Abstract
Purpose
We analyzed the Exome Aggregation Consortium (ExAC) data set for the presence of individuals with pathogenic genotypes implicated in Mendelian pediatric disorders.
Methods
ClinVar likely/pathogenic variants supported by at least one peer-reviewed publication were assessed within the ExAC database to identify individuals expected to exhibit a childhood disorder based on concordance with disease inheritance modes: heterozygous (for dominant), homozygous (for recessive) or hemizygous (for X-linked recessive conditions). Variants from 924 genes reported to cause Mendelian childhood disorders were considered.
Results
We identified ExAC individuals with candidate pathogenic genotypes for 190 previously published likely/pathogenic variants in 128 genes. After curation, we determined that 113 of the variants have sufficient support for pathogenicity and identified 1,717 ExAC individuals (~2.8% of the ExAC population) with corresponding possible/disease-associated genotypes implicated in rare Mendelian disorders, ranging from mild (e.g., due to SCN2A deficiency) to severe pediatric conditions (e.g., due to FGFR1 deficiency).
Conclusion
Large-scale sequencing projects and data aggregation consortia provide unprecedented opportunities to determine the prevalence of pathogenic genotypes in unselected populations. This knowledge is crucial for understanding the penetrance of disease-associated variants, phenotypic variability, somatic mosaicism, as well as published literature curation for variant classification procedures and predicted clinical outcomes.
Keywords: Mendelian pediatric disorder, phenotypic variability, population, somatic mosaicism, variant classification
Introduction
Next-generation sequencing technology (NGS) has revolutionized the discovery of gene defects causing rare Mendelian disease.1, 2, 3 The estimate for the total number of rare genetic disorders is currently approximately 7,000.1 Based on the pace at which new gene–disease associations are established (~250/year),4 the genetic basis for most monogenic Mendelian disorders is expected to be uncovered in the next decade. However, the major challenge and limiting step of NGS remains in data interpretation, variant classification, and identification of true pathogenic variant(s) relevant to clinical phenotype from amongst hundreds of thousands of variants.
The American College of Medical Genetics and Genomics (ACMG) and Association of Molecular Pathology have provided an important set of standards and guidelines5 for the interpretation of sequence variants with the intent to improve and standardize pathogenicity classification by assessing and weighting the strength of available genetic evidence. The prioritization of candidate variants based on occurrence in a population of unselected individuals is assessed in several ACMG categories5 and is a crucial step in bioinformatics gene-discovery pipelines and pathogenicity classification. When investigating fully penetrant pediatric Mendelian disorders a genotype-centric approach has been recommended. The presence of an individual with a candidate “pathogenic genotype” in a “healthy” population is considered as strong evidence that the candidate variant is benign.5, 6 However, given the unique aspect of certain genes, unknown penetrance, and phenotypic variability of a variant in different genetic backgrounds, the control population may contain individuals with milder phenotypes and/or resilient individuals,7 resulting in misclassification of potentially pathogenic variants.
Understanding the prevalence of disease-associated genotypes implicated in rare Mendelian disorders in a general population is important for the effective use of genomics in variant classification procedures and prediction of clinical outcomes. The Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/)8 compiled whole-exome sequencing data on 60,706 unrelated individuals from 17 disease-specific and population genetic studies, excluding individuals affected by a severe pediatric disease. The ExAC provides information about different human populations, as well as the presence of heterozygous, homozygous, and hemizygous (X-linked recessive conditions) individuals, and has proven to be an invaluable resource for the efficient filtering of candidate disease-causing variants and variant interpretation in patients with rare Mendelian diseases.8
The ExAC data set was evaluated as a representative control for analysis and classification of sequence variants across a subset of 19 genes.9 To further understand the ExAC population, we assessed the sequence and genotype data of the ExAC data set for the presence of individuals with “pathogenic genotypes” previously described in Mendelian pediatric disorders. We analyzed variants spanning a set of 924 genes. Candidate pathogenic genotypes were identified in ExAC individuals for 190 previously published and classified as likely/pathogenic variants in ClinVar10 in 128 genes. After reassessment of variant pathogenicity, 113 variants with possible/disease-associated genotypes implicated in pediatric disease were determined to be present in presumably “healthy” individuals in ExAC.
Materials and methods
The ExAC data set8 was profiled, using custom scripts available upon request, for variants classified as likely pathogenic or pathogenic in a set of 924 genes believed to cause Mendelian childhood disorders using version 0.3 (13 January 2015) of the ExAC database and the ClinVar10 February 2016 data set. The 924 genes (Supplementary Table S1A online) were selected based on the panel of 874 genes recently described in the Resilience Project (Supplementary Table S1B)7 and an additional 50 genes (Supplementary Table S1C) considered to be candidates in our Treatable Intellectual Disability Endeavor in British Columbia (TIDE-BC) neurometabolic gene discovery project3; 25 candidate genes in the TIDE-BC study were already included in the Resilience Project gene panel (Supplementary Table S1C). The ExAC VCF file (ExAC.r0.3.sites.vep.vcf.gz), based on the genome build GRCh37/hg19, was downloaded from the ExAC homepage (http://exac.broadinstitute.org/downloads), while the annotated ClinVar VCF file (version clinvar_20160203.vcf.gz, clinvar_20160203.vcf.gz.tbi) was downloaded from the hg19 folder on the ClinVar FTP site (ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37/). The VCF files were processed using the VariantAnnotation R-package.11 The ExAC and ClinVar variants within the genes of interest were extracted from the raw VCF files using the genomic coordinate ranges, and subsequently position-sorted via vcftool12 and indexed.13 The ClinVar variants were further filtered to include only variants annotated at least once as likely pathogenic (coded as 4) or pathogenic (coded as 5). Subsequently, the ExAC variants were compared against the selected set of ClinVar variants classified as likely/pathogenic to identify the variants present in ExAC, of which the variant annotation (e.g., allele information, genotype frequency) was retrieved from ExAC. The variants were further filtered based on the expected gene/disease-specific inheritance pattern: heterozygous (for dominant conditions), homozygous (for recessive conditions), or hemizygous (for X-linked recessive conditions). The final set of candidate variants was narrowed to include only likely/pathogenic variants supported by at least one PubMed ID (PMID) record (i.e., a peer-reviewed publication) and manually reviewed.
Results
Identification of pathogenic genotypes associated with Mendelian pediatric disorders
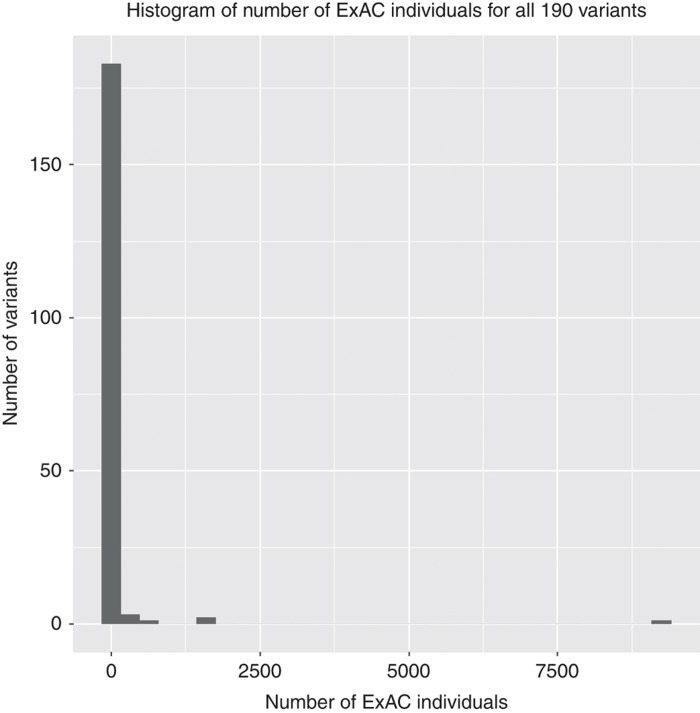
We analyzed the ExAC data set for the presence of likely/pathogenic genotypes for well-characterized and highly penetrant diseases (Supplementary Table S1 for complete list of 924 genes). We identified 190 (Supplementary Table S2A) previously published variants that were classified as likely pathogenic or pathogenic in the ClinVar database (February 2016 released). The variants corresponded to the appropriate genotypes based on the published inheritance patterns across 128 genes: 99 variants were associated with autosomal dominant inheritance, six with X-linked dominant inheritance, 73 with autosomal recessive inheritance, and 12 with X-linked recessive inheritance (Table 1; Supplementary Table S2A). Although the majority of the variants occurred in a small number of individuals (with median at 1) (Figure 1; Supplementary Table S2A), some variants are prohibitively frequent to be truly pathogenic (e.g., rs2266782 observed in 9,246 homozygous individuals in ExAC).
Table 1. Characteristics of the 190 likely/pathogenic genotypes.
| Characteristics | Number of variants (%) |
|---|---|
| Inheritance patterns of the entire set of likely/pathogenic variants | 190 |
| Autosomal dominant | 99 (52%) |
| X-linked dominant | 6 (3%) |
| Autosomal recessive | 73 (39%) |
| X-linked recessive | 12 (6%) |
| High-frequency variants (evidence supporting disease association) | 48 |
| Disease-associated | 5 (10%) |
| Benign and/or obvious but not life-threatening trait | 3 |
| Severe with variable clinical outcome, adult onset or mild | 2 |
| Possibly disease-associated | 9 (19%) |
| Benign and/or obvious but not life-threatening trait | 1 |
| Insufficient evidence to lead to a disease in homozygotes | 4 |
| Susceptibility allele | 3 |
| Severe with variable clinical outcome, adult onset or mild | 1 |
| Insufficient evidence | 34 (71%) |
| Rare variants (evidence supporting disease association) | 142 |
| Disease-associated | 43 (30%) |
| Severe, highly penetrant, early onset | 9 |
| Severe with variable clinical outcome, adult onset or mild | 26 |
| Benign and/or obvious but not life-threatening trait | 8 |
| Possibly disease-associated | 56 (40%) |
| Severe, highly penetrant, early onset | 3 |
| Severe with variable clinical outcome, adult onset or mild | 18 |
| Benign and/or obvious but not life-threatening trait | 16 |
| Insufficient evidence to lead to a disease in homozygotes | 8 |
| Susceptibility allele | 8 |
| Modifier allele | 3 |
| Insufficient evidence | 43 (30%) |
Figure 1.
Histogram plot of the number of ExAC individuals with 190 likely/pathogenic variants corresponding to candidate genotypes. This plot shows the distribution of the entire set of 190 variants in terms of the number of ExAC individuals corresponding to each variant (bin size of histogram=30). The y-axis represents the number of variants with number of ExAC individuals equal to the corresponding value on the x-axis. According to the five-number summary of all 190 variants: sample minimum = 1, lower quartile = 1, median = 1, upper quartile = 6, sample maximum = 9,246.
High-frequency variants associated with Mendelian pediatric disorders
With large-scale sequencing projects using NGS data, some of the previously published variants deemed to be Mendelian disease-causing have been shown to be more common in a population, and thus the evidence for pathogenicity of those variants has been challenged.8, 14, 15, 16 Recently, MacArthur et al. manually inspected evidence of pathogenicity for 192 variants with an allele frequency >1% that were previously classified as disease mutations in the Human Gene Mutation Database (HGMD).8 The majority (134 of 192) of these variants have since been retired or reclassified in HGMD.8 Only nine variants had sufficient data to support disease association.8 To assess the 190 likely/pathogenic variants identified here, we grouped them into two sets: frequent variants (Table 1; Supplementary Table S2B) and rare variants (with ExAC highest population frequency of <1% for recessive and <0.1% for dominant disorders) (Table 1; Supplementary Table 2C).
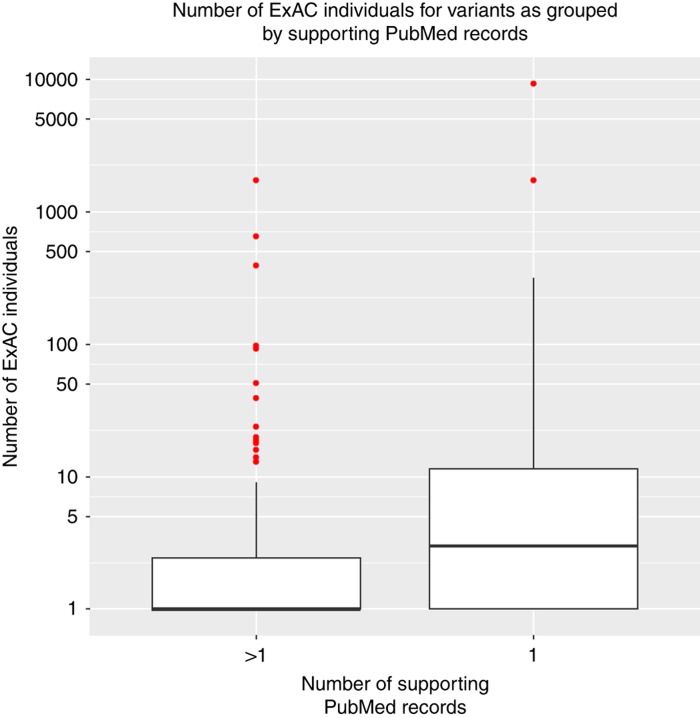
The frequent variant set contained 48 likely/pathogenic variants, 38 in recessive disorders and 10 in dominant disorders (Supplementary Table S2B). Only 6 of the 192 variants reported by Lek et al.,8 which were classified as disease mutations in the HGMD, were also classified as likely/pathogenic by ClinVar10 and were identified in our set of 48 frequent variants. The remaining 42 frequent likely/pathogenic variants found in ExAC have not been reported.8 We observed that these frequent variants tend to be supported by less evidence for pathogenicity classification (only one PMID record) compared to the rare variant set (tends to be supported by more than one PMID record) (Figure 2). We then performed a literature review and assessed the available support for variant pathogenicity and found that 34 out of 48 (71%) frequent variants had insufficient evidence (IE) for pathogenicity given the assessed genotypic configuration (Table 1; Supplementary Table S2B), further supporting the need for reclassification of the variants in databases.8, 14, 15, 16, 17, 18 In agreement with Lek et al.,8 we found that the 6 overlapping frequent variants (rs11555096, rs397514333, rs13078881, rs28937880, rs373002889, and rs35312232) had insufficient evidence to be disease-causing in homozygotes. The 14 variants with medium to strong support for pathogenicity (disease-associated (DA) or possibly disease-associated (PDA)) were either linked to benign and/or non-life-threatening traits, mild and/or variable clinical outcomes, increased susceptibility rather than causal, or had evidence to cause a disease when inherited in compound heterozygous state but with insufficient evidence to lead to a disease in homozygotes (Table 1; Supplementary Table S2B).
Figure 2.
Box plot of the number of ExAC individuals with 190 likely/pathogenic variants grouped according to the number of PMID support. This plot shows distribution of the entire set of 190 variants. The y-axis represents a log10 scale, while the x-axis reflects grouping of the variants based on the level of the published support (PMID=1 vs. PMID>1). Outliers are represented by a dot. It shows that the PMID = 1 group has a higher number of ExAC individuals than the PMID >1 group.
Rare pathogenic variants with variable penetrance and clinical outcomes
The rare variant set contained 142 likely/pathogenic variants (Table 1); 47 corresponded to recessive disorders and 95 corresponded to dominant disorders (Supplementary Table S2C). Our literature review and assessment revealed that 43 (30%) of the rare variants have insufficient evidence for pathogenicity (Table 1; Supplementary Table S2C). However, unlike the frequent variants, the majority of the rare variants (n=99) did have evidence supporting either the disease association (n=43) or possible disease association (n=56) (Table 1; Supplementary Table S2C). The analysis of phenotypic severity of these 99 variants revealed that the majority (n=87) have been described to contribute to variable and/or milder (even benign) clinical manifestations (Table 1). For example, Gaucher disease (MIM 230800) is an autosomal recessive inborn error of metabolism caused by mutations in GBA. There is variable age of onset and different combinations of multiorgan involvement.19 One of the ExAC individuals is homozygous for the well-established Gaucher disease pathogenic variant N370S (NM_001005741.2; c.1226A>G (p.N409S)); however, the age of onset and clinical variability have been shown to be the highest in patients homozygous for the N370S variant20 suggesting that this ExAC individual may have been unaffected or only mildly affected at the time of recruitment. Furthermore, two ExAC individuals are homozygous for the nonsense Y176* (NM_174878:c.528T>G (p.Y176*)) variant in CLRN1, which has been reported in unrelated individuals with Usher syndrome (MIM 276902).21 Usher syndrome is characterized by progressive sensorineural hearing loss, vestibular dysfunction, and retinitis pigmentosa, which is an obvious but non-life-threatening condition. These examples imply that ExAC contains individuals who may have had a mild-to-no clinical presentation or a presentation of a non-life-threatening condition at the time of recruitment.
Rare pathogenic variants implicated in severe Mendelian pediatric disorders
In addition to rare genotypes that may be attributed to variable, milder clinical manifestations of pediatric disorders, we identified 18 ExAC individuals with possibly/disease-associated genotypes implicated in severe Mendelian early-onset disorders (Table 2) with high penetrance and low variability based on current knowledge. These included five reported homozygotes for recessive disorders (glycogen storage disease (MIM 232200), pontocerebellar hypoplasia (MIM 612389), hyaline fibromatosis syndrome (MIM 228600), congenital dyserythropoietic anemia (MIM 224100) and mitochondrial complex III deficiency (MIM 124000)); one hemizygote for a X-linked recessive disorder (chondrodysplasia punctate (MIM 302950)); and 12 heterozygotes for autosomal dominant disorders (Pfeiffer syndrome (MIM 101600), Apert syndrome (MIM 101200), cardiofaciocutaneous syndrome (MIM 115150), Schuurs–Hoeijmakers syndrome (MIM 615009) and Bohring–Opitz syndrome (MIM 605039)) (Table 2).
Table 2. 18 ExAC individuals with pathogenic genotypes implicated in severe Mendelian early-onset disorders.
| Gene | Phenotype | rsID | Variant | Zygosity | Variant support | ExAC | Severity |
|---|---|---|---|---|---|---|---|
| CAP Set | |||||||
| G6PC | Glycogen storage disease Type Ia | rs1801175 (DA) | c.247C>T; p.R83C (NM_000151.2) | Hom | 156 (100%) | 1 | Severe hypoglycemia and hepatomegaly manifesting as growth retardation; autosomal recessive |
| TSEN2 | Pontocerebellar hypoplasia | rs113994149 (PDA) | c.926A>G; p.Y309C (NM_025265.3) | Hom | 63 (98%) | 1 | Severe early childhood disorder characterized by an abnormally small cerebellum and brainstem; progressive microcephaly from birth combined with extrapyramidal dyskinesia and chorea, epilepsy, and normal spinal cord findings; autosomal recessive |
| FGFR1 | Pfeiffer syndrome | rs121909627 (DA) | c.755G>C; P252R (NM_023110.2) | Het | 35 (43%) | 1 | Severe autosomal dominant craniosynostosis syndrome with characteristic anomalies of the hands and feet |
| Other | |||||||
| ANTXR2 | Hyaline fibromatosis syndrome | rs312262690 (PDA) | c.1073dupC; p.A359Cfs (NM_058172.5) | Hom | Possibly low-quality site | 1 | Severe disorder characterized by abnormal growth of hyalinized fibrous tissue usually affecting subcutaneous regions on the scalp, ears, neck, face, hands, and feet; autosomal recessive |
| SEC23B | Dyserythropoietic anemia, congenital, type II | rs121918221 (DA) | c.325G>A; p.E109K (NM_032985.4) | Hom | 46 (100%) | 1 | Severe disorder characterized by ineffective erythropoiesis, presence of bi- and multinucleated erythroblasts in bone marrow, progressive splenomegaly, gallstones and iron overload, liver cirrhosis or cardiac failure; autosomal recessive |
| BCS1L | Mitochondrial complex III deficiency, nuclear type 1 | rs121908578 (DA) | c.550C>T; p.R184C (NM_004328.4) | Hom | 43 (100%) | 1 | Severe disorder characterized by early onset (at birth) lactic acidosis, hypotonia, hypoglycemia, failure to thrive, encephalopathy, and delayed psychomotor development. Many patients die in early childhood, but some may show longer survival; autosomal recessive |
| ARSE | Chondrodysplasia punctata | rs28935474 (PDA) | c.1732C>T; p.P578S (NM_000047.2) | Hemi | Possibly low-quality site | 1 | Severe disorder characterized by punctiform calcification of the bones; X-linked recessive |
| BRAF | Cardiofaciocutaneous syndrome | rs121913355 (DA) | c.1406G>A; p. G469E (NM_004333.4) | Het | 30 (16%) | 1 | Severe disorder characterized by multiple congenital anomalies, distinctive facial appearance, heart defects, and intellectual disability; autosomal dominant |
| BRAF | Cardiofaciocutaneous syndrome | rs397507473 (DA) | c.1403T>C; p.F468S (NM_004333.4) | Het | 161 (37%) | 1 | Severe disorder characterized by multiple congenital anomalies, distinctive facial appearance, heart defects, and intellectual disability; autosomal dominant |
| FGFR2 | Apert syndrome | rs79184941 (DA) | c.755C>G; p.S252W (NM_022970.3) | Het | Unknown | 1 | Severe disorder characterized by craniosynostosis, midface hypoplasia, and syndactyly of the hands and feet with a tendency to fusion of bony structures; autosomal dominant |
| PACS1 | Schuurs–Hoeijmakers syndrome | rs398123009 (DA) | c.607C>T; p.R203W (NM_018026.3) | Het | 30 (19%) | 1 | Severe disorder characterized by mental retardation, distinct craniofacial features, and variable additional congenital abnormalities; all to date reported individuals have the same de novo R203W variant; autosomal dominant |
| ASXL1 | Bohring–Opitz syndrome | rs373145711 (DA) | c.1210C>T; p.R404* (NM_015338.5) | Het | 43 (41%) 16 (21%) 15 (23%) 63 (43%) 18 (19%) Unknown Unknown | 7 | Severe disorder characterized by severe intrauterine growth retardation, poor feeding, profound mental retardation, trigonocephaly, prominent metopic suture, exophthalmos, nevus flammeus of the face, upslanting palpebral fissures, hirsutism, and flexion of the elbows and wrists with deviation of the wrists and metacarpophalangeal joints; autosomal dominant |
CAP, core allele panel; DA, disease-associated; Hemi, hemizygous; Het, heterozygous; Hom, homozygous; PDA, possibly disease-associated.
One explanation for the presence of the individuals with genotypes implicated in severe Mendelian early-onset disorders may be that the called genotypes are not the true genotypes of these individuals. The ExAC database offers an interactive visualization of reads through the Integrative Genomics Viewer.22 We inspected the reads (depth and alternative variant read support) and evidence for homozygosity and heterozygosity calls (Table 2). Three of the called homozygotes/hemizygotes for recessive disorders may not be true homozygotes (two due to possibly low-quality sites (ANTXR2 and ARSE) and one due to one read supporting a reference base (TSEN2), Table 2), while three had good (100% alternative variant) support and may truly exemplify incomplete penetrance or resilience (Table 2). The inspection of six heterozygous variants (12 individuals) revealed potential evidence of somatic mosaicism in five individuals (Table 2).
As a part of the Resilience Project, established with the goal to search for and identify presumably healthy individuals with completely penetrant Mendelian disease-causing mutations, a core allele panel comprised of 674 well-established and well-annotated pathogenic variants representing 125 severe, early-onset diseases was compiled7. We tested our set of likely/pathogenic variants (Supplementary Table S2A) and identified six of the alleles from the core allele panel (Supplementary Table S2A). Three of the variants were grouped in the “severe with variable clinical outcome, adult onset or mild” category (Supplementary Table S2C). The R76W variant (ADA: NM_000022.2: c.226C>T; (p.R76W)) was observed in two ExAC homozygotes and is associated with partial adenosine deaminase deficiency (MIM 102700), the Y63C (PTPN11: NM_002834.3:c.188A>G (p.Y63C)) and N308D (PTPN11: NM_002834.3:c.922A>G (p.N308D)) variants were each observed in one ExAC heterozygote each, and both have been associated with Noonan syndrome (MIM 163950). The remaining three variants from the core allele panel are believed to result in severe, highly penetrant, early-onset Mendelian disorders (Table 2). One heterozygous variant (NM_023110.2: c.755C>G; (p.P252R)) in FGFR1 has also been identified in one of the 13 first-tier candidate individuals proposed to be resilient to severe Mendelian childhood diseases, in this case for Pfeiffer syndrome (MIM 101600).7 Importantly, the 17 additional ExAC individuals identified here have neither been reported in the first-tier (n=13) nor the second-tier candidate resilient individuals (n=111).7
Variant interpretation using ExAC in rare disease discovery
In our TIDE-BC project, we perform NGS in well-characterized patients with unexplained neurometabolic phenotypes to identify genetic causes of intellectual developmental disorders (IDDs) that may be treatable (i.e., due to underlying inborn errors of metabolism).3 As a part of our semiautomated gene-discovery pipeline,3 we utilize the ExAC database in the variant prioritization and classification process. However, we do take into account that ExAC contains individuals with possible/disease-associated genotypes (Supplementary Table S2D).
From October 2012 through January 2016, more than 150 probands meeting the TIDE-BC eligibility criteria were subjected to genome and exome sequencing. The results of our efforts were recently published on a subset (n=47) of these probands.3 As outlined below, in three probands (Table 3) variants were identified in candidate genes previously implicated in human disease that were also present in at least one individual in the ExAC database.
Table 3. TIDE-BC patients with established pathogenic genotypes reported in the ExAC individuals.
| Gene | Phenotype | rsID | Variant | Zygosity | ExAC | TIDE patient phenotype |
|---|---|---|---|---|---|---|
| GJB2 | Deafness, autosomal recessive | rs72474224 (DA_frequent) | c.109G>A; p.V37I (NM_004004.5) | Hom | 39 | Moderate stable sensorineural hearing loss |
| CBL | Noonan syndrome–like disorder | rs267606706 (DA_rare) | c.1111T>C; p.Y371H (NM_005188.3) | Het | 2 | Mild IDD, ADHD, dysmorphic features, splenomegaly, thrombocytopenia |
| PACS1 | Schuurs–Hoeijmakers syndrome | rs398123009 (DA_rare) | c.607C>T; p.R203W (NM_018026.3) | Het | 1 | Severe IDD, microcephaly, facial dysmorphisms, myopia, bifid uvula and submucous cleft, progressive ataxia and cerebellar atrophy, dysplastic pulmonary and aortic valves, failure to thrive |
ADHD, attention deficit hyperactivity disorder; Het, heterozygous; Hom, homozygous; IDD, intellectual development disorder.
Sensorineural hearing loss
A proband with moderate but stable sensorineural hearing loss was found to be homozygous for the V37I variant in GJB2,3 a known cause of hearing impairment (deafness, autosomal recessive 1A (MIM 220290)) with reduced penetrance (Table 3).23 The V37I variant is a frequent, disease-associated variant (Supplementary Table S2B) with 39 homozygous individuals reported in ExAC. Given that there is reported evidence for reduced penetrance, and that the trait is obvious but not life-threatening, the V37I variant is considered to be diagnostic in this TIDE-BC patient.
Noonan syndrome–like disorder
A proband with mild IDD, attention deficit hyperactivity disorder, dysmorphic features, splenomegaly, and thrombocytopenia was found to be heterozygous for a de novo variant in CBL (Table 3).3 The Y371H variant is a rare variant that was previously reported in multiple unrelated patients with Noonan syndrome–like disorder and onset of juvenile myelomonocytic leukemia (MIM 613563) in the first years of life (Supplementary Table S2C).24, 25, 26 The variant is found in two ExAC individuals (one with potential evidence for somatic mosaicism (20% variant support) and one with good evidence for heterozygosity (48% variant support)). However, the potential somatic mosaicism and the reported variable clinical outcome with possibly milder presentation and/or adult onset may explain the occurrence of the individuals with pathogenic genotypes in the ExAC database. Thus, the Y371H variant is considered to be diagnostic in our patient.
Schuurs–Hoeijmakers syndrome
A proband with severe IDD, microcephaly, facial dysmorphisms, and failure to thrive was found to be heterozygous for a de novo variant affecting PACS1 (Table 3).3, 27 The R203W variant is a rare variant that has been reported in all the unrelated patients reported with Schuurs–Hoeijmakers syndrome (MIM 615009).27, 28 The clinically recognizable phenotype of the R203W variant led to diagnosis of the Schuurs–Hoeijmakers syndrome in our patient, despite the presence of one ExAC individual with the same called genotype. Furthermore, the 19% read support for the variant may suggest somatic mosaicism rather than heterozygosity in the ExAC individual. Alternatively, if there were a possibility to confirm the ExAC individual as a true heterozygote and not showing the phenotypic features, she/he may hold an important clue to unknown modifier(s) that alleviate(s) effect of the R203W variant, and potential preventive and/or therapeutic avenues for patients with Schuurs–Hoeijmakers syndrome.
Discussion
Until now, the focus of NGS technology in rare disorders has been predominantly placed on the discovery of underlying primary pathogenic variant(s) affecting a single gene that explains observed clinical phenotype(s) of a patient or a cohort of patients. The availability of large-scale sequencing data sets from data aggregation consortia7, 8 offers novel possibilities to assess the prevalence of disease-associated genotypes implicated in rare Mendelian disorders in an unselected population.8, 29 This is an unprecedented research avenue that holds promise to reveal new knowledge on penetrance, phenotypic variability, somatic mosaicism, genetic modifiers, and interpretation of variant pathogenicity in context of individual genetic backgrounds, not unlike personalized medicine.
Here we assess the ExAC resource8 for the presence of individuals with pathogenic genotypes implicated in Mendelian pediatric disorders and uncover 15,698 ExAC individuals with candidate pathogenic genotypes based on likely/pathogenic classification of 190 variants in the ClinVar database.10 We show that this unexpectedly large number is mainly due to 77 variants (n=13,981 individuals) that currently have insufficient evidence to be classified as pathogenic and/or likely pathogenic. These findings further illustrate the power of large-scale sequencing projects and underscore the importance of continued data sharing for systematic reassessment of the reported disease–variant associations in centralized databases like ClinVar.8, 14, 15, 16, 17, 18
Importantly, we identify 1,717 individuals (2.8% of the ExAC population) with possible (n=1,535) disease-associated (n=182) genotypes implicated in rare Mendelian disorders. The assessment of the published evidence on their phenotypic severity revealed that the majority of those genotypes may (i) either have variable clinical outcome presenting as mild form of the disease (e.g., partial adenosine deaminase deficiency (MIM 102700)) and/or appear with later adult onset (e.g., Alexander disease (MIM 203450)), (ii) result in benign (e.g., benign familial hematuria (MIM 141200)) and/or obvious but not life-threatening trait (e.g., deafness), (iii) have insufficient evidence to cause a disease in homozygotes (e.g., R395W variant in NCF2), (iv) confer susceptibility (e.g., R726L in the AR and susceptibility to prostate cancer), or (v) are considered to be modifying alleles (e.g., R488H variant in SLC22A5). Therefore, considering a potential milder effect of these variants, it may be possible that deeper phenotyping and follow-up would reveal signs of the disease-associated phenotypes in those individuals. Unfortunately, clinical phenotype data is not available for most of the ExAC individuals8 or individuals in other control population databases,7 further stressing the need for future efforts to combine both clinical and genetic data to better interpret and understand human disease.30
Beyond the genotypes that may be attributed to variable, milder clinical manifestations of pediatric disorders, we identified ExAC individuals expected to exhibit a severe Mendelian early-onset disorder. For example, Bohring–Opitz syndrome (MIM 605039) is a rare severe pediatric condition characterized by profound IDD and multiple congenital malformations.31 In all the individuals described to date, de novo nonsense or frameshift variants in ASXL1 were identified.31, 32 One of those variants, the R404*, has been observed in seven presumably heterozygous ExAC individuals (Table 2). Although craniofacial dysmorphism with reduced penetrance was demonstrated in Asxl1 heterozygous null mice,33 reduced penetrance in patients with Bohring–Opitz syndrome has not been reported. Considering the severity and rarity of this seemingly fully penetrant condition in humans, as well as the fact that R404* variant truncates the protein early in the protein sequence, we do expect that some if not all of the seven ExAC individuals may be due to somatic mosaicism.34, 35, 36 Indeed, available data of five ExAC individuals revealed evidence of somatic mosaicism in at least three of the individuals (percent of reads supporting the variant is 19, 21, and 23% rather than the expected ~50% Table 2). Unfortunately, the DNA samples of these individuals are not available to us for testing via Sanger sequencing or pyrosequencing to confirm true genotypes, which is another important aspect to be taken into consideration in future large-scale population sequencing projects. Nonetheless, the R404* pathogenic variant serves as an important advocate for careful use of frequencies in population databases in variant classification procedures. Although the exact prevalence of Bohring–Opitz syndrome is unknown, there are fewer than 60 children in the world diagnosed with this syndrome, suggesting that ExAC frequency of the pathogenic p.R404* variant is higher than expected. The ASXL1 p.R404* variant exemplifies that hard frequency cutoffs based on prevalence of a disorder may further complicate variant interpretation and lead to misclassification of true pathogenic variants.
Recently Chen et al.17 identified 13 candidate resilient individuals spanning eight diseases as part of the Resilience Project effort.7 The P252R heterozygous variant in the FGFR1 (Pfeiffer syndrome (MIM 101600)) that we identified in one ExAC individual is also observed in one of the confirmed Resilience Project heterozygotes.7 Interestingly, this heterozygous European (non-Finnish) individual was identified from the SWE-SCZ data set (Swedish schizophrenia cohort), which is also one of the ExAC contributing projects,8 suggesting that this may be the same individual. Thus, the ExAC data set does contain individuals with true pathogenic genotypes who may hold clues to the better understanding of penetrance in rare Mendelian disorders.
The important limitation of our study is in the unavailability of additional information (e.g., age and clinical phenotype) and patient samples for further assessment. However, this information is generally unavailable to clinicians who rely on ExAC and other currently available population databases in variant classification procedures. Accurate interpretation of genetic variants based on population frequencies has a direct impact on patient welfare both from the perspective of a variant misclassified as pathogenic (e.g., due to insufficient knowledge of different ancestry backgrounds),18 as well as a variant classified as likely/benign due to lack of understanding of a control population and true clinical etiology of the genetic disease. While recent ACMG gudelines5 have advanced the variant classification procedures, these tend to be stringent in the recommended use of five specific categories (pathogenic, likely pathogenic, uncertain significance, likely benign, and benign)5 and tailored for assumed highly penetrant Mendelian disorders.5 Here we show that up to 2.8% of the ExAC population does have likely/pathogenic genotypes reported in ClinVar, which is important information to be considered when interpreting variants in known as well as yet to be discovered rare Mendelian disorders. Our results show that particular caution needs to be exercised when applying the recommended genotype-centric approach.5, 6 The possibilities of (i) genotypes being different than reported (e.g., due to somatic mosaicism, sequencing, or alignment errors), (ii) unknown penetrance, and (iii) unknown phenotypic variability (e.g., milder clinical representation) need to be taken into consideration. Continued data aggregation efforts that combine clinical and genetic data of diverse populations will play a crucial role in better understanding of variability and penetrance of both known and novel rare disease variants in the context of different genetic and nongenetic backgrounds.30 As these new discoveries continue to broaden our knowledge of rare diseases genetics, there is a growing need to expand the recommended five-category ACMG gudelines.5 Currently, the “likely pathogenic” and “likely benign” variants are defined as having greater than 90% probability of being disease-causing or benign, according to the ACMG gudelines,5 while the uncertain significance (VUS) category is a default classification for an unknown, an entire spectrum of variants that given the current lack of knowledge have insufficient evidence to adequately estimate their clinical significance. Recently, a seven-point scale has been proposed where the VUS category has been further subcategorized to include three subclasses: VUS-suggesting pathogenic, VUS, and VUS-suggesting benign.37 The proposed scale adds much needed granularity in the classification procedure where VUS is equal to unknown, while possibly disease-causing and possibly benign variants are recognized in addition to probable (>90%) disease-causing or benign variants. With the new discoveries of rare diseases genetics, including the role of somatic mosaicism, incomplete penetrance, and variable expressivity, guidelines for variant interpretation are expected to be updated and refined to enhance our ability to diagnose patients.
Acknowledgments
This work was supported by funding from Genome Canada Bioinformatics and Computational Biology (255ONT), Canadian Institutes of Health Research (CIHR): Bioinformatics and Computational Biology (BOP-149430), BC Children’s Hospital Foundation (Treatable Intellectual Disability Endeavour in British Columbia: 1st Collaborative Area of Innovation http://www.tidebc.org), Genome Canada/Genome BC/CIHR Large Scale Applied Research Grant ABC4DE project (174CDE), and the Ministry of Education and Science of the Republic of Serbia (ON173052). We thank D. Arenillas and M. Hatas for systems support, and D. Pak for research management support.
ADDENDUM
While this manuscript was In Press others also demonstrated presence of somatic ASXL1 variants in reference databases.38
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/gim
The authors declare no conflict of interest.
Supplementary Material
References
- Boycott KM, Vanstone MR, Bulman DE et al, Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet 2013;14:681–691. [DOI] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F et al, Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Shyr C, Ross CJ et al, Exome sequencing and the management of neurometabolic disorders. N Engl J Med 2016;374:2246–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger AM, Guturu H, Bernstein JA et al, Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med 2016;19:209–214. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al, Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J-L, Conley ME, Seligman SJ et al, Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med 2014;211:2137–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J et al, Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol 2016;34:531–538. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV et al, Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Gardner SA, Hovhannisyan H et al, Exploring the landscape of pathogenic genetic variation in the ExAC population database: insights of relevance to variant classification. Genet Med 2015;18:850–854. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M et al, ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44(D1):D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenchain V, Lawrence M, Carey V et al, VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics 2014;30:2076–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G et al, The variant call format and VCFtools. Bioinformatics 2011;27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A et al, The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Dinwiddie DL, Miller NA et al, Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 2011;3:65ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Chen Y, Ayub Q et al, Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet 2012;91:1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Redin C, Mandel J-L. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet 2013;93:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan-Müller C, Ahlberg G, Ghouse J et al, Integration of 60,000 exomes and ACMG guidelines question the role of catecholaminergic polymorphic ventricular tachycardia associated variants. Clin Genet 2016;91:63–72. [DOI] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL et al, Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016;375:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato D, Stachiw T, Clarke JTR et al, Gaucher disease: variability in phenotype among siblings. J Inherit Metab Dis 2004;27:659–669. [DOI] [PubMed] [Google Scholar]
- Grabowski GA, Zimran A, Ida H. Gaucher disease types 1 and 3: phenotypic characterization of large populations from the ICGG Gaucher Registry. Am J Hematol 2015;90(suppl 1):S12–S18. [DOI] [PubMed] [Google Scholar]
- Joensuu T, Hämäläinen R, Yuan B et al, Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet 2001;69:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W et al, Integrative genomics viewer. Nat Biotechnol 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak A, Skórka A, Mueller-Malesińska M et al, M34T and V37I mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. Am J Med Genet A 2007;143A:2534–2543. [DOI] [PubMed] [Google Scholar]
- Loh ML, Sakai DS, Flotho C et al, Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood 2009;114:1859–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli S, De Luca A, Stellacci E et al, Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am J Hum Genet 2010;87:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez B, Mechinaud F, Galambrun C et al, Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomonocytic leukaemia. J Med Genet 2010;47:686–691. [DOI] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers JHM, Oh EC, Vissers LELM et al, Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am J Hum Genet 2012;91:1122–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers JHM, Landsverk ML, Foulds N et al, Clinical delineation of the PACS1-related syndrome—report on 19 patients. Am J Med Genet A 2016;170:670–675. [DOI] [PubMed] [Google Scholar]
- Natarajan P, Gold NB, Bick AG et al, Aggregate penetrance of genomic variants for actionable disorders in European and African Americans. Sci Transl Med 2016;8:364ra151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrai AK, Ioannidis JPA, Kohane IS. Clinical genomics: from pathogenicity claims to quantitative risk estimates. JAMA 2016;315:1233–1234. [DOI] [PubMed] [Google Scholar]
- Hoischen A, van Bon BWM, Rodríguez-Santiago B et al, De novo nonsense mutations in ASXL1 cause Bohring–Opitz syndrome. Nat Genet 2011;43:729–731. [DOI] [PubMed] [Google Scholar]
- Magini P, Della Monica M, Uzielli MLG et al, Two novel patients with Bohring–Opitz syndrome caused by de novo ASXL1 mutations. Am J Med Genet A 2012;158A:917–921. [DOI] [PubMed] [Google Scholar]
- Abdel-Wahab O, Gao J, Adli M et al, Deletion of AsXL1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med 2013;210:2641–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE et al, Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J et al, Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J et al, Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbassi I, Maston GA, Love A et al, A standardized DNA variant scoring system for pathogenicity assessments in Mendelian disorders. Hum Mutat 2016;37:127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlston CM, O’Donnell-Luria AH, Underhill HR et al, Pathogenic ASXL1 somatic variants in reference databases complicate germline variant interpretation for Bohring-Opitz Syndrome. Hum Mutat 2017;38:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.