Abstract
Background:
Pancreatic carcinoma (PC) is an aggressive malignancy that lacks strategies for early detection. This study aimed to develop a coherent, high-throughput and non-discriminatory pipeline for the novel clinical biomarker discovery of PC.
Methods:
We combined mass spectrometry (MS)-intensive methods such as isobaric tags for relative and absolute quantitation with two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2DLC-MS/MS), 1D-targeted LC-MS/MS, prime MRM (P-MRM) and stable isotope dilution-based MRM (SID-MRM) to analyse serum samples from healthy people (normal control, NC), patients with benign diseases (BD) and PC patients to identify novel biomarkers of PC.
Results:
On the basis of the newly developed pipeline, we identified >1000 proteins, verified 142 differentially expressed proteins and finally targeted four proteins for absolute quantitation in 100 serum samples. The novel biomarker panel of apolipoprotein E (APOE), inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3), apolipoprotein A-I (APOA1), apolipoprotein L1 (APOL1), combining with CA19-9, statistically-significantly improved the sensitivity (95%) and specificity (94.1%), outperforming CA19-9 alone, for the diagnosis of PC.
Conclusions:
We developed a highly efficient pipeline for biomarker discovery, verification and validation, with each step systematically informing the next. A panel of proteins that might be clinically relevant biomarkers for PC was found.
Keywords: pancreatic cancer, serum, biomarker, mass spectrometry, LC-MS, MRM
Pancreatic cancer (PC) has a dismal prognosis and is currently the fourth leading cause of cancer-related mortality, and it is expected to become the second within the next 20 years (Li et al, 2004; Bardeesy and DePinho, 2002; Ryan et al, 2014). Owing to its asymptomatic nature and high metastatic potential, the diagnosis of PC is only possible for those in an advanced state, and the prognosis of PC remains the worst of the major malignancies (Ghaneh et al, 2007; Xu et al, 2015). The medium survival rate of PC after diagnosis is <6% and the 5-year survival rate has remained at ∼5–7% for decades (Yachida et al, 2010; Wolfgang et al, 2013; Siegel et al, 2014). The ability to diagnose PC in asymptomatic patients would allow many patients to be actively treated, thereby greatly improving their prognosis (Kaur et al, 2017).
Many researchers have aimed to identify effective biomarkers for the early detection of PC (Brand et al, 2011). In our previous work, novel prognostic predictors of PC and PC-associated diabetes mellitus were investigated based on the analysis of surgically resected fresh PC tissues and adjacent non-tumour tissues (Wang et al, 2013a, 2013b, and so on). Ideally, a blood-based biomarker or biomarker panel would be more optimal as it would be more feasible and minimally invasive. The Food and Drug Administration-approved blood-based biomarker CA 19–9 has demonstrated only modest effectiveness for the diagnosis of PC, with variable sensitivity (SN, 60–90%) and specificity (SP, 68–91%) (Locker et al, 2006; Goonetilleke et al, 2007; Goonetilleke and Siriwardena, 2007; Kaur et al, 2012). It also showed false negative results in the Lewis negative phenotype (5–10%) (Locker et al, 2006) and false positive results in the presence of obstructive jaundice (10–60%) (Duraker et al, 2007). These limitations of CA 19-9 have led to the urgent search for alternative biomarkers. The development of new methodologies for the discovery of biomarkers is an ongoing endeavor (Chen et al, 2016).
A typical proteomics-based biomarker pipeline starts with a discovery stage, followed by verification and validation of the candidate biomarker for its intended clinical use (Rifai et al, 2006). Discovery proteomics analyses have rapidly developed to detect and comprehensively quantitate proteins expressed in complex biological systems, generating hundreds of candidate biomarkers of differential abundance (Ross et al, 2004; Zhou et al, 2013; Zhang et al, 2014; Cancer Genome Atlas Research Network, 2014). Immunoassays and stable isotope dilution-multiple reaction monitoring mass spectrometry (SID-MRM) offers robust, high-throughput, and absolute quantification of targeted peptide(s) across different samples (Addona et al, 2009; Keshishian et al, 2009; Picotti and Aebersold, 2012). However, lacking of commercial available antibodies for specific proteins and posttranslational modifications hinders the development of newly discovered biomarkers (Larkin et al, 2016). Besides, it is both time consuming and expensive to verify dozens or hundreds of candidate biomarkers for both antibody-based and SID-MRM assays. Additionally, because of the wide dynamic range of protein content in serum samples, high-abundant protein depletion strategies and extensive separation of enzymatically digested peptides are utilised for better coverage of protein identification in the discovery stage. Therefore, a general approach is needed to verify and prioritise the subset of candidate biomarkers that are detectable in the whole serum sample using one-dimensional liquid chromatography MRM-MS (1D LC-MRM-MS) analyses, which can validate biomarkers with high throughput and high efficiency.
Herein, we employed isobaric tags for relative and absolute quantitation (iTRAQ)-based comparative proteomics analysis, 1D targeted LC-MS/MS, a prime MRM without SIS peptides and SID-MRM in an integrated workflow for biomarker candidate discovery, verification and validation, respectively. The data from each stage can systematically inform the next stage without discrimination. To the best of our knowledge, this is the first construction of a coherent and MS-intensive pipeline for biomarker development in PC. A total of 150 serum samples from healthy people (normal control, NC), patients with benign diseases (BD) and PC patients were analysed, and a new panel of candidate biomarkers consisting of apolipoprotein E (APOE), inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3), apolipoprotein A-I (APOA1) and apolipoprotein L1 (APOL1) showed significant differences between PC vs NC and BD groups. The combined diagnosis of the four proteins and CA19-9 outperformed CA19-9 alone in the diagnosis of PC and thus could serve as a potential predictive biomarker panel.
Materials and methods
Patients and specimens
A total of 150 blood samples were recruited (using informed consent) at the Zhongshan Hospital between June 2010 and January 2012, and categorised as follows: NC (n=40), BD (n=30, pancreatitis (4), pancreatic cysts (13), benign tumours (13)), and PC (n=80). The research followed the tenets of the Declaration of Helsinki and was approved by the Ethics Committee of the Fudan University Shanghai Zhongshan hospital. Blood samples were collected in the morning after an overnight fast using Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ, USA) without anticoagulant and allowed to clot at room temperature for 1 h before centrifugation at 1500 g for 10 min. The serum was removed, immediately aliquoted in sterile centrifuge tubes and stored at −80 °C for future analysis.
2D LC-MS/MS analysis of iTRAQ-labelled peptides
In the discovery stage, every 10 serum samples were pooled together in each group for subsequent analysis. The high-abundant proteins from the pooled serum sample were depleted using a Human 14 Multiple Affinity Removal System Column (Agilent Technologies, Santa Clara, CA, USA). Proteins were then digested (Wisniewski et al, 2009), followed by iTRAQ labelling, according to the manufacture’s instructors. As a consequence, two sets of iTRAQ 8-Plex (NC with 113 and 114 tags, BD with 115 and 116 tags, and PC with 117, 118, 119 and 121 tags) and 1 set of iTRAQ 5-Plex (NC with 113 tags, BD with 115 and 116 tags, and PC with 117 tags) were constructed to provide multiple biological replicates. The labelled peptides were then fractionated with high pH reversed-phase liquid chromatography on a UPLC system (Waters, Milford, MA, USA).
Nano-LC-MS/MS analyses were performed on a Nanoeasy system with a 50- cm-long column (75 um id × 50- cm-long,C18, Thermo Fisher Scientific, San Jose, CA, USA) connected to a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific), and a 1D Plus nano LC system (Eksigent of Sciex, Framingham, MA, USA) coupled with the Triple TOF 5600 system (Sciex, Framingham, MA, USA). The detailed methods may be found in the Supplementary section.
Protein identification and quantitation analysis were performed with Proteome Discovery (v.1.3, Thermo Fisher Scientific) and ProteinPilot (version 4.5, Sciex). All the data were searched against the Swiss-Prot human database (20,238 entries) with MS tolerance set at 20 ppm, and MS/MS tolerance set at 0.1 Da.
In this study, a false discovery rate (FDR) lower than 1% was used to control protein level identification based on the target-decoy strategy. Proteins with at least one unique peptide with confidence higher than 95% were used for quantitation. Student's t-test (PC vs control (NC and BD)) was applied to compare the protein expression levels between the PC group and the control group. The mean value of the ratio of each group was used to calculate the fold change. Proteins with a fold change larger than 1.2 or less than 0.8 with a Student's t-test P-value <0.05 were selected as differently expressed proteins. A total of 142 proteins met these criteria.
1D-targeted LC-MS/MS
Referring to the spectrums generated above, the precursor-ion intensities of all the unique peptides of the 142 proteins were analysed. The six most intense unique peptides of each protein were selected and their m/z values were set as the inclusion list for 1D-targeted LC-MS/MS detection on a Triple TOF 5600 system. A total of 2 μg of enzymatically digested crude serum sample without high abundant protein depleted was used for the analysis. A short list of 49 proteins was identified with high confidence (FDR<1%, peptide confidence >95%).
P-MRM analysis
For the P-MRM analysis, a total of 96 crude unique peptides corresponding to the 49 proteins were synthesised and used to optimise the transition selection, method building, retention time scheduling for the MRM assay development. An exogenous peptide was added to each digested crude serum sample working as an internal standard. In total, 1 pmol of each crude peptide and 2 μg of digested peptides from 52 crude serum samples were analysed separately.
The MRM analyses were performed on a 6500 QTRAP hybrid triple quadrupole/liner ion trap mass spectrometer (Sciex) interfaced with a UPLC system (Eksigent of Sciex) using a 15-cm-long column (75 μm id × 150, C18). The MRM data were processed using Skyline software (v 3.1) resulting in 4 significantly changed candidate biomarkers (P<0.05%, comparing PC with NC and BD groups; fold change >1.2 or fold change <0.8).
SID-MRM analysis
For the SID-MRM analysis, four stable isotope-labelled peptides corresponding to the best performing peptides of the targeted proteins were synthesised (Bankpeptide, Ltd., China). SIS peptide of 4–5 orders of magnitude were added to the digested serum proteins and tested in triplicate to construct a standard curve (Supplementary Information S8). A certain concentration of SIS peptide were spiked in each sample and the absolute quantitation of interested peptides were carried out in 100 serum samples (34 NCs, 26 BDs and 40 PCs). The concentrations of the endogenous peptides were calculated as follows: Cendogenous=CSIS × peak areaendogenous/peak areaSIS. Detailed information can be found in the Supplementary Methods Section.
Immunoassay measurement of CA19-9 level
CA19-9 levels of the 100 serum samples tested in SID-MRM analysis were determined by electrochemiluminescence immunoassay (double-antibody sandwich ELISA) on a Roche cobas e 602 module according to the manufacturer’s instructions (Roche Diagnostics, Mannheim, Germany).
Statistical construction of a diagnostic model
The quantitative results from the P-MRM and SID-MRM analyses were compared and visualised using Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA). The peptide concentrations of APOE, ITIH3, APOA1, APOL1 and the expression level of CA19-9 in the serum of the NC group (34 cases), BD (26 cases) and the PC group (40 cases) were used to construct the diagnostic model. The statistical analyses were performed using SPSS (v24.0, IBM, Armonk, NY, USA), and P<0.05 was considered statistically significant. Receiver operation characteristic curves (ROCs) were calculated to determine the specificity and sensitivity, as well as to compare the area under the curve (AUC) of single candidate biomarkers and their combinations using a binary logistic regression analysis (DeLong et al, 1988).
Immunohistochemistry
A total of 4 μm of whole formalin-fixed and paraffin-embedded tissue section samples (cancer and para-cancer tissues of pancreas) were prepared. The samples were deparaffinised with xylene, followed by rehydration in a series of four graded alcohols (70, 80, 90 and 100%). Rabbit polyclonal antibodies, anti-APOL1 (1 : 300 dilution, Proteintech, Wuhan, China), anti-APOA1 (1 : 50 dilution, Proteintech), anti-ITIH3 (1 : 50 dilution, Proteintech), and anti-APOE (1 : 1000 dilution, Proteintech) were incubated for 1 h at room temperature, detected with ImmPRESS-HRP anti-rabbit IgG reagent (Beyotime, China) and visualised using DAB+substrate (Dako). An Aperio Scanscope XT (Leica Biosystems, Vista, CA, USA) was used to digitally scan the slides.
Results
Discovery MS
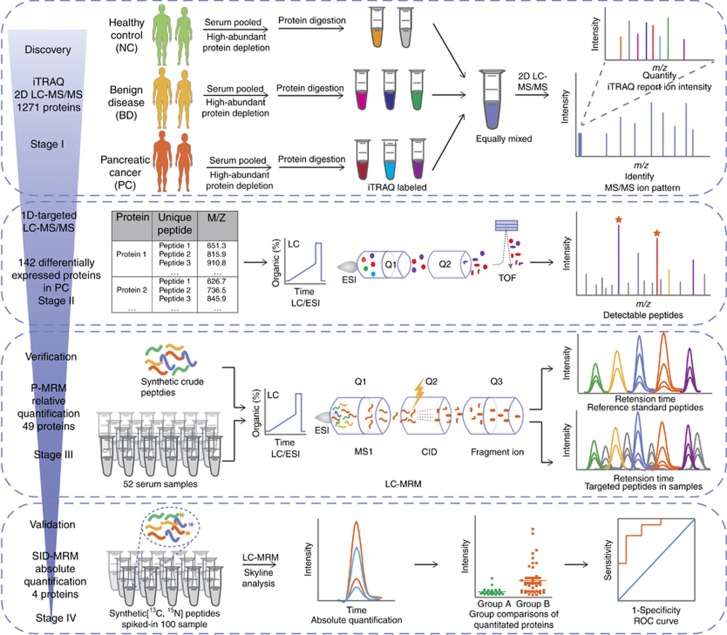
Figure 1 shows the overall workflow for the discovery, verification and validation of the candidate biomarkers for PC. In the whole pipeline, a series of MS-based methods were applied as follows: in stage I, iTRAQ-2DLC-MS/MS was applied to analyse the expression level for up to a thousand proteins in the serum samples (with highly abundant proteins depleted) from the NC, BD and PC groups. In stage II, 1D-targeted LC-MS/MS was utilised to ascertain the detection of the 142 altered protein in 1D LC-MS/MS, resulting in a shorten list of 49 interesting proteins. For cost savings and non-discriminant selection of candidate biomarkers, in stage III, the 49 proteins were further verified using P-MRM in 52 crude serum samples, resulting in a prioritised panel of proteins that were further absolutely quantitated and validated by SID-MRM in 100 serum samples in stage IV. The performance of the biomarker candidates was evaluated using the ROC curves, which was based on the quantitated concentration of these proteins.
Figure 1.
The overall scheme of the MS-based pipeline for the development of novel pancreatic cancer biomarkers. The pipeline utilised a serial of MS-based methods such as iTRAQ-2DLC-MS/MS Strategy, 1D-targeted LC-MS/MS, P-MRM and SID MRM to construct a coherent, high-throughput and non-discriminatory pipeline with strict quantity control. The performance of newly developed biomarkers was statistically evaluated by ROC curves. ROC = receiver operation characteristic.
iTRAQ 2D-LC-MS/MS enables an unbiased quantitative comparison of the expression levels of proteins in different samples and is widely used in biomarker discovery. However, protein content of serum samples has a wide dynamic range which spans >12 orders of magnitude (Tonack et al, 2013). High abundant proteins such as albumin masks or sequesters the detection of lower abundant proteins (Larkin et al, 2016). To extensively identify serum proteome, we depleted high abundant proteins prior to comparative proteomics analysis. Besides, we used extremely high pressures in LC and a long column packed with small particles to improve the separation efficiency (Yin et al, 2014). In this study, a total of 1,217 proteins were identified with a FDR of <1%, of which 142 proteins were differentially expressed according to the criteria mentioned above (Supplementary Information S1–S3). Among these, 78 proteins were increased >1.20-fold in serum samples from PC group compare with NC and BD groups with P-value <0.05, and 64 proteins were decrease <0.8-fold in the PC group (Supplementary Information S4). All the up and down regulated proteins were applied for further analysis.
1D-targeted LC-MS/MS
To ascertain which of the proteins discovered in a iTRAQ- 2D-LC-MS/MS method could also be detected in 1D-LC-MS/MS analysis of crude serum sample without high abundant protein depletion, a 1D-targeted LC-MS/MS analysis was incorporated in the pipeline. As a result, a total of 49 proteins were identified; other proteins were not detected due to the weak signal and ion suppression that resulted from the high dynamic range of the crude serum proteins (Supplementary Information S4).
Relative quantification of candidate proteins using the P-MRM assay
To ensure the quality of P-MRM method analysis, crude peptides corresponding to the 49 proteins were used for the assay development and another exogenous peptide was monitored as an internal standard. The relative quantitation and comparison of each peptide was based on the integration of the areas of the chromatography peaks of the transitions for each peptide (Supplementary Information S5). The coefficient of variation (CV) of the summed area of transitions of the internal standard peptide was 14% in all 52 samples, which indicated that it is reliable to approximately evaluate the relative amount of peptides in the different samples according to the P-MRM results.
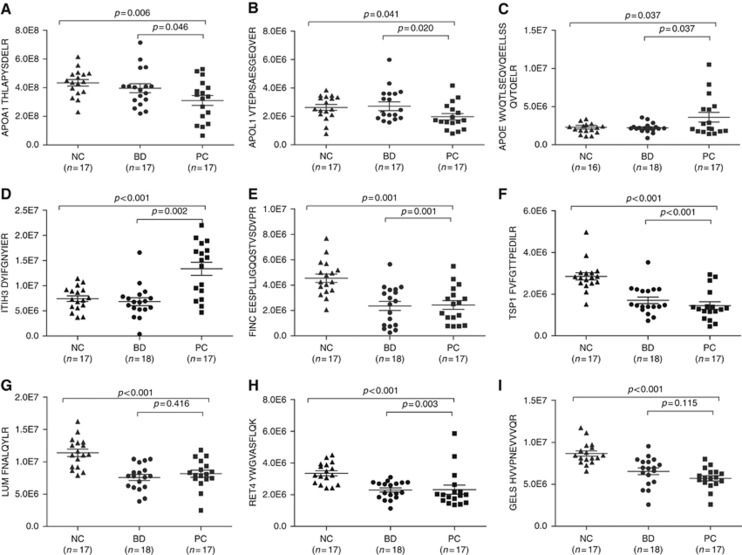
According to the results, 47 peptides corresponding to 27 proteins showed significant changes between the PC group and NC group (Supplementary Information S6). Some of the proteins, such as fibronectin (FINC, P=0.001), thrombospondin-1 (TSP1, P<0.001), lumican (LUM, P<0.001), retinol-binding protein 4 (RET4, P<0.001), and gelsolin (GELS, P<0.001) (Figure 2), showed marked differences between the PC and NC groups (p-values as above). However, the concentrations of these proteins were not significantly different comparing PC with BD groups. These proteins can help us to distinguish patients with pancreatic disorders from healthy people, but they are not appropriate biomarkers for PC.
Figure 2.
Plots of determined concentrations of 9 differentially expressed proteins using P-MRM. The concentration of AOPA1 (A), APOL1 (B), APOE (C), ITIH3 (D), FINC (E), TSP1 (F), LUMA (G), RET4 (H) and GELs (I) in different groups are illustrated. The mean value and standard deviations of the serum proteins from the NC, BD, and PC groups are shown. Student’s t-test was used for the pairwise comparisons of the concentrations of proteins from the NC, BD, and PC groups. A P-value <0.05 was considered statistically significant. BD = benign diseases; NC = normal control; PC = pancreatic carcinoma.
Of all the results, APOE and ITIH3 expression was significantly increased in PC (Figure 2C and D), whereas APOA1 and APOL1 expression was apparently decreased in PC compared with that in the controls (BD and NC groups) (Figure 2A and B), which was consistent with the iTRAQ findings. These four proteins were selected as candidates for further confirmation and absolute quantification using the SID-MRM assay.
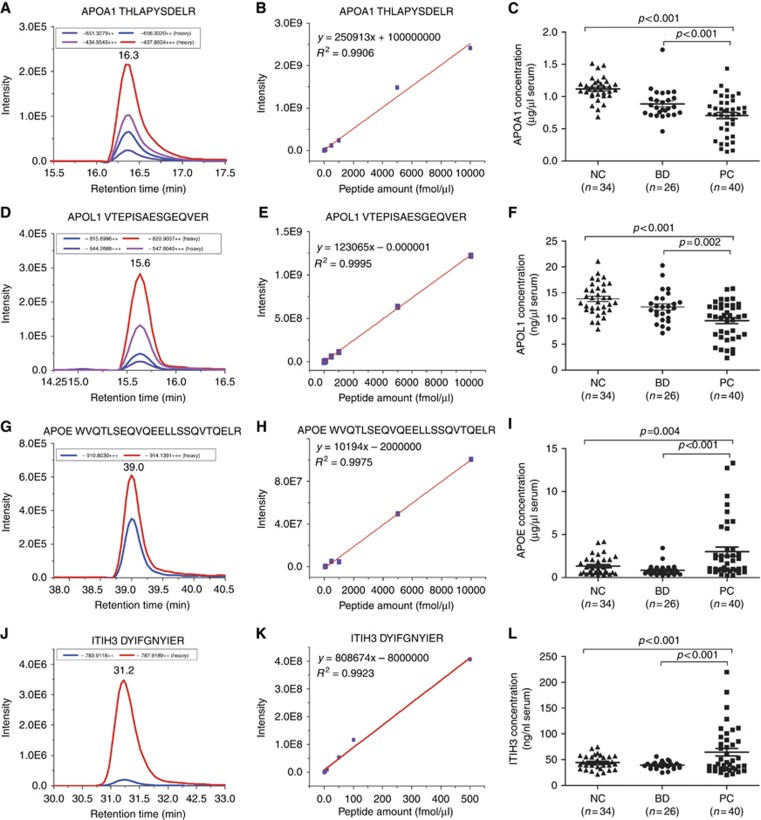
Absolute quantification of 4 candidate proteins using SID-MRM
On the basis of the relative quantification results, four SIS peptides corresponding to the 4 selected proteins (APOE, ITIH3, APOA1, and APOL1) were synthesised for absolute quantification. The details of the confirmed peptides are shown in Supplementary Information S7. Standard curves were tested based on the SIS peptides. The correlation coefficients of the weighted calibration curves of the four SIS peptides ranged from 0.9 to 1 (Figure 3A, D, G and J). The lower limit of quantitation (LLOQ, S/N>10) of the 4 SIS peptides was also determined (Supplementary Information S8). The good linearity and reproducibility of the 4 SIS peptides (Figure 3B, E, H and K) proved the reliability of the SID-MRM method we developed. Group comparisons were performed according to the concentration of each endogenous peptide (Figure 3C, F, R, L, Supplementary Information S10). The results showed that the concentrations of APOA1, APOL1, APOE, and ITIH3 were significantly different (P<0.004) between the PC vs NC and BD groups (Supplementary Information S9).
Figure 3.
Absolute quantification of APOE, ITIH3, APOA1, and APOL1 in the NC, BD and PC groups using the SID-MRM method. Extracted ion chromatograms (A, D, G, J), Standard curves (B, E, H, K) and Group comparison of the concentrations (C, F, I, L) of the 4 peptides in the NC, BD and PC groups are illustrated. The mean value and standard deviations of the serum proteins from the NC, BD, and PC groups are shown. Student’s t-test was used for the pairwise comparisons of the concentrations of proteins from the NC, BD, and PC groups. A P-value <0.05 was considered statistically significant. BD = benign diseases; NC = normal control; PC = pancreatic carcinoma.
Immunoassay measurement of CA19-9 level
The result of CA19-9 expression level of the 100 serum samples are shown in Supplementary Information S9.
Combination biomarker models outperform CA19-9 alone
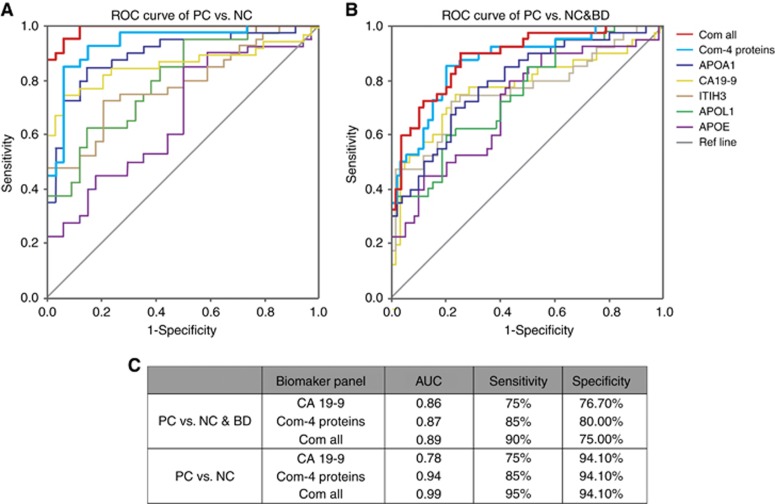
To gain a further insight to the utility of these markers, binary logic regression was performed to produce predictive models that were then analysed by ROC curves. Figure 4 shows the performance, in terms of the area under the curve (AUC), sensitivity, specificity values, of the serum factors such as APOE, ITIH3, APOA1, APOL1, CA19-9. The assessment of the combination of the 4 newly discovered proteins (Com-4 proteins) and the panel of the combination of all the five elements (Com-all) were carried out as well.
Figure 4.
ROC analysis of the newly developed biomarker panels. ROC Curves of APOA1, APOL1, ITIH3, APOE, CA19-9, the combinations of the 4 novel protein biomarkers (Com-4 proteins) and the combination all five elements (Com-all) are shown for differentiating PC vs NC group (A), and PC vs NC & BD groups (B). The AUC, sensitivity and specify of CA19-9 alone, Com-4 proteins and Com-all are demonstrated (C). BD = benign diseases; NC = normal control; PC = pancreatic carcinoma.
For analysis that uses all the samples, including 34 NCs, 26 BDs and 40 PCs, we set NC and BD group together as the control. To differentiate PC from the control group, the plot demonstrates a significant improvement of AUC and Youden Index for the Com-4 proteins and Com-all compare with CA19-9 alone (Figure 4B). The Com-4 proteins and Com-all panels outperformed CA19-9 alone for the differentiation of PC vs NC &BD group.
Comparing PC with NC group, the AUC values for APOE, ITIH3, APOA1, APOL1, CA19-9 were 0.669 (P=0,013), 0.784 (P<0.001), 0.896 (P<0.001), 0.803 (P<0.001), and 0.78(P<0.001), respectively. The Com-4 proteins robustly increased the AUC to 0.937 (P<0.001), and the sensitivity and specificity were 85.0 and 94.1% (Figure 4A). Incorporating with CA19-9, the multi-marker panels named Com-all, remarkably elevated the AUC to 0.99 with a sensitivity of 95% and specificity of 94.1%. The combination of proteins discovered in our analysis and CA19-9 proved to be highly discriminatory between the PC and NC groups.
Biomarker validation by immunohistochemistry in the tissue samples
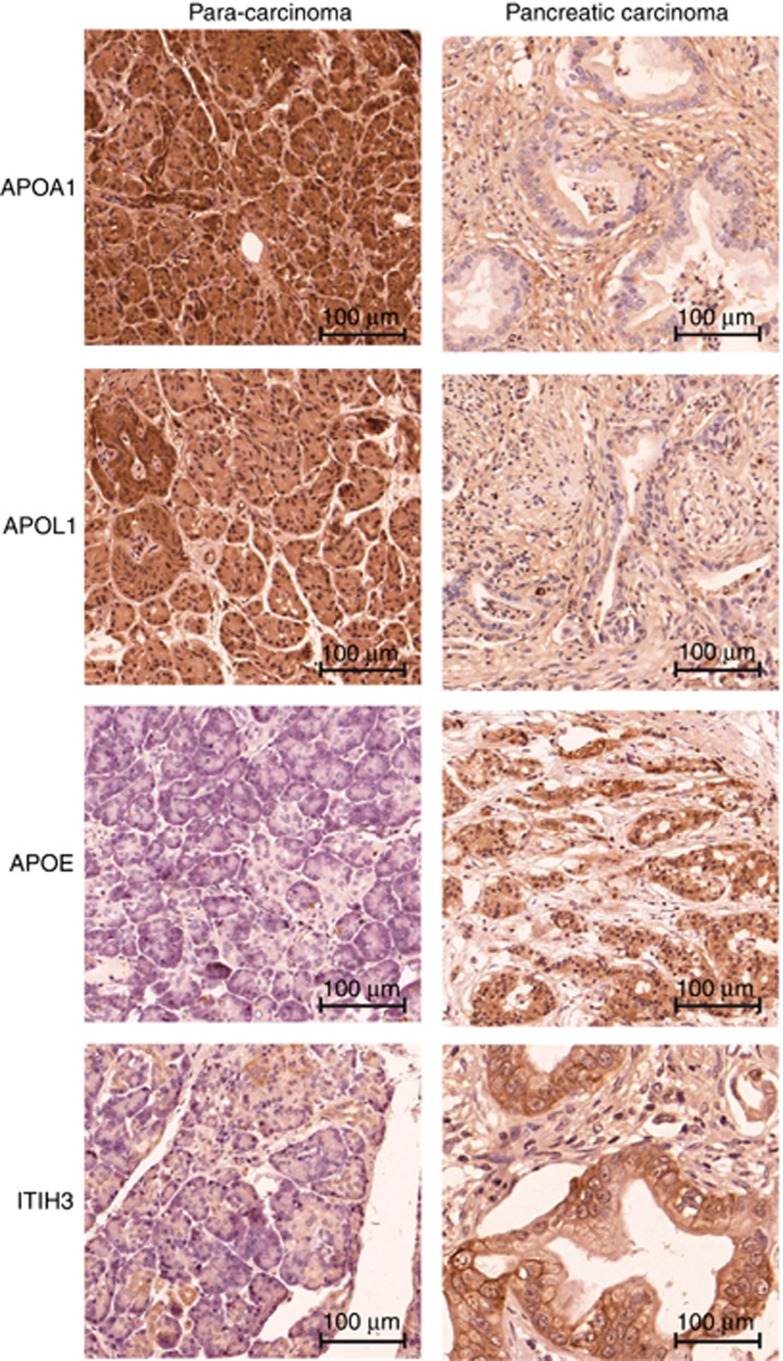
Biomarker profiles of a specific cancer are factors generated by the tumour itself or by the systemic response to the growing and progressing tumour. The new panel of biomarkers identified in the serum samples was further validated by assessing the expression level using immunohistochemistry. The antibody staining demonstrated that APOA1 and APOL1 expression was strong in para-carcinoma tissues. In contrast, APOE and ITIH3 expression was higher in PC tissues (Figure 5). These results were consistent with the differential expression levels of the four proteins in the serum samples.
Figure 5.
APOA1, APOL1, APOE and ITIH3 expression in pancreatic tissue samples. Representative immunohistochemistry images of tissues stained with APOA1, APOL1, APOE and ITIH3 antibodies.
Discussion
A pipeline consisting of an extensive discovery stage followed by a timely verification and validation of altered proteins is becoming increasingly essential for the putative discovery of candidate biomarkers. However, the lack of a highly efficient verification method for the evaluation of multiple altered proteins has hindered the clinical application of candidate biomarkers identified through research (Chambers et al, 2014). Due to its multiplexing capability and antibody independence, robust and high-throughput MRM assays can be developed to verify and quantify hundreds of targeted proteins across large sample sets. The combination of large-scale proteome screens and the high-throughput MRM evaluation of interesting proteins show the potential to increase the efficiency of biomarker development.
On the basis of the MS methods mentioned above, in the current work, we developed a high-throughput and non-discriminatory pipeline for biomarker discovery, verification and validation, where each step systematically informed the next stage. The following aspects were monitored carefully to ensure the functionality of the pipeline. First, several approaches were applied for high resolution biomarker discovery. For example, most of the high-abundant proteins in the serum samples were selectively depleted using affinity columns; a long column (50 cm) was utilised to improve of the separation efficiency of the peptides; and three subsets of iTRAQ–2DLC-MS/MS experiments were analysed separately. The combination of these three approaches highly increased the number of proteins identified in the serum samples. In the discovery stage, a total of 1217 serum proteins were identified, among which 142 circulating proteins were revealed to be differentially expressed in PC compared with the controls. For cost savings and non-discriminant selection of biomarker candidates, 1D-targeted LC-MS/MS was used to confirm the peptide detection; P-MRM was conducted to relative quantification of the targeted proteins, thus bridging the gap between the high-throughput discovery stage and the large-scale targeted validation of samples. Strict quantity control was employed in the whole MRM analysis. Fragmentation properties (such as the retention time and chromatography traces of fragment ions) of the synthesised peptides were used as a constraint for the correct detection of the targeted peptides in the complex background. An exogenous peptide was added to each sample to monitor the reproducibility of the MRM runs and to normalise the results of each sample. A standard curve was constructed to ensure the high performance of the final SID-MRM assays. Thus, reliable relative quantification of the targeted proteins was achieved, which helped to prioritise the candidates for further validation. Finally, the significantly altered proteins were absolutely quantitated and evaluated in large-scale serum samples using SID-MRM. Based on the newly developed pipeline and excellent management of each step, a panel of proteins was observed and the results of the ROC analysis highlight the superiority of the newly developed 4 proteins and the combination of the new panel with CA19-9 for the diagnosis of PC.
Our results also indicate that PC is associated with circulating alterations in a number of proteins that represent a diverse set of biological families, particularly proteins with functions related to retinoid regulation (Bleul et al, 2015), inflammation and multi-molecular metabolism. For example, APOE (2.27 times higher in PC vs NC) is a highly abundant protein in serum and is essential for the normal catabolism of triglyceride-rich lipoprotein constituents (Poersch et al, 2016). Previous studies revealed that during tumour progression, APOE is overexpressed in ovarian carcinomas to maintain cell growth and prevent apoptosis (Poersch et al, 2016). ITIH3 (1.68 times higher in PC vs NC) can be found in the extracellular matrix of various organs as well as in the blood circulation. One study proposed that the ITIH family acts an important factor to stabilise hyaluronic acid on the extracellular matrix. When tumours grow, the epithelial hyaluronic complex increases in size (Ivancic et al, 2013); thus, ITIH3 may play an important role in extracellular matrix remodelling during tumour progression. APOA1 (1.59 times lower in PC vs NC) is a key component of the reverse cholesterol transport pathway, binding to prion inflammatory phospholipids, thereby giving it anti-inflammatory properties (Edelson, 2010). Furthermore, APOL1 (1.34 times lower in PC vs NC) possesses both extra- and intra-cellular functions that are crucial in host defense and cellular homeostatic mechanisms (Hu et al, 2012).
Although we developed a powerful and high-throughput pipeline for biomarker development, there are still some limitations. As mentioned above, not all of the candidate biomarkers discovered using 2D LC-MS/MS can be detected with 1D LC-MRM. Some efforts could be made to improve the assays developed, such as special enrichment of interested proteins with very low abundance prior to MRM analysis (Ahn et al, 2012, 2015; Kennedy et al, 2016). Nonetheless, in terms of time and cost, this MS-intensive pipeline may still be one of the most powerful analytical approaches for biomarker discovery, with high throughput and high efficiency. The panel of these 4 proteins discovered based on this pipeline was identified to have a high predictive value with good sensitivity (85%) and specificity (94.1%),when combining with CA19-9, the sensitivity significantly increased to 95%, which outperformed CA19-9 alone for highly discriminate the PC group from the NC group. These proteins have potential value as novel predictive circulating biomarkers for PC.
Acknowledgments
We thank the funding supported by the Special Project on Precision Medicine under the National Key R&D Program (SQ2017YFSF090210), the National Key Research and Development Program of China (2017YFA0505100), National Basic Research Program of China (2013CB910802), the National High Technology Research and Development Program of China (2014AA020902), the National Natural Science Foundation of China (21675033) and the China Postdoctoral Science Foundation (2015M570324).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJL, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol 27: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn YH, Kim JY, Yoo JS (2015) Quantitative mass spectrometric analysis of glycoproteins combined with enrichment methods. Mass Spectrom Rev 34: 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn YH, Shin PM, Oh NR, Park GW, Kim H, Yoo JS (2012) A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J Proteomics 75: 5507–5515. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, DePinho RA (2002) Pancreatic cancer biology and genetics. Nat Rev Cancer 2: 897–909. [DOI] [PubMed] [Google Scholar]
- Bleul T, Ruhl R, Bulashevska S, Karakhanova S, Werner J, Bazhin AV (2015) Reduced retinoids and retinoid receptors' expression in pancreatic cancer: A link to patient survival. Mol Carcinogen 54: 870–879. [DOI] [PubMed] [Google Scholar]
- Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM, Langmead CJ, Landsittel DP, Whitcomb DC, Grizzle WE, Lokshin AE (2011) Serum biomarker panels for the detection of pancreatic cancer. Clin Cancer Res 17: 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AG, Percy AJ, Simon R, Borchers CH (2014) MRM for the verification of cancer biomarker proteins: recent applications to human plasma and serum. Expert Rev Proteomics 11: 137–148. [DOI] [PubMed] [Google Scholar]
- Chen WC, Yuan JS, Xing Y, Mitchell A, Mbong N, Popescu AC, McLeod J, Gerhard G, Kennedy JA, Bogdanoski G, Lauriault S, Perdu S, Merkulova Y, Minden MD, Hogge DE, Guidos C, Dick JE, Wang JCY (2016) An integrated analysis of heterogeneous drug responses in acute myeloid leukemia that enables the discovery of predictive biomarkers. Cancer Res 76: 1214–1224. [DOI] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44: 837–845. [PubMed] [Google Scholar]
- Duraker N, Hot S, Polat Y, Hobek A, Gencler N, Urhan N (2007) CEA, CA 19-9, and CA 125 in the differential diagnosis of benign and malignant pancreatic diseases with or without jaundice. J Surg Oncol 95: 142–147. [DOI] [PubMed] [Google Scholar]
- Edelson S (2010) Going APOA1 in cancer. SciBX 3: 19–20. [Google Scholar]
- Ghaneh P, Costello E, Neoptolemos JP (2007) Biology and management of pancreatic cancer. Gut 56: 1134–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke KS, Mason JM, Siriwardana P, King NK, France MW, Siriwardena AK (2007) Diagnostic and prognostic value of plasma tumor M2 pyruvate kinase in periampullary cancer- evidence for a novel biological marker of adverse prognosis. Pancreas 34: 318–324. [DOI] [PubMed] [Google Scholar]
- Goonetilleke KS, Siriwardena AK (2007) Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol 33: 266–270. [DOI] [PubMed] [Google Scholar]
- Hu CA, Klopfer EI, Ray PE (2012) Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett 586: 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivancic MM, Huttlin EL, Chen XD, Pleiman JK, Irving AA, Hegeman AD, Dove WF, Sussman MR (2013) Candidate serum biomarkers for early intestinal cancer using N-15 metabolic labeling and quantitative proteomics in the apcmin mouse. J Proteome Res 12: 4152–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Baine MJ, Jain M, Sasson AR, Batra SK (2012) Early diagnosis of pancreatic cancer: challenges and new developments. Biomark Med 6: 597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Smith LM, Patel A, Menning M, Watley DC, Malik SS, Krishn SR, Mallya K, Aithal A, Sasson AR, Johansson SL, Jain M, Singh S, Guha S, Are C, Raimondo M, Hollingsworth MA, Brand RE, Batra SK (2017) A Combination of MUC5AC and CA19-9 improves the diagnosis of pancreatic cancer: a multicenter study. Am J Gastroenterol 112: 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy JJ, Yan P, Zhao L, Ivey RG, Voytovich UJ, Moore HD, Lin C, Pogosova-Agadjanyan EL, Stirewalt DL, Reding KW, Whiteaker JR, Paulovich AG (2016) Immobilized metal affinity chromatography coupled to multiple reaction monitoring enables reproducible quantification of phospho-signaling. Mol Cell Proteomics 15: 726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshishian H, Addona T, Burgess M, Mani DR, Shi X, Kuhn E, Sabatine MS, Gerszten RE, Carr SA (2009) Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics 8: 2339–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin SET, Johnston HE, Jackson TR, Jamieson DG, Roumeliotis TI, Mockridge CI, Michael A, Manousopoulou A, Papachristou EK, Brown MD, Clarke NW, Pandha H, Aukim-Hastie CL, Cragg MS, Garbis SD, Townsend PA (2016) Detection of candidate biomarkers of prostate cancer progression in serum: a depletion-free 3D LC/MS quantitative proteomics pilot study. Br J Cancer 115: 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Xie K, Wolff R, Abbruzzese JL (2004) Pancreatic cancer. Lancet 363: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC (2006) ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol 24: 5313–5327. [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 9: 555–566. [DOI] [PubMed] [Google Scholar]
- Poersch A, Grassi ML, Carvalho VPd, Lanfredi GP, Palma CdS, Greene LJ, de Sousa CB, Carrara HHA, Candido dos Reis FJ, Faça VM (2016) A proteomic signature of ovarian cancer tumor fluid identified by highthroughput and verified by targeted proteomics. J Proteomics 145: 226–236. [DOI] [PubMed] [Google Scholar]
- Rifai N, Gillette MA, Carr SA (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol 24: 971–983. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169. [DOI] [PubMed] [Google Scholar]
- Ryan DP, Hong TS, Bardeesy N (2014) Pancreatic Adenocarcinoma REPLY. New Engl J Med 371: 2140–2141. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ma JM, Zou ZH, Jemal A (2014) Cancer Statistics, 2014. Ca-Cancer J Clin 64: 9–29. [DOI] [PubMed] [Google Scholar]
- Tonack S, Jenkinson C, Cox T, Elliott V, Jenkins RE, KitteringhamNR, Greenhalf W, Shaw V, Michalski CW, Friess H, Neoptolemos JP, Costello E (2013) iTRAQ reveals candidate pancreatic cancer serum biomarkers: influence of obstructive jaundice on their performance. Br J Cancer 108: 1846–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WS, Liu XH, Liu LX, Jin DY, Yang PY, Wang XL (2013. a) Identification of proteins implicated in the development of pancreatic cancer-associated diabetes mellitus by iTRAQ-based quantitative proteomics. J Proteomics 84: 52–60. [DOI] [PubMed] [Google Scholar]
- Wang WS, Liu XH, Liu LX, Lou WH, Jin DY, Yang PY, Wang XL (2013. b) iTRAQ-based quantitative proteomics reveals myoferlin as a novel prognostic predictor in pancreatic adenocarcinoma. J Proteomics 91: 453–465. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362. [DOI] [PubMed] [Google Scholar]
- Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH (2013) Recent progress in pancreatic cancer. Ca-Cancer J Clin 63: 318–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Chang RX, Peng ZY, Wang YM, Ji WW, Guo JY, Song LL, Dai C, Wei W, Wu YJ, Wan XJ, Shao CH, Zhan LX (2015) Loss of polarity protein AF6 promotes pancreatic cancer metastasis by inducing Snail expression. Nat Commun 6: 7184. [DOI] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA (2010) Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467: 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XY, Liu XH, Zhang Y, Yan GQ, Wang F, Lu HJ, Shen HL, Yang PY (2014) Rapid and sensitive profiling and quantification of the human cell line proteome by LC-MS/MS without prefractionation. Proteomics 14: 2008–2016. [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJC, Carr SA, Tabb DL, Coffey RJ, Slebos RJC, Liebler DC, Cptac N (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lu Y, Ficarro SB, Adelmant G, Jiang WY, Luckey CJ, Marto JA (2013) Genome-scale proteome quantification by DEEP SEQ mass spectrometry. Nat Commun 4: 2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.