Balish A, Yu HJ, Katz JM, Klimov A, Cox N, Li DX, Wang Y, Guo YJ, Yang WZ, Donis RO, Shu YL. Development of a new candidate H5N1 avian influenza virus for pre-pandemic vaccine production. Influenza Other Respir Viruses. 2009;3:287–295. doi: 10.1111/j.1750-2659.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle TM, Li C, Bucher DJ, Hashem AM, Van Domselaar G, Wang J, Farnsworth A, She YM, Cyr T, He R, Brown EG, Hurt AC, Li X. A monoclonal antibody targeting a highly conserved epitope in influenza B neuraminidase provides protection against drug resistant strains. Biochem Biophys Res Commun. 2013;441:226–229. doi: 10.1016/j.bbrc.2013.10.041. [DOI] [PubMed] [Google Scholar]
- Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng NY, Andrews S, Huang M, Qu X, Huang Y, Salgado-Ferrer M, Ho IY, Taylor W, Hai R, Wrammert J, Ahmed R, Garcia-Sastre A, Palese P, Krammer F, Wilson PC. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Investig. 2015;125:1255–1268. doi: 10.1172/JCI74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberger MC, Wan H. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- Farooqui A, Liu W, Zeng T, Liu Y, Zhang L, Khan A, Wu X, Wu R, Wu S, Huang L, Cai Y, Kelvin AA, Paquette SG, Hu K, Zheng N, Chen H, Xu S, Lin C, Sun P, Yao Z, Wang J, Ma H, Zhu Z, Lin P, Chen W, Fang X, Bermejo-Martin JF, Leon AJ, Kelvin DJ. Probable hospital cluster of H7N9 influenza infection. N Engl J Med. 2016;374:596–598. doi: 10.1056/NEJMc1505359. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations, F. H7N9 Situation Update. 2016. [Google Scholar]
- Gulati U, Hwang CC, Venkatramani L, Gulati S, Stray SJ, Lee JT, Laver WG, Bochkarev A, Zlotnick A, Air GM. Antibody epitopes on the neuraminidase of a recent H3N2 influenza virus (A/Memphis/31/98) J Virol. 2002;76:12274–12280. doi: 10.1128/JVI.76.23.12274-12280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solorzano A, Garcia-Sastre A, Palese P, Bouvier NM. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun. 2013;4:2854. doi: 10.1038/ncomms3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Zhang X, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet (London, England) 2013;381:2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Shichinohe S, Nakayama M, Igarashi M, Ishii A, Ishigaki H, Ishida H, Kitagawa N, Sasamura T, Shiohara M, Doi M, Tsuchiya H, Nakamura S, Okamatsu M, Sakoda Y, Kida H, Ogasawara K. Emergence of H7N9 influenza a virus resistant to neuraminidase inhibitors in nonhuman primates. Antimicrob Agents Chemother. 2015;59:4962–4973. doi: 10.1128/AAC.00793-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, Dekker CL, Zheng NY, Huang M, Sullivan M, Wilson PC, Greenberg HB, Davis MM, Fisher DS, Quake SR. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med. 2013;5:171ra119. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson BE, Bucher DJ, Kilbourne ED. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J Virol. 1989;63:1239–1246. doi: 10.1128/jvi.63.3.1239-1246.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver WG, Colman PM, Webster RG, Hinshaw VS, Air GM. Influenza virus neuraminidase with hemagglutinin activity. Virology. 1984;137:314–323. doi: 10.1016/0042-6822(84)90223-x. [DOI] [PubMed] [Google Scholar]
- Lentz MR, Air GM, Laver WG, Webster RG. Sequence of the neuraminidase gene of influenza virus A/Tokyo/3/67 and previously uncharacterized monoclonal variants. Virology. 1984;135:257–265. doi: 10.1016/0042-6822(84)90135-1. [DOI] [PubMed] [Google Scholar]
- Malby RL, Tulip WR, Harley VR, McKimm-Breschkin JL, Laver WG, Webster RG, Colman PM. The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody. Struct Lond Engl. 1994;1993(2):733–746. doi: 10.1016/s0969-2126(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Monto AS, Kendal AP. Effect of neuraminidase antibody on Hong Kong influenza. Lancet (London, England) 1973;1:623–625. doi: 10.1016/s0140-6736(73)92196-x. [DOI] [PubMed] [Google Scholar]
- Murphy BR, Kasel JA, Chanock RM. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med. 1972;286:1329–1332. doi: 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, Xu K, Shi C, Zhu FC, Zhou MH, Wang H. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ Clin Res ed. 2013;347:f4752. doi: 10.1136/bmj.f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Taylor G, Laver WG, Kawaoka Y, Webster RG. Antigenicity of the N8 influenza A virus neuraminidase: existence of an epitope at the subunit interface of the neuraminidase. J Virol. 1994;68:1790–1796. doi: 10.1128/jvi.68.3.1790-1796.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild GC. Antibody against influenza A2 virus neuraminidase in human sera. J Hyg. 1969;67:353–365. doi: 10.1017/s0022172400041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science. 2013;342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- Shoji Y, Chichester JA, Palmer GA, Farrance CE, Stevens R, Stewart M, Goldschmidt L, Deyde V, Gubareva L, Klimov A, Mett V, Yusibov V. An influenza N1 neuraminidase-specific monoclonal antibody with broad neuraminidase inhibition activity against H5N1 HPAI viruses. Hum Vaccin. 2011;7(Suppl):199–204. doi: 10.4161/hv.7.0.14595. [DOI] [PubMed] [Google Scholar]
- Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg Infect Dis. 2013;19:1521–1524. doi: 10.3201/eid1909.130724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, Krammer F. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 2016;12:e1005578. doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K, Subramanian V, Viswanathan K, Sloan S, Yen HL, Barnard DL, Leung YH, Szretter KJ, Koch TJ, Delaney JC, Babcock GJ, Wogan GN, Sasisekharan R, Shriver Z. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc Natl Acad Sci U S A. 2015;112:10890–10895. doi: 10.1073/pnas.1502374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosh PK, Jacobson RM, Poland GA. Influenza vaccines: from surveillance through production to protection. Mayo Clin Proc. 2010;85:257–273. doi: 10.4065/mcp.2009.0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulip WR, Varghese JN, Laver WG, Webster RG, Colman PM. Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J Mol Biol. 1992a;227:122–148. doi: 10.1016/0022-2836(92)90687-f. [DOI] [PubMed] [Google Scholar]
- Tulip WR, Varghese JN, Webster RG, Laver WG, Colman PM. Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface. J Mol Biol. 1992b;227:149–159. doi: 10.1016/0022-2836(92)90688-g. [DOI] [PubMed] [Google Scholar]
- Varghese JN, Colman PM, van Donkelaar A, Blick TJ, Sahasrabudhe A, McKimm-Breschkin JL. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc Natl Acad Sci U S A. 1997;94:11808–11812. doi: 10.1073/pnas.94.22.11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol. 2013;87:9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J, Eichelberger MC. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat Commun. 2015;6:6114. doi: 10.1038/ncomms7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Air GM, Metzger DW, Colman PM, Varghese JN, Baker AT, Laver WG. Antigenic structure and variation in an influenza virus N9 neuraminidase. J Virol. 1987;61:2910–2916. doi: 10.1128/jvi.61.9.2910-2916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Brown LE, Laver WG. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology. 1984;135:30–42. doi: 10.1016/0042-6822(84)90114-4. [DOI] [PubMed] [Google Scholar]
- Wilson JR, Guo Z, Tzeng WP, Garten RJ, Xiyan X, Blanchard EG, Blanchfield K, Stevens J, Katz JM, York IA. Diverse antigenic site targeting of influenza hemagglutinin in the murine antibody recall response to A(H1N1)pdm09 virus. Virology. 2015;485:252–262. doi: 10.1016/j.virol.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Tzeng WP, Spesock A, Music N, Guo Z, Barrington R, Stevens J, Donis RO, Katz JM, York IA. Diversity of the murine antibody response targeting influenza A(H1N1pdm09) hemagglutinin. Virology. 2014;458–459:114–124. doi: 10.1016/j.virol.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold TJ, Chromikova V, Tan GS, Meade P, Amanat F, Comella P, Hirsh A, Krammer F. Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against lethal H10N8 influenza virus infection in mice. J Virol. 2016;90:851–861. doi: 10.1128/JVI.02275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Bi Y, Vavricka CJ, Sun X, Zhang Y, Gao F, Zhao M, Xiao H, Qin C, He J, Liu W, Yan J, Qi J, Gao GF. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res. 2013;23:1347–1355. doi: 10.1038/cr.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol. 2008;82:10493–10501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney P, Stevens J. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr. 2010;2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol. 2013;87:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Nguyen HT, Carney PJ, Guo Z, Chang JC, Jones J, Davis CT, Villanueva JM, Gubareva LV, Stevens J. Structural and functional analysis of surface proteins from an A(H3N8) influenza virus isolated from New England harbor seals. J Virol. 2015;89:2801–2812. doi: 10.1128/JVI.02723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, Zhang Y, Shi Y, Yang L, Zhu W, Bai T, Qin K, Lan Y, Zou S, Guo J, Dong J, Dong L, Zhang Y, Wei H, Li X, Lu J, Liu L, Zhao X, Li X, Huang W, Wen L, Bo H, Xin L, Chen Y, Xu C, Pei Y, Yang Y, Zhang X, Wang S, Feng Z, Han J, Yang W, Gao GF, Wu G, Li D, Wang Y, Shu Y. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499:500–503. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]